Abstract
Research increasingly relies on interrogating large-scale data resources. The NIH National Heart, Lung, and Blood Institute developed the NHLBI BioData CatalystⓇ (BDC), a community-driven ecosystem where researchers, including bench and clinical scientists, statisticians, and algorithm developers, find, access, share, store, and compute on large-scale datasets. This ecosystem provides secure, cloud-based workspaces, user authentication and authorization, search, tools and workflows, applications, and new innovative features to address community needs, including exploratory data analysis, genomic and imaging tools, tools for reproducibility, and improved interoperability with other NIH data science platforms. BDC offers straightforward access to large-scale datasets and computational resources that support precision medicine for heart, lung, blood, and sleep conditions, leveraging separately developed and managed platforms to maximize flexibility based on researcher needs, expertise, and backgrounds. Through the NHLBI BioData Catalyst Fellows Program, BDC facilitates scientific discoveries and technological advances. BDC also facilitated accelerated research on the coronavirus disease-2019 (COVID-19) pandemic.
Keywords: data-driven science, cloud computing, data analysis, reproducibility of results, team science
BACKGROUND AND SIGNIFICANCE
For decades, the National Heart, Lung, and Blood Institute (NHLBI) has supported the generation of rich data resources for heart, lung, blood, and sleep (HLBS) conditions.1 These resources underpin ambitious translational research programs, including NHLBI Trans-Omics for Precision Medicine (TOPMed),2 the Cure Sickle Cell Initiative,3 and multiple coronavirus disease-2019 (COVID-19) efforts.4 Originally collecting clinical, epidemiological, and imaging data, studies increasingly included genomics, proteomics, and other omics data.2 Mining these data resources advanced our understanding of health and disease, leading to novel diagnostics, therapies, and prevention strategies. However, researchers often lack the data access, computational expertise, or local resources to effectively interrogate this growing wealth of data.5
OBJECTIVE
To address these challenges, the NHLBI established the NHLBI BioData CatalystⓇ (BDC),6,7 a community-driven ecosystem, democratizing data and computational access, implementing data science solutions, and advancing biomedical research. BDC users can find, access, share, store, and compute on large-scale datasets within secure workspaces. User requests, testing, and feedback have driven technical development innovations, including easy data search and exploration; queries to refine hypotheses and cohort-building for analysis; secure cloud workspaces; integrating annotations for rare variant genomic association analysis; rapid medical imaging data ingestion for viewing, annotation, and sharing, as well as machine learning; and tools for enhancing and promoting the FAIRness (Findable, Accessible, Interoperable, Reusable) of data and analyses and setting the stage for future expectations for similar accessibility norms.8
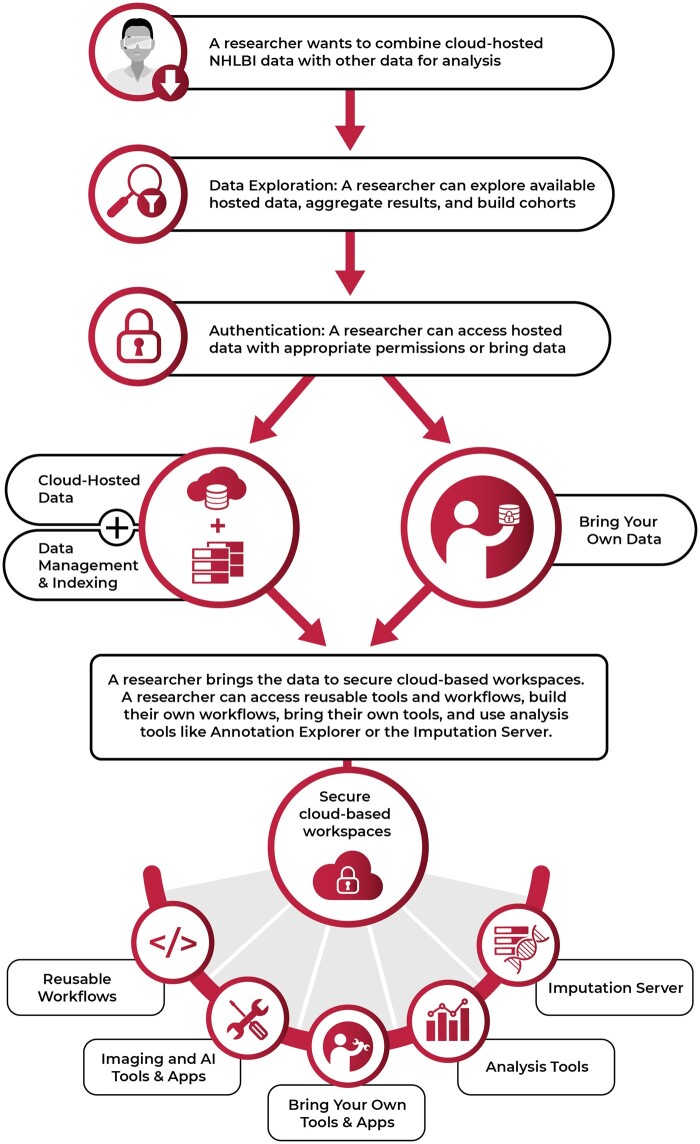
Research increasingly relies on interrogating large-scale data resources. BDC provides a broad, diverse community accelerated access to large-scale datasets and computational resources that support precision medicine for HLBS conditions, leveraging separately developed and managed platforms to maximize flexibility based on researcher needs, expertise, and backgrounds. The integrated ecosystem of platforms provides secure, cloud-based workspaces, user authentication and authorization, search, tools and workflows, applications, and innovative features to address community needs (Figure 1). Today, BDC contains over 3 petabytes of available data,9,10 with more released regularly. Researchers across institutions and with diverse domain expertise, including bench and clinical scientists, statisticians, and algorithm developers, securely collaborate and communicate in real-time; configure tools, workflows, and analysis environments; and bring their own data and tools to co-analyze or expand on provided datasets. Finally, end-to-end provenance and workflow versioning makes analyses easily reproducible and shareable.
Figure 1.
BDC is designed for user flexibility, exploring, and collaborating across data collections in a secure cloud workspace, and creating or using existing tools and workflows.
BDC was initiated as an institute-specific iteration of the NIH Data Commons Pilot Phase Consortium, which was funded by the NIH Office of the Director, Office of Strategic Coordination.11 NIH solicited competitive proposals for that program, which was intended to accelerate “new biomedical discoveries by developing and testing a cloud-based platform where investigators could store, share, access, and interact with digital objects.”12 The pilot phase spanned September 2017–January 2019, funding 12 multi-institutional teams at over 30 institutions.13 NHLBI requested that some teams from that cohort propose developing innovative computing solutions to meet NHLBI’s specific scientific needs: enhancing data sharing, access, and NHLBI-generated data resource value. NHLBI selected private, public, and nonprofit institutions with expertise delivering production-quality biomedical data discovery systems and, with these institutions, collaboratively developed a plan to support these goals. This collaborative decision making continues as the ecosystem evolves to meet the needs of a diverse research community.
While this article will not catalog the entire collection of BDC components, it will provide a basic overview tailored for users looking to access HLBS data and tools. Here, we seek to provide insight into the initial vision for this system and information about basic practicalities, as well as discussion of technical issues and early successes. Future publications will discuss the system in more detail and seek to provide more depth on the strategic capabilities.
MATERIALS AND METHODS
Exploratory data analysis
Rapid expansion of NHLBI data resources made data discovery more challenging, with complicated data access approvals, data from disparate sources and in different formats, and inadequate computing resources. BDC enables users to access data, analysis, and workflows without requiring separate data downloads, providing highly secure and cost-effective computation (Table 1). Prior to requesting access, users can search all hosted phenotypic data and view aggregate results in the cloud, ensuring available data meets their needs. Hosted data are updated on a regular basis and currently includes data from the NHLBI TOPMed program, many NHLBI-funded longitudinal cohort studies and clinical trials, and some COVID-19 initiatives.9,10 BDC is data type agnostic and provides the ability to co-analyze phenotypic, molecular (genomic, transcriptomic, proteomic, etc), as well as imaging and sensor (polysomnography, actigraphy, electrophysiology, etc) data.
Table 1.
Researchers are using the BDC ecosystem to access large-scale datasets and computational resources that support precision medicine for HLBS conditions
| BDC occurrence | Number |
|---|---|
| Average number of unique logins per month | 385 |
| Registered users (Seven Bridges/Dockstore) | 1400+ |
| Years of total computation time across all users to date (Seven Bridges) | 86 |
| Publicly available workflows | 1850 |
Note: Metrics in this table were captured in September 2022.
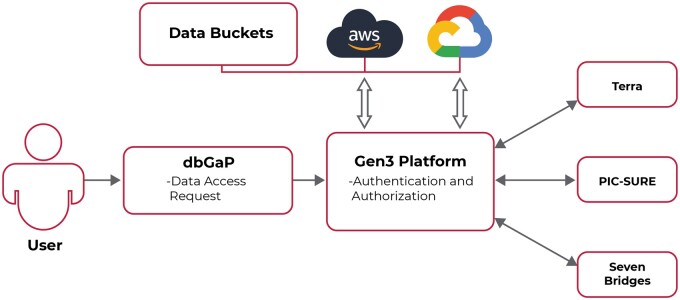
BDC supports data access and analysis through 4 interoperable platforms (outlined in Figure 2):
Figure 2.
BDC supports data analysis and access through 4 interoperable platforms, providing researchers the ability to use a wide variety of techniques to store, share, compute on, and analyze their data, while also providing opportunities for collaboration and cohort building. Copyright 2023 by NHLBI BioData Catalyst. Reprinted from our website,9 with our own permission.
BDC Powered by Gen3 (BDC-Gen3): an interactive data exploration platform for searching and filtering study-specific clinical and genomic data, while managing data resources and user access.
BDC Powered by PIC-SURE (BDC-PIC-SURE): an interactive search with advanced exploratory features, allowing users to browse available hosted phenotypic and genomic data at the variable-value level, build cohorts, access participant-level data, aggregate counts of participants or distributions of biomedical markers, and export participant-level clinical and/or genomic data into workspaces in a statistically-ready tabular format. BDC-PIC-SURE also enables researchers to search all clinical and genomic data at the variable-value and variant levels and bypass file manipulation and data decoding to easily search variables (such as “gender”) and values (such as “female”) of interest, apply filters, and build cohorts across phenotypic and genomic data to include participants that meet the query criteria.
BDC Powered by Seven Bridges (BDC-Seven Bridges): a collaborative, secure, cloud-based computational workspace providing cloud cost optimization of tools and workflows, described in Common Workflow Language (CWL), and a visual editor to support diverse user profiles.
BDC Powered by Terra (BDC-Terra): a collaborative, cloud-based computational workspace, using Workflow Description Language (WDL) and Galaxy, for secure collaboration and cross-dataset analysis, which is particularly well suited to rapidly develop new workflows.14
Hosting controlled-access data requires policies and technical controls to protect data privacy and security and respect study participant consent. All platforms are certified at a FISMA or FedRAMP moderate level, providing strong protection for controlled and private data.15,16 Users request access to these studies via dbGaP Data Access Requests, and dataset permissions are programmatically reflected in BDC platforms. These workspaces provide bioinformatics analysis at scale, allowing researchers to share workflows, perform interactive analysis, and create visualizations of results via cloud-based, interactive analysis tools such as Jupyter, Jupyterlab notebooks, RStudio, and SAS. Moreover, researchers can compute on Amazon Web Services (AWS) or Google Cloud Platform (GCP). BDC-Seven Bridges is hosted on AWS, but users can configure their data storage and computation for AWS or GCP to combine existing private or public data and hosted data without egress costs. BDC-Terra is hosted on GCP, allowing researchers to leverage GCP accounts to support computation and data storage costs. It should be noted, given that there are commercial facilities available in this ecosystem, those facilities may be subject to constraints in the future.
BDC-Gen3 17 supports BDC by assigning globally unique persistent identifiers (GUIDs) to data collections, specifying metadata, and providing data indexing to ensure data collection FAIRness.8 It also allows dbGAP authorized users to access approved NHLBI data, with flexibility to manage manually generated access control lists. Researchers can browse file- and variable-level data. BDC-Gen3 also supports interoperability for large and complex data sets in the cloud through APIs and data exchange formats. BDC-Gen3’s technical implementation of authentication and authorization, along with policies outlined in the BDC Security Statement,18 protect the confidentiality, integrity, provenance, and availability of hosted data. Further, BDC’s “Bring Your Own Data” functionality allows users to analyze their datasets in isolation or combine them with hosted datasets.
Genomic tools
The advent of rapid DNA sequencing technology caused an explosion in discovery of rare variants with potential roles in human health and disease.2,19 Generating sufficient statistical power to detect their effect on complex traits, however, requires combining multiple genomic datasets, using significant computational time and resources. To assist, BDC uses two unique systems.
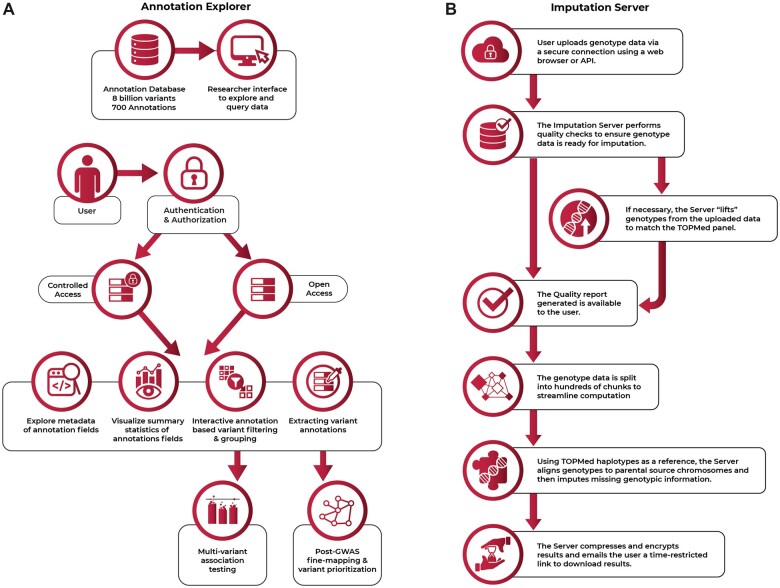
Annotation Explorer, by the University of Washington Genetic Analysis Center (GAC) and BDC-Seven Bridges, is a high-performance annotation database with optimized resource availability, hosted controlled-access and open-access datasets, billions of variants, hundreds of annotations, and a graphical user interface for interactively exploring, querying, visualizing, and storing outputs. Annotation Explorer allows users to interactively select, aggregate, and filter billions of variants (Figure 3A). Users can explore and evaluate multiple custom grouping and filtering options in real time, a task that once took hours, and identify subsets for further analysis. As proof of principle, the GAC used BDC to generate multiple annotation-informed variant sets. Members of multiple phenotype-centric TOPMed working groups utilized these variant sets in analyses, leading to discovery of rare variants associated with phenotypes such as blood cell count, fasting glucose, and lung function.20
Figure 3.
(A and B) Annotation Explorer and the Imputation Server are powerful, cloud-based tools for genomic analysis.
The TOPMed Imputation Server by the TOPMed Informatics Research Center enables genotype imputation with a reference panel, based on TOPMed sequencing. Imputation compares short stretches of individual genomes to previously characterized stretches and reconstructs missing data. It is a key component of genetic association studies, increases statistical power, facilitates meta-analysis, and aids in interpretation of signals. The Server provides web-based access to imputation reference panels and allows users to request imputation of their samples and securely download results (Figure 3B). The TOPMed panel includes over 300 million variants derived from 97 256 multiethnic participants, significantly improving imputation accuracy compared to other publicly available panels.2,21
Reproducibility
Science faces a reproducibility crisis, and data science is no exception.22,23 Reproducibility allows other researchers to evaluate analytical robustness by applying new methods to the same data; replication conducts the same analysis on new data. BDC seeks to foster and facilitate reproducible science. For example, BDC-PIC-SURE enabled researchers to use data from the PETAL Network: Outcomes Related to COVID-19 Treated With Hydroxychloroquine Among Inpatients With Symptomatic Disease (ORCHID) Trial to reproduce the original JAMIA study analysis.24 Notebooks used for this study are public projects, available on BDC-Seven Bridges and BDC-Terra.
BDC tools and resources, including unique identifiers, assist other users in conducting reproducible science, leveraging container-based tools and workflow description standards, such as CWL and WDL, which allow for reproducing complex analyses across different compute environments. Using sample Jupyter notebooks as a starting point to recreate an existing analysis, users can further modify code to conduct new analyses for their own research questions, demonstrating FAIR scientific data and accelerating scientific development and innovations.25
These platforms integrate with Dockstore, a centralized catalog of thousands of public, scientific tools and workflows following FAIR principles.26 Dockstore provides features to find, reuse, test, vet, cite, and publish tools and workflows to promote integrity of data science methodology. BDC users leverage Dockstore to share analyses as reproducible methods including, for example, genome-wide association testing, rare variant analysis, gene by environment interactions, and structural variant calling (tools and workflows are shared at https://dockstore.org/organizations/bdcatalyst).
Interoperability
Several cloud-based data analysis platforms across NIH host large, high-value datasets and tools. Research questions cross these platforms, necessitating co-analysis of disparate datasets from multiple NIH institutes and centers. For example, a researcher may utilize TOPMed data on BDC and GTEx data on National Human Genome Research Institute (NHGRI) AnVIL platform.27 Currently, this requires separate accounts, resulting in siloed research that increases costs28 and decreases efficiency.
BDC addresses this challenge as part of the NIH Cloud Platform Interoperability project.29 This effort includes instantiation of a single sign-on, via the NIH Researcher Auth Service, to authenticate and authorize users across multiple platforms with a search capability to promote data FAIRness. Empowering end-user analyses across participating platforms helps realize a trans-NIH, federated data ecosystem to harness data science approaches that support precision medicine for HLBS conditions and beyond. For example, authorized researchers can find relevant datasets on AnVIL, BDC, Cancer Research Data Commons, or Kids First and seamlessly combine them within a BDC workspace for analysis, thanks to BDC’s investments in Global Alliance for Genomics and Health (GA4GH) Data Repository Service (DRS), minimal metadata attributes, and common manifest transmission standards. Already, this interoperability helped researchers perform joint calling on the Pediatric Cardiac Genomics Consortium dataset in the cloud, which is split between NHLBI30 and Kids First.31 Prior to the interoperability work, this call would not have been possible and was a key pain point for researchers. NIH funded distinct projects to build on existing interoperation between Seven Bridges and Terra workspaces, allowing tools, data, and WDL and CWL workflows to be accessed and utilized by users across platforms.
Users benefit from separate but interoperable workspaces. While BDC-Seven Bridges and BDC-Terra share core data access, management, and analysis capabilities, BDC-Seven Bridges provides visual interfaces to compose and edit workflows, without coding, as well as advanced user management capabilities suited to large consortia and working groups like the TOPMed and The Collaborative Cohort of Cohorts for COVID-19 Research (C4R) working groups. BDC-Terra provides rich capabilities for advanced users, such as directly leveraging their own Google account and supporting rich data visualization and data portal development. While users can encode workflows in CWL or WDL, WDL enables rapid prototyping and iteration for code-savvy researchers, whereas CWL supports programmatic and visual construction and workflow editing.
RESULTS, DISCUSSION, AND CONCLUSION
Since its launch, BDC’s development has focused on end-user needs, emphasizing data, tools, training, and other resources to accelerate research and advance scientific insights for improved patient outcomes. BDC is a people-centric endeavor, building a community to collaboratively solve technical and scientific challenges. Community building efforts include an active help desk,32 peer-to-peer mentoring, interest groups, a community forum,33 and monthly community hours34 with researcher showcases and virtual meet and greets. Additionally, BDC teams regularly solicit feedback from researchers, users, data generators, an external expert panel, and other stakeholders for planning future development (Table 2). BDC teams and program leadership review and prioritize needs and add top priority development items to the upcoming year’s roadmap.
Table 2.
BDC is a community-driven ecosystem, with user requests, testing, and feedback driving technical development innovations
| BDC ecosystem events as of September 2022 | Number |
|---|---|
| Institutions represented at Community Hours | 109 |
| Unique attendees at Community Hours (of 523 total) | 266 |
| People reached with live training at workshops | 350+ |
| Conference presentations or booths | 20+ |
| Publications and pre-prints referencing BDC | 60+ |
Note: Continual tracking, through researcher submissions and internal BDC efforts, shows a strong record of ecosystem successes.
To strengthen the BDC community and facilitate initial research efforts, NHLBI offers many new users cloud credits that cover costs associated with accessing, analyzing, and storing data. Costs on the ecosystem may include cloud data storage and computation. Analysis of whole genome sequencing data, for example, can cost $5–$10 from alignment through variant calling. After using pilot funds, researchers can connect their STRIDES-enabled Google account on BDC-Terra or receive the STRIDES discount through BDC-Seven Bridges. Both platforms provide extensive cost optimization capabilities. For example, BDC-Seven Bridges’ workflow execution metrics let developers select the most time- and cost-effective instances for each type of analysis, while optimized workflows help new researchers cost effectively execute common analysis. Detailed common workflow benchmarking information supports cloud costs planning and interactive calculators are planned.
The nature of cloud computing means that the cost of using BDC is variable and is currently covered by NIH. NIH will provide training to ensure researchers are able to estimate the cost of cloud resource use, to allow researchers to understand potential costs before research begins. The NIH will continue to provide limited cloud credits to allow an investigator to conduct preliminary research and estimate costs that can then be included in a grant application budget.
Researchers requiring significant computational resources for novel use cases are needed to continue building on BDC’s success in advancing understanding of HLBS disorders. Accordingly, NHLBI funded the NHLBI BDC Fellows Program, which supports early-career scientists in their novel and innovative data science and data-focused research. These users informed data ingest decisions, developed or requested high-utility tools and workflows, and provided insight to streamline BDC for future researchers. Early success stories include Dr. Jean Monlong, who developed a “pangenome short read mapper” called Giraffe, allowing his team to genotype 2000 samples from the Multi-Ethnic Study of Atherosclerosis (MESA) cohort in four days, and 3202 samples from a high-coverage dataset in 6 days.35
Because BDC is flexible and designed to meet changing research needs, it pivoted to support accelerated collaborative research on COVID-19. As part of NHLBI's multi-pronged COVID-19 research strategy,36 BDC is rapidly aggregating and ingesting COVID-19 datasets, creating shared workspaces to promote collaborations among COVID researchers, and providing mechanisms for demonstrating reproducibility. The goal is to leverage ecosystem resources to better understand Severe acute respiratory syndrome-associated coronavirus (SARS-CoV-2) biology and identify, develop, and evaluate new therapies to improve public health outcomes. Adding new datasets will bring together researchers from disparate domains, focused on COVID-19, leading to new tool and workflow development to meet these researchers’ needs.
As exemplified by NIH-led COVID-19 projects,37,38 competing scientists and organizations are increasingly collaborating on large-scale research.39 While projects spanning multiple institutions can result in “delayed, misinterpreted, or nonexistent communication … imped[ing] discovery, trust, productivity, consensus, and division of labor,”40 BDC consortium members developed communications and workflows at program inception that allow room for multiple voices from potentially competing interests while focusing on programmatic goals. This approach, plus allowing competitors to develop distinct but interoperable components, allows the ecosystem to support a wide range of use cases, truly democratizing data access and analysis, while meeting a broad range of potential users and user needs despite rapidly evolving technology. Furthermore, engaging those collaborating on standards, who may, where appropriate, compete on implementation standards, increases ways users can interact with data.
BDC operates for, and because of, the potential for scientific collaborations to tackle some of the world’s greatest medical questions. It leverages emerging opportunities in data science to open new frontiers in HLBS research, leading to discovery of biological, social, and behavioral determinants associated with HLBS health and disease to improve public health outcomes.
ACKNOWLEDGMENTS
Support for this work was provided by the National Institutes of Health, National Heart, Lung, and Blood Institute, through the BioData Catalyst program (award 1OT3HL142479-01, 1OT3HL142478-01, 1OT3HL142481-01, 1OT3HL142480-01, 1OT3HL147154-01) and the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). Any opinions expressed in this document are those of the author(s) and do not necessarily reflect the views of NHLBI, individual BioData Catalyst team members, or affiliated organizations and institutions.
Contributor Information
Stan Ahalt, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Paul Avillach, Department of Biomedical Informatics, Harvard Medical School, Boston, Massachusetts, USA.
Rebecca Boyles, RTI International, Triangle Park, North Carolina, USA.
Kira Bradford, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA; RTI International, Triangle Park, North Carolina, USA.
Steven Cox, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Brandi Davis-Dusenbery, Velsera, Boston, Massachusetts, USA.
Robert L Grossman, University of Chicago, Chicago, Illinois, USA.
Ashok Krishnamurthy, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Alisa Manning, The Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Benedict Paten, UC Santa Cruz Genomics Institute, University of California, Santa Cruz, Santa Cruz, California, USA.
Anthony Philippakis, The Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Ingrid Borecki, Independent Consultant, BioData Catalyst Steering Committee Chair, St. Louis, Missouri, USA.
Shu Hui Chen, National Heart, Lung, and Blood Institute, NIH, Bethesda, Maryland, USA.
Jon Kaltman, National Heart, Lung, and Blood Institute, NIH, Bethesda, Maryland, USA.
Sweta Ladwa, Axle Informatics, Rockville, Maryland, USA.
Chip Schwartz, Coresoft, LLC, Gaithersburg, Maryland, USA.
Alastair Thomson, National Heart, Lung, and Blood Institute, NIH, Bethesda, Maryland, USA.
Sarah Davis, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Alison Leaf, Velsera, Boston, Massachusetts, USA.
Jessica Lyons, Department of Biomedical Informatics, Harvard Medical School, Boston, Massachusetts, USA.
Elizabeth Sheets, UC Santa Cruz Genomics Institute, University of California, Santa Cruz, Santa Cruz, California, USA.
Joshua C Bis, Department of Medicine, University of Washington, Seattle, Washington, USA.
Matthew Conomos, Department of Biostatistics, University of Washington, Seattle, Washington, USA.
Alessandro Culotti, The Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Thomas Desain, Department of Biomedical Informatics, Harvard Medical School, Boston, Massachusetts, USA.
Jack Digiovanna, Velsera, Boston, Massachusetts, USA.
Milan Domazet, Velsera, Boston, Massachusetts, USA.
Stephanie Gogarten, Department of Biostatistics, University of Washington, Seattle, Washington, USA.
Alba Gutierrez-Sacristan, Department of Biomedical Informatics, Harvard Medical School, Boston, Massachusetts, USA.
Tim Harris, UC Santa Cruz Genomics Institute, University of California, Santa Cruz, Santa Cruz, California, USA.
Ben Heavner, Department of Biostatistics, University of Washington, Seattle, Washington, USA.
Deepti Jain, Department of Biostatistics, University of Washington, Seattle, Washington, USA.
Brian O'Connor, Nimbus Informatics, San Jose, California, USA.
Kevin Osborn, UC Santa Cruz Genomics Institute, University of California, Santa Cruz, Santa Cruz, California, USA.
Danielle Pillion, Department of Biomedical Informatics, Harvard Medical School, Boston, Massachusetts, USA.
Jacob Pleiness, Department of Biostatistics, University of Michigan School of Public Health, Ann Arbor, Michigan, USA.
Ken Rice, Department of Biostatistics, University of Washington, Seattle, Washington, USA.
Garrett Rupp, Velsera, Boston, Massachusetts, USA.
Arnaud Serret-Larmande, Department of Biomedical Informatics, Harvard Medical School, Boston, Massachusetts, USA.
Albert Smith, Department of Biostatistics, University of Michigan School of Public Health, Ann Arbor, Michigan, USA.
Jason P Stedman, Department of Biomedical Informatics, Harvard Medical School, Boston, Massachusetts, USA.
Adrienne Stilp, Department of Biostatistics, University of Washington, Seattle, Washington, USA.
Teresa Barsanti, Nimbus Informatics, San Jose, California, USA.
John Cheadle, RTI International, Triangle Park, North Carolina, USA.
Christopher Erdmann, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Brandy Farlow, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Allie Gartland-Gray, RTI International, Triangle Park, North Carolina, USA.
Julie Hayes, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Hannah Hiles, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Paul Kerr, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Chris Lenhardt, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Tom Madden, RTI International, Triangle Park, North Carolina, USA.
Joanna O Mieczkowska, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Amanda Miller, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Patrick Patton, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Marcie Rathbun, RTI International, Triangle Park, North Carolina, USA.
Stephanie Suber, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Joe Asare, RTI International, Triangle Park, North Carolina, USA.
FUNDING
This work was supported by the National Institutes of Health, National Heart, Lung, and Blood Institute through the BioData Catalyst program (award 1OT3HL142479-01, 1OT3HL142478-01, 1OT3HL142481-01, 1OT3HL142480-01, 1OT3HL147154-01) to SA, PA, RB, KB, SC, BND, RG, AK, AKM, BP, AP, IB, SD, AL, JL, ES, AC, TD, JD, MD, AG, TH, BO, KO, DP, JP, GR, AS, AVS, JS, JA, TB, JC, CE, AG, JH, HH, PK, CL, TM, JM, AM, PP, MR, and SS; and National Institutes of Health, National Heart, Lung, and Blood Institute through the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I) to JB, MC, SG, BH, DJ, KR, and AS.
AUTHOR CONTRIBUTIONS
All listed authors made substantial contributions to the work, revised it for important intellectual content, approved the final version for publication, and agree to be accountable for all aspects of the work. SA, PA, RB, KB, SC, BD-D, RG, AK, AM, BP, and AP are principal investigators for the ecosystem. IB, SHC, JK, SL, CS, and AT provide project oversight. SD, BF, AL, JL, and ES provided key support in writing this manuscript in addition to developing the ecosystem. JCB, MC, AC, TDS, JDG, MD, SG, AG-S, TH, BH, DJ, BO’C, KO, DP, JP, KR, GR, AS-L, AS, JPS, and AS are key developers of the ecosystem. JA, TB, JC, CE, AG-G, JH, HH, PK, CL, TM, JOM, AM, PP, MR, and SS provide ecosystem coordination.
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare.
DATA AVAILABILITY
No new data were generated or analyzed in support of this research. We are featuring a system that enables better analysis.
REFERENCES
- 1. U.S. Department of Health and Human Services. NHLBI's COVID-19 Research Strategy. National Heart Lung and Blood Institute, 2021. https://www.nhlbi.nih.gov/coronavirus/research-strategy. Accessed March 31, 2023. [Google Scholar]
- 2. Taliun D, Harris DN, Kessler MD,. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature2021; 590 (7845): 290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. U.S. Department of Health and Human Services. National Heart, Lung, and Blood Institute. Cure sickle cell initiative: advancing research in sickle cell disease: cure sickle cell. Cure Sickle Cell Initiative | Advancing research in sickle cell disease | Cure Sickle Cell. 2023. https://curesickle.org/. Accessed March 31, 2023.
- 4. U.S. Department of Health and Human Services. Collaborating Network of Networks for Evaluating COVID-19 and Therapeutic Strategies (CONNECTS). National Heart Lung and Blood Institute, n.d. https://www.nhlbi.nih.gov/science/collaborating-network-networks-evaluating-covid-19-and-therapeutic-strategies-connects. [Google Scholar]
- 5. Powell K. The broken promise that undermines human genome research. Nature News. February 10, 2021. https://www.nature.com/articles/d41586-021-00331-5. [DOI] [PubMed]
- 6. U.S. Department of Health and Human Services. National Heart Lung and Blood Institute, 2023. https://biodatacatalyst.nhlbi.nih.gov/. Accessed March 31, 2023.
- 7. U.S. Department of Health and Human Services. BioData Catalyst. National Heart Lung and Blood Institute, 2023. https://www.nhlbi.nih.gov/science/biodata-catalyst. Accessed March 31, 2023.
- 8. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016; 3 (1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.NHLBI BioData CatalystⓇ Studies Available Throughout the BioData Catalyst Ecosystem, https://biodatacatalyst.nhlbi.nih.gov/resources/data/. Accessed March 31, 2023.
- 10.NHLBI BioData CatalystⓇ Powered by Gen3 Dictionary, https://gen3.biodatacatalyst.nhlbi.nih.gov/DD. Accessed March 31, 2023.
- 11.NIH RM-17-026_CommonsPilotPhase. https://commonfund.nih.gov/sites/default/files/RM-17-026_CommonsPilotPhase.pdf. Accessed March 31, 2023.
- 12.NIH Office of Strategic coordination: The common Fund. https://commonfund.nih.gov/commons. Accessed March 31, 2023.
- 13.NIH RePORTER. https://reporter.nih.gov/. Accessed March 31, 2023.
- 14. Birger C, Hanna M, Salinas E, et al. FireCloud, a scalable cloud-based platform for collaborative genome analysis: Strategies for reducing and controlling costs [published online ahead of print 2017]. BioRxiv. doi: 10.1101/209494, preprint: not peer reviewed. [DOI]
- 15. U.S. Department of Homeland Security. Federal Information Security Modernization Act. Cybersecurity and Infrastructure Security Agency CISA, 2014. https://www.cisa.gov/federal-information-security-modernization-act. Accessed March 31, 2023. [Google Scholar]
- 16. General Services Administration. The Federal Risk and Authorization Management Program. Technology Transformation Services, 2022. https://www.fedramp.gov/. Accessed May 9, 2022. [Google Scholar]
- 17. Grossman RL, Lakes D.. Clouds and commons: a review of platforms for analyzing and sharing genomic data. Trends Genet 2019; 35: 223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NHLBI BioData CatalystⓇ Ecosystem Security Statement, https://bdcatalyst.gitbook.io/biodata-catalyst-documentation/community/request-for-comments/nhlbi-biodata-catalyst-ecosystem-security-statement. Accessed March 31, 2023.
- 19. Lee S, Abecasis GR, Boehnke M, Lin X.. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet 2014; 95 (1): 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mikhaylova AV, McHugh CP, Polfus LM, et al. Whole-genome sequencing in diverse subjects identifies genetic correlates of leukocyte traits: The NHLBI TOPMed program. Am J Hum Genet 2021; 108 (10): 1836–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kowalski MH, Qian H, Hou Z, et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet 2019; 15 (12): e1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baker M. 1,500 scientists lift the lid on reproducibility. Nature 2016; 533 (7604): 452–4. [DOI] [PubMed] [Google Scholar]
- 23. Stupple A, Singerman D, Celi LA.. The reproducibility crisis in the age of digital medicine. NPJ Digit Med 2019; 2 (1). doi: 10.1038/s41746-019-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Self WH, Semler MW, Leither LM,. et al. Effect of hydroxychloroquine on clinical status at 14 days in hospitalized patients with COVID-19: A randomized clinical trial. JAMA2020; 324 (21): 2165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. St. Martin A, Hebert KM, Serret-Larmande A, et al. Long-term survival after hematopoietic cell transplant for sickle cell disease compared to the United States population. Transplant Cell Ther 2022. doi: 10.1016/j.jtct.2022.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yuen D, Cabansay L, Duncan A, et al. The Dockstore: enhancing a community platform for sharing reproducible and accessible computational protocols. Nucleic Acids Res 2021; 49 (W1): W624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schatz MC, Philippakis AA, Afgan E, et al. Inverting the model of genomics data sharing with the NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-space. Cell Genom 2022; 2 (1): 100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krissaane I, De Niz C, Gutiérrez-Sacristán A, et al. Scalability and cost-effectiveness analysis of whole genome-wide association studies on Google Cloud Platform and Amazon Web Services. JAMIA 2020; 27 (9): 1425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.NIH Cloud Platform Interoperability Effort, https://datascience.nih.gov/nih-cloud-platform-interoperability-effort. Accessed March 31, 2023.
- 30.Bench to Bassinet Program, https://benchtobassinet.com/. Accessed March 31, 2023.
- 31.European Genome-Phenome Archive, https://ega-archive.org/studies/phs001194. Accessed March 31, 2023.
- 32.NHLBI BioData CatalystⓇ Help Desk, https://biodatacatalyst.nhlbi.nih.gov/contact. Accessed March 31, 2023.
- 33.NHLBI BioData CatalystⓇ Community Forum, https://bdcatalyst.freshdesk.com/support/discussions. Accessed March 31, 2023.
- 34.NHLBI BioData CatalystⓇ YouTube Channel. https://www.youtube.com/channel/UCGkmY5oNK8uFZzT8vV_9KgQ/videos. Accessed March 31, 2023.
- 35. Sirén J, Monlong J, Chang X, et al. Pangenomics enables genotyping of known structural variants in 5202 diverse genomes. Science 2021; 374 (6574): abg8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rising to the Challenge of COVID-19: The NHLBI Community Response (2020). https://www.nhlbi.nih.gov/directors-messages/coronavirus-covid-19-nhlbi-response. Accessed March 31, 2023.
- 37.The COVID MUSIC study: long-term outcomes after the multisystem inflammatory syndrome in children. https://covidmusicstudy.com/. Accessed March 31, 2023.
- 38.RECOVER: researching COVID to enhance recovery. https://recovercovid.org/. Accessed March 31, 2023.
- 39. Wu L, Wang D, Evans JA.. Large teams develop and small teams disrupt science and technology. Nature 2019; 566 (7744): 378–82. [DOI] [PubMed] [Google Scholar]
- 40. Robasky K, Boyles R, Bradford K, et al. How to launch transdisciplinary research communication [published online ahead of print 2020]. RTI Press; 2020. doi: 10.3768/rtipress.2020.rb.0022.2004. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analyzed in support of this research. We are featuring a system that enables better analysis.