Abstract
Genetic testing for susceptibility genes through next-generation sequencing (NGS) has become a widely used technique. Using this, a number of genetic variants have been identified, several of which are variants of unknown significance (VUS). These VUS can either be pathogenic or benign. However, since their biological effect remains unclear, functional assays are required to classify their functional nature. As the use of NGS becomes more mainstream as a diagnostic tool in clinical practice, the number of VUS is expected to increase. This necessitates their biological and functional classification. In the present study, a VUS was identified in the BRCA1 gene (NM_007294.3:c.1067A>G) in two women at risk for breast cancer, for which no functional data has been reported. Therefore, peripheral lymphocytes were isolated from the two women and also from two women without the VUS. DNA from all samples were sequenced by NGS of a breast cancer clinical panel. Since the BRCA1 gene is involved in DNA repair and apoptosis, the functional assays chromosomal aberrations, cytokinesis-blocked micronucleus, comet, γH2AX, caspase and TUNEL assays were then conducted on these lymphocytes after a genotoxic challenge by ionizing radiation or doxorubicin to assess the functional role of this VUS. The micronucleus and TUNEL assays revealed a lower degree of DNA induced-damage in the VUS group compared with those without the VUS. The other assays showed no significant differences between the groups. These results suggested that this BRCA1 VUS is likely benign, since the VUS carriers were apparently protected from deleterious chromosomal rearrangements, subsequent genomic instability and activation of apoptosis.
Keywords: next-generation sequencing, breast cancer, variants of unknown significance (VUS), BRCA1, genotoxic challenge, functional analysis of VUS
Introduction
The development of targeted diagnostic clinical panels based on next-generation sequencing (NGS) has allowed the identification of various pathogenic variants in high penetrant genes associated with different types of cancer (1–3). This is especially the case in those involving hereditary syndromes. For example, pathogenic variants in the BRCA genes (BRCA1 and BRCA2) have been reported to be associated with a higher lifetime risk of developing breast cancer in women. They account for ~20% all familial breast cancers and >10% patients with early-onset triple-negative breast cancer.
The Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) consortium has received to date >6,000 submissions of unique variants of unknown significance (VUS) identified in >13,000 families from >17 countries (http://www.enigmaconsortium.org/). These figures are expected to increase with the increased use of gene sequencing techniques, especially NGS. NGS is capable of also covering untranslated and deeper intronic regions (4). For certain types of BRCA gene variants, generation of functional evidence is essential before a variant can be clearly classified to be pathogenic, VUS or benign. How the multiple functions of the BRCA1 and BRCA2 proteins are associated with cancer predisposition remains poorly understood (5).
Some of the high penetrant genes involved in hereditary cancers include genes of the DNA damage response (DDR) pathway. DNA repair serves a critical role in preventing the development of cancer. A number of genes in the DDR pathway have been documented to be mutated in hereditary cancers, such as BRCA1, BRCA2 and ATM. In response to DNA damage, cells activate the DDR pathway and arrest cell-cycle progression, allowing time for DNA repair or, depending on the extent of damage, activation of apoptosis (6–9). The complex and multi-layered process of DNA repair is critical in response to DNA damage and subsequent cancer cell survival (10–13). Double-stranded DNA breaks (DSBs) are amongst the major threats to genomic integrity. DSBs are repaired by either one of the following two mechanistically distinct pathways: Homologous recombination (HR), which is a conservative form; and non-homologous end-joining (NHEJ), which is a non-conservative form (13,14). BRCA1/2 proteins are crucial for HR repair (15,16). BRCA1 interacts with tumor suppressors, DNA repair proteins and cell cycle regulators through its numerous functional domains. Therefore, it can serve a role in a multitude of DNA repair pathways and checkpoint regulation during DDR (17). Accordingly, cells carrying BRCA1 mutations are particularly sensitive to DNA-damaging agents (18). This enhanced sensitivity to DNA-damaging agents provides an opportunity to assess the response of BRCA1 variants to genotoxic agents in vitro. This can then be compared with that of non-pathogenic BRCA1 variants (18).
Sequence variants that disrupt the interaction of BRCA1/2 with its binding partners are associated with increased risks of developing breast and ovarian cancer (17,19). Although some sequence variants can be pathogenic, such as non-synonymous variants, other variants can result in amino acid changes that do not alter the network of BRCA1/2 interactions, even if they are non-synonymous. However, BRCA1/2 gene products are involved in multiple processes during various stages of the cell cycle, each of which serves a specific function (20). Therefore, the simple diagnosis of a sequence variant by NGS is typically insufficient to directly predict its putative role in breast cancer. To overcome this hurdle, functional analysis of VUS could be highly beneficial, even if it is time-consuming, labor intensive and not necessarily conclusive (21–23).
During the course of routine NGS assessment of families at risk for breast cancer, a VUS was identified in the BRCA1 gene of two women, without information on its pathogenicity. Therefore, the aim of the present study was to perform a functional in vitro analysis of the identified BRCA1 VUS using peripheral blood lymphocytes from these women. This was performed through the assessment of cellular responses to genotoxic challenge induced by γ-radiation and the chemotherapeutic agent doxorubicin.
Material and methods
Patient target population
Women at high risk of familial cancer were genotyped through NGS with a costum-made panel of high-risk genes, namely BRCA1, BRCA2, PTEN, TP53, BRCA1-interacting helicase 1, RAD51C, RAD51D, ATM, partner and localizer of BRCA2, checkpoint kinase 2 and cadherin-1, which are described to be essential in the clinical guidelines for breast cancer studies (21,23,24). All exons and exon-intron boundaries had 100% coverage. In addition, two healthy, non-carrier controls (NC) with no VUS identified after NGS for the same clinical panel were also included in this study.
The participants enrolled were selected from a collaboration with the company Ophiomics Precision Medicine. All participants were informed about the present study and the collection of blood samples by venous puncture were preceded by the signing of an informed consent form agreeing to the use of their blood samples for research. Detailed family history of oncological diseases for each patient/participant was also collected. All personal data was anonymized, and the samples were coded. The present study was approved by the National Commission of Data Protection (approval no. 10637/2016) for the use of samples for research and also by the Ethical Commission of NOVA Medical School/Faculdade Ciências Médicas (NMS/FCM; approval no. 54/2018/CEFCM).
DNA extraction
Genomic DNA extraction from total peripheral blood cells was performed with the GeneJET Whole Blood Genomic DNA Purification Mini kit (Thermo Fisher Scientific, Inc.) according to manufacturer's instructions, with a minor change: the final elution volume was 75 µl. The quantification and the quality of the extracted DNA was evaluated with the electrophoresis system Agilent 2200 TapeStation System (Agilent Technologies, Inc.) using the Agilent Genomic DNA ScreenTape and Reagents kit (Agilent Technologies, Inc.) according to manufacturer's instructions.
Next-generation sequencing (NGS) for variant detection
Genomic DNA extracted from peripheral blood (200 µl) of patients was evaluated for DNA concentration and integrity; DNA isolated from each sample was quantified in 2200 TapeStation using the Genomic DNA ScreenTape (Agilent Technologies, Inc.). Genomic DNA libraries were prepared using the Ion Ampliseq Library kit (2.0) using the custom Ion Ampliseq panel described above and quantified by quantitative PCR with the Ion Library Quantification kit (Thermo Fisher Scientific, Inc.). The emulsion PCR of amplified libraries was performed using Ion Chef (Thermo Fisher Scientific, Inc.). Sequencing runs were performed with Ion personal machine using 316 Chips (Thermo Fisher Scientific, Inc.) aiming for a mean sequencing depth coverage of 100×. Variant annotation was performed in reference to Human Genome version GRCh38 and based on information contained in the databases ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), DGVa (https://www.ebi.ac.uk/dgva/), dbSNP (https://www.ncbi.nlm.nih.gov/snp/), HGMD-PUBLIC (https://www.hgmd.cf.ac.uk/ac/index.php), EBI Variation HomoSapiens (https://www.ebi.ac.uk/eva/). The bioinformatics algorithms used to predict the functional impact of variants were: PolyPhen (http://genetics.bwh.harvard.edu/pph2/), SIFT (https://sift.bii.a-star.edu.sg), LoF (http://aloft.gersteinlab.org), Condel (https://bbglab.irbbarcelona.org/fannsdb/help/condel.html), BLOSUM62 scoring matrix used in BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and CAROL (https://www.sanger.ac.uk/tool/carol/).
Detection of copy number variants for BRCA1 and BRCA2 genes was performed by MLPA with the panels MLPA® Salsa® P002-BRCA1 and MLPA® Salsa® P090-BRCA2 (MRC-Holland BV), respectively. The Portuguese founder mutation (c_156_157 inserção Alu) BRCA2 (OMIM:600185) was also screened by PCR. All reported variants classified as pathogenic or unknown significance, occurring in coding regions and at frequencies >10% were validated by Sanger sequencing (ABI3100 Avant; Thermo Fisher Scientific, Inc.).
Sanger sequencing
PCR reactions to prepare samples for Sanger sequencing were performed according to Platinum® PCR SuperMix High Fidelity (Invitrogen; Thermo Fisher Scientific, Inc.) manufacturer's instructions. Briefly, a total volume of 20 µl was used, containing 18 µl of Platinum® PCR SuperMix High Fidelity (Invitrogen; Thermo Fisher Scientific, Inc.), 0.2 µM of forward (5′-AATGATAGGCGGACTCCCAG-3′) and reverse (5′-GAGGCTTGCCTTCTTCCGAT-3′) primers. High quality genomic DNA (5–50 ng) extracted from peripheral blood cells was added to the PCR mix and the PCR reactions were performed in a Veriti™ 96-Well Thermal Cycler (Thermo Fisher Scientific, Inc.). The cycling conditions employed were initial denaturation at 94°C, 2 min, 30 cycles of amplification at 94°C, 15 sec, 55°C, 15 sec, 68°C, 1 min and held at 10°C. The size and quantity of each fragment analyzed was evaluated with the electrophoresis system Agilent 2200 TapeStation System (Agilent Technologies, Inc.) using the kit DNA ScreenTape and Reagents (Agilent Technologies, Inc.) according to manufacturer's instructions. Each sample was sequenced with both forward and reverse primers in an Applied Biosystems 3130 Genetic Analyzer (Thermo Fisher Scientific, Inc.) according to manufacturer's instructions.
In vitro γ-irradiation
Blood samples were irradiated in vitro using a 60Co radiation source in a Precisa 22 irradiator at the Ionizing Radiation Installations at Center for Nuclear Sciences and Technologies-Instituto Superior Técnico (C2TN-IST) in Lisbon. Each donor sample was irradiated with a dose of 2 Gy and a non-irradiated control (0 Gy) was included. A total of ~4 ml whole blood was isolated from each donor and subject to irradiation after which each assay was performed as described below. To perform the comet assay, lymphocytes were isolated and then distributed into 4-ml glass tubes for irradiation.
CA assay
After irradiation, blood samples were cultured in triplicates or quadruplicates for each donor. The experiments were performed as previously described (25–27) with minor modifications. Briefly, 500 µl irradiated and non-irradiated whole blood was added to 4.5 ml RPMI-1640 medium with L-Glutamine (MilliporeSigma), supplemented with 25% FBS (MilliporeSigma), 1.5% penicillin-streptomycin (Pen-Strep), 0.5% sodic heparin (5,000 UI/ml; B. Braun Medical Inc.) and 2.5% phytohemagglutinin (Gibco; Thermo Fisher Scientific, Inc.). Cultures were maintained in an incubator at 37°C and 5% CO2 at an 40° angle for 48 h. After 24 h, colcemid (0.08 µg/ml; Gibco; Thermo Fisher Scientific, Inc.) was added to the culture. At the end of the 48 h, cultures were centrifuged at 400 × g for 5 min at RT (RT). The pellet was then resuspended with mild stirring before 10 ml KCl solution [0.56% (p/v)] previously warmed to 37°C was added and homogenization by inversion. The tubes were incubated at 37°C for 20 min to promote hypotonic shock and then centrifuged at 400 × g for 5 min at RT. The cells were fixed under stirring with 5 ml fixative mixture of methanol:acetic acid [3:1 (v/v)] previously cooled at −20°C, before being centrifuged at 400 × g for 5 min at RT. These two steps of fixation and subsequent centrifugation were repeated two or three times, until the supernatant became clear. Finally, 10 ml fixative mixture was added to each tube and the samples were stored at −20°C.
Samples held at −20°C were centrifuged at 400 × g for 5 min at RT following which the supernatant was removed and the suspension homogenized by gentle tapping. Glass slides were washed and immersed in distilled water at 4°C and a few drops of the cell suspension were spread onto each slide. Once well dried for 24 h at RT, the slides were stained with Giemsa's solution 4% (v/v) in 0.01 M phosphate buffer (pH 6.8) for 10 min. Excess dye was then washed off under running water. Once well dried again, permanent slides were prepared using a mounting medium [Entellan® (MilliporeSigma)]. Slides were then scored using an optical microscope at ×1,000 magnification. Scoring was performed in 200 complete metaphases (46 chromosomes) by two independent evaluators (100 each), according to the criteria described by Rueff et al (28) and following the recommendations of the International Atomic Energy Agency (IAEA) (29). Each metaphase was analyzed according to the following criteria: The presence of chromosomal aberrations, namely chromatid with gaps or breaks; chromosomes with gaps or breaks; excess of acentric fragments; dicentric chromosomes DIC; and rings. The metaphases containing ≥1 chromosome aberration except gaps were accounted for the frequency (%) of aberrant cells excluding gaps [chromosomal aberration excluding gaps (CAEG)].
Cytokinesis-blocked micronucleus assay (CBMN)
For the CBMN assay, blood samples were cultured after irradiation in triplicate or quadruplicate for each donor (26,30,31). Briefly, 0.5 ml irradiated whole blood was added into each tube containing 4.5 ml RPMI-1640 medium with L-Glutamine, supplemented with 25% FBS, 1.5% of Pen-Strep, 0.5% of sodic heparin and 2.5% of phytohemagglutinin. Cultures were maintained at 37°C, 5% CO2 and at an angle of 40° for ~72 h. After 44 h, cytochalasin-B (6 µg/ml; MilliporeSigma) was added. At the end of the 72 h, cultures were centrifuged at 110 × g for 10 min at RT. After discarding the supernatant, cells were washed twice with 5 ml washing solution [RPMI-1640 medium with L-Glutamine and NaHCO3 (0.1 g/l), supplemented with 2% FBS] and centrifuged at 110 × g for 7 min at RT. Mild hypotonic treatment was then performed by adding 5 ml 4:1 distilled water: RPMI-1640 medium with L-Glutamine (pH 7.2) and NaHCO3 (0.1 g/l), supplemented with 2% FBS, followed by centrifugation at 110 × g for 5 min at RT. After concentrating the pellet by discarding most of the supernatant, a drop of cell suspension was placed onto each glass slide and a smear was performed.
Once the glass slides were completely dry, they were fixed with pre-cooled 5 ml methanol:acetic acid solution [3:1 (v/v)] for 20 min at −20°C. The slides were then dried and stained with Giemsa's solution in 0.01 M phosphate buffer (pH 6.8) for 8 min at RT. The permanent slides were prepared as aforementioned with Entellan® mounting medium. Slides were imaged and scored using an optical microscope at ×400 magnification. For each donor and dose, 2,000 binucleated cells were scored by two independent scorers (1,000 each) according to the IAEA criteria (29). The number of micronucleated binucleated cells were recorded.
Single-cell gel electrophoresis (comet assay)
Peripheral blood mononuclear cells (PBMCs) were prepared from fresh blood samples and isolated through density gradient centrifugation using Histopaque-1077 (MilliporeSigma) according to the manufacturer's protocols. Briefly, blood was diluted with an equal volume of PBS before 5 ml of this diluted blood was carefully added to a canonical centrifuge tube, which contains 3.5 ml Histopaque-1077, before being centrifuged at 700 × g for 30 min at RT. PBMCs were then harvested from the interface and washed with PBS and centrifuged again at 200 × g for 10 min at RT. The pellet was suspended in RPMI-1640 medium supplemented with 25% FBS and 1.5% Pen-Strep. For irradiation, part of the cell suspension (~4 ml) was used, whereas the rest was used as control. Cell suspensions were held on ice until the single-cell gel electrophoresis assay was performed. For chemical exposure, 1×106 PBMCs were cultured in 12-well plates and exposed for 2 h with doxorubicin (BioAustralis) at 37°C at 5% CO2. The samples were then centrifuged at 200 × g for 5 min at RT, washed in 1 ml PBS and centrifuged again. The pellet was then resuspended in 100 µl 0.5% low-melting point agarose.
The comet assay (SCGE) was performed as previously described (32) with slight modifications. Briefly, the cell suspensions were spread on glass microscope slides previously coated with 1% normal-melting point agarose and kept at 4°C for 20 min. The slides were then left overnight in a cold lysis buffer (2.5 M NaCl, 10 mM Tris, 100 mM EDTA and 1% Triton, pH 10). After the overnight lysis, slides were washed with previously cooled double-distilled water and remained immersed for 10 min at 4°C. They were then immersed in cold electrophoresis buffer (10 M NaOH and 200 mM EDTA, pH >13) for 20 min at 4°C. Electrophoresis was conducted for 20 min at 25 V (400 mA) before the slides were neutralized three times with neutralization buffer (0.4 M Tris, pH 7.5) at 5 min each, dried with ethanol (50, 75 and 100%; 5 min each) and stained with 3X GelRed (Biotium, Inc.). Slides were scored using a fluorescent microscope (Zeiss Z2; Carl Zeiss AG) at ×200 magnification, before ~200 cells were selected and images captured. The cell images captured were then examined using the CometScore V1.5 Software, which calculated the % DNA in the tail.
Blood cell culture and chemical treatment
The functional assays through chemical exposure were performed for all samples carrying the VUS and for NC controls. In total. three different concentrations of doxorubicin were chosen (0.1, 1.0 and 5.0 µM). The duration of chemical exposure varied according to the assay performed. Treated samples were incubated at 37°C for specific periods of time.
Functional assays
γH2A histone family member X (γH2AX) assay
Following doxorubicin treatment for 2 h at 37°C with 5% CO2, the samples were centrifuged at 200 × g for 5 min at RT. RPMI-1640 medium (1 ml) supplemented with 25% of FBS and 1.5% of Pen-Strep was then added to each sample and incubated for 30 min at 37°C with 5% CO2. Samples were centrifuged at 200 × g at RT and 1 ml PBS and 1 µl violet fluorescent reactive dye (LIVE/DEAD™ Fixable Violet Dead Cell Stain kit; Thermo Fisher Scientific, Inc.) was added to the respective sample and incubated for 30 min at RT protected from light. The samples were then centrifuged again at 200 × g for 5 min at RT and the pellet was washed with 1 ml PBS, followed by centrifugation at 200 × g for 5 at RT min again. This was followed by fixation in 500 µl 2% formaldehyde for 15 min on ice. The samples were centrifuged again at the same speed and time at 4°C, and each pellet was resuspended in 500 µl 70% cold ethanol in PBS before being kept overnight at 4°C. The next day, the samples were centrifuged at 200 × g for 5 min at 4°C and each pellet was resuspended in 1 ml blocking buffer (containing 4% BSA in PBS, 4% goat serum and 0.25% Triton X-100) and were centrifuged further in the same conditions as before. In total, 1:500 antibody [Phospho-Histone H2A.X (Ser139) Monoclonal Antibody (CR55T33), PE, eBioscience; Thermo Fisher Scientific, Inc.] was added to the respective pellet, followed by 2 h incubation at RT protected from light. Cells were washed with 1.5 ml 1% BSA and centrifuged at 200 × g for 5 min at RT. Each pellet was resuspended in 200 µl 0.1% BSA. The samples were analyzed by flow cytometry using a BD FACSCanto II Cytometer (BD Biosciences), where 20,000 events were counted. Image analysis were performed using the FlowJo v10 software (FlowJo LLC).
Caspase activity assay
Caspase assays were performed using commercially available kits, specifically by CaspaTag™ Caspase-3/7 In Situ Assay kit and CaspaTag™ Caspase-9 In Situ Assay kit (Thermo Fisher Scientific, Inc.). The methodology was performed according to the manufacturer's protocols, with minor alterations. The methodology used for both assays was the same, with the main difference being the FLICA concentration specific for each one, according to the manufacturer's instructions. Briefly, after treatment for 2 h (or overnight for Caspase-9) at 37°C with 5% CO2, samples were centrifuged at 200 × g for 5 min at RT before the pellet was resuspended in 200 µl PBS. In total, 10 µl 6X FLICA for Caspases 3/7 and 15X FLICA for Caspase 9 were added to the respective tubes. They were then incubated for 1 h at 37°C with 5% CO2 protected from light. Tubes were gently swirled three times, before 1 ml 1X wash buffer (10X wash buffer provided in the kit) was added and centrifuged at 400 × g for 5 min at RT. The pellet was resuspended in 1 ml 1X wash buffer and centrifuged again at 400 × g for 5 min at RT. The pellet was resuspended in 400 µl 1X wash buffer and 2 µl propidium iodide (provided in kit) was added to the respective tubes. Samples were analyzed by flow cytometry using a BD FACSCanto II Cytometer (BD Biosciences) and 20,000 events were counted. Image analysis were performed using the FlowJo v10 software (FlowJo LLC).
TUNEL assay
TUNEL assay was performed using the APO-BrdU™ TUNEL Assay kit (Thermo Fisher Scientific, Inc.). The methodology was performed according to the manufacturer's protocols with minor alterations. Briefly, after treatment for 4 h at 37°C with 5% CO2, samples were centrifuged at 200 × g for 5 min at RT. In total, 500 µl PBS was added to the samples before PBMCs were pelleted (300 × g for 5 min at RT) followed by fixation in 500 µl 2% formaldehyde for 15 min on ice. Samples were then centrifuged at 300 × g for 5 min at 4°C and each pellet was resuspended in 1 ml PBS and centrifuged again at 300 × g for 5 min at 4°C. The pellet was resuspended in 500 µl PBS and 1 ml 70% cold ethanol in PBS before being incubated for 30 min on ice. Samples were centrifuged (300 × g for 5 min at 4°C) and the pellet was resuspended and centrifuged twice with 1 ml wash buffer (provided in kit). Each pellet was then resuspended in 50 µl DNA-labeling solution, which contains reaction buffer, TdT enzyme, BrdUTP and ddH2O, before being kept overnight at 22–24°C. The next day, samples were resuspended and centrifuged twice in same conditions as before (300 × g for 5 min at RT) with 1 ml rinse buffer (provided in kit), 106 cells were added to 100 µl diluted solution, which contains the Alexa Fluor™ 488-conjugated anti-BrdU mouse monoclonal antibody PRB-1 (provided in kit) and rinse buffer. They were then incubated for 30 min at RT protected from light. Samples were analyzed by flow cytometry using a BD FACSCanto II Cytometer (BD Biosciences), where 20,000 events were counted. Image analysis were performed using FlowJo v10 software (FlowJo LLC).
Statistical analysis
All graphs were plotted using the GraphPad Prism 9 software (Dotmatics). Data were presented as the means ± standard deviation. All graphs were obtained for the grouped samples according to their genetic status: NC carriers or VUS carriers. Statistical analysis was performed using GraphPad Prism 9 taking into account the pooled samples. For the CA and MN assays, χ2 or Fisher's exact tests was applied, where P<0.05 was considered to indicate a statistically significant association. For the SCGE or comet assays, Kolmogorov-Smirnov normality test was performed to examine if samples followed a Gaussian distribution. If this was not observed, then non-parametric tests were used to analyze the data. To compare controls (0 Gy) and irradiated samples (2 Gy), Wilcoxon signed rank test was applied. The non-parametric Mann-Whitney U test was performed to compare the different groups of samples. P<0.05 was considered to indicate a statistically significant difference.
Results
Study summary
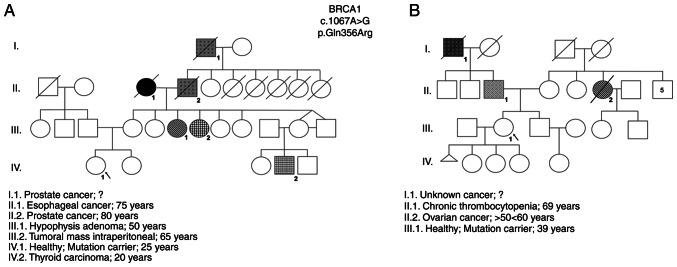
The results obtained by NGS revealed two women with a VUS in the BRCA1 gene (NM_007294.3:c.1067A>G) ambiguously defined as probably pathogenic according to the PolyPhen2 database and as benign in the ClinVar database were identified (Table I). These VUS-carriers were female individuals with no identified tumors, belonging to two distinct families with high incidence of oncologic diseases and the same VUS (rs1799950). The familial history of each woman and their pedigrees were constructed and are shown in Fig. 1.
Table I.
Characterization of participants carrying the VUS in the BRCA1 gene.
| Characterization | ||||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Sample ID | Age (years) | Cancer | Gene | Variant ID | EBI amino/genomic | rs ID | PolyPhen | ClinVar |
| VUS_BRCA1_ 1 | 25 | Healthy | BRCA1 | NM_007294.3 | ENSP00000418960.2: | rs1799950 | Probably | Benign |
| c.1067A>G | p.Gln356Arg 17:g | damaging | ||||||
| 43094464T>C | ||||||||
| VUS_BRCA1_2 | 39 | |||||||
VUS, variants of unknown significance.
Figure 1.
Pedigree of women enrolled in the study that carry the rs1799950 genetic variant. (A) VUS_BRCA1_1 genealogy (IV.1). (B) VUS_BRCA1_2 genealogy (III.1).
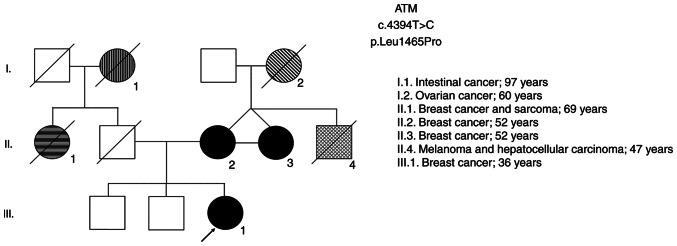
In order to validate the sensitivity of the functional assays to verify the pathogenicity of the gene variant, and since it was not possible to include a pathogenic BRCA1 variant selected by NGS in the present study, two women also with high-risk genealogy harboring variants in the ATM gene that are likely to be pathogenic were included in the study (Table II). These carriers of ATM mutations are first degree-relatives (mother and daughter). The two were diagnosed with breast cancer in a family with a relevant pedigree history of oncological diseases (Fig. 2). In addition, this variant was also identified in a third relative in this family (case II.3), who was also diagnosed with breast cancer along with her twin sister (case II.2; ATM2) and her niece (case III.1; ATM1), emphasizing the high probability of this being a pathogenic variant. Together with the BRCA1 gene, the ATM gene is also involved in the DDR pathway and is important in the cellular response to genotoxic agents.
Table II.
Characterization of participants carrying the probable pathogenic variant in the ATM gene.
| Characterization | ||||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Sample ID | Age | Cancer | Gene | Variant ID | EBI amino/genomic | rs ID | PolyPhen | ClinVar |
| ATM 1 | 36 | Breast | ATM | NM_000051.3: | ENSP00000278616.4: | rs730881391 | Probably | Likely |
| c.4394T>C | p.Leu1465Pro | damaging | Pathogenic | |||||
| NC_000011.10:g | ||||||||
| 108289759T>C | ||||||||
| ATM 2 | 52 | |||||||
Figure 2.
Genealogy of ATM carriers: III.1 corresponds to ATM 1; II.2 corresponds to ATM 2.
DNA damage was induced by γ-radiation and chemical exposure to doxorubicin, a DNA damaging agent that is also used as a first line chemotherapeutic for several cancers. The present study was intended to be exploratory and a proof of concept. The genotoxic and functional studies performed between NC carriers and carriers of VUS and ATM mutation are described below and were chosen with the objective of measuring the extent of DNA damage and several apoptosis end-points.
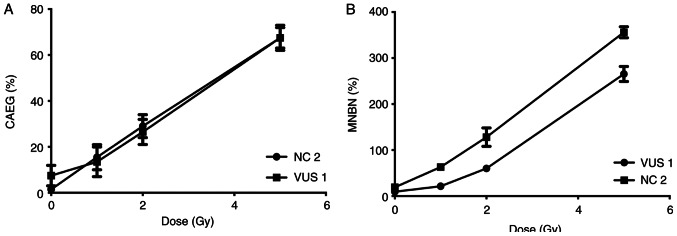
Samples from two participants, VUS_BRCA1_1 and NC 2, were first used to establish a dose response curve for the present study. Dose-response curves at 0, 1, 2 and 5 Gy were performed for the MN and CA assays (Fig. 3). The radiation dose chosen (2 Gy) was previously described (33) and is used in biological dosimetry requirements.
Figure 3.
Dose-response curve. Observed frequencies of (A) CAEG and (B) MNBN are represented as solid marks. Two samples were selected to evaluate the irradiation dose-response (a VUS carrier and a NC control). CAEG, chromosomal aberrations excluding gaps; MNBN, micronucleated binucleated cells; VUS, variants of unknown significance; NC, non-carrier.
CA assay results
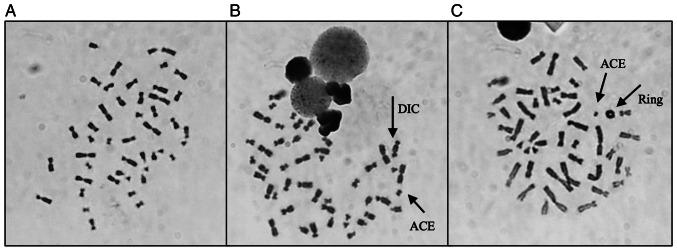
Table III shows the results obtained after analysis, where it is possible to observe a global increase in the frequency of CAEG globally following radiation exposure. The most frequent CA present were acentric fragments and DIC which, apart from rings, are the main CAs identified following γ-radiation exposure. Fig. 4 shows representative images of metaphases showing these structures that were observed during analysis. The results obtained individually for each woman were then grouped to evaluate the effect attributed to the presence of each genetic variant. The frequency of CAEG observed is shown in Fig. 5. No significant differences could be observed among the NC carriers, VUS_BRCA1 carriers and ATM carriers.
Table III.
Chromosomal aberrations distribution in cells, and the frequency of cells with at least one chromosomal aberration excluding gaps (CAEG %).
| DIC Distribution | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample name | Dose (Gy) | Total Cells | CTG | CHG | CTB | CHB | Excess ACE | DIC |
|
CAEG (%) | |||||
| 0 | 1 | 2 | 3 | 4 | Ring | ||||||||||
| NC 1 | 0 | 200 | 0 | 0 | 5 | 0 | 2 | 1 | 199 | 1 | 0 | 0 | 0 | 0 | 3.50 |
| 2 | 200 | 0 | 0 | 2 | 2 | 34 | 21 | 179 | 21 | 0 | 0 | 0 | 2 | 25.50 | |
| NC 2 | 0 | 200 | 0 | 0 | 5 | 1 | 3 | 3 | 197 | 3 | 0 | 0 | 0 | 1 | 6.00 |
| 2 | 200 | 1 | 0 | 3 | 3 | 45 | 28 | 172 | 22 | 3 | 0 | 0 | 1 | 32.50 | |
| Global | 0 | 400 | 0 | 0 | 10 | 1 | 5 | 4 | 396 | 4 | 0 | 0 | 0 | 1 | 4.75 |
| 2 | 400 | 1 | 0 | 5 | 5 | 79 | 49 | 351 | 43 | 3 | 0 | 0 | 3 | 29.00 | |
| VUS_BRCA1_1 | 0 | 200 | 5 | 0 | 6 | 1 | 3 | 0 | 200 | 0 | 0 | 0 | 0 | 0 | 4.50 |
| 2 | 200 | 0 | 1 | 6 | 0 | 26 | 22 | 178 | 22 | 0 | 0 | 0 | 4 | 25.00 | |
| VUS_BRCA1_ 2 | 0 | 200 | 0 | 0 | 4 | 1 | 0 | 1 | 199 | 1 | 0 | 0 | 0 | 0 | 3.00 |
| 2 | 200 | 2 | 0 | 7 | 3 | 30 | 30 | 170 | 22 | 4 | 0 | 0 | 3 | 30.50 | |
| Global | 0 | 400 | 5 | 0 | 10 | 2 | 3 | 1 | 399 | 1 | 0 | 0 | 0 | 0 | 3.75 |
| 2 | 400 | 2 | 1 | 13 | 3 | 56 | 52 | 348 | 44 | 4 | 0 | 0 | 7 | 27.75 | |
| ATM 1 | 0 | 200 | 3 | 0 | 5 | 1 | 0 | 0 | 200 | 0 | 0 | 0 | 0 | 1 | 3.50 |
| 2 | 200 | 2 | 1 | 3 | 1 | 39 | 33 | 167 | 21 | 6 | 0 | 0 | 2 | 29.00 | |
| ATM 2 | 0 | 200 | 0 | 0 | 1 | 1 | 9 | 0 | 200 | 0 | 0 | 0 | 0 | 0 | 5.00 |
| 2 | 200 | 2 | 0 | 2 | 3 | 31 | 27 | 173 | 27 | 0 | 0 | 0 | 2 | 28.00 | |
| Global | 0 | 400 | 3 | 4 | 6 | 2 | 9 | 0 | 400 | 0 | 0 | 0 | 0 | 1 | 5.50 |
| 2 | 400 | 4 | 1 | 5 | 4 | 70 | 60 | 340 | 48 | 6 | 0 | 0 | 4 | 28.50 | |
CTG, chromatid gap; CHG, chromosome gap; CTB, chromatid break; CHB, chromosome break; ACE, acentric fragment; DIC, dicentric chromosome; CAEG, chromosomal aberration excluding gaps; NC, non-carrier; VUS, variants of unknown significance.
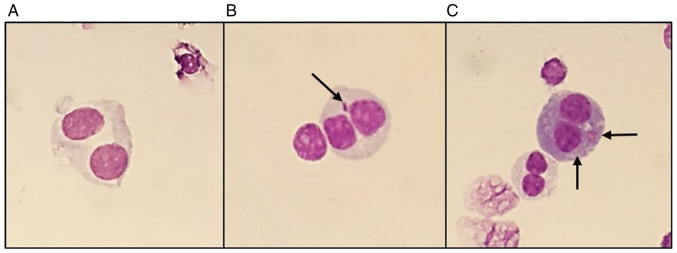
Figure 4.
Representative images of metaphases obtained during analysis, with the characteristic structures associated with γ-radiation exposure. (A) Representation of a normal metaphase with 46 chromosomes (0 Gy); (B) representation of a metaphase with 46 chromosomes, containing one dicentric chromosome and one acentric fragment (2 Gy); (C) representation of a metaphase with 46 chromosomes, containing one ring and one acentric fragment (2 Gy). Magnification, ×1,000. DIC, dicentric chromosomes; ACE, acentric fragment.
Figure 5.
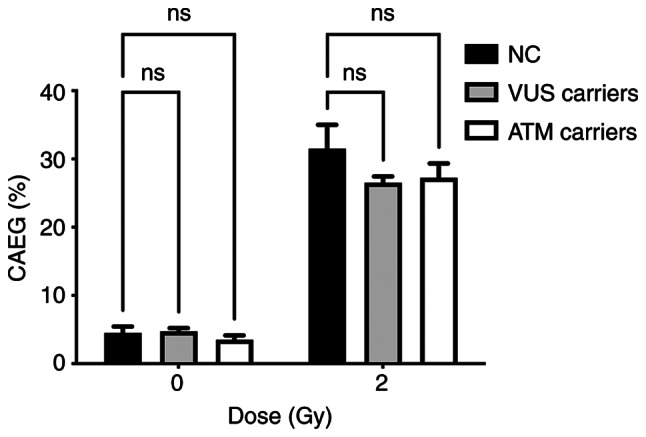
CAEG frequency distribution for each group of genetic variants. CAEG, chromosomal aberrations excluding gaps; NC, non-carrier; VUS, variants of unknown significance; ns, no significance.
CBMN assay
Micronuclei slides were then analyzed. For each participant, 1,000 binucleated cells were counted and independently analyzed by two independent evaluators. The data obtained for the micronuclei distribution in each participant are shown in Table IV. Fig. 6 shows representative images captured during the data analysis, with the results observed. An overall analysis of the results showed an increase in the rates of binucleated cells with micronuclei (MNBN) and total micronuclei (TMN) following exposure to a dose of 2 Gy, compared with those in the control group of 0 Gy. Furthermore, the occurrence of ≥2 micronuclei after radiation exposure (Table III) was observed across all samples.
Table IV.
Micronuclei distribution in binucleated cells, frequency of binucleated cells with micronuclei, total micronuclei (TMN) and nuclear division index (NDI) for each volunteer.
| MN Distribution | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample name | Dose (Gy) | Total BN |
|
MNBN (‰) | TMN (‰) | NDI | ||||
| 0 MN | 1 MN | 2 MN | 3 MN | 4 MN | ||||||
| NC 1 | 0 | 2,000 | 1,981 | 15 | 2 | 0 | 0 | 8.50 | 9.50 | 1.58 |
| 2 | 2,000 | 1,626 | 263 | 43 | 7 | 1 | 157.00 | 187.00 | 1.54 | |
| NC 2 | 0 | 2,000 | 1,961 | 35 | 2 | 0 | 0 | 18.50 | 19.50 | 1.72 |
| 2 | 2,000 | 1,629 | 288 | 37 | 3 | 0 | 164.00 | 185.50 | 1.60 | |
| Global | 0 | 4,000 | 3,942 | 50 | 4 | 0 | 0 | 13.50 | 14.50 | |
| 2 | 4,000 | 3,255 | 551 | 80 | 10 | 1 | 160.50 | 186.25 | ||
| VUS_BRCA1_1 | 0 | 2,000 | 1,984 | 16 | 0 | 0 | 0 | 8.00 | 8.00 | 1.84 |
| 2 | 2,000 | 1,753 | 176 | 29 | 3 | 1 | 104.50 | 123.50 | 1.81 | |
| VUS_BRCA1_2 | 0 | 2,000 | 1,991 | 9 | 0 | 0 | 0 | 4.50 | 4.50 | 1.82 |
| 2 | 2,000 | 1,733 | 209 | 26 | 2 | 0 | 118.50 | 133.50 | 1.71 | |
| Global | 0 | 4,000 | 3,975 | 25 | 0 | 0 | 0 | 6.25 | 6.25 | |
| 2 | 4,000 | 3,486 | 385 | 55 | 5 | 1 | 111.50 | 128.50 | ||
| ATM 1 | 0 | 2,000 | 1,992 | 8 | 0 | 0 | 0 | 4.00 | 4.00 | 1.71 |
| 2 | 2,000 | 1,834 | 126 | 20 | 0 | 0 | 73.00 | 83.00 | 1.77 | |
| ATM 2 | 0 | 2,000 | 1,980 | 18 | 1 | 0 | 0 | 9.50 | 10.00 | 1.23 |
| 2 | 2,000 | 1,745 | 192 | 27 | 3 | 0 | 111.00 | 127.50 | 1.17 | |
| Global | 0 | 4,000 | 3,972 | 26 | 1 | 0 | 0 | 6.75 | 7.00 | |
| 2 | 4,000 | 3,579 | 318 | 47 | 3 | 0 | 92.00 | 105.25 | ||
BN, binucleated cells; MN, micronuclei; NC, non-carrier; VUS_BRCA1_1 and VUS_BRCA1_2, VUS, variants of unknown significance carriers.
Figure 6.
Representative images of binucleated cells obtained during analysis, showing the micronuclei structures, associated with γ-radiation exposure: (A) Normal binucleated cell (0 Gy dose); (B) binucleated cells containing one micronuclei (2 Gy dose); and (C) binucleated cells containing two micronuclei (2 Gy dose). Micronuclei indicated by arrows. Magnification, ×400.
The main aim of the present study was to functionally characterize the genetic variant by assessing the cellular response to γ-radiation. Given the limited number of samples analyzed, the data were grouped according to the presence of each genetic variant: VUS carriers, ATM carriers and NC carriers. Fig. 7 represents the distribution of MNs in each group. Analysis of the MN frequency distribution revealed a significant decrease in the MNBN frequency in VUS and ATM carriers compared with NC carriers.
Figure 7.
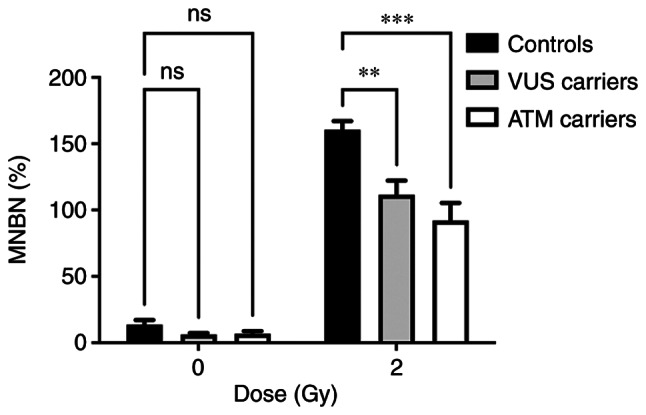
MN frequency distribution for each group of genetic variants (**P<0.01; ***P<0.001). MN, micronuclei; MNBN, micronucleated binucleated cells; VUS, variants of unknown significance; ns, no significance.
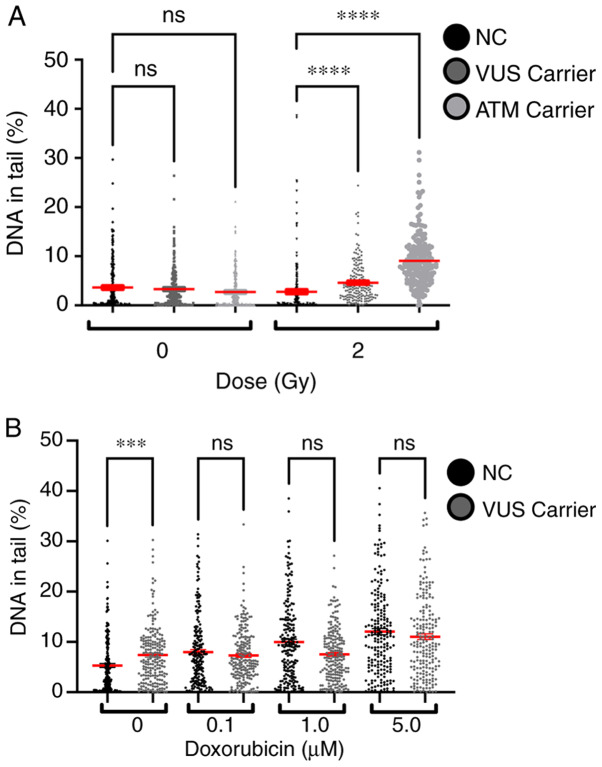
Single-cell gel electrophoresis (comet assay)
The % DNA in tail for all samples was measured to evaluate the effect of γ-radiation and chemical exposure to doxorubicin. A dose-dependent effect was observed in both experiments, with higher doses inducing more lesions (Fig. 8). Comparing the effects of 2 Gy on NC and VUS carriers, an increase in the number of DNA lesions was observed in the VUS carriers, with an even more significant increase in ATM carriers, compared with the NC carriers. However, this effect was not observed after doxorubicin exposure except for the basal level, where VUS carriers have significantly more basal DNA damage than NC carriers (Fig. 8B).
Figure 8.
DNA lesions measured by the comet assay following (A) exposure to γ-radiation and (B) chemical exposure to doxorubicin (***P<0.001; ****P<0.0001). NC, non-carrier; VUS, variants of unknown significance; ns, no significance.
γH2AX assay
The γH2AX assay is one functional method that can be used to detect DSBs following chemical exposure. The data obtained was grouped into NC and VUS carriers. The results obtained demonstrated a dose-dependent effect in both NC and VUS carriers (Fig. 9). However, the differences between VUS carriers and NC carriers were non-significant.
Figure 9.
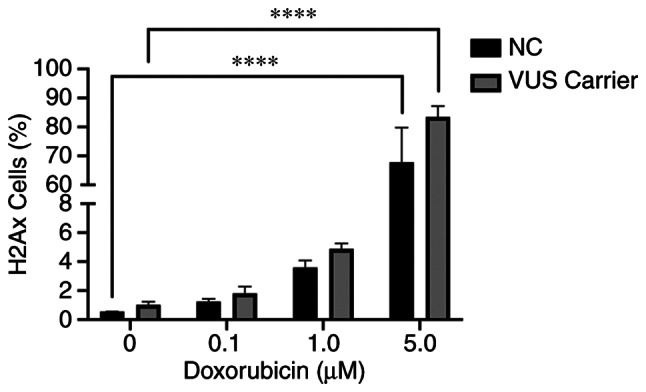
DSB-induced by chemical exposure measured by flow-cytometry of γH2AX assay (****P<0.0001). DSB, double-stranded DNA breaks; NC, non-carrier; VUS, variants of unknown significance; ns, no significance.
Caspase 3–7 and 9 assays
The caspase signaling cascade is responsible for the activation of apoptosis and inflammatory processes, which allow cells to maintain their genomic stability whilst controlling programed cell death. In the present study, two caspases with two different functions in the apoptosis pathway were assessed; caspase 9 (initiator caspase) and caspases 3–7 (executioner caspases) (34). These caspases operate in the intrinsic pathway, which is triggered in response to death stimuli generated by DNA damage. Chemical exposure was found to slightly activate caspase 9, while almost no activation could be detected for caspase 3–7 (Fig. 10). Caspases 3–7 activity exhibited only slight variations at the different doxorubicin concentrations. However, the activity of caspase 9 increased sharply at higher doxorubicin concentrations, suggesting that activation of the initiator caspase cascade occurred due to DNA damage.
Figure 10.
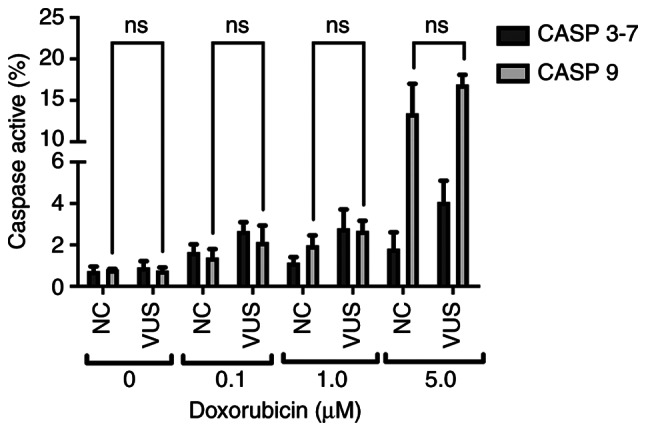
Representation of Caspase activation signal measured by flow cytometry. NC, non-carrier; VUS, variants of unknown significance; ns, no significance.
TUNEL assay
To the best of the authors' knowledge, cells undergoing apoptosis exhibit several changes in nuclear morphology, especially during the later stages of programmed cell death or apoptosis. These features include DNA fragmentation and DNA strand breaks, both of which are relevant when evaluating the biological role of DNA repair genes. The damage inflicted in PBMCs by doxorubicin exposure was also evaluated using the TUNEL assay as quantified by flow cytometry. This assay measures DNA fragments resulting from the apoptotic process. A significant difference at higher concentrations of doxorubicin between NC carriers and VUS carriers was observed (Fig. 11), with NC carriers being more sensitive to DNA fragmentation, consistent with the results obtained for MN (Fig. 4).
Figure 11.
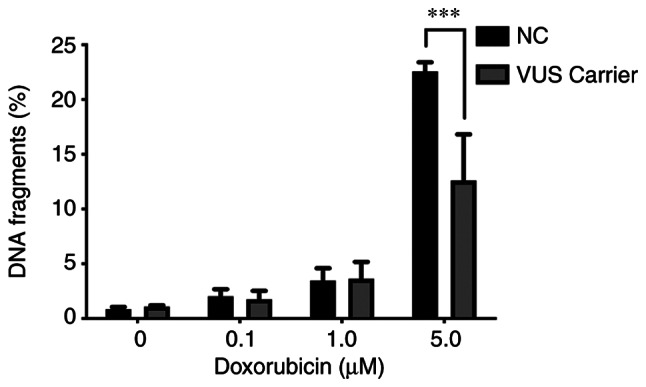
Representation of % DNA fragmentation for TUNEL measured by flow cytometry (***P<0.001). NC, non-carrier; VUS, variants of unknown significance.
Discussion
One of the limitations in studying the clinical significance of VUS in human samples is the rarity of their occurrence, restricting the number of individuals available for functional studies which can be performed in peripheral lymphocytes. BRCA1 plays a major role in DNA repair and is broadly expressed in a wide variety of cells, including lymphocytes (https://www.proteinatlas.org/ENSG00000012048-BRCA1/tissue), which justifies the use of patients' lymphocytes as a surrogate tissue for breast tissue. The BRCA1 protein is involved in repairing damaged DNA, produced either endogenously or by exogenous factors or when chromosomes exchange genetic material in preparation for cell division, replication fork protection, cell cycle regulation and gene transcription regulation (35). The BRCA1 protein interacts with several other proteins to repair DNA strand breaks that also occur when chromosomes exchange genetic material in preparation for cell division. Thus, BRCA1 acts as a tumor suppressor. Mutations in BRCA1 have long been associated with increased risk of breast cancer in men and women, as well as several other types of cancer and increased DSB, indicating a defect in DNA repair (36). To date, there is no evidence of cell-type specific differences in the activity of DSB repair pathways and in other BRCA1-specific interactions.
Identification of a pathogenic germline variant is crucial for the correct clinical management of families with increased risk for hereditary breast cancer. This would enable the early identification of individuals most at-risk and those who require increased surveillance and/or prophylactic interventions (1). The increasingly common application of NGS has identified a number of variants in genes suspected to be involved in cancer predisposition, in particular for breast cancer. Several of these genes are associated with DNA repair and have been reported in female and male breast cancer patients (36). However, the increased identification of variants of high penetrant genes through NGS has led to considerable difficulties in the adequate classification of their pathogenicity. This assessment therefore relies mostly on co-segregation with disease, co-occurrence with known pathogenic variants and family history of cancer. Therefore, understanding the impact of VUS on protein function is critical for understanding the functional consequences and potential therapy responses (37–39).
The c.1067A>G (rs1799950) BRCA1 missense variant results in the replacement of glutamine with arginine at codon 356 of the BRCA1 gene. Missense mutations that do not lead to the complete disruption of protein function may slightly alter the structures of domains important for protein function. The effect of these mutations may be estimated according to their position and the type of altered amino acid using specific software, which is measured by the probability of disrupting a particular protein function increasing the disease risk. For the present study, analysis using two different in silico prediction tools revealed two distinct prognostic results for this VUS. PolyPhen2 identified this VUS as likely damaging (0.998) (PolyPhen 2, 2020), whereas ClinVar classified this as benign (ClinVar-NCBI, 2020). Therefore, this substitution cannot be classified as benign or pathogenic with confidence, since in silico prediction tools could only provide a theoretical prediction of the effects of this variant on protein structure and function. In addition, given its rarity, data on the role of this variant on cancer risk are scarce.
The role of DNA repair genes in breast cancer has been extensively studied, to an extent that clinical panels integrating the most relevant genes for breast cancer progression have been reported, emphasizing their importance. The challenge in the present study was to establish a set of assays that allowed the evaluation and characterization of genetic variants that may affect the DNA repair mechanisms. Therefore the decision was centered on approaches that facilitate the measurement of DNA lesions induced by genotoxic agents (radiation and doxorubicin) with the added advantage of studying human samples. One of the methodologies selected is the CA assay, which is considered to be the ‘gold standard’ for radiation biodosimetry. This approach allows for the microscopic visualization of features of DNA damage, such as DSB (40). The most representative lesion caused by radiation exposure is dicentric chromosomes, as discussed in previous studies (Fig. 4B) (29,33). High frequencies of CA in peripheral blood lymphocytes have been associated with significantly elevated risks of cancer development (40,41). Results in the present study showed a clear dose-dependent increase in the rates of CA after radiation exposure. These results are consistent with those from previous biodosimetry studies (33). However, no statistical difference could be observed between NC and VUS carriers.
An alternative method to the CA assay that also allows for the detection and evaluation of DNA damage induced by genotoxic agents is the CBMN assay (29,42). The MN results from the present study also revealed a dose-dependent effect. In particular, increases in the frequency of MNBN and TMN were observed at higher doses. However, the VUS carrier group exhibited lower levels of DNA lesions in response to radiation compared with those in the NC group. Previous studies have associated higher frequencies of MN with an increased risk of cancer (43–45). Therefore, if this VUS is a potentially pathogenic variant in the BRCA1 gene, then higher levels of DNA damage should have been observed.
The comet assay has also been previously applied to evaluate both DNA damage and repair. In addition, it is a well-known technique for assessing DNA damage after radiation and chemical exposure (46–48). According to this assay, the present study showed a global increase in DNA damage after exposure. The differences between the exposure types were also evident, showing statistical differences, specifically higher sensitivity to DNA damage in BRCA1 VUS carriers. The effect observed for irradiated samples demonstrates that this assay is a viable method for evaluating primary DNA lesions (49), suggesting a potential role of this genetic variant in hampering the repair mechanisms.
To measure the DSB repair sites induced by chemical exposure, a H2AX assay was also performed. Although this assay can also evaluate DNA damage, it could not reveal a significant difference between NC and VUS (Fig. 9). This assay is likely to be more beneficial for evaluating primary DNA lesions but showed substantial limitations over longer time-scales, due to the rapid signal decline (48). Nevertheless, the data suggest that the VUS carriers displays a higher trend of doxorubicin-induced γH2AX foci.
The relationship between DNA repair and apoptotic pathways remains poorly understood. However, it is clear that both radiation and chemotherapy exposure can activate apoptotic pathways. Different programed cell death pathways can be activated depending on the stimulus and the damage level of the cells. According to the three main activation pathways, results from the present study appear to indicate activation of the intrinsic pathway, which mainly involves the formation of apoptosomes, followed by the activation of caspase-9 (50). Once activated, caspase-9 initiates a caspase activation cascade by processing caspases-3 and −7 (34). The present study showed a clear dose-dependent signal in caspase-9 activity, but no significant difference could be observed in the signals for caspases-3 and −7, even at increasing doxorubicin concentrations. However, these data could not be correlated with the presence of the VUS.
Later stage of apoptosis can be measured through the detection and quantification of apoptotic DNA fragmentation using the TUNEL assay (50). In agreement with the results from MN assay, a significant difference between the NC and VUS carriers was observed, with the former displaying less DNA fragmentation. Theoretically, the similarity between the MN and TUNEL results suggests a benign effect attributed to the presence of this VUS in the BRCA1 gene.
BRCA1 associates with the MRE11/RAD50/NBS1 complex of proteins, which acts as a DSB sensor and signals for the recruitment of downstream DNA repair pathway components. This process typically favors HR rather than NHEJ (19). This association is mediated by RAD50 with residues 341–748 of the BRCA1 protein. The VUS (rs1799950; T>C) analyzed in the present study occurs in residue 356 of the BRCA1 protein, leading to an arginine instead of a glutamine. Although this change was within the RAD50 interacting region, it did not show clear divergent results compared with NC carriers even though glutamine has an uncharged R group and arginine is a positively charged amino acid. This suggested that this amino acid change did not affect its interaction with RAD50.
DNA repair mechanisms serve a crucial role in maintaining genome stability and integrity. The presence of a single VUS in the BRCA1 gene may modulate its role in protecting the cell population from DSBs that lead to chromosomal rearrangements by decreasing micronuclei formation. This in turn reduces genomic instability and leads to the activation of programmed cell death. Thus, the present study suggested that this VUS is probably benign. Supporting this, recent meta-analyzes described this VUS to be a low-risk variant for breast cancer, demonstrating its possible protective behavior (51,52).
However, further studies should be performed to understand the underlying mechanisms, due to the exploratory nature of the present study. Genome-wide sequencing technologies will continue to identify novel VUS that will urgently require functional characterization. This can be achieved using assaying techniques applied in the present study. In particular, a clear classification is of utmost importance in the clinical setting and for planning resulting actions.
The strategy followed by the present study, which mainly assessed DNA-damage endpoints as readout for investigating the effects of putative modifications on the DNA repair capacity of this BRCA1 VUS, may shed light on the possible functional consequences of sequence variants. This is because VUS may not result in pathogenicity through a direct effect on protein-protein interaction due to amino acid changes. Several regulatory variants discovered by expression quantitative trait loci mapping have already been validated. It was assumed that these regulatory variants may act by affecting transcription factors binding sites to interfere with the function of the protein. For example, they may operate through a regulatory mechanism that lowers or even abort BRCA1 expression (53).
In conclusion, the results obtained suggested that this VUS is benign or highly likely to be benign. Although the present study was exploratory, the strategy can be successfully used to study other variants.
Acknowledgements
The authors would like to do acknowledge in particular all participants of this study. The authors also acknowledge all the technical support by Dr Ana Catarina Antunes and the irradiation of samples by Dr Pedro Santos (Centro de Ciências e Tecnologias Nucleares).
Funding Statement
The present study was funded by Terry Fox Grant 2017 from Liga Portuguesa Contra o Cancro and by Fundação de Ciência e Tecnologia (FCT; grant nos. UID/BIM/0009/2020 and UIDP/00009/2020.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
Conceptualization was mainly performed by SNS, JR, JBPL and JC. Collection of patient clinical data and family history was performed by TL. SNS, JBPL and JC confirm the authenticity of all the raw data. RAL and ML performed the experiments, and OG designed the methodology and performed the experiments. Validation of proceedings was performed by SNS and OG. Formal analysis was performed by SNS. Investigation was mainly performed by RAL and ML. Resources were acquired in collaboration with Ophiomics-Precision Medicine by SNS, JR, JBPL and JC. JC and JBPL were responsible for the recruitment of families to be included in the present study and performed the NGS sequencing analysis. ASR analyzed and interpreted data, and critically revised the manuscript. Data curation was performed by JBPL, JC and SNS. Writing of the original draft was by SNS and reviewing and editing was by SNS, JR and ASR. Supervision of the present study was by SNS and JR. Project administration funding acquisition was performed by SNS. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the National Commission of Data Protection (approval no. 10637/2016) for the use of samples for research and also by the Ethical Commission of NMS/FCM (approval no. 54/2018/CEFCM). All regulations were respected and were in agreement with the Declaration of Helsinki. Informed consent was obtained from all participants, after their being informed about the usage of their blood samples were used for scientific research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kar SP, Beesley J, Amin Al Olama A, Michailidou K, Tyrer J, Kote-Jarai Z, Lawrenson K, Lindstrom S, Ramus SJ, Thompson DJ, et al. Genome-wide meta-analyses of breast, ovarian, and prostate cancer association studies identify multiple new susceptibility loci shared by at least two cancer types. Cancer Discov. 2016;6:1052–1067. doi: 10.1158/2159-8290.CD-15-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulo P, Pinto P, Peixoto A, Santos C, Pinto C, Rocha P, Veiga I, Soares G, Machado C, Ramos F, Teixeira MR. Validation of a next-generation sequencing pipeline for the molecular diagnosis of multiple inherited cancer predisposing syndromes. J Mol Diagn. 2017;19:502–513. doi: 10.1016/j.jmoldx.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Pinto P, Paulo P, Santos C, Rocha P, Pinto C, Veiga I, Pinheiro M, Peixoto A, Teixeira MR. Implementation of next-generation sequencing for molecular diagnosis of hereditary breast and ovarian cancer highlights its genetic heterogeneity. Breast Cancer Res Treat. 2016;159:245–256. doi: 10.1007/s10549-016-3948-z. [DOI] [PubMed] [Google Scholar]
- 4.Breast Cancer Association Consortium, corp-author. Dorling L, Carvalho S, Allen J, González-Neira A, Luccarini C, Wahlström C, Pooley KA, Parsons MT, Fortuno C, et al. Breast cancer risk genes-association analysis in more than 113,000 women. N Engl J Med. 2021;384:428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eccles DM, Mitchell G, Monteiro ANA, Schmutzler R, Couch FJ, Spurdle AB, Gómez-García EB, ENIGMA Clinical Working Group BRCA1 and BRCA2 genetic testing-pitfalls and recommendations for managing variants of uncertain clinical significance. Ann Oncol. 2015;26:2057–2065. doi: 10.1093/annonc/mdv278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasparini P, Lovat F, Fassan M, Casadei L, Cascione L, Jacob NK, Carasi S, Palmieri D, Costinean S, Shapiro CL, et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci USA. 2014;111:4536–4541. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 8.Prasad CB, Prasad SB, Yadav SS, Pandey LK, Singh S, Pradhan S, Narayan G. Olaparib modulates DNA repair efficiency, sensitizes cervical cancer cells to cisplatin and exhibits anti-metastatic property. Sci Rep. 2017;7:12876. doi: 10.1038/s41598-017-13232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ratanaphan A. A DNA repair BRCA1 estrogen receptor and targeted therapy in breast cancer. Int J Mol Sci. 2012;13:14898–14916. doi: 10.3390/ijms131114898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conde J, Silva SN, Azevedo AP, Teixeira V, Pina JE, Rueff J, Gaspar JF. Association of common variants in mismatch repair genes and breast cancer susceptibility: A multigene study. BMC Cancer. 2009;9:344. doi: 10.1186/1471-2407-9-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva SN, Costa B, Rueff J, Gaspar JF. DNA repair perspectives in thyroid and breast cancer: The role of DNA repair polymorphisms. In: Vengrova S, editor. DNA Repair and Human Health. InTech; 2011. [Google Scholar]
- 12.Silva SN, Moita R, Azevedo AP, Gouveia R, Manita I, Pina JE, Rueff J, Gaspar J. Menopausal age and XRCC1 gene polymorphisms: Role in breast cancer risk. Cancer Detect Prev. 2007;31:303–309. doi: 10.1016/j.cdp.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Silva SN, Tomar M, Paulo C, Gomes BC, Azevedo AP, Teixeira V, Pina JE, Rueff J, Gaspar JF. Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer Epidemiol. 2010;34:85–92. doi: 10.1016/j.canep.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Gomes BC, Silva SN, Azevedo AP, Manita I, Gil OM, Ferreira TC, Limbert E, Rueff J, Gaspar JF. The role of common variants of non-homologous end-joining repair genes XRCC4, LIG4 and Ku80 in thyroid cancer risk. Oncol Rep. 2010;24:1079–1085. [PubMed] [Google Scholar]
- 15.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 17.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenblick A, de Azambuja E, Azim HA, Jr, Piccart M. An update on PARP inhibitors-moving to the adjuvant setting. Nat Rev Clin Oncol. 2015;12:27–41. doi: 10.1038/nrclinonc.2014.163. [DOI] [PubMed] [Google Scholar]
- 19.Christou C, Kyriacou K. BRCA1 and Its network of interacting partners. Biology (Basel) 2013;2:40–63. doi: 10.3390/biology2010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma B, Kaur RP, Raut S, Munshi A. BRCA1 mutation spectrum, functions, and therapeutic strategies: The story so far. Curr Probl Cancer. 2018;42:189–207. doi: 10.1016/j.currproblcancer.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Colas C, Golmard L, de Pauw A, Caputo SM, Stoppa-Lyonnet D. ‘Decoding hereditary breast cancer’ benefits and questions from multigene panel testing. Breast. 2019;45:29–35. doi: 10.1016/j.breast.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Calò V, Bruno L, La Paglia L, Perez M, Margarese N, Di Gaudio F, Russo A. The clinical significance of unknown sequence variants in BRCA genes. Cancers (Basel) 2010;2:1644–1660. doi: 10.3390/cancers2031644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beitsch PD, Whitworth PW, Hughes K, Patel R, Rosen B, Compagnoni G, Baron P, Simmons R, Smith LA, Grady I, et al. Underdiagnosis of hereditary breast cancer: Are genetic testing guidelines a tool or an obstacle? J Clin Oncol. 2019;37:453–460. doi: 10.1200/JCO.18.01631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S, van Leeuwen FE, Milne RL, Andrieu N, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 25.Martins V, Antunes AC, Cardoso J, Santos L, Gil OM. Influence of age and gender in response to γ-radiation in Portuguese individuals using chromosomal aberration assay-preliminary findings. Radiat Meas. 2011;46:1000–1003. doi: 10.1016/j.radmeas.2011.05.002. [DOI] [Google Scholar]
- 26.Gil OM, Oliveira NG, Rodrigues AS, Laires A, Ferreira TC, Limbert E, Léonard A, Gerber G, Rueff J. Cytogenetic alterations and oxidative stress in thyroid cancer patients after iodine-131 therapy. Mutagenesis. 2000;15:69–75. doi: 10.1093/mutage/15.1.69. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues AS, Oliveira NG, Gil OM, Léonard A, Rueff J. Use of cytogenetic indicators in radiobiology. Radiat Prot Dosimetry. 2005;115:455–460. doi: 10.1093/rpd/nci072. [DOI] [PubMed] [Google Scholar]
- 28.Rueff J, Brás A, Cristóvão L, Mexia J, Sá da Costa M, Pires V. DNA strand breaks and chromosomal aberrations induced by H2O2 and 60Co gamma-radiation. Mutat Res. 1993;289:197–204. doi: 10.1016/0027-5107(93)90070-V. [DOI] [PubMed] [Google Scholar]
- 29.Cytogenetic Dosimetry, corp-author. International Atomic Energy Agency; Vienna: 2011. Applications in preparedness for and response to radiation emergencies. [Google Scholar]
- 30.Antunes AC, Martins V, Cardoso J, Santos L, Monteiro Gil O. The cytokinesis-blocked micronucleus assay: Dose estimation and inter-individual differences in the response to γ-radiation. Mutat Res Genet Toxicol Environ Mutagen. 2014;760:17–22. doi: 10.1016/j.mrgentox.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Oliveira NG, Neves M, Rodrigues AS, Monteiro Gil O, Chaveca T, Rueff J. Assessment of the adaptive response induced by quercetin using the MNCB peripheral blood human lymphocytes assay. Mutagenesis. 2000;15:77–83. doi: 10.1093/mutage/15.1.77. [DOI] [PubMed] [Google Scholar]
- 32.Pingarilho M, Oliveira NG, Martins C, Fernandes AS, de Lima JP, Rueff J, Gaspar JF. Genetic polymorphisms in detoxification and DNA repair genes and susceptibility to glycidamide-induced DNA damage. J Toxicol Environ Health A. 2012;75:920–933. doi: 10.1080/15287394.2012.690709. [DOI] [PubMed] [Google Scholar]
- 33.Martins V, Antunes AC, Monteiro Gil O. Implementation of a dose-response curve for γ-radiation in the Portuguese population by use of the chromosomal aberration assay. Mutat Res. 2013;750:50–54. doi: 10.1016/j.mrgentox.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Cullen SP, Martin SJ. Caspase activation pathways: Some recent progress. Cell Death Differ. 2009;16:935–938. doi: 10.1038/cdd.2009.59. [DOI] [PubMed] [Google Scholar]
- 35.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 36.Silva SN, Gomes BC, André S, Félix A, Rodrigues AS, Rueff J. Male and female breast cancer: The two faces of the same genetic susceptibility coin. Breast Cancer Res Treat. 2021;188:295–305. doi: 10.1007/s10549-021-06159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guidugli L, Carreira A, Caputo SM, Ehlen A, Galli A, Monteiro AN, Neuhausen SL, Hansen TV, Couch FJ, Vreeswijk MP, ENIGMA consortium Functional assays for analysis of variants of uncertain significance in BRCA2. Hum Mutat. 2014;35:151–164. doi: 10.1002/humu.22478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boonen RACM, Rodrigue A, Stoepker C, Wiegant WW, Vroling B, Sharma M, Rother MB, Celosse N, Vreeswijk MPG, Couch F, et al. Functional analysis of genetic variants in the high-risk breast cancer susceptibility gene PALB2. Nat Commun. 2019;10:5296. doi: 10.1038/s41467-019-13194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Millot GA, Carvalho MA, Caputo SM, Vreeswijk MP, Brown MA, Webb M, Rouleau E, Neuhausen SL, Hansen TVO, Galli A, et al. A guide for functional analysis of BRCA1 variants of uncertain significance. Hum Mutat. 2012;33:1526–1537. doi: 10.1002/humu.22150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obe G, Pfeiffer P, Savage JRK, Johannes C, Goedecke W, Jeppesen P, Natarajan AT, Martínez-López W, Folle GA, Drets ME. Chromosomal aberrations: Formation, identification and distribution. Mutat Res. 2002;504:17–36. doi: 10.1016/S0027-5107(02)00076-3. [DOI] [PubMed] [Google Scholar]
- 41.Terzoudi GI, Pantelias GE. Cytogenetic methods for biodosimetry and risk individualisation after exposure to ionising radiation. Radiat Prot Dosimetry. 2006;122:513–520. doi: 10.1093/rpd/ncl509. [DOI] [PubMed] [Google Scholar]
- 42.Sommer S, Buraczewska I, Kruszewski M. Micronucleus assay: The state of art, and future directions. Int J Mol Sci. 2020;21:1534. doi: 10.3390/ijms21041534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murgia E, Ballardin M, Bonassi S, Rossi AM, Barale R. Validation of micronuclei frequency in peripheral blood lymphocytes as early cancer risk biomarker in a nested case-control study. Mutat Res. 2008;639:27–34. doi: 10.1016/j.mrfmmm.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Cardinale F, Bruzzi P, Bolognesi C. Role of micronucleus test in predicting breast cancer susceptibility: A systematic review and meta-analysis. Br J Cancer. 2012;106:780–790. doi: 10.1038/bjc.2011.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott D, Hu Q, Roberts SA. Dose-rate sparing for micronucleus induction in lymphocytes of controls and ataxia-telangiectasia heterozygotes exposed to 60Co gamma-irradiation in vitro. Int J Radiat Biol. 1996;70:521–527. doi: 10.1080/095530096144725. [DOI] [PubMed] [Google Scholar]
- 46.Gunasekarana V, Raj GV, Chand P. A comprehensive review on clinical applications of comet assay. J Clin Diagn Res. 2015;9:GE01–GE05. doi: 10.7860/JCDR/2015/12062.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kopjar N, Garaj-Vrhovac V, Milas I. Assessment of chemotherapy-induced DNA damage in peripheral blood leukocytes of cancer patients using the alkaline comet assay. Teratog Carcinog Mutagen. 2002;22:13–30. doi: 10.1002/tcm.1035. [DOI] [PubMed] [Google Scholar]
- 48.Vinnikov V, Hande MP, Wilkins R, Wojcik A, Zubizarreta E, Belyakov O. Prediction of the acute or late radiation toxicity effects in radiotherapy patients using ex vivo induced biodosimetric markers: A review. J Pers Med. 2020;10:285. doi: 10.3390/jpm10040285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaur S, Sangeeta, Galhna KK, Gautam N. Assessment of radiation induced DNA damage in human peripheral blood lymphocytes using COMET assay. Int J Life Sci Scienti Res. 2017;3:1208–1214. doi: 10.21276/ijlssr.2017.3.4.17. [DOI] [Google Scholar]
- 50.Majtnerová P, Roušar T. An overview of apoptosis assays detecting DNA fragmentation. Mol Biol Rep. 2018;45:1469–1478. doi: 10.1007/s11033-018-4258-9. [DOI] [PubMed] [Google Scholar]
- 51.Brignoni L, Cappetta M, Colistro V, Sans M, Artagaveytia N, Bonilla C, Bertoni B. Genomic diversity in sporadic breast cancer in a Latin American population. Genes (Basel) 2020;11:1272. doi: 10.3390/genes11111272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu GP, Zhao Q, Wang D, Xie WY, Zhang LJ, Zhou H, Chen SZ, Wu LF. The association between BRCA1 gene polymorphism and cancer risk: A meta-analysis. Oncotarget. 2018;9:8681–8694. doi: 10.18632/oncotarget.24064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majewski J, Pastinen T. The study of eQTL variations by RNA-seq: From SNPs to phenotypes. Trends Genet. 2011;27:72–79. doi: 10.1016/j.tig.2010.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.