Duchenne muscular dystrophy (DMD) (MIM: 310200) is a severe and frequent neuromuscular disorder (incidence of ∼1 in 5,500 boys) caused by mutations in the vast X-linked DMD gene (∼2.2 Mb), whose largest product, the long rod-shaped 427-kDa dystrophin isoform, is a key structural component of the striated musculature.1 Contributing to the urgency in the development of currently inexistent DMD therapies is the observation that ∼1/3 of cases arise de novo through germline mutations, often intragenic deletions that disrupt the mRNA reading frame. Critically, naturally occurring DMD gene deletions resulting in in-frame transcripts coding for internally truncated, yet partially functional, dystrophins cause the milder Becker muscular dystrophy (MIM: 300376). Hence, DMD gene manipulations yielding Becker-like dystrophins via direct coding sequence reframing or exon skipping have the potential of offering long-lasting therapeutic effects.1 Toward this end, among other technologies, CRISPR-Cas9 nucleases and adeno-associated viral (AAV) vectors are being investigated for rescuing dystrophin expression upon double-strand DNA break (DSB) formation and ensuing chromosomal end-joining.1 These experiments demonstrate that AAV/CRISPR-Cas9-based dystrophin restoration can improve striated muscle function in mice; however, a potentially insidious outcome is the identification of prevalent capture of Cas9-encoding AAV at nuclease target sites, including at Dmd exons 51 and 53.2,3 Moreover, programmable nucleases can trigger other untoward effects, e.g., locus- or chromosome-wide rearrangements.4 There is, therefore, a pressing need to expand candidate DMD genetic therapies to those based on DSB-independent genome editing systems. In a timely study published in Molecular Therapy – Nucleic Acids, Chai and coworkers5 identify adenine base editors (ABEs) and guide RNAs (gRNAs) (Figure 1) that, after implementing single base-pair substitutions (i.e., A⋅T-to-G⋅C transitions) at splicing motifs, a process that they name “single-swap” editing, lead to genotype-specific DMD repair through exon skipping. Next, the authors assemble a dual AAV ABE trans-splicing system to demonstrate in dystrophin-defective mice the amelioration of dystrophic traits at the cellular and organismal levels upon intramuscular or systemic administrations. This study identifies ABE:gRNA complexes compatible with ∼30% of DMD-causing genotypes and, notwithstanding its inherent complexity, establishes dual AAV ABE trans-splicing as a DSB-free DMD gene correction option.
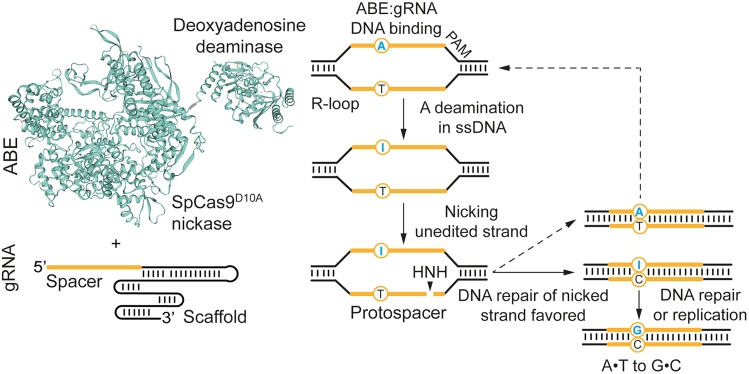
Figure 1.
Adenine base editing
Adenine base editors (ABEs) catalyze A⋅T-to-G⋅C substitutions and consist of a fusion product between a disabled or nicking Cas9, or ortholog protein, and an evolved deoxyadenosine deaminase, e.g., Escherichia coli tRNA adenosine deaminase (TadA) derivatives. Upon PAM binding, ABE:gRNA complexes form an R-loop at a gRNA-defined target sequence exposing a region of single-stranded DNA. A nucleotides in this single-stranded protospacer “bubble” become targets for the ABE effector domain that converts A nucleotides to inosine (I) intermediates preferentially within an “activity window.” Subsequently, nicking of the unedited strand induces DNA repair that installs C nucleotides opposite I intermediates with additional DNA repair events (or replication) establishing the final A⋅T-to-G⋅C transitions.
Chai and colleagues start by testing in HEK293T cells and DMD iPSCs, ABE:gRNA complexes that, depending on their ABE component, i.e., ABE8e6 or ABEe-NG, recognize, respectively, canonical NGG or NG PAMs. DNA sequencing assays in DMD iPSCs identified ABE:gRNA complexes yielding high-frequency target-base editing at DMD exons 51 and 45 splice acceptor (SA) motifs (71.6% and 79.3%–83.3%, respectively). As a result, robust expression of Becker-like dystrophins was detected in cardiomyocytes differentiated from base-edited DMD iPSCs. Interestingly, it was found that DMD exon 51 targeting serendipitously established an in-frame 11-nucleotide deletion instead of the intended exon skipping, presumably due to internal cryptic splice site usage. This finding per se stresses the importance of carefully assessing gene-edited products even when using subtle DSB-independent systems. Moreover, various amounts of bystander A⋅T-to-G⋅C transitions were also detected (range: 3.3%–91%). These bystander changes might have limited consequences as they map to either spliced-out intron or, if accompanied with the intended SA edits, to skipped exon sequences. Further investigations will be, nonetheless, necessary to probe for slight (or otherwise) splicing alterations in different cell types or contexts. Regardless, base editors with narrower “editing windows” should facilitate more favorable target-to-bystander ratios.
AAV vectors are attractive in vivo gene-editing tool delivery vehicles owing to their lack of viral genes and serotype diversity of their parental viruses. Indeed, packaging vector genomes in AAV serotype capsids with a strong tropism for certain cell types (pseudotyping) facilitates tissue-directed transductions. However, the limited AAV packaging capacity (<4.7 kb) permits delivering neither base editing nor other large constructs. To obviate this limitation, researchers are developing base editors with compact architectures,7 testing alternative delivery systems or applying dual AAV strategies in which split constructs linked to N- and C-terminal intein domains are packaged in different AAV vectors (Figure 2). Upon target cell co-transductions, intein-mediated protein trans-splicing results in the in situ assembly of full-length proteins. Indeed, dual AAV-vectored base editor trans-splicing is currently undergoing intense investigation for tackling various disease-causing mutations, including DMD mutations.8,9 In Chai et al.,5 a dual AAV ABE trans-splicing system is assembled to address frequent DMD deletions through exon skipping modulation. By exploiting high human-murine conservation over intron 44 to exon 45 junctions and guided by their earlier in vitro experiments, the authors apply dual AAVs to deliver a split version of an ABE8e variant (i.e., ABE.TadA-8eV106W), selected for its reduced off-target nucleic acid deaminase activities,6 together with a gRNA targeting the exon 45 SA region (Figure 2). To favor base editing in striated muscles over non-target organs of Dmd exon 44-deleted mice, the vector constructs were packaged in AAV serotype-9 capsids with the split ABE.TadA-8eV106W moieties being expressed through the tissue-specific CK8e promoter. Intramuscular dual AAV co-administrations of 1 × 1011 total vector genomes (VGs) per tibialis anterior (TA) led to 29.5% ± 2.7% A⋅T-to-G⋅C edits with minimal indel formation (i.e., 0.2% ± 0.1%), as determined by deep sequencing at 3 weeks post injection. Systemic dual AAV co-administrations of 1.5 × 1014 VG kg−1 and 3 × 1014 VG kg−1 via the temporal facial veins of postnatal day 2 (P2) mice yielded, in TA muscles, 5.5% ± 1.2% and 8.1% ± 3.0% edits and, in hearts, 22.0% ± 2.2% and 26.2% ± 4.4% edits, respectively, with <0.1% indels detected at 8 weeks post injection. Critically, a general dose-dependent improvement of disease-associated molecular, cellular, and functional endpoints is reported. In this regard, up to 31% and 60% of wild-type dystrophin levels estimated in treated TA and heart muscles corresponded to over 76% and 95% of dystrophin-positive myofibers and cardiomyocytes, respectively. Partial dystrophin rescue translated, in turn, in noticeable reduction of myofiber central nucleation, diameter distribution, and fibrosis, all hallmarks of muscle degeneration. Finally, Dmd exon 44-deleted mice systemically treated at P2 with low and high doses of dual AAV ABE trans-splicing particles registered 31% and 41% grip strength augmentation, respectively, when compared with their untreated counterparts. Follow-up studies will be instructive to determine the long-term effects of the local and systemic gene-editing procedures in the treated animals. Of notice, despite the aforementioned transductional and transcriptional targeting measures, significant base editing was detected in the liver (i.e., 11.1% ± 5.9%), which correlated with high VG copy numbers present specifically in this organ. These data confirm the importance of developing liver de-targeting protocols and strictly myotropic AAV capsids.10 Indeed, as dose-dependent toxicity and immunological constrains have emerged during AAV clinical applications, optimization of tissue tropism and expression will be particularly important for dual AAV trans-splicing approaches due to the necessarily higher particle amounts required for maximizing co-expression and full-length protein assembly. Moreover, besides seeking the elimination of the observed editing at two of five top-ranked in silico-predicted candidate off-target sites,5 unbiased genome- and transcriptome-wide assessments of off-target effects, including gRNA-independent deamination, will complement the safety profile of DMD-targeting ABE:gRNA complexes. Concluding, Chai and colleagues demonstrate that AAV-vectored ABE trans-splicing can induce robust synthesis of Becker-like dystrophins in striated muscles of dystrophic mice upon DSB-free splice site knockout and exon skipping.5 The resulting improvement in pathological traits measured at the molecular, tissue, and functional levels validates the use of this platform for the efficacious testing and optimization of the growing number of ABE reagents in vivo and expands the range of potential treatment modalities for DMD patients.
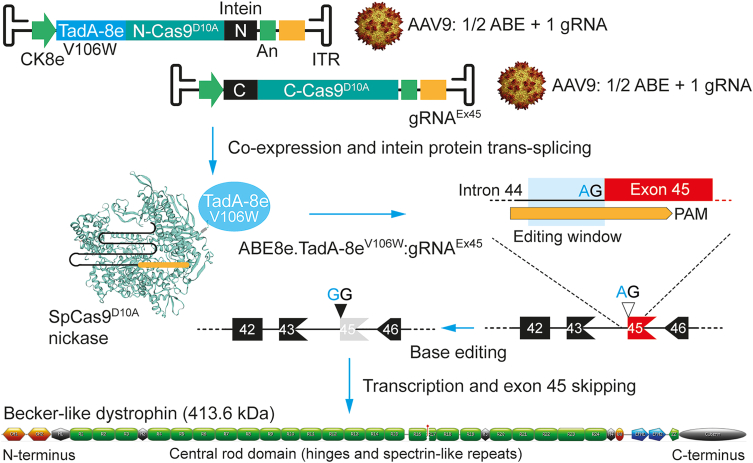
Figure 2.
Dual AAV ABE trans-splicing system for dystrophin repair
Two AAV serotype-9 vectors expressing separated portions of an adenine base editor (ABE.TadA-8eV106W) and a gRNA (gRNAEx45) lead to intein-mediated assembly of complete ABE:gRNA complexes. Base editing involving A·T-to-G·C transitions at the splice acceptor site of exon 45 establishes permanent exon 45 skipping in striated muscle cells. In dystrophin-defective muscle cells lacking exon 44, exon 45 skipping restores the reading of mature transcripts that code for a truncated Becker-like dystrophin with therapeutic potential for DMD patients. ITR, T-shaped hairpin-structured AAV serotype-2 inverted terminal repeats (cis-acting elements needed for vector DNA replication and packaging in producer cells). CK8e and An, synthetic striated muscle-specific promoter and polyadenylation signal, respectively. The dystrophin diagram was generated via: http://edystrophin.genouest.org/index.php?page=home.
Acknowledgments
Research in our laboratory is supported by the Prinses Beatrix Spierfonds (W.OR21-01), the Duchenne Parent Project NL, the Dutch Research Council (NWO) – Open Technology Program, and EU Marie Skłodowska-Curie Doctoral Network Actions. Z.L. holds a Ph.D. fellowship from the China Scholarship Council – Leiden University Joint Scholarship Program. Authors of this work are members of the European Reference Network – Neuromuscular diseases (ERN EURO-NMD).
Declaration of interests
The authors declare no competing interests.
References
- 1.Choi E., Koo T. CRISPR technologies for the treatment of Duchenne muscular dystrophy. Mol. Ther. 2021;29:3179–3191. doi: 10.1016/j.ymthe.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson C.E., Wu Y., Gemberling M.P., Oliver M.L., Waller M.A., Bohning J.D., Robinson-Hamm J.N., Bulaklak K., Castellanos Rivera R.M., Collier J.H., et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanlon K.S., Kleinstiver B.P., Garcia S.P., Zaborowski M.P., Volak A., Spirig S.E., Muller A., Sousa A.A., Tsai S.Q., Bengtsson N.E., et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 2019;10:4439. doi: 10.1038/s41467-019-12449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chai A.C., Chemello F., Li H., Nishiyama T., Chen K., Zhang Y., Sánchez-Ortiz E., Alomar A., Xu L., Liu N., et al. Single-swap editing for the correction of common Duchenne muscular dystrophy mutations. Mol. Ther. Nucleic Acids. 2023;32:522–535. doi: 10.1016/j.omtn.2023.04.009. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W., Wilson C., Koblan L.W., Zeng J., Bauer D.E., et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis J.R., Wang X., Witte I.P., Huang T.P., Levy J.M., Raguram A., Banskota S., Seidah N.G., Musunuru K., Liu D.R. Efficient in vivo base editing via single adeno-associated viruses with size-optimized genomes encoding compact adenine base editors. Nat. Biomed. Eng. 2022;6:1272–1283. doi: 10.1038/s41551-022-00911-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chemello F., Chai A.C., Li H., Rodriguez-Caycedo C., Sanchez-Ortiz E., Atmanli A., Mireault A.A., Liu N., Bassel-Duby R., Olson E.N. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci. Adv. 2021;7:eabg4910. doi: 10.1126/sciadv.abg4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L., Zhang C., Li H., Wang P., Gao Y., Mokadam N.A., Ma J., Arnold W.D., Han R. Efficient precise in vivo base editing in adult dystrophic mice. Nat. Commun. 2021;12:3719. doi: 10.1038/s41467-021-23996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Koay T.W., Maiakovska O., Zayas M.L., Grimm D. Progress in bioengineering of myotropic Adeno-associated viral (AAV) gene therapy vectors. Hum. Gene Ther. 2023 doi: 10.1089/hum.2023.057. Online ahead of print. [DOI] [PubMed] [Google Scholar]