Abstract
Lung cancer is the leading cause of cancer death all over the world. The majority (80-85 %) of lung cancer cases are classified as non-small cell lung cancer (NSCLC). Within NSCLC, adenocarcinoma (AC) and squamous cell carcinoma (SCC) are the most often recognized. The histological and immunohistochemical examination of NSCLC is a basic diagnostic tool, but insufficient for comprehensive therapeutic decisions. In some NSCLC patients, mainly adenocarcinoma, molecular alterations in driver genes, like EGFR, KRAS, HER2, ALK, MET, BRAF, RET, ROS1, and NTRK are recognized. The frequency of some of those changes is different depending on race, and between smokers and non-smokers. The molecular diagnostics of NSCLC using modern methods, like next-generation sequencing, is essential in estimating targeted, personalized therapy. In recent years, a breakthrough in understanding the importance of molecular studies for the precise treatment of NSCLC has been observed. Many new drugs were approved, including tyrosine kinase and immune checkpoint inhibitors. Clinical trials testing novel molecules like miRNAs and trials with CAR-T cells (chimeric antigen receptor - T cells) dedicated to NSCLC patients are ongoing.
Keywords: non-small cell lung cancer, driver genes, molecular alterations, targeted therapies
Introduction
According to data from the World Health Organization from 2020, lung cancer is the second most frequently diagnosed cancer, with 12,2 % of new cases a year. Regarding mortality, lung cancer is responsible for 18,2 % of cancer-related deaths (Global Cancer Observatory https://gco.iarc.fr/, accessed 20 April 2023). The high death ratio may be caused by 70 % of lung cancer cases being diagnosed in an advanced or metastatic stage where radical therapy (surgery or radiotherapy) is impossible to implement (Lemjabbar-Alaoui et al., 2015[47]). According to clinical classification, lung cancer is divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), where the latter accounts for about 80-85 % of all lung cancer cases. The majority of NSCLC morphological recognition is based on a small amount of tissue (fine-needle aspiration or bronchoscopic biopsies) and two main subtypes can be distinguished: adenocarcinoma (AC) and squamous cell carcinoma (SCC). ACC accounts for 40-50 %, and SCC about 20-30 % of NSCLC cases (Osmani et al., 2018[63]; Zheng, 2016[100]). ACC and SCC show positive immunohistochemical reactions (IHC) for specific markers (Table 1(Tab. 1); Reference in Table 1: Zheng, 2016[100]). These IHC reactions demonstrate high sensitivity and specificity (Kriegsmann et al., 2019[46]). Figure 1(Fig. 1) shows the results of some IHC reactions in ACC and SCC. Some NSCLC cases could not be diagnosed only with regard to morphological features and remained NSCLC-NOS (NOS- not otherwise specified). IHC investigation of these samples leads to a more precise diagnosis (Righi et al., 2014[75]).
Table 1. IHC markers used in NSCLC diagnostics (Zheng, 2016, changed).
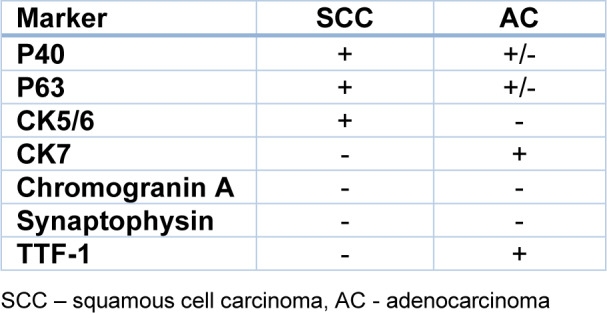
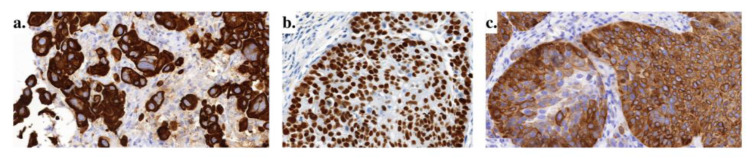
Figure 1. Positive IHC reactions in NSCLC. a) Adenocarcinoma - the tumor cells stain strongly for cytokeratin CK7, 400x; b) Squamous cell carcinoma - the tumor cells are positive for p40 and c) positive for cytokeratin CK5/6, 400x. The tissue samples were obtained from archived paraffin blocks collected in the Department of Oncological Pathology University Clinical Hospital in Poznań (photos and IHC reactions A. Grodzka, K. Kowalska, M. Krzyżaniak).
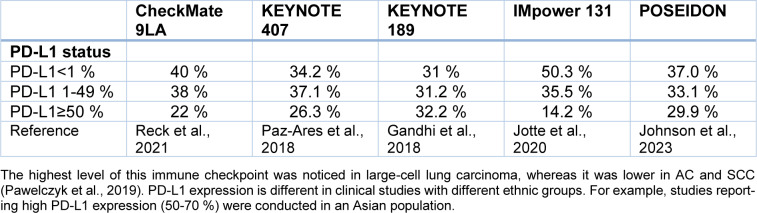
IHC is also used to examine PD-L1 status in NSCLC patients. This protein is a ligand of one of the immune checkpoints. Based on Keynote 001 lung cohort results, two cutoff points (1 % and 50 %) were established (Garon et al., 2015[28]). The higher one (50 %) was recognized as the predictive marker of first Pembrolizumab and further Atezolizumab and Cemiplimab monotherapy (Reck et al., 2016[73]). Many evaluations were performed to assess PD-L1 prevalence. In the Polish study, IHC PD-L1 expression in ≥1 % of cancer cells was observed in 32,5 % of NSCLC patients (Pawelczyk et al., 2019[65]). High expression in ≥50 % of tumor cells were detected in approximately 23 to 28 % of advanced NSCLC in the Pembrolizumab registration trial (Reck et al., 2016[73]). The data from large clinical trials are summarized in Table 2(Tab. 2) (References in Table 2: Gandhi et al., 2018[26]; Johnson et al., 2023[40]; Jotte et al., 2020[42]; Pawelczyk et al., 2019[65]; Paz-Ares et al., 2018[66]; Reck et al., 2021[72]).
Table 2. PD-L1 status in NSCLC patients according to five clinical trials.
For example, studies in which high PD-L1 expression (50-70 %) was reported were conducted on the Asian population. Unfortunately, clinical series that correlated PD-L1 expression with clinicopathologic and/or molecular variables and/or survival have reported conflicting results. Probably, not only differences in ethnicity and/or histologic types but also differences in the PD-L1 IHC method can be responsible for this (Mino-Kenudson, 2016[59]).
NSCLC patients, compared to SCLC, show a higher frequency of driver gene alterations (Vollbrecht et al., 2015[88]). It is known that their accumulation successively increases uncontrolled cell proliferation (Tomasetti et al., 2017[84]). Regarding the complex genetics of NSCLC, primary attention is paid to molecular diagnostics. The alterations in EGFR and KRAS genes are the most frequently observed in NSCLC. ALK, ROS1, HER2, BRAF, MET, and RET or NRTK are other driver genes involved in NSCLC oncogenesis. Molecular alterations within these genes are mainly point mutations, amplifications, chromosome rearrangement, or protein overexpression (Ren et al., 2022[74]; Zhu et al., 2017[102]).
Molecular Diagnostics in NSCLC and the Driver Genes
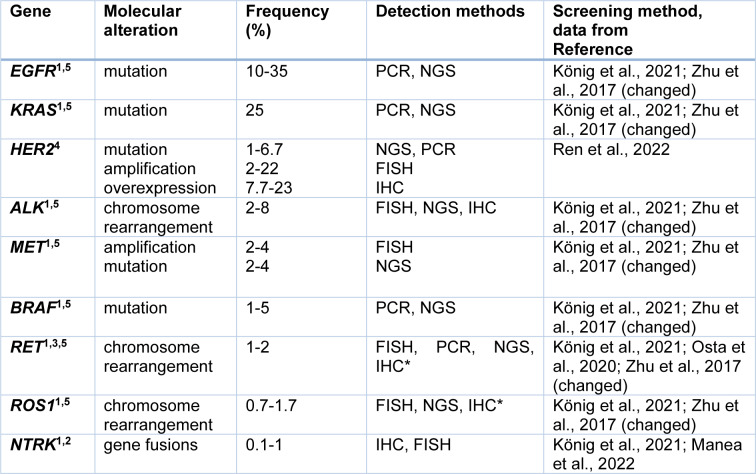
Many methods, like PCR, qPCR, RT-PCR, FISH, and NGS, are involved in assessing NSCLC molecular alterations (Table 3(Tab. 3); References in Table 3: König et al., 2021[45]; Manea et al., 2022[56]; Osta et al., 2020[64]; Ren et al., 2022[74]; Zhu et al., 2017[102]). The role of IHC is limited only to the evaluation of protein overexpression or expression of pathologic one. FISH is helpful mainly for detecting chromosome rearrangements. PCR methods identify chosen DNA sequences. NGS enables the simultaneous evaluation of specific genes using a cancer gene panel. This method can also be used for transcriptomic and epigenetics analysis (Dong et al., 2019[20]). However, according to an international survey conducted by the International Association for the Study of Lung Cancer (IASLC), the adoption of molecular testing for lung cancer is suboptimal. Most patients with molecular alterations were only tested for EGFR and ALK. Unfortunately, there are still many limitations, like testing cost and access (Smeltzer et al., 2020[77]).
Table 3. The main driver mutations in NSCLC and the methods used in their diagnostics.
The material for genetic studies is usually the tissue sample or the cell aspirate from a biopsy, but if the sample is insufficient, or the risk of biopsy is too high, a liquid biopsy emerges as a reasonable option (Smolle et al., 2021[78]). DNA in the blood comes from circulating tumor cells, exosomes, or circulating cell-free material (Ansari et al., 2016[3]; Chen and Zhao, 2019[11]). Liquid biopsy is not only a diagnosis of genetic alterations, but it also has the potential to monitor the response to the treatment and the development of acquired resistance (new driver mutations in cancer cells).
About 50 % of NSCLC patients do not show any known molecular alterations (Stencel et al., 2021[81]). The driver mutations are recognized mainly in AC cases (Joshi et al., 2021[41]). Only 4-5.8 % of SCC patients in Asia show EGFR mutations and 1-1.7 % KRAS molecular changes (Gou and Wu, 2014[29]; Joshi et al., 2021[41]). For AC, significant differences in the frequency of EGFR and KRAS genetic alterations are observed among Caucasian and East Asian populations (Joshi et al., 2021[41]). KRAS mutations are the most common in the Caucasian group (25-50 %), while EGFR gene mutations dominate in East Asians (27-62 %).
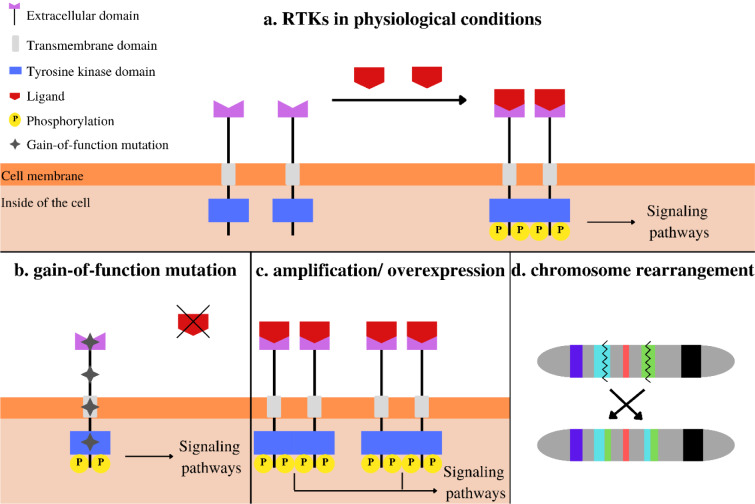
Many molecular driver alterations in NSCLC involve receptor tyrosine kinases (RTKs) genes. They are proteins responsible for controlling a wide range of biological processes by involvement in cell-to-cell communication. For example, they regulate cell growth, differentiation, and metabolism and are associated with oncogenesis. The main components of RTKs are an extracellular ligand binding domain, a single transmembrane helix, and an intracellular region - a tyrosine kinase domain. Its activation is a result of ligand binding and receptor dimerization and/or oligomerization (Figure 2a(Fig. 2); References in Figure 2: Du and Lovly; 2018[23]; Gandhi et al., 2015[27]; Noor et al., 2020[62]). It tends to conformational changes and autophosphorylation. The next step is an activation of cascades of other intracellular proteins of signaling pathways. Under normal physiological conditions, the number of RTKs and their activity is strictly regulated. Their dysregulation in cancer cells can be the result of different molecular alterations, like activation by gain-of-function mutations (Figure 2b(Fig. 2)), RTKs proteins overexpression- following gene amplification (Figure 2c(Fig. 2)), or chromosomal rearrangements (Figure 2d(Fig. 2)) (Du and Lovly, 2018[23]).
Figure 2. Schematic illustration of RTKs action and their changes; (a) physiological conditions; (b) gain-of-function mutations; (c) gene amplification/receptor overexpression; (d) chromosomal rearrangement (Du and Lovly; 2018; Gandhi et al., 2015; Noor et al., 2020).
EGFR
EGFR (ErbB1) is a transmembrane tyrosine kinase receptor, which belongs, like HER2/neu (ERBB2), HER3 (ERBB3), and HER-4 (ERBB4), to the ErbB family (Hsu et al., 2019[31]). EGFR gene mutations in cancer cells have been found in 2004 (Lynch et al., 2004[55]). They were the first targetable and predictive oncogenic driver alteration discovered in lung cancer. A deletion in exon 19 (del19) and point mutation in exon 21 of the EGFR gene (L858R) is the most frequent (König et al., 2021[45]). It results in receptor hyperactivation, and then cell proliferation is observed. EGFR mutations have significantly higher rates in never-smokers, east Asians race, females' gender, and younger age (Couraud et al., 2012[15]). The study of Warth et al. (2014[90]) shows that this alteration is almost doubled in women (21.7 % women vs 11.7 % men). EGFR IHC is useless as a diagnostic tool because targetable EGFR mutations do not influence the extent of EGFR expression at the cell surface. That is why genotyping is a golden standard for EGFR (Yang et al., 2022[96]).
NSCLC cell lines expressing mutant EGFRs show low expression of some negative regulators for EGFR (Yang et al., 2015[95]). One of them is tumor suppressor CD82, which is upregulated by wild type of EGFR but downregulated by mutant EGFRs. This change can be critical for elevated tumorigenic activity triggered by EGFR mutations. EGFR gene amplification and protein overexpression were also found in NSCLC patients but rather in SCC (Du and Lovly, 2018[23]; Hirsch et al., 2003[30]). Those alterations are not clinically meaningful.
KRAS
KRAS is a member of the rat sarcoma oncogenes family (RAS). The RAS gene encodes a low molecular weight G protein with GTPase activity that acts as molecular signal transduction of cell growth and differentiation (Xie et al., 2021[92]). KRAS mutation is one of the most frequent alterations in human cancers and NSCLC (König et al., 2021[45]; De Maglio et al., 2022[18]). It leads to the constitutive activation of the KRAS protein and the subsequent signal transduction (De Maglio et al., 2022[18]). The majority of mutations are located in codon 12, and point mutation variant G12C (glycine replaced by cysteine at codon 12) is the most frequent (Michelotti et al., 2022[58]). KRAS mutation is substantially associated with smoking status. Most patients with NSCLC harboring KRAS G12C mutation were current (40.7 %) or former (50 %) smokers. This mutation has prognostic value (Finn et al., 2021[25]). It is significantly associated with poorer prognosis. The risk of death is higher for KRAS G12C mutated patients compared with KRAS-nonmutated or KRAS other mutations by 32 % and 39 %, respectively.
HER2
Human epidermal growth factor receptor 2 (HER2 or ErbB2) is a RTK that belongs to the same family as EGFR. These receptors consist of a ligand-binding extracellular domain and an intracellular tyrosine kinase domain. Ligand binding induces a homo- or heterodimerization with other family members. HER2 heterodimerizes with other HER receptors and entails activation of downstream signaling through PI3K/AKT and RAS/MAP/ MEK pathways (Pillai et al., 2017[68]). There are three types of molecular alterations of HER2 in NSCLC: activating mutations, gene amplification, and protein overexpression (Ren et al., 2022[74]). In breast cancer, HER2 overexpression often occurs concurrently with amplification. In lung cancer, significant correlation exists between HER2 gene copy number, and protein overexpression. HER2 amplification and HER2 mutations are mutually exclusive (Uy et al., 2022[86]).
HER2 overexpression in NSCLC is a complex phenomenon with distinct molecular features making this alteration a weak biomarker in NSCLC. Its commonness is described with a wide range (Table 3(Tab. 3)), probably because of the lack of consensus on defining HER2 overexpression using IHC in NSCLC. The poor association between HER2 amplification and HER2 overexpression in NSCLC, compared to breast cancer, is probably the reason for the low effectiveness of HER2-targeted therapies in NSCLC. Although the overlap between IHC 3+ staining and HER2 amplification was found, the IHC low/negative probes were FISH-positive (Yu et al., 2022[97]). Currently, only mutation is recognized as a valid biomarker for therapeutic decisions (Li et al., 2022[50]). Mutations are mainly found in females (62.4 %), never-smokers (60.4 %), and patients with AC (Mazières et al., 2016[57]). The rates of HER2 alteration cases in NSCLC differ by country (Ren et al., 2022[74]). For the USA, mutations are observed in 3 % of cases, amplification in 3 %, and overexpression in 0 %, whereas in China, mutations are equal to 4.8 %, amplification to 15 %. Interestingly, HER2 gene alterations can coexist with TP53 mutation (Xu et al., 2020[93]).
ALK
ALK in physiological conditions is expressed in neural tissue, the small intestine, but it is not present in healthy lungs (König et al., 2021[45]). ALK regulates signaling pathways shared with other RTKs, like MAPK, PI3K-AKT, and JAK-STAT. Several fusions of the ALK gene were discovered. The most common is rearrangement with echinoderm microtubule-associated protein-like 4-four genes (EML4). Both are located on chromosome 2 but ALK at P23 and EML4 at P21. More than 21 forms of ALK-EML4 have been reported (Dong et al., 2019[20]; Liu et al., 2019[54]). The fusion is a consequence of inversion on the short arm of chromosome 2. EML4 is joined to the intracellular tyrosine kinase domain of ALK, and thus it promotes dimerization and oligomerization, inducing constitutive activation of the ALK kinase (König et al., 2021[45]; Liu et al., 2019[54]). FISH is the golden standard for detecting ALK gene rearrangements (Dong et al., 2019[20]). However, because of a strong association between ALK gene rearrangement and ALK protein expression, IHC is also a good tool for pre-screening or preliminary tests (Yang et al., 2022[96]).
This molecular alteration, with a frequency of less than 10 % of NSCLC cases, is more frequent in non-smokers, younger age patients, and AC. For example, according to data from the European Thoracic Oncology Platform Lungscape iBiobank, 5.4 % of patients with NSCLC are ALK-positive, among them 79.2 % were AC (Letovanec et al., 2018[49]). Analysis of almost 20 thousand patients with NSCLC from the USA revealed that only 2.6 % of cases were ALK- positive, most at 18-44. Moreover, non-smokers had the most significant mutation rate, and ALK alteration was very rare if EGFR, ROS1, KRAS, or BRAF changes were present (Allen et al., 2020[2]).
MET
MET proto-oncogene also encodes one of the RTKs. Hepatocyte growth factor as a ligand leads to receptor dimerization and autophosphorylation of tyrosine residues. Via signaling pathways, like MAPK or PI3K, cell proliferation, migration, invasion, angiogenesis, and the epithelial-to-mesenchymal transition can be activated (Drilon et al., 2017[21]). MET alterations found in NSCLC patients can include gene amplification, mutation, or protein overexpression leading to aberrant activation of downstream pathways (Michelotti et al., 2022[58]). MET exon 14 skipping mutation is observed in 2-4 % of cases, resulting in a decreased degradation of the MET protein and increased activation of downstream signaling pathways. METex14 mutation is recognized mainly in patients with AC (68.8 %), over 65 years old (79 %), and females (60.4 %) (Schrock et al., 2016[76]; Yang et al. 2022[96]). This molecular alteration can be identified by DNA-based sequencing in most cases. However, some METex14 skipping mutations might be caused by alterations in deep intronic regions which could not be detected by assay limited to exonic and canonical splice site sequences. METex14 skipping mutations usually coexist with overexpression of this gene but MET IHC has minimal utility as a diagnostic tool for METex14 skipping alterations (Yang et al., 2022[96]). MET changes detected by liquid biopsy occur in more patients, than MET alterations found in tissue (Ikeda et al., 2018[34]). MET alterations, mainly gene amplification, strongly impact the therapy with RTK inhibitors in EGFR-mutant NSCLC patients (Zhang et al., 2019[99]).
BRAF
BRAF, full name v-Raf murine sarcoma viral oncogene homolog B is a serine/threonine kinase, a part of the MAP/ERK signaling pathway. This pathway might be deregulated due to activating point mutation of the BRAF gene. BRAF mutations in NSCLC are rare (Table 3(Tab. 3)) and commonly occur in never-smokers, women with AC. Recently three classes of BRAFV600 mutations have been distinguished: class I- including RAS-independent kinase-activating V600 functioning as monomers; class II- RAS-independent kinase-activating nonV600 dimers; class III- RAS-dependent kinase-inactivating nonV600 heterodimers (König et al., 2021[45], Yan et al., 2022[94]). 50-80 % of BRAF mutations in NSCLC are nonV600 and belong to class II or III (Bracht et al., 2019[7]; Yan et al., 2022[94]). BRAF mutations in lung cancer can coexist with EGFR and KRAS molecular alterations (Li et al., 2014[51]). It is not common to analyze BRAF gene mutation separately, but it is encouraged to include it in the gene panel strategy (Yan et al., 2022[94]).
RET
Among NSCLC patients, RET gene alterations are infrequent (Table 3(Tab. 3)). The gene was discovered in 1985 (Takahashi et al., 1985[83]). This protooncogene on chromosome 10q11.2 encodes a transmembrane glycoprotein receptor tyrosine kinase with a ligand that belongs to the factors of the glial cell line-derived neurotrophic family (Choudhry and Drilon, 2020[13]). It can be activated by two mechanisms: RET fusions and RET point mutations (Osta et al., 2020[64]). Chromosomal rearrangement of RET can be the effect of the fusion of the 3`coding region for RET kinase domain on chromosome 10 with a 5` upstream partner gene with domain coiled-coil or LIS1 homology (Choudhry and Drilon, 2020[13]). Intrachromosomal rearrangements are found in genes such as KIF5B (72 %) and CCDC6 (23 %) (Osta et al., 2020[64]). Chimeric fusion proteins, produced by rearrangement, can cause ligand-independent constitutive activation of RET, promoting cancer cell proliferation (Drusbosky et al., 2021[22]). However, not all RET structural variants result in oncogenic fusion proteins because some are not associated with RET kinase fusions. Considering this, DNA- and RNA-based sequencing methods need to be used to discover the significance of particular structural variants (Yang et al., 2022[96]). RET fusions, discovered in 2012, are reported mainly in young, never smokers, with AC (Choudhry and Drilon, 2020[13]).
ROS1
ROS1 is a member of the insulin receptor family, and its extracellular domain is one of the largest among all human RTKs (Bubendorf et al., 2016[9]; König et al., 2021[45]). Its ligands in humans remain unknown, although, in studies with mice, chickens, and rats, ROS1 expression has been detected in the epithelial cells of the kidneys, male reproductive organs, small intestine, heart, and lungs. ROS1 activates downstream oncogenic pathways, like STAT3, PI3K/AKT/mTOR, and RAS-MAPK/ERK, which controls cell proliferation (Zhu et al., 2015[101]). Similar to ALK, ROS1 gene alteration in NSCLC is a chromosome rearrangement (Lin and Shaw, 2017[52]). The ROS1 gene is located at q21 of chromosome 6, and rearrangement is mainly concentrated in exons 32-36 (Dong et al., 2019[20]). Fourteen fusion partners have been found: the most frequent is CD74. In contrast to the ALK gene, ROS1 fusion partners do not provide dimerization domains that induce constitutive kinase activation (Lin and Shaw, 2017[52]). IHC might distinguish patients with ROS1 rearrangements, but because of low specificity, the diagnosis must be confirmed by FISH (Yang et al., 2022[96]). The molecular alteration of ROS1 is relatively rare in NSCLC patients (only approximately 1 %), and it is detected mostly in AC (86 % of NSCLC cases with ROS1 rearrangements). ROS1 alterations are slightly more frequent among women (Cui et al., 2020[17]), and non-smokers (Song et al., 2017[80]).
NTRK
The neurotrophic tyrosine receptor kinase family (NTRK or TRK) comprises NTRK1, NTRK2, and NTRK3 genes. They encode proteins of the tropomyosin receptor kinase (TRK) family, transmembrane receptor tyrosine kinases responsible for neuronal development (Manea et al., 2022[56]). Alterations of NTRK genes can be involved in carcinogenesis in neurogenic and non-neurogenic cells. NTRKs may become a part of fusion oncogenic proteins in different types of tumors but rarely in NSCLC (König et al., 2021[45]). NTRK1 and NTRK2 are preferentially expressed in SCC. It has been found that the presence of NTRK2 in SCC cases is correlated with a good prognosis (Liu et al., 2022[53]). Gene fusions affect different genomic rearrangements, but the 3' sequences of the NTRK gene are always fused to the 5' sequence of a fusion partner gene. It eventually leads to persistent activation of downstream signaling pathways involved in cell growth, proliferation, differentiation, survival, and apoptosis prevention (Liu et al., 2022[53]). Numerous fusion partners for the NTRK gene in NSCLC have been found: MPRIP, CD74, QSTM1, TPR, IRF2BP2, BCL9, LMNA, and PHF20 (Liu et al., 2022[53]). The existence of NTRK fusions and other molecular alterations characteristic of NSCLC is mutually exclusive (Manea et al., 2022[56]).
Targeted Therapies in NSCLC and Perspectives
Driver mutation genes found in NSCLC patients are the excellent target in precise and personalized therapies with tyrosine kinase inhibitors (TKIs), immune checkpoint inhibitors (ICIs), or CAR-T cells. TKIs are the first targeted drugs for NSCLC patients (Michelotti et al., 2022[58]). During the last several years, ICIs were also approved for immunotherapy of NSCLC patients (Jiang et al., 2019[38]). Many clinical trials are focused on testing different drug combinations. Studies of CAR-T cell therapy applications in solid tumors, including NSCLC, are conducted. Also, epigenetic targets, like miRNAs, can be involved in developing new therapeutic strategies in NSCLC (Ahn et al., 2020[1]; Wang et al., 2019[89]).
Tyrosine kinase inhibitors (TKIs)
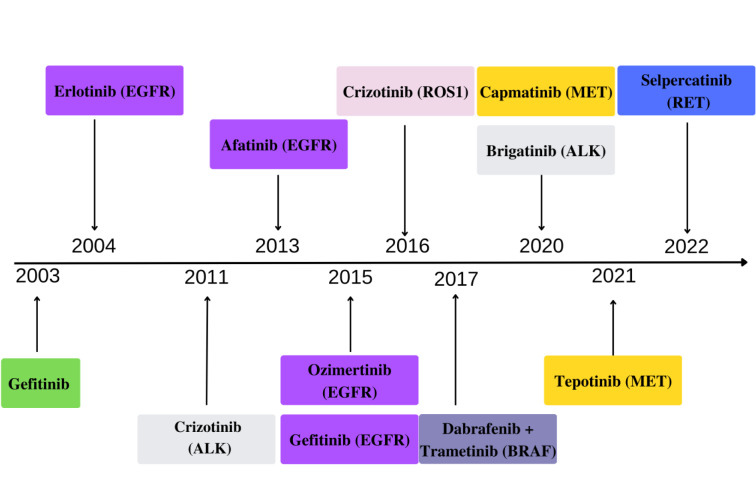
The first TKI approved by FDA for NSCLC patients was Gefitinib (Figure 3(Fig. 3); References in Figure 3: Cohen et al., 2003[14]; Michelotti et al., 2022[58]) as a treatment for patients with locally advanced or metastatic disease after failure of platinum-based and docetaxel chemotherapies (Cohen et al., 2003[14]), without indication of any gene mutation. The next was Erlotinib, whose mechanism of action was based on reverse binding to the cytoplasmic domain of EGFR as a target. During the following years, other TKIs were approved, including Afatinib and Gefitinib (EGFR mut). Osimertinib, approved in 2015, is the third-generation TKI against EGFR, both frontline and after treatment with first- or second-generation EGFR-TKIs (Zhang et al., 2019[99]). Crizotinib was the first-generation TKI for patients with ALK rearrangement. Second-generation ALK inhibitors are Ceritinib, Alectinib, and Brigatinib, followed by third-generation Lorlatinib (Michelotti et al., 2022[58]). ROS1 alterations are highly sensitive to Crizotinib and BRAFV600 to a combination of Dabrafenib with Trametinib. However, NSCLC patients with BRAF non-V600 mutations show much less sensitivity than V600 to these drugs, and novel BRAF kinase inhibitors are tested in clinical trials (Bracht et al., 2019[7]; Yan et al., 2022[94]). Over the last two years, Tepotinib (MET inhibitor) and Selpercatinib (RET inhibitor) were approved. The regular approval of Selpercatinib was in 2022. However, the first approval as an accelerated was based on the LIBRETTO-001 trial (NCT 3157128) in 2020 (Michelotti et al., 2022[58]).
Figure 3. Timeline progress in NSCLC targeted therapy with FDA-approved TKIs (Cohen et al., 2003, Michelotti et al., 2022; U.S. Food & Drug Administration https://www.fda.gov/, accessed 20 April 2023, changed).
Targeted therapies with TKIs prolong the overall survival of NSCLC patients. However, during therapy, the acquired resistance to these inhibitors is developed. More than 50 % of the resistance to the first-generation of EGFR-TKIs results from EGFR exon 20 T790M mutations (Benedettini et al., 2010[6]). It was found that amplification of MET also can be responsible for acquired resistance to EGFR TKIs, e.g., Osimertinib. MET amplification is diagnosed in 5-22 % of EGFR-mutated NSCLC patients with acquired resistance to EGFR-TKIs (Michelotti et al., 2022[58]; Zhang et al., 2019[99]). These patients should be treated simultaneously with EGFR and MET inhibitors (Zhang et al., 2019[99]). MET amplification is not connected with EGFR exon 20 T790M mutation in some patients. According to data for the Korean NSCLC patients' group (Ji et al., 2013[37]), the coexistence of T790M mutation and MET amplification was found in 11.5 % of patients, and almost the same result was obtained for the group with only MET alteration.
The mechanisms responsible for the development of acquired resistance to EGFR-TKIs can be divided into three main groups: 1) secondary mutations on EGFR or EGFR amplifications, 2) activation of new signaling pathways or different gene amplification (e.g. MET), and 3) phenotypic plasticity and epithelial-mesenchymal transition or transformation to small-cell lung cancer (Delahaye et al., 2022[19]).
Concerning the contribution of MET in EGFR-TKI resistance, many clinical trials were conducted with molecules acting as inhibitors of MET. For example, Capmatinib was tested in GEOMETRY mono-1 and Tepotinib in VISION (Wolf et al., 2020[91]). The FDA approved these drugs in 2020 and 2021, respectively (Michelotti et al. 2022[58]).
A driver oncogene RET can also be developed as a mechanism of acquired resistance to EGFR mutation during therapy with EGFR inhibitors. Most RET fusions involved in the resistance are CCDC6 (58 %) and NCOA4 (26 %) (Osta et al., 2020[64]). Selpercatinib and Pralsetinib are new RTKIs for RET fusion-positive NSCLC patients (Michelotti et al., 2022[58]). According to the results of clinical trials, Selpercatinib and Pralsetinib can be used as potent and selective inhibitors of RET fusions and mutations, irrespective of the tissue of origin (Drusbosky et al., 2021[22]). These RET-TKIs cross the blood-brain barrier and show tolerable toxicity profiles. It was described that acquired resistance to Selpercatinib can be developed during long-term RET inhibition. RETG810 mutations have been found in circulating tumor DNA and patient-xenograft model (Solomon et al., 2020[79]). However, the resistance can also be RET-independent, such as acquired MET or KRAS amplification. Currently, several novel agents targeting RET fusions in NSCLC are being tested. TAS0953/HM06 and TPX-0046 are in phase 1/2, while RXDX-105, and BOS172738 are in phase 1 (Michelotti et al., 2022[58]).
In 2020, a new inhibitor- Sotorasib was approved to treat NSCLC patients with KRAS mutation (Michelotti et al., 2022[58]). This drug is not a typical TKI inhibitor, but it is an inhibitor of the RAS GTPase family and targets a specific mutation, G12C, in the protein K-Ras. Concerning a high frequency of KRAS mutations, in many European countries, the test for KRAS as a biomarker in molecular diagnostics of NSCLC was recommended (Kerr et al., 2021[44]). KRAS mutated gene is a target of clinical trials NCT04504669 and NCT03101839 with antisense oligonucleotides (Bartolucci et al., 2022[5]). These molecules can be an emerging class of biotherapeutics for a new era of precision anti-cancer medicine.
The excellent response to inhibitors is observed not only in NSCLC patients with known driver mutations, like EGFR, KRAS, or ALK rearrangement. The results of Hu et al. (2019[32]), obtained for NSCLC Chinese patients indicate that the effect of EGFR-TKIs is also good in patients with mutations of the HER2 gene (Michelotti et al., 2022[58]; Uy et al., 2022[86]) and germline BRCA mutations (Hu et al., 2019[32]). For HER2-altered NSCLC, clinical trials are conducted with antibody-drug conjugates, like T-DXd (Trastuzumab Deruxtecan), T-DM1 (Trastuzumab Emtansine), but also with TKIs, like Pyrotinib and Poziotinib (Uy et al., 2022[86]). In NSCLC patients, several clinical trials were also conducted with PARP inhibitors. For example, Olaparib versus placebo monotherapy was tested in a multicenter, randomized, controlled phase 2 (Fennel et al., 2022[24]). A clinical trial (phase 2) has been conducted with Niraparib plus immune checkpoint inhibitor Pembrolizumab (Ramalingam et al., 2022[71]).
Immune checkpoint inhibitors (ICIs)
PD-1/PD-L1 inhibitors are promising immunotherapeutic agents approved for many cancer types, including NSCLC (Jiang et al., 2019[38]). ICIs therapies give a chance to many NSCLC patients who do not show any driver gene mutations and are excluded from the targeted therapy. Based on the Chen et al. (2021[12]) meta-analysis, it can be found that ICIs give an excellent objective response rate and duration of response. The first ICI in NSCLC was approved in March 2015. It was Nivolumab, in the second-line treatment of advanced disease stage. A few months later, Pembrolizumab was approved, and in 2016 Atezolizumab as well (Jain et al., 2018[35]). Since 2017, Pembrolizumab can be applied as first-line systemic therapy for patients with PD-L1 expression in cancer cells >50 % or as a second-line systemic therapy after progression on first-line chemotherapy, with at least 1 % PD-L1 expression in tumor cells. High PD-L1 expression is also a predictive biomarker for Atezolizumab in monotherapy (Jassem et al., 2021[36]). Recent data led to the approval of combining a single ICI (Pembrolizumab) with chemotherapy (Gandhi et al., 2018[26]).
The association between PD-L1 expression and clinicopathological features is not clear. In the study of Pawelczyk et al. (2019[65]) conducted on 866 samples of NSCLC in TMA (tissue microarray) probes, it was found that the patients with low PD-L1 expression had prolonged overall survival compared to the group with high expression, but only in AC. According to the results of Pawelczyk et al., as well as of Igarashi et al. (2016[33]), higher expression of PD-L1 is noticed in G2 and G3 in AC patients, compared to G1. PD-L1 positivity is higher in males, smokers, positive blood vessels, and lymphatic invasion (Miyazawa et al. 2019[60]).
There was a concept that high tumor mutation burden (TMB) could be one of the predictors of the response to immunotherapy with ICIs. The higher the TMB level, the more new antigens are present and the more likely the immune system is to be activated against cancer cells (Dong et al., 2019[20]). Among NSCLC patients, those with HER2 mutations show the lowest levels of PD-L1 expression. Hence, the effect of ICIs will be relatively weak (Vathiotis et al., 2021[87]).
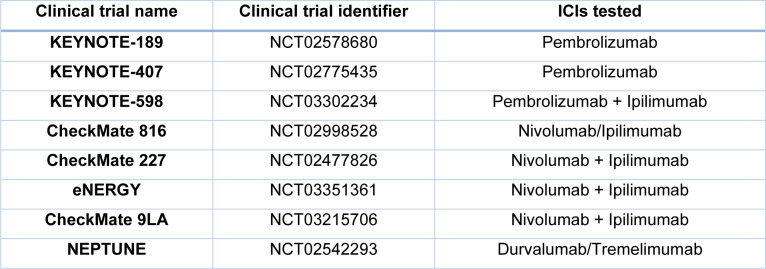
A pan-tumor retrospective analysis of participants with advanced solid tumors, including NSCLC, showed that TMB ≥175 mutations are associated with improvement in the effectiveness of Pembrolizumab monotherapy vs. chemotherapy (Cristescu et al., 2022[16]). However, large clinical trials did not confirm TMB as a useful predictive tool. In recent years, clinical trials have also been conducted on ICIs combinations (e.g., with chemotherapeutic agents and other ICIs) (Table 4(Tab. 4); References in Table 4: Paz-Ares et al., 2022[67], Pinto et al., 2019[69]; Zerdes et al., 2018[98]). Some clinical trials are focused on Ipilimumab, an anti-CTLA-4 ICI (Paz-Ares et al., 2022[67]). It was the first ICI approved in cancer immunotherapy (Tomasini et al., 2012[85]). However, this agent has shown limited efficacy as a single agent in lung cancer, compared to those obtained in melanoma. New Ipilimumab combinations with chemotherapeutic agents or other ICIs, like Nivolumab, show better effects (Lena et al., 2022[48]).
Table 4. Phase 3 trials of ICIs or their combinations in NSCLC (Zerdes et al., 2018; Pinto et al., 2019; Paz-Ares et al., 2022, changed).
CAR-T cells
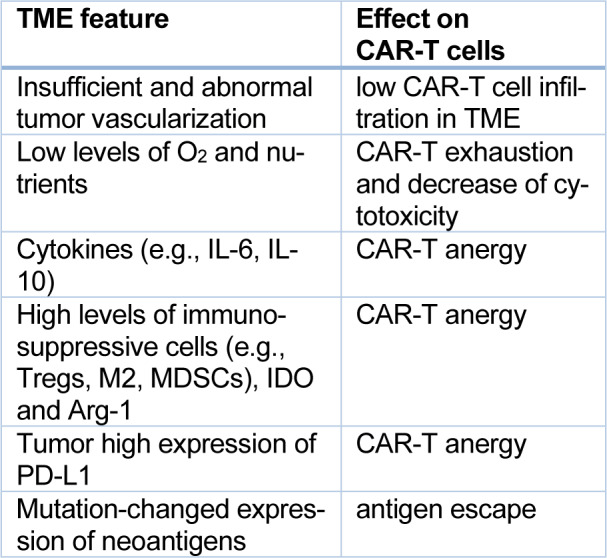
CAR-T cell therapy can be another promising way for many NSCLC patients. T lymphocytes can be genetically modified to recognize antigens of NSCLC cells, like EGFR, PD-L1, mesothelin, mucin 1, and CEA (Qu et al., 2021[70]; Chen et al., 2022[10]). So far, the CAR-T cell procedure has been dedicated to hematological diseases, and the first CAR-T cell drugs, Kymriah and Yescarta, were approved in 2017 for patients with acute lymphoblastic leukemia and diffuse large B-cell lymphoma (Styczyński, 2020[82]). Many clinical trials are conducted with CAR-T cell therapy in solid tumors, including NSCLC (Barros et al., 2022[4]; Chen et al., 2022[10]). However, few severe obstacles exist in developing this strategy in solid tumors. Among them is a low level of tumor-specific antigens, T-cells infiltration, and high tumor microenvironment immunosuppression (Qu et al., 2021[70]). CAR-T cells are applicated intravenously. However, as seen in Table 5(Tab. 5) (References in Table 5: Chen et al., 2022[10]; Johnson et al., 2022[39]), impaired tumor vasculature and other tumor microenvironment (TME) features are responsible for the immunosuppressive influence on CAR-T cells (Johnson et al., 2022[39]). Despite these difficulties, many preclinical and clinical studies have been conducted with the aim to improve CAR-T cell access in solid tumors (Nguyen et al., 2022[61]). One of the clinical trials is the NCT04153799 study of CXCR5-modified EGFE chimeric antigen receptor autologous T-cells in EGFR-positive patients with advanced NSCLC (phase 1).
Table 5. Immunosuppressive influence of TME on CAR-T cells (Chen et al., 2022; Johnson et al., 2022 changed).
miRNAs
Lung cancer cells show not only genetic but also epigenetic changes. There can be upregulation of oncogenic miRNAs, and downregulation of tumor suppressive miRNAs. Oncogenic miRNAs can be intensely involved in tumor growth, angiogenesis, epithelial-mesenchymal transition, or immune escape (Ahn et al., 2020[1]; Wang et al., 2019[89]). A great attention is paid to non-coding RNAs (miRNAs, circRNAs) in cancer stem cells responsible for cancer metastasis and drug resistance (Bryl et al., 2022[8]). Non-coding RNAs can be the new weapon in developing targeted therapies to combat cancer. For example, applying tumor suppressive miRNA mimetics or reducing oncogenic miRNAs in cancer cells seems to be a good perspective for a new therapeutic strategy. In recent years, several in vivo lung cancer models with miR-7, miR-34a, or miR-200c were tested (Ahn and Ko, 2020[1]). So far, two clinical trials with miRNAs have been conducted (Wang et al., 2019[89]). For example, in NCT02369198, a mimetic of miR16 was tested (phase 1). This potential drug consists of three components: 1) miR-16-based miRNA mimic, 2) drug delivery vehicle composed of non-living bacterial minicells (about 400 nm in size) which allow efficient drug packaging, 3) targeting moiety of nanoparticles to EGFR-expressing cancer cells with an anti-EGFR bispecific antibody (Kara et al., 2022[43]).
Conclusions
Significant correlations between driver gene alterations and therapeutic outcomes of NSCLC patients show that precise molecular diagnostics is crucial, and there is an urgent clinical need to applicate effective, targeted therapies. Hence, molecular diagnostic methods, like NGS, should become a daily practice. The unresolved problem in NSCLC patients under protein inhibitors' treatment is the acquired resistance to the drugs. Current studies should be concentrated on the mechanisms of the resistance and addressing them, as well as on looking for novel targets and tools. Therapies with ICIs, and their combinations with chemotherapy are also of great clinical interest. The novel treatment with CAR-T cell therapy or the application of miRNA-based drugs can also be a promising direction for NSCLC patients.
Notes
Anna Grodzka and Agnieszka Knopik-Skrocka contributed equally as first author.
Declaration
Conflict of interest
Maciej Bryl COI: Honoraria, consulting or advisory role and travel, accommodations, expenses: Boehringer Ingelheim, Roche/Genentech, MSD, Bristol-Myers Squibb, AstraZeneca, Takeda, Novartis, Pfizer. Other authors declare no conflict of interest.
Acknowledgments
The authors would like to thank Ms. Oliwia Piwocka from Radiobiology Laboratory, Department of Medical Physics, Greater Poland Cancer Center, Poznań, for professional language assistance.
References
- 1.Ahn YH, Ko YH. Diagnostic and therapeutic implications of microRNAs in Non-Small Cell Lung Cancer. Int J Mol Sci. 2020;21(22):8782. doi: 10.3390/ijms21228782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen T, Xiao Y, Abraham A, Redpath S, Engstrom-Melnyk J, Croix D, et al. Prevalence of ALK mutations in advanced NSCLC patients in the United States: Analysis with A real world oncology database. J Clin Oncol. 2020;38(15_Suppl):e21586. [Google Scholar]
- 3.Ansari J, Yun JW, Kompelli AR, Moufarrej YE, Alexander JS, Herrera GA, et al. The liquid biopsy in lung cancer. Genes Cancer. 2016;7:355–367. doi: 10.18632/genesandcancer.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barros LRC, Couto SCF, da Silva Santurio D, Paixão EA, Cardoso F, da Silva VJ, et al. Systematic review of available CAR-T Cell trials around the world. Cancers (Basel) 2022;14(11):2667. doi: 10.3390/cancers14112667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartolucci D, Pession A, Hrelia P, Tonelli R. Precision anti-cancer medicines by oligonucleotide therapeutics in clinical research targeting undruggable proteins and non-coding RNAs. Pharmaceutics. 2022;14(7):1453. doi: 10.3390/pharmaceutics14071453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benedettini E, Sholl LM, Peyton M, Reilly J, Ware C, Davis L, et al. Met activation in non-small cell lung cancer is associated with de novo resistance to EGFR inhibitors and the development of brain metastasis. Am J Pathol. 2010;177:415–423. doi: 10.2353/ajpath.2010.090863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bracht JWP, Karachaliou N, Bivona T, Lanman RB, Faull I, Nagy RJ, et al. BRAF mutations classes I, II, and III in NSCLC patients included in the SLLIP trial: the need for a new pre-clinical treatment rationale. Cancers (Basel) 2019;11(9):1381. doi: 10.3390/cancers11091381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryl R, Piwocka O, Kawka E, Mozdziak P, Kempisty B, Knopik-Skrocka A. Cancer stem cells - the insight into non-coding RNAs. Cells. 2022;11(22):3699. doi: 10.3390/cells11223699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bubendorf L, Büttner R, Al-Dayel F, Dietel M, Elmberger G, Kerr K, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch. 2016;469:489–503. doi: 10.1007/s00428-016-2000-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Chen F, Li J, Pu Y, Yang C, Wang Y, et al. CAR-T cell therapy for lung cancer: Potential and perspective. Thorac Cancer. 2022;13:889–899. doi: 10.1111/1759-7714.14375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13(1):34. doi: 10.1186/s40246-019-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Zhang Z, Zheng X, Tao H, Zhang S, Ma J, et al. Response efficacy of PD-1 and PD-L1 inhibitors in cflinical trials: a systematic review and meta-analysis. Front Oncol. 2021;11:562315. doi: 10.3389/fonc.2021.562315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhry NJ, Drilon A. Decade in review: a new era for RET-rearranged lung cancers. Transl Lung Cancer Res. 2020;9:2571–2580. doi: 10.21037/tlcr-20-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen MH, Williams GA, Sridhara R, Chen G, Pazdur R. FDA drug approval summary: gefitinib (ZD1839) (Iressa) tablets. Oncologist. 2003;8:303–306. doi: 10.1634/theoncologist.8-4-303. [DOI] [PubMed] [Google Scholar]
- 15.Couraud S, Zalcman G, Milleron B, Morin F, Souquet PJ. Lung cancer in never smokers - a review. Eur J Cancer. 2012;48:1299–1311. doi: 10.1016/j.ejca.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Cristescu R, Aurora-Garg D, Albright A, Xu L, Liu XQ, Loboda A, et al. Tumor mutational burden predicts the efficacy of pembrolizumab monotherapy: a pan-tumor retrospective analysis of participants with advanced solid tumors. J Immunother Cancer. 2022;10(1):e003091. doi: 10.1136/jitc-2021-003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui M, Han Y, Li P, Zhang J, Ou Q, Tong X, et al. Molecular and clinicopathological characteristics of ROS1-rearranged non-small-cell lung cancers identified by next-generation sequencing. Mol Oncol. 2020;14:2787–2795. doi: 10.1002/1878-0261.12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Maglio G, Pasello G, Dono M, Fiorentino M, Follador A, Sciortino M. The storm of NGS in NSCLC diagnostic-therapeutic pathway: How to sun the real clinical practice. Crit Rev Oncol Hematol. 2022;169:103561. doi: 10.1016/j.critrevonc.2021.103561. [DOI] [PubMed] [Google Scholar]
- 19.Delahaye C, Figarol S, Pradines A, Favre G, Mazieres J, Calvayrac O. Early steps of resistance to targeted therapies in non-small-cell lung cancer. Cancers (Basel) 2022;14(11):2613. doi: 10.3390/cancers14112613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong J, Li B, Lin D, Zhou Q, Huang D. Advances in targeted therapy and immunotherapy for non-small cell lung cancer based on accurate molecular typing. Front Pharmacol. 2019;10:230. doi: 10.3389/fphar.2019.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drilon A, Cappuzzo F, Ou SI, Camidge DR. Targeting MET in lung cancer: will expectations finally be MET? J Thorac Oncol. 2017;12:15–26. doi: 10.1016/j.jtho.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drusbosky LM, Rodriguez E, Dawar R, Ikpeazu CV. Therapeutic strategies in RET gene rearranged non-small cell lung cancer. J Hematol Oncol. 2021;14(1):50. doi: 10.1186/s13045-021-01063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer. 2018 Feb 19;17(1):58. doi: 10.1186/s12943-018-0782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fennel DA, Porter C, Lester J, Danson S, Blackhall F, Nicolson M, et al. Olaparib maintenance versus placebo monotherapy in patients with advanced non-small cell lung cancer (PIN): A multicentre, randomised, controlled, phase 2. eClinicalMedicine. 2022;52:101595. doi: 10.1016/j.eclinm.2022.101595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn SP, Addeo A, Dafni U, Thunnissen E, Bubendorf L, Madsen LB, et al. Prognostic impact of KRAS G12C mutation in patients with NSCLC: results from the European Thoracic Oncology Platform Lungscape Project. J Thorac Oncol. 2021;16:990–1002. doi: 10.1016/j.jtho.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Gandhi L, Rodriguez-Abren D, Gadgeel S, Esteban E, Felip E, De Angelis F. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378:2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 27.Gandhi S, Chen H, Zhao Y, Dy GK. First-line treatment of advanced ALK-positive non-small-cell lung cancer. Lung Cancer (Auckl) 2015;6:71–82. doi: 10.2147/LCTT.S63491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 29.Gou LY, Wu YL. Prevalence of driver mutations in non-small-cell lung cancers in the People's Republic of China. Lung Cancer (Auckl) 2014;5:1–9. doi: 10.2147/LCTT.S40817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Di Maria MV, Veve R, Bremmes RM, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 31.Hsu PC, Jablons DM, Yang CT, You L. Epidermal Growth Factor Receptor (EGFR) pathway, Yes-Associated Protein (YAP) and the regulation of Programmed Death-Ligand 1 (PD-L1) in Non-Small Cell Lung Cancer (NSCLC) Int J Mol Sci. 2019;20(15):3821. doi: 10.3390/ijms20153821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu X, Yang D, Li Y, Li L, Wang Y, Chen P, et al. Prevalence and clinical significance of pathogenic germline BRCA1/2 mutations in Chinese non-small cell lung cancer patients. Cancer Biol Med. 2019;16:556–564. doi: 10.20892/j.issn.2095-3941.2018.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igarashi T, Teramoto K, Ishida M, Hanaoka J, Daigo Y. Scoring of PD-L1 expression intensity on pulmonary adenocarcinomas and the correlations with clinicopathological factors. ESMO Open. 2016;1(4):e000083. doi: 10.1136/esmoopen-2016-000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda S, Schwaederle M, Mohindra M, Fontes Jardim DL, Kurzrock R. MET alterations detected in blood-derived circulating tumor DNA correlate with bone metastases and poor prognosis. J Hematol Oncol. 2018;11(1):76. doi: 10.1186/s13045-018-0610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain P, Jain C, Velcheti V. Role of immune-checkpoint inhibitors in lung cancer. Ther Adv Respir Dis. 2018;12:1753465817750075. doi: 10.1177/1753465817750075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jassem J, de Marinis F, Giaccone G, Vergnenegre A, Barrios CH, Morise M. Updated overall survival analysis from IMpower110: atezolizumab versus platinum-based chemotherapy in treatment-naive programmed death-ligand 1–selected NSCLC. J Thorac Oncol. 2021;16:1872–1882. doi: 10.1016/j.jtho.2021.06.019. [DOI] [PubMed] [Google Scholar]
- 37.Ji W, Choi CM, Rho JK, Jang SJ, Park YS, Chun SM, et al. Mechanisms of acquired resistance to EGFR-tyrosine kinase inhibitor in Korean patients with lung cancer. BMC Cancer. 2013;13:606. doi: 10.1186/1471-2407-13-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, Chen M, Nie H, Yuan Y. PD-1 and PD-L1 in cancer immunotherapy: clinical implications and future considerations. Hum Vaccin Immunother. 2019;15:1111–1122. doi: 10.1080/21645515.2019.1571892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson A, Townsend M, O'Neill K. Tumor microenvironment immunosuppression: a roadblock to CAR T-cell advancement in solid tumors. Cells. 2022;11(22):3626. doi: 10.3390/cells11223626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson ML, Cho BC, Luft A, Alatorre-Alexander J, Geater SL, Laktionov K, et al. Durvalumab with or without tremelimumab in combination with chemotherapy as first-line therapy for metastatic non-small-cell lung cancer: The Phase III POSEIDON Study. J Clin Oncol. 2023;41:1213–1217. doi: 10.1200/JCO.22.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi A, Mishra R, Desai S, Chandrani P, Kore H, Sunder R, et al. Molecular characterization of lung squamous cell carcinoma tumors reveals therapeutically relevant alterations. Oncotarget. 2021;12:578–588. doi: 10.18632/oncotarget.27905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jotte R, Cappuzzo F, Vynnychenko I, Stroyakovskiy D, Rodríguez-Abreu D, Hussein M, et al. Atezolizumab in combination with carboplatin and nab-paclitaxel in advanced squamous NSCLC (IMpower131): results from a randomized phase III trial. J Thorac Oncol. 2020;15:1351–1360. doi: 10.1016/j.jtho.2020.03.028. [DOI] [PubMed] [Google Scholar]
- 43.Kara G, Arun B, Calin GA, Ozpolat B. miRacle of microRNA-driven cancer nanotherapeutics. Cancers (Basel) 2022;14(15):3818. doi: 10.3390/cancers14153818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerr KM, Bibeau F, Thunnissen E, Botling J, Ryška A, Wolf J, et al. The evolving landscape of biomarker testing for non-small cell lung cancer in Europe. Lung Cancer. 2021;154:161–175. doi: 10.1016/j.lungcan.2021.02.026. [DOI] [PubMed] [Google Scholar]
- 45.König D, Savic Prince S, Rothschild SI. Targeted therapy in advanced and metastatic non-small cell lung cancer. an update on treatment of the most important actionable oncogenic driver alterations. Cancers (Basel) 2021;13(4):804. doi: 10.3390/cancers13040804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kriegsmann K, Cremer M, Zgorzelski C, Harms A, Muley T, Winter H, et al. Agreement of CK5/6, p40, and p63 immunoreactivity in non-small cell lung cancer. Pathology. 2019;51:240–245. doi: 10.1016/j.pathol.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Lemjabbar-Alaoui H, Hassan OU, Yang YW, Buchanan P. Lung cancer: Biology and treatment options. Biochim Biophys Acta. 2015;1856:189–210. doi: 10.1016/j.bbcan.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lena H, Monnet I, Bylicki O, Audigier-Valette C, Falchero L, Vergnenegre A, et al. Randomized phase III study of nivolumab and ipilimumab versus carboplatin-based doublet in first-line treatment of PS 2 or elderly (≥ 70 years) patients with advanced non–small cell lung cancer (energy-GFPC 06-2015 study) J Clin Oncol. 2022;40(16_Suppl):9011. [Google Scholar]
- 49.Letovanec I, Finn S, Zygoura P, Smyth P, Soltermann A, Bubendorf L, et al. Evaluation of NGS and RT-PCR methods for ALK rearrangement in European NSCLC patients: results from the European Thoracic Oncology Platform Lungscape Project. J Thorac Oncol. 2018;13:413–425. doi: 10.1016/j.jtho.2017.11.117. [DOI] [PubMed] [Google Scholar]
- 50.Li BT, Smit EF, Goto Y, Nakagawa K, Udagawa H, Mazières J, et al. Trastuzumab deruxtecan in HER2-mutant non-small-cell lung cancer. N Engl J Med. 2022;386:241–251. doi: 10.1056/NEJMoa2112431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, Li L, Zhu Y, Huang C, Qin Y, Liu H, et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer. 2014;110:2812–2820. doi: 10.1038/bjc.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin JJ, Shaw AT. Recent advances in targeting ROS1 in lung cancer. J Thorac Oncol. 2017;12:1611–1625. doi: 10.1016/j.jtho.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu F, Wei Y, Zhang H, Jiang J, Zhang P, Chu Q. NTRK fusion in non-small cell lung cancer: diagnosis, therapy, and TRK inhibitor resistance. Front Oncol. 2022;12:864666. doi: 10.3389/fonc.2022.864666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Ye X, Yu Y, Lu S. Prognostic significance of anaplastic lymphoma kinase rearrangement in patients with completely resected lung adenocarcinoma. J Thorac Dis. 2019;11:4258–4270. doi: 10.21037/jtd.2019.09.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 56.Manea CA, Badiu DC, Ploscaru IC, Zgura A, Bacinschi X, Smarandache CG, et al. A review of NTRK fusions in cancer. Ann Med Surg (Lond) 2022;79:103893. doi: 10.1016/j.amsu.2022.103893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazières J, Barlesi F, Filleron T, Besse B, Monnet I, Beau-Faller M, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: results from the European EUHER2 cohort. Ann Oncol. 2016;27:281–286. doi: 10.1093/annonc/mdv573. [DOI] [PubMed] [Google Scholar]
- 58.Michelotti A, de Scordilli M, Bertoli E, De Carlo E, Del Conte A, Bearz A. NSCLC as the paradigm of precision medicine at its finest: the rise of new druggable molecular targets for advanced disease. Int J Mol Sci. 2022;23(12):6748. doi: 10.3390/ijms23126748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mino-Kenudson M. Programmed cell death ligand-1 (PD-L1) expression by immunohistochemistry: could it be predictive and/or prognostic in non-small cell lung cancer? Cancer Biol Med. 2016;13:157–170. doi: 10.20892/j.issn.2095-3941.2016.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyazawa T, Marushima H, Saji H, Kojima K, Hoshikawa M, Takagi M, et al. PD-L1 Expression in non-small-cell lung cancer including various adenocarcinoma subtypes. Ann Thorac Cardiovasc Surg. 2019;25(1):1–9. doi: 10.5761/atcs.oa.18-00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen DT, Ogando-Rivas E, Liu R, Wang T, Rubin J, Jin L, et al. CAR T cell locomotion in solid tumor microenvironment. Cells. 2022;11(12):1974. doi: 10.3390/cells11121974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noor ZS, Cummings AL, Johnson MM, Spiegel ML, Goldman JW. Targeted therapy for non-small cell lung cancer. Semin Respir Crit Care Med. 2020;41:409–434. doi: 10.1055/s-0039-1700994. [DOI] [PubMed] [Google Scholar]
- 63.Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103–109. doi: 10.1016/j.semcancer.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osta BE, Ramalingam SS. RET Fusion: joining the ranks of targetable molecular drivers in NSCLC. JTO Clin Res Rep. 2020 May 13;1(3):100050. doi: 10.1016/j.jtocrr.2020.100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pawelczyk K, Piotrowska A, Ciesielska U, Jablonska K, Gletzel-Plucinska N, Grzegrzolka J, et al. Role of PD-L1 expression in non-small cell lung cancer and their prognostic significance according to clinicopathological factors and diagnostic markers. Int J Mol Sci. 2019;20(4):824. doi: 10.3390/ijms20040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. 2018;379:2040–2051. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 67.Paz-Ares LG, Ramalingam SS, Ciuleanu TE, Lee JS, Urban L, Caro RB. First-line nivolumab plus ipilimumab in advanced NSCLC: 4-year outcomes from the randomized, open-label, Phase 3 CheckMate 227 Part 1 trial. J Thorac Oncol. 2022;17:289–308. doi: 10.1016/j.jtho.2021.09.010. [DOI] [PubMed] [Google Scholar]
- 68.Pillai RN, Behera M, Berry LD, Rossi MR, Kris MG, Johnson BE, et al. HER2 mutations in lung adenocarcinomas: A report from the Lung Cancer Mutation Consortium. Cancer. 2017;123:4099–4105. doi: 10.1002/cncr.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pinto JA, Raez LE, Oliveres H, Rolfo CC. Current knowledge of Ipilimumab and its use in treating non-small cell lung cancer. Expert Opin Biol Ther. 2019;19:509–515. doi: 10.1080/14712598.2019.1610380. [DOI] [PubMed] [Google Scholar]
- 70.Qu J, Mei Q, Chen L, Zhou J. Chimeric antigen receptor (CAR)-T-cell therapy in non-small-cell lung cancer (NSCLC): current status and future perspectives. Cancer Immunol Immunother. 2021;70:619–631. doi: 10.1007/s00262-020-02735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramalingam SS, Thara E, Awad MM, Dowlat A, Haque B, Stinchcombe TE. JASPER: Phase 2 trial of first- line niraparib plus pembrolizumab in patients with advanced non–small cell lung cancer. Cancer. 2022:65–74. doi: 10.1002/cncr.33885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reck M, Ciuleanu TE, Cobo M, Schenker M, Zurawski B, Menezes J, et al. First-line nivolumab plus ipilimumab with two cycles of chemotherapy versus chemotherapy alone (four cycles) in advanced non-small-cell lung cancer: CheckMate 9LA 2-year update. ESMO Open. 2021;6(5):100273. doi: 10.1016/j.esmoop.2021.100273.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 74.Ren S, Wang J, Ying J, Mitsudomi T, Lee DH, Wang Z, et al. Consensus for HER2 alterations testing in non-small-cell lung cancer. ESMO Open. 2022;7(1):100395. doi: 10.1016/j.esmoop.2022.100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Righi L, Vavalà T, Rapa I, Vatrano S, Giorcelli J, Rossi G, et al. Impact of non-small-cell lung cancer-not otherwise specified immunophenotyping on treatment outcome. J Thorac Oncol. 2014;9:1540–1546. doi: 10.1097/JTO.0000000000000271. [DOI] [PubMed] [Google Scholar]
- 76.Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL, et al. Characterization of 298 patients with lung cancer harboring MET Exon 14 skipping alterations. J Thorac Oncol. 2016;11:1493–1502. doi: 10.1016/j.jtho.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 77.Smeltzer MP, Wynes MW, Lautuejoul S, Soo R, Ramalingam SS, Varell-Garcia M. The International Association for the Study of Lung Cancer Global Survey on Molecular Testing in Lung Cancer. J Thorac Oncol. 2020;15:1434–48. doi: 10.1016/j.jtho.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 78.Smolle E, Taucher V, Lindenmann J, Pichler M, Smolle-Juettner FM. Liquid biopsy in non-small cell lung cancer-current status and future outlook-a narrative review. Transl Lung Cancer Res. 2021;10:2237–2251. doi: 10.21037/tlcr-21-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Solomon BJ, Tan L, Lin JJ, Wong SQ, Hollizeck S, Ebata K, et al. RET solvent front mutations mediate acquired resistance to selective RET inhibition in RET-driven malignancies. J Thorac Oncol. 2020;15:541–549. doi: 10.1016/j.jtho.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song Z, Zheng Y, Wang X, Su H, Zhang Y, Song Y. ALK and ROS1 rearrangements, coexistence and treatment in epidermal growth factor receptor-wild type lung adenocarcinoma: a multicenter study of 732 cases. J Thorac Dis. 2017;9:3919–3926. doi: 10.21037/jtd.2017.09.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stencel K, Chmielewska I, Milanowski J, Ramlau R. Non-small-cell lung cancer: new rare targets-new targeted therapies-state of the art and future directions. Cancers (Basel) 2021;13(8):1829. doi: 10.3390/cancers13081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Styczyński J. A brief history of CAR-T cells: From laboratory to the bedside. Acta Haematol Pol. 2020;51(1):2–5. doi: 10.2478/ahp-2020-0002. [DOI] [Google Scholar]
- 83.Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell. 1985;42:581–588. doi: 10.1016/0092-8674(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 84.Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355(6331):1330–1334. doi: 10.1126/science.aaf9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tomasini P, Khobta N, Greillier L, Barlesi F. Ipilimumab: its potential in non-small cell lung cancer. Ther Adv Med Oncol. 2012;4(2):43–50. doi: 10.1177/1758834011431718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uy NF, Merkhofer CM, Baik CS. HER2 in non-small cell lung cancer: a review of emerging therapies. Cancers (Basel) 2022;14(17):4155. doi: 10.3390/cancers14174155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vathiotis IA, Charpidou A, Gavrielatou N, Syrigos KN. HER2 aberrations in non-small cell lung cancer: from pathophysiology to targeted therapy. Pharmaceuticals (Basel) 2021;14(12):1300. doi: 10.3390/ph14121300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vollbrecht C, Werner R, Walter RF, Christoph DC, Heukamp LC, Peifer M, et al. Mutational analysis of pulmonary tumours with neuroendocrine features using targeted massive parallel sequencing: a comparison of a neglected tumour group. Br J Cancer. 2015;113:1704–1711. doi: 10.1038/bjc.2015.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang WT, Han C, Sun YM, Chen TQ, Chen YQ. Noncoding RNAs in cancer therapy resistance and targeted drug development. J Hematol Oncol. 2019;12(1):55. doi: 10.1186/s13045-019-0748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Warth A, Penzel R, Lindenmaier H, Brandt R, Stenzinger A, Herpel E, et al. EGFR, KRAS, BRAF and ALK gene alterations in lung adenocarcinomas: patient outcome, interplay with morphology and immunophenotype. Eur Respir J. 2014;43:872–883. doi: 10.1183/09031936.00018013. [DOI] [PubMed] [Google Scholar]
- 91.Wolf J, Seto T, Han JY, Reguart N, Garon EB, Groen HJM, et al. Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer. N Engl J Med. 2020;383:944–957. doi: 10.1056/NEJMoa2002787. [DOI] [PubMed] [Google Scholar]
- 92.Xie M, Xu X, Fan Y. KRAS-mutant non-small cell lung cancer: an emerging promisingly treatable subgroup. Front Oncol. 2021;11:672612. doi: 10.3389/fonc.2021.672612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu F, Yang G, Xu H, Yang L, Qiu W, Wang Y. Treatment outcome and clinical characteristics of HER2 mutated advanced non-small cell lung cancer patients in China. Thorac Cancer. 2020;11:679–685. doi: 10.1111/1759-7714.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yan N, Guo S, Zhang H, Zhang Z, Shen S, Li X. BRAF-mutated non-small cell lung cancer: current treatment status and future perspective. Front Oncol. 2022;12:863043. doi: 10.3389/fonc.2022.863043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang CH, Chou HC, Fu YN, Yeh CL, Cheng HW, Chang IC, et al. EGFR over-expression in non-small cell lung cancers harboring EGFR mutations is associated with marked down-regulation of CD82. Biochim Biophys Acta. 2015;1852:1540–1549. doi: 10.1016/j.bbadis.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 96.Yang SR, Schultheis AM, Yu H, Mandelker D, Ladanyi M, Buttner R. Precision medicine in non-small cell lung cancer: Current applications and future directions. Semin Cancer Biol. 2022;84:184–198. doi: 10.1016/j.semcancer.2020.07.009. [DOI] [PubMed] [Google Scholar]
- 97.Yu X, Ji X, Su C. HER2-altered non-small cell lung cancer: biology, clinicopathologic features, and emerging therapies. Front Oncol. 2022;12:860313. doi: 10.3389/fonc.2022.860313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zerdes I, Matikas A, Bergh J, Rassidakis GZ, Foukakis T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene. 2018;37:4639–4661. doi: 10.1038/s41388-018-0303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Z, Yang S, Wang Q. Impact of MET alterations on targeted therapy with EGFR-tyrosine kinase inhibitors for EGFR-mutant lung cancer. Biomark Res. 2019;7:27. doi: 10.1186/s40364-019-0179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng M. Classification and pathology of lung cancer. Surg Oncol Clin N Am. 2016;25:447–468. doi: 10.1016/j.soc.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 101.Zhu Q, Zhan P, Zhang X, Lv T, Song Y. Clinicopathologic characteristics of patients with ROS1 fusion gene in non-small cell lung cancer: a meta-analysis. Transl Lung Cancer Res. 2015;4:300–309. doi: 10.3978/j.issn.2218-6751.2015.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu QG, Zhang SM, Ding XX, He B, Zhang HQ. Driver genes in non-small cell lung cancer: Characteristics, detection methods, and targeted therapies. Oncotarget. 2017;8:57680–57692. doi: 10.18632/oncotarget.17016. [DOI] [PMC free article] [PubMed] [Google Scholar]