Abstract
Despite decades of research, human cytomegalovirus (CMV) continues to contribute to significant morbidity and mortality in transplant settings and remains the leading cause of viral congenital infections. Clinical diagnosis of CMV infection and/or reactivation under these settings is completed using real time quantitative polymerase chain reaction (RT-qPCR). This assay performs well but is hampered by poor sensitivity and a lack of standardization among testing facilities. A point-of-care rapid diagnostic to determine CMV viremia could address these issues and improve patient care. In this manuscript, we introduce clustered regularly interspaced short palindromic repeats (CRISPR)-Cas12a technology to design and validate a rapid diagnostic for CMV. This system was tested using CMV spiked human saliva and urine samples. Sensitivity of the assay was ~10 infectious units (IU)/mL. Specificity of the assay was robust and failed to detect other herpesviruses. Collectively, we have designed and validated a rapid diagnostic for CMV that overcomes limitations of the current standard diagnostic. This assay has the potential to be used as a point-of-care screening tool in transplant and neonatal settings.
Keywords: Cytomegalovirus, CMV, CRISPR-Cas12a, gRNA, Rapid diagnostic, Virus, Congenital CMV
1. Introduction
Cytomegalovirus (CMV) is a double-stranded DNA betaherpesvirus exhibiting seroprevalence of 50–80% in the United States and greater than 95% in developing countries (Cannon et al., 2010; Bate et al., 2010). In immunocompetent individuals, primary infection is controlled by the immune system, but the virus is never cleared resulting in lifelong infection. CMV undergoes periodic reactivation characterized by the detection of viral DNA or CMV-specific IgM or IgG in the blood, increasing in frequency with age (Parry et al., 2016; van Boven et al., 2017). Despite the widespread availability of effective antivirals, CMV continues to adversely affect transplantation populations and is the leading infectious cause of congenital infections (Dollard et al., 2007). Quantitative detection of CMV from bodily fluids is completed using real time quantitative polymerase chain reaction (RT-qPCR). This assay is the gold standard for CMV detection. The significant limitations of this test include moderate sensitivity (~100–400 copies/mL) (altona-diagnostics. RealStar CMV PCR Kit, 2017; testguide.labmed.uw.edu. CMV Quantitative by PCR, 2022) and poor standardization between testing sites, despite the 2010 implementation of a World Health Organization (WHO) International Standard (Preiksaitis et al., 2016). The lack of standardization has resulted in establishing thresholds for preemptive therapy that varies between distinct centers (Preiksaitis et al., 2016; Hayden et al., 2017). Further, the assay requires expensive equipment and trained laboratory personnel to complete. Standard sample analysis requires 24–48 h before results are returned to physicians (testguide.labmed.uw.edu. CMV Quantitative by PCR, 2022; Aruplab. Cytomegalovirus by Quantitative PCR, 2022).
Under immunosuppressed conditions, such as transplant settings, CMV can reactivate resulting in high levels of viremia (Vallejo et al., 2022; Duan et al., 2022). During transplantation procedures, CMV reactivation is directly linked to severe end-organ disease (Pneumonia, colitis, retinitis, etc.) and has been associated with decreased overall survival and increased non-relapse mortalilty (Teira et al., 2016). In solid organ transplant settings, CMV is associated with significant morbidity and patients are at high risk for CMV complications during the 3–6 months post-transplantation due to immunosuppressive therapies (Fishman, 2007). Antiviral therapy is effective but is limited by toxicity and antiviral resistance (Krosky et al., 1998; Imlay and Kaul, 2021). The ability to monitor CMV-associated viremia quickly and quantitatively could minimize the impact of toxicity and resistance in transplant settings.
Congenital CMV (cCMV) infection is the leading pathogenic cause of fetal loss, infant neurologic deficits, and birth defects (Zhang et al., 2022; Li et al., 2021). The birth prevalence of cCMV is estimated to be ~0.7% worldwide (Kenneson and Cannon, 2007; Fowler and Boppana, 2006). Pregnant individuals exhibiting or suspected of having a CMV infection can undergo prenatal CMV antibody screening, but use of this assay for clinical decisions is controversial (Leruez-Ville et al., 2020). Suspicion of cCMV may be identified during fetal imaging as intracranial calcifications and intrauterine growth restriction (Ito et al., 2013). Suspected cCMV cases are referred to a maternal-fetal medicine specialist who confirms a cCMV diagnosis using PCR to identify CMV in amniotic fluid via amniocentesis (Leruez-Ville et al., 2020; Rawlinson et al., 2017). This assay is high risk to the fetus and may jeopardize a healthy pregnancy. Only 10% of cCMV cases are symptomatic resulting in neurological sequelae (Boppana and Fowler, 2017). In asymptomatic cases, 10–15% of infants develop progressive sensoneural hearing loss and cognitive dysfunction (Fowler and Boppana, 2018; Foulon et al., 2019). Recent data has emphasized the importance of early antiviral intervention to improve hearing and developmental outcomes in neonates with symptomatic cCMV (Kimberlin et al., 2015; Nishida et al., 2016; Yamada et al., 2020). Detection of cCMV in neonates is confirmed by RT-qPCR in urine or saliva during the first 3 weeks of life. This test is ordered based on clinical suspicion, resulting in up to 90% of cases being undiagnosed at birth (Sorichetti et al., 2016). As most cases are asymptomatic, a universal screening approach is needed to identify the majority of cCMV cases.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein (Cas) system is being explored as a pathogen detection system (Huang et al., 2021; Xu et al., 2020; Kaminski et al., 2020a). CRISPR sequences are repeat and spacer sequences that can match viral DNA. The Cas protein has helicase and nuclease properties that cut DNA upstream of the CRISPR-Cas locus. The CRISPR-Cas system exhibits high specificity and sensitivity. CRISPR-Cas12a identifies thymine-rich double-stranded DNA protospacer adjacent motifs (PAM). The sticky ends of the staggered cut offered by Cas12a allow enhanced target specific DNA assembly compared to traditional restriction enzyme approaches (Kim et al., 2017). Cas12a cleaves DNA 18–24 base pairs downstream of the PAM site, resulting in sequence recognition after repair, enabling numerous rounds of cleavage. This attribute allows Cas12a to be utilized as a diagnostic, in which one side is guided to the target by a designed guide RNA (gRNA) and the other end cleaves an oligonucleotide fluorescent probe, allowing for quantification. This property is central to the model and is leveraged here to develop a CMV assay, similar to previous work (Huang et al., 2021; Broughton et al., 2020; Nguyen et al., 2020). An overview of the assay design is provided in Fig. 1. Collectively, the need for a low cost, point-of-care rapid diagnostic for CMV is critical to change how we think about and treat CMV-associated pathology. This work describes the design, optimization and validation a CRIPSR-Cas12a CMV rapid diagnostic.
Fig. 1. Overview of the CRISPR-Cas12a assay used in this manuscript.
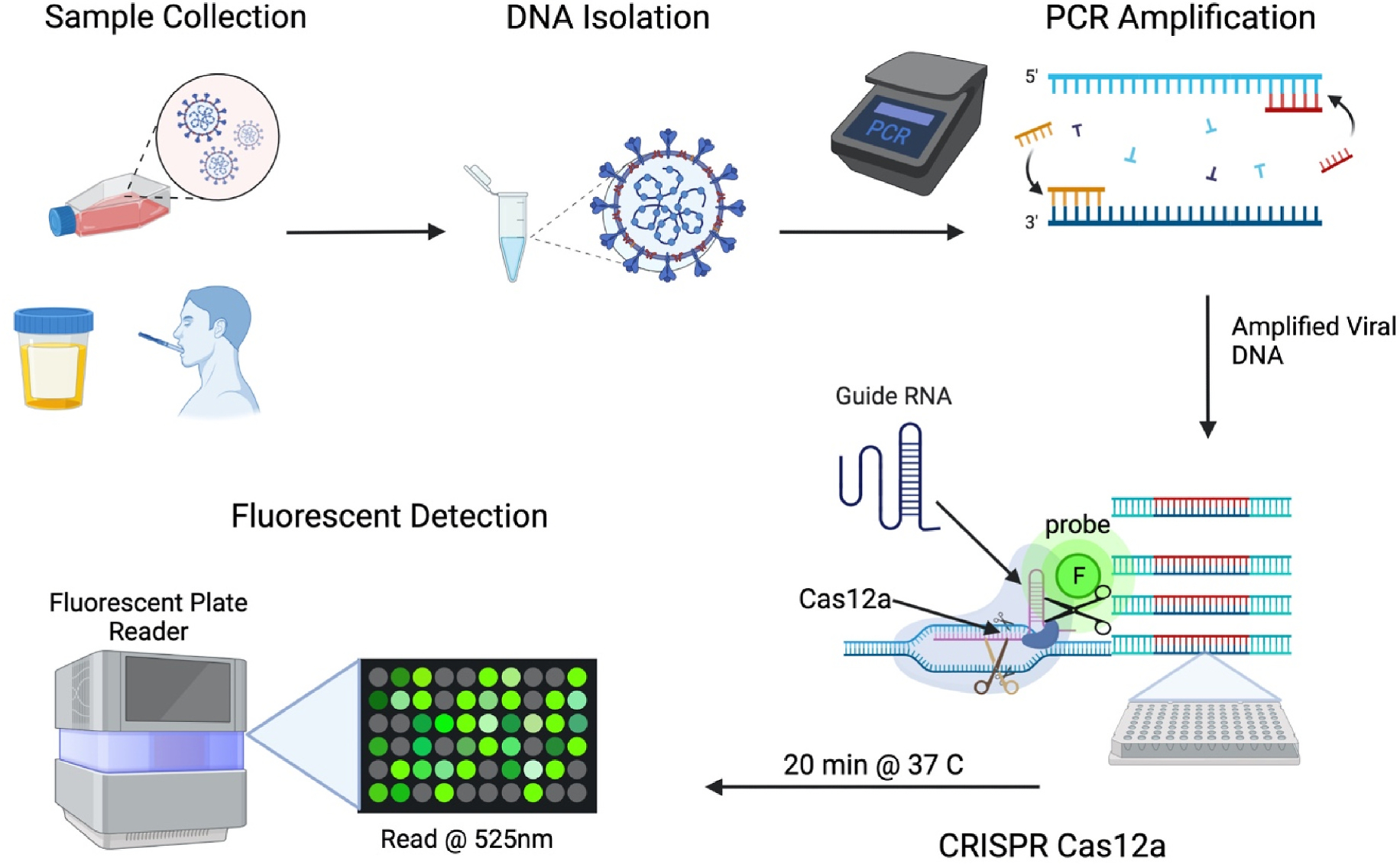
Urine or saliva was spiked with HCMV. Viral DNA was isolated using commercially available kits. HCMV DNA target sequences were amplified using a thermocycler. Amplified HCMV target sequences were mixed with CRISPR-Cas12a reagents and incubated for 20 min. A fluorescent plate reader was used to detect HCMV. Created with BioRender.com.
2. Methods
2.1. gRNA and primer design
The CMV Merlin strain sequence (ref: GenBank: AY446894.2) was used to design gRNA for CMV detection. The CMV genes UL123 and US28 were scanned for CRISPR-Cas12a PAM sequence(s) TTTV (V indicating any base but T). The gRNA sequence is the immediate 20–24 base pairs after the PAM sequence. CMV Merlin-specific primer sets were designed using NCBI Primer-Blast. Premier biosoft.com qOligo was used to analyze the forward and reverse primers for GC content, melting point, GC clamp, dimers and hairpins. Primers and gRNA were ordered from Integrated DNA Technologies (IDT).
2.2. Viral DNA propagation and isolation
CMV Towne (ATCC VR-977), Toledo (generous gift from Dr. John Sinclair), and TR (generous gift from Dr. Andrew Yurochko) were propagated in Human Foreskin Fibroblasts (HFF) (ATCC SCRC-1041) using DMEM (Gibco) supplemented with 10% FBS and 1X Glutamax (Gibco). HHV-6A strain HST (acquired from HHV-6 foundation) was propagated in HSB-2 cells (ATCC CCL-120.1) using IMDM media (Gibco) supplemented with 10% FBS. HHV-6B strain Z29 (acquired from HHV-6 foundation) was propagated using Molt-3 cells (ATCC CRL-1552) using RPMI-1640 media (Gibco) + 10% FBS. Human simplex virus 1 (HSV-1) (generous gift from Dr. Shitao Li) was propagated in Vero cells (ATCC CCL-81) grown in DMEM + 10% FBS. All viral strains were purified through centrifugation and titered as previously described (Britt, 2010; Combs et al., 2019). Each 100 μl viral sample was heat inactivated at 60 °C for 30 min and stored at −80 °C after use. Viral DNA was isolated using a Quick-DNA/RNA Viral Kit (Zymo Research) following manufacturers recommendations. Isolated CMV DNA was stored in nuclease-free water at −20 °C until use.
2.3. Human samples
Human saliva and urine samples from three distinct healthy donors were a generous gift from Dr. Elizabeth Norton. Deidentified samples (Tulane IRB#15–727936) were collected between 2015 and 2016, aliquoted and stored at −80 °C. Samples were thawed and 90 μL were aliquoted for serial dilution and spiked with 10 μL of titered heat- inactivated CMV Towne, TR, or Toledo resulting in a 105 to 10−1 viral IU/mL concentration range. DNA/RNA shield was added to the samples and DNA was isolated using Quick-DNA/RNA Viral Kit (Zymo Research).
2.4. PCR
PCR reactions were performed for each targeted viral region by combining 5 μL of template and 15 μL of PCR mastermix containing 0.5 μL forward primer (10 μM), 0.5 μL reverse primer (10 μM) (IDT), 0.2 μL of AccuPrime Taq DNA Polymerase, 2 μL of AccuPrime PCR Buffer (10x) (ThermoFisher) and 11.8 μL of nuclease-free water (Invitrogen). Sample DNA was amplified using a T100 thermocycler (Bio-Rad) for each 20 μL reaction [denaturation (2 min at 94 °C), amplification (38 cycles: 10 s at 98 °C, 10 s at 60 °C, 15 s at 72 °C) and elongation (5 min at 72 °C)]. Amplified product was used immediately or stored at −20 °C.
2.5. CRISPR-Cas12a fluorescent detection of target regions
All CRISPR-Cas12a fluorescent detection conditions and concentrations have previously been optimized and used at a molar ratio of 1:7.5 (Cas12a/gRNA: fluorescent probe) for an RNA virus (Huang et al., 2020). This assay used a 1:3 M ratio (Cas12a/gRNA: fluorescent probe) to adjust for a DNA virus. Each 32 μL reaction contained end concentrations of 2 μL of RT-PCR product, 10 μM gRNA, 6.7 μM of oligonucleotide fluorescent probe (IDT), 10 μM LbCas12a (New England Biolabs), 3 μL of 10x NEBuffer 2.1 (New England Biolabs), and 11.8 μL of nuclease-free water. Triplicates were loaded on a 96-well opaque half area black flat bottom microplate (Corning) and incubated in the dark for 20 min at 37 °C. A Synergy H1 Microplate reader (BioTek) was used to measure fluorescence at 525 nm in intervals of 5 min. A positive cutoff was determined to be the mean of the no template control (NTC) + 3 times the standard deviation (SD) (3*SD).
2.6. Cross-reactivity and specificity
In silico testing for specificity was completed using the NCBI BLAST tool to assess the gRNA/primer sets for cross-reactivity with any other organism (except taxonomy IDs of CMV strains) in the nonredundant nucleotide database. In vitro testing for specificity was completed by testing HSV-1, HHV-6A (strain HST), and HHV-6B (strain Z29) against each CMV-targeted gRNA/primer set. gRNA/primer sets exhibiting cross-reactivity were excluded from further use. Cross-reactivity was defined by a positive value (3*SD).
2.7. Limit of detection
gRNA/primer sets without cross reactivity were tested for limit of detection (LoD) using serial dilutions of isolated viral DNA. Isolated DNA dilutions of each sample type (purified and spiked) and strain, ranging from 105 to 10−1 IU/mL, were used to create a standard curve that was tested using the CRISPR-Cas12a platform with each gRNA/primer set. A positive sample was defined as the arithmetic mean of the NTC ± (3*SD). This approach was used to determine the lowest viral DNA dilution detectable for each gRNA/primer set and sample type/viral strain. This process was repeated in triplicate and from three distinct biological donors (described in section 2.3).
2.8. Statistical analysis
Data were expressed as means and standard deviations. One-way ANOVA was used to compare intensity responses for sensitivity and specificity thresholds by strain type. Multiple comparisons were performed using Dunnett’s multiple comparison test. Statistical analysis was performed using GraphPad Prism (Version 9, GraphPad Software Inc.). The type I error threshold was set at 5%. Cartoons created with BioRender.com.
3. Results
3.1. Limit of detection using a CRISPR-Cas12a CMV rapid diagnostic
Using the CMV Merlin strain as the standard CMV genome, we identified PAM sequences and gRNA within the UL123, UL122, and US28 gene regions (Fig. 2). To validate that the identified targets could be detected using a CRISPR-Cas12a system, we added CMV into nuclease-free water resulting in a viral concentration range of 105 to 10−1 infectious units (IU)/mL. Viral DNA was then extracted from the CMV spiked water. The limit of detection using CMV TR was significant at 10 IU/mL (Fig. 3A) and at 100 IU/mL for Toledo and Towne (Fig. 3B and C). UL123 recorded a limit of detection as low as 0.1 IU/mL using the CMV TR and Toledo strains. CMV Towne could be detected at 10 IU/mL. US28 exhibited similar limits of detection with values detected at 0.1 IU/mL in TR, Toledo, and Towne (Fig. 3D and E) achieving significance at 100 IU/mL for TR, 10 IU/mL for Toledo, and 1 IU/mL for Towne. We were unable to detect UL122 regardless of the CMV strain used (Supplemental Figs. 1A–C). These results validate the design and approach of a CRISPR-Cas12a system under laboratory conditions.
Fig. 2. Primer and guide RNA (gRNA) sequences for CRISPR-Cas12a detection of CMV.
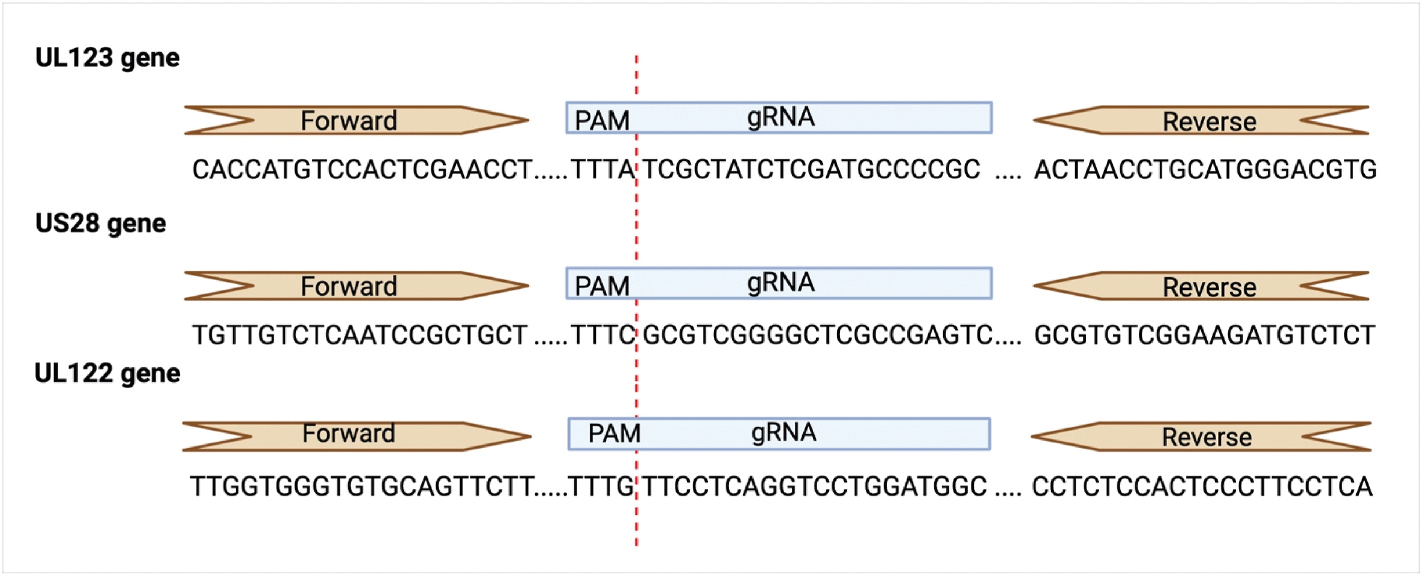
Forward and reverse primer and guide RNA (gRNA) sequences for UL123, US28, and UL122 CMV gene targets were identified and validated. PAM sequences were identified as TTT(V) (V being any base pair except T). Primers were designed against the gRNA region following the PAM sequence. Created with BioRender.com.
Fig. 3. Limit of Detection using a CRISPR-Cas12a CMV assay.
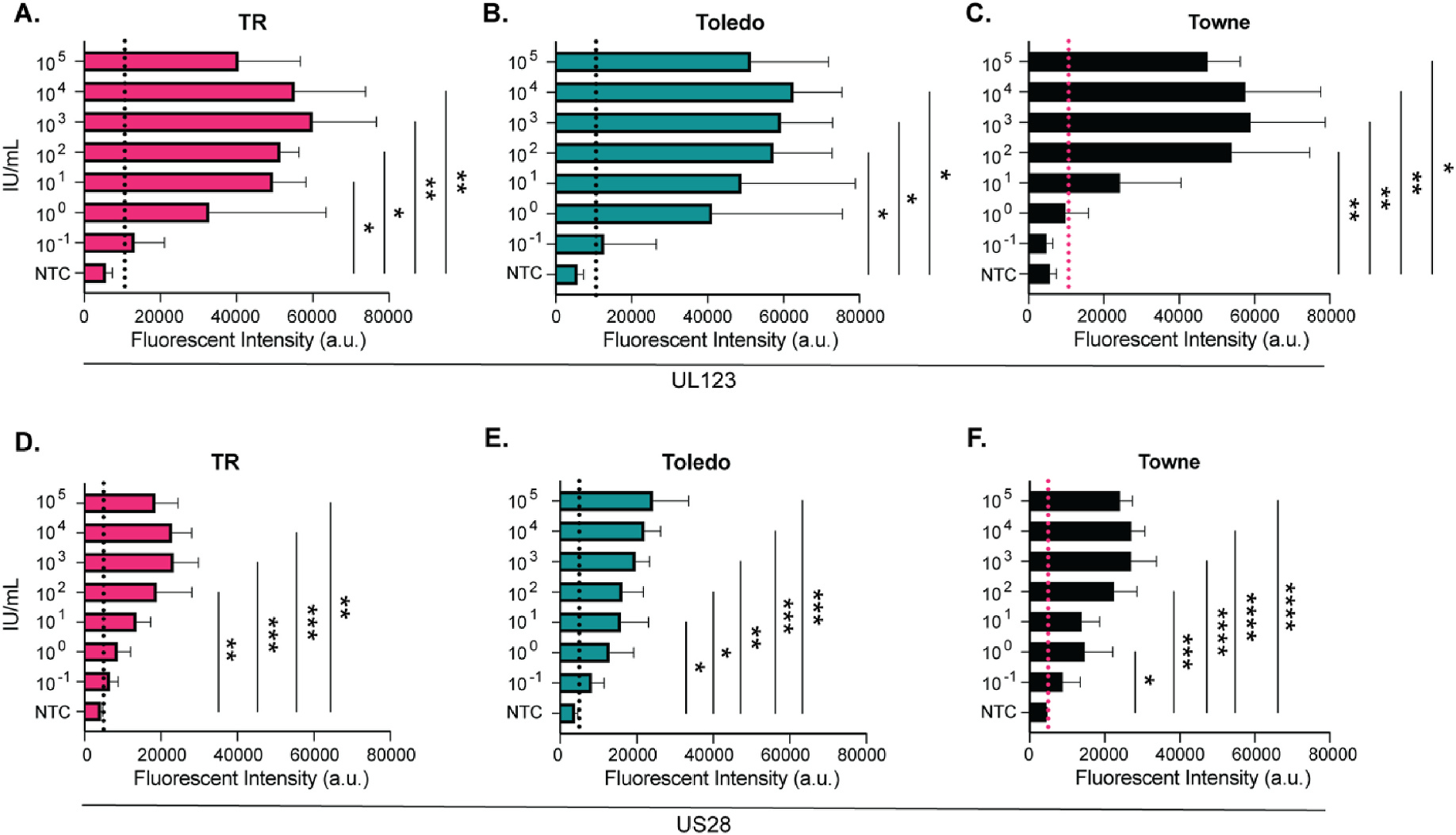
The CMV strains Towne, TR, and Toledo were serially diluted (IU/mL) into nuclease free water, prior to DNA extraction and PCR amplification. Resultant samples were assayed using a CRISPR-Cas12a based fluorescence detection assay. The limit of detection of the CMV gene UL123 using the CMV strain A) TR, B) Toledo, or C) Towne. The limit of detection of the CMV gene US28 using the CMV strain D) TR, E) Toledo, or F) Towne. N = 3 for all experiments. Positive is calculated by the mean of the NTC + (3*SD) and is indicated by the dotted line.*(P < 0.05), **(P < 0.01), ***(P < 0.001), ****(P < 0.0001), determined using a one-way ANOVA with a Dunnett’s multiple comparison test single pooled variance. NTC = no template control. a.u. = arbitrary units. IU = infectious units.
3.2. Validation of specificity
We next addressed the issue of viral specificity by testing the ability of the CRISPR-Cas12a CMV rapid diagnostic to detect other closely related human herpesviruses. Fig. 4A shows the CMV UL123 target is detected using the CMV strains TR, Toledo and Towne but the alphaherpesvirus human simplex virus 1 (HSV-1), the betaherpesviruses human herpesvirus 6A (HHV-6A) and HHV-6B were not detected. These observations were repeated using US28 targets (Fig. 4B). Positive values were considered greater than the mean of the NTC + (3*SD). Together, these results showed that other closely related herpesviruses do not generate false positives with this CRISPR-Cas12a CMV rapid diagnostic.
Fig. 4. Viral specificity of a CRISPR-Cas12a assay.
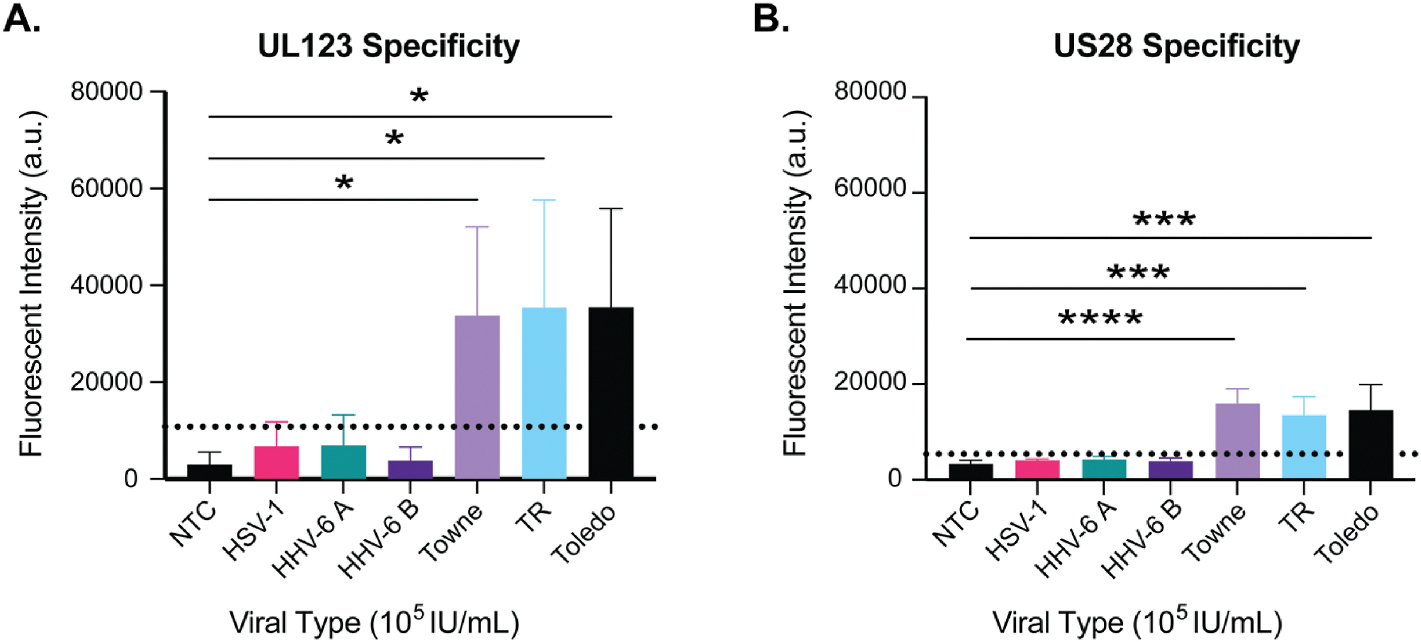
The CMV targets UL123 and US28 were tested for specificity against HSV-1, HHV-6A, HHV-6B and the CMV strains Towne, TR, and Toledo. Virus was spiked in nuclease free water at 105 IU/mL followed by DNA extraction and PCR amplification. Samples were then tested using a CRISPR-Cas12a assay. Specificity using A) UL123 and B) US28 were assayed. N = 3. Positive is calculated by the mean of the NTC + (3*SD) and is indicated by the dotted line. *(P < 0.05), ***(P < 0.001), ****(P < 0.001) statistical significance determined using a one-way ANOVA with a Dunnett’s multiple comparison test single pooled variance. IU = infectious units.
3.3. Evaluation of CMV-spiked human saliva samples
As CMV is secreted at high levels in saliva, we tested the sensitivity of this diagnostic in this fluid. Saliva samples spiked with 105 to 10−1 IU/mL of CMV were tested using the UL123 and US28 targets. Detection of UL123 was significant at 0.1 IU/mL using TR and Toledo strains (Fig. 5A and B). Towne UL123 was detected at 0.1 IU/mL but was significant at 100 IU/mL (Fig. 5C). The limit of detection using US28 was less robust. TR and Toledo were detected at 0.1 IU/mL with significance occurring at 1 IU/mL for the Toledo strain and 100 IU/mL using TR (Fig. 5D and E). The Towne strain was detected at 1 IU/mL; however significance was not achieved (Fig. 5F). The results indicate that this CRIPSR-Cas12a CMV rapid diagnostic can detect CMV in saliva samples without additional preparation or processing of saliva.
Fig. 5. The limit of detection in saliva using a CRISPR-Cas12a CMV assay.
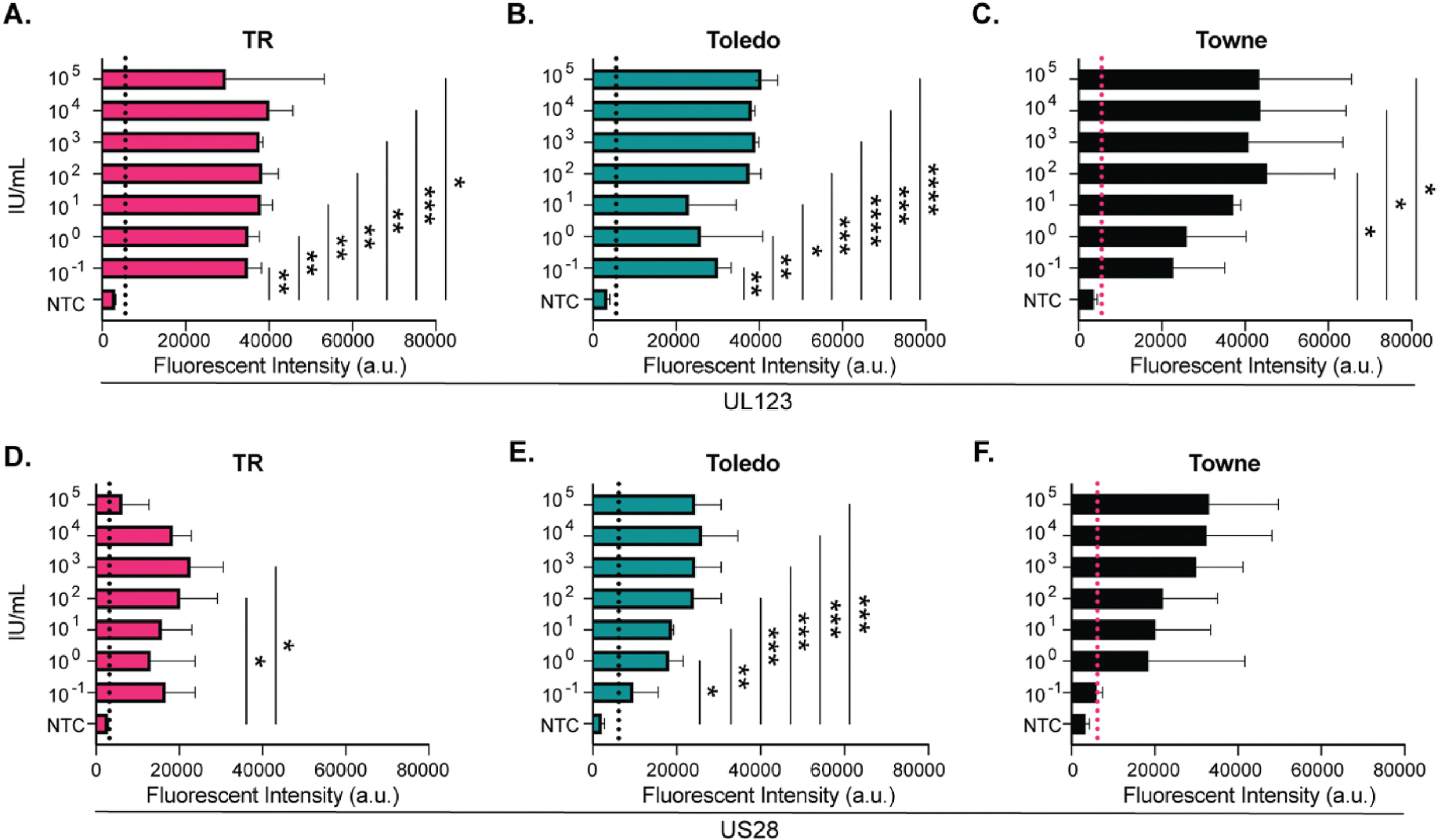
Saliva samples were spiked with CMV followed by DNA isolation and amplification. The limit of detection for UL123 is reported in A) TR, B) Toledo and C) Towne. Comparison of limit of detection against US28 targets using D) TR, E) Toledo, and F) Towne CMV strains. N = 3, Positive is calculated by the mean of the NTC + (3*SD) and is indicated by the dotted line. *(P < 0.05), **(P < 0.01), ***(P < 0.001), **** (P < 0.001). Significance determined using a one-way ANOVA with a Dunnett’s multiple comparison test single pooled variance. NTC = no template control. a.u. = arbitrary units. IU = infectious units.
3.4. Evaluation of spiked human urine samples
CMV can be detected in urine from infected neonates, so sensitivity of this assay was determined using CMV spiked urine samples. UL123 and US28 targets were tested. UL123 had a limit of detection in urine of 0.1 IU/mL for all CMV strains achieving significance at 0.1 IU/mL for TR, 100 IU/mL for Toledo, and 1 IU/mL for Towne (Fig. 6A–C). US28 exhibited similar results with the limit of detection being lowest using TR and Towne strains (0.1 IU/mL) (Fig. 6D and F). Toledo was detected at 10 IU/mL (Fig. 6E). Significance was observed at 100, 10, and 100,000 IU/mL for TR, Toledo, and Towne respectively.
Fig. 6. Limit of detection in urine.
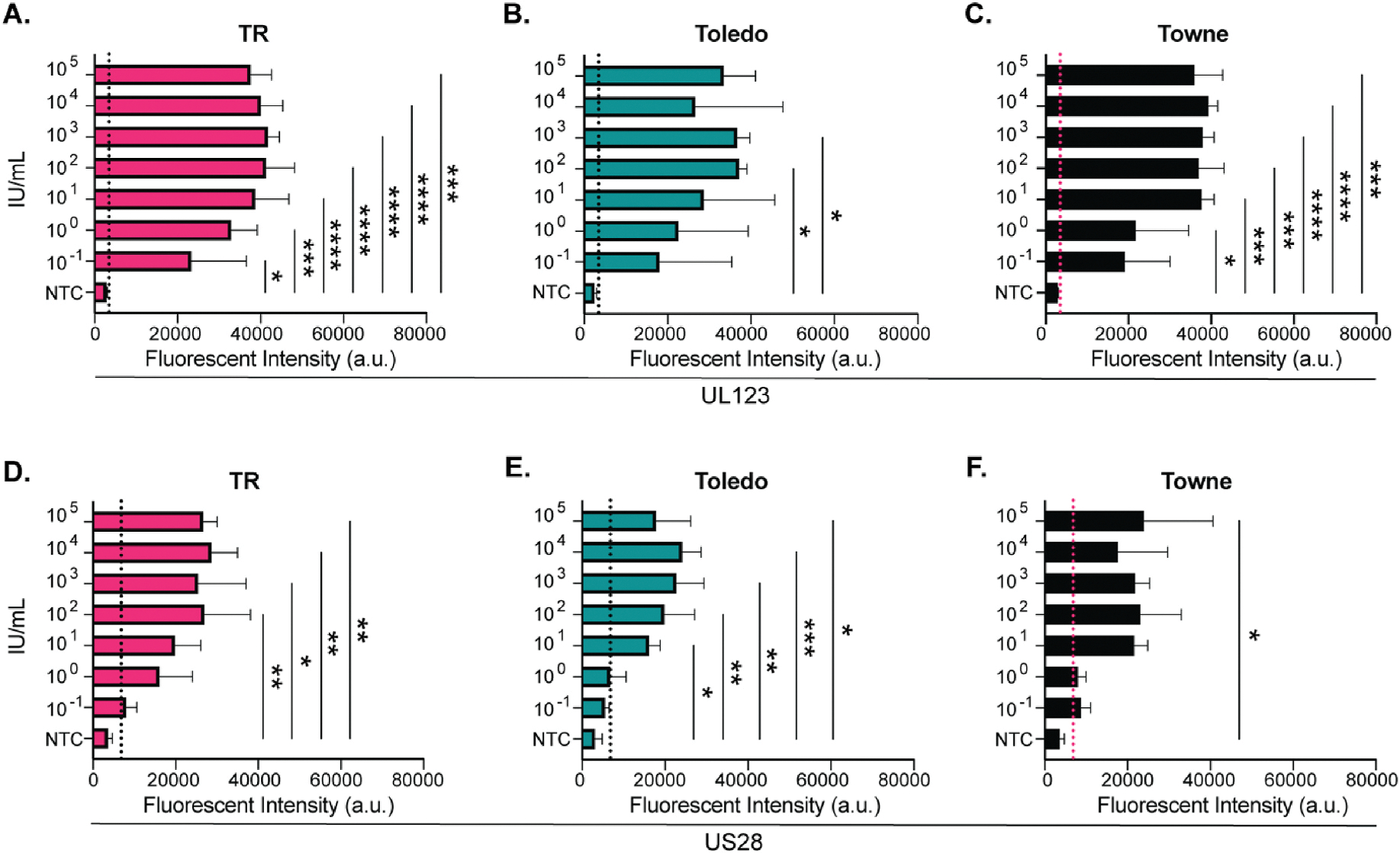
CMV was serially diluted (IU/mL) into human urine, followed by DNA extraction and PCR amplification before CRISPR-based fluorescence detection. Limit of detection of UL123 targets using A) TR, B) Toledo, or C) Towne. Limit of detection of US28 targets using D) TR, E) Toledo, or F) Towne. N = 3. Positive is calculated by the mean of the NTC + (3*SD) and is indicated by the dotted line. *(P < 0.05), **(P < 0.01), ***(P < 0.001), ****(P < 0.001). Significance determined using a one-way ANOVA with a Dunnett’s multiple comparison test single pooled variance. NTC = no template control. a.u. = arbitrary units. IU = infectious units.
3.5. Evaluation of detection time
Last, we characterized how fast the CRISPR-Cas12a reaction was able to detect CMV. Again, using a range of CMV IU (105 to 10−1 IU/mL), detection of CMV as measured by fluorescent intensity was plotted against time. Consistent with the limit of detection results (Fig. 3), we were able to consistently detect CMV at 101 IU/mL within 15 min after addition of the amplified sample (Fig. 7A–L) using UL123 and US28 CMV targets. The total length of this assay is approximately 90 min: 5 min for DNA extraction, 30 min of PCR amplification, 15 min for CRISPR-Cas12a fluorescent detection, and 30 min for sample preparation between steps. Strain-specific differences and CMV target-specific differences were observed. Regardless, this data supports the potential for rapid analysis using CRISPR-Cas12a detection after target amplification. This finding further substantiates the potential of this assay to detect diverse CMV strains using various bodily fluids.
Fig. 7. Rapid detection of CMV products using CRISPR-Cas12a assay.
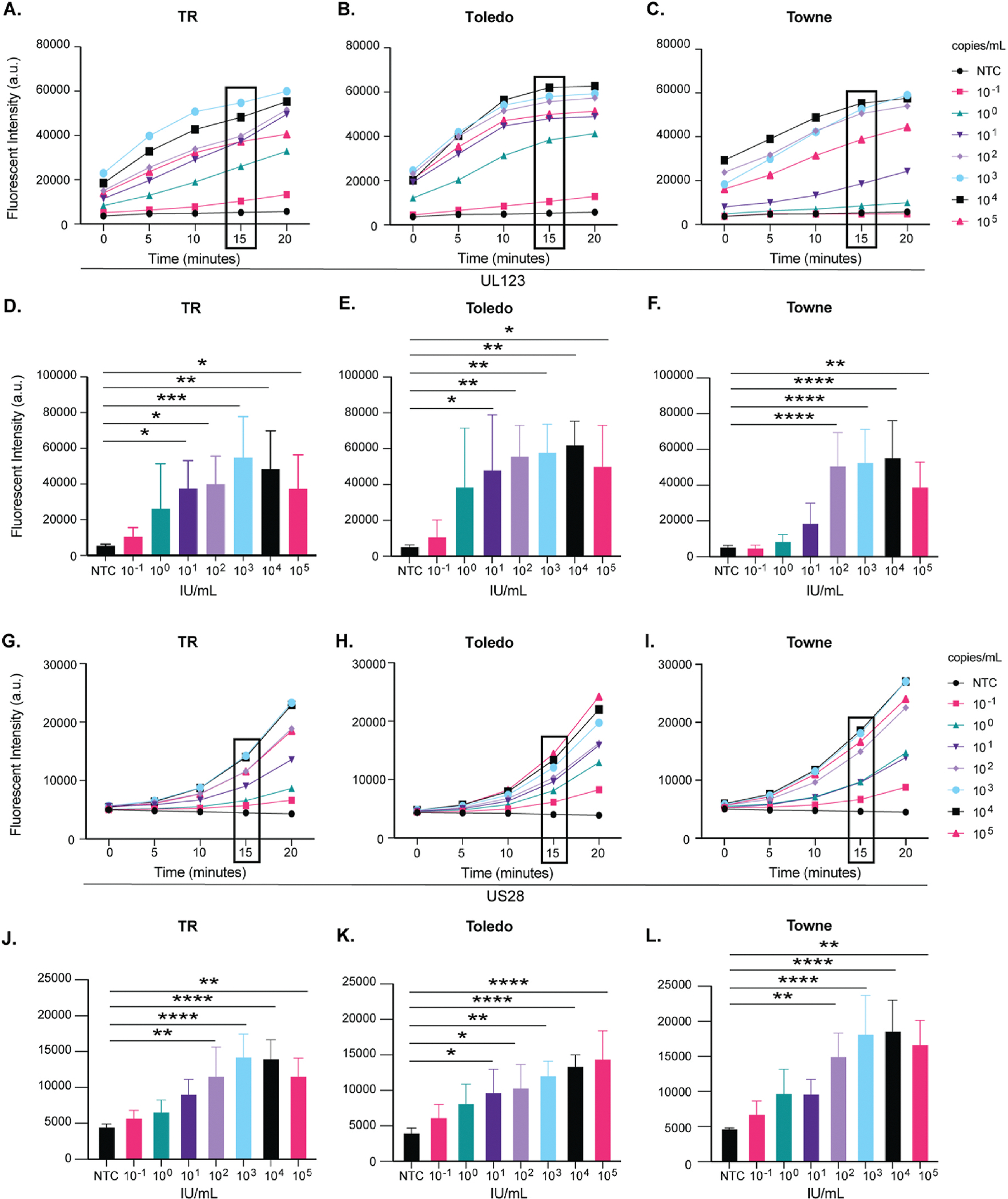
Fluorescent intensity progression at 5-min intervals over 20 min showing UL123 detection in A) TR, B) Toledo, C) Towne and US28 in G) TR, H) Toledo, and I) Towne. (N = 3). The 15-min timepoint of UL123 detection (black box) is displayed with significance in D) TR, E) Toledo, and F) Towne and US28 detection (black box) for J) TR, K) Toledo, and L) Towne respectively. *(P < 0.05), **(P < 0.01), ***(P < 0.001), ****(P < 0.001). NTC = no template control. a.u. = arbitrary units. IU = infectious units.
4. Discussion
Implementation of a robust, quantitative, cost-effective point-of-care diagnostic for CMV has the potential to significantly reduce CMV-associated morbidity and mortality. Rapid results or universal screening can prevent or reduce CMV associated pathologies, improving patient outcomes and lessening the enormous economic burden resulting from CMV-associated disease. In this study, we describe the design, optimization, and validation of a CRISPR-Cas12a CMV rapid diagnostic. The assay was tested using human saliva and urine in which CMV is commonly detected. Collectively, these results provide a path toward the development of a rapid diagnostic for CMV that can be used to minimize the severity of CMV-associated congenital infections. A direct comparison between this assay and the current standard assay using neonatal urine and saliva patient samples must be completed prior to definitive conclusions.
We validated this rapid diagnostic test using biologically relevant samples. Urine samples have been found to be reliable indicators of cCMV. Significantly higher viral loads are observed in infant urine samples than umbilical cord blood or cerebral spinal fluid (CSF) (Halwachs-Baumann et al., 2002). This observation may suggest that urine is representative of early CMV infection. Additionally, difficulty with plasma CMV isolation has previously been observed (Kaminski et al., 2020b). Urine collection is simple and requires no medical training, making it ideal for universal CMV screening in a medical facility or monitoring in an at home setting. Urine poses unique obstacles such as PCR inhibition due to high urea concentrations (>50 mM) while testing for CMV (Khan et al., 1991). Inhibition of amplification complicated by a high urea content in urine samples may display a lower limit of detection, which prompted us to test how this might hinder this assay.
We also tested saliva in parallel and results were similar to previously reported CMV detection sensitivity (Yamamoto et al., 2006; Ross et al., 2014). Previous publications using RT-PCR to detect cCMV in saliva report a limit of detection of approximately 200 copies/mL or failed to report quantification (Ross et al., 2014), (eurofins-viracor. Cytomegalovirus, 2022). It should be noted that a study by Ross et al., did not include a viral DNA extraction step (Ross et al., 2014). This may explain lower sensitivity versus the results reported here. This approach also provides insight into improving the CRISPR-Cas12a assay and transitioning to a true point-of-care assay. Saliva provides a simple and less invasive collection process than blood and urine making it the preferred sample type for this assay.
There are two key limitations to this study. This assay was not directly used on fresh clinical samples and this study did not validate the use of the CRISPR-Cas12a assay with whole blood. Expanded use of this assay to other relevant clinical cohorts, such as transplantation patients, would require experimental confirmation that results reported in this manuscript can be replicated using blood. Clinical samples may have a lower sensitivity due to PCR-inhibitory molecules in the sample that interfere with amplification (Sidstedt et al., 2018). In clinical validation, we plan to use the WHO standard for CMV to standardize these results using international units and to directly compare to current standard of care assays. Without this standardization, we have chosen to report the results in infectious units/mL, as it more accurately represents the samples used in this study. Unfortunately, infectious units/mL is determined by viral titering, which adds an additional factor of error and could impact the limit of detection. Also, similar to PCR based assays, this assay does not distinguish between infectious virus and viral DNA. This is important when interpreting results from patients on letermovir as they may have DNAemia but not infectious virus. Understanding the limitations of this assay using a range of clinical samples will provide insights into areas of improvement or reassessment of specificity and/or limit of detection. We are currently validating the CRIPSR-Cas12a system on transplant patient samples, directly comparing the rapid diagnostic results to the standard of care (RT-qPCR) results. An area of interest that would benefit this assay is the elimination of the in vitro amplification of nucleic acid, currently included in this protocol. The current protocol for this assay does not eliminate the need for PCR-based amplification. We are currently testing a number of methods to eliminate this step from the protocol, enabling this diagnostic to be a true rapid, point-of-care diagnostic.
Under transplantation settings, the current standard of care for CMV includes blood draws and RT-qPCR analysis for CMV viremia. Antiviral concentrations are adjusted accordingly after the return of results. An assay that can return results immediately could increase antiviral efficacy and shorten antiviral schedules, while also decreasing antiviral toxicity and viral resistance concerns. Current diagnostic kits cite an in vitro limit of detection of 0.238 International Units/μL (approximately 400 copies/mL) (altona-diagnostics. RealStar CMV PCR Kit, 2017), with most commercially available tests displaying a limit of detection of 100 copies/mL or higher (Aruplab. Cytomegalovirus by Quantitative PCR, 2022). Some assays can detect 10 copies/mL but specificity is limited as a result (94%) (Waggoner et al., 2012; fda.gov. Abbott RealTime CMV, 2017). A similar PCR-based assay used the CMV UL123 region for detection reported a limit of detection of 300 International Units/mL (534 copies/mL) (testguide.labmed.uw.edu. CMV Quantitative by PCR, 2022). Validation of this CRISPR-Cas12a assay using blood, could potentially permit integration of this assay with current diagnostics. This could increase specificity and the limit of detection of current assays.
The aim in this manuscript was to provide a proof-of-concept design to validate further exploration using CRISPR-Cas12a technology as a rapid diagnostic for CMV. There were unexpected results during this study. The CRISPR-Cas12a targets were designed using an identical approach but yield a wide variation in results. We are uncertain why the UL122 failed to produce a signal. We also do not have a strong explanation for the difference in sensitivity between UL123 and US28. Experiments are continuing to further define biochemical factors that may impact CRISPR-Cas12a design. An advantage of the CRISPR-Cas12a system, is that multiple targets can be integrated into a single assay. We are testing other CMV targets that could be included in future tests. Alternatively, different CMV panels may be developed to provide results for distinct clinical requirements. The results show high specificity to CMV, high sensitivity with accurate measurements below 100 IU/mL, and detection of CRISPR-Cas12a tagged targets from a pre-amplified product in under 20 min. These results were completed using saliva and urine, further showing the promise of future development of a point-of-care CMV diagnostic. The goal is to validate this CRISPR-Cas12a CMV rapid diagnostic in clinical trials in the near future after continued improvement and validation.
In summary, this study shows the promise of a CRISPR-Cas12a rapid diagnostic for the detection of CMV in saliva and urine. This assay can be applied to other body fluids such as blood or ocular fluid to detect CMV infection. This assay is low-cost, easy to use and quantitative, permitting repeated testing during CMV-associated complications. We are further validating this assay in various human clinical samples including neonatal samples to prove the feasibility and affordability of a universal neonatal CMV screen. Achieving an at-home version would lessen the requirement of a visit to the doctor’s office and is of great interest in future work.
Supplementary Material
Acknowledgements
We appreciate the generous support of Dr. Elizabeth Norton for providing urine samples used in this manuscript. This work was supported in part by NIH grants 1R21AI169582–01A1 (B.N.), U54GM104940, P20GM103629 (K.J.Z), 1R01HD107790 (K.J.Z), HHV-6 Foundation Pilot Grant (K.J.Z), U54CA260581 (K.J.Z & J.G.S).
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2023.105624.
Data availability
Data will be made available on request.
References
- Altona-diagnostics. RealStar CMV PCR kit 1.0. https://www.altona-diagnostics.com/files/public/Content%20Homepage/-%2002%20RealStar/MAN%20-%20CE%20-%20EN/RealStar%20CMV%20PCR%20Kit%201.0_WEB_CE_EN-S02.pdf, 2017.
- Aruplab. Cytomegalovirus by Quantitative PCR, 2022. https://ltd.aruplab.com/Tests/Pub/0051813.
- Bate SL, Dollard SC, Cannon MJ, 2010. Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988–2004. Clin. Infect. Dis. 50, 1439–1447. 10.1086/652438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boppana SB, Fowler KB, 2017. Insight into long-term neurodevelopmental outcomes in asymptomatic congenital CMV infection. Pediatrics 140. 10.1542/peds.2017-2526. [DOI] [PubMed] [Google Scholar]
- Britt WJ, 2010. Human cytomegalovirus: propagation, quantification, and storage. Curr Protoc Microbiol Chapter 14. 10.1002/9780471729259.mc14e03s18. Unit 14E.13. [DOI] [PubMed] [Google Scholar]
- Broughton JP, et al. , 2020. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38, 870–874. 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon MJ, Schmid DS, Hyde TB, 2010. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 20, 202–213. 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- Combs JA, et al. , 2019. Human cytomegalovirus alters host cell mitochondrial function during acute infection. J. Virol. 10.1128/JVI.01183-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollard SC, Grosse SD, Ross DS, 2007. New estimates of the prevalence of neurological and sensory sequelae and mortality associated with congenital cytomegalovirus infection. Rev. Med. Virol. 17, 355–363. 10.1002/rmv.544. [DOI] [PubMed] [Google Scholar]
- Duan Z, et al. , 2022. Risk factors and survival of refractory cytomegalovirus reactivation after allogeneic peripheral blood stem cell transplantation. J Glob Antimicrob Resist 31, 279–285. 10.1016/j.jgar.2022.10.009. [DOI] [PubMed] [Google Scholar]
- Eurofins-Viracor. Cytomegalovirus (CMV) Saliva Real-Time PCR, 2022. https://www.eurofins-viracor.com/clinical/test-menu/5571-cytomegalovirus-cmv-saliva-real-time-pcr/.
- fda.gov. Abbott RealTime CMV. https://www.accessdata.fda.gov/cdrh_docs/pdf16/P160044B.pdf.
- Fishman JA, 2007. Infection in solid-organ transplant recipients. N. Engl. J. Med. 357, 2601–2614. 10.1056/NEJMra064928. [DOI] [PubMed] [Google Scholar]
- Foulon I, et al. , 2019. Hearing loss with congenital cytomegalovirus infection. Pediatrics 144. 10.1542/peds.2018-3095. [DOI] [PubMed] [Google Scholar]
- Fowler KB, Boppana SB, 2006. Congenital cytomegalovirus (CMV) infection and hearing deficit. J. Clin. Virol. 35, 226–231. 10.1016/j.jcv.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Fowler KB, Boppana SB, 2018. Congenital cytomegalovirus infection. Semin. Perinatol. 42, 149–154. 10.1053/j.semperi.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Halwachs-Baumann G, et al. , 2002. Human cytomegalovirus load in various body fluids of congenitally infected newborns. J. Clin. Virol. 25, 81–87. 10.1016/S1386-6532(02)00188-9. [DOI] [PubMed] [Google Scholar]
- Hayden RT, et al. , 2017. Progress in quantitative viral load testing: variability and impact of the WHO quantitative international standards. J. Clin. Microbiol. 55, 423–430. 10.1128/JCM.02044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, et al. , 2020. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 164, 112316 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, et al. , 2021. Sensitive tracking of circulating viral RNA through all stages of SARS-CoV-2 infection. J. Clin. Invest. 131 10.1172/jci146031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay HN, Kaul DR, 2021. Letermovir and Maribavir for the Treatment and Prevention of Cytomegalovirus Infection in Solid Organ and Stem Cell Transplant Recipients, vol. 73. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, pp. 156–160. 10.1093/cid/ciaa1713. [DOI] [PubMed] [Google Scholar]
- Ito Y, et al. , 2013. Risk factors for poor outcome in congenital cytomegalovirus infection and neonatal herpes on the basis of a nationwide survey in Japan. Pediatr. Int. 55, 566–571. 10.1111/ped.12122. [DOI] [PubMed] [Google Scholar]
- Kaminski MM, et al. , 2020a. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nat Biomed Eng 4, 601–609. 10.1038/s41551-020-0546-5. [DOI] [PubMed] [Google Scholar]
- Kaminski MM, et al. , 2020b. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nature Biomedical Engineering 4, 601–609. 10.1038/s41551-020-0546-5. [DOI] [PubMed] [Google Scholar]
- Kenneson A, Cannon MJ, 2007. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev. Med. Virol. 17, 253–276. 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- Khan G, Kangro HO, Coates PJ, Heath RB, 1991. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J. Clin. Pathol. 44, 360–365. 10.1136/jcp.44.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, et al. , 2017. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8, 14406 10.1038/ncomms14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin DW, et al. , 2015. Valganciclovir for symptomatic congenital cytomegalovirus disease. N. Engl. J. Med. 372, 933–943. 10.1056/NEJMoa1404599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosky PM, et al. , 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72, 4721–4728. 10.1128/jvi.72.6.4721-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leruez-Ville M, Foulon I, Pass R, Ville Y, 2020. Cytomegalovirus infection during pregnancy: state of the science. Am. J. Obstet. Gynecol. 223, 330–349. 10.1016/j.ajog.2020.02.018. [DOI] [PubMed] [Google Scholar]
- Li M, et al. , 2021. Non-human primate models to investigate mechanisms of infection-associated fetal and pediatric injury, teratogenesis and stillbirth. Front. Genet. 12, 680342 10.3389/fgene.2021.680342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Smith BM, Jain PK, 2020. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 11, 4906. 10.1038/s41467-020-18615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, et al. , 2016. Neurological outcomes in symptomatic congenital cytomegalovirus-infected infants after introduction of newborn urine screening and antiviral treatment. Brain Dev. 38, 209–216. 10.1016/j.braindev.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Parry HM, et al. , 2016. Cytomegalovirus viral load within blood increases markedly in healthy people over the age of 70 years. Immun. Ageing 13, 1. 10.1186/s12979-015-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiksaitis JK, et al. , 2016. Are we there yet? Impact of the first international standard for cytomegalovirus DNA on the harmonization of results reported on plasma samples. Clin. Infect. Dis. 63, 583–589. 10.1093/cid/ciw370. [DOI] [PubMed] [Google Scholar]
- Rawlinson WD, et al. , 2017. Congenital cytomegalovirus infection in pregnancy and the neonate: consensus recommendations for prevention, diagnosis, and therapy. Lancet Infect. Dis. 17, e177–e188. 10.1016/S1473-3099(17)30143-3. [DOI] [PubMed] [Google Scholar]
- Ross SA, et al. , 2014. Detection of congenital cytomegalovirus infection by real-time polymerase chain reaction analysis of saliva or urine specimens. J. Infect. Dis. 210, 1415–1418. 10.1093/infdis/jiu263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidstedt M, et al. , 2018. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal. Bioanal. Chem. 410, 2569–2583. 10.1007/s00216-018-0931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorichetti B, et al. , 2016. Symptomatic congenital cytomegalovirus infection is underdiagnosed in British columbia. J. Pediatr. 169, 316–317. 10.1016/j.jpeds.2015.10.069. [DOI] [PubMed] [Google Scholar]
- Teira P, et al. , 2016. Early cytomegalovirus reactivation remains associated with increased transplant-related mortality in the current era: a CIBMTR analysis. Blood 127, 2427–2438. 10.1182/blood-2015-11-679639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- testguide.labmed.uw.edu. CMV Quantitative by PCR. https://testguide.labmed.uw.edu/view/CMVQN?, 2022.
- Vallejo M, et al. , 2022. Risk prediction of CMV reactivation after allogeneic stem cell transplantation using five non-HLA immunogenetic polymorphisms. Ann. Hematol. 101, 1567–1576. 10.1007/s00277-022-04841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boven M, et al. , 2017. Infectious reactivation of cytomegalovirus explaining age- and sex-specific patterns of seroprevalence. PLoS Comput. Biol. 13, e1005719 10.1371/journal.pcbi.1005719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner J, Ho DY, Libiran P, Pinsky BA, 2012. Clinical significance of low cytomegalovirus DNA levels in human plasma. J. Clin. Microbiol. 50, 2378–2383. 10.1128/jcm.06800-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, et al. , 2020. CRISPR-assisted DNA detection: a novel dCas9-based DNA detection technique. CRISPR J 3, 487–502. 10.1089/crispr.2020.0041. [DOI] [PubMed] [Google Scholar]
- Yamada H, et al. , 2020. A cohort study of the universal neonatal urine screening for congenital cytomegalovirus infection. J. Infect. Chemother. 26, 790–794. 10.1016/j.jiac.2020.03.009. [DOI] [PubMed] [Google Scholar]
- Yamamoto AY, et al. , 2006. Is saliva as reliable as urine for detection of cytomegalovirus DNA for neonatal screening of congenital CMV infection? J. Clin. Virol. 36, 228–230. 10.1016/j.jcv.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. , 2022. The epidemiology and disease burden of congenital TORCH infections among hospitalized children in China: a national cross-sectional study. PLoS Neglected Trop. Dis. 16, e0010861 10.1371/journal.pntd.0010861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


