Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is one of the deadliest and most aggressive hematological malignancies, but its pathological mechanism in controlling cell survival is not fully understood. Oculocerebrorenal syndrome of Lowe is a rare X-linked recessive disorder characterized by cataracts, intellectual disability, and proteinuria. This disease has been shown to be caused by mutation of oculocerebrorenal syndrome of Lowe 1 (OCRL1; OCRL), encoding a phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] 5-phosphatase involved in regulating membrane trafficking; however, its function in cancer cells is unclear. Here, we uncovered that OCRL1 is overexpressed in T-ALL cells, and knockdown of OCRL1 results in cell death, indicating the essential role of OCRL in controlling T-ALL cell survival. We show OCRL is primarily localized in the Golgi and can translocate to plasma membrane (PM) upon ligand stimulation. We found OCRL interacts with oxysterol-binding protein–related protein 4L, which facilitates OCRL translocation from the Golgi to the PM upon cluster of differentiation 3 stimulation. Thus, OCRL represses the activity of oxysterol-binding protein–related protein 4L to prevent excessive PI(4,5)P2 hydrolysis by phosphoinositide phospholipase C β3 and uncontrolled Ca2+ release from the endoplasmic reticulum. We propose OCRL1 deletion leads to accumulation of PI(4,5)P2 in the PM, disrupting the normal Ca2+ oscillation pattern in the cytosol and leading to mitochondrial Ca2+ overloading, ultimately causing T-ALL cell mitochondrial dysfunction and cell death. These results highlight a critical role for OCRL in maintaining moderate PI(4,5)P2 availability in T-ALL cells. Our findings also raise the possibility of targeting OCRL1 to treat T-ALL disease.
Keywords: Ca2+ homeostasis; oculocerebrorenal syndrome of Lowe 1; OSBP-related protein 4L; phosphatidylinositol 4,5-bisphosphate; T-cell acute lymphoblastic leukemia
The gene responsible for Lowe syndrome was identified by positional cloning of X chromosome breakpoints, designated oculocerebrorenal syndrome of Lowe 1 (OCRL1) (1), and the mutations of this gene are associated with both the oculocerebrorenal syndrome of Lowe and Dent disease (2). OCRL was originally described as a Golgi-localized protein in fibroblasts and epithelial cells (3). Subsequent reports suggested that OCRL is localized to the Golgi (4, 5), PM (6), early endosomes (7, 8), lysosomes (9), and clathrin-coated trafficking intermediates (4, 10). OCRL can also be detected in the PM when quiescent cells are stimulated with growth factors, suggesting that it translocates to the PM. This may be mediated by Rac1, a member of the Rho GTPase family, which forms a stable complex with the C-terminal Rho GAP-like domain of OCRL (5, 6). Furthermore, there are two clathrin-binding sites and a clathrin adaptor AP-2 binding site present in OCRL (11), which play important roles in clathrin-mediated endocytosis defects (12). The interaction of OCRL with clathrin regulates protein trafficking between the Golgi and endosomes (4).
Human inositol 5-phosphatases form a family of 10 members, and their catalytic substrates include inositol 1,4,5-trisphosphate (IP3), inositol 1,3,4,5-tetrakisphosphate, phosphatidylinositol 3,4,5-trisphosphate, and phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] (13). OCRL has a marked preference for PI(4,5)P2 compared with the other inositol 5-phosphatases, suggesting that it is mainly a PI(4,5)P2 5-phosphatase, that produces phosphatidylinositol 4-phosphate [PI(4)P] (14). In addition, PI(4,5)P2 is a precursor of IP3 and diacylglycerol after phosphoinositide phospholipase C (PLC) activation, which is essential for early signaling of cell surface receptors. Among them, IP3 from PLC-mediated inositol phosphate breakdown induces Ca2+ release from the endoplasmic reticulum (ER) (15). PLC-mediated activation of PI(4,5)P2 hydrolysis and the Ca2+ is a key intracellular messenger that controls different cellular functions but causes cell death when the homeostasis is broken (16). Ca2+ released from the ER is taken up by mitochondria through the mitochondrial Ca2+ uniporter (MCU) (17). Furthermore, stimulation of mitochondrial oxidative metabolism by Ca2+ is generally considered important for the control of cellular ATP homeostasis (18). Excessive Ca2+ taken up by MCU into mitochondria results in mitochondrial Ca2+ overload, disrupting mitochondrial permeability transition pore opening, release of cytochrome c and activation of caspase3/7 (19). Thus, the content of PI(4,5)P2 in the PM used for PLC hydrolysis should be controlled to maintain Ca2+ homeostasis.
Oxysterol-binding protein (OSBP)-related protein 4 (ORP4; also known as OSBP2) is a member of the oxysterol-binding protein–related protein (ORP) family, which are described as lipid-binding/transfer proteins with variable tissue expression, ligand specificity, and subcellular localization and function (20). It has three recognized variants, OSBP-related protein 4L (ORP4L), ORP4M, and ORP4S. All together 23 exons are required to encode the full-length ORP4L protein, including the OSBP-related ligand-binding domain, pleckstrin homology domain, and a motif designated two phenylalanines in an acidic tract, to facilitate lipid transfer and signaling activities (21). Our previous study showed that ORP4L promotes phospholipase C β3 (PLCβ3) translocation from the nucleus to the PM (22) and forms a PLCβ3-Gαq/11-CD3ε complex that activates PLCβ3 upon anti-cluster of differentiation 3 (CD3) stimulation, regulating IP3 production and ER Ca2+ release in T-ALL cells (23). ORP4L extracts PI(4,5)P2 from the PM and presents it to PLCβ3, enabling IP3 generation, and subsequent Ca2+-dependent bioenergetics and cell survival (24).
The CD3 molecule is an important marker of the surface of T cells, consisting of five polypeptide chains of γ, δ, ε, ζ, and η, which are involved in the assembly, stabilization and signal transduction of the TCR–CD3 complex (25). Anti-human CD3 monoclonal antibodies recognize ε chains of CD3 molecules on the TCR-CD3 complex, thereby enhancing the activation and proliferation of T lymphocytes (26). In normal T cells, upon stimulation with anti-CD3, LAT (pp36) is heavily tyrosine phosphorylated and subsequently binds to PLCγ1 to increase IP3 and intracellular Ca2+ in a G protein–independent manner (27). This process is required for the recruitment of the CD3ζ chain to active ZAP70, which plays a central role in CD3 signaling (28). However, CD3ζ chain expression is defective in T-ALL cells (29). Thus, CD3 signaling is swapped to Gαq/11 in the presence of ORP4L and sequentially activates Gαq/11 and PLCβ3, which becomes the dominant enzyme for IP3 generation and intracellular Ca2+ homeostasis in T-ALL cells (23).
In this study, we show that OCRL directly binds to ORP4L which stimulates OCRL translocation from the Golgi to the PM, maintaining moderate PI(4,5)P2 consumption by PLCβ3 and Ca2+ homeostasis for T-ALL cell survival.
Results
Elevated OCRL1 expression is essential for T-ALL cell survival
As OCRL catalyzes hydrolysis of PI(4,5)P2, the same substrate that PLCβ3 uses, we explored the relationship between OCRL and ORP4L. As we previously reported (23), ORP4L mRNA and protein are undetectable in normal T-cells, but they are upregulated in primary T-ALL cells and two T-ALL cell lines, Jurkat T-cells and Molt-4 cells (Fig. 1, A and B). OCRL1 is expressed in normal T-cells, but it is significantly elevated in T-ALL cells (Fig. 1, A and B). By analyzing 1036 human cancer cell lines in CCLE database (https://portals.broadinstitute.org/ccle), we found OCRL1 was expressed in T-ALL cell lines (Fig. S1A). To further determine OCRL1 expression in T-ALL cells, the assessment of human leukemia databases (GSE48558) showed a significant increase in OCRL1 expression in patients’ T-ALL cells and T-ALL cell lines compared with that in normal T-cells (Fig. S1B). Thus, we identified upregulation of OCRL1 in T-ALL cells.
Figure 1.
Elevated OCRL1 expression in T-ALL cells maintains cell survival.A, qRT-PCR analysis of ORP4L (left) and OCRL1 (right) mRNA expression in normal human T-cells, primary T-ALL cells, and two T-ALL cell lines, Jurkat T-cells and Molt-4 cells. The mRNA expression level is calculated relative to actin expression. Values are shown as the mean ± S.D. of three repeats folded over the T-ALL #1 group (for ORP4L) or T-cell #1 group (for OCRL1), respectively. B, Western blot analysis of ORP4L and OCRL protein expression in normal human T-cells, primary T-ALL cells, and two T-ALL cell lines. C, Western blot showing OCRL expression in Jurkat T-cells (upper) and Molt-4 cells (lower) subjected to control (shNT), OCRL1 knockdown (shOCRL1) alone, or combined with OCRL1 rescued expression (shOCRL1+OCRL1). Cells were transduced with lentivirus expressing control nontargeting shRNA or OCRL1-specific shRNA for 48 h before transduction with lentivirus expressing OCRL1. After additional 48 h of culture, the cells were used for analysis. D, cell proliferation analysis of Jurkat T-cells and Molt-4 cells subjected as in panel C. Cells were incubated with EdU for 1 h and analyzed by flow cytometer. E, cell death analysis of Jurkat T-cells and Molt-4 cells subjected as in panel C. Cells were stained by LIVE/DEAD Fixable Dead Cell Stain Kits and analyzed by flow cytometer. The data represent mean ± SD (n = 3). ∗∗∗p < 0.001; Student's t test. EdU, ethynyl-deoxyuridine; OCRL, oculocerebrorenal syndrome of Lowe 1; ORP4L; OSBP-related protein 4L; OSBP, oxysterol-binding protein; T-ALL, T-cell acute lymphoblastic leukemia.
To investigated the role of OCRL in T-ALL cell survival, we transduced Jurkat and Molt-4 cells with lentivirus carrying shOCRL1 alone or combined with OCRL1 cDNA to rescue its expression. OCRL1 expression was significantly knockdown by shRNA but can rescue after cotransduction of OCRL1 cDNA, which were confirmed by Western blot (Fig. 1C). By performing cell proliferation experiments, we found that cells with knockdown of OCRL1 display a reduced percentage of cells with ethynyl-deoxyuridine (EdU) in newly synthesized DNA, indicating decreased cell proliferation (Fig. 1D). Simultaneously, cell death analysis revealed that knockdown of OCRL1 increased cell death (Fig. 1E), whereas, these phenotypes were abolished in OCRL1 expression rescued cells (Fig. 1, D and E). To avoid the off-target effect of shRNA, we confirmed the effects of OCRL1 knockdown by using an independent shRNA (Fig. S1, C–E). Take together, we evidenced that elevated OCRL1 expression is essential for T-ALL cell survival.
ORP4L is a novel binding partner of OCRL in T-ALL cells
ORP4L was important for T-ALL cell Ca2+ homeostasis and cell survival (23). As both OCRL and ORP4L regulate PM PI(4,5)P2 and require for T-ALL cell survival, we further study their relationship in Jurkat T-cells. We first tested whether they interact with each other by a series of experiments. In a pull-down assay using lysates from OCRL1-overexpressing Jurkat T-cells, the OCRL protein was pulled down by glutathione S-transferase (GST)-ORP4L but not by GST (Fig. 2A). We carried out immunoprecipitation experiments in Jurkat T-cells and detected an interaction between endogenous OCRL and ORP4L proteins (Fig. 2B). Bimolecular fluorescence complementation (BiFC) provides the potential for direct visualization of protein interactions in living cells (30). To investigate the interaction between OCRL and ORP4L in living cells, expression vectors encoding the Venus fusion protein fragments were cotransfected into Jurkat T-cells. After 36 h, heterodimer fluorescence was observed at the PM of the cells transfected with OCRL1/pVn-N1 and ORP4L/pVc-C1 (Fig. 2C). Furthermore, confocal fluorescence microscopy analysis revealed a largely Golgi distribution of OCRL, but some of OCRL also localized at the PM, overlapping with the ORP4L staining in Jurkat T-cells (Fig. 2D). These results evidenced that OCRL interacts with ORP4L in T-ALL cells.
Figure 2.
The interaction of OCRL and ORP4L.A, OCRL interacts with ORP4L in GST-pulldown assays. Lysates derived from Jurkat T-cells were incubated with glutathione-agarose beads preloaded with the GST or GST-ORP4L fusions. The retained proteins were eluted, fractionated by 10% SDS/PAGE, and immunoblotted with anti-OCRL antibody. Approximately equal amounts of different GST fusions were immobilized on glutathione-agarose beads in these assays, as verified by immunoblotted with anti-GST antibody. B, coimmunoprecipitation of endogenous OCRL and ORP4L in Jurkat T-cells; precipitation was carried out with 5 μg OCRL antibody or control-IgG followed by Western blot analysis with antibodies against ORP4L or OCRL. C, visualization of interaction among OCRL and ORP4L in living cells using BiFC analysis. Fluorescence images of Jurkat T-cells transfected with plasmid encoding OCRL1 or ORP4L fused to the Venus protein fragments as indicated were acquired 36 h after transfection. Scale bar, 10 μm. D, colocalization of ORP4L (red) and OCRL (green) in plasma membrane of Jurkat T-cells. Nucleus, blue. Scale bar, 10 μm. E, the full-length OCRL1 in bait vector pGBKT7 and truncated fragments of ORP4L in prey vector pGADT7 cotransformed into the yeast were grown on SD/2-; 53/T is a positive control, and Lam/T is a negative control; the interaction was monitored by growth on SD/4- and the use of X-gal assay. F, the full-length ORP4L in bait vector pGBKT7 and truncated fragments of OCRL1 in prey vector pGADT7 cotransformed into the yeast were grown on SD/2-; 53/T is a positive control, and Lam/T is a negative control; the interaction was monitored by growth on SD/4- and the use of X-gal assay. G, predicted binding mode of ORP4L (yellow) to OCRL (blue) and the site view show the detailed maps of key interaction residues of ORP4L (yellow) and OCRL (blue) complex. H and I, OCRL interacts with ORP4L and ORP4L(mut) (H), or ORP4L interacts with OCRL and OCRL (mut) (I) in GST-pulldown assays. Lysates derived from Jurkat T-cells were incubated with glutathione-agarose beads preloaded with the indicated GST fusions. The retained proteins were eluted, fractionated by 10% SDS/PAGE, and immunoblotted with anti-OCRL antibody (H) or anti-ORP4L (I). Approximately equal amounts of different GST fusions were immobilized on glutathione-agarose beads in these assays, as verified by immunoblotted with anti-GST antibody. Bimolecular fluorescence complementation. BiFC, bimolecular fluorescence complementation; GST, glutathione S-transferase; OCRL, oculocerebrorenal syndrome of Lowe 1; ORP4L; OSBP-related protein 4L; OSBP, oxysterol-binding protein.
In order to map which region of ORP4L is involved in the interaction with OCRL, deletion constructs of ORP4L were inserted in the prey plasmid pGADT7, and their interactions with OCRL were evaluated by using the yeast 2-hybrid assay (Fig. 2E). The positive control 53/T and negative control Lam/T demonstrated the reliability of these experiments. None of the deletion constructs of ORP4L grew on SD/4- plates or activated the X-gal reporter gene when combined with the empty plasmid pGBKT7, indicating that ORP4L did not autoactivate the reporter. The prey constructs containing full-length ORP4L or the segment containing aa 391 to 455 could grow on SD/4- plates and activated the X-gal reporter, indicating that this region in ORP4L interacts with OCRL. Conversely, to identify the region of OCRL that is sufficient for interaction with ORP4L, truncated fragments of OCRL1 were inserted in the prey plasmid pGADT7. When combined with the empty plasmid pGBKT7, none of the deletion constructs of OCRL1 grew on SD/4- plates or activated the X-gal reporter. The prey constructs containing full-length OCRL1 or the segment containing aa 564 to 901 grew on SD/4- plates and activated the X-gal reporter, suggesting that aa 564 to 901 of OCRL are sufficient for the interaction with ORP4L (Fig. 2F).
To further determine the key amino acids mediating the binding between OCRL and ORP4L, we predicted the binding mode of ORP4L to OCRL by molecular docking. Asp743/Trp739 of OCRL are in contact with Arg383 of ORP4L, and Lys713 of OCRL is in contact with Arg396 of ORP4L. Furthermore, Lys991 of OCRL is in contact with Phe417 of ORP4L (Fig. 2G). We further mutated the predicted binding site in ORP4L (the Arg383, Arg396, Phe417) and OCRL (Asp743, Trp739, Lys713, Lys691). In the pull-down assay, as compared to wildtype ORP4L, the OCRL protein pulled down by GST-ORP4L (mut) was dramatically reduced (Fig. 2H), and the ORP4L protein pulled down by GST-OCRL (mut) was also decreased (Fig. 2I), indicating the requirement of the Arg383, Arg396, and Phe417 site of ORP4L and the Asp743, Trp739, Lys713, and Lys691 site of OCRL for their interaction.
In order to use this OCRL (mut) for functional study, we next detected its localization and phosphatase activity. For these purposes, we overexpressed Xpress-tagged OCRL1 or OCRL1(mut) in Jurkat T-cells, then isolated the PM and Golgi fractionations to analyze their localization. We verified PM and Golgi purification by the corresponding marker proteins (Fig. S2A). We found the equal amount of OCRL and OCRL (mut) protein in PM and Golgi (Fig. S2B). Immunofluorescence staining provided further evidence that OCRL (mut) did not alter its intracellular localization (Fig. S2C). For in vitro phosphatase activity, we purified recombinant OCRL and OCRL (mut) proteins and incubated them with PI(4,5)P2 containing PM and then measured the conversion of PI(4,5)P2 to PI(4)P. Both the OCRL and OCRL (mut) increased the PI(4)P production and reduced the PI(4,5)P2 levels (Fig. S2D) with the similar activity, indicating that this OCRL1 mutation did not affect its phosphatase activity.
OCRL is translocated from the Golgi to the PM upon anti-CD3 stimulation in ORP4L-dependent manner
It has been found that after Rac activation, OCRL is translocated to the PM and accumulates specifically in membrane ruffles (6). As a protein that directly interacts with OCRL, ORP4L has been shown to have the ability of assisting in protein translocation to the PM (23). So, we explored whether OCRL can translocate to the PM in T-ALL cells and whether ORP4L plays a role in this process.
Confocal imaging analyses showed that endogenous OCRL clustered in Golgi of unstimulated Jurkat T-cells, with a small amount of OCRL distribution at the PM where it colocalized with ORP4L. However, anti-CD3 stimulation induced a striking OCRL translocation from the Golgi to the PM and increased its colocalization with ORP4L (Fig. 3A). In contrast, translocation of OCRL was not observed in ORP4L-depleted cells, and the percentage of cells that responds to anti-CD3 stimulation was significantly reduced (Fig. 3A).
Figure 3.
Analysis of OCRL translocation in Jurkat T-cells upon anti-CD3 stimulation.A, confocal microscopy analysis of endogenous OCRL (green) and ORP4L (red) localization in shNT- or shORP4L-transfected Jurkat T-cells. Cells were stimulated with or without anti-CD3 (10 μg ml−1) for 5 min. Scale bars, 10 μm. The histograms represent the percentage of cells with OCRL translocation predominant PM localization. The percentage of cells that represent OCRL distribution upon anti-CD3 stimulation was quantified from >50 cells of each group. The histograms showed the results from three independent experiments. B, cells with wildtype OCRL1 or OCRL1 (mut) overexpression, confocal microscopy analysis of OCRL (by Xpress) and ORP4L in Jurkat T-cells after 5-min anti-CD3 stimulation (10 μg ml−1). Scale bars, 10 μm. The histograms represent the percentage of cells with OCRL translocation predominant PM localization. The percentage of cells that represent OCRL1 distribution upon anti-CD3 stimulation was quantified from >50 cells of each group. The histograms showed the results from three independent experiments. C, BiFC analysis between OCRL1/pVn-N1 and ORP4L/pVc-C1 or OCRL1 (mut)/pVn-N1 and ORP4L/pVc-C1 in Jurkat T-cells. After transfected with the fragments for 24 h, cells were stimulated with or without anti-CD3 (10 μg ml−1) for 5 min and analyzed. Scale bars, 10 μm. The histograms showed the relative BiFC intensity from 10 cells. D, OCRL protein levels in PM, Golgi, and total lysate of Jurkat T-cells with ORP4L knockdown. Cells were stimulated with 10 μg ml−1 anti-CD3 for 5 min. The histograms showed the relative OCRL protein levels in PM (upper) and Golgi (lower) from three independent experiments. E, OCRL protein levels in PM, Golgi, and total lysate of Jurkat T-cells with OCRL1 or OCRL1 (mut) overexpression. Cells were stimulated with 10 μg ml−1 anti-CD3 for 5 min. OCRL were detected by Xpress antibody. The histograms showed the relative OCRL protein levels in PM (upper) and Golgi (lower) from three independent experiments. ∗∗p < 0.01; ∗∗∗p < 0.001; Student's t test. BiFC, bimolecular fluorescence complementation; CD3, cluster of differentiation 3; OCRL, oculocerebrorenal syndrome of Lowe 1; OSBP; oxysterol-binding protein, ORP4L; OSBP-related protein 4L; PI(4,5)P2, phosphatidylinositol 4,5-diphosphate; PM, plasma membrane.
Next, we examined the translocation of exogenous overexpressed Xpress-tagged OCRL upon anti-CD3 stimulation in Jurkat T-cells. The results showed that anti-CD3 stimulation similarly induced exogenous OCRL translocation from the Golgi to the PM and increased its colocalization with ORP4L (Fig. 3B). However, this translocation was not observed for the OCRL (mut) incapable of binding to ORP4L (Fig. 3B). In BiFC assay, the OCRL and ORP4L can evoked a BiFC signal, which was enhanced upon anti-CD3 stimulation; however, these signals were undetectable in OCRL (mut) and ORP4L-transfected cells (Fig. 3C). Cell fractionation followed by the Western blot analysis provided further evidence for the translocation of OCRL from the Golgi to the PM upon anti-CD3 stimulation (Fig. 3, D and E). Anti-CD3 stimulation increased the OCRL protein level in PM, decreased it in Golgi, but these phenotypes were abolished in ORP4L knockdown cells (Fig. 3E). Similarly, anti-CD3 stimulation results in increase of exogenous OCRL, but not OCRL (mut) protein translocation from Golgi to PM (Fig. 3E). These results suggested that with the assistance of ORP4L, anti-CD3 stimulation activated OCRL translocation from the Golgi to the PM.
OCRL mediates PI(4,5)P2 consumption at the PM by binding ORP4L
PI(4,5)P2 is a major inositol 5-phosphatase substrate that shows signaling properties, and its content in membranes could be affected by PI phosphatase activities (13). Since ORP4L-mediated PLCβ3 translocation also triggered the hydrolysis of PI(4,5)P2, we further analyzed the changes of PI(4,5)P2 after OCRL translocation to the PM in Jurkat T-cells.
In Jurkat T-cells with OCRL1 knockdown, wildtype OCRL1 or OCRL1 (mut) were re-overexpressed for rescue experiments (Fig. 4A). We analyzed the PI(4,5)P2 contents in PM by using PI(4,5)P2 indicator GFP-PHPLCδ1 domain (31). In the resting cells, OCRL1 knockdown resulted in PI(4,5)P2 abundantly accumulated at the PM. Anti-CD3 stimulation decreased the PI(4,5)P2 level in control cells, but this decrease were reduced in OCRL1 knockdown cells (Fig. 4, B and C). In rescue experiments, OCRL1 re-overexpression abolish the effects on PI(4,5)2 accumulation, whereas no rescue was observed with OCRL1(mut) (Fig. 4, B and C). As a second method, the PMs were isolated for dot-blot to detect PI(4,5)P2 contents. We found the similar results as analysis by PI(4,5)P2 indicator (Fig. S3A).This observation indicated that OCRL consumed PI(4,5)P2 at the PM dependent on its binding to ORP4L.
Figure 4.
OCRL-ORP4L interaction is required for PI(4,5)P2consumption in the PM.A, Western blot showing OCRL expression in Jurkat T-cells expressing PI(4,5)P2 probe GFP-PHPLCδ1. Cells were subjected to control (shNT), OCRL1 knockdown (shOCRL1) alone, or combined with OCRL1 rescued expression (shOCRL1+OCRL1) or combined with OCRL1(Mut) rescued expression (shOCRL1+OCRL1(Mut)). Cells were transduced with lentivirus expressing shNT or shOCRL1 for 48 h, before transduction with lentivirus expressing OCRL1 or OCRL1(Mut). After additional 48 h of culture, the cells were used for analysis. B, PI(4,5)P2 levels at the PM as detected by PI(4,5)P2 probe GFP-PHPLCδ1 in Jurkat T-cells treated as panel (A). Cells were stimulated with 10 μg ml−1 anti-CD3 for 5 min before analyzed. The cells with OCRL1 knockdown or rescue expression were considered for analysis, and the relative PI(4,5)P2 levels expressed as fold change are shown in C. The data represent mean ± SD (n = 10 cells). Scale bar, 10 μm. D, Western blot showing ORP4L expression in Jurkat T-cells expressing PI(4,5)P2 probe GFP-PHPLCδ1. The cells subjected to control (shNT), ORP4L knockdown (shORP4L) alone, or combined with ORP4L rescued expression (shORP4L+ORP4L) or combined with ORP4L (Mut) rescued expression (shORP4L + ORP4L (Mut)). Cells were transduced with lentivirus expressing shNT or shORP4L for 48 h, before transduction with lentivirus expressing ORP4L or ORP4L (Mut). After additional 48 h of culture, the cells were used for analysis. E, PI(4,5)P2 levels at the PM as detected by PI(4,5)P2 probe GFP-PHPLCδ1 in Jurkat T-cells treated as panel (D). Cells were stimulated with 10 μg ml−1 anti-CD3 for 5 min before analyzed. The cells with ORP4L knockdown or rescue expression were considered for analysis, and the relative PI(4,5)P2 levels expressed as fold change are shown in F. The data represent mean ± SD (n = 10 cells). Scale bar, 10 μm. ∗∗∗p < 0.001; Student's t test. CD3, cluster of differentiation 3; OCRL, oculocerebrorenal syndrome of Lowe 1, OSBP, oxysterol-binding protein; ORP4L, OSBP-related protein 4L; PI(4,5)P2, phosphatidylinositol 4,5-diphosphate; PLC; phosphoinositide phospholipase C; PM, plasma membrane.
Similarly, in Jurkat T-cells with ORP4L knockdown, wildtype ORP4L or ORP4L (mut) were re-overexpressed (Fig. 4D). ORP4L knockdown completely prevented OCRL and PLCβ3 translocation, therefore we found an intense PI(4,5)P2 accumulation at the PM in the ORP4L knockdown cells with or without anti-CD3 stimulation. Re-overexpression of the wildtype ORP4L abolished the PI(4,5)P2 accumulation, while the ORP4L (mut) only partly rescued this phenotype (Figs. 4, E and F and S3B). These results suggested that PI(4,5)P2 at the PM was consumed by OCRL and PLCβ3, both of them dependent of ORP4L.
OCRL is essential for cytosol and mitochondria Ca2+ homeostasis of Jurkat T-cells
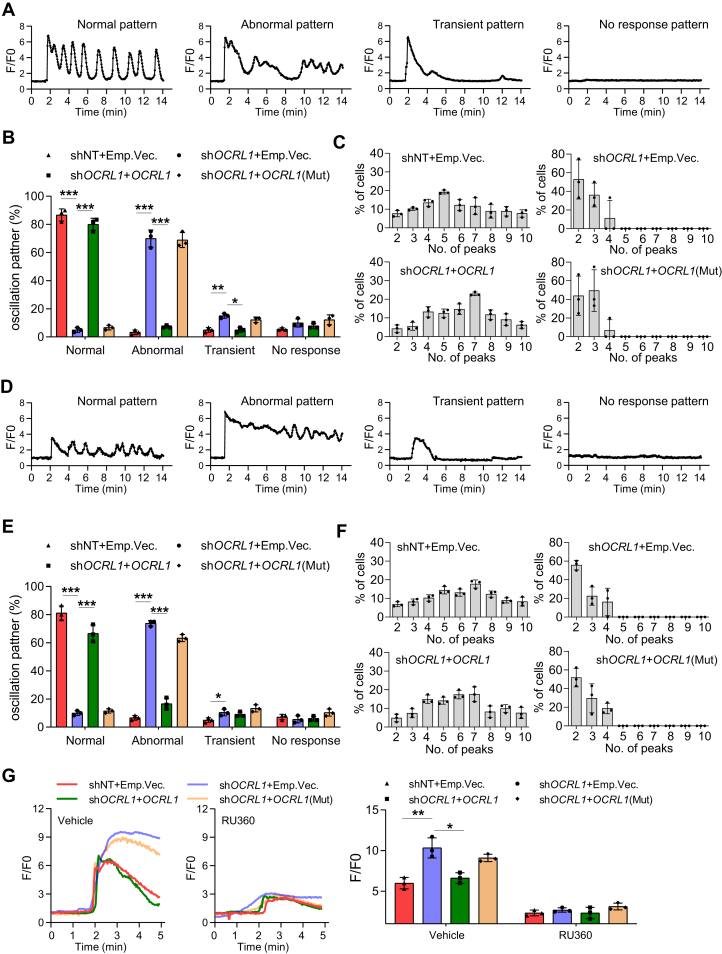
Ca2+ signaling has been closely associated with PLC-mediated PI(4,5)P2 breakdown, namely through IP3-induced Ca2+ release from the ER (15). Therefore, we measured the cytosol Ca2+ oscillation in OCRL1 knockdown Jurkat T-cells after low concentration of anti-CD3 stimulation. To analyze results quantitatively, we classified intracellular Ca2+ signal patterns into four groups (Fig. 5A): normal oscillation pattern consists of repetitive spike-shape Ca2+ rises which interval are about 2 min; abnormal oscillation consists of a combination of continuous Ca2+ rise and oscillation; transient pattern consists of one transient Ca2+ rises; no-response pattern consists of no Ca2+ rises during observation periods. We found most of the control cells displayed normal oscillation pattern upon anti-CD3 stimulation, and OCRL1 knockdown decreased the normal pattern responding cells, but increased the abnormal and transient pattern responding cells (Fig. 5B). In rescue experiments, OCRL1 but not OCRL1(mut) re-overexpression abolished the reduced normal pattern resulting from OCRL1 knockdown (Fig. 5B). We quantified frequency of normal pattern oscillation. The number of peaks seen during stimulation was 2 to 10 peaks in control cells, but there were only 2 to 4 peaks in OCRL1 knockdown cells, the reduction can be rescued by OCRL1, but not the OCRL1(mut) (Fig. 5C).
Figure 5.
OCRL modulates cytosolic and mitochondrial Ca2+homeostasis in Jurkat T-cells.A, the four types of cytosolic calcium oscillation patterns in Jurkat T-cells upon low concentration of anti-CD3 (1 μg ml−1) stimulation. B, the percentage of cells represent cytosolic calcium oscillation patterns from three independent experiments. Changes in [Ca2+] were recorded as F/F0 ratio using Fluo-4-AM in confocal Ca2+ imaging. For each experiment, >30 cells of each group were quantified. C, the percentage of cells represent different number of calcium peak in cells with normal oscillation pattern. D, the four types of mitochondrial calcium oscillation patterns in Jurkat T-cells upon low concentration of anti-CD3 (1 μg ml−1) stimulation. Changes in [Ca2+] were recorded as F/F0 ratio using Rhod-2 in confocal Ca2+ imaging. E, the percentage of cells represent mitochondrial calcium oscillation patterns from three independent experiments. For each experiment, >30 cells of each group were quantified. F, the percentage of cells represent different number of calcium peak in cells with normal oscillation pattern. G, high concentration of anti-CD3 (10 μg ml−1)-induced [Ca2+] peaks in mitochondrial of Jurkat T-cells with or without MCU inhibitor RU360 pretreatment (5 μM, 30 min). Changes in [Ca2+] were recorded as the F:F0 ratio using Rhod-2 in confocal Ca2+ imaging. Average [Ca2+] peak amplitudes were shown from three independent experiments. For each experiment, >30 cells of each group were quantified. The data represent mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; Student's t test. CD3, cluster of differentiation 3; MCU, mitochondrial Ca2+ uniporter; OCRL, oculocerebrorenal syndrome of Lowe 1.
Constitutive Ca2+ oscillations released periodically from the ER are taken up by mitochondria via the MCU (17), and T-ALL cells had mitochondrial Ca2+ oscillation follow the same patterner as cytosol Ca2+ oscillation (32). We next analyzed the mitochondrial Ca2+ oscillation patterns. OCRL1 knockdown also decreased the mitochondrial normal pattern and the number of peaks, which can be rescued by OCRL1, but not the OCRL1(mut) (Fig. 5, D–F). To study whether the amount of PI(4,5)P2 affects the Ca2+ signal patterns, we pretreated Jurkat T-cells with exogenous PI(4,5)P2, the moderate addition of PI(4,5)P2 increased the normal oscillation pattern and the number of peaks. Whereas, the excess PI(4,5)P2 produces opposite effect and results in decreased normal pattern and number of peaks but increased abnormal pattern (Fig. S4, A and B).
Excessive Ca2+ taken up by MCU into mitochondria results in mitochondrial Ca2+ overload. To strengthen the link between OCRL function and mitochondrial, we next assayed Ca2+ uptake by mitochondrial by MCU. OCRL1 knockdown markedly increased the Ca2+ peak amplitude in mitochondria (Fig. 5G) in high concentration of anti-CD3–stimulated cells, whereas OCRL1 but not OCRL1 (mut) re-overexpression reduced this amplitude (Fig. 5G). To test whether OCRL1 knockdown induces mitochondrial Ca2+ overloading via MCU complex, we blocked the pore with the MCU inhibitor Ruthenium 360 (Ru360). The highly peak amplitude of OCRL1 knockdown cells was significantly reduced in Ru360 pretreated cells (Fig. 5G). Altogether, these results suggest that OCRL1 knockdown leads to excess PI(4,5)P2 accumulation in PM and cytosol and mitochondrial Ca2+ dyshomeostasis in T-ALL cells.
OCRL sustains mitochondrial function and T-ALL cell survival in vivo
Mitochondrial Ca2+ overload results in mitochondrial permeability transition pore opening, release of cytochrome c, activation of caspase3/7, and ATP synthesis inhibition (19). We next asked whether the Ca2+ overloading in mitochondrial have a detrimental effect and lead to cell death. OCRL1 knockdown decreased the mitochondrial membrane potential as measured by JC-1 (Fig. 6A), decreased ATP production and oxygen consumption rate (Fig. 6, B and C), and also increased the Caspase 3 activity and accelerated cell death (Fig. 6, D and E). These phenotypes were all rescued by wildtype OCRL1 but not OCRL1 (mut) re-overexpression (Fig. 6, A–E).
Figure 6.
OCRL regulates mitochondrial function and cell survival in vivo.A, flow cytometry analysis of mitochondrial membrane potential by JC-1 staining in Jurkat T-cells transduced with the indicated lentiviruses. The histograms expressed as ratio of JC-1 aggregates (red) and monomer (green) (Red/Green). B–E, the ATP levels (B), OCR (C), caspase-3 activity (D) and cell death rate (E) of Jurkat-T cells transduced with the indicated lentiviruses. F and G, flow cytometric analysis of engrafted Jurkat T-cells in the bone marrow (F) and spleen (G) of NSG mice. Jurkat T-cells transduced with the indicated lentiviruses were injected via the tail vein, and after 6 weeks, the mice were sacrificed for analysis. The data represent mean ± SD (n = 3 for A–E, n = 4 for F and G). ∗∗∗p < 0.001; Student's t test. OCR, oxygen consumption rate; OCRL, oculocerebrorenal syndrome of Lowe 1.
A xenograft model was established in NSG mice to determine the contribution of OCRL to T-ALL cell survival in vivo. Mice were injected with Jurkat T-cells with OCRL1 gene modifications. At 6 weeks postinjection, the mice were scarified, and the engraftment of T-ALL cells were detected. We found the percentage of engrafted cells in bone marrow and spleen was significantly lower in mice injected with OCRL1 knockdown cells. Whereas, this reduction was abolished only in mice injected with OCRL1 re-overexpression cells, but not the OCRL1(mut) re-overexpression cells (Fig. 6, F and G).
Discussion
Previous studies have shown that OCRL in Lowe syndrome patient cells translocates from the Golgi to the PM after Rac activation, and its deficiency causes PI(4,5)P2 to accumulate at the PM (6). This is a new perspective on Lowe syndrome pathophysiology. It demonstrates that the Golgi/trans-Golgi network is not the only organelle affected by loss of OCRL function and raises the question of the role of OCRL-induced PM perturbation in the pathophysiology of Lowe syndrome. According to this finding, an anomalous mode of OCRL action may exist in some diseases in which OCRL1 expression is misregulated. In this study, we found that OCRL interacts specifically with ORP4L, and anti-CD3 stimulation enhances the colocalization between OCRL and ORP4L, facilitating more OCRL to be translocated from the Golgi to the PM in T-ALL cells. Confocal imaging showed the redistribution of OCRL and colocalization with ORP4L at the PM. This phenomenon occurs only when the interaction sites of OCRL and ORP4L remain intact. These findings suggest that OCRL anchors to ORP4L upon translocation from the Golgi to the PM.
As an inositol polyphosphate 5-phosphatase that dephosphorylates PI(4,5)P2, OCRL plays an important role in controlling the PI(4,5)P2 content of membranes. Translocation of OCRL increases hydrolysis of PI(4,5)P2 at the membrane, converting it to PI(4)P, thereby maintaining the balance between PI(4,5)P2 and PI(4)P (33). When OCRL1 was knocked down, the balance was broken and PI(4,5)P2 abundantly accumulated at the PM. In addition, PI(4,5)P2 is a precursor of IP3 and diacylglycerol after PLC activation, which is essential for early signaling of cell surface receptors. ORP4L plays an important role in this process (24). PLC is activated upon anti-CD3 stimulation (23), and the accumulation of PI(4,5)P2 results in a corresponding increase in the second messenger IP3. Similar to our observation in OCRL1 knockdown cells, glutamate regulates Ca2+ oscillations in a concentration-dependent manner, low concentration increases oscillation frequency, but high concentration disrupts oscillation and results in single high amplitude (34). Mitochondrial Ca2+ is a double-edged sword. The low levels of Ca2+ is required for ATP production, but extreme level Ca2+ leads to loss of mitochondrial. Thus, the moderate amount of PI(4,5)P2 in PM used for hydrolysis is important for IP3/Ca2+ signaling and cell survival. OCRL1 knockdown resulting in excessive PI(4,5)P2 hydrolysis by PLCβ3 leads to excessive ER Ca2+ release and disruption of constitutive Ca2+ oscillation pattern and cell death.
In recent years, great progress has been made in understanding the role played by OCRL in cellular metabolism. OCRL appears to regulate many processes within the cell, most of which depend on the coordination of membrane dynamics with actin cytoskeleton remodeling (35). Here, we identify a previously unknown translocation mechanism of OCRL in T-ALL cells that may influence T-ALL progression. This finding is novel, but further studies are needed to determine the importance of OCRL in T-ALL cells and the uniqueness of this translocation mechanism of OCRL in T-ALL cells. Because of the difference in expression between normal T-cells and T-ALL cells, we connected OCRL to another protein, ORP4L, which plays an important role in T-ALL and confirmed their interaction. Another question is whether there is an alternative enzyme to OCRL in T-ALL cells. The inositol polyphosphate 5-phosphatase INPP5B is closely related to OCRL, sharing a similar substrate specificity, domain organization, overlapping subcellular distribution and similar interaction partners (36). In particular, OCRL and INPP5B are the only human 5-phosphatases with a RhoGAP-like domain, both widespread in vertebrates (37). Determining whether INPP5B affects the physiologic effects of OCRL depletion is one of the necessary topics of follow-up research. Finally, as OCRL appears as a new factor regulating energy metabolism in T-ALL, its possible use as a target for T-ALL treatment needs further verification by in vivo experiments.
In conclusion, in this study, we revealed that OCRL controls the activity of ORP4L upon translocation from the Golgi to the PM, by consuming PI(4,5)P2 together with PLCβ3 to maintain Ca2+ homeostasis. The role of OCRL in T-ALL cells survival suggests that it may be a viable therapeutic target for T-ALL, providing a new idea to the treatment of this deadly malignancy.
Experimental procedures
Human leukocyte specimens and cell lines
The human leukocytes and cell lines used were cultured as described in our previous study (23). Briefly, peripheral blood mononuclear cells from healthy human donors and T-ALL patients were isolated by Ficoll-Hypaque gradient centrifugation, followed by purified using an Enhanced Human T Cell Recovery Column Kit (Cedarlane). The Jurkat and Molt-4 cells were maintained in RPMI1640 containing 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37 °C in a humidified incubator with 5% CO2.
Cell transfection
High-titer lentivirus (>109 transduction units per mL) expressing nontargeting shRNA (shNT), OCRL1-specific shRNA (shOCRL1, shOCRL1#2), ORP4L shRNA (shORP4L), or the OCRL1 cDNA, OCRL1(mut) cDNA, ORP4L cDNA, and the GFP-PHPLCδ1 domain cDNA were packaged by Shanghai GenePharma (Shanghai,China). The shRNA targeting sequences used were: ORP4L: 5′-TCAGAGTCAAGCTCAGGTGTA-3′; OCRL1: 5′-GGTTCCCTGCCATTTTTCA-3′; OCRL1#2: 5′-TGAGAGAGCGCCGCTTTGA-3′. For single lentivirus transduction, cells were transduced and culture for 72 h before used. For two separate lentivirus transductions, cells were transduced with the shRNA lentivirus and cultured for 48 h, followed by the cDNA lentivirus transduction and culturing for additional 48 h. For lentivirus infection, 1 × 105 cells were resuspended in 100 μl of medium supplemented with polybrene (5 μg/ml) and containing lentivirus [multiplicity of infection = 100] in 24-well culture plates. Spinfection was conducted by spinning the plates at 250 g for 1 h at 25 °C. Then 400 μl of medium was added dropwise on top of each well and cultured at 37 °C.
Quantitative RT-PCR
Total RNA was isolated with TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. RNA samples were reverse transcribed using random hexamer primers in the presence of RNase Inhibitor (Takara Bio). Quantitative RT-PCR was performed with SYBR Premix EX Taq (Takara Bio) using the CFX96 Sequence Detection System (Bio-Rad Laboratories). A relative quantification analysis was performed using the ΔΔ cycle threshold method, and the expression levels were normalized using actin as endogenous control. Relative gene expression is presented as log (2−ΔΔ cycle threshold). The primer sequences are as following: ORP4L (sense 5′-CCCTTCACTAAGGCCGCATC-3′, anti-sense 5′-GAACCCCAAGAGGAGTCTTCG-3′); OCRL1 (sense 5′-CACTGACCTGGGATCTTTG-3′, anti-sense 5′-CCAGCTGAATCCGAAATCC-3′); Actin (sense 5′-GGCATCCTCACCCTGAAGTA-3′, anti-sense 5′-AGGTGTGGTGCCAGATTTTC-3′).
Cell proliferation and cell death assay
Cell proliferation was analyzed by using the Click-iT Plus EdU Alexa Fluor 488 Flow Cytometry Assay Kit. Cells were treated with 10 μM EdU for 1 h and stained with Invitrogen Alexa Fluor 488 picolyl azide, according to the protocol for the Invitrogen Click-iT Plus EdU Alexa Fluor 488 Flow Cytometry Assay Kit. Cells were then analyzed by flow cytometry using 488 nm excitation (for Click-iT EdU Alexa Fluor 488 dye).
Cell death was analyzed by LIVE/DEAD Fixable Dead Cell Stain Kits (Life Technologies) according to the manufacturer's instructions. Briefly, cells were washed once with PBS and then incubated with LIVE/DEAD Fixable Dead Cell Stain in PBS for 30 min at room temperature in the dark. After washing with PBS with 1% FBS, cells were resuspended in PBS with 1% FBS and analyzed using flow cytometer (FACSAriaTM, BD), and the data were analyzed by FlowJo_V10 software.
Coimmunoprecipitation
Cells were washed twice with ice-cold PBS and incubated on ice for 30 min with 1 ml of lysis buffer (50 mM Tris-Cl, 150 mM NaCl, 0.5 mM MgCl2, 10% glycerol, and 0.5% Triton X-100, pH 8.0) supplemented with protease inhibitor mixture (Roche Applied Science). Cell lysates were centrifuged for 15 min at 15,000g. The supernatant was preabsorbed with 50 μl of Protein G-agarose (Invitrogen) for 1 h at 4 °C. The recovered supernatant was incubated overnight with OCRL1 or control antibody at 4 °C. 50 μl of Protein G-agarose was added to the lysate–antibody mixture and incubated for 2 h at 4 °C on a roller. The beads were washed four times with lysis buffer and boiled in SDS-PAGE loading buffer. Samples were resolved on 10% SDS-polyacrylamide gels and subjected to Western blot analysis with antibodies.
GST pulldown assay
The wildtype ORP4L, OCRL1, OCRL1(mut), or ORP4L(mut) cDNA were cloned into pGEX-4T-1 vector. These constructs were transformed into E. coli RosettaTM (DE3) (Novagen) and cultured at 37 °C to A600 = 0.5 to 1.0, followed by induction with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside to induced protein expression for 16 to 18 h at 18 °C. The cells were collected, and crude bacterial lysates were prepared by sonication in lysis buffer 1 (50 mM Tris-Cl, 150 mM NaCl, 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride, pH 8.0) in the presence of the protease inhibitor mixture (Roche Applied Science). Bacterial lysates were centrifuged for 20 min at 12,000g, the soluble protein supernatants were collected, and the protein concentration detected by using BCA Protein Assay Kit (Beyotime). Approximately equal amounts of different GST fusions were incubated with glutathione-Sepharose 4B beads (GE Healthcare) for 1 h at 4 °C, then the samples were centrifuged at 1300 g for 1 min to collect GST protein-beads complexes. For pulldown, Jurkat T-cells were washed twice with cold PBS, lysed in lysis buffer, and shaken for 30 min on ice, and the lysate was cleared by a 10-min centrifugation at 12,000 rpm in a microcentrifuge. Jurkat T-cells lysates were then added and incubated with GST protein–beads complexes at 4 °C overnight. Then the beads were washed three times with lysis buffer, resuspended into 2 × SDS-PAGE loading buffer at 98 °C for 5 min, and resolved on 10% SDS-polyacrylamide gels for Western blotting. Approximately equal amounts of different GST fusions used were verified by immunoblotting with anti-GST antibody.
Immunofluorescence microscopy
For protein staining, cells were seeded onto coverslips, stimulated with anti-CD3 for the indicated times, and then fixed with 4% paraformaldehyde for 30 min at room temperature followed by permeabilization with 0.1% Triton X-100 for 5 min and blocking with 10% FBS for 30 min at room temperature. Cells were then incubated with primary antibodies in 5% FBS at 4 °C overnight. After washing three times (10 min each) with PBS, cells were incubated with fluorescent secondary antibody conjugates at 37 °C for 30 min followed by staining with Hoechst 33342 at room temperature for 10 min. Antibodies are used as following: Rabbit anti-ORP4L (Sigma-Aldrich, 1:200), mouse anti-OCRL (Santa Cruz, 1:50), Alexa Fluor 488-Goat Anti-Mouse IgG (H + L) secondary antibody (Thermo Fisher Scientific, 1:200), and Alexa Fluor 546 Goat Anti-Rabbit IgG (H + L) secondary antibody (Thermo Fisher Scientific, 1:200). The specimens were analyzed using Olympus FV3000 laser-scanning confocal microscope system.
Yeast 2-hybrid analysis
To identify the interaction regions of OCRL and ORP4L, full-length OCRL1 or ORP4L in bait vector pGBKT7 and the truncated fragments of ORP4L or OCRL1 in prey vector pGADT7 (Takara Bio) were cotransformed into the yeast stain AH109 before plating on synthetic defined medium without Leu, Trp (SD/2-) and without Ade, His, Leu, and Trp (SD/4-) plates and culturing at 30 °C. The colonies that appeared were then transferred onto membrane for X-gal assay. The interaction was monitored as growth on SD/4- and by X-gal cleavage assay. The vectors pGBKT7-53 and pGADT7-T were used as positive controls because the murine p53 protein (53) can interact with SV40 large T-antigen (T) in yeast, while pGBKT7-Lam and pGADT7-T were used as negative controls because human lamin C protein (Lam) cannot interact with T in yeast. The empty plasmid pGBKT7 was used to exclude self-activating activity.
Homology modeling and docking simulation
The structure prediction of partial ORP4L was generated using Rosetta (38, 39). Hydrogen atoms were added to the protein using the MOE (MOE 2015.10. Chemical Computing Group, Inc.) before carrying out the docking studies. The structures of ORP4L and OCRL (PDB: 2QV2) were subjected to an energy minimization protocol. The atomic partial charges were calculated with the Amber12:EHT force field, and all possible ionization states were generated at pH 7.0 using the MOE suite. ORP4L was docked into OCRL using MOE software. The top scoring conformation was used for the binding model analysis.
In vitro phosphatase activity assays
Cells were incubated with 1 μM soluble short-chain diC8-PI(4,5)P2 (Echelon Biosciences) for 10 min at room temperature in culture medium, then the PM was prepared by using Plasma Membrane Protein Isolation and Cell Fractionation Kit (Invent Biotechnologies, Inc.) according to the manufacturer’s instructions. The assay mixture consisted of 50 μl of assay buffer (50 mM Hepes, pH 7.0, 100 mM KCl, 6 mM MgCl2, 0.6 mM CaCl2, 2 mM EGTA), and 10 μg of PM was incubated on ice for 10 min. The reaction was started by the addition of 5 μg of OCRL or OCRL(mut) recombinant proteins and incubation at 37 °C for 10 min. After the reaction, The PM pellet was centrifuged and collected for analysis of PI(4)P and PI(4,5)P2 levels by dot blot.
BiFC assay
BiFC constructs using fragments derived from Venus were generated, and wildtype or mutated ORP4L and OCRL1 cDNAs were inserted into pVn-N1, pVc-N1, pVn-C1 and pVc-C1 vectors (40). The OCRL1/pVn-N1 and ORP4L/pVc-N1, OCRL1/pVn-C1 and ORP4L/pVc-C1, OCRL1/pVn-N1 and ORP4L/pVc-C1, OCRL1/pVc-N1 and ORP4L/pVn-C1, or OCRL1(Mut)/pVn-N1 and ORP4L/pVc-C1 vectors were cotransfected into Jurkat T-cells by using an 4D-Nucleofector System (Lonza) according to the manufacturer’s instructions. After cultured for 24 h, cells were stimulated with or without 10 ug/ml anti-CD3 for 5 min and then fixed with 4% paraformaldehyde for 15 min at 4 °C. The BiFC fluorescence was detected by using Olympus FV3000 confocal Microscope.
Isolation of Golgi and PM fractions
PM and Golgi fractions were prepared by using Plasma Membrane Protein Isolation and Cell FractionationKit (Invent Biotechnologies, Inc) and Golgi Apparatus Enrichment Kit (Invent Biotechnologies, Inc) according to the manufacturer’s instructions.
Dot blots for PI(4,5)P2 and PI(4)P levels
PI(4,5)P2 and PI(4)P of PMs were released by incubation with lysis buffer (50 mM Tris, pH 7.5, 300 mM NaCl, 5 mM EGTA, 20 mM DTT, 2% Triton X-100 and 50 mM NaF) at room temperature for 30 min. Dot blots were conducted as described (41). Briefly, after centrifugation, 1 μl of suspension was spotted onto nitrocellulose membrane (Bio-Rad Laboratories), probed with PI(4,5)P2 antibody (1:500, Echelon Biosciences), PI(4)P antibody (1:500, Echelon Biosciences), or PM internal loading control pan-cadherin antibody (Santa Cruz), and detected using an HRP-conjugated secondary antibody and enhanced chemiluminescence. Experiments were repeated at least three times.
Ca2+ oscillation and amplitude measurement
Cells (1 × 105) were incubated with 1 μM Fluo-4 AM (for cytosolic Ca2+ measurement) or 2 μM Rhod-2 AM (for mitochondrial Ca2+ measurement) for 15 min at 37 °C in extracellular calcium buffer (130 mM NaCl, 5 mM KCl, 1.5 mM CaCl2, 1 mM MgCl2, 25 mM Hepes, pH 7.5, 1 mg/ml BSA, and 5 mM glucose) in dark, after which they were collected and resuspended in extracellular calcium buffer for an additional incubation at 25 °C for 30 min to permit dye de-esterification. Then, cells were plated onto a glass-bottomed dish and excited with low-intensity 488-nm laser excitation (for Fluo-4 AM) or 516-nm laser excitation (for Rhod-2 AM). Images were acquired at 3-s intervals under time-lapse mode by Olympus FV3000 confocal Microscope. For oscillation measurement, fluorescence was collected for 1 min before the low concentration of anti-CD3 (1 ug/ml) was added into suspension. For amplitude measurement, fluorescence was collected for 1 min before the high concentration of anti-CD3 (10 ug/ml) was added into suspension. Image data were subsequently analyzed using ImageJ (National Institutes of Health) and are presented as a ratio of F/F0 in final results, where F0 represents baseline fluorescence intensity in each cell.
Mitochondrial membrane potential measurement
The mitochondrial membrane potential was detected by using JC-1 dye (Beyotime). 5 × 105 cells were suspended in 0.5 ml PBS containing 1 μM JC-1 and incubated for 15 min at 37 °C in dark. After washed once with PBS, the green and red fluorescence were detected by flow cytometer (FACSAriaTM, BD). The green fluorescence represents JC-1 monomers, whereas red fluorescence represents JC-1 aggregates.
Oxygen consumption assay
Oxygen consumption was analyzed by using MitXpress-Xtra-HS (Cayman Chemical). 2 × 105 cells were transferred into fresh culture medium containing 1% FBS, then 10 μl of porphyrin-based phosphorescent oxygen-sensitive probe was added, and the cells were equilibrated at 37 °C. The assay was read by a microplate reader. The maximal rate of oxygen consumption is proportional to the change in probe fluorescence during the linear phase of the assay.
ATP measurement
ATP levels were measured using ATP Assay Kit (Abcam). All the reagents such as ATP standard dilution, ATP reaction mix, and Background reaction mix were prepared according to manufacturer’s protocol. ATP standards were prepared to obtain standard curve. 1 × 106 cells were seeded into 6-well plates and lysed with lyolysis on ice. The samples were centrifuged for 5 min at 13,000g to remove insoluble material. The supernatants were collected and transferred to new tubes. Before loading on 96-well plate, volume of all samples was adjusted to 50 μl with ATP Assay Buffer. Reaction mix (50 μl) was added into each standard and sample well, and 50 μl of background reaction mix was added into the background control sample wells. The plate was incubated at room temperature for 30 min protected from light. Absorbance was detected at 570 nm using a microplate reader. The results are presented as the ratio between the test and control values.
Fluorogenic caspase 3 activity assay
1 × 106 cells were washed with PBS and lysis by cell lysis buffer (Cell Signaling Technology). After centrifuged at 4 °C for 10 min at 12,000g, cell lysates were diluted to a concentration of approximately 5 μg/μl. Caspase activity assay (Cell Signaling Technology) was performed according to the manufacturer’s instructions. Fluorescence was detected by excitation of 380 nM and emission of 460 nm and expressed in relative fluorescence units.
Human T-ALL cell xenograft in vivo
The 4- to 6-weeks-old female NSG mice (Beijing Biocytogen) were kept under pathogen-free conditions in the Laboratory Animal Center, Jinan University. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Laboratory Animal Ethics Committee at Jinan University. To establish an engraftment model, Jurkat T-cells were transfected with lentivirus and cultured for 48 h, then injected (2 × 107 cells/0.2 ml of PBS) via the tail vein. Weekly monitoring of mice for circulating leukemia cells in peripheral blood was performed by analysis of human CD45 expression with flow cytometry. At 6 weeks postengraftment, mice were killed. Cells were then isolated from spleens by mechanical disaggregation and red blood cell lysis and collected from bone marrow by flushing of femurs with PBS. The leukemia cells burden in spleens and bone marrow were staining with CD45 antibody and assessed by flow cytometer (FACSAriaTM, BD).
Western blot analysis
Cells were suspended in lysis buffer (50 mM Tris-Cl, pH8.0, 150 mM NaCl, 0.5 mM MgCl2, 10% glycerol, 1% Triton X-100, 0.1% SDS) with protease inhibitor cocktail (Roche Diagnostics) on ice for 30 min. Samples were centrifuged at 4 °C for 10 min at 12,000g. The supernatants were collected, and protein concentrations were measured using the BCA Protein Assay Kit (Beyotime) according to the manufacturer’s instructions. Protein extracts were run on a 10 or 12% SDS polyacrylamide gel before they were transferred to a polyvinylidene difluoride membrane. Membranes were blocked with 5% milk for 1 h and incubated with primary antibodies overnight, followed by incubation with the secondary antibodies for 1 h at room temperature. Antibodies are used as following: anti-ORP4L (Sigma-Aldrich, 1:1000), anti-OCRL (Santa Cruz, 1:200), anti-pan-cadherin (Santa Cruz, 1:200), anti-GM130 (Thermo Fisher Scientific, 1:3000), anti-Canexin (Cell signaling, 1:1000), anti-Cytochrome c (Santa Cruz, 1:200), anti-p-PDH (Novus Biologicals, 1:100), anti-PDH (Cell signaling, 1:1000), anti-actin (Proteintech Group, 1:3000), Goat anti-rabbit IgG (H + L), HRP conjugate antibody (Proteintech Group, 1:3000), Goat anti-mouse IgG (H + L), and HRP conjugate antibody (Proteintech Group, 1:3000).
Statistical analyses
The data were expressed as mean ± SD from at least three independent experiments. All comparisons between groups were made by unpaired two-tailed Student's t test. p values of <0.05 were considered statistically significant.
Data availability
All data generated for this study are included within this article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
H. C., C. L., Y. T., J. Z., M. X., J. X., S. L., Z. T., and C. L. investigation. W. Z. and D. Y. conceptualization; W. Z. and D. Y. supervision; W. Z., V. M. O., and D. Y. funding acquisition; H. C. and W. Z. writing-original draft. V. M. O., M. W.-B., and M. L. writing-review and editing.
Funding and additional information
This work was supported by grants from the NSFC for Young Scientists of China (Grant 31900548 to W. Z.), NSFC of China (Grant 32071280 to D. Y.), the Academy of Finland (Grants 285223 and 322647 to V. M. O.), the Sigrid Jusélius Foundation, and the Magnus Ehrnrooth Foundation (V. M. O.).
Reviewed by members of the JBC Editorial Board. Edited by George M. Carman
Contributor Information
Daoguang Yan, Email: tydg@jnu.edu.cn.
Wenbin Zhong, Email: winbeyz@163.com.
Supporting information
References
- 1.Attree O., Olivos I.M., Okabe I., Bailey L.C., Nelson D.L., Lewis R.A., et al. The Lowe's oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 1992;358:239–242. doi: 10.1038/358239a0. [DOI] [PubMed] [Google Scholar]
- 2.Hichri H., Rendu J., Monnier N., Coutton C., Dorseuil O., Poussou R.V., et al. From Lowe syndrome to dent disease: correlations between mutations of the OCRL1 gene and clinical and biochemical phenotypes. Hum. Mutat. 2011;32:379–388. doi: 10.1002/humu.21391. [DOI] [PubMed] [Google Scholar]
- 3.Dressman M.A., Olivos-Glander I.M., Nussbaum R.L., Suchy S.F. Ocrl1, a PtdIns(4,5)P(2) 5-phosphatase, is localized to the trans-Golgi network of fibroblasts and epithelial cells. J. Histochem. Cytochem. 2000;48:179–190. doi: 10.1177/002215540004800203. [DOI] [PubMed] [Google Scholar]
- 4.Choudhury R., Diao A., Zhang F., Eisenberg E., Saint-Pol A., Williams C., et al. Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol. Biol. Cell. 2005;16:3467–3479. doi: 10.1091/mbc.E05-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faucherre A., Desbois P., Satre V., Lunardi J., Dorseuil O., Gacon G. Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum. Mol. Genet. 2003;12:2449–2456. doi: 10.1093/hmg/ddg250. [DOI] [PubMed] [Google Scholar]
- 6.Faucherre A., Desbois P., Nagano F., Satre V., Lunardi J., Gacon G., et al. Lowe syndrome protein Ocrl1 is translocated to membrane ruffles upon Rac GTPase activation: a new perspective on Lowe syndrome pathophysiology. Hum. Mol. Genet. 2005;14:1441–1448. doi: 10.1093/hmg/ddi153. [DOI] [PubMed] [Google Scholar]
- 7.Erdmann K.S., Mao Y., McCrea H.J., Zoncu R., Lee S., Paradise S., et al. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell. 2007;13:377–390. doi: 10.1016/j.devcel.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vicinanza M., Di Campli A., Polishchuk E., Santoro M., Di Tullio G., Godi A., et al. OCRL controls trafficking through early endosomes via PtdIns4,5P(2)-dependent regulation of endosomal actin. EMBO J. 2011;30:4970–4985. doi: 10.1038/emboj.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Leo M.G., Staiano L., Vicinanza M., Luciani A., Carissimo A., Mutarelli M., et al. Autophagosome-lysosome fusion triggers a lysosomal response mediated by TLR9 and controlled by OCRL. Nat. Cell Biol. 2016;18:839–850. doi: 10.1038/ncb3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ungewickell A., Ward M.E., Ungewickell E., Majerus P.W. The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13501–13506. doi: 10.1073/pnas.0405664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhury R., Noakes C.J., McKenzie E., Kox C., Lowe M. Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J. Biol. Chem. 2009;284:9965–9973. doi: 10.1074/jbc.M807442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nandez R., Balkin D.M., Messa M., Liang L., Paradise S., Czapla H., et al. A role of OCRL in clathrin-coated pit dynamics and uncoating revealed by studies of Lowe syndrome cells. Elife. 2014;3 doi: 10.7554/eLife.02975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramos A.R., Ghosh S., Erneux C. The impact of phosphoinositide 5-phosphatases on phosphoinositides in cell function and human disease. J. Lipid Res. 2019;60:276–286. doi: 10.1194/jlr.R087908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Jefferson A.B., Auethavekiat V., Majerus P.W. The protein deficient in Lowe syndrome is a phosphatidylinositol-4,5-bisphosphate 5-phosphatase. Proc. Natl. Acad. Sci. U. S. A. 1995;92:4853–4856. doi: 10.1073/pnas.92.11.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berridge M.J., Irvine R.F. Inositol phosphates and cell signalling. Nature. 1989;341:197–205. doi: 10.1038/341197a0. [DOI] [PubMed] [Google Scholar]
- 16.Bagur R., Hajnoczky G. Intracellular Ca(2+) sensing: its role in calcium homeostasis and signaling. Mol. Cell. 2017;66:780–788. doi: 10.1016/j.molcel.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foskett J.K., Philipson B. The mitochondrial Ca(2+) uniporter complex. J. Mol. Cell Cardiol. 2015;78:3–8. doi: 10.1016/j.yjmcc.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyman L., Karbowski M., Lederer W.J. Regulation of mitochondrial ATP production: ca(2+) signaling and quality control. Trends Mol. Med. 2020;26:21–39. doi: 10.1016/j.molmed.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvo-Rodriguez M., Hou S.S., Snyder A.C., Kharitonova E.K., Russ A.N., Das S., et al. Increased mitochondrial calcium levels associated with neuronal death in a mouse model of Alzheimer's disease. Nat. Commun. 2020;11:2146. doi: 10.1038/s41467-020-16074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietrangelo A., Ridgway N.D. Bridging the molecular and biological functions of the oxysterol-binding protein family. Cell Mol. Life Sci. 2018;75:3079–3098. doi: 10.1007/s00018-018-2795-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C., JeBailey L., Ridgway N.D. Oxysterol-binding-protein (OSBP)-related protein 4 binds 25-hydroxycholesterol and interacts with vimentin intermediate filaments. Biochem. J. 2002;361:461–472. doi: 10.1042/0264-6021:3610461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan G., Cao X., Liu B., Li C., Li D., Zheng J., et al. OSBP-related protein 4L promotes phospholipase Cbeta3 translocation from the nucleus to the plasma membrane in Jurkat T-cells. J. Biol. Chem. 2018;293:17430–17441. doi: 10.1074/jbc.RA118.005437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong W.B., Yi Q., Xu B., Li S.Q., Wang T., Liu F.P., et al. ORP4L is essential for T-cell acute lymphoblastic leukemia cell survival. Nat. Commun. 2016;7:12702. doi: 10.1038/ncomms12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong W., Xu M., Li C., Zhu B., Cao X., Li D., et al. ORP4L extracts and presents PIP2 from plasma membrane for PLCbeta3 catalysis: targeting it eradicates leukemia stem cells. Cell Rep. 2019;26:2166–2177.e2169. doi: 10.1016/j.celrep.2019.01.082. [DOI] [PubMed] [Google Scholar]
- 25.Guy C.S., Vignali D.A.A. Organization of proximal signal initiation at the TCR:CD3 complex. Immunol. Rev. 2009;232:7–21. doi: 10.1111/j.1600-065X.2009.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Transy C., Moingeon P.E., Marshall B., Stebbins C., Reinherz E.L. Most anti-human CD3 monoclonal antibodies are directed to the CD3 epsilon subunit. Eur. J. Immunol. 1989;19:947–950. doi: 10.1002/eji.1830190525. [DOI] [PubMed] [Google Scholar]
- 27.Park D.J., Rho H.W., Rhee S.G. CD3 stimulation causes phosphorylation of phospholipase C-gamma 1 on serine and tyrosine residues in a human T-cell line. Proc. Natl. Acad. Sci. U. S. A. 1991;88:5453–5456. doi: 10.1073/pnas.88.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deindl S., Kadlecek T.A., Brdicka T., Cao X., Weiss A., Kuriyan J. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129:735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 29.Torelli G.F., Paolini R., Tatarelli C., Soriani A., Vitale A., Guarini A., et al. Defective expression of the T-cell receptor-CD3 zeta chain in T-cell acute lymphoblastic leukaemia. Br. J. Haematol. 2003;120:201–208. doi: 10.1046/j.1365-2141.2003.04044.x. [DOI] [PubMed] [Google Scholar]
- 30.Kerppola T.K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varnai P., Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao X., Chen J., Li D., Xie P., Xu M., Lin W., et al. ORP4L couples IP(3) to ITPR1 in control of endoplasmic reticulum calcium release. FASEB J. 2019;33:13852–13865. doi: 10.1096/fj.201900933RR. [DOI] [PubMed] [Google Scholar]
- 33.Prosseda P.P., Luo N., Wang B., Alvarado J.A., Hu Y., Sun Y. Loss of OCRL increases ciliary PI(4,5)P2 in Lowe oculocerebrorenal syndrome. J. Cell Sci. 2017;130:3447–3454. doi: 10.1242/jcs.200857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X., Yang Z., Han L., Li X., Feng M., Zhang T., et al. Raloxifene neutralizes the adverse effects of glutamate on cultured neurons by regulation of calcium oscillations. Mol. Med. Rep. 2015;12:6207–6214. doi: 10.3892/mmr.2015.4191. [DOI] [PubMed] [Google Scholar]
- 35.Mehta Z.B., Pietka G., Lowe M. The cellular and physiological functions of the Lowe syndrome protein OCRL1. Traffic. 2014;15:471–487. doi: 10.1111/tra.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams C., Choudhury R., McKenzie E., Lowe M. Targeting of the type II inositol polyphosphate 5-phosphatase INPP5B to the early secretory pathway. J. Cell Sci. 2007;120:3941–3951. doi: 10.1242/jcs.014423. [DOI] [PubMed] [Google Scholar]
- 37.Ooms L.M., Horan K.A., Rahman P., Seaton G., Gurung R., Kethesparan D.S., et al. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 2009;419:29–49. doi: 10.1042/BJ20081673. [DOI] [PubMed] [Google Scholar]
- 38.Raman S., Vernon R., Thompson J., Tyka M., Sadreyev R., Pei J.M., et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77:89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song Y., DiMaio F., Wang R.Y., Kim D., Miles C., Brunette T., et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong W., Zhou Y., Li S., Zhou T., Ma H., Wei K., et al. OSBP-related protein 7 interacts with GATE-16 and negatively regulates GS28 protein stability. Exp. Cell Res. 2011;317:2353–2363. doi: 10.1016/j.yexcr.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 41.Li J.L., Tanhehco E.J., Russell B. Actin dynamics is rapidly regulated by the PTEN and PIP2 signaling pathways leading to myocyte hypertrophy. Am. J. Physiol-heart C. 2014;307:H1618–H1625. doi: 10.1152/ajpheart.00393.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated for this study are included within this article.