Abstract
The use of modified nucleosides is an important approach to mitigate the intrinsic immunostimulatory activity of exogenous mRNA and to increase its translation for mRNA therapeutic applications. However, for vaccine applications, the intrinsic immunostimulatory nature of unmodified mRNA could help induce productive immunity. Additionally, the ionizable lipid nanoparticles (LNPs) used to deliver mRNA vaccines can possess immunostimulatory properties that may influence the impact of nucleoside modification. Here we show that uridine replacement with N1-methylpseudouridine in an mRNA vaccine encoding influenza hemagglutinin had a significant impact on the induction of innate chemokines/cytokines and a positive impact on the induction of functional antibody titers in mice and macaques when MC3 or KC2 LNPs were used as delivery systems, while it impacted only minimally the titers obtained with L319 LNPs, indicating that the impact of nucleoside modification on mRNA vaccine efficacy varies with LNP composition. In line with previous observations, we noticed an inverse correlation between the induction of high innate IFN-α titers in the macaques and antigen-specific immune responses. Furthermore, and consistent with the species specificity of pathogen recognition receptors, we found that the effect of uridine replacement did not strictly translate from mice to non-human primates.
Keywords: MT: Oligonucleotides: Therapies and Applications, mRNA vaccine, lipid nanoparticle, N1-methylpseudouridine, mRNA modification, delivery system, ionizable lipids, innate immunity, immune response
Graphical abstract
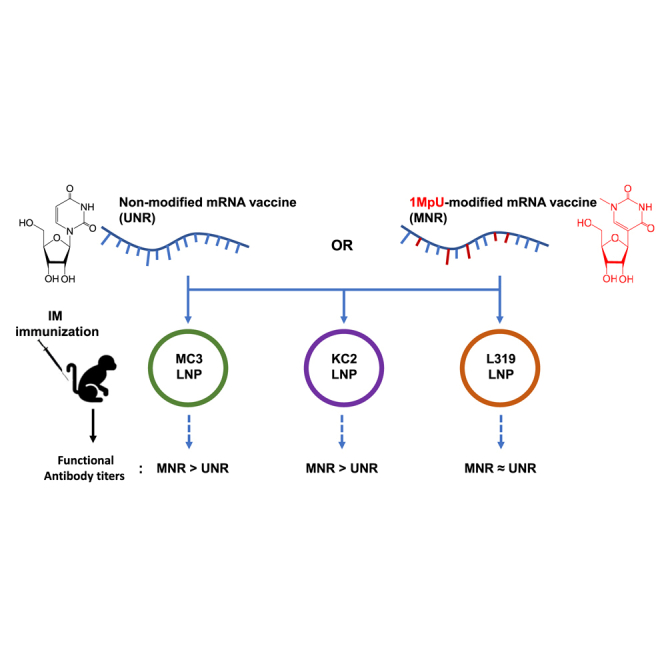
Haensler and colleagues compared the immune responses evoked by a native and N1-methylpseudouridine (1MpU)-modified influenza mRNA vaccine in mice and macaques when delivered using three different lipid nanoparticles (LNPs). They observed that the beneficial impact of uridine replacement with 1MpU varied with the ionizable lipid component of the LNPs.
Introduction
The use of nucleoside-modified mRNA for the development of mRNA vaccines remains an outstanding question with important implications in terms of intellectual property and industrial development. For instance, both licensed mRNA vaccines against Covid-19 (Pfizer-BioNTech’s BNT162b2; Comirnaty and Moderna’s mRNA-1273; Spikevax) contain N1-methylpseudouridine (1MpU) instead of uridine to preserve antigen expression by protecting against the activation of innate immune mechanisms that are deleterious to mRNA stability and translation.1,2,3 Moreover, the use of unmodified mRNA in Curevac’s SARS-CoV-2 vaccine candidate (CVnCoV) that uses the same lipid nanoparticle (LNP) delivery system as Comirnaty was thought to play a role in the lack of effectiveness of this vaccine (47% effectiveness at preventing Covid-19 in phase 3 trials versus >93% for the licensed vaccines of Pfizer-BioNTech and Moderna).2,4 While Curevac was able to improve its Covid-19 vaccine candidate to the level of marketed benchmarks in preclinical models by using mRNA sequence optimization without introducing modified nucleosides,5,6 in recent clinical studies on Covid-19 and influenza mRNA vaccines, the use of modified mRNA resulted into a substantially improved reactogenicity profile and offered a broader dose range compared to unmodified mRNA. This has finally led Curevac to select modified mRNA for further vaccine development.7 Indeed, exogenous mRNA produced by in vitro transcription, as in the case of mRNA vaccines, has been shown to stimulate innate immune mechanisms through various pathogen recognition receptors such as Toll-like receptor (TLR)3,8 TLR7, and TLR8,9 the retinoic acid-inducible gene I (RIG-I),10 and melanoma differentiation-associated 5 (MDA5)11 receptors, leading to type I interferon induction, RNase L activation, translation inhibition, and inflammation.12,13 To attenuate this effect, modified nucleosides such as pseudouridine, N1-methylpseudouridine, and other nucleoside analogs that are specific to endogenous mRNAs are typically incorporated.14,15,16,17,18,19,20,21,22 The application of modified nucleosides to mRNA immunosilencing has been extensively reviewed elsewhere.23,24,25,26
The licensed mRNA vaccines and most mRNA vaccines in clinical and preclinical development use LNPs to deliver the mRNA in vivo.27,28,29 The most critical component of LNPs is an ionizable amino lipid that is required for mRNA compaction and intracellular delivery.30,31,32 Since cationic and ionizable amino lipids can as well effectively activate innate immune mechanisms33,34,35,36,37,38 to a level that some of them have been used as adjuvants for protein vaccines,39,40,41,42,43,44,45 we speculated that the composition of the LNP that is used to deliver the mRNA vaccine could influence the impact of nucleoside modification on mRNA vaccine-induced immune responses. Therefore, in the present work, we aimed to compare in mice and macaques three well-established LNPs—MC3, KC2, and L31946—for the delivery of both an unmodified (UNR) and a 1MpU-modified (MNR) version of a model mRNA vaccine encoding an influenza H1N1 strain hemagglutinin (HA-H1).
Results
Our goal was to study if mRNA base modification could differently impact mRNA/LNP vaccine performance depending on the ionizable amino lipid component of the LNP delivery system. In this aim, we encapsulated influenza strain A/Netherlands/602/2009 (H1N1) HA-encoding mRNA, with or without 1MpU modification, into three LNPs differing solely by their ionizable lipid component. The ionizable lipids chosen for this study were DLin-MC3-DMA (MC3), which is a well-known ionizable lipid47 that is present in the marketed siRNA drug product Onpattro (Patisiran from Alnylam),48 L319 that comprises the same dimethyl amino ionizable head group as MC3 but a different lipid tail with hydrolysable ester bonds,49 and DLin-KC2-DMA that comprises the same dilinoleoyl lipid tail and same dimethyl amino head group as MC3 but a dioxolane linker instead of an ester linker between the head and tail.50 The structures of the three lipids are shown in Table 1. For LNP formulation, each ionizable lipid was combined with the same helper lipids, 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DMPE-PEG2000) at a molar ratio of 50:10:38.5:1.5, respectively, and LNPs were prepared at an ionizable lipid to RNA charge ratio (N/P) of 6 by using a microfluidic mixing device (NanoAssemblr; Precision Nanosystems, Vancouver, BC), knowing from the literature that these compositions and conditions would be appropriate to yield homogeneous LNPs.46,49,51 These LNPs were characterized by dynamic light scattering to determine particle sizes and by using a Ribogreen dye accessibility assay to determine the rate of mRNA encapsulation. Their apparent pKas determined by the 2-(p-toluidino)-6-naphthalene sulfonic acid (TNS) binding assay were taken from the literature.47,49 Table 1 shows the characteristics of MC3, KC2, and L319 LNPs loaded with the different mRNAs of the present study, namely 5-methoxyuridine-modified, cyanine-5-labeled CleanCapmRNA encoding firefly luciferase (FLuc mRNA) and unmodified (UNR) or 1MpU-modified (MNR) mRNA encoding influenza H1N1 hemagglutinin (HA-H1 mRNA). Note that the particle sizes of MC3 and L319 LNPs varied in opposite ways when switching from FLuc mRNA to HA-H1 mRNA, probably due to the cyanine-5 molecules attached to the FLuc mRNA.
Table 1.
Characterization of LNPs used in preclinical studies
| Ionizable amino lipid | pKa | mRNA | pH | Osmolality mOsmol/kg | Z average (nm) | Encaps. (%) |
|---|---|---|---|---|---|---|
MC3 
|
6.4 | Cya-5-Fluc | 7.38 | 280 | 136 | 89 |
| HA-H1 UNR | 7.16 | 287 | 87 | 96 | ||
| HA-H1 MNR | 7.20 | 289 | 88 | 98 | ||
KC2 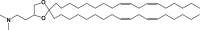
|
6.7 | Cya-5-Fluc | 7.36 | 281 | 71 | 90 |
| HA-H1 UNR | 7.21 | 287 | 87 | 98 | ||
| HA-H1 MNR | 7.19 | 290 | 64 | 98 | ||
L319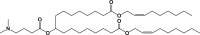
|
6.4 | Cya-5-Fluc | 7.37 | 284 | 90 | 100 |
| HA-H1 UNR | 7.19 | 289 | 155 | 98 | ||
| HA-H1 MNR | 7.16 | 288 | 163 | 97 |
Functional in vivo mRNA delivery by using MC3, KC2, and L319 LNPs
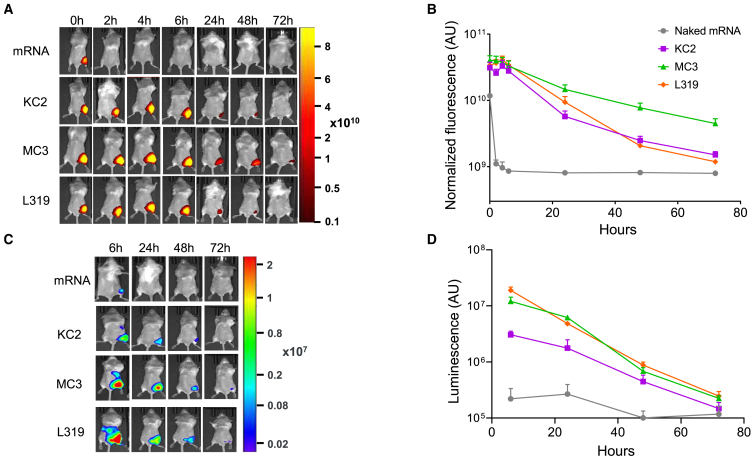
In a preliminary step, we used the FLuc mRNA as a reporter system to confirm that the three LNPs were effective at protecting and delivering an active mRNA construct in vivo by using in vivo fluorescence and bioluminescence techniques. In mice injected i.m. in the right quadriceps with 5 μg of naked FLuc mRNA, the fluorescence signal associated with the mRNA disappeared within 2 h following the injection, consistent with a rapid degradation of the mRNA in vivo. Conversely, when the labeled FLuc mRNA was encapsulated within MC3, KC2, or L319 LNPs, fluorescence could be detected in the injected muscle for at least 48 h following the injection and even longer (72 h) with MC3 LNPs (Figures 1A and 1B). Encapsulation into LNPs also afforded stronger luciferase expression from the injected FLuc mRNA as assessed by using the bioluminescence modality of the IVIS spectrum bioimaging system (Figures 1C and 1D). In this experiment, the strongest luciferase expression was obtained from L319 and MC3 LNPs, with a signal detected not only around the site of injection but also at distal sites in the mouse abdomen area (liver, spleen). In an independent experiment, using the same cyanine-5-labeled FLuc mRNA in L319 LNPs, the injected mice were sacrificed after 6 or 24 h to perform ex vivo imaging on harvested organs (muscle, popliteal and axillary lymph nodes, spleen, liver, lung, heart, kidneys, and intestines). Thus, the luciferase-expressing organs could be identified as being the injected muscle, the draining lymph nodes, the liver, and the spleen (data not shown). At 72 h post injection, luciferase signal was detected only in the injected muscle.
Figure 1.
Kinetics of Cy5-Fluc mRNA fluorescence and expression after i.m. delivery via different LNPs
(A) Representative IVIS images of cyanine-5 fluorescence signal in mice injected with 5 μg of naked cyanine-5-labeled, 5- methoxyuridine-modified CleanCap FLuc mRNA or the same mRNA encapsulated within MC3, KC2, or L319 LNPs. (B) Quantification of the fluorescence signal by total radiance efficiency (AU = (ph/s)/(μW/cm2)) in the injected site at 0, 2, 4, 6, 24, 48 and 72 h post injection. (n = 4 to 5 mice per group). Data are presented as mean values +/− SEM. (C) Representative bioluminescence images of luciferase expression in the Cy5-Fluc mRNA-injected mice. (D) Quantification of the bioluminescence signal (AU = photons/s) in the injected site at 6, 24, 48 and 72 h post injection. (n = 4 to 5 mice per group). Data are presented as mean values +/− SEM.
Influence of 1MpU modification on immune responses induced by mRNA in MC3, KC2, and L319 LNPs
After having confirmed functional mRNA delivery in vivo, we wanted to compare the three LNPs for their ability to induce innate chemokines and cytokines and functional antibody and T cell responses with the unmodified (UNR) and 1MpU-modified (MNR) versions of HA-H1 mRNA in cynomolgus macaques and in mice. For the sake of interspecies translational analysis, the mice and macaques were immunized the same day with the same mRNA/LNP preparations, the monkeys receiving 500 μL-doses (50 μg mRNA) and the mice receiving 50 μL-doses (5 μg mRNA) of the same preparations by the intramuscular (i.m.) route.
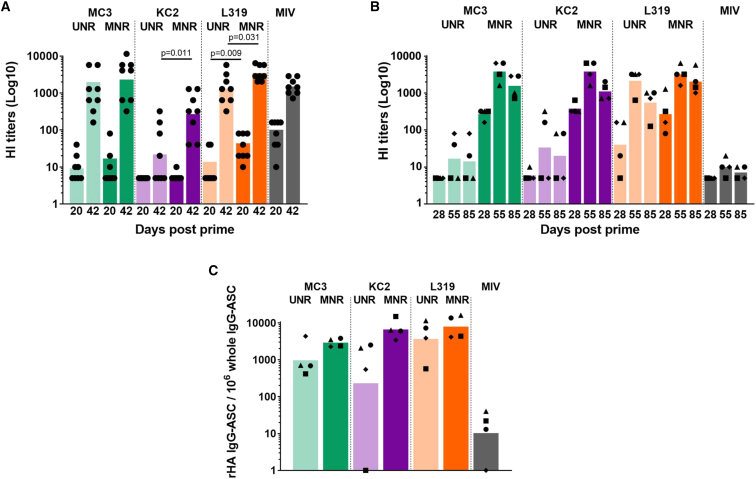
In mice, there was a general trend toward an increase in functional antibody responses (hemagglutination-inhibiting [HI] titers) when substituting uridine with 1MpU (Figure 2A). MC3 and L319 LNPs, which both induced robust HI titers with UNR, benefited only modestly from 1MpU modification with a 2- to 3-fold increase in HI titers (Figure 2A). This increase was found to be statistically significant for L319 LNPs post prime and post boost (p = 0.009 and p = 0.031, respectively) but not for MC3 LNPs. Conversely, KC2 LNPs, which induced low seroconversion and the lowest HI titers when used with UNR, benefited much more from the switch to MNR with a 100% increase of seroconversion rate and over 10-fold increase in average HI titers post boost (p = 0.011). However, despite this 10-fold increase, the level of HI titers induced by KC2 LNPs with MNR did not reach that of MC3 and L319 LNPs (p < 0.001). Of note, two injections were needed to induce substantial seroconversion in naive mice with this mRNA, independently of base modification and of LNP delivery vehicle. 3 weeks after a single injection of 5 μg of HA-H1 mRNA, only a few responding mice (1–4 out of 8) with low HI titers were detected in the different groups (Figure 2A).
Figure 2.
Comparison of MC3, KC2, and L319 LNPs in mice and NHPs with unmodified and 1MpU-modified mRNA
(A) Individual and mean HI titers measured in sera collected from Balb/c mice (n = 8/group) after a single immunization (D20) and two immunizations (D42) with 5 μg of a monovalent A/California/07/2009 (H1N1) split influenza vaccine (Vaxigrip; MIV) or with 5 μg of unmodified mRNA (UNR) or 1MpU-modified mRNA (MNR) encoding full-length hemagglutinin (HA) of closely related influenza virus strain A/Netherlands/602/2009 (H1N1). The mRNA was encapsulated into MC3 (green), KC2 (purple), or L319 (orange) LNPs. Immunization was performed by 2 i.m. injections into the quadriceps given 3 weeks apart (D0, D21) under a final volume of 50 μL of PBS. For each lipid, UNR versus MNR comparison was performed by ANOVA after log transformation of the HI titers. When difference was significant, p-value was reported directly on the graph (B) 500 μL fractions of the mRNA vaccines used for the immunization of mice (i.e., 50 μg of UNR or MNR) were used to immunize cynomolgus macaques. The macaques (n = 4/group) were immunized by 2 i.m. injections into the deltoid given 4 weeks apart (D0, D28). A control group received 15 μg of the monovalent A/California/07/2009 Vaxigrip (MIV) following the same schedule. Individual and mean HI titers measured after a single immunization (D28) and after two immunizations (D55 and D85) are shown. A given symbol was attributed to each individual macaque from the different groups and kept the same throughout the graphs. (C) 8 weeks following the second immunization (D85), PBMCs were collected from the macaques for the determination of antibody-secreting cells (ASCs). The percentage of antigen-specific ASCs to the total IgG + ASCs is shown for each individual macaque represented by its symbol. The bar represents the mean for the group.
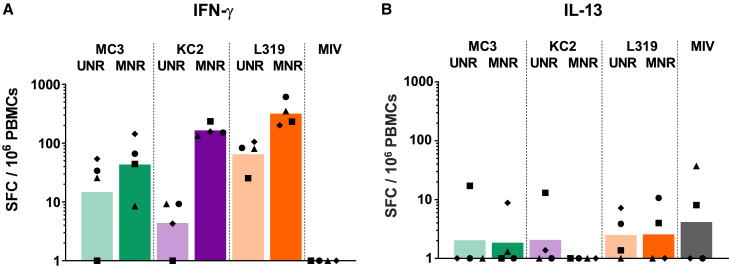
In macaques, an even more striking effect of 1MpU modification was observed on HI titers since all three LNPs, including KC2 LNPs, brought about similarly strong HI titers when loaded with MNR (Figure 2B). The peak titers achieved by MC3, KC2, and L319 LNPs loaded with MNR (mean titers of 2,600–3,400) were 100-fold above those induced by MC3 and KC2 LNPs loaded with UNR (mean titers of 20–40). In the L319 LNP group, similar HI peak titers were measured with UNR and MNR at D55, 4 weeks following the second administration (mean titers of 1,600 versus 3,200 with UNR and MNR respectively), but with MNR, the titer was higher at D28 (after a single injection) and appeared to decline less rapidly compared with those obtained with UNR (Figure 2B). Note also that whatever the LNP in this study, two injections were required to induce substantial seroconversion rates and HI titers with UNR in the macaques, while MNR induced robust HI titers comprised between 80 and 640 at D28, 4 weeks after a single injection. In accordance with the HI responses, the cellular responses obtained with MNR were more robust than those obtained with UNR, independently of the LNP composition, as evidenced by the frequencies of HA-specific IgG-secreting B cells (Figure 2C) and IFN-γ-secreting T cells (Figure 3A) determined respectively 2 months (D85) and 1 month (D55) following the second injection. Note that no HA-specific IgM-secreting B cells (not shown) and IL-13-secreting T cells were detected in this experiment (Figure 3B). KC2 and L319 LNPs tended to induce higher cellular responses than MC3 LNPs with MNR, but due to the low number of animals per group and the heterogeneity of the responses in some of the groups, it is hard to rank the formulations based on these cellular responses. Note also that only modest HI responses and no cellular immune responses were induced by i.m. injection of two 15-μg doses of monovalent influenza split vaccine Vaxigrip in the macaques. This observation was in accordance with previous observations indicating that an adjuvant was required for the induction of significant immune responses with Vaxigrip in the macaques.41
Figure 3.
T cell responses in NHPs immunized with MC3, KC2, and L319 LNPs loaded with unmodified and 1MpU-modified mRNA
4 weeks following the second immunization (D55), PBMCs were collected from the macaques and analyzed for (A) IFN-γ and (B) IL-13 secreting cells by FluoroSpot assay. Cell counts from each individual macaque are shown after subtracting counts obtained with PBMCs collected from the same animal pre-immunization. Each macaque can be identified on the graph with its symbol attributed in Figure 2. The bar represents the mean for the group.
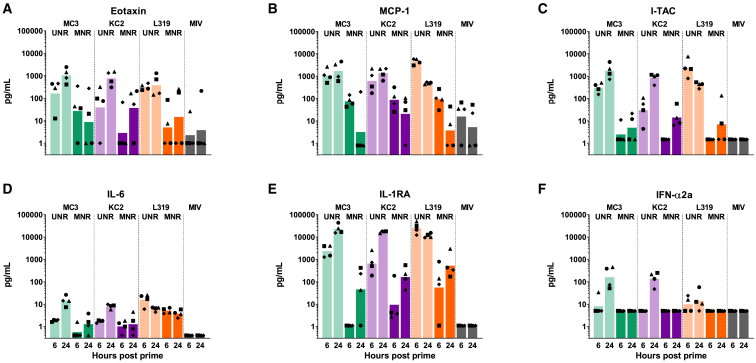
Influence of 1MpU modification on innate chemokines/cytokines responses induced by mRNA in MC3, KC2, and L319 LNPs in macaques
The different vaccines were also tested for the induction of innate cytokines and chemokines in the macaques by using a multiplexing assay for IL-1RA, IL-1β, IL-6, IL-8 (CXCL8), IL-17, IFN-α2a, I-TAC (CXCL11), MCP-1 (CCL2), TNF-α, and eotaxin. The induction of innate chemokines/cytokines measured in the macaque serum 6 and 24 h post immunization is represented in Figure 4 for each individual animal and each of the induced chemokines/cytokines. With UNR, the three LNPs induced similarly high levels of I-TAC and IL1-RA and moderate levels of MCP1 and eotaxin (Figures 4A, 4B, 4C, and 4E). However, while MC3 and KC2 LNPs induced also high levels of IFN-α, the level of IFN-α induced by L319 LNPs was much lower, indicating that high IFN-α induction resulted from a combined action of UNR and LNP amino lipid on the innate immune system (Figure 4F). Furthermore, the L319 group also stood out with a faster induction of IL-1RA, I-TAC, and MCP1, with highest levels measured in this group with UNR at 6 h. When formulated with MNR, the induction of innate chemokines/cytokines was dramatically reduced in all animals whatever the LNP composition, but IL-1RA could still be detected in animals immunized with MNR (Figure 4E). Although it is hard to draw definitive conclusions from a study comprising only four monkeys per group, the strong reduction in innate chemokine/cytokine induction when switching from UNR to MNR was in line with the expected effects of mRNA modification. It is noteworthy that there was no significant induction of major inflammatory cytokines—TNF-α, IL-1β, and IL-6—and no significant induction of IL-8 and IL-17A in any of the immunized macaques across all groups (not shown).
Figure 4.
Innate responses in NHPs immunized with MC3, KC2, and L319 LNPs loaded with unmodified and 1MpU-modified mRNA
6 and 24 h following the first immunization, serum samples from macaques of Figure 2 were tested for innate chemokines/cytokines by MSD assay. Measured levels of (A) eotaxin, (B) MCP-1, (C) I-TAC, (D) IL-6, (E) IL-1RA, and (F) IFN-α2a are represented in pg/mL after subtracting background levels measured pre-immunization for each individual macaque identified by its symbol attributed in Figure 2. The bar represents the mean for the group. Levels of IL-1β, IL-8, and IL-17A did not increase over background and are not represented.
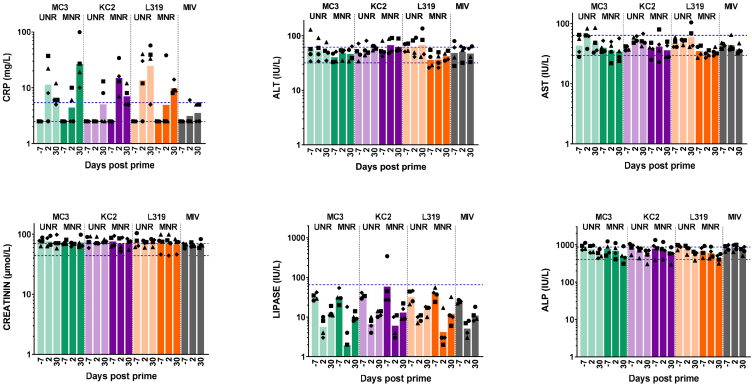
Influence of 1MpU modification on reactogenicity or blood parameters induced by mRNA in MC3, KC2, and L319 LNPs in mice and macaques
In terms of reactogenicity, the vaccinated mice did not display any systemic or local reactions, and their body weight increased regularly over the study in all groups.
Likewise, none of the vaccinated macaques experienced clinical symptoms, except one animal in the KC2/UNR group and one in the L319/UNR group, which both developed a site of injection rash, which resolved within 2 days, following the first injection. These local reactions did not occur after the second injection. Bleeding was sometimes observed immediately after administration due to the needle stick with the occurrence of scab, edema, hematoma, or redness that resolved within the following days. There were no adverse variations in food consumption, rectal temperature, and body weight throughout the study. The animals were also tested for a typical set of blood biomarkers—C-reactive protein (CRP), lipase, aspartate transaminase (AST), alanine transaminase (ALT), alkaline phosphatase (ALP), and creatinine—before immunization and 2, 5, and 7 days following each immunization. Representative values detected before injection (D-7) and 2 days after each injection (D2, D30) are depicted in Figure 5. The mRNA vaccines, as well as the monovalent Vaxigrip, did not induce major variations in the blood biomarker levels compared with baseline levels, with values mostly within the normal range (as described in materials and methods). The only exception was for CRP values, which increased in some animals from the mRNA/LNP groups, whether UNR or MNR was used, on D2 and/or D30 and rapidly returned to normal levels at the next tested time points. These findings, reflective of a transient systemic inflammation in the immunized macaques, could not be linked to a specific formulation or to an increase/decrease of specific immune responses. Note that CRP concentration values below 23 mg/L in serum are typically considered as a normal inflammation status in young cynomolgus macaques according to the monkey breeder.
Figure 5.
Blood biomarkers in NHPs immunized with MC3, KC2, and L319 LNPs loaded with unmodified and 1MpU-modified mRNA
7 days before immunization (D-7) and 2 days following each immunization (D2, D30), plasma samples from the macaques were analyzed for standard blood parameters: alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine, lipase, and C-reactive protein (CRP). Protein concentrations are shown for each individual macaque identified by its symbol attributed in Figure 2. Values are expressed in IU/L for ALP, ALT, AST, and lipase, in μmol/L for creatinine, and in mg/L for CRP. The bars represent the mean from each group. Lower and upper dashed lines in each graph represent the fifth and 95th percentile, respectively, of reference pre-dose values commonly observed in cynomolgus macaques based on historical data. The fifth to 95th percentile range covers 90% of these pre-dose data.
Discussion
For mRNA therapeutics, the use of modified nucleosides is an important approach to mitigate the intrinsic immunostimulatory activity of exogenous mRNA and to increase its translation. However, this was not always found to be the case,52,53 and in a recent study, Melamed and coworkers demonstrated that the benefit of base modification in systemically injected mRNA/LNPs could vary with the composition of the LNPs and the nature of their target organs/cells.54 In the mRNA/LNP vaccine space, the importance of nucleoside modification remains a matter of debate since increased innate immunity activation, although potentially detrimental to mRNA translation, could be beneficial for the induction of productive immune responses.55 This led us to further speculate that some mRNA/LNP vaccines could benefit more than others from nucleoside modification, depending on their cationic lipid component and immunostimulatory potential. Therefore, in the present work, we aimed to compare in mice and macaques three well-established LNPs—MC3, KC2 and L31946—for the delivery of both an unmodified (UNR) and a 1MpU-modified (MNR) version of an mRNA vaccine encoding influenza HA. First, we observed that switching from MC3 or KC2 to L319 impacted LNP particle sizes since L319 LNPs (Ca. 160 nm) were about twice as big as MC3 and KC2 LNPs (Ca. 80 nm) (note that process parameters, nature, and ratio of helper lipids and N/P ratio remained constant in the different formulations). We then observed that, depending on the ionizable amino lipid in the LNP, 1MpU modification could also affect LNP sizes since the size of KC2 LNPs decreased from 87 nm to 64 nm when switching from UNR to MNR, while the sizes of MC3 and L319 LNPs remained essentially unaffected (Table 1). We believe this may be the result of potential effects of 1MpU on the HA-H1 mRNA hydrophobicity and secondary structure, which in turn may impact its compaction by the cationic lipid. More importantly, we found that depending on the nature of the cationic amino lipid used for LNP formulation, switching from UNR to MNR had either a neutral or positive effect on the level of functional HI antibodies induced by the mRNA/LNP vaccine; there was no case, however, where the switch had a negative effect, which is in line with Curevac’s recent clinical observations.7 In macaques, we observed strong HI responses when using MNR, with all three tested LNPs, but L319 LNPs brought about similarly high HI titers with UNR, indicating that robust HI can be obtained with UNR in the monkeys. These titers were in the same range as those reported by Lutz and colleagues and by Chivukula and colleagues after i.m. immunization of macaques with LNP-formulated UNR encoding influenza HA.55,56 In the present study, the better performance of L319 LNPs with UNR compared to KC2 and MC3 LNPs may be linked to the larger particle size of the L319 LNPs (Ca. 160 nm vs. Ca. 80 nm). Indeed, according to recent studies, the larger particles, which are susceptible to contain and deliver a larger amount of mRNA copies per particle and target different cell types, could induce better responses, provided the particle size remains within the limits of lymphatic drainage.57,58,59 Furthermore, the better performance of L319 LNPs with UNR seems to be linked to the lack of strong IFN-α induction by this formulation since we observed a negative correlation between the induction of high HI titers and high innate IFN-α production. With L319 LNPs, there was no or little IFN-α detected, even when loaded with UNR, and antibody and T cell responses were high with both UNR and MNR. With MC3 and KC2 LNPs, IFN-α was induced when the LNPs were loaded with UNR but not with MNR and antibody, and T cell responses were strongly increased with MNR. The negative correlation between strong IFN-α induction and mRNA vaccine efficacy had already been observed by others60,61 and is in line with the reported effects of IFN-α on the upregulation and activation of enzymes involved in translation inhibition and mRNA degradation.62 IFN-α is typically induced by the stimulation of innate immune receptors such as TLR7 and RIG-I.10,13 In the present study using highly purified HA mRNA, induction of significant IFN-α secretion with UNR was somehow modulated by the ionizable cationic lipid and abrogated by the use of MNR. With UNR, there was clearly less IFN-α secreted with L319 LNPs than with KC2 and MC3 LNPs (Figure 4F). Interestingly, with L319 LNPs, there was also a faster induction of IL-1RA, as detected 6 h post first immunization (Figure 4E). In a recent study,63 Tahtinen et al. demonstrated that the IL-1 pathway was critical in triggering mRNA vaccine-associated innate signaling, with a key role of IL-1RA in negatively regulating inflammation. Noteworthy, the authors reported that the immunostimulatory activity of a modified mRNA was highly dependent on the lipid formulation: LNPs formulated with SM-102 ionizable lipid (used in the Moderna COVID-19 vaccine, mRNA-1273)29 elicited a robust increase in IL-1β in mRNA-LNP-treated human peripheral blood mononuclear cells (PBMCs), while IL-1RA levels remained low, consistent with the reactogenicity observed in mRNA-1273-vaccinated individuals.64 In contrast, LNPs formulated with MC3 lipid were much less potent at stimulating IL-1β release in this system, with IL-1RA levels even lower than with SM-102. In our study, although IL-1β was undetectable in the macaque serum samples, it might have been locally induced in vivo and rapidly counteracted by the anti-inflammatory IL-1RA cytokine. Hence, the capacity of L319 LNPs with UNR to elicit a faster induction of IL-1RA—acting as a regulator of IL-1 pathway-mediated inflammation—and lower levels of innate IFN-α may have contributed to its better performance. Whether this resulted from the larger particle size of the L319 LNPs, from a different distribution or cellular uptake of these LNPs, from the chemistry of the L319 lipid itself, which contains hydrolysable ester bonds designed to accelerate biodegradation,49 or from the nature of the interaction between L319 and UNR remains to be studied.
In the macaques, 1MpU modification had also a positive effect on the development of specific T cell responses dominated by IFN-γ-secreting T cells, in accordance with the ability of mRNA vaccines to induce potent Th-1 responses.61 Regarding Th-2 type responses, we were not able to detect specific IL-13-secreting cells in the immunized macaques, whatever the formulation. This result is aligned with previously published observations by our group.56,65
Interestingly, although 1MpU modification had a positive impact on the induction of HI titers in both mice and macaques with all three tested LNPs, the fact that MC3 and KC2 would be as effective as L319 for inducing functional antibodies in the macaques when used with MNR could not be predicted from the results obtained in mice. Although these results may be influenced also by the quality and purity of the mRNA, the testing in mice may be considered insufficient to assess the real performance of an mRNA vaccine. This is in line with the species specificity of pathogen recognition receptors (PRRs) such as TLR7/8 and others.9 Hence, as for vaccine adjuvants,66 and immunomodulatory products in general, one recommendation would be to rapidly validate the preclinical work on mRNA vaccine sequence design and formulation optimization in small phase I clinical trials.
Regarding safety/reactogenicity, the products used in the described studies were generally well tolerated by mice and macaques. However, two macaques administered with UNR and none in the MNR groups developed a transient injection site rash following the first administration. This observation, although from a small sample group, speaks in favor of MNR and is aligned with clinical observations.7 While the switching from UNR to MNR generally strongly reduced the levels of circulating innate cytokines/chemokines (Figure 4), major inflammatory cytokines (IL-6, TNF-α, and IL-1β) did not increase above background levels even in macaques injected with UNR, although this does not preclude these cytokines from being secreted locally, at least to some degree. No safety signal was detected from the monitoring of standard blood biomarkers; all values remained within normal ranges67,68 in the immunized macaques (Figure 5). Although we may have missed an increase of these blood biomarkers at early time points following injection (before 2 days), none of the macaques experienced any systemic reactions (fever, body weight loss, reduced food consumption) throughout the study period.
Other interesting observations come from the study using the three LNPs encapsulating cyanine-5-labeled FLuc mRNA, although this experiment was essentially designed as a control experiment to simply demonstrate in vivo mRNA expression from each of the three LNPs. First, we observed that the fluorescent signal emitted by the labeled FLuc mRNA at the site of administration disappeared rapidly, within 2 h, following i.m. injection of naked mRNA, which is in line with a rapid degradation of extracellular mRNA in vivo.69 Consistent with the protective effect of LNPs, the encapsulation of the FLuc mRNA in MC3, KC2, or L319 LNPs led to an increase of its stability in vivo as reflected by fluorescence detection for at least 24 h at the site of administration following injection, with MC3 LNPs promoting a brighter and longer lasting signal compared to L319 and KC2 LNPs. The increased signal observed with MC3/Fluc mRNA at the site of injection may be due to the larger particle size of the MC3 LNPs when loaded with the cyanine-5-labeled Fluc mRNA (136 nm versus 90 and 71 nm for, respectively, L319 and KC2 LNPs) or from an increased uptake or protection of mRNA from degradation promoted by MC3 LNPs in the muscle. Some of this may be further explored by conducting in vitro uptake and stability assays on HSkMC and C2C12 muscle cells for instance. Next, we observed that luciferase expression was predominant at the site of administration, but all three LNPs were able to induce some expression in distal organs such as draining lymph nodes, liver, and spleen 6 h following i.m. injection, as already observed by others.70 Note that we also obtained some luciferase expression in the muscle injected with naked cyanine-5-labeled Fluc mRNA at early time points, which was consistent with the pioneering findings of Wolff and coworkers.71 With LNP-encapsulated mRNA, MC3 and L319 LNPs brought about similar levels of luciferase expression, and these were stronger than that obtained from KC2 LNPs. In this regard, there was reasonable alignment between the level of in vivo luciferase expression and immune responses obtained with the three different LNPs after i.m. injection into mice, although the luciferase and vaccine mRNAs had different modifications (5-methoxyuridine and cyanine-5-uridine for Fluc and 1MpU for HA-H1 mRNA) and were obtained from different suppliers. This correlation, however, seems to be incidental, as our general experience reveals an inconsistent correlation between the level of mRNA expression and the induced immune responses from a given LNP formulation, as already reported previously65 and by others.32 Concerning distribution of Fluc expression, it is not clear from this study whether distribution could be affected by the cyanine-5 label and 5-methoxyuridine modification on the Fluc mRNA.
Conclusion
In line with the recent report of Melamed and coworkers showing that the effect of mRNA base modification in systemically injected mRNA/LNPs could vary with the LNP composition,54 our study illustrates the interplay between 1MpU mRNA modification and LNP ionizable lipid in intramuscularly delivered mRNA/LNP vaccines. Consistent with the ability of IFN-α to counteract the antigen-specific immune responses induced by mRNA vaccines,60,61 we found that KC2 and MC3 LNPs that induced high levels of IFN- α when used with UNR were the ones that benefited most from the uridine replacement with 1MpU. Conversely, with L319 LNPs, IFN-α production was lower, anti-inflammatory IL-1RA induction was faster, the immune responses obtained with the UNR-based vaccine were high, and mRNA modification with 1MpU had only a modest improvement on immune responses. The underlying mechanisms of lower IFN-α induction, the earlier production of IL1-RA, the increased potency of L319 LNPs loaded with UNR, and the potential role played by LNP lipid chemistry, particle sizes, distribution, and uptake remain to be studied in detail. Work to understand why some mRNA/LNP vaccine formulations could benefit more than others from the UNR to MNR switching is currently under way in our group.
Materials and methods
Lipids
di((Z)-non-2-en-1-yl) 9-((4-(dimethylamino)butanoyl)oxy)heptadecanedioate (L319) was synthesized as described by Maier et al.,49 heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino)butanoate (DLin-MC3-DMA; MC3) as described by Jayaraman et al.,47 and 2,2-dilinoleyl-4-(2-dimethylaminoethyl)-[1,3]-dioxolane (DLin-KC2-DMA; KC2) as described by Semple et al.50 and obtained at a purity >90% from SAI Life Sciences (Hyderabad, India).
1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC; ref. 850365), 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000](14:0 PEG2000-PE; ref. 880150) and cholesterol (Chol; ref. 700100) were obtained from Avanti Polar Lipids (Alabaster, AL, USA).
mRNAs
High purity, custom-synthesized mRNA with and without N1-methyl pseudouridine (1MpU) substitution and encoding the hemagglutinin (HA) of the influenza strain A/Netherlands 2009 (H1N1) was obtained from Amptec. The mRNAs were prepared by in vitro transcription (IVT) with T7 RNA polymerase starting with a linear DNA template generated by PCR and a mixture of all natural nucleosides (UNR) or with 1MpU instead of U (MNR). The mRNAs were capped (Cap0) with anti-reverse cap analog introduced into the nucleotide triphosphate mix used for the IVT reaction. The mRNAs were tailed with a 120-A-long polyA tail encoded directly in the DNA template. The same 5′ and 3′ UTRs, specific to the vendor, were used in UNR and MNR. mRNAs were treated with phosphatase to remove the triphosphate group from the 5′ end of uncapped products and purified to high degree of purity by using spin column purification. Certificates of analysis showed single peaks for both UNR and MNR by using Agilent 2100 Bioanalyzer and A260/A280 ratios of 2.1 and 1.9, respectively, for UNR and MNR.
The 5-methoxyuridine-modified CleanCapmRNA encoding firefly luciferase and labeled with cyanine 5 for direct visualization was obtained from TriLink (catalog number: L-7702). Cyanine 5-UTP and 5-methoxy-UTP were used at a ratio of 1:3. Substitution of uridine in this ratio results in mRNA that is easily visualized and can still be translated in cell culture (note that translation efficiency correlates inversely with cyanine 5-UTP substitution).
LNP formulation and characterization
L319, MC3, and KC2 were formulated into LNPs by using a NanoAssemblr (Precision Nanosystems, Vancouver, BC) as previously described.51 In brief, lipids were first dissolved in ethanol at molar ratios of 50:10:38.5:1.5 (ionizable lipid/DSPC/Chol/DMPE-PEG2000). The lipid mix at 20 mg/mL in ethanol and the mRNA at 0.305 mg/mL in 50 mM citrate buffer (pH 4.0) were then injected into the NanoAssemblr at a flow rate ratio of 1:3 respectively with a combined final flow rate of 4 mL/min and an N/P ratio of 6 (cationic nitrogen groups from the ionizable lipid over anionic phosphate groups from the mRNA). Formulations were then dialyzed using 10-kDa MWCO cassettes (Spectrum Labs, Rancho Dominguez, CA) against 50 mM citrate buffer (pH 4.0) for at least 4 h followed by phosphate buffered saline (pH 7.4; PBS) for 24 h. Obtained LNP suspensions were filtered on 0.22-μm PES membranes and stored liquid at 4°C under inert atmosphere.
The size of the LNPs was characterized by dynamic light scattering using the Malvern Zetasizer NanoZS (Malvern, Worcestershire, UK), and mRNA encapsulation efficiency was determined by mRNA accessibility to Ribogreen using the QuantiTRibogreen RNA assay (Life Technologies, Burlington, ON).
Animal studies and ethics statements
All experiments were included in a program approved by the French Ministry of Higher Education, Research and Innovation and were conducted in accordance with the European Directive 2010/63/UE as published in the French Official Journal of February 7th, 2013.
BALB/c ByJ mice (Balb/c) were purchased from Charles River (Saint-Germain-Nuelles, France) and cynomolgus macaques (Macaca fascicularis) from Noveprim (Camarney, Spain). Mice and macaques were housed in animal facilities accredited by AAALAC International. Mouse study protocols were reviewed by the Ethics Committee #11 of Sanofi Pasteur. The macaque study protocol was also reviewed by the Animal Welfare Body of Cynbiose and the Ethics Committee of VetAgro-Sup (Marcy l’Étoile, France) and approved under number 1633-V3 (MESR number: 2016071517212815).
Bioimaging studies
8-week-old female Balb/c mice were anesthetized with 2% isoflurane in oxygen (Alcyon, France; ref. 1818290) and injected into the right quadriceps with 5 μg of cyanine-5-labeled, 5- methoxyuridine-modified CleanCap FLuc mRNA either naked or encapsulated within L319, MC3, or KC2 LNPs under a final volume of 50 μL PBS.
Bioimaging acquisitions were performed using IVIS Spectrum CT imaging system (PerkinElmer). The fluorescence signal of cyanine-5-FLuc mRNA was acquired at the time of injection (T0), 2, 4, 6, 24, 48, and 72 h post injection. The level of FLuc mRNA expression was assessed at 6, 24, 48, and 72 h post injection by bioluminescence imaging, in anesthetized mice, 15 min after intraperitoneal injection of 150 mg/kg of D-luciferin (Invitrogen). In an independent experiment, using the same labeled Fluc mRNA in L319 LNPs, the injected mice were sacrificed after 6 or 24 h to perform ex vivo imaging on harvested organs by using the IVIS system.
All data were analyzed using the Living Image software provided by PerkinElmer and the quantification was performed over a region of interest (ROI) applied to the injected area. The fluorescence signal was expressed as total radiant efficiency [(photon/s)/(μW/cm2)] and normalized by subtracting the background signal measured in the PBS-injected group. Bioluminescence was quantified by measuring the total radiance (photon/s) of the ROI.
Immunization studies
Groups of four cynomolgus macaques (three females and one male weighing from 2.4 to 4.4 kg in each group) were immunized at D0 and D28 with 10 μg of a monovalent A/California/7/09 (H1N1) split influenza vaccine or with MC3, KC2, or L319 LNPs loaded with 50 μg of unmodified mRNA (UNR) or 1MpU-modified mRNA (MNR) encoding full-length HA of closely related influenza virus strain A/Netherlands/602/2009 (H1N1). The immunogens in PBS were injected into the animals’ right (first inj.) and left (second inj.) deltoid under a final volume of 500 μL. Blood samples were collected pre-immunization (D-22 and D-7) and at different time points following immunization for the analysis of functional antibody responses by HI titration (D28, D55, D85), HA-specific B and T cell responses by FluoroSpot (D20, D55, D85), innate cytokines by Meso Scale Diagnostics (MSD) assay (6 and 24 h post injection), and standard blood biochemistry markers (D2, D5, D7, D30, D33, D35). For functional antibody and innate cytokine titration in serum, blood was collected in dry tubes containing clot activator and serum separator (Becton-Dickinson, Vacumed; ref. 42711). After an overnight incubation at 5°C ± 3°C, blood samples were centrifuged at 2000 x g for 20 min, and sera were stored at −80°C until analysis. For T cell responses, blood was collected in Sodium-Heparin tubes (Becton-Dickinson Vacutainer; ref. 267876) for PBMC isolation. PBMCs were stored frozen in liquid nitrogen until analysis. For blood biochemistry markers, blood was collected in lithium-heparin tubes with separator gel (Greiner Bio-One, Vacuette; ref. 454089). Tubes were stored at 5°C ± 3°C for 60 ± 10 min prior performing the analyses. Clinical observations were performed daily over a period of 5 days after each injection. The presence of symptoms such as decrease in food intake, limited mobility, polypnea, systemic or local reactions were noted during the whole period of the study. Animals were weighed and their body temperature monitored once weekly.
In parallel, 50-μL fractions of the vaccine formulations used for macaque immunization (i.e., 5 μg of mRNA) were used to immunize Balb/c mice. Mice (eight per group) were injected twice, 3 weeks apart (D0, D21) by i.m. route into the quadriceps. Blood samples were collected in pre-immunized mice (D-9) and 3 weeks following each injection (D20, D42) for HI titration. Clinical observations were performed daily over a period of 5 days after each injection. Systemic and local reactions were monitored over the whole study period. Animals were weighed before immunization (D-9, D-1) and 3 weeks following each injection (D20, D42).
Hemagglutination inhibition assay
The level of functional antibodies was determined by HI titration of individual sera against 4 hemagglutination units (HAU) of an A/California/07/09 (H1N1) influenza virus (strain closely related to A/Netherlands/602/2009 encoded by the mRNA vaccine) using chicken red blood cells (cRBCs). Individual sera collected at defined time points were first treated with Receptor Destroying Enzyme and absorbed on 10% cRBCs in PBS. The HI titer of a given serum was defined as the reciprocal of its last dilution preventing hemagglutination. A value of 5, corresponding to half of the initial dilution (1/10), was arbitrarily given to a serum with no HI activity to perform statistical analysis.
Quantification of IFN-γ- and IL-13-secreting T cells
IFN-γ- and IL-13-secreting T cells were determined by using, respectively, the Monkey IFN- γ FluoroSpot BASIC (550) kit (Mabtech; Ref. FS1-21-550) and the Monkey IL-13 ELISpot BASIC (HRP) kit (Mabtech; ref. 3470M-2H) by following the kits’ instructions. In brief, Multiscreen 96-well IPFL plates (Mabtech; ref. 3654-FL-10) were coated by adding 100 μL/well of anti-monkey IFN- γ (Mabtech; ref. MT126L) or anti-monkey IL-13 capture mAb (Mabtech; Ref. IL13-I) at 0.5 mg/mL and incubated overnight at 4°C. After washing with PBS and plate blocking with RPMI medium containing 200 mM glutamine, 10 mg/mL streptomycin, 10000U/mL penicillin, and 10% heat-inactivated fetal calf serum, purified PBMCs (200,000 per well) were cultured for 48 h with either 1 μg per well of recombinant influenza hemagglutinin (rHA) from A/California/07/2009 strain (Sanofi, Protein Sciences) or with medium alone as a negative control. Positive controls were obtained by stimulating PBMCs with 100 ng anti-CD3 (Mabtech; Ref. CD3-1) for IFN- γ or 2 μg phytohemagglutinin (PHA) for IL-13. After overnight culture and washing with PBS-0.25% BSA, biotinylated anti-IFN- γ (7-B6-1; Mabtech) or anti-IL13 (IL-13-3; Mabtech) detection mAb was added and incubated at room temperature, followed by streptavidin-550 (Mabtech; Ref. SA-550) for IFN- γ or streptavidin PE (SouthernBiotech; ref. 7100-09L) for IL-13 detection. IFN- γ and IL-13-secreting cells were counted using a plate reader (Microvision Instruments, Evry, France). Data shown were corrected by subtracting counts from negative control wells containing PBMCs collected from the same animal prior to immunization.
Memory B cell FluoroSpot assay
Freshly thawed PBMCs were first stimulated for 5 days in RPMI medium containing 200 mM glutamine, 10 mg/mL streptomycin, 10,000 U/mL penicillin, and 10% heat-inactivated fetal calf serum supplemented with R848 (1 μg/mL) plus IL-2 (10 ng/mL) to promote B cell differentiation into antibody-secreting cells (ASCs). ASC frequency was then measured by the Human IgG/IgM FluoroSpot kit (Mabtech; Ref. FS-05R17G-10). Specifically, Multiscreen 96-well IPFL plates were prepared as described above for the T cell assays and coated with 4 μg of recombinant influenza hemagglutinin (rHA) from A/California/07/2009 strain (Sanofi, Protein Sciences).
After washing with PBS and blocking with complete medium, the plates were seeded with the pre-stimulated PBMCs. After 5 h of incubation at 37°C, the plates were washed in PBS-0.05% Tween 20 and PBS and incubated with respectively Cy3-labeled anti-human IgG-550 mAbs (Mabtech; Ref. MT78/145) or FITC-labeled anti-human IgM-490 mAb (Mabtech; Ref. MT22). After washing with PBS, fluorescent spots were enumerated with a spot reader equipped with filters for Cy3 and FITC (Microvision). Data shown were corrected by subtracting FluoroSpot counts from negative control wells containing PBMCs collected from the same animal prior to immunization.
Innate cytokine assay
10 cytokines (Eotaxin, IFN-α2a; IL-1β; IL-1RA, IL-6, IL-8, IL-17A; I-TAC, MCP-1, TNFα) were determined in macaque serum samples collected 7 days prior to immunization, 6 and 24 h following the first immunization by using an MSD U-plex kit (MSD; Ref. K15068L-2), and following kit instructions. Amounts were expressed in pg/mL after subtracting background levels measured pre-immunization for each individual macaque.
Blood biomarkers
Plasma levels of ALP, ALT, AST, creatinine, lipase, and CRP were measured before immunization and 2, 5, and 7 days following each injection using standard protocols applied for blood parameter determination by Biovelys (Marcy l’Etoile, France). Blood protein concentrations were expressed in IU (International Units)/L for ALP, ALT, AST, and lipase, and in μmol/L for creatinine and in mg/L for CRP for each individual macaque. When CRP was below the lower limit of quantification (5 mg/L), an arbitrary value of 2.5 mg/L was assigned. Other blood parameters could be quantified at each time point and for each animal.
Normal range values for cynomolgus macaques were provided by Biovelys based on their own data library (ALP, ALT, AST, creatinine) or derived from the literature (lipase, CRP).67,68 From these reference values, the fifth percentile (lowermost edge) and the 95th percentile (uppermost edge) were calculated, and the fifth to the 95th percentile range was represented, covering 90% of the most common pre-dose data.
Statistical analyses
No statistical analysis was performed on the monkey study due to the low number of animals per group. For the mouse study, the HI titers were log transformed and an analysis of variance (ANOVA) with LNP and mRNA as fixed factors was performed for group comparisons. For non-responders, an arbitrary minimum value of 5 HI units was attributed. Heterogeneity between the groups was considered. The model’s residuals were studied to test the model’s validity (normality, extreme individuals, etc.). All analyses were done on SAS v9.4. A margin of error of 5% was used for effects of the main factors.
Data availability
Research data are not shared.
Acknowledgments
We gratefully thank Julie Piolat for statistical analyses, Frank DeRosa, Sudha Chivukula, and Brian Schanen for helpful discussions and critical review of this manuscript, and Sanofi for the funding of this work.
Author contributions
Conceptualization and methodology: M.-C.B., F.B., B.R., and J.H. Investigations: M.-C.B., E.B., N.P., K.L., S.C., C.V., S.M., V.P., and M.R. Writing – original draft: M.-C.B. K.L., and J.H. Writing – review & editing: J.H., K.L., S.R., F.B., and B.R. Supervision: M.-C.B., M.G., F.B., B.R., and J.H.
Declaration of interests
All authors are Sanofi employees or were under contract with Sanofi (K.L., V.P., and M.R.) at the time of the study and may hold shares and/or stock options in the company.
References
- 1.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen J., van der Meulen Rodgers Y. What went wrong with CureVac's mRNA vaccine? Science. 2021;16:1381–1395. [Google Scholar]
- 3.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. The dawn of mRNA vaccines: the COVID-19 case. J. Contr. Release. 2021;333:511–520. doi: 10.1016/j.jconrel.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machado B.A.S., Hodel K.V.S., Fonseca L.M.S., Mascarenhas L.A.B., Andrade L.P., Rocha V.P.C., Soares M.B.P., Berglund P., Duthie M.S., Reed S.G., Badaró R. The importance of RNA-based vaccines in the fight against COVID-19: an overview. Vaccines. 2021;9:1345. doi: 10.3390/vaccines9111345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth N., Schön J., Hoffmann D., Thran M., Thess A., Mueller S.O., Petsch B., Rauch S. CV2CoV, an enhanced mRNA-based SARS-CoV-2 vaccine candidate, supports higher protein expression and improved immunogenicity in rats. bioRxiv. 2021 doi: 10.1101/2021.05.13.443734. Preprint at. [DOI] [Google Scholar]
- 6.Gebre M.S., Rauch S., Roth N., Yu J., Chandrashekar A., Mercado N.B., He X., Liu J., McMahan K., Martinot A., et al. Optimization of non-coding regions for a non-modified mRNA COVID-19 vaccine. Nature. 2022;601:410–414. doi: 10.1038/s41586-021-04231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curevac conference call & webcast of January 6th, 2023 – CureVac announces positive data on joint COVID-19 and flu mRNA vaccine development programs. Presentation in Events - CureVac (Last Accessed March 7th, 2023). https://www.curevac.com/en/newsroom/events.
- 8.Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. mRNA is an endogenous ligand for toll-like receptor 3. J. Biol. Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 9.Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 10.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 11.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Haro C., Méndez R., Santoyo J. The eIF-2α kinases and the control of protein synthesis. Faseb. J. 1996;10:1378–1387. doi: 10.1096/fasebj.10.12.8903508. [DOI] [PubMed] [Google Scholar]
- 13.Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 14.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Nallagatla S.R., Bevilacqua P.C. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2’-5′-Oligoadenylate Synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karikó K., Muramatsu H., Keller J.M., Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andries O., McCafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Contr. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 20.Durbin A.F., Wang C., Marcotrigiano J., Gehrke L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio. 2016;7 doi: 10.1128/mBio.00833-16. e00833008333-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parr C.J.C., Wada S., Kotake K., Kameda S., Matsuura S., Sakashita S., Park S., Sugiyama H., Kuang Y., Saito H. N 1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 2020;48:e35. doi: 10.1093/nar/gkaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 24.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linares-Fernández S., Lacroix C., Exposito J.Y., Verrier B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 2020;26:311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Mu X., Hur S. Immunogenicity of in vitro-transcribed RNA. Acc. Chem. Res. 2021;54:4012–4023. doi: 10.1021/acs.accounts.1c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhary N., Weissman D., Whitehead K.A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021;20:817–838. doi: 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J., Eygeris Y., Gupta M., Sahay G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021;170:83–112. doi: 10.1016/j.addr.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buschmann M.D., Carrasco M.J., Alishetty S., Paige M., Alameh M.G., Weissman D. Nanomaterial delivery systems for mRNA vaccines. Vaccines. 2021;9:65. doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hafez I.M., Maurer N., Cullis P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 31.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filion M.C., Phillips N.C. Toxicity and immunomodulatory activity of liposomal vectors formulated with cationic lipids toward immune effector cells. Biochim. Biophys. Acta. 1997;1329:345–356. doi: 10.1016/s0005-2736(97)00126-0. [DOI] [PubMed] [Google Scholar]
- 34.Yan W., Chen W., Huang L. Reactive oxygen species play a central role in the activity of cationic liposome based cancer vaccine. J. Contr. Release. 2008;130:22–28. doi: 10.1016/j.jconrel.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Lonez C., Vandenbranden M., Ruysschaert J.M. Cationic liposomal lipids: from gene carriers to cell signaling. Prog. Lipid Res. 2008;47:340–347. doi: 10.1016/j.plipres.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Lonez C., Vandenbranden M., Ruysschaert J.M. Cationic lipids activate intracellular signaling pathways. Adv. Drug Deliv. Rev. 2012;64:1749–1758. doi: 10.1016/j.addr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Lonez C., Bessodes M., Scherman D., Vandenbranden M., Escriou V., Ruysschaert J.M. Cationic lipid nanocarriers activate toll-like receptor 2 and NLRP3inflammasome pathways. Nanomedicine. 2014;10:775–782. doi: 10.1016/j.nano.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Yun C.H., Bae C.S., Ahn T. Cargo-free nanoparticles containing cationic lipids induce reactive oxygen species and cell death in HepG2 cells. Biol. Pharm. Bull. 2016;39:1338–1346. doi: 10.1248/bpb.b16-00264. Erratum in: Biol Pharm Bull. 2018. 41, 285. [DOI] [PubMed] [Google Scholar]
- 39.Pizzuto M., Bigey P., Lachagès A.M., Hoffmann C., Ruysschaert J.M., Escriou V., Lonez C. Cationic lipids as one-component vaccine adjuvants: a promising alternative to alum. J. Contr. Release. 2018;287:67–77. doi: 10.1016/j.jconrel.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 40.Alameh M.G., Tombácz I., Bettini E., Lederer K., Sittplangkoon C., Wilmore J.R., Gaudette B.T., Soliman O.Y., Pine M., Hicks P., et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity. 2021;54:2877–2892.e7. doi: 10.1016/j.immuni.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guy B., Pascal N., Françon A., Bonnin A., Gimenez S., Lafay-Vialon E., Trannoy E., Haensler J. Design, characterization and preclinical efficacy of a cationic lipid adjuvant for influenza split vaccine. Vaccine. 2001;19:1794–1805. doi: 10.1016/s0264-410x(00)00386-8. [DOI] [PubMed] [Google Scholar]
- 42.Hartikka J., Bozoukova V., Yang C.K., Ye M., Rusalov D., Shlapobersky M., Vilalta A., Wei Q., Rolland A., Smith L.R. Vaxfectin, a cationic lipid-based adjuvant for protein-based influenza vaccines. Vaccine. 2009;27:6399–6403. doi: 10.1016/j.vaccine.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 43.Christensen D., Korsholm K.S., Rosenkrands I., Lindenstrøm T., Andersen P., Agger E.M. Cationic liposomes as vaccine adjuvants. Expert Rev. Vaccines. 2007;6:785–796. doi: 10.1586/14760584.6.5.785. [DOI] [PubMed] [Google Scholar]
- 44.Alving C.R., Beck Z., Matyas G.R., Rao M. Liposomal adjuvants for human vaccines. Expert Opin. Drug Deliv. 2016;13:807–816. doi: 10.1517/17425247.2016.1151871. [DOI] [PubMed] [Google Scholar]
- 45.Swaminathan G., Thoryk E.A., Cox K.S., Smith J.S., Wolf J.J., Gindy M.E., Casimiro D.R., Bett A.J. A Tetravalent sub-unit dengue vaccine formulated with ionizable cationic lipid nanoparticle induces significant immune responses in rodents and non-human primates. Sci. Rep. 2016;6:34215. doi: 10.1038/srep34215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam Y.Y.C., Chen S., Cullis P.R. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics. 2013;5:498–507. doi: 10.3390/pharmaceutics5030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 49.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W.Y., Stebbing D., Crosley E.J., Yaworski E., et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 51.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melamed J.R., Hajj K.A., Chaudhary N., Strelkova D., Arral M.L., Pardi N., Alameh M.G., Miller J.B., Farbiak L., Siegwart D.J., et al. Lipid nanoparticle chemistry determines how nucleoside base modifications alter mRNA delivery. J. Contr. Release. 2022;341:206–214. doi: 10.1016/j.jconrel.2021.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lutz J., Lazzaro S., Habbeddine M., Schmidt K.E., Baumhof P., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Heidenreich R., Fotin-Mleczek M. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chivukula S., Plitnik T., Tibbitts T., Karve S., Dias A., Zhang D., Goldman R., Gopani H., Khanmohammed A., Sarode A., et al. Development of multivalent mRNA vaccine candidates for seasonal or pandemic influenza. NPJ Vaccines. 2021;6:153. doi: 10.1038/s41541-021-00420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hassett K.J., Higgins J., Woods A., Levy B., Xia Y., Hsiao C.J., Acosta E., Almarsson Ö., Moore M.J., Brito L.A. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Contr. Release. 2021;335:237–246. doi: 10.1016/j.jconrel.2021.05.021. [DOI] [PubMed] [Google Scholar]
- 58.Alishetty S., Carrasco M., Alameh M.G., Paige M., Said H., Wright L., Narayanan A., Alem F., Hernandez K., Gillevet P., et al. Novel lipid nanoparticle provides potent SARS-CoV-2 mRNA vaccine at low dose with low local reactogenicity, high thermostability and limited systemic biodistribution. Res Sq. 2021 doi: 10.21203/rs.3.rs-798453/v1. [DOI] [Google Scholar]
- 59.Cui L., Hunter M.R., Sonzini S., Pereira S., Romanelli S.M., Liu K., Li W., Liang L., Yang B., Mahmoudi N., Desai A.S. Mechanistic studies of an automated lipid nanoparticle reveal critical pharmaceutical properties associated with enhanced mRNA functional delivery in vitro and in vivo. Small. 2022;18:e2105832. doi: 10.1002/smll.202105832. [DOI] [PubMed] [Google Scholar]
- 60.Pollard C., Rejman J., De Haes W., Verrier B., Van Gulck E., Naessens T., De Smedt S., Bogaert P., Grooten J., Vanham G., De Koker S. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 2013;21:251–259. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Röhss J., John S., Hassett K., Yuzhakov O., Bahl K., et al. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tahtinen S., Tong A.-J., Himmels P., Oh J., Paler-Martinez A., Kim L., Wichner S., Oei Y., McCarron M.J., Freund E.C., et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat. Immunol. 2022;23:532–542. doi: 10.1038/s41590-022-01160-y. [DOI] [PubMed] [Google Scholar]
- 64.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-Cov-2-preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ripoll M., Bernard M.-C., Vaure C., Bazin E., Commandeur S., Perkov V., Lemdani K., Nicolaï M.C., Bonifassi P., Kichler A., et al. An imidazole modified lipid confers enhanced mRNA-LNP stability and strong immunization properties in mice and non-Human primates. Biomaterials. 2022;286:121570. doi: 10.1016/j.biomaterials.2022.121570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alving C.R. Design and selection of vaccine adjuvants: animal models and human trials. Vaccine. 2002;20:S56–S64. doi: 10.1016/s0264-410x(02)00174-3. [DOI] [PubMed] [Google Scholar]
- 67.Verlangieri A.J., DePriest J.C., Kapeghian J.C. Normal serum biochemical, hematological and EKG parameters in anesthetized adult male Macaca fascicularis and Macaca arctoides. Lab. Anim. Sci. 1985;35:63–66. [PubMed] [Google Scholar]
- 68.Abee C.R., Mansfield K., Tardif S., Morris T., editors. Nonhuman Primates in Biomedical Research: Diseases. Second edition. Volume 2. Elsevier; 2012. [Google Scholar]
- 69.Tsui N.B.Y., Ng E.K.O., Lo Y.M.D. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin. Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 70.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Contr. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data are not shared.