Abstract
Scoparone (SCO) is a compound found in the stems and leaves of Artemisia capillaris. The pharmacological uses of SCO include significant hypotensive, cholagogic, anti-inflammatory, analgesic, lipid-lowering, anti-asthmatic and anti-coagulant effects. The present study aimed to verify the anticancer potential of SCO in breast cancer (BC) cells and its underlying molecular mechanism. Cell Counting Kit-8 and flow cytometry were used to analyze the effects of SCO on cell viability and apoptosis. Nucleocytoplasmic separation was used to analyze the location of the long non-coding RNA (lncRNA) small nucleolar RNA host gene 12 (SNHG12) in BC cells. Reverse transcription-quantitative PCR was used to analyze the effect of SCO on the expression levels of SNHG12, microRNA (miRNA/miR)-140-3p and tumor necrosis factor receptor associated factor 2 (TRAF2). Western blotting was used to analyze the protein expression levels of TRAF2 and downstream nuclear factor κB (NF-κB) signaling pathways. The results demonstrated that SCO had a time- and dose-dependent inhibitory effect on the viability of BC cells, and that the upregulated lncRNA SNHG12 in BC cells was inhibited by SCO. SNHG12, which was primarily expressed in the cytoplasm, acted as a competing endogenous RNA, sponged miR-140-3p and inhibited the expression of miR-140-3p. The transcriptional activity and translational level of TRAF2, a downstream target of miR-140-3p, decreased following the SCO-mediated suppression of SNHG12 expression. As an upstream effector, TRAF2 activity reduction mediated the inhibition of NF-κB signaling, decreased the viability and migration of BC cells, and promoted BC cell apoptosis. In conclusion, SCO-induced inhibition of viability and promotion of apoptosis in BC cells are achieved through the inhibition of NF-κB signaling, which is associated with regulation of the SNHG12/miR-140-3p/TRAF2 axis. This understanding provides new drug candidates for the treatment of BC and a theoretical basis for biology.
Keywords: SCO, BC, SNHG12, miR-140-3p, TRAF2, NF-κB
Introduction
Breast cancer (BC) is the most common cancer in women globally (1). However, the early stages of BC are considered curable (2); therefore, early screening and diagnostic marker detection are critical for effective BC treatment. As BC progresses, the growth of cancer cells accelerates and causes distant migration, which greatly shortens the survival expectancy of BC patients and increases the difficulty of establishing effective treatment strategies (3). Given the current limited treatment options, new drugs are urgently required to treat BC. The primary aim of this study was to explore the therapeutic effect and internal mechanism of scoparone (SCO; 6,7-dimethoxycoumarin) in BC.
Traditional Chinese medicine (TCM) has the remarkable ability to reduce the side effects of chemotherapy alongside being used to prevent and treat cancer (4). The anticancer effects of Chinese herbal medicines have also been verified against BC. For example, plantamajoside can inhibit BC cell growth and pulmonary metastasis by decreasing the activity of matrix metalloproteinase-9 and -2(5). Liew et al (6) determined that treatment with Chinese herbal medicine during chemotherapy reduced fatigue, nausea, and anorexia in BC patients; therefore, TCM can improve quality of life in cancer patients. SCO is the primary component of the Chinese herb Artemisia capillaris; this component specifically has antioxidant, lipid-lowering, and anti-inflammatory effects (7,8). Recent studies have demonstrated that scopolamine (SCO) can play a significant role in cancer treatment. For example, SCO shows significant anti-tumor effects against prostate cancer cells by inhibiting the activity of signal transducer and activator of transcription 3(9). SCO also reduces migration and induces apoptosis of laryngeal cancer cells in a dose-dependent manner (10). However, no systematic study has been conducted on the potential anti-tumor effect of SCO in BC.
Long noncoding RNAs (lncRNAs) are important players in the progression of BC (11), and their dysregulation may alter intercellular signal transduction and affect the growth of BC cells (12). In addition, lncRNAs can act as competing endogenous RNA (ceRNAs) that regulate the transcriptional activity and translation level of downstream mRNA by adsorbing microRNAs (miRNAs/miRs). For example, LINC00673 acts as a ceRNA that adsorbs miR-515-5p to regulate microtubule affinity-regulating kinase 4, thereby affecting the transduction of Hippo signaling and the growth of BC cells (13). Small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation, migration, and invasion, and inhibits BC cell apoptosis through the miR-15a-5p/spalt-like transcription factor 4 axis (14). In addition, miRNAs regulate the transcriptional activity and translation level of downstream mRNA by binding to the 3'-untranslated region (UTR) of mRNA. For example, the expression of miR-140-3p is reduced in BC, which promotes the expression of tripartite motif containing 28 and accelerates the viability, migration, and invasion of BC cells (15). Additionally, tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) is upregulated in BC; nonetheless, miR-502-5p can bind to the 3'-UTR of TRAF2, thereby inhibiting TRAF2 and reducing the progression of BC (16). TRAF2 also promotes the proliferation and metastasis of BC cells through the nuclear factor κB (NF-κB) pathway by directly interacting with various TNF receptors (17,18).
Overall, this study aims to investigate the effect and mechanism of SCO on BC cells and provides a theoretical basis for its potential clinical application in BC treatment.
Materials and methods
Cell culture and treatment
All cell lines used in this study were obtained from the Procell (Wuhan, China). These cell lines included the normal human mammary epithelial cell line MCF-10A (culture conditions: Dulbecco's modified eagle medium [DMEM]/nutrient mixture F-12 containing 5% horse serum) and the human BC cell lines MCF-7 (culture conditions: minimum essential medium containing 10% fetal bovine serum [FBS]), MDA-MB-231 (culture conditions: DMEM containing 5% FBS), ZR-75-1 (culture conditions: Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% FBS), HCC1937 (culture conditions: RPMI-1640 containing 10% FBS), and HEK293 (culture conditions: DMEM containing 10% FBS). All cells were seeded in 6-well plates at a density of 2x105; gradient concentrations of SCO (0, 100, 200, 500, and 1,000 µM) were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used to treat cells for 0, 24, 48, or 72 h.
Cell transfection
Initially, ov-SNHG12 (50 nM; pcDNA3.1), miR-140-3p mimic/inhibitor (100 nM), and three TRAF2-targeting small interfering RNAs (si-TRAF2) (50 nM) were transfected into MCF-7 and MDA-MB-231 cells (density of 3x105) using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc., Massachusetts, USA), according to the manufacturer's instructions. ov-SNHG12, miR-140-3p mimic/inhibitor, si-TRAF2, and their negative controls (NCs) were purchased from Sangon Biotech (Shanghai, China); the corresponding sequences are listed in Table SI.
Cell viability assay
MCF-10A and BC cells were seeded in 96-well plates at a density of 3x103 cells/well and cultured for 12 h. Optical density values were measured at 450 nm for 60 min at 37˚C; samples were taken at 24 h intervals across 72 h and measurements were conducted using a Cell Counting Kit-8 (CCK8; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and an enzyme labeling instrument (Thermo Fisher Scientific).
Nucleocytoplasmic separation and reverse transcription-quantitative PCR (RT-qPCR) assay
The Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, Ontario, Canada) was used to isolate RNA from the cells according to the manufacturer's instructions; total RNA was extracted from BC cells using TRIzol (Invitrogen). After centrifugation at 12,000 x g (4˚C, 10 min) in a high-speed refrigerated centrifuge (JIDI-17RS; Guangzhou JiDi Instrument Co., Ltd., Guangzhou, China), the precipitate was resuspended in diethyl pyrocarbonate water (Invitrogen); aliquots of these precipitates were then reverse-transcribed to complementary DNA using a PrimeScript RT kit (Takara Bio, Kyoto, Japan). The SYBR Premix Ex Taq II kit (Invitrogen) was used to perform RT-qPCR analysis using an ABI 7500 Real-Time PCR system (Applied Biosystems, California, USA). PCR amplification was performed under the following conditions: 95˚C for an initial 30 sec, followed by 40 cycles at 95˚ for 5 sec and 60˚C for 30 sec. The sequences of the primer pairs used for amplification are listed in Table SII. SNHG12, miR-140-3p, and TRAF2 RNA levels were normalized to those of the housekeeping genes β-actin or U6 and were determined using the 2-ΔΔCq method (19).
RNA pull-down assay
Biotin-labeled SNHG12 and miR-140-3p were transfected into BC cells. After 24 h of transfection, cells were lysed with RIPA lysis buffer and incubated with Dynabeads M-280 Streptavidin (Invitrogen) for 15 min, followed by RT-qPCR analysis. Biotinylated RNA was obtained from Sangon Biotech.
Flow cytometry (FCM) analysis of apoptosis
Apoptosis was detected using the Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (BD Biosciences, New Jersey, USA) according to the manufacturer's instructions. After incubation with annexin V-FITC (5 µl) and propidium iodide (PI; 10 µl) in the dark (15 min, 25˚C), cell apoptosis was assessed by FCM (FACSCanto II; BD FACSChorus software, version: 1.0; BD Biosciences).
Transwell migration and invasion assays
For the Transwell migration assay, BC cells were transferred to the upper chamber of a Transwell plate (Corning, New York, USA) containing 100 µl of culture medium without FBS. The lower chamber contained 600 µl of culture medium containing FBS. After 48 h of incubation, BC cells were immobilized with 4% paraformaldehyde for 15 min, washed with PBS, and stained with 0.1% crystal violet for 10 min. The migrated cells were counted in five randomly selected visual fields. For the Transwell invasion assay, the experimental procedure was similar to that of the Transwell migration assay, except for the replacement of the Transwell migration plate with the Transwell invasion plate (Corning).
Western blot analysis
A Beyotime Biotechnology kit (Shanghai, China) was used to detect the protein concentration in each sample. We mixed 40 µg protein with SDS buffer (5X), boiled the sample for 10 min, and added these samples to 10% SDS-PAGE gels for electrophoresis. Proteins were then transferred from the SDS-PAGE gel to a PVDF membrane (Millipore, Billerica, Massachusetts, USA). The membrane was blocked with 5% skim milk powder in Tris-buffered saline with Tween 20 (0.1%, v/v; Solarbio) for 1 h at 25˚C. The membrane was then incubated overnight at 4˚C, according to the optimum conditions of the primary antibodies. After washing thrice, the membrane was incubated with the appropriate horseradish peroxidase secondary antibody for 1 h. Finally, enhanced chemiluminescence (Millipore) was used to detect the blots. Antibodies against TRAF2 (1:1,000, ab126758), NF-κB (1:100, ab16502), and p-NF-κB (1:1,000, ab76302) were obtained from Abcam (Cambridge, UK).
Dual luciferase assays
The SNHG12 or TRAF2 3'-UTR sequences were inserted into the psi-CHECK2 construct and co-transfected into HEK293 cells with miR-140-3p mimic/inhibitor using Lipofectamine 3000 at 37˚C for 4 h. After 48 h of culture, the Dual-Luciferase Reporter assay system (Promega Corp., Wisconsin, USA) was used to measure luciferase activity at 490 nm. The ratio of firefly to Renilla luciferase activity was used to normalize the firefly luciferase values.
Statistical analysis
Three separate experiments were conducted for each measurement. The corresponding results are expressed as mean ± standard deviation. Statistical analysis was performed using an unpaired Student's t-test or one-way analysis of variance followed by Tukey's post hoc test using SPSS (Chicago, USA). Statistical significance was set at P<0.05.
Results
SCO inhibits BC cell growth and SNHG12 expression
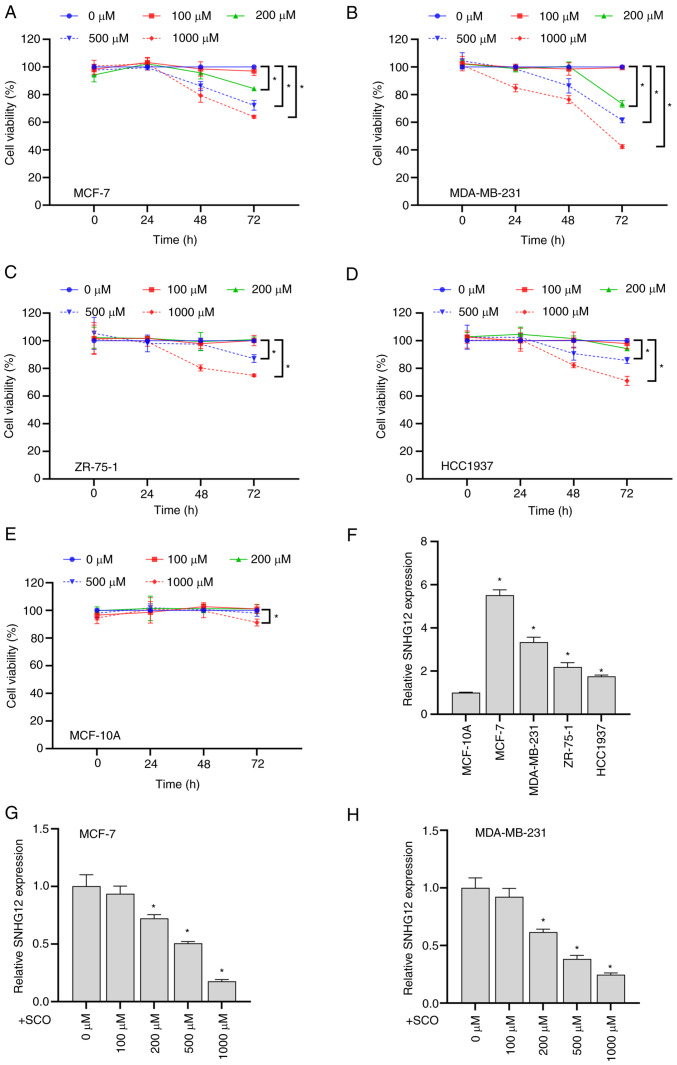
Overall, SCO exhibited time- and dose-dependent inhibitory effects on the viability of the four BC cell types to varying degrees (Fig. 1A-D). However, when the SCO dose reached 1000 µM, the viability of MCF-10A cells significantly decreased (Fig. 1E). Therefore, we used a relatively safe dose of 500 µM in subsequent experiments to explore the molecular mechanism by which SCO inhibits BC cell viability. Compared with the normal mammary epithelial cell line MCF-10A, lncRNA SNHG12 expression in BC cells was upregulated to varying degrees (Fig. 1F). Among these cell lines, the highest expression of SNHG12 was observed in MCF-7 and MDA-MB-231 cells. Therefore, these two cell lines were used in subsequent experiments to study cellular function. In addition, the expression of SNHG12 gradually decreased with increasing SCO concentrations (Fig. 1G and H). These results suggested that the inhibition of BC cell viability by SCO may be involved in corresponding inhibition of SNHG12 expression.
Figure 1.
SCO inhibits BC cell growth and SNHG12 expression. CCK8 assay to analyze the effects of different doses of SCO on the cell viability of (A) MCF-7, (B) MDA-MB-231, (C) ZR-75-1, (D) HCC1937 and (E) MCF-10A cells. *P<0.05. (F) RT-qPCR analysis of the differential expression of lncRNA SNHG12 in MCF-7, MDA-MB-231, ZR-75-1 and HCC1937 cells compared to MCF-10A. *P<0.05 vs. MCF-10A. RT-qPCR analysis of the differing expression levels of lncRNA SNHG12 in (G) MCF-7 and (H) MDA-MB-231 cells under different SCO doses. *P<0.05 vs. 0 µM. CCK8, Cell Counting Kit-8; SCO, scoparone; BC, breast cancer; lncRNA, long non-coding RNA; SNHG12, small nucleolar RNA host gene 12; RT-qPCR, reverse transcription-quantitative PCR.
Overexpression of SNHG12 alleviated the SCO-mediated growth inhibition of BC cells
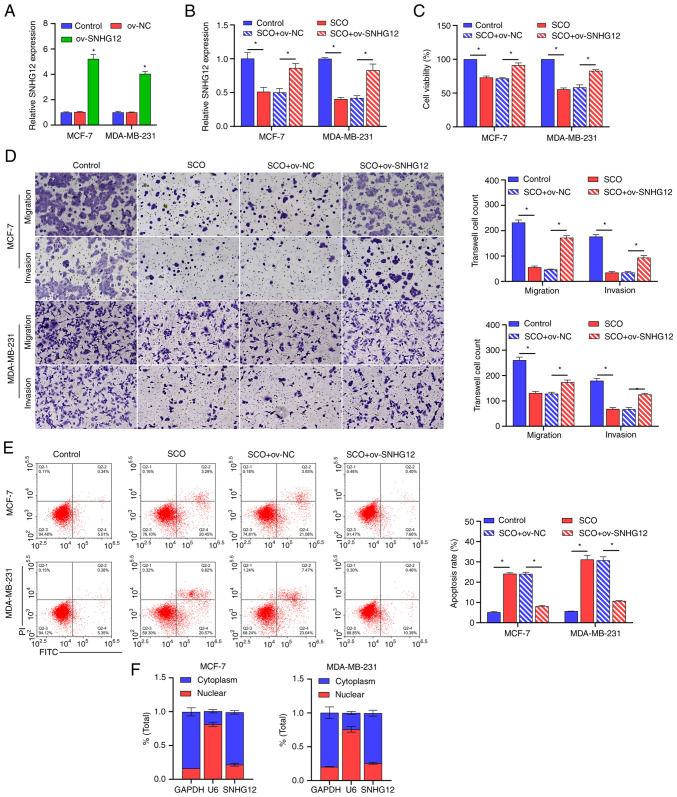
We constructed a SNHG12 overexpression plasmid and transfected it into BC cells. The corresponding results demonstrated that the expression of SNHG12 was significantly upregulated in ov-SNHG12 compared to that in ov-NC (Fig. 2A). Additionally, SCO inhibited SNHG12 expression, cell viability, and migration/invasion ability, and increased apoptosis, all of which were reversed by overexpression of SNHG12 (Fig. 2B-E). To further understand the functional principle of SNHG12, we conducted a nuclear plasma separation experiment. Similar to the positive control GAPDH, SNHG12 was highly expressed in the cytoplasm, whereas the positive control U6 was highly expressed in the nucleus (Fig. 2F). This finding suggests that SNHG12 exerts its downstream effects through post-transcriptional epigenetic regulation.
Figure 2.
Overexpression of SNHG12 alleviates the SCO-mediated growth inhibition of BC cells. (A) RT-qPCR analysis verifying the effectiveness of SNHG12 overexpression plasmids. *P<0.05 vs. ov-NC. (B) RT-qPCR analysis of the effect of SNHG12 overexpression on SNHG12 expression in SCO-treated cells. (C) CCK8 analysis of the effect of SNHG12 overexpression on BC cell viability in SCO-treated cells. (D) Transwell analysis of the effect of SNHG12 overexpression on BC cell migration/invasion in SCO-treated cells. Magnification, x200. (E) FCM analysis of the effect of SNHG12 overexpression on apoptosis in SCO-treated cells. (F) Nucleocytoplasmic separation analysis of the localization of SNHG12 in MCF-7 and MDA-MB-231 cells. *P<0.05. CCK8, Cell Counting Kit-8; SCO, scoparone; BC, breast cancer; lncRNA, long non-coding RNA; SNHG12, small nucleolar RNA host gene 12; ov, overexpression; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; FCM, flow cytometry; FITC, fluorescein isothiocyanate; PI, propidium iodide.
SNHG12 acts as a ceRNA to adsorb miR-140-3p
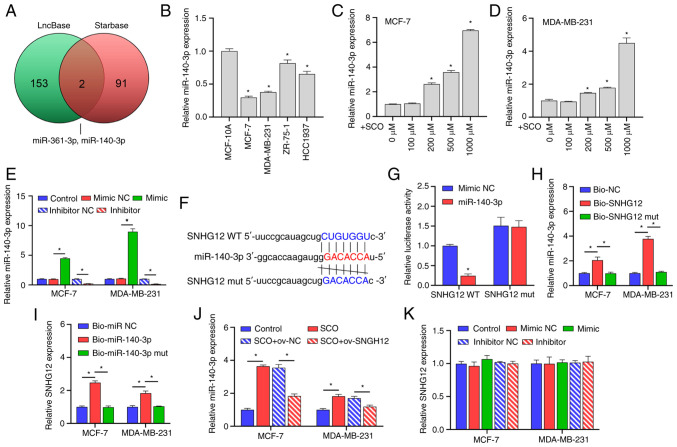
Through joint analysis of the LncBase and Starbase databases, we determined that miR-361-3p and miR-140-3p may be potential targets of SNHG12 (Fig. 3A). Studies have shown that miR-140-3p acts as a tumor suppressor gene in BC and when this gene is suppressed, BC progression is promoted (20,21); however, it has been established that miR-361-3p may be unfavorable for the treatment and prognosis of BC (22,23). Further, miR-140-3p expression was observed to be downregulated in both MCF-7 and MDA-MB-231 cells compared with that in the normal mammary epithelial cell line MCF-10A (Fig. 3B). However, with an increasing concentration of SCO, the miR-140-3p expression gradually increased (Fig. 3C and D). In this study, the miR-140-3p mimic/inhibitor was synthesized and transfected into MCF-7 and MDA-MB-231 cells. The corresponding expression of miR-140-3p was up-regulated in the mimic-transfected group and downregulated in the inhibitor-transfected group, suggesting that the synthesis of the miR-140-3p mimic/inhibitor was effective (Fig. 3E). Bioinformatic analysis demonstrated that there were potential binding sites between SNHG12 and miR-140-3p (Fig. 3F). Subsequent dual luciferase results showed that the fluorescence activity of the WT SNHG12 + miR-140-3p mimic co-transfected group was significantly decreased compared with that of the WT SNHG12 + miR-140-3p mimic NC co-transfected group. Nonetheless, there was no significant difference observed between the fluorescence activity of the mutant SNHG12 + miR-140-3p inhibitor NC co-transfection group and the mutant SNHG12 + miR-140-3p inhibitor co-transfection group (Fig. 3G). Subsequent RNA pull-down assays showed that miR-140-3p and SNHG12 were enriched in the miR-140-3p and SNHG12 groups, further establishing the binding efficiency between SNHG12 and miR-140-3p (Fig. 3H and I). In addition, ov-SNHG12 inhibited the expression of miR-140-3p compared with that of the o-NC group (Fig. 3J). However, changes in miR-140-3p expression had no significant effect on the expression of SNHG12 (Fig. 3K).
Figure 3.
SNHG12 acts as a ceRNA to adsorb miR-140-3p. (A) LncBase and Starbase joint analysis of potential targets for SNHG12. (B) RT-qPCR analysis of the differential expression of miR-140-3p in MCF-7, MDA-MB-231, ZR-75-1 and HCC1937 cells compared to MCF-10A. *P<0.05 vs. MCF-10A. RT-qPCR analysis of the difference in expression of miR-140-3p in (C) MCF-7 and (D) MDA-MB-231 cells under different SCO doses. *P<0.05 vs. 0 µM. (E) RT-qPCR analysis verifying the effectiveness of the miR-140-3p mimic/inhibitor. *P<0.05. (F) Bioinformatic analysis of the potential binding sites between SNHG12 and miR-140-3p. (G) Dual luciferase analysis of the binding between SNHG12 and miR-140-3p. *P<0.05 vs. mimic NC. RNA pull-down analysis was used to examine the binding between (H) bio-SNHG12 and miR-140-3p, as well as (I) bio-miR-140-3p and SNHG12, respectively, in MCF-7 and MDA-MB-231 cells. (J) RT-qPCR analysis of miR-140-3p expression under the combined action of SCO and ov-SNHG12. (K) RT-qPCR analysis of the effect of miR-140-3p mimic/inhibitor on SNHG12 expression. *P<0.05. SCO, scoparone; lncRNA, long non-coding RNA; SNHG12, small nucleolar RNA host gene 12; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; miR, microRNA; Bio, biotinylated; ceRNA, competing endogenous RNA.
miR-140-3p directly targets TRAF2 to regulate the NF-κB signaling pathway
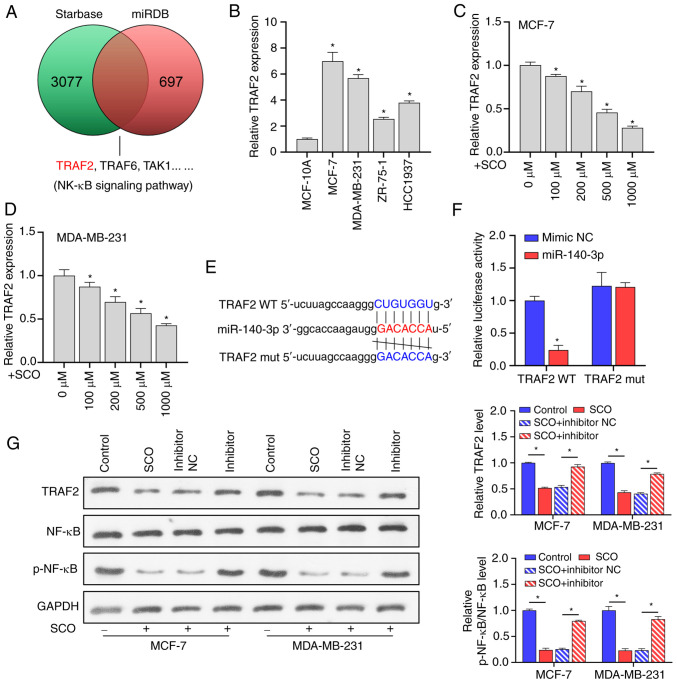
miRNAs are typically used to bind to the 3'-UTR of the downstream target to inhibit transcription and translation, thereby affecting signal transduction between BC cells (24,25). Using the miRDB and Starbase databases, we found that many putative target genes of miR-140-3p, such as TRAF2, are upstream regulators of NF-κB signaling (Fig. 4A). Prior studies have shown that TRAF2 is involved in the activation of NF-κB (26) and induces the growth and distal migration of breast cancer cells (27). In the current study, TRAF2 expression was upregulated in both MCF-7 and MDA-MB-231 cells compared to that in the normal mammary epithelial cell line MCF-10A (Fig. 4B). However, as the concentration of SCO increased, TRAF2 expression gradually decreased (Fig. 4C and D). Bioinformatic analysis indicated that there were potential binding sites between miR-140-3p and TRAF2 (Fig. 4E). The dual luciferase assay results showed that the fluorescence activity of the WT TRAF2 + miR-140-3p mimic co-transfected group was significantly decreased compared with that of the WT TRAF2 + miR-140-3p mimic NC co-transfected group. Nonetheless, there was no significant difference observed between the fluorescence activity of the mutant TRAF2 + miR-140-3p inhibitor NC co-transfection and mutant TRAF2 + miR-140-3p inhibitor co-transfection groups (Fig. 4F). In addition, SCO inhibited TRAF2 protein levels, and NF-κB activation was partially reversed by the miR-140-3p inhibitor. Overall, miR-140-3p negatively regulated TRAF2 expression (Fig. 4G).
Figure 4.
miR-140-3p directly targets TRAF2 to regulate the NF-κB signaling pathway. (A) miRDB and Starbase database joint analysis of potential targets for miR-140-3p. (B) RT-qPCR analysis of the differential expression of TRAF2 in MCF-7, MDA-MB-231, ZR-75-1 and HCC1937 cells compared to MCF-10A. *P<0.05 vs. MCF-10A. RT-qPCR analysis of TRAF2 expression in (C) MCF-7 and (D) MDA-MB-231 cells under different SCO doses. *P<0.05 vs. 0 µM. (E) Bioinformatic analysis of the potential binding sites between TRAF2 and miR-140-3p. (F) Dual luciferase analysis of the binding between TRAF2 and miR-140-3p. *P<0.05 vs. mimic NC. (G) Western blot analysis of the effect of SCO combined with miR-140-3p inhibitor on protein levels of TRAF2, NF-κB, and p-NF-κB. *P<0.05. SCO, scoparone; TRAF2, receptor-associated factor 2; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; miR, microRNA; Bio, biotinylated; ceRNA, competing endogenous RNA; NF-κB, nuclear factor κB.
miR-140-3p is negatively correlated with TRAF2 expression
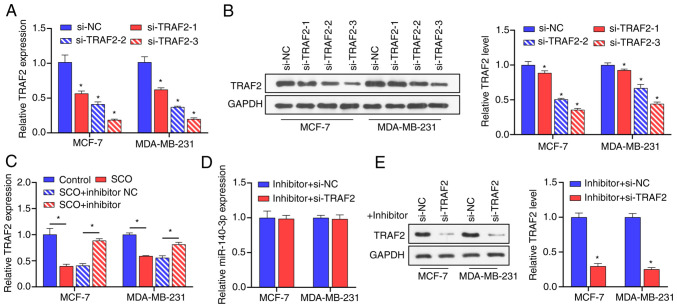
To confirm the negative correlation between miR-140-3p and TRAF2, we constructed three siRNAs that targeted TRAF2 and transfected MCF-7 and MDA-MB-231 cells with these siRNAs. Corresponding TRAF2 gene expression and protein levels were downregulated in BC cells after siRNA transfection (Fig. 5A and B). Among these siRNAs, si-TRAF2-3 possessed the best inhibition efficiency; therefore, si-TRAF2-3 was used in subsequent experiments. Subsequent RT-qPCR results demonstrated that, compared with the control group, the miR-140-3p inhibitor upregulated the expression of TRAF2 (Fig. 5C). In the presence of SCO, inhibition of TRAF2 expression had no significant effect on the expression of miR-140-3p (Fig. 5D). Nonetheless, in the presence of SCO and miR-140-3p inhibitor, the inhibition of TRAF2 gene expression reduced corresponding TRAF2 protein levels (Fig. 5E). These results indicated that miR-140-3p is negatively correlated with the expression of the downstream target gene TRAF2.
Figure 5.
miR-140-3p is negatively correlated with the expression of TRAF2. (A) RT-qPCR analysis verifying the effectiveness of 3 siRNAs against TRAF2 in MCF-7 and MDA-MB-231 cells. *P<0.05 vs. si-NC. (B) Western blot analysis verifying the effectiveness of 3 siRNAs against TRAF2 in MCF-7 and MDA-MB-231 cells. *, si-TRAF2-1, si-TRAF2-2,si-TRAF2-3 groups vs. si-NC group. (C) RT-qPCR analysis of the effect of SCO combined with miR-140-3p inhibitor on the expression of TRAF2. *P<0.05. (D) RT-qPCR analysis of the effect of si-TRAF2 on miR-140-3p expression. (E) Western blot analysis of the effect of si-TRAF2 on protein levels of TRAF2 in the presence of miR-140-3p inhibitor. *P<0.05 vs. inhibitor + si-NC. SCO, scoparone; TRAF2, receptor-associated factor 2; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; miR, microRNA; Bio, biotinylated; ceRNA, competing endogenous RNA; si, small interfering.
SCO regulates the NF-κB signaling pathway through the SNHG12/miR-140-3p/TRAF2 axis to inhibit BC cell growth
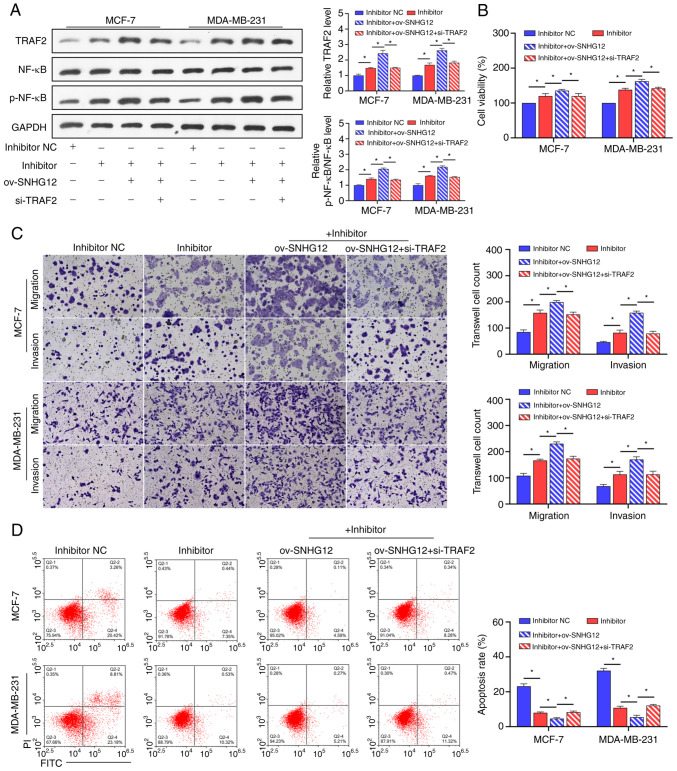
To understand the mechanism of the SNHG12/miR-140-3p/TRAF2/NF-κB axis in SCO treatment of BC, ov-SNHG12 was co-transfected with an miR-140-3p inhibitor and si-TRAF2 in BC cells in the presence of SCO. Overall, we found that the promotion of TRAF2 protein levels and NF-κB activation by the miR-140-3p inhibitor was enhanced by ov-SNHG12. The combined effect of ov-SNHG12 and the miR-140-3p inhibitor was partially mitigated in the presence of si-TRAF2 (Fig. 6A). In addition, the miR-140-3p inhibitor-mediated promotion of activity, migration/invasion ability, and apoptosis inhibition in BC cells was enhanced by overexpression of SNHG12. Further, inhibition of TRAF2 expression partially prevented the combined effect of the ov-SNHG12/miR-140-3p inhibitor (Fig. 6B-D).
Figure 6.
SCO regulates the NF-κB signaling pathway through the SNHG12/miR-140-3p/TRAF2 axis to inhibit BC cell growth. (A) Western blot analysis of the effect of the interaction between ov-SNHG12, miR-140-3p inhibitor, and si-TRAF2 on protein levels of TRAF2, NF-κB, and p-NF-κB in the presence of SCO. (B) CCK8 analysis of the effect of the interaction between ov-SNHG12, miR-140-3p inhibitor, and si-TRAF2 on BC cell viability in the presence of SCO. (C) Transwell analysis of the effect of the interaction between ov-SNHG12, miR-140-3p inhibitor, and si-TRAF2 on BC cell migration/invasion in the presence of SCO. Magnification, x200. (D) FCM analysis of the effect of the interaction between ov-SNHG12, miR-140-3p inhibitor, and si-TRAF2 on BC cell apoptosis in the presence of SCO. *P<0.05. SCO, scoparone; BC, breast cancer; lncRNA, long non-coding RNA; SNHG12, small nucleolar RNA host gene 12; ov, overexpression; TRAF2, receptor-associated factor 2; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; miR, microRNA; Bio, biotinylated; ceRNA, competing endogenous RNA; si, small interfering; NF-κB, nuclear factor κB; FCM, flow cytometry; FITC, fluorescein isothiocyanate; PI, propidium iodide.
Discussion
Prior studies have shown that TCM can effectively increase treatment efficiency, improve survival rates, and reduce the side effects of chemotherapy in patients with BC (28,29). SCO is a major component of the Chinese herbal medicine A. capillaris (30) and plays an important role in cancer therapy (7,31). In the current study, our results confirmed that SCO inhibited BC cell viability and promoted apoptosis, aligning with those of a previous study (9). This indicated that SCO had a significant anti-BC effect. Notably, our data also indicated that SCO is less toxic to normal mammary epithelial cells and is suitable for the treatment of BC.
Our finding that lncRNA SNHG12 is upregulated in BC cells and plays an oncogenic role in BC is consistent with a previous study (32). Furthermore, SCO inhibited the expression of SNHG12 in a dose-dependent manner, whereas the overexpression of SNHG12 reversed the effect of SCO on BC cells. Therefore, we determined that SCO inhibits the growth of BC cells through SNHG12. Cytoplasmic lncRNA acts as a ceRNA; therefore, we first conducted a nucleocytoplasmic separation experiment and confirmed that SNHG12 is predominantly located in the cytoplasm of BC cells, which is consistent with a previous study (33). We then demonstrated for the first time that SNHG12 adsorbs miR-140-3p, a tumor suppressor gene in BC (20), thereby downregulating the expression of miR-140-3p. Therefore, SCO-mediated inhibition of SNHG12 expression promotes the expression of miR-140-3p, leading to a reduction in BC cell growth.
It is well understood that TRAF2 acts as an oncogene in various cancers, such as gastric cancer (34) and prostate cancer (35), and plays a key role in the promotion of cell viability. Additionally, high TRAF2 expression promotes the growth and invasion of BC cells (27,36). The current study confirmed, for the first time, the direct binding relationship between miR-140-3p and TRAF2. TRAF2 is highly expressed in BC, and the growth and migration of BC cells promoted by the miR-140-3p inhibitor were reversed by si-TRAF2. Prior studies have demonstrated that abnormal NF-κB regulation contributes to autoimmune diseases, chronic inflammation, and many cancers (37,38). In BC progression, NF-κB activation is an important contributor to tumor development (39). As an upstream effector of NF-κB, the main pathway through which TRAF2 mediates BC progression is via NF-κB activation (40). In the present study, we demonstrated that SCO suppresses NF-κB signaling, but this signaling was partially restored by the miR-140-3p inhibitor. The promoting effect of the miR-140-3p inhibitor on NF-κB signaling was enhanced by overexpression of SNHG12 and decreased by treatment with si-TRAF2. Thus far, we have confirmed that SCO inhibits the NF-κB signaling pathway through the SNHG12/miR-140-3p/TRAF2 axis, inhibits BC cell viability, and promotes apoptosis.
Nonetheless, this study had some limitations. First, we did not analyze the therapeutic effect of SCO in BC patients or the correlation of clinical data. Second, the development of BC involves many RNA and protein interactions and signaling pathway alterations, which have not been assessed here. Third, SCO directly or indirectly affects the stability or processing of SNHG12, potentially through post-transcriptional modification or through interactions with RNA-binding proteins or other regulatory factors; however, these mechanisms were not explored within this study. Finally, it is unknown whether SCO can increase the susceptibility of BC cells to drug resistance. Overall, these research areas remain unclear; therefore, we aim to focus on these factors in future research of SCO.
In conclusion, our study demonstrated that SCO inhibits BC cell survival by inhibiting intercellular NF-κB signaling, which is related to the regulation of the SNHG12/miR-140-3p/TRAF2 axis. These results indicated that SCO may be a promising anti-BC therapeutic drug, which provides a strong theoretical basis for the treatment of BC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CE designed experiments. XW and XL performed experiments and drafted the manuscript. JL, XZ and YC collected and analyzed the data. CE and JL confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Azamjah N, Soltan-Zadeh Y, Zayeri F. Global trend of breast cancer mortality rate: A 25-year study. Asian Pac J Cancer Prev. 2019;20:2015–2020. doi: 10.31557/APJCP.2019.20.7.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 3.Libson S, Lippman M. A review of clinical aspects of breast cancer. Int Rev Psychiatry. 2014;26:4–15. doi: 10.3109/09540261.2013.852971. [DOI] [PubMed] [Google Scholar]
- 4.Tang JL, Liu BY, Ma KW. Traditional Chinese medicine. Lancet. 2008;372:1938–1940. doi: 10.1016/S0140-6736(08)61354-9. [DOI] [PubMed] [Google Scholar]
- 5.Pei S, Yang X, Wang H, Zhang H, Zhou B, Zhang D, Lin D. Plantamajoside, a potential anti-tumor herbal medicine inhibits breast cancer growth and pulmonary metastasis by decreasing the activity of matrix metalloproteinase-9 and -2. BMC Cancer. 2015;15(965) doi: 10.1186/s12885-015-1960-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liew AC, Peh KK, Tan BS, Zhao W, Tangiisuran B. Evaluation of chemotherapy-induced toxicity and health-related quality of life amongst early-stage breast cancer patients receiving Chinese herbal medicine in Malaysia. Support Care Cancer. 2019;27:4515–4524. doi: 10.1007/s00520-019-04724-1. [DOI] [PubMed] [Google Scholar]
- 7.Jung SH, Lee GB, Ryu Y, Cui L, Lee HM, Kim J, Kim B, Won KJ. Inhibitory effects of scoparone from chestnut inner shell on platelet-derived growth factor-BB-induced vascular smooth muscle cell migration and vascular neointima hyperplasia. J Sci Food Agric. 2019;99:4397–4406. doi: 10.1002/jsfa.9674. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Wang M, Chen B, Shi J. Scoparone attenuates high glucose-induced extracellular matrix accumulation in rat mesangial cells. Eur J Pharmacol. 2017;815:376–380. doi: 10.1016/j.ejphar.2017.09.039. [DOI] [PubMed] [Google Scholar]
- 9.Kim JK, Kim JY, Kim HJ, Park KG, Harris RA, Cho WJ, Lee JT, Lee IK. Scoparone exerts anti-tumor activity against DU145 prostate cancer cells via inhibition of STAT3 activity. PLoS One. 2013;8(e80391) doi: 10.1371/journal.pone.0080391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kielbus M, Skalicka-Wozniak K, Grabarska A, Jeleniewicz W, Dmoszynska-Graniczka M, Marston A, Polberg K, Gawda P, Klatka J, Stepulak A. 7-substituted coumarins inhibit proliferation and migration of laryngeal cancer cells in vitro. Anticancer Res. 2013;33:4347–4356. [PubMed] [Google Scholar]
- 11.Zhao Z, Guo Y, Liu Y, Sun L, Chen B, Wang C, Chen T, Wang Y, Li Y, Dong Q, et al. Individualized lncRNA differential expression profile reveals heterogeneity of breast cancer. Oncogene. 2021;40:4604–4614. doi: 10.1038/s41388-021-01883-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkatesh J, Wasson MD, Brown JM, Fernando W, Marcato P. LncRNA-miRNA axes in breast cancer: Novel points of interaction for strategic attack. Cancer Lett. 2021;509:81–88. doi: 10.1016/j.canlet.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Qiao K, Ning S, Wan L, Wu H, Wang Q, Zhang X, Xu S, Pang D. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38(418) doi: 10.1186/s13046-019-1421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan JH, Li WX, Hu C, Zhang B. Upregulation of SNHG12 accelerates cell proliferation, migration, invasion and restrain cell apoptosis in breast cancer by enhancing regulating SALL4 expression via sponging miR-15a-5p. Neoplasma. 2020;67:861–870. doi: 10.4149/neo_2020_190808N731. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Y, Wang B, Wang Y, Chen G, Lian Q, Wang H. miR-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol Lett. 2019;17:3835–3841. doi: 10.3892/ol.2019.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun LL, Wang J, Zhao ZJ, Liu N, Wang AL, Ren HY, Yang F, Diao KX, Fu WN, Wan EH, Mi XY. Suppressive role of miR-502-5p in breast cancer via downregulation of TRAF2. Oncol Rep. 2014;31:2085–2092. doi: 10.3892/or.2014.3105. [DOI] [PubMed] [Google Scholar]
- 17.Taminiau A, Draime A, Tys J, Lambert B, Vandeputte J, Nguyen N, Renard P, Geerts D, Rezsohazy R. HOXA1 binds RBCK1/HOIL-1 and TRAF2 and modulates the TNF/NF-κB pathway in a transcription-independent manner. Nucleic Acids Res. 2016;44:7331–7349. doi: 10.1093/nar/gkw606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang KW, Lee KH, Kim SH, Jin T, Choi EY, Jeon HJ, Kim E, Han YS, Chung JH. Ubiquitin ligase CHIP induces TRAF2 proteasomal degradation and NF-κB inactivation to regulate breast cancer cell invasion. J Cell Biochem. 2011;112:3612–3620. doi: 10.1002/jcb.23292. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Onyeisi JOS, Greve B, Espinoza-Sanchez NA, Kiesel L, Lopes CC, Gotte M. microRNA-140-3p modulates invasiveness, motility, and extracellular matrix adhesion of breast cancer cells by targeting syndecan-4. J Cell Biochem. 2021;122:1491–1505. doi: 10.1002/jcb.30071. [DOI] [PubMed] [Google Scholar]
- 21.Dou D, Ren X, Han M, Xu X, Ge X, Gu Y, Wang X, Zhao S. Circ_0008039 supports breast cancer cell proliferation, migration, invasion, and glycolysis by regulating the miR-140-3p/SKA2 axis. Mol Oncol. 2021;15:697–709. doi: 10.1002/1878-0261.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamarbide Losada JN, Sulpice E, Combe S, Almeida GS, Leach DA, Choo J, Protopapa L, Hamilton MP, McGuire S, Gidrol X, et al. Apoptosis-modulatory miR-361-3p as a novel treatment target in endocrine-responsive and endocrine-resistant breast cancer. J Endocrinol. 2023;256(e220229) doi: 10.1530/JOE-22-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua B, Li Y, Yang X, Niu X, Zhao Y, Zhu X. MicroRNA-361-3p promotes human breast cancer cell viability by inhibiting the E2F1/P73 signalling pathway. Biomed Pharmacother. 2020;125(109994) doi: 10.1016/j.biopha.2020.109994. [DOI] [PubMed] [Google Scholar]
- 24.Jin T, Zhang Y, Zhang T. MiR-524-5p Suppresses migration, invasion, and EMT progression in breast cancer cells through targeting FSTL1. Cancer Biother Radiopharm. 2020;35:789–801. doi: 10.1089/cbr.2019.3046. [DOI] [PubMed] [Google Scholar]
- 25.Cheng S, Huang Y, Lou C, He Y, Zhang Y, Zhang Q. FSTL1 enhances chemoresistance and maintains stemness in breast cancer cells via integrin β3/Wnt signaling under miR-137 regulation. Cancer Biol Ther. 2019;20:328–337. doi: 10.1080/15384047.2018.1529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu M, Zhou W, Chen J, Zhao Y, Xie C, Zhou Z. TRAF2 gene silencing induces proliferation and represses apoptosis of nucleus pulposus cells in rats with intervertebral disc degeneration. Life Sci. 2021;279(119670) doi: 10.1016/j.lfs.2021.119670. [DOI] [PubMed] [Google Scholar]
- 27.Yao Y, Zhao K, Yu Z, Ren H, Zhao L, Li Z, Guo Q, Lu N. Wogonoside inhibits invasion and migration through suppressing TRAF2/4 expression in breast cancer. J Exp Clin Cancer Res. 2017;36(103) doi: 10.1186/s13046-017-0574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li QW, Zhang GL, Hao CX, Ma YF, Sun X, Zhang Y, Cao KX, Li BX, Yang GW, Wang XM. SANT, a novel Chinese herbal monomer combination, decreasing tumor growth and angiogenesis via modulating autophagy in heparanase overexpressed triple-negative breast cancer. J Ethnopharmacol. 2021;266(113430) doi: 10.1016/j.jep.2020.113430. [DOI] [PubMed] [Google Scholar]
- 29.Qi F, Zhao L, Zhou A, Zhang B, Li A, Wang Z, Han J. The advantages of using traditional Chinese medicine as an adjunctive therapy in the whole course of cancer treatment instead of only terminal stage of cancer. Biosci Trends. 2015;9:16–34. doi: 10.5582/bst.2015.01019. [DOI] [PubMed] [Google Scholar]
- 30.Liu B, Deng X, Jiang Q, Li G, Zhang J, Zhang N, Xin S, Xu K. Scoparone improves hepatic inflammation and autophagy in mice with nonalcoholic steatohepatitis by regulating the ROS/P38/Nrf2 axis and PI3K/AKT/mTOR pathway in macrophages. Biomed Pharmacother. 2020;125(109895) doi: 10.1016/j.biopha.2020.109895. [DOI] [PubMed] [Google Scholar]
- 31.Li N, Yang F, Liu DY, Guo JT, Ge N, Sun SY. Scoparone inhibits pancreatic cancer through PI3K/Akt signaling pathway. World J Gastrointest Oncol. 2021;13:1164–1183. doi: 10.4251/wjgo.v13.i9.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Cheng G, Huang Z, Bao L, Liu J, Wang C, Xiong Z, Zhou L, Xu T, Liu D, et al. Long noncoding RNA SNHG12 promotes tumour progression and sunitinib resistance by upregulating CDCA3 in renal cell carcinoma. Cell Death Dis. 2020;11(515) doi: 10.1038/s41419-020-2713-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. 2018;495:1822–1832. doi: 10.1016/j.bbrc.2017.12.047. [DOI] [PubMed] [Google Scholar]
- 34.Ye Y, Ye F, Li X, Yang Q, Zhou J, Xu W, Aschner M, Lu R, Miao S. 3,3'-diindolylmethane exerts antiproliferation and apoptosis induction by TRAF2-p38 axis in gastric cancer. Anticancer Drugs. 2021;32:189–202. doi: 10.1097/CAD.0000000000000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei B, Liang J, Hu J, Mi Y, Ruan J, Zhang J, Wang Z, Hu Q, Jiang H, Ding Q. TRAF2 is a valuable prognostic biomarker in patients with prostate cancer. Med Sci Monit. 2017;23:4192–4204. doi: 10.12659/msm.903500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiao C, Chen W, Tan X, Liang H, Li J, Yun H, He C, Chen J, Ma X, Xie Y, Yang BB. Ganoderma lucidum spore oil induces apoptosis of breast cancer cells in vitro and in vivo by activating caspase-3 and caspase-9. J Ethnopharmacol. 2020;247(112256) doi: 10.1016/j.jep.2019.112256. [DOI] [PubMed] [Google Scholar]
- 37.Poma P. NF-κB and disease. Int J Mol Sci. 2020;21(9181) doi: 10.3390/ijms21239181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-kappaB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct Target Ther. 2020;5(209) doi: 10.1038/s41392-020-00312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan Y, Sun R, Liu L, Yang D, Xiang Q, Li L, Tang J, Qiu Z, Peng W, Wang Y, et al. Tumor suppressor DRD2 facilitates M1 macrophages and restricts NF-κB signaling to trigger pyroptosis in breast cancer. Theranostics. 2021;11:5214–5231. doi: 10.7150/thno.58322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang L, Yu L, Zhang X, Lei F, Wang L, Liu X, Wu S, Zhu J, Wu G, Cao L, et al. miR-892b silencing activates NF-κB and promotes aggressiveness in breast cancer. Cancer Res. 2016;76:1101–1111. doi: 10.1158/0008-5472.CAN-15-1770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.