Abstract
Machine learning is a powerful tool that is increasingly being used in many research areas, including neuroscience. The recent development of new algorithms and network architectures, especially in the field of deep learning, has made machine learning models more reliable and accurate and useful for the biomedical research sector. By minimizing the effort necessary to extract valuable features from datasets, they can be used to find trends in data automatically and make predictions about future data, thereby improving the reproducibility and efficiency of research. One application is the automatic evaluation of micrograph images, which is of great value in neuroscience research. While the development of novel models has enabled numerous new research applications, the barrier to use these new algorithms has also decreased by the integration of deep learning models into known applications such as microscopy image viewers. For researchers unfamiliar with machine learning algorithms, the steep learning curve can hinder the successful implementation of these methods into their workflows. This review explores the use of machine learning in neuroscience, including its potential applications and limitations, and provides some guidance on how to select a fitting framework to use in real-life research projects.
Keywords: Artificial intelligence, Data science, Image analysis, Machine learning, Micrographs
CHALLENGES IN MICROGRAPH ANALYSIS
Digital microscopy is an important tool for basic and clinical research. While the technical instruments available to obtain ever more accurate images of tissue slices have steadily improved over the last years, accurate analysis of the images is key to validate hypotheses. Two of the main challenges of manual microscopy image analysis are that it is prone to human errors and it is time consuming. When relying on human raters to manually analyze images, there can be significant interrater variability (1,2). Priming effects can subconsciously influence evaluation of images, as faster and more efficient microscopes enable us to gather more data in a shorter amount of time; the limiting factor becomes the time needed to analyze those large quantities of data. As researchers must manually select representative images to analyze, this can introduce selection biases. Leaving the selection process to humans can therefore become problematic even when not considering the additional errors and biases in the actual analysis workflow. Using fully automated analysis software solutions could help by significantly reducing the time needed to perform image analysis and allowing for a larger amount of data to be analyzed in the same amount of time. Using algorithms to extract the features needed for statistical evaluation to confirm or reject the null hypothesis also reduces the risk of subconsciously favoring some images over others. The research community has been trying to deal with this problem for centuries, using blinding, randomization, and anonymization techniques to evaluate clinical and research data without prejudice, but in practice this is oftentimes hard to achieve.
Another challenge, which is especially hard for more inexperienced researchers and students, is the learning curve when analyzing datasets. As most tasks are problem specific (e.g. counting axons, nuclei, or other subcellular components), raters could adapt their judgment over the course of the rating process, resulting in different evaluation standards for images that were evaluated later in the study compared to those rated earlier (3).
Reproducibility through deterministic algorithms
According to a survey of 1576 researchers conducted by Nature, more than 70% of researchers have failed to reproduce other scientists’ experiments (4). Many preclinical studies cannot be translated into the clinical field and findings cannot be reproduced by other laboratories. A team at Bayer HealthCare in Germany reported that only about 25% of published preclinical studies could be validated to the point at which projects could continue because there were significant inconsistencies between published data and in-house data when investigators tried to reproduce the results of the articles (5). Discrepancies between published data and validation data can happen for a number of reasons, including the use of different protocols and equipment, lack of identifiability of research resources (6), and the use of a p value as the only marker of significance (7). A lack of reproducibility is also impacted by subjective analysis and interpretation of data by human researchers. To overcome this challenge, multiple approaches are needed to generate more reliable and reproducible results. One approach could be the use of modern computer vision algorithms, which have become increasingly reliable for tasks such as cell detection, segmentation, and feature analysis, for example of heterogeneity, which can be used as prognostic factors of disease (8,9). Algorithms and software for the analysis of microscopic images have been used since the 1950s and have been considerably improved and developed (10). New algorithms are developed constantly, with ever higher accuracy and less segmentation errors, using state-of-the-art machine learning methods that are able to compete using the latest statistical segmentation methods (11).
One of the main advantages of using computer vision applications and machine learning classifiers for the analysis of microscopy images is their deterministic nature. Once trained, a convolutional neural network (CNN) will consistently produce the same results for a given set of data (12). Furthermore, the increasing open-source community and the possibility to easily share source code, algorithms, and configurations with each other allow for a rapid transferability of newly developed algorithms between research labs (13). This will allow researchers to re-run the experiments of their colleagues to better understand their findings and to investigate how the proposed algorithms perform on their own datasets, which may not be publicly available. Leveraging new technologies to iterate through multiple analysis approaches, using different kinds of architectures and algorithm designs without the need to implement every single one from scratch, can significantly speed up the research process.
Computer-aided digital microscopy has already been shown to significantly reduce intra- and interobserver variabilities (14). The addition of machine learning algorithms to micrograph analysis software could further improve this effect. The analysis of large-scale datasets could be made possible through the use of deep learning-based algorithms, which have proven to be able to tackle complex tasks such as differentiating complex structures in immunohistochemical-stained images (15).
Another advantage of learning methods is their inherent ability to adapt to new data, as a trained classifier can be retrained or expanded once new data for training are available. In the best-case scenario, this could lead to permanently improving an algorithm that dynamically adapts to shifts and changes in the data as it becomes more and more “familiar” with the new input.
MACHINE LEARNING IN MICROSCOPY
Review of the machine learning methodology
This review does not strive to give an extensive explanation for the diverse and complex machine learning algorithms and model architectures that are continuously improved and invented, representing an entirely separate topic. Multiple articles have been published that explain the theory and mathematical foundations as well as the possible use cases of machine learning and deep learning as applied in medical research (16–18). This review will focus on providing a conceptual overview of the main concepts to provide a better understanding of real-use cases.
Machine learning is a term used to describe any computing strategy that allows a program to build an analytical model on its own when only provided with a dataset without any human expert knowledge (12). In recent years, the design of new algorithms and the acceleration of computations by better hardware have had an impact on many fields of research including neuroscience by identifying predictive variables or model complex structures (19). In microscopy analyses, machine learning algorithms help to automate image computing to study and detect new aspects in computational neuroscience by analyzing patterns and modeling complex structures (20).
One application is the automated identification of cell-type-specific genes in the mouse brain using images of in situ hybridization expression patterns matched with gene datasets (21). This application provides quantitative data that facilitate more accurate computational modeling of the results than other methods.
One subfield of machine learning is deep learning, which utilizes artificial neural networks. Deep learning algorithms have a special architecture, that is neural networks, which are comprised of different layers composed of multiple nodes called neurons. In densely connected networks, which form the basis of deep learning architectures, each neuron of each layer is connected to each neuron in the next layer; this allows them to learn complex relationships of different features of the input data. The organization of the neurons in the networks can be summed up into 3 main components: an input layer, 1 or more hidden layers, and an output layer (22); processing happens in the hidden layers. To understand how machine learning algorithms differ from more traditional, statistical rule-based approaches to image segmentation and classification, we must first understand the task at hand. If we consider the challenge of image segmentation, the goal is to identify a distinct set of features that characterize the objects of interest inside the obtained image (23). Traditional segmentation methods relied on a mixture of different features, for example intensity, texture, and the detection of edges of objects, to segment images. These features can be combined to form a set of rules to semantically segment images. This mixture of different features can be regarded as a function that transforms the input data (e.g. a microscopy image), to the desired output (e.g. the full segmentation of the image into subregions such as cells). While rule-based systems are designed by hand by researchers using their subject-specific knowledge, machine learning approaches extract this function autonomously once they are properly trained. The selection and curation of training data by humans is critical for the performance and reliability of machine learning models. The quality and representativeness of the training data directly impact the accuracy and generalizability of the resulting model. Training data that are incomplete, biased, or of poor quality can lead to poor performance and inaccurate predictions even if the machine learning algorithm is well-designed. Therefore, it is essential that human experts carefully curate and annotate the training data to ensure that they are representative of the real-world data and cover a wide range of relevant scenarios. This is particularly important in the field of microscopy image analysis in which the complexity and variability of the data can pose significant challenges for machine learning algorithms. The best possibly achievable performance of a model is most often dependent on the quality of the data.
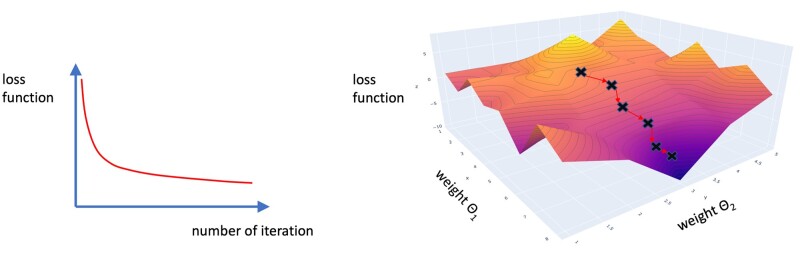
The “learning” property of machine learning models is achieved by implementing 2 functions into the algorithm: a loss function and an optimization function. The loss function describes how far off the machine learning algorithm performance is from the task at hand. In the example of image segmentation, it computes a numerical value for the difference between the desired segmentation and the actual output of the algorithm. This “loss” is then subject to minimization by the optimization function. The optimization function tries to find the combination of different parameters of the algorithm (also called weights), which results in the smallest loss (Fig. 1). The resulting architecture is a continuous iteration between computing the loss of the algorithm by the loss function, adapting its parameters by the optimization function, and then running the new, improved algorithm again and evaluating its loss again; this is called Backpropagation (24).
Figure 1.
Illustrations of the loss function as function of the number of iterations (left) and as function of the weight of the network (right). The loss of the machine learning algorithm decreases with more iterations (i.e. more training time). The network tries to find the best combination of weights (also called parameters) that result in the smallest loss function. With every iteration, the network moves further toward the global minimum. The x- and y-axes represent the weights of the network. For visualization purposes, only 2 parameters are displayed. When dealing with a deeper neural network and simultaneously optimizing all parameters during the training, this multivariate optimization problem can take on much higher dimensions. The z-axis illustrates the cumulative loss function. It is possible for the machine learning algorithm to get stuck in local minima. Multiple solutions to this problem exist, depending on the architecture of the algorithm.
The step-wise adaption of parameters to find the global minimum of the loss function is based on the principle of gradient descent (25). Machine learning algorithms can either learn supervised or unsupervised. Unsupervised learning deals with data without labels provided by humans. Its most common uses are to find relationships between the data points in large datasets, to subsequently find clusters in the dataset, or to extract the most important features of the dataset. The loss function here is based in finding parameters that allow for minimal differences between the data points in each subgroup. Supervised learning requires a set of data with the optimal solution for the task, called training data. This training set is used to compute the loss function in the initial learning phase of the algorithm to optimize its parameters. After the training phase, it can take in new data without the solution and compute the solution itself based on the previously learning parameters.
Recent improvements in software algorithms (for the loss and optimization functions) (25) and hardware architecture, such as graphical processing units and the introduction of tensor processing units (26), have enabled a significant advancement in the field of machine learning-related research areas. By increasing speed and while simultaneously decreasing computing costs and the effort required for the setup, machine learning research has become feasible for research areas outside of mathematics and computer science.
The barrier to include machine learning algorithms efficiently into research workflows decreases further as time progresses with the invention of new tools that require less and less coding experience (27). While training and designing deep learning models from scratch for specific tasks is a complex and challenging task, there are already a number of pretrained models for many tasks such as cell segmentation and pathology classification available, which can be used out-of-the-box with the appropriate software. Some approaches to this task are presented later in this review.
Dataset curation
The curation of a high-quality training dataset is essential for developing accurate and reliable machine learning models for microscopy image analysis. In this section, we discuss the key steps that are necessary for the curation of a training dataset.
Identify the relevant data: the first step in curating a training dataset is to identify the relevant data that will be used for model training. This may include a range of microscopy image types, including brightfield, fluorescence, and confocal images, among others. It is important to ensure that the data are representative of the real-world scenarios for which the model is intended to be used.
Data cleaning and preprocessing: once the relevant data have been identified, the next step is to clean and preprocess the data. This may include tasks such as removing irrelevant images or correcting errors in the metadata associated with the images.
Preprocessing is an essential step in preparing microscopy images for machine learning, as it can help to improve the quality and consistency of the data and can enhance the accuracy and performance of machine learning models. Some of the most important preprocessing steps for microscopy images in machine learning include the following: (1) Image normalization: normalization is a technique used to standardize the intensity values of the pixels in an image. This can help to reduce the effects of variation in illumination and can improve the consistency of the data. Most machine learning frameworks provide this functionality. (2) Filtering: image filtering can be used to remove noise and improve the quality of the image. There are many different types of filters that can be used, including median filters, Gaussian filters, and bilateral filters, which are implemented in most image viewer platforms. (3) Image segmentation: segmentation is the process of dividing an image into different regions or objects. This can be useful for identifying specific features or structures within the image and can be used to extract quantitative information for further analysis. For example, in microscopy images, 1 step could be to divide the image into foreground and background, to be able to operate more efficiently. (4) Image registration: image registration is the process of aligning multiple images to a common coordinate system. This can be useful for creating composite images or when comparing images over time. This step is not always needed for microscopy image analysis but, for example it is necessary for Magnetic Resonance Imaging (MRI) scans of organs that need to be aligned to each other. (5) Image augmentation: image augmentation is the process of creating new training images by applying random transformations to existing images. This can help to increase the diversity of the training data and improve the generalizability of the machine learning model, particularly when there is not much training data available.
Annotation of data: if the goal is to develop a new model, the next step is to annotate the data by adding labels or tags to the images. This can be done manually or using automated tools that are implemented in most image viewer software (e.g. level tracing and grow from seeds). It is important to ensure that the annotation is accurate and consistent and that the labels reflect the true nature of the image content. The quality of the training data will be responsible for the final performance and accuracy of the model to a significant degree.
Selection of training and validation data: the dataset must be split into training and validation datasets. The training data are used to train the model, while the validation data are used to evaluate the performance of the model. It is important to ensure that the split is representative and that the training data are not biased toward certain types of images. A very important point is to avoid any leakage of test data into the training dataset. If we happen to train our model on test data, we will produce a falsely elevated accuracy when evaluating the performance of our model. Keeping the training a testing data separate is therefore a key step in dataset preparation.
Balancing the dataset: in some cases, the dataset may be unbalanced, with a disproportionate number of images in 1 class compared to others. It is important to balance the dataset by adding or removing images to ensure that each class is equally represented. If we are training a model to classify images as pathologic or healthy but training our model on a dataset of mostly healthy images, it may learn that predicting “healthy” for every image results on average on a high score and incentivize the model to produce false negatives.
Quality control: the final step is to perform quality control on the dataset to ensure that it meets the desired quality standards. This may involve reviewing the annotations, checking the balance of the dataset, and evaluating the performance of the model on the validation dataset.
In conclusion, the curation of a training dataset is a critical step in developing accurate and reliable machine learning models for microscopy image analysis. By following the steps outlined above, it is possible to create a high-quality dataset that can be used to train and evaluate machine learning models for a wide range of applications in the field of medical microscopy.
Deep learning algorithms
Deep learning architectures include a variety of models, which mainly differ in architecture and optimization and loss functions. Currently, there are several main architectures of neural networks, namely multilayer perceptron, recurrent neural networks, and CNN.
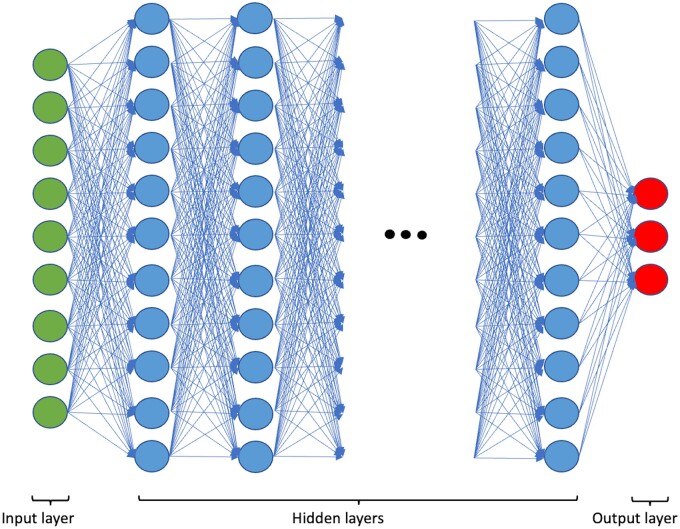
A multilayer perceptron is a feedforward artificial neural network model that maps sets of input data onto a set of appropriate outputs (Fig. 2) (28). It is composed of multiple layers of nodes in a directed graph, with each layer fully connected to the next one. Each node uses a nonlinear activation function, which means it uses all its input values to return a value between 0 and 1 that gets forwarded to each neuron in the next layer. The concept of linking multiple nodes together and combining their inputs to obtain the activation for the next layer enables the network to learn complex functions to process data on its own. It can be thought of as the blueprint for a complicated function in which parameters get dynamically adapted over time to find the best combination for the task at hand.
Figure 2.
Basic neural network architecture. The Input layer represents the data which is handed to the network as pixel values. The hidden layers can have an arbitrary number, the depth of the networks correlates with the computational burden. The dynamic adaption of weights during the training process happens in the hidden layers. Each neuron represents a covariable of the final transformation function learning by the network. The output layer is the output of the final computation, for example the classification of data into different labels. In the case of classification problems, it outputs a single label, while in segmentation problems it outputs pixels with a label assigned to each individually, which are then reconstructed into the final image.
Recurrent neural networks (RNNs) are a class of artificial neural networks that use a directed cyclic graph to process sequences of data items (29). They are like simple feedforward artificial neural networks but contain cycles that enable them to exhibit dynamic temporal behavior. RNNs are designed to process variable-length sequences. Subtypes of this architecture are mostly used for tasks such as natural language processing and time series prediction as they are designed to process and extract patterns from sequential data thereby profiting from their ability to consider previous states (30,31). Neural networks have been successfully used to predict Alzheimer disease progression by extracting patterns from temporal data obtained over the course of the disease from its training data (31,32). Using similar approaches with other network architecture and careful data selection could lead to detect patterns of disease and progression markers in a number of neurological conditions.
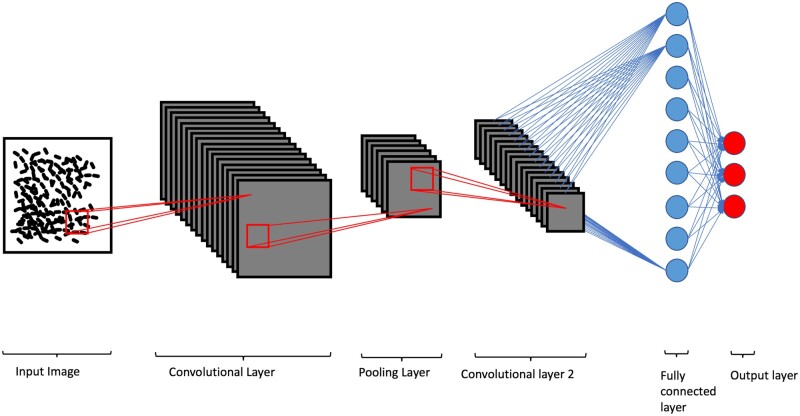
CNNs are composed of alternating convolutional and pooling layers (Fig. 3) (33). They are designed to process data that have a grid-like topology, such as digital images, films, and sound, and can extract spatial features from them in a process similar to that in the human visual cortex. They are most widely used for image classification, object detection, and image segmentation. Convolutional layers consist of a set of feature detectors that are applied to small regions of the input. Each detector uses a small set of parameters; the weights are shared among all locations and scales at which the detector is applied. This means that a single weight matrix is used to compute the output of a convolutional layer. By using convolutions, which can be also thought of as kernels, the network is able to learn basic features in images such as borders, edges, and shapes, depending on the depth of the network. We can observe a spatial hierarchy in CNNs in which simple patterns learned by the first couple of layers will be combined to form more complex patterns in the following layers. Convolution layers are often followed by subsampling layers. The subsampling layers reduce the spatial resolution of the input and thus decrease the size of the feature vectors. This effectively reduces the computational burden of the network and forces the network to learn a more invariant representation of the input. The decrease in the computational burden is important because the number of learnable parameters in a CNN increases much faster with its depth when compared to traditional neural networks. Multiple adaptions in its architecture were invented to deal with this problem, such as the addition of “skip connections,” which bypass some hidden layers and connect directly into layers further down the network (34).
Figure 3.
The architecture of a convolutional neural network. The input image is processed by multiple kernels which result in several feature maps learned by the network in the convolutional layers. The pooling layers act to decrease the computational burden and emphasize the primary features of the learned patterns. The last layers are fully connected to end in an output layer which again outputs the desired result of the computation, for example a classification of the picture or a label map of the image. This architecture is specifically designed for dealing with imaging data, as the spatial relationships between neighboring pixel groups can be preserved and learned by the network.
The CNN architecture can be trained to detect different types of objects in images, such as the presence of a road sign on a highway, a brain tumor in an MRI or distinct cell lines in microscopy images. This makes them versatile and capable of adapting to many different requirements of different specialties in medical research. The adaptability of these networks to different challenges has led to various applications in neuroscience, such as glial cell segmentation (35), segmentation of neuronal membranes (36), and subcortical brain structure segmentation (37). The main advantage of CNNs is their ability to automatically learn robust features from large training datasets when trained accordingly. By using data augmentation methods during training, they are able to learn features that are invariant to rotations and translations by extracting geometric features in an image; this helps to reduce the amount of manual feature engineering by human experts (29). The main disadvantages of CNNs are that they require large amounts of training data and computational power to train and they are notoriously difficult to optimize (Table 1). This is because the network depth and the number of tunable parameters are usually large, even though there are some recent advances in dealing with overfitting problems (38,39).
Table 1.
Comparison of advantages and disadvantages of convolutional neural networks
| Advantages | Disadvantages |
|---|---|
| High accuracy | Requires large amounts of data |
| Designed to deal with image data | High computational costs |
| Can extract spatial features in a hierarchical matter | Hard to optimize due to large parameter size |
The displayed arguments can be seen as a tradeoff between accuracy and complexity, which results in an increased need for time and computational costs.
The U-Net is a special kind of CNN that was developed for use in medical image segmentation and proposed by a research team from the University of Freiburg in 2015 (40). This architecture is characterized by a symmetric configuration of a contracting sequence of layers, which reduce the size of the input image and extract features from it, and a symmetric, expansive path, which up-samples the segmented image to match the input shape and therefore produce an accurate segmentation of the image. This symmetric contracting-expanding architecture inspired the name “U-Net” because the shape is reminiscent of a “U.” This network takes in an image and outputs its complete segmentation into the labels it was trained on. The segmentations each model can produce therefore largely depend on the training set and labels on which it was originally trained, again highlighting the need for high-quality, curated datasets. The efficiency of the U-Net has been demonstrated in several studies; this has recently resulted in the design of the “nnU-Net,” short for “no-new-U-Net,” which is a framework developed by the Medical Image Computing Team in Heidelberg (41). This design has won numerous segmentation challenges and is currently considered state-of-the-art for most medical segmentation tasks (42).
The nnU-Net aims to remove most of the preprocessing and architectural design, which are major barriers for new researchers coming to the field of machine learning. It works in a completely automatic fashion and designs the entire training and validation pipeline autonomously when it is provided with adequate training data. Many research laboratories have already adapted the framework and conducted studies on its performance on multiple tasks, such as brain tumor segmentation (43) and fetal brain tissue segmentation (44). Most of the studies have studied the performance of nnU-Net on radiologic images, but there are several networks specifically designed for histology images, such as StarDist (11). A more detailed look at networks specifically designed for micrograph analysis follows.
Classification versus segmentation problems
Classification tasks for image analysis describe the purpose of predicting a discrete class label from a set of classes; for example, if the tissue seen in the image contains pathological tissue or not (45,46). Each input image is assigned a label. Possible use cases are the detection of alteration in tissue slices, such as scarring, tumor cell infiltration, or abnormal growth patterns.
Segmentation describes the process of dividing an image into multiple segments that are continuous sets of pixels, also known as superpixels. Each segmented region is homogeneous and represents a distinct component of the image. Examples for segmentation problems are outlines of single cells, defining tumor borders or finding distinct patterns in images (47). Segmentation tasks can be thought of as a “pixel-wise” classification problem, answering the question of classification for each pixel separately. While there are already multiple rule-based segmentation methods that are heavily used to facilitate microscopy image analysis, data-driven implementation can provide several advantages. A study in 2020 showed that data-driven algorithms do not only hold up and even beat the accuracy of rule-based systems but at the same time are much faster once trained (48). This is crucial in a dynamic field of biomedical research in which reiterating through numerous approaches to solve a problem is needed.
Current challenges and their possible solutions
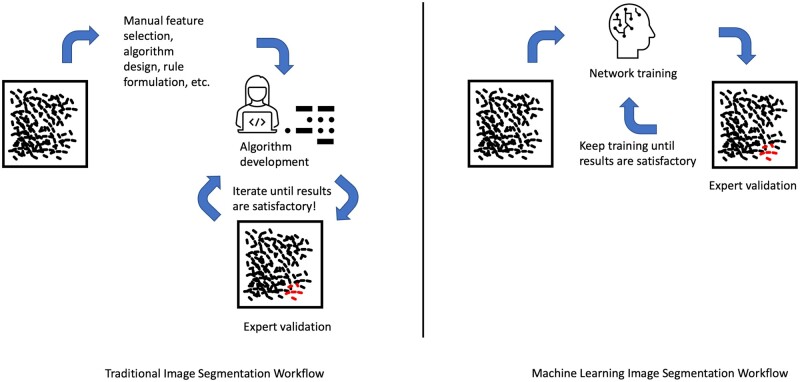
As far as modern machine learning algorithms and research in the fields of computer science and mathematics have come, there are still some significant challenges in the attempt to translate these insights into the healthcare/research sector. Furthermore, the authors want to emphasize that machine learning approaches to data analysis problems are not necessarily a substitute for the use of statistical algorithms or rule-based models. They should be seen as another tool to automate and advance the image analysis process and as a new way to gather insights into the growing amount of data we are able to gather with new technologies. By combining statistical and machine learning-based algorithms we can profit from concepts and tools from both toolsets (Fig. 4).
Figure 4.
Comparison between traditional image segmentation workflows and machine learning workflows. Left: the traditional approach, where the statistical algorithm to solve the classification or segmentation problem is designed by manually finding rules in the dataset which are then applied and tested on the whole dataset. If no satisfying outcome is observed, the process is repeated with another choice of parameters. Right: the machine learning approach. Researchers must decide on network architecture and hyperparameters, such as depth, learning rate and optimization algorithm. Once the network is designed, it is fed with training data to learn accurate representations and transformative functions on its own. If the results are not satisfying, we can either keep training the network or adapt its architecture.
Articles published in life science journals do oftentimes not provide enough information about the exact machine learning algorithms and tech stack used to enable other researchers to recreate the experiment (49). Only 6% of research articles on artificial intelligence release their code, leaving many questions about the correct and exact implementations of algorithms open (50). Furthermore, the training of models depends heavily on the abundance of high-quality datasets. Models trained on a subset of data acquired at 1 institution may not be transferable to another due to the use of different protocols, different equipment, and different analysis approaches, all of which may have an influence on the performance of the model. Public datasets are rare, which carries the risk of unintentional overfitting the data when training multiple machine learning algorithms frequently on the same datasets. This demonstrates the need for open, publicly available datasets in addition to open-sourced algorithm to further advance research in the biomedical imaging domain. The work on those archives has already started, with a great example being The Cancer Imaging Archive (TCIA) (51). This effort by the National Cancer Institute aims to provide curated and open datasets to help researchers build generalizable machine learning and data analysis models. The importance of archives like the TCIA is illustrated by the fact that only 51% of research articles in the field of machine learning for human health care use public datasets (50). Developing machine learning algorithms on private datasets prevents other researchers from independently evaluating the integrity and generalizability of the model and can lead to unintentional overfitting of the data to the oftentimes much smaller, private datasets.
The key to successful application of machine learning is to identify the classes of data that can be collected in a reliable and consistent manner. In biomedicine, we often deal with highly complex, multifaceted data, which makes data collection and preparation more difficult. One of the challenges in this field is how to ensure that the data collected are truly representative of the underlying system.
If large amounts of data are collected efficiently, the amount of data needed to analyze and discover patterns and relationships will grow accordingly. This will put a strain on the computational resources research institutions have available and necessitates a cooperative environment between computer scientists and researchers. It is notable that dataset size is not the only or even the most important factor for the performance of machine learning algorithms in the medical domain. A 2021 study found that the choice of the classifier and how closely the distribution of the dataset represents the true distribution of data are among the most important factors (52). This further confirms the notion that the overall design of machine learning studies should be picked deliberately and with sufficient knowledge of the different parameters in mind.
Another different kind of challenge is the additional task for researchers in the biomedical fields to keep up with the advances in the development of new algorithms and best practices in the bioinformatics sector; selecting the best algorithm for the problem at hand and configuring it accordingly can significantly improve its performance (53). The possible solution is to outsource the tasks of providing computing power, disk space, and server maintenance to cloud-based systems. However, this is especially problematic in the field of medical research, where data often fall under very strict security laws and restrictions.
File types and sizes
Another challenge for the consistent implementation of machine learning-based software solutions for micrograph analysis is the variety of file types that have been established over the years. There are several file types that are commonly used, including TIFF (tagged image file format), JPEG (joint photographic experts group), PNG (portable network graphics) (54), BMP (bitmap), DICOM (55), and proprietary file formats such as CZI (56). Among these, TIFF is considered the gold standard for microscopy image analysis due to its high resolution and support for multichannel images. TIFF files can store uncompressed data or lossless compressed image data that preserve the quality of the original image (57). JPEG is another commonly used file type, particularly for web-based image sharing due to its relatively small file size. However, JPEG compression can result in the loss of image quality, which can limit its usefulness in some microscopy applications. PNG is another lossless file format that is becoming more popular due to its support for transparency and its ability to store multichannel images. BMP and GIF (graphics interchange format) are older file formats that are less commonly used in microscopy image analysis today but may still be encountered in some contexts. An example for a popular proprietary file format is the CZI file format, used by Zeiss microscopes, which is becoming increasingly popular in microscopy image analysis. The CZI format can store multiple images, metadata, and annotations in a single file, making it useful for storing large amounts of microscopy data (57). In addition, the CZI format can store multichannel images, time-lapse sequences, and 3D stacks, which is useful for a variety of applications. However, since the CZI format is proprietary, it may pose challenges in data sharing and interoperability with other software packages that do not support this format. Interoperability is key for conducting high-quality research, as other laboratories and research groups should be able to seamlessly integrate new insights into their own work and comprehend the steps described in each study. Another important file format, especially in the clinical setting, is the DICOM (digital imaging and communications in medicine) file format, which is the standard format used in medical imaging, including microscopy imaging. DICOM files can store a variety of image types and metadata, such as patient information, imaging parameters, and image processing history. DICOM files are widely used in medical settings due to their compatibility with various imaging modalities and equipment, as well as their support for interoperability between different healthcare systems (56), However, the DICOM format can be more complex and less flexible than other image file formats and may require specialized software for analysis and visualization.
Micrograph images can differ greatly in size depending on the microscopy technique used, the imaging parameters, and the resolution required. For instance, a typical light microscopy image of a single cell can range from a few hundred kilobytes to several gigabytes in size. Similarly, electron microscopy images of cellular organelles or tissues can range in size from a few hundred kilobytes to several gigabytes. Particularly, when dealing with whole slide images in a high resolution, the size can grow quickly and this can pose challenges for image viewers in terms of storage, retrieval, and visualization.
To deal with large image sizes, image viewers can use various techniques. One common technique is image compression, which reduces the size of the image file by removing redundant or irrelevant data. Lossless compression methods can be used to compress image files without losing any information, while lossy compression methods such as JPEG can be used to reduce file size at the cost of some loss of image quality (58).
Another approach is to use image viewers that can handle large image sizes, such as specialized software packages like Fiji/ImageJ or Orbit, which have been specifically designed for this. These software packages can handle large image sizes by loading only a portion of the image into memory at a time, which allows users to navigate and visualize large datasets efficiently.
Lastly, cloud-based image viewers or image hosting services such as OMERO or Cytomine can also be used to store and share large microscopy datasets with collaborators across different locations. These platforms provide web-based interfaces that allow users to view, annotate, and share microscopy data without the need for large file transfers (59).
Deep learning for micrograph analysis
There are special networks adapted for micrograph image analysis that have gained momentum over the last years, with research labs around the globe exploring new ways of developing and testing different network configuration, setups, and integration into existing software to produce more reliable and usable deep learning models. One notable contribution of the open-source researcher community to these efforts has been the creation of ZeroCostDL4Mic (60). This platform aims to simplify the process of using these sophisticated networks and apply them to actual research problems by providing access to pretrained models, which are already embedded in the appropriate setup to be used out-of-the-box without excessive need for configuration. If researchers want to train their own models using the published architecture of the model instead of using the pretrained weights, there is also the possibility of connecting their local dataset to the environment to train the model for themselves. In most cases, a GoogleColab (61) environment is utilized to provide quick access for researchers to test out the networks and evaluate them for their specific needs.
This helps translating the research on deep learning architectures into the field of their actual implementation in micrograph analysis. They also provide tutorials and references to the papers where the networks have been published for the first time.
As for the different networks, there have been numerous different approaches to the already mentioned problems of segmentation and classification, but also for further tasks such as denoising, that is artificially increasing the resolution of images and translating image types into each other (62) (e.g. brightfield to IHC). A very practical overview can be found in the work of Xing et al (63), which gives an overview about the different tasks that have already been solved using deep learning networks, which networks the teams used, and what the advantages and disadvantages of each study were. Most research conducted with neural networks for image analysis is based on the same network architectures discussed above. Therefore, one of the main tasks to build a deep learning framework fitting the specific needs of research laboratories is to identify the correct model for the problem at hand. To provide a useful overview on how to apply different networks to different problems, we have summarized some of the most important networks and their use cases in Table 2.
Table 2.
Overview of the most commonly used, open-source deep learning models designed for the task of microscopy image analysis
| Project | Reference | Model architecture | Use cases | Advantages | Open-source code |
|---|---|---|---|---|---|
| U-Net | (76) | U-Net | Segmentation of various structures, very adaptive |
|
Various |
| StarDist | (11) | Modified U-Net + polygonal representation of objects | 2D and 3D object detection and segmentation, designed for complex shapes. |
|
https://github.com/stardist/stardist |
| Cellpose | (60) | Modified U-Net + vector representation of objects | 2D and 3D object detection and segmentation, continuously improved by the community training |
|
https://github.com/MouseLand/cellpose |
| SplineDist | (77) | Modified U-Net (extended StarDist) | Extension of the StarDist network, segmentation of objects with more complex shapes and curves |
|
https://github.com/uhlmanngroup/splinedist |
| EmbedSeg | (78) | ERFNet (convolutional network with residual connections) | Instance segmentation of cells and nuclei in microscopy |
|
https://github.com/juglab/EmbedSeg |
| CellSeg | (79) | R-CNN (region based convolutional network) | Segmentation on highly multiplexed fluorescence images |
|
https://github.com/michaellee1/CellSeg |
Every model can be used for the task it was originally trained for or retrained from scratch, given enough training data is present. All of the listed models are freely and publicly available, some are even provided with hands on tutorials to help new researchers get started using them. While this list is not exhaustive, the authors would recommend to start out with one of the models listed above, due to their ease of use and already proven accuracy at the task of micrograph analysis.
The StarDist network has proven to be a highly accurate and reliable network choice for segmentation tasks at the cellular level (11). StarDist is a deep learning method that uses a CNN for the detection and segmentation of cells in microscopy images. The network is based on the concept of star-convex polygons, which are a generalization of star-convex shapes. During inference, the network predicts a set of star-convex polygons that fit the cells in the image. The star-convex polygons are defined as the intersection of multiple disks and are chosen to provide a good tradeoff between fitting the cell shape and being computationally efficient. StarDist is also available as an open-source software package, making it readily accessible to researchers and practitioners in the field (https://github.com/stardist/stardist).
Another very popular choice of network is Cellpose, which has been trained on a large training dataset of cells of varying size and shape (64). Cellpose is based on a combined approach that uses simulated diffusion to produce topological maps of the cells and a U-Net to predict gradients along these maps. By following the gradients, which converge to the same center, single cells can be tracked. This unique approach significantly increases the accuracy of segmentation in comparison to a plain U-Net, probably due to the complex nature of segmenting cells with different shapes and sizes.
Other open-source models specifically designed for microscopy image analysis and approaches to address the challenge of differentiation objects of such varying shape as cells and subcellular structures are listed in Table 4.
Table 4.
Examples of different applications of machine learning applications for different fields of neuropathological challenges
| Name of study | Reference | Model | Outcome | Relevant field |
|---|---|---|---|---|
| 3D segmentation of glial cells using fully convolutional networks and k-terminal cut | (37) | CNN | High accuracy (F1 0.89) in 3D segmentation of glial cells | Neuropathology, basic research |
| Code-free machine learning for classification of central nervous system histopathology images | (80) | CNN | High precision in detecting various brain tumor entities (e.g. glioma, subtypes of glioma, metastasis) | Neuropathology, neurooncology |
| Deep neural networks segment neuronal membranes in electron microscopy images | (38) | CNN | Superhuman pixel error rate (60 × 10−3) on segmenting neuronal membranes for studying connectomes | Basic research |
| Artificial intelligence in neuropathology: deep learning-based assessment of tauopathy | (81) | CNN | High accuracy in detecting neurofibrillary tangles for diagnosis of tauopathies | Neuropathology, neurodegeneration |
| Automated brain histology classification using machine learning | (82) | CNN | Low- versus high-grade glioma classification of brain histology slides | Neuropathology, neurooncology |
| Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks | (83) | CNN | Intraoperative diagnosis of brain tumors from hematoxylin and eosin-stained (H&E) specimens | Neurooncology, neurosurgery, neuropathology |
The papers listed here show some of the diverse use cases of neural networks in supporting researchers and pathologists in their work and research projects.
CNN, convolutional neural network.
PRACTICAL OPEN-SOURCE SOFTWARE SOLUTIONS FOR RESEARCHERS
With an ever-increasing amount of data, the need for convenient, reliable, and efficient data analysis software is greater than ever. Open-source software is an important component of this effort as it provides a way to share and collaborate on software. It allows researchers from different institutions around the world to integrate seamlessly, add to existing solutions, and learn from and share their software with the community (65). The availability of open-source software helps to keep data analysis costs low and accelerate the development process in institutions in developing countries. There are already several well-validated and sophisticated open-source image analysis applications that specialize in providing tools for scientific imaging. We present an overview of those solutions as well as a short comparison between them in Table 3. Moreover, recent advances in code-free machine learning systems enable researchers with no or little coding knowledge background to leverage machine learning resources with so-called “code-free” machine learning platforms (27). While these will help with widespread adoption, a thorough understanding of the underlying algorithms and background is recommended to ensure the use of these algorithms to their full potential. The section below will give an overview of the most commonly used tools for implementing machine learning approaches into the workflow of digital histopathology analysis. It is notable that there are many more efficient and highly developed commercial software solutions that provide functionalities for the analysis of micrograph images. An overview of the most commonly used Open-Source Viewers, meaning that the source code is open to the public and freely available, follows.
Table 3.
Comparison table between advantages, disadvantages, and the used programming language for development of different open-source microscopy viewers, namely QuPath, orbit, ImageJ/Fiji, and napari
| Viewer | Advantages | Disadvantages | Language | Operating system | WSI support |
|---|---|---|---|---|---|
| QuPath |
|
|
Java | macOS, Windows, Linux | Yes |
| ImageJ/Fiji |
|
|
Java | macOS, Windows, Linux | Plugin required |
| Orbit |
|
|
Java | macOS, Windows, Linux | Yes |
| napari |
|
|
Python | macOS, Windows, Linux | Plugin required |
| BioImageXD | Many native functions for common image processing steps implemented from well validated frameworks |
|
Python, C++ | macOS, Windows, Linux | Yes |
| Cytomine |
|
|
Java | Web based | Yes |
| Icy |
|
|
Java | MacOS, Windows, Linux | Yes |
| Cellprofiler |
|
|
Python | macOS, Windows | No |
This list is not exhaustive but provides an overview about the different arguments a research department might consider when choosing a software for building image analysis workflows. Different requirements have different solutions, where each of the viewer software solutions was built with different intentions.
WSI, whole slide image.
QuPath
QuPath is an open-source, multiplatform, software for image processing and analysis designed for scientists to analyze and annotate large volumes of histopathological image data (66). It is based on Java and runs on Windows-, macOS-, and Linux-based systems. Its main advantages are its ease of use software as it comes with a comprehensive and intuitive graphical user interface. It offers an integrated, powerful application programming interface for custom algorithms to develop problem-specific workflows inside the application. Experienced users can write their own scripts in the scripting language “groovy” to automate workflow inside the software. It comes with a large number of preinstalled algorithms and support for popular machine learning frameworks, most notably OpenCV, which is one of the most popular computer vision libraries currently available (67). The ready-to-use implementation makes it easy to try out different algorithms for the same problem; however, it limits the possibilities for researchers to implement other frameworks and algorithm into QuPath when compared to Fiji.
ImageJ2/Fiji
ImageJ2 is a medical computing software designed to analyze images and the rewrite of the original ImageJ software, optimized for dealing with multidimensional image data to focus on scientific imaging (68). Fiji, which stands for “Fiji is just ImageJ” is a distribution of ImageJ2 that already bundles a lot of the most used plugins developed for ImageJ into a single installment. It is used in many areas of medicine including biomedical imaging, pathology, molecular biology, neuroscience, and dentistry and is accessible to users of all levels of expertise. The software is available for all operating systems and can integrate with a variety of other tools and viewers. In September 2015, the ImageJ website reported that the software was downloaded over 1 000 000 times, with more than 30 000 registered users contributing to the community. Its widespread adoption in the medical community has led to an extensive network of developers that create and maintain ready-to-use plugins for various tasks such as segmentation, classification, and feature extraction. Thanks to the numerous contributions of its large community and the active development, Fiji offers custom solutions to nearly every common task in medical imaging and many specialized problems as indicated by its over 2000 plugins available on its update site.
Orbit
Orbit is another open-source whole slide image analysis tool written in Java (69). It was designed to be used with a tight integration with existing solutions in mind. One outstanding feature is therefore the possibility to connect to existing image servers like the OMERO server architecture (60). This can shorten the time it takes for new researchers to get started with analyzing their images by reducing the setup time and specific knowledge to set up data directories. Orbit can connect to local servers or cloud clusters and is able to connect with ImageJ to use the wider plugin options. Orbit was designed to handle whole-slide scans that are challenging to analyze due to their oftentimes very large size. Similar to QuPath, experienced users can create their own scripts in the scripting language “groovy.”
napari
napari is the newest software of the image viewer applications having been released in 2020 as an alpha-build (70). It is still under heavy development but has attracted many researchers due to its native integration with the scientific programming stack of the Python programming language. This image viewer is completely written in Python, which established itself as the main programming language of the scientific community in recent years. This tight integration with the Python programming framework and its associated scientific libraries results in a modular platform that can be easily extended and modified, provided that some knowledge of the Python programming language is present. The napari image viewer can also be directly started from Jupyter notebooks, which are a document format specifically developed for researchers of all fields to publish their code and results in a readable and executable format (71). napari aims to integrate seamlessly with the current landscape of research using Python frameworks and libraries. Currently, however, there are multiple features still missing that are available by other software solutions, such as easy intuitive ways to process multiple images at once or design workflows without programming knowledge.
BioImageXD
BioImageXD is an open-source bioimage informatics platform that provides a range of advanced image processing functionalities for 3D and 4D microscopy data (72). One of the strengths of BioImageXD is its ability to implement other well-established frameworks such as VTK (the visualization toolkit) and ITK (the insight toolkit), which further extend its capabilities in image processing and analysis. This makes it possible to utilize various algorithms and methods for filtering, segmentation, registration, and visualization, which are useful for various microscopy applications.
In addition to the implementation of other frameworks, BioImageXD has a large selection of native image processing functionalities that are designed to enable efficient and effective processing of 3D and 4D microscopy data. These include the ability to perform complex segmentation tasks, 3D rendering and animation, and tracking of cells in time-lapse images, among others. BioImageXD also offers the ability to combine different processing methods and workflows to create customized image analysis pipelines, which can be used to perform sophisticated analysis on complex microscopy data.
However, 1 potential disadvantage of BioImageXD is that its user interface may be less intuitive compared to other software packages, which can make it more challenging for some users to navigate and use effectively. This may also be one of the reasons why the community support around BioImageXD is significantly less pronounced than for most of the other image viewers. For this reason, it may require a learning curve for some researchers to fully utilize its capabilities. Even though it provides many functions without the need for programming skills, the lack of good tutorials and learning materials can pose an obstacle to implementing this software solution into the workflow of laboratories.
Cytomine
Cytomine is an open-source software package designed for collaborative analysis and management of large-scale microscopy imaging data. Cytomine provides a web-based interface that allows users to access and manage microscopy data, as well as collaborate with other researchers in the analysis and annotation of the data. One of the key strengths of Cytomine is its ability to handle large datasets because it uses a distributed architecture that can scale to thousands of images and terabytes of data. This also solves the problems of having less computationally capable hardware available because the main setup and configuration as well as storage and computation are all handled in the cloud. Additionally, Cytomine provides a variety of image analysis tools, including segmentation, classification, and annotation, which can be used to perform detailed and accurate analysis of microscopy data.
Another advantage of Cytomine is the support of a wide range of image formats, including proprietary formats from various microscopy equipment manufacturers, which can facilitate data sharing and interoperability.
However, 1 potential disadvantage of Cytomine is that the client is solely web based. This necessitates uploading the data to the Cytomine cloud. Even though Cytomine was designed with data security and privacy in mind, this may pose a challenge for many laboratories. Some researchers may be subject to data protection regulations that require them to store and process their data within certain jurisdictions, which may limit their ability to use cloud-based software that operates outside those jurisdictions. As such, researchers should carefully consider their data protection obligations and seek legal advice, if necessary, before using cloud-based software for microscopy image analysis. Nonetheless, Cytomine is a powerful tool for collaborative analysis and management of large microscopy imaging data, particularly for those who require machine learning and deep learning capabilities.
Icy
Icy is an open-source software package designed for bioimaging, including microscopy image analysis (73). One advantage of Icy is its user-friendly interface that provides easy access to a wide range of analysis tools and plugins. Icy also supports a variety of microscopy image data types, including multidimensional time-lapse images, 3D stacks, and high-content screening data. Moreover, Icy provides a flexible and modular architecture that enables users to develop and integrate their own plugins and algorithms into the software.
One disadvantage of Icy is that it can be slow and memory intensive when working with large datasets, which may limit its scalability. Additionally, while Icy provides a wide range of analysis tools, some users may require additional plugins or algorithms that are not readily available in the software. Moreover, while user friendly, Icy may lack some of the advanced features and customizability that are available as plugins of other software packages such as ImageJ/Fiji due to the comparatively smaller plugin offer.
CellProfiler/cell profiler analyst
CellProfiler is an open-source software package designed specifically for high-throughput microscopy image analysis (74). CellProfiler also includes a wide range of prebuilt image analysis modules and algorithms, as well as the ability to create customized workflows for specific analysis tasks. Moreover, CellProfiler has a user-friendly interface that makes it accessible to researchers with varying levels of programming expertise. CellProfiler allows users to create a pipeline of analysis modules to be applied to multiple images in a batch-processing mode, thereby allowing for efficient processing of large datasets.
This batch-processing mode is particularly useful for high-throughput microscopy applications where thousands or even millions of images need to be analyzed. The predefined workflow also enables users to apply standardized and reproducible analysis methods, reducing variability in the results and facilitating comparison between different datasets.
Once the images have been processed using CellProfiler, the CellProfiler Analyst software provides a unique way to quickly analyze and visualize the results obtained by the image analysis workflow. This tight integration can be used to perform data analysis, including visualization, statistical analysis, and machine learning in 1 step, further simplifying the process of analyzing large datasets and extracting useful features. The Analyst software is a user-friendly interface for exploring and visualizing large datasets and for identifying correlations and trends within the data.
The ability to automate image analysis and data extraction with CellProfiler and subsequently analyze the data using CellProfiler Analyst is a unique and powerful feature that makes it possible to perform sophisticated image and data analyses on large datasets in a standardized and reproducible manner. This is particularly important for high-throughput microscopy applications, where the volume and complexity of the data can make manual analysis impractical or error prone.
A disadvantage of CellProfiler is that it can be computationally intensive, requiring high processing power and storage capacity to handle large datasets. Additionally, the learning curve for CellProfiler can be steep, particularly for researchers without prior programming experience, even though the software provides a way to create workflows completely from a graphical interface without the need to write any code. However, there is no native support for whole slide images. To use whole slide images, the extension of CellProfiler with other image viewer software solutions like QuPath is necessary.
The Python programming framework (including the most used libraries)
Python is an object-oriented, open-source programming language that was designed to be both readable and easy to learn (Python Software Foundation. Python Language Reference, version 3.10.7. Available at http://www.python.org). Python is suited to rapid application development, with clear syntax and an extensive standard library. As of 2022, it is the most widely used programming language on GitHub, the largest developer platform to date. It also includes several standard modules that allow for the processing of scientific data, such as NumPy, SciPy, and matplotlib (often referred to as the SciPy stack, short for scientific Python). Used together, Python and SciPy provide some of the most powerful tools to analyze complex datasets and create statistical and machine learning models and visualizations. Due to the above-mentioned features, the Python programming framework has seen a rise in adaption among the research community in the recent decade. Its intuitive syntax allows for a fast adaption and easy learning curve, which enables researchers with little to no programming background to still develop data analysis and visualization workflows within a reasonable amount of time. Its popularity as a beginner-friendly language has also resulted in an extensive and growing set of openly available tutorial for various recurring tasks, ranging from computer vision and visualizations to natural language processing and workflow optimization. The importance of Python as the main development language for machine learning and artificial intelligence applications is visualized by the fact that the 2 most widely used frameworks for deep learning applications, namely TensorFlow (75) Google and PyTorch by Meta (76), are both primarily aimed toward Python developers. There are also dedicated frameworks and libraries for image processing tasks in the Python programming language, such as scikit-Image (77) and, more recently, the deep learning PyTorch-based Medical Open Network for Artificial Intelligence (MONAI) framework which is actively developed by many research institutions and commercial companies like NVIDIA (78).
Conclusion
Machine learning algorithms can have a positive impact on the research workflows of neuroscience labs dealing with micrograph images. They can be used to automatically segment and classify images and extract quantitative information from them. By automating tasks that were traditionally done manually, they can help to increase reproducibility, objectivity, and efficiency in the research group. While many network architectures are actively developed and improved, the most used and best-tested implementations can be accessed easily by their integration into modern open-source microscopy viewers. When choosing a research framework for the image analysis workflow, care must be taken to choose the right tools and networks for the challenge at hand. While more mature solutions have a larger community and plugin collection, newer viewers can provide possibilities for researchers to integrate the latest implementations of machine learning models and algorithms into their work. Using machine learning in neuroscience research does require a basic understanding of the underlying concepts to be able to identify possible shortcomings and challenges and choose the right algorithms. Doing so can improve the quality of research and offer new ways to gather insights into the data.
FUNDING
Funding was generously provided by the National Center for Advancing Translational Sciences (NCATS) UL1 TR002377 and TL1 TR002380, the Bowen Foundation, the Nemitz Foundation, the Morton Cure Paralysis Fund, and the Mayo Clinic Benefactor Funded Career Development Award – Regenerative Medicine Initiative.
Contributor Information
Frederic Thiele, Department of Neurology, Mayo Clinic, Rochester, Minnesota, USA; Department of Neurosurgery, Medical Center of the University of Munich, Munich, Germany.
Anthony J Windebank, Department of Neurology, Mayo Clinic, Rochester, Minnesota, USA.
Ahad M Siddiqui, Department of Neurology, Mayo Clinic, Rochester, Minnesota, USA.
CONFLICT OF INTEREST
The authors have no duality or conflicts of interest to declare.
REFERENCES
- 1. Eriksson H, Frohm-Nilsson M, Hedbled MA, et al. Interobserver variability of histopathological prognostic parameters in cutaneous malignant melanoma: Impact on patient management. Acta Derm Venereol 2013;93:411–6 [DOI] [PubMed] [Google Scholar]
- 2. Aldape K, Simmons ML, Davis RL, et al. Discrepancies in diagnoses of neuroepithelial neoplasms: The San Francisco Bay Area Adult Glioma Study. Cancer 2000;88:2342–9 [PubMed] [Google Scholar]
- 3. Schmitz SK, Hjorth JJJ, Joemai RMS, et al. Automated analysis of neuronal morphology, synapse number and synaptic recruitment. J Neurosci Methods 2011;195:185–93 [DOI] [PubMed] [Google Scholar]
- 4. Baker M, Penny D.. Is there a reproducibility crisis? Nature 2016;533:452–4 [DOI] [PubMed] [Google Scholar]
- 5. Prinz F, Schlange T, Asadullah K.. Believe it or not: How much can we rely on published data on potential drug targets? Nat Rev Drug Discov 2011;10:712. [DOI] [PubMed] [Google Scholar]
- 6. Vasilevsky NA, Brush MH, Paddock H, et al. On the reproducibility of science: Unique identification of research resources in the biomedical literature. PeerJ 2013;1:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ioannidis JPA. Why most published research findings are false. PLoS Med 2005;2:e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ali R, Gooding M, Szilágyi T, et al. Automatic segmentation of adherent biological cell boundaries and nuclei from brightfield microscopy images. Mach Vi Appl 2012;23:607–21 [Google Scholar]
- 9. Yuan Y, Failmezger H, Rueda OM, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med 2012;4:157ra143. [DOI] [PubMed] [Google Scholar]
- 10. Meijering E. Cell segmentation: 50 years down the road [life sciences]. IEEE Signal Process Mag 2012;29:140–5. [Google Scholar]
- 11. Schmidt U, Weigert M, Broaddus C, Myers G.. Cell detection with star-convex polygons. In: Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); vol. 11071; 2018:265–73. [Google Scholar]
- 12. Janiesch C, Zschech P, Heinrich K.. Machine learning and deep learning. Electron Markets 2021;31:685–95 [Google Scholar]
- 13. Li R, Sharma V, Thangamani S, et al. Open-source biomedical image analysis models: A meta-analysis and continuous survey. Front Bioinform 2022;2:912809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gavrielides MA, Gallas BD, Lenz P, et al. Observer variability in the interpretation of HER2/neu immunohistochemical expression with unaided and computer-aided digital microscopy. Arch Pathol Lab Med 2011;135:233–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koga S, Ghayal NB, Dickson DW.. Deep learning-based image classification in differentiating tufted astrocytes, astrocytic plaques, and neuritic plaques. J Neuropathol Exp Neurol 2021;80:306–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cao C, Liu F, Tan H, et al. Deep learning and its applications in biomedicine. Genomics Proteomics Bioinformatics 2018;16:17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ravi D, Wong C, Deligianni F, et al. Deep learning for health informatics. IEEE J Biomed Health Inform 2017;21:4–21 [DOI] [PubMed] [Google Scholar]
- 18. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88 [DOI] [PubMed] [Google Scholar]
- 19. Glaser JI, Benjamin AS, Farhoodi R, et al. The roles of supervised machine learning in systems neuroscience. Prog Neurobiol 2019;175:126–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peng H, Roysam B, Ascoli GA.. Automated image computing reshapes computational neuroscience. BMC Bioinformatics 2013;14:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J, Ranka S, Sahni S.. Automated identification of cell-type-specific genes in the mouse brain by image computing of expression patterns. BMC Bioinformatics 2014;15:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goodfellow I, Bengio Y, Courville A.. Deep learning. Genet Program Evolvable Mach 2017;19:305–7 [Google Scholar]
- 23. Shapiro LG, Stockman GC.. Computer Vision. 1st ed.; 2001, p. 580. [Google Scholar]
- 24. Rumelhart DE, Hinton GE, Williams RJ.. Learning representations by back-propagating errors. Nature 1986;323:533–6 [Google Scholar]
- 25. Ruder S. An overview of gradient descent optimization algorithms, September 15, 2016. Available at: https://arxiv.org/abs/1609.04747.
- 26. Jouppi NP, Young C, Patil N, et al. In-datacenter performance analysis of a tensor processing unit. SIGARCH Comput Archit News 2017;45:1–12 [Google Scholar]
- 27. Jungo P, Hewer E.. Code-free machine learning for classification of central nervous system histopathology images. J Neuropathol Exp Neurol 2023;82:221–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lecun Y, Bengio Y, Hinton G.. Deep learning. Nature 2015;521:436–44 [DOI] [PubMed] [Google Scholar]
- 29. Zeiler MD, Fergus R.. Visualizing and understanding convolutional networks. In: Computer Vision—ECCV 2014; vol. 8689; 2014:818–33. arXiv:13112901v3 [csCV]. November 28, 2013. Available at: https://link.springer.com/chapter/10.1007/978-3-319-10590-1_53 [Google Scholar]
- 30. Schmidt RM, Recurrent neural networks (RNNs): A gentle introduction and overview, November 23, 2019. Available at: https://arxiv.org/abs/1912.05911
- 31. Wang T, Qiu RG, Yu M.. Predictive modeling of the progression of Alzheimer’s disease with recurrent neural networks. Sci Rep 2018;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen M, He T, An L, et al. ; Alzheimer's Disease Neuroimaging Initiative. Predicting Alzheimer’s disease progression using deep recurrent neural networks. Neuroimage 2020;222:117203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krizhevsky A, Sutskever I, Hinton GE.. ImageNet classification with deep convolutional neural networks. Commun ACM 2017;60:84–90 [Google Scholar]
- 34. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, December 2016; 2015:770–8. Available at: https://arxiv.org/abs/1512.03385
- 35. Yang L, Zhang Y, Guldner IH, et al. 3D segmentation of glial cells using fully convolutional networks and k-terminal cut. In: Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); vol. 9901; 2016:658–66. Available at: https://dl.acm.org/doi/abs/10.1007/978-3-319-46723-8_76 [Google Scholar]
- 36. Ciresan D, Giusti A, Gambardella LM, et al. Deep neural networks segment neuronal membranes in electron microscopy images. In: Pereira F, Burges CJ, Bottou L, eds. Advances in Neural Information Processing Systems 25 (NIPS 2012). Available at: http://www.idsia.ch/. Accessed October 14, 2022. [Google Scholar]
- 37. Shaken M, Tsogkas S, Ferrante E, et al. Sub-cortical brain structure segmentation using F-CNN’S. In: Proceedings—International Symposium on Biomedical Imaging, June 2016; 2016:269–72. Available at: https://ieeexplore.ieee.org/document/7493261
- 38. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, December 2016; 2016:770–8. Available at: https://arxiv.org/abs/1512.03385
- 39. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, June 7–12, 2015; 2015:1–9. Available at: https://ieeexplore.ieee.org/document/7298594
- 40. Ronneberger O, Fischer P, Brox T.. U-net: Convolutional networks for biomedical image segmentation. In: Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); vol. 9351; 2015:234–41. [Google Scholar]
- 41. Isensee F, Jaeger PF, Kohl SAA, et al. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203–11 [DOI] [PubMed] [Google Scholar]
- 42. Antonelli M, Reinke A, Bakas S, et al. The medical segmentation decathlon. Nat Commun 2022;13:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bakas S, Reyes M, Jakab A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. 2018;124. Available at: https://arxiv.org/abs/1811.02629 [Google Scholar]
- 44. Karimi D, Rollins CK, Velasco-Annis C, et al. Learning to segment fetal brain tissue from noisy annotations, March 25, 2022. Available at: https://arxiv.org/abs/2203.14962 [DOI] [PMC free article] [PubMed]
- 45. Litjens G, Sánchez CI, Timofeeva N, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 2016;6:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cruz-Roa A, Basavanhally A, González F, et al. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. Available at: https://engineering.case.edu/centers/ccipd/sites/ccipd.case.edu/files/Automatic_detection_of_invasive_ductal_carcinoma_in_whole.pdf
- 47. Bindhu V. Biomedical image analysis using semantic segmentation. J Real Time Image Process 2019;2:91–101 [Google Scholar]
- 48. Rueckert D, Schnabel JA.. Model-based and data-driven strategies in medical image computing. Proc IEEE 2020;108:110–24. Available at: https://ieeexplore.ieee.org/document/8867900 [Google Scholar]
- 49. McDermott MBA, Wang S, Marinsek N, et al. Reproducibility in machine learning for health. In: RML@ICLR 2019 Workshop—Reproducibility in Machine Learning, July 2, 2019. Available at: https://arxiv.org/abs/1907.01463
- 50. Gundersen OE, Kjensmo S. State of the art: Reproducibility in artificial intelligence. Available at: https://ojs.aaai.org/index.php/AAAI/article/view/11503. Accessed August 7, 2022.
- 51. Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J Digit Imaging 2013;26:1045–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Althnian A, AlSaeed D, Al-Baity H, et al. Impact of dataset size on classification performance: An empirical evaluation in the medical domain. Appl Sci 2021;11:796 [Google Scholar]
- 53. Olson RS, La Cava W, Mustahsan Z, et al. Data-driven advice for applying machine learning to bioinformatics problems. In: Pacific Symposium on Biocomputing; vol. 212669; 2018:192–203. Available at: https://arxiv.org/abs/1708.05070 [PMC free article] [PubMed] [Google Scholar]
- 54. Adler M, Boutell T, Bowler J, et al. Portable Network Graphics (PNG) Specification. 2nd ed. Available at: https://www.w3.org/TR/2003/REC-PNG-20031110/. Accessed April 15, 2023.
- 55.PS3.1. Available at: https://dicom.nema.org/medical/dicom/current/output/html/part01.html. Accessed April 15, 2023.
- 56.CZI Image File Format. Available at: https://www.zeiss.com/microscopy/en/products/software/zeiss-zen/czi-image-file-format.html. Accessed April 15, 2023.
- 57. Kabachinski J. TIFF, GIF, and PNG: Get the picture? Biomed Instrum Technol 2007;41:297–300. [DOI] [PubMed] [Google Scholar]
- 58. Guarneri F, Vaccaro M, Guarneri C.. Digital image compression in dermatology: Format comparison. Telemed J E Health 2008;14:666–70 [DOI] [PubMed] [Google Scholar]
- 59. Allan C, Burel JM, Moore J, et al. OMERO: Flexible, model-driven data management for experimental biology. Nat Methods 2012;9:245–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. von Chamier L, Laine RF, Jukkala J, et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat Commun 2021;12:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bisong E, Google colaboratory. In: Building Machine Learning and Deep Learning Models on Google Cloud Platform; 2019:59–64. Available at: https://scholar.google.com/scholar?q=doi:10.1007/978-1-4842-4470-8_7&hl=en&as_sdt=0&as_vis=1&oi=scholart
- 62. Isola P, Zhu JY, Zhou T, et al. Image-to-image translation with conditional adversarial networks. In: Proceedings—30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, January 2017; 2016:5967–76. Available at: https://ieeexplore.ieee.org/document/8100115
- 63. Xing F, Xie Y, Su H, et al. Deep learning in microscopy image analysis: A survey. IEEE Trans Neural Netw Learning Syst 2018;29:4550–68 [DOI] [PubMed] [Google Scholar]
- 64. Stringer C, Wang T, Michaelos M, et al. Cellpose: A generalist algorithm for cellular segmentation. Nat Methods 2021;18:100–6 [DOI] [PubMed] [Google Scholar]
- 65. Cardona A, Tomancak P.. Current challenges in open-source bioimage informatics. Nat Methods 2012;9:661–5 [DOI] [PubMed] [Google Scholar]
- 66. Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. The OpenCV Library. Available at: https://www.drdobbs.com/open-source/the-opencv-library/184404319. Accessed November 9, 2022.
- 68. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods 2012;9:676–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stritt M, Stalder AK, Vezzali E.. Orbit Image Analysis: An open-source whole slide image analysis tool. PLoS Comput Biol 2020;16:e1007313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sofroniew N, Lambert T, Evans K, et al. napari: A multi-dimensional image viewer for Python, May 31, 2022. Available at: https://zenodo.org/record/6598542#.ZFqXvi_MLLs
- 71. Kluyver T, Ragan-Kelley B, Pérez F, et al. Jupyter Notebooks—A publishing format for reproducible computational workflows. In: Positioning and Power in Academic Publishing: Players, Agents and Agendas—Proceedings of the 20th International Conference on Electronic Publishing, ELPUB 2016, 2016:87–90. Available at: https://eprints.soton.ac.uk/403913/
- 72. Kankaanpää P, Paavolainen L, Tiitta S, et al. BioImageXD: An open, general-purpose and high-throughput image-processing platform. Nat Methods 2012;9:683–9 [DOI] [PubMed] [Google Scholar]
- 73. De Chaumont F, Dallongeville S, Chenouard N, et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat Methods 2012;9:690–6 [DOI] [PubMed] [Google Scholar]
- 74. Stirling DR, Swain-Bowden MJ, Lucas AM, et al. CellProfiler 4: Improvements in speed, utility and usability. BMC Bioinformatics 2021;22:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Abadi M, Agarwal A, Barham P, et al. TensorFlow: Large-scale machine learning on heterogeneous distributed systems, March 14, 2016. Available at: https://arxiv.org/abs/1603.04467.
- 76. Paszke A, Gross S, Massa F, et al. PyTorch: An imperative style, high-performance deep learning library. Adv Neural Inf Process Syst 2019;32. Available at: https://arxiv.org/abs/1912.01703. [Google Scholar]
- 77. Van Der Walt S, Schönberger JL, Nunez-Iglesias J, et al. ; scikit-image contributors. Scikit-image: Image processing in python. PeerJ 2014;2:e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cardoso MJ, Li W, Brown R, et al. MONAI: An open-source framework for deep learning in healthcare, November 4, 2022. Available at: https://arxiv.org/abs/2211.02701.
- 79. Lee MY, , BediaJS, , Bhate SS,. et al. CellSeg: a robust, pre-trained nucleus segmentation and pixel quantification software for highly multiplexed fluorescence images. BMC Bioinformatics 2022;23:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jungo P, , Hewer E.. Code-free machine learning for classification of central nervous system histopathology images. J Neuropathol Exp Neurol 2023;82:221–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Signaevsky M, , PrastawaM, , Farrell K,. et al. Artificial intelligence in neuropathology: deep learning-based assessment of tauopathy. Lab Invest 2019;99:1019–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ker J, , Ba Y, , Lee HY,. et al. Automated brain histology classification using machine learning. J Clin Neurosci 2019;66:239–45 [DOI] [PubMed] [Google Scholar]
- 83. Hollon TC, , PandianB, , Adapa AR,. et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med 2020;26:52–8 [DOI] [PMC free article] [PubMed] [Google Scholar]