Abstract
Human members of the ELAV family, referred to as ELAV-like proteins (ELPs), include HuC, HuD, Hel-N1 and HuR. These proteins bind to AU-rich elements in the 3′-untranslated regions (3′-UTRs) of many growth-related mRNAs, including c-myc and VEGF, and may participate in regulating the stability of these transcripts. Here, I have developed an enzyme-linked immunosorbent assay (ELISA) which can rapidly assess the RNA–protein-binding properties of ELPs. With this assay, I demonstrate that HuC and HuD bind to the VEGF 3′-UTR regulatory segment (VRS) and to the c-myc 3′-UTR in a specific and concentration-dependent pattern, with both proteins showing a greater affinity for the VRS. Further analysis of the VRS indicated that the binding affinity was greater for the 3′-end where the majority of AU motifs reside. Binding to the VRS could be competed by both proteins as well as a poly(U) ribohomopolymer. The binding could not be competed by other ribohomopolymers or serum from patients with high titer anti-HuD antibodies. In summary, this assay provides a rapid analysis of ELP–RNA binding which can be utilized for further characterization of RNA-binding properties and for identification of competitor molecules for in vivo functional analysis of ELPs.
INTRODUCTION
AU-rich elements (ARE) are present within the 3′-untranslated regions (3′-UTRs) of many protooncogene, cytokine and lymphokine mRNAs and has been linked to the stability of these transcripts (1,2). The ELAV family of RNA-binding proteins is a prime candidate for participation in this level of gene regulation because of its avid and specific binding to the ARE (3–6). The four human members, HuR, Hel-N1, HuC and HuD, referred to as ELAV-like proteins (ELP), have been shown by immunoprecipitation or gel mobility shift assay to bind c-myc, c-fos, Id, N-myc, GM-CSF, VEGF and GAP-43 mRNAs (3,4,6–10). Although the biological consequence of this binding remains to be elucidated, recent evidence suggests that HuR may participate in the stabilization of VEGF and c-fos mRNAs (11–13). While the basic requirement of an ARE for ELAV protein binding is clear, many questions remain unanswered. Different members, for example, have been shown to bind to the same 3′-UTRs, such as c-fos and c-myc (3,4,6), but do some family members bind preferentially to certain 3′-UTRs? Are there other requirements for binding in addition to the AU motifs within the ARE? Are there competitor molecules that can disrupt the RNA–protein binding and thus be utilized in vivo to investigate the biological function of ELPs? Here I describe an enzyme-linked immunosorbent assay (ELISA)-based RNA-binding assay which can rapidly delineate the RNA-binding properties of ELPs. Using the 3′-UTRs of VEGF and c-myc as model RNA targets, I demonstrate several novel binding characteristics of two neural-specific ELPs, HuC and HuD. This assay will facilitate future efforts to characterize the ELP–RNA interaction and to identify competitor molecules which can inhibit this interaction.
MATERIALS AND METHODS
Synthesis of RNA probes
The human VEGF 3′-UTR was cloned by reverse transcription–PCR as follows. Total RNA was extracted from the human glioma cell line U251 using Trizol (Gibco BRL, Gaithersburg, MD) and reverse transcribed with random hexamers as previously described (14). The following oligodeoxynucleotides were then used to amplify the VEGF regulatory segment (VRS) within the VEGF 3′-UTR shown previously to bind HuR (11): 5′-TTCAGTATTCTTGGTTAATA-3′ (upstream) and 5′-GA-ATTTAATTCTGATCTCAAAG-3′ (downstream). The PCR product was subcloned into the pCRTopo vector (InVitrogen, San Diego, CA) and verified by sequencing. For c-myc 3′-UTR, a human genomic clone was kindly provided by Dr David Jones. The following oligonucleotides were used to amplify the 3′-UTR: 5′-GGAAAAGTAAGGAAAACGA-3′ (upstream) and 5′-TT-TATTTTTTCTAAAAACAATAG-3′ (downstream). The PCR reaction conditions were as previously described (14). The product was then subcloned into the SmaI site of pBluescript KS (Stratagene, La Jolla, CA). The negative control RNA consisted of pBluescript SK linearized with PvuII, which yields a transcript of 332 nt with T7 RNA polymerase. The RNA probes were transcribed as follows. Two micrograms of linearized template were added to transcription buffer, which consisted of 40 mM Tris–HCl, pH 7.5, 6 mM MgCl2, 10 mM DTT, 2 mM spermidine, 10 mM NaCl, 1 mM ATP, CTP and GTP, 6.5 mM UTP, 4.0 mM biotin-16-UTP (Boehringer Mannheim, Indianapolis, IN), 10 U RNasin (Promega, Madison, WI) and 20 U T7 RNA polymerase (Promega, Madison, WI). The reactions were carried out for 2 h at 37°C. Three units of RQ1 DNase I (Promega, Madison, WI) were added to the samples and incubated for an additional 15 min at 37°C. The probes were precipitated in 0.4 M LiCl2 and then analyzed on a denaturing acrylamide gel. The probes were quantitated using a commercial assay (BioWorld, Dublin, OH).
Preparation of recombinant proteins and ELISA plates
HuC and HuD were expressed as fusion proteins with a 6-histidine tag as previously described (15). Recombinant kinesin, fused to the same tag, was kindly provided by Dr Steven Rosenfeld. The proteins were quantitated with a commercial assay (Bio-Rad, Hercules, CA) using bovine serum albumin (BSA) as the standard. All proteins were diluted in phosphate-buffered saline (PBS) and incubated overnight at 4°C in a high-binding ELISA plate (Costar, Cambridge, MA) in a total volume of 50 µl. The following day, just prior to the binding assay, the plates were washed four times in diethylpyrocarbate (DEPC)-treated PBS.
RNA-binding assay
All binding reactions were carried out in 50 µl of 25 mM HEPES, pH 7.5, 0.5 mM EGTA, 100 mM NaCl, 0.5 mM DTT, 4 mM MgCl2, 20 mM KCl, 0.05% NP-40, 0.5 mg/ml yeast tRNA, 0.05 mg/ml poly(A)+ RNA, 0.125 mg/ml BSA, 0.4 mM vanadyl ribonucleoside complex, 80 U/ml RNasin, 5% glycerol. The probe/buffer mix was added to the wells and then incubated for 30 min at room temperature. The wells were washed four times in DEPC-treated PBS and drained completely. The wells were incubated with alkaline phosphatase-linked streptavidin (Pierce, Rockford, IL), diluted to 9.6 µg/ml in 50 µl of DEPC-treated PBS. After a 30 min incubation at room temperature, the wells were washed four times in DEPC-treated PBS and then incubated with 75 µl of p-nitrophenyl phosphate (Sigma, St Louis, MO) for times ranging from 10 to 30 min. The plates were analyzed in a Titertek Multiskan MCC/340 Reader (ICN, Costa Mesa, CA) using a 405 nm filter. All binding reactions were done in duplicate and the average OD was taken and expressed as arbitrary units. For RNA kinetics, 0.1 pmol of VRS probe and 300 ng of HuD were utilized for each binding reaction. Binding curves were estimated using DeltaGraph software (SSPS, Chicago, IL). RNA-binding curves for the VRS and c-myc probes as well as all competition assays were done in duplicate or triplicate and found to be highly reproducible.
Competition assays
For binding in the presence of competitor ELPs, 100 ng of HuD or HuC was adsorbed to the ELISA well as described above. A fixed amount of VRS (1 pmol) probe was used in the binding reaction. Competitor protein (HuD or HuC) was added to the RNA-binding buffer which was then immediately added to the ELISA well. For homoribopolymer competitions, 300 ng of HuD or HuC protein was used in the ELISA well and 1.25 pmol of RNA probe (VRS or c-myc 3′-UTR) in the binding reaction. Poly(A), poly(C) and poly(U) ribohomopolymers (Pharmacia, Piscataway, NJ) were added to the binding buffer (containing the probe) which was then immediately added to the ELISA well. For the anti-Hu antibody experiments, 2 µl of patient or normal control serum was added just prior to the binding reaction. All competitions were carried out simultaneously in the same ELISA plate.
RESULTS
The Hu proteins bind to the c-myc 3′-UTR and the VRS in a concentration-dependent pattern
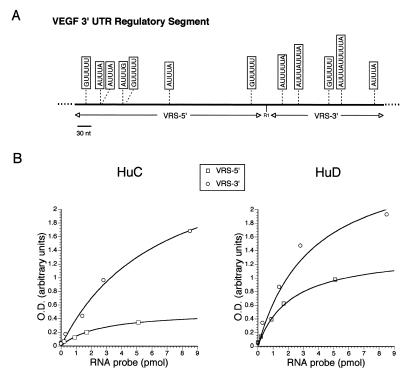
Probes for VEGF and c-myc were derived from 3′-UTR sequences previously shown to bind ELPs by immunoprecipitation or gel shift mobility assays (Fig. 1) (3,11). The VEGF probe represents a portion of the 3′-UTR referred to as the VEGF regulatory segment (VRS) (4). This segment has been implicated in the post-transcriptional regulation of VEGF mRNA (16). Motifs that have been associated with ELP–RNA binding are underlined in Figure 1 (3–5). HuD, HuC and control proteins were incubated on the same ELISA plate and all RNA-binding reactions were carried out simultaneously to minimize any variation in conditions or incubation times. As shown in Figure 2A and B, HuD and HuC bound to the VRS and the c-myc 3′-UTR probes in a concentration-dependent pattern. Recombinant kinesin (biosynthesized in the same way as recombinant HuC and HuD) and BSA did not bind to the RNA probes, indicating that the ELP binding was specific. As a control for the RNA, a riboprobe transcribed from pBluescript vector sequence was incubated with all proteins (in parallel with the VRS and c-myc reactions) and did not bind to any of the proteins over a similar range of probe concentrations (Fig. 2C). The OD values for the VRS were overall substantially higher than those for c-myc with both HuD and HuC. Although this difference may reflect an increased number of uridine residues in the VRS probe (165 versus 98 for c-myc), the apparent Kd was nearly 10-fold less than for c-myc with HuD (0.2 versus 2.0 pmol) and 5-fold less with HuC (0.4 versus 2.0 pmol). These findings indicate that both ELPs have a higher affinity for the VRS. This difference may be related to the number of AU motifs in the two probes. Compared to c-myc, the VRS probe contains an additional three AUUUA motifs (total of seven) and two longer motifs, AUUUUA and AUUUUUA, which have been identified as high affinity sites for another ELP, HuR (Fig. 1) (4). The c-myc 3′-UTR only contained one of these longer motifs. The VRS also contained a higher number of two other motifs (GUUUU and AUUUG) which had been identified in an immunoprecipitation-based RNA-binding assay with Hel-N1, another ELP (3). To determine further whether the binding affinity was governed by the number of AU motifs, the VRS was truncated into 5′- and 3′-fragments using an internal EcoRI site (Fig. 1). The 5′-fragment contained three AUUUA motifs whereas the 3′-segment contained four motifs (two being in tandem). The latter fragment also contained the two variant motifs, AUUUUA and AUUUUUA. Both probes were tested simultaneously in the same ELISA plate. As shown in Figure 3, HuC and HuD bound more avidly to the 3′-segment, with respect to maximal binding (~2-fold greater with HuD and 6-fold greater with HuC). This difference cannot be explained by the discrepancy in labeling as the 5′-fragment has 68% more uridine residues than the 3′-fragment. Moreover, the 5′-fragment contained three GUUUU pentamers versus one in the 3′-fragment, indicating that this motif contributes less to the binding affinity of HuC and HuD for the VRS. These findings suggest that the degree of binding to HuC and HuD is related to the number of AU motifs within the RNA sequence.
Figure 1.
Sequence from the 3′-UTRs of c-myc and VEGF utilized as probes in the ELISA-based RNA-binding assay. The VEGF probe represents the portion of the VEGF 3′-UTR which encompasses the VRS (11,16). Motifs linked to ELAV-like protein binding are underlined (3–5). The boxed sequence is the nonamer motif associated with mRNA degradation (22,23). The arrowhead indicates the EcoRI restriction site used to create truncated VRS probes.
Figure 2.
RNA-binding curves for HuC, HuD and control proteins (kinesin and BSA) generated by the ELISA assay. Three biotinylated riboprobes (VRS, c-myc 3′-UTR and Bluescript) were analyzed at the concentrations shown.
Figure 3.
RNA-binding analysis of HuD and HuC with truncated portions of the VRS. (A) Schematic diagram of the VRS highlighting the motifs which have been linked to ELP–RNA binding (3–5). The truncated VRS probes are shown below the diagram. (B) RNA-binding curves for HuC and HuD to the truncated VRS probes.
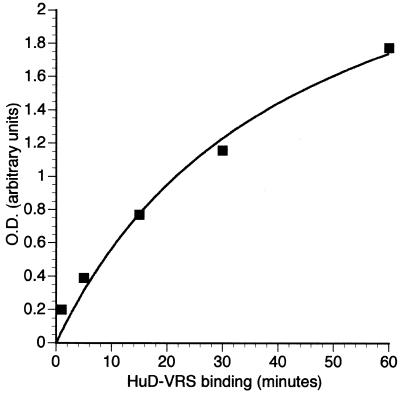
The kinetics of HuD and VRS binding were further analyzed in a series of timed reactions. Unlike gel mobility shift or immunoprecipitation assays, where the inter-sample lag time for washing or gel loading is significant, all reactions in the ELISA assay for RNA kinetics were initiated and terminated simultaneously. As shown in Figure 4, the steepest slope in the curve was between 0 and 30 min. These kinetics were similar to those of HuD and the c-fos 3′-UTR where the largest increase in binding for complex II occurred between the 10 and 30 min intervals (5). Interestingly, those binding experiments were done at 37°C (versus 25°C in this study). This assay can therefore accurately assess RNA-binding kinetics and may be used for future analyses of wild-type and mutant ELPs in a variety of environmental conditions, such as hypoxia or acidosis.
Figure 4.
Analysis of RNA-binding kinetics with HuD and the VRS. The probe was incubated in the ELISA well with adsorbed HuD for varying time intervals and the OD values were obtained as described in Materials and Methods.
HuD and HuC can compete with each other for binding to the VRS
HuD or HuC protein was added to the RNA-binding buffer at varying excess amounts (relative to the amount of protein affixed to the ELISA well) immediately prior to the binding reaction. For these experiments, less binding protein (100 ng) was used in the ELISA well to reduce the amount of exogenous competitor protein required. As shown in Figure 5, the binding of HuC and HuD to VRS was reduced in a concentration-dependent manner using either protein as a competitor. As little as 10-fold addition of competitor maximally reduced binding. Both proteins were equally effective in their capacity to abrogate RNA binding. The overall lower OD values reflect the reduced amount of binding protein used in the experiment. No competition was seen when equal amounts of BSA were added (not shown). Although the competitor protein in solution may have an advantage over the affixed protein for RNA binding, these findings underscore the specificity of the RNA–protein binding and suggest that the ELPs can effectively compete with each other for RNA binding.
Figure 5.
RNA-binding results with the VRS and ELPs in the presence of varying concentrations of competitor ELPs (indicated in the figure). The primary ELP adsorbed to the ELISA well is indicated above each graph.
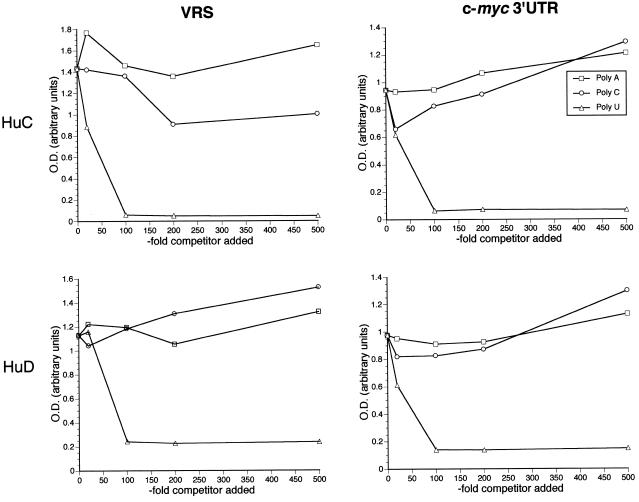
A poly(U) homoribopolymer can compete with HuC and HuD binding to the c-myc 3′-UTR and the VRS
The binding reaction was carried out in the presence of three ribohomopolymers, poly(A), poly(C) and poly(U), at various amounts in excess of the VRS and c-myc probes. As shown in Figure 6, there was a marked and consistent inhibition of both VRS and c-myc binding with the poly(U) homoribopolymer, occurring between 10- and 100-fold excess of probe. This range of inhibitory concentrations was similar to that observed by gel mobility shift assay with the Xenopus homolog of HuR (elr A) and the UUUUUAU-type cytoplasmic polyadenylation element (17). With the exception of HuC and VEGF, the addition of poly(A) and poly(C) did not inhibit binding. In contrast to poly(U), there was even enhancement of binding at higher concentrations (e.g. 500-fold excess of probe). A modest inhibition of HuC and VRS binding by poly(C) (Fig. 6A, right) was observed at higher concentrations (200- and 500-fold excess probe).
Figure 6.
RNA-binding results with the VRS (A) or the c-myc 3′-UTR (B) and HuC or HuD in the presence of varying excess concentrations of ribohomopolymers (shown as fold excess of RNA probe).
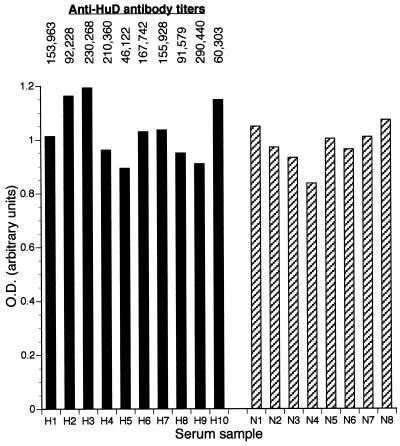
VRS binding to HuD in the presence of anti-Hu antibodies
Patients with paraneoplastic encephalomyelitis/sensory neuronopathy develop high titer polyclonal antibodies to ELPs (3,7,15,18). These patients suffer from various central nervous system signs and symptoms ranging from sensory neuronopathy to limbic encephalitis (19,20). Although the pathophysiology of this disease is unknown, one possibility is that the polyclonal immune response may disrupt binding of the ELP to target RNAs (6). As an initial step toward testing that possibility, the ELISA assay was used to evaluate the ability of HuD to bind to RNA in the presence of anti-Hu antibodies. The VRS was chosen as the target RNA because of its robust binding to HuD. As shown in Figure 7, there was no significant inhibition of RNA binding in the presence of anti-Hu sera compared to control sera. While a fixed volume of patient serum was used in each binding experiment, the anti-HuD titer, as determined by ELISA (15), varied from 1:46 122 to 1:230 268, thus providing a range of antibody doses. Preincubating the serum in the ELISA well 2 h prior to adding the RNA probe also did not show any inhibitory effect on RNA binding (not shown). It should be stressed, however, that the in vivo concentrations of antibody within the central nervous system are substantially higher (21) and thus a critical antibody dosage may not have been obtained here.
Figure 7.
RNA binding with HuD and the VRS in the presence of anti-HuD (H) or normal (N) serum. The corresponding anti-HuD titer for each serum sample (as measured elsewhere; 15) is shown above the graph.
DISCUSSION
In this paper I have demonstrated a novel, ELISA-based assay to analyze the RNA-binding properties of HuC and HuD using the c-myc 3′-UTR and the VRS as model RNA targets. The biological relevance of these RNA–protein interactions has recently come to light when HuR, another ELP, was shown to participate in stabilization of the VEGF transcript under hypoxic conditions (11). The ELPs, however, have been shown to bind the 3′-UTRs of many other growth-related genes, almost interchangeably. This assay will expedite both the analysis and comparison of binding by individual ELPs to these many possible target RNAs. Furthermore, it will help define additional cis elements which participate in the specificity of binding among ELPs.
Previous RNA-binding analyses with ELPs, including the initial one linking them to AU-rich RNA sequences, have centered on the standard techniques of immunoprecipitation, gel mobility shift, UV crosslinking or filter binding (3,4,6,8,9,11). In each of these assays, both the protein and target RNA remain in solution during the binding phase. The assay described here, however, indicates that the RNA-binding capacity of ELPs is preserved following adsorption to an ELISA well. The ELISA assay permits a rapid assessment (requiring <2 h) of RNA-binding properties as shown with the c-myc 3′-UTR and the VRS (Fig. 2). For each protein, a concentration-dependent binding curve is generated, from which an apparent Kd can be calculated. The lack of any signal with control proteins or RNA, moreover, indicated that the RNA–protein binding within the ELISA well was highly specific. The assay also allowed an accurate assessment of binding kinetics, indicating that near maximal binding of HuD to the VRS occurred within 60 min. In addition to a shortened time of analysis, the advantage of this assay is that a large number of individual RNA-binding experiments can be performed simultaneously. With the use of a multi-pipettor and washer, for example, 40–60 binding reactions can be initiated and stopped within 1 min. This number of samples would be prohibitive for the standard techniques of gel shift analysis or immunoprecipitation. Likewise, the assay utilizes biotinylated probes which obviates the need for radioisotopes.
With this assay, the first observation with HuC and HuD was the substantially higher affinity of both proteins for the VRS compared to the c-myc 3′-UTR (Fig. 2). The apparent Kd, for example, was from 4- to 10-fold higher for the VRS. To explore potential explanations for this discrepancy, both sequences were analyzed with respect to motifs linked to ELP binding. In the original RNA-binding study with ELPs, Hel-N1 was shown by immunoprecipitation to bind three U-rich motifs, including AUUUG, GUUUUU and the canonical motif for RNA stability, AUUUA (1–3). Since then, two other high affinity binding sites have been identified for HuD and HuR (in addition to the AUUUA motif): AUUUUA and AUUUUUA (4,5). The higher affinity of binding to the VRS probe observed here may relate to the greater number of AU motifs within this sequence. Notably, the VRS probe contains a higher number of nearly all the motifs which have been linked to ELP binding (Table 1). The c-myc probe also lacks one of the two high affinity sites identified for HuR by RNase T1 mapping (4). The importance of these motifs was demonstrated by analyzing the truncated segments of the VRS (Fig. 3). When these motifs are not present, as with the VRS 5′ construct, binding of both HuD and HuC was markedly reduced (Fig. 3). This finding is similar to that for HuD and c-fos ARE binding, where deletion of two uridine stretches of 4–7 nt (with flanking adenosines) completely abrogated binding (5). Interestingly, both the VRS 5′ and 3′ constructs had a similar number of AUUUA motifs and the 5′ construct had a higher number of GUUUUs, which further emphasizes the importance of longer uridine stretches in binding affinity. The effective inhibition of ELP–VRS binding by the poly(U) homoribopolymer further supports this observation. The absolute number of uridine residues, however, does not predict ELP binding as the VRS 3′ construct contained 59% fewer residues than the 5′ construct. Interestingly, the 3′ construct contained the nonamer motif UUAUUUAUU, which has been linked to mRNA degradation (22,23). The importance of this motif in ELP binding will require additional mutational analyses.
Table 1. Summary of the Elav-like protein binding motifs within the c-myc 3′UTR and the VEGF 3′UTR regulatory segment.
aPreviously linked to Elav-like protein binding (3–5).
bVEGF 3′UTR regulatory segment.
cVRS segment 3′ to the EcoRI site in Figure 1.
dVRS segment 5′ to the EcoRI site in Figure 1.
This assay also effectively demonstrates the capacity of ELPs to compete with each other for RNA binding. The competition for binding to the VRS occurred with similar concentrations of either protein (10-fold excess), which is consistent with the similar binding affinities observed in Figure 2. This finding suggests that the ELPs could potentially be used as inhibitors to further analyze the biological roles of individual family members in certain cell systems. Overexpression of HuD in gliomas, for example, could effectively compete with HuR for VEGF binding and block the adaptive responses to hypoxia (11). Polyclonal anti-ELP antibodies, on the other hand, did not compete for VRS binding over a range of titers. This finding raises the possibility that the dominant epitopes for the polyclonal antibodies are away from the RNP motifs where RNA binding is thought to occur (24,25). The antibodies, however, may participate in the pathogenesis of paraneoplastic encephalomyelitis by other mechanisms, such as inhibiting ELP interactions with other proteins.
In summary, the ELISA-based RNA-binding assay provides a rapid assessment of ELPs and their capacity to bind AREs. Since a large number of growth-related genes, including cytokine, lymphokine and other protooncogene genes, contain AREs in the 3′-UTR (2), the assay will permit a rapid analysis and ranking of the ELPs and their capacity to bind these mRNAs. Furthermore, this assay will provide an efficient screening process to identify potential competitor molecules (peptides, nucleic acids or antibodies) that can disrupt RNA binding. The in vivo inhibition of HuR binding to the VRS, for example, may destabilize the mRNA and down-regulate its expression (11–13,26). Such an inhibition could have therapeutic benefits in cancers where neovascularization has been linked to the stabilization of VEGF mRNA (27–29).
Acknowledgments
ACKNOWLEDGEMENTS
I would like to thank Dr Steven Hajduk for his thoughtful comments in the preparation of this manuscript. This work was supported by a grant from the American Cancer Society (AC RPG-97-111-01-CCE). Oligonucleotides were provided by the Oligo Core Facility at UAB with support from NIH grant 5P50 CA13148.
REFERENCES
- 1.Shaw G. and Kamen,R. (1986) Cell, 46, 659–667. [DOI] [PubMed] [Google Scholar]
- 2.Chen C.A. and Shyu,A. (1995) Trends Biochem. Sci., 20, 465–470. [DOI] [PubMed] [Google Scholar]
- 3.Levine T.D., FenBiao,G., King,P.H., Andrews,L.G. and Keene,J.D. (1993) Mol. Cell. Biol., 13, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma W., Cheng,S., Campbell,C., Wright,A. and Furneaux,H. (1996) J. Biol. Chem., 271, 8144–8151. [DOI] [PubMed] [Google Scholar]
- 5.Chung S., Jiang,L., Cheng,S. and Furneaux,H. (1996) J. Biol. Chem., 271, 11518–11524. [DOI] [PubMed] [Google Scholar]
- 6.Liu J., Dalmau,J., Szabo,A., Rosenfeld,M., Huber,J. and Furneaux,H. (1995) Neurology, 45, 544–550. [DOI] [PubMed] [Google Scholar]
- 7.Szabo A., Dalmau,J., Manley,G., Rosenfeld,M., Wong,E., Henson,J., Posner,J.B. and Furneaux,H.M. (1991) Cell, 67, 325–333. [DOI] [PubMed] [Google Scholar]
- 8.King P.H., Levine,T.D., Fremeau,R.T. and Keene,J.D. (1994) J. Neurosci., 14, 1943–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chagnovich D., Fayos,B.E. and Cohn,S.L. (1996) J. Biol. Chem., 271, 33587–33591. [DOI] [PubMed] [Google Scholar]
- 10.Chung S., Eckrich,M., Perrone-Bizzozero,N., Kohn,D.T. and Furneaux,H. (1997) J. Biol. Chem., 272, 6593–6598. [DOI] [PubMed] [Google Scholar]
- 11.Levy N., Chung,S., Furneaux,H. and Levy,A. (1998) J. Biol. Chem., 273, 6417–6423. [DOI] [PubMed] [Google Scholar]
- 12.Peng S.S.-Y., Chen,C.-Y.A., Xu,N. and Shyu,A.-B. (1998) EMBO J., 17, 3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan X.C. and Steitz,J.A. (1998) EMBO J., 17, 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King P.H. (1994) Gene, 151, 261–265. [DOI] [PubMed] [Google Scholar]
- 15.King P.H., Redden,D., Palmgren,J.S., Nabors,L.B. and Lennon,V.A. (1999) J. Autoimmun., 13, 435–443. [DOI] [PubMed] [Google Scholar]
- 16.Levy A.P., Kevy,N.S. and Goldberg,M.A. (1996) J. Biol. Chem., 271, 2746–2753. [DOI] [PubMed] [Google Scholar]
- 17.Wu L., Good,P.J. and Richter,J.D. (1997) Mol. Cell. Biol., 17, 6402–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dropcho E.J. and King,P.H. (1994) Ann. Neurol., 36, 200–205. [DOI] [PubMed] [Google Scholar]
- 19.Dalmau J., Graus,F., Rosenblum,M.K. and Posner,J.B. (1992) Medicine, 71, 59–72. [DOI] [PubMed] [Google Scholar]
- 20.Lucchinetti C.F., Kimmel,D.W. and Lennon,V.A. (1998) Neurology, 50, 652–657. [DOI] [PubMed] [Google Scholar]
- 21.Furneaux H.M., Reich,L. and Posner,J.B. (1990) Neurology, 40, 1085–1091. [DOI] [PubMed] [Google Scholar]
- 22.Zubiaga A.M., Belasco,J.G. and Greenberg,M.E. (1995) Mol. Cell. Biol., 15, 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagnado C.A., Brown,C.Y. and Goodall,G.J. (1994) Mol. Cell. Biol., 14, 7984–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenan D.J., Query,C.C. and Keene,J.D. (1991) Trends Biochem. Sci., 16, 214–220. [DOI] [PubMed] [Google Scholar]
- 25.Nagai K., Oubridge,C., Jessen,T.H., Li,J. and Evans,P.R. (1990) Nature, 348, 515–520. [DOI] [PubMed] [Google Scholar]
- 26.Myer V.E., Fan,X.C. and Steitz,J.A. (1997) EMBO J., 16, 2130–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda E., Achen,M.G., Breier,G. and Risau,W. (1995) J. Biol. Chem., 270, 19761–19766. [DOI] [PubMed] [Google Scholar]
- 28.Damert A., Machein,M., Breier,G., Fujita,M.Q., Hanahan,D., Risau,W. and Plate,K.H. (1997) Cancer Res., 57, 3860–3864. [PubMed] [Google Scholar]
- 29.Stein I., Neeman,M., Shweiki,D., Itin,A. and Keshet,E. (1995) Mol. Cell. Biol., 15, 5363–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]