Abstract
Aim
This cross-sectional survey aimed to identify aerobic bacteria, antimicrobial resistance, and multi-drug resistance profiles of bacteria isolated from different wound infections among a group of Egyptian patients.
Results
Of 120 positive samples, 170 isolates were identified. Polymicrobial infections were determined in 55% of samples. The dominant Gram-positive isolated strains were Staphylococcus aureus, especially from wound infections because of accidents (71.8%). Piperacillin, methicillin, ampicillin/sulbactam, and amoxicillin/clavulanic acid were all highly resistant to S. aureus and Coagulase-negative Staphylococci. The prevalence of methicillin-resistant S. aureus in wound infections was 89.9%. S. aureus showed superior sensitivity to vancomycin (85.3%) and linezolid (81.3%). The highest prevalence of Gram-negative isolates was for Pseudomonas aeruginosa (40%), which was highly sensitive to ciprofloxacin (79.2%) and highly resistant to levofloxacin (83.3%). Several isolates revealed a multi-drug resistance profile (52.4%). The overall MDR rate of Gram-positive and Gram-negative isolates were 50% and 54.9%, respectively.
Conclusion
The prevalence of MRSA isolated from various wound infections and MDR is a warning issue in Upper Egypt. It should implement a health education strategy and hygiene measures to prevent the spread of wound infection-causing organisms in the community.
Keywords: Egypt, Prevalence, Wound infection, S. aureus, P. aeruginosa, MRSA, MDR
Introduction
Skin provides a good medium for pathologic bacteria to proliferate. Subsequently, the chance of a skin wound being infected is boosted, and interferes with the healing process [1]. The origin of wounds varies, from acute postsurgical wounds (i.e., surgical site infections), post-traumatic wounds (i.e., wounds following an accident or burns), or chronic wounds such as those associated with diabetes mellitus (e.g. diabetic foot ulcers) [2].
The rate of MDR bacteria elucidates a worldwide increase as announced by the Centers for Disease Control and Prevention (CDC) [3]. This phenomenon has negative implications for the healthcare system and increases the threat of antibiotic failure, which raises the mortality rates [4]. More than 90% of S. aureus are resistant to penicillin, and that remains a global issue [5]. Despite that, MRSA strains have the tendency to expand quickly within a health facility via colonized or infected patients or health professionals, as well as contaminated sites within the facility [6]. Normally, antibiotic resistance gradually emerges. The injudicious administration of broad-spectrum topical and systemic antibiotics hastens the emergence of resistant bacteria [7]. Unfortunately, in developing countries, the unwise use problem of antibiotics is exacerbated by the absence of strict precautions to dispense the antibiotics without a medical prescription [8].
Regardless the type of wound infection, it is important to monitor the dynamic changes in the prevalence of sensitivity profiles and MDR profiles over time. This is important in detecting a structured therapeutic strategy to prevent microbial proliferation while avoiding side effects [9]. Regarding the MDR of bacteria isolated from different kinds of wound infection in Upper Egypt, the current survey was conducted to identify aerobic bacteria isolated from a group of Egyptian patients with different types of wounds and burn infections attend Minia University Hospital. Furthermore, to detect the antimicrobial resistance profile and MDR profile of different bacterial isolates.
Materials and methods
Design and setting
The study designed as a cross-sectional design that has been carried out from November 2019 to September 2021. The wound samples were collected from the patients who attended the Department of Plastic and Reconstructive Surgery, Minia University Hospital (MUH). The hospital provides its services to a geographical area of approximately 6 million people in Upper Egypt. The hospital included 330 beds.
Demographic data of the study population
The study included males and females aged 1 month to 60 years. Wound infection is suspected if a wound was not healing well, getting bigger, and exudation of pus or fluids. Samples were obtained from various wound types including burn wounds, surgical wounds from different anatomical sites, and abscesses. Specimens were properly labeled, indicating the source, gender, and age of patients.
Sample size
A total of 146 randomly selected wound swabs were collected from MUH. Wound infection is suspected when a wound is not healing properly, grows in size, or exudes pus or fluids. The specimens were collected on sterile cotton swabs and the wounds were cleaned before collection to avoid surface contamination. Swabs are transferred immediately, within 2 h to the laboratory. In the laboratory, the specimens were registered, and swabs were cultured, streaked, and incubated at 37 °C for 24 h. After incubation, plates were checked for bacterial growth. Plates with negative bacterial growth were additionally incubated for another 24 h.
Ethical consideration
The hospital collects samples from different clinical sources on a daily basis from patients and sends them to the hospital’s laboratories for analysis. Samples were collected from hospital laboratories without dealing with patients directly. Upon getting permission from the Head of Plastic and Reconstructive Surgery Department, Faculty of Medicine, Minia University, the collection of laboratory samples has begun.
Isolation and identification of wound bacterial isolates
The microorganisms were identified by Gram stain, culturing, biochemical reactions, and motility testing as per standard guidelines [10]. Culture plates of Nutrient agar, Muller Hinton agar, nutrient broth, Cetrmide agar, Mannitol salt agar (MSA), Brain heart infusion broth, Triple sugar iron (TSI) agar and DNase agar (Oxoid, England). Simmon citrate agar (CONDA). MacConkey agar, Eosin methylene blue, and Sulphide indo motility test (SIM), all produced by Himedia Laboratories, India. All media were prepared according to the instructions of the manufacturers. The media were sterilized by autoclaving at 121ºC for 15 min. Subcultures were then made into plates of nutrient agar, MSA, and MacConkey agar and incubated for another 24 h. The primary identification of the bacterial isolates was based on colonial appearance, pigmentation, morphology, and Gram staining characteristics using a light microscope. Biochemical tests were performed to identify the isolates. Biochemical tests were the standard catalase test, coagulase (tube and slide) test, DNase test, triple sugar iron test, Simmon citrate test, Indole, and sulphide production motility. Colonies were maintained by storing them at -80˚C in stocks with 2.5 M glycerol.
Antibiotic sensitivity testing
Antimicrobial sensitivity was determined by the Kirby Bauer agar disc diffusion method. A small inoculum of each pure bacterial isolate was emulsified in 2 mL sterile normal saline. The turbidity of the cell suspension was adjusted to correspond to 0.5 MCfarland standard (1.5 × 108 CFU/mL). The inoculum was dispensed on the surface of Mueller-Hinton agar plate and ramified with a sterile metallic wire loop and the plates were allowed to dry for 3–5 min. Antibiotic discs were used with the following concentrations: linezolid (30 µg, Bioanalyse limited -Turkey), tetracycline (30 µg, Himedia India), chloramphenicol (30 µg, Bioanalyse limited -Turkey), rifampin (5 µg, Himedia India), piperacillin (100 µg, Bioanalyse limited -Turkey), amoxicillin/ clavulanic acid (30 µg, Bioanalyse limited -Turkey), ampicillin/ sulbactam (20 µg, Bioanalyse limited -Turkey), levofloxacin (5 µg, Himedia India), gentamicin (10 µg, Himedia India), vancomycin (30 µg, Himedia India), cefoxitin (30 µg, Sigma USA), ciprofloxacin (5 µg, Bioanalyse limited -Turkey), cefuillin/tazobactam (75/10 µg, Himedia India), cefuroxime (30 µg, Himedia India), ceftazidime( 30 µg, Himedia India ), tigecycline( 15 µg, Himedia India), cefazolin (30 µg, Himedia India ), trimethoprim-sulfamethoxazole (1.25/23.75 µg, Himedia India), and imipinem (10 µg, Himedia India). Antibiotic discs were applied on the surface of the plates at least 15 mm apart from the edges of the plates to prevent overlapping of inhibition zones. The plates were incubated at 37oC for 24 h and the diameters of zones of inhibition were measured in mm and the results are compared to those from the Clinical Laboratory Standard Institute (CLSI) (CLSI, 2018). S. aureus isolates were considered MRSA when the diameter of inhibition zone of cefoxitin disc is ≤ 21 mm according to CLSI (CLSI, 2018).
Statistical analysis
Statistical analyses were performed using the chi-square or Fisher’s exact test using SPSS software version 16 (SPSS, Inc., Chicago, IL, USA). The results were considered statistically significant when the P value was less than 0.05.
Results
Participant’s demographic characteristics and wound samples
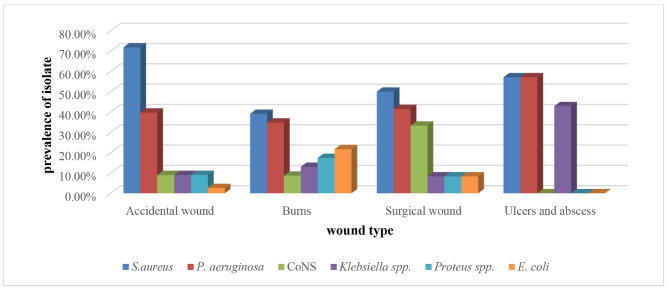
Of 146 samples, bacterial growth was evident in 120 (82.2%) samples, and 26 cultures (17.8%) were clear. The majority of participants were males, representing 73.5% (n = 88) and the rest was females represented 26.7% (n = 32). The ages of the patients ranged from 1 month to 60 years, with an average of 25.28 ± 16.21. The frequencies of samples isolated from different wound infections were shown in Fig. 1.
Fig. 1.
Prevalence of isolates from patients suffering from different types of wound infections. % was calculated out of number of samples obtained from a certain kind of wound
Bacterial isolate identification and distribution among wound infections
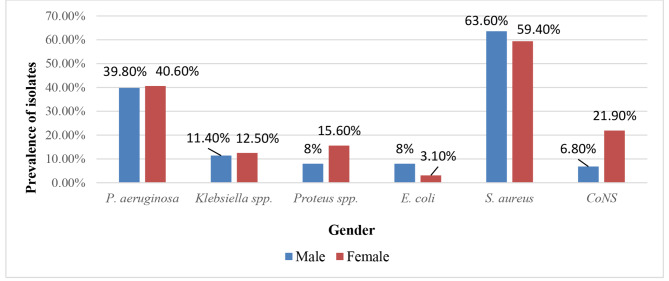
Out of 120 samples, 170 isolates were identified. Poly-microbial infections were determined in 66 (55%) samples, while mono-microbial infections were detected in 54 (45%) samples. There were 88 Gram-positive isolates (51.8%) and 82 Gram-negative isolates (48.2%). The dominant strains isolated from all types of wound infections were S. aureus (62.5%) followed by P. aeruginosa (40%), Klebsiella spp. (11.6%), CoNS (10.8%) Protues spp. (10%), and E. coli (6.67%) (Fig. 1). Figure 2 shows the prevalence of bacteria isolated from different types of wound infections in relation to gender. Regarding isolates, 121 isolates (71.2%) were isolated from males. While 49 isolates (28.8%) were isolated from females. The prevalence of S. aureus and E. coli was greater in males. While the prevalence of CoNS, P. aeruginosa, Klebsiella spp., and Proteus spp., was greater in females.
Fig. 2.
Prevalence of bacteria isolated from different types of wound infections in relation to gender. % of bacterial isolates was calculated out of total number of males or females
Antibiotic sensitivity and resistance profiles of Gram-positive isolates
Both S. aureus and CoNS showed high resistance to piperacillin, cefoxitin, ampicillin/sulbactam, and amoxicillin/clavulanic acid. MRSA was detected in 89.9% of S. aureus isolates. The details of sensitivity and resistance profiles are mentioned in Table 1. The association between susceptibility or resistance to various antibiotics among S. aureus or CoNS was significant in case of rifampin, piperacillin, gentamicin, levofloxacin and ciprofloxacin (p < 0.05).
Table 1.
Antimicrobial sensitivity and resistance profile of Gram-positive isolated from wound and burn infections
| Antibiotic |
S. aureus
(N = 75) |
CoNS (N = 13) |
P value* | ||||
|---|---|---|---|---|---|---|---|
| S% | I (%) | R% | S% | I (%) | R% | ||
| Linezolid | 61 (81.3%) | 0 (0) | 14 (18.6%) | 11 (84.6%) | 0 (0) | 2 (15.4%) | 0.777 |
| Tetracycline | 22 (29.3%) | 7 (9.3) | 46 (61.3%) | 5 (38.5%) | 3 (23.1) | 5 (38.5%) | 0.273 |
| Chloramphenicol | 52 (69.3%) | 14 (18.6) | 9 (12%) | 11(84.6%) | 1 (7.7) | 1 (7.7%) | 0.554 |
| Rifampin | 43 (57.3%) | 3 (4) | 29 (38.7%) | 9 (69.2%) | 1 (7.7) | 3 (23.1%) | 0.018 |
| Piperacillin | 1 (1.3%) | 0 (0) | 74 (98.7%) | 2 (15.4%) | 0 (0) | 11 (84.6%) | 0.010 |
| Ampicillin / Sulbactam | 8 (10.7%) | 0 (0) | 67 (89.3%) | 1 (7.7%) | 0 (0) | 12 (92.3%) | 0.744 |
| Amoxicillin/ Clavulanic | 8 (10.7%) | 0 (0) | 67 (89.3%) | 1 (7.7%) | 0 (0) | 12 (92.3%) | 0.744 |
| Cefoxitin | 8 (10.7%) | 0 (0) | 67 (89.3%) | 1 (7.7%) | 0 (0) | 12 (92.3%) | 0.744 |
| Gentamycin | 37 (49.3%) | 11(14.7) | 27 (36%) | 12 (92.3%) | 0 (0) | 1 (7.7%) | 0.018 |
| Levofloxacin | 44 (58.67%) | 5 (0.7) | 26 (34.67%) | 12 (92.3%) | 0 (0) | 1 (7.7%) | 0.002 |
| Ciprofloxacin | 41 (54.67%) | 8 (10.7) | 26 (34.67%) | 12 (92.3%) | 0 (0) | 1 (7.7%) | 0.030 |
| Vancomycin | 64 (85.3%) | 0 (0) | 11 (14.67%) | 11 (84.6%) | 0 (0) | 2 (15.4%) | 0.976 |
* X2 test was used for statistical analysis and P values are significant at < 0.05
Antibiotic sensitivity and resistance profiles of Gram-negative isolates
Data in Table 2 shows various susceptibility and resistance profiles to different antibiotics. For instance, P. aeruginosa was sensitive to ciprofloxacin (79.2%) and highly resistant to cefazolin (100%) and levofloxacin (83.3%). Klepsiella spp. were sensitive to gentamicin (78.6%) and highly resistant to cefazolin (100%) and ceftazidime (85%). Protius spp. was highly resistant to ampicillin/sulbactam (100%), cefazolin (100%) and ceftazidime (91.9%). Finally, E. coli showed absolute resistance against cefuroxime and cefazolin (100%) and high resistance against ampicillin/sulbactam (87.5%). In the case of levofloxacin and ciprofloxacin, the association between susceptibility or resistance to various antibiotics was significant (p < 0.05).
Table 2.
Antimicrobial sensitivity and resistance profile of Gram-negative organisms isolated from wound and burn infections
| Antibiotic |
P. aeruginosa
(N = 48) |
Klebsiella spp. (N = 14) |
Proteus spp. (N = 12) |
E. coli
(N = 8) |
P value* | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S% | I (%) | R% | S% | I (%) | R% | S% | I (%) | R% | S% | I (%) | R% | ||
| Chloramphenicol | 21 (43.8%) | 10 (20.8) | 17 (35.4%) | 6 (42.9%) | 2 (14.3) | 6 (42.9%) | 8 (66.7%) | 3 (25) | 1 (8.3%) | 5 (62.5%) | 0 (0) | 3 (37.5%) | 0.262 |
| Tigycyclin | 17 (35.4%) | 18 (37.5) | 13 (27.1%) | 1 (7.1%) | 9 (64.3) | 4 (28.6%) | 4 (33.3%) | 8 (66.7) | 0 (0%) | 2 (25%) | 6 (75) | 0 (0%) | 0.186 |
| Gentamycin | 29 (60.4%) | 3 (6.3) | 16 (33.3%) | 11 (78.6) | 1 (7.1) | 2 (14.3%) | 6 (50%) | 1 (8.3) | 5 (41.7%) | 4 (50%) | 0 (0) | 4 (50%) | 0.318 |
| Levofloxacin | 8 (16.7%) | 0 (0) | 40 (83.3%) | 9 (64.3%) | 1 (7.1) | 4 (28.6%) | 7 (58.3%) | 1 (8.3) | 4 (33.3%) |
5 (62.5) |
1 (12.5) |
2 (25%) |
< 0.0001 |
| Ciprofloxacin | 38 (79.2%) | 2 (4.2) | 8 (16.7%) | 6 (42.9%) | 4 (28.6) | 4 (28.6%) | 7 (58.3%) | 1 (8.3) | 4 (33.3%) | 2 (25%) | 2 (25) | 4 (50%) | 0.042 |
| Ampicillin / Sulbactam | 4 (8.3%) | 8 (16.7) | 36 (75%) | 1 (7.1%) | 6 (42.9) | 7 (50%) | 0 (0%) | 0 (0) | 12 (100%) | 0 (0%) | 1 (12.5) | 7 (87.5%) | 0.532 |
| Amoxicillin/ Clavulanic | 11 (23%) | 5 (10.4) | 32 (66.7%) | 3 (21.4%) | 4 (28.6) | 7 (50%) | 0 (0%) | 6 (50) | 6 (50%) | 0 (0%) | 5 (62.5) | 3 (37.5%) | 0.189 |
|
Ticracillin /tazobactam |
15 (31.3%) | 15 (31.3) | 18 (37.5%) | 2 (14.3%) | 5 (35.7) | 7 (50%) | 0 (0%) | 6 (50) | 6 (50%) | 4 (50%) | 1 (12.5) | 3 (37.5%) | 0.090 |
| Cefuroxime | 9 (18.8%) | 9 (18.8) | 30 (62.5’%) | 3 (21.4%) | 2 (14.3) | 9 (64.3%) | 2 (16.7%) | 3 (25) | 7 (58.3%) | 0 (0%) | 0 (0) | 8 (100%) | 0.497 |
| Cefazolin | 0 (0%) | 0 (0) | 48 (100%) | 0 (0%) | 0 (0) | 14 (100%) | 0 (0%) | 0 (0) | 12 (100%) | 0 (0%) | 0 (0) | 8 (100%) | N/A |
| Ceftazidime | 6 (12.5%) | 7 (14.6) | 35 (73%) | 2 (14.3%) | 0 (0) | 12 (85.%) | 0 (0%) | 1 (8.3) | 11 (91.7) | 2 (25%) | 0 (0) | 6 (75%) | 0.445 |
| Imipineme | 24 (50%) | 8 (16.7) | 16 (33.3%) | 7 (50%) | 2 (14.3) | 5 (35.%) | 1 (8.3%) | 7 (58.3) | 4 (33.3%) | 4 (50%) | 1 (12.5) | 3 (37.5%) | 0.472 |
| Trimethoprim/ sulfamethoxazole | 19 (39.6%) | 8 (16.7) | 21 (43.8%) | 11 (78.6) | 0 (0) | 3 (21.%) | 7 (58.3%) | 0 (0) | 5 (41.7%) | 2 (25%) | 0 (0) | 6 (75%) | 0.053 |
* X2 test was used for statistical analysis and P values are significant at < 0.05
Gender and age group correlations with the antibiotic resistance profile
Gender was not significantly correlated with the antibiotic resistance profile among Gram-positive samples (67 patients) and Gram-negative samples (53 patients) (Tables 3 and 4, respectively). The correlations between age group and antibiotic resistance profile among Gram-positive and Gram-negative samples were illustrated in Tables 5 and 6, respectively. No statistically significant difference was detected between the resistance profiles of tested antibiotics among both Gram-positive and Gram-negative samples and the participant’s age group (p > 0.05).
Table 3.
Correlation between gender and antibiotic resistance profile among Gram-positive samples
| Antibiotic | Resistance profile | P-value* | ||
|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | ||
| Linezolid | ||||
|
Male Female |
38 15 |
0 0 |
9 5 |
0.547 |
| Tetracycline | ||||
|
Male Female |
9 3 |
9 0 |
29 17 |
0.079 |
| Chloramphenicol | ||||
|
Male Female |
31 14 |
9 3 |
7 3 |
0.919 |
| Rifampin | ||||
|
Male Female |
25 10 |
2 2 |
20 8 |
0.662 |
| Piperacillin | ||||
|
Male Female |
1 0 |
0 0 |
46 20 |
0.511 |
| Cefoxitin | ||||
|
Male Female |
3 1 |
0 0 |
44 19 |
0.827 |
| Gentamycin | ||||
|
Male Female |
22 8 |
7 4 |
18 8 |
0.828 |
| Ampicillin/Sulbactam | ||||
|
Male Female |
3 1 |
0 0 |
44 19 |
0.827 |
| Levofloxacin | ||||
|
Male Female |
3 1 |
0 0 |
44 19 |
0.827 |
| Ciprofloxacin | ||||
|
Male Female |
25 10 |
3 2 |
19 8 |
0.872 |
| Amoxicillin/Clavulanic | ||||
|
Male Female |
3 1 |
0 0 |
44 19 |
0.827 |
| Vancomycin | ||||
|
Male Female |
3 1 |
0 0 |
44 19 |
0.827 |
*Chi square test; P-value was set to 0.05
Table 4.
Correlation between gender and antibiotic resistance profile among Gram-negative samples
| Antibiotic | Resistance profile | P-value* | ||
|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | ||
| Ticracillin /Tazobactam | ||||
|
Male Female |
5 0 |
13 7 |
23 5 |
0.168 |
| Tigycyclin | ||||
|
Male Female |
9 2 |
21 5 |
11 5 |
0.314 |
| Chloramphenicol | ||||
|
Male Female |
17 4 |
7 1 |
17 7 |
0.547 |
| Cefuroxime | ||||
|
Male Female |
2 4 |
6 1 |
33 7 |
0.063 |
| Cefazolin | ||||
|
Male Female |
0 0 |
0 0 |
41 12 |
N/A |
| Imipineme | ||||
|
Male Female |
12 7 |
10 0 |
19 5 |
0.076 |
| Gentamycin | ||||
|
Male Female |
21 5 |
4 1 |
16 6 |
0.794 |
| Ampicillin/Sulbactam | ||||
|
Male Female |
3 1 |
3 2 |
35 9 |
0.609 |
| Amoxicillin/Clavulanic | ||||
|
Male Female |
3 3 |
9 0 |
29 9 |
0.073 |
| Ceftazidime | ||||
|
Male Female |
1 0 |
5 2 |
35 10 |
0.803 |
| Levofloxacin | ||||
|
Male Female |
27 8 |
3 0 |
11 4 |
0.601 |
| Ciprofloxacin | ||||
|
Male Female |
24 6 |
4 2 |
13 4 |
0.772 |
| Trimethoprim/Sulfamethoxazole | ||||
|
Male Female |
11 5 |
6 3 |
24 4 |
0.304 |
*Chi square test; P-value was set to 0.05; N/A: not applicaple
Table 5.
Correlation between age group and antibiotic resistance profile among Gram-positive samples
| Antibiotic | Resistance profile | P-value* | ||
|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | ||
| Linezolid | ||||
|
1–20 20–40 41–60 |
25 21 7 |
0 0 0 |
10 3 1 |
0.270 |
| Tetracycline | ||||
|
1–20 20–40 41–60 |
7 4 1 |
4 4 1 |
24 16 6 |
0.964 |
| Chloramphenicol | ||||
|
1–20 20–40 41–60 |
23 16 6 |
8 3 1 |
4 5 1 |
0.742 |
| Rifampin | ||||
|
1–20 20–40 41–60 |
20 12 3 |
2 1 1 |
23 11 4 |
0.804 |
| Piperacillin | ||||
|
1–20 20–40 41–60 |
1 0 0 |
0 0 0 |
34 24 8 |
0.629 |
| Cefoxitin | ||||
|
1–20 20–40 41–60 |
4 0 0 |
0 0 0 |
31 24 8 |
0.143 |
| Gentamycin | ||||
|
1–20 20–40 41–60 |
18 9 3 |
7 3 1 |
10 12 4 |
0.513 |
| Ampicillin / Sulbactam | ||||
|
1–20 20–40 41–60 |
4 0 0 |
0 0 0 |
31 24 8 |
0.143 |
| Levofloxacin | ||||
|
1–20 20–40 41–60 |
4 0 0 |
0 0 0 |
31 24 8 |
0.143 |
| Ciprofloxacin | ||||
|
1–20 20–40 41–60 |
22 9 4 |
2 2 1 |
11 13 3 |
0.400 |
| Amoxicillin/ Clavulanic | ||||
|
1–20 20–40 41–60 |
4 0 0 |
0 0 0 |
31 24 8 |
0.143 |
| Vancomycin | ||||
|
1–20 20–40 41–60 |
4 0 0 |
0 0 0 |
31 24 8 |
0.143 |
*Chi square test; P-value was set to 0.05
Table 6.
Correlation between age group and antibiotic resistance profile among Gram-negative samples
| Antibiotic | Resistance profile | P-value* | ||
|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | ||
| Ticracillin / Tazobactam | ||||
|
1–20 20–40 41–60 |
2 0 3 |
13 4 3 |
8 11 9 |
0.065 |
| Tigycyclin | ||||
|
1–20 20–40 41–60 |
3 2 6 |
11 10 5 |
9 3 4 |
0.157 |
| Chloramphenicol | ||||
|
1–20 20–40 41–60 |
7 7 7 |
4 2 2 |
12 6 6 |
0.838 |
| Cefuroxime | ||||
|
1–20 20–40 41–60 |
4 0 2 |
3 3 1 |
16 12 12 |
0.448 |
| Cefazolin | ||||
|
1–20 20–40 41–60 |
0 0 0 |
0 0 0 |
23 15 15 |
N/A |
| Imipineme | ||||
|
1–20 20–40 41–60 |
10 4 5 |
3 2 5 |
10 9 5 |
0.369 |
| Gentamycin | ||||
|
1–20 20–40 41–60 |
12 5 9 |
2 2 1 |
9 8 5 |
0.677 |
| Ampicillin / Sulbactam | ||||
|
1–20 20–40 41–60 |
3 0 1 |
4 0 1 |
16 15 13 |
0.185 |
| Amoxicillin/ Clavulanic | ||||
|
1–20 20–40 41–60 |
3 1 2 |
2 2 5 |
18 12 8 |
0.309 |
| Ceftazidime | ||||
|
1–20 20–40 41–60 |
0 0 1 |
5 1 1 |
18 14 13 |
0.285 |
| Levofloxacin | ||||
|
1–20 20–40 41–60 |
19 8 8 |
0 2 1 |
4 5 6 |
0.171 |
| Ciprofloxacin | ||||
|
1–20 20–40 41–60 |
17 5 8 |
1 4 1 |
5 6 6 |
0.077 |
| Trimethoprim / Sulfamethoxazole | ||||
|
1–20 20–40 41–60 |
6 5 3 |
5 3 3 |
12 8 8 |
0.942 |
*Chi square test; P-value was set to 0.05; N/A: not applicaple
MDR, XDR, and PDR profiles of isolates
MDR, XDR, and PDR percentages of each bacteria were calculated out of the total number of isolates of each bacteria. Table 7 shows that 52.4% of total isolates elucidated a MDR profile (i.e. resistance to at least 3 antibiotic classes). The overall MDR of Gram-positive isolate was 50% (56% for S. aureus and 15.4% for CoNS). Total MDR rate of Gram-negative isolates was 54.9% with the highest prevalence pattern of E. coli (87.5%). For other Gram-negative species, the MDR rates were as follows: P. aeruginosa (54.2%), Klebsiella spp. (50%), and Proteus spp. (41.7%). Both Proteus spp. and E. coli had significant distributions of resistance profiles across antibiotic classes (p < 0.05). Only 4.7% and 2.9% of all isolates showed XDR and PDR profile, respectively. XDR rate of Gram-positive isolate was 2.3% and that of Gram-negative isolates was 7.3%, while the PDR rates of Gram-positive and Gram-negative isolates were 3.4% and 2.4%, respectively.
Table 7.
Prevalence of MDR, XDR and PDR profile of wound infections isolates
| Organism | R0 % |
R1 % |
R2 % |
R3 % |
R4 % |
R5 % |
R6 % |
R7 % |
R % % |
P value* | MDR | XDR | PDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. aureus (n = 75) |
0 (0%) | 3 (4%) | 3 (4%) | 1 (1.3%) | 10 (13.3%) | 18 (24%) | 7 (9.3%) | 8 (10.7%) | 25 (33.3%) | 0.997 | 42 (56%) | 2 (2.7%) | 2 (2.7%) |
|
CoNS (n = 13) |
1 (7.7%) | 0 (0%) | 0 (0%) | 1 (7.7%) | 5 (38.5%) | 4 (30.8%) | 0 (0%) | 0 (0%) | 2 (15.4%) | 0.437 | 2 (15.4%) | 0 (0%) | 1 (7.7%) |
|
P. aeruginosa (n = 48) |
0 (0%) | 0 (0%) | 3 (6.3%) | 7 (14.6%) | 5 (10.4%) | 6 (12.5%) | 6 (12.5%) | 4 (8.3%0 | 16 (33.3%) | 0.981 | 26 54.2% | 4 (8.3%) | 1 (2.1%) |
|
Klebsiella spp. (n = 14) |
0 (0%) | 0 (0%) | 0 (0%) |
3 (21.4%) |
4 (28.6%) | 1 (7.1%) | 2 (14.3%) | 0 (0%) | 4 (28.6%) | 0.437 | 7 (50%) | 1 (7.1%) | 1 (7.1%) |
|
Protues spp. (n = 12) |
0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (25%) | 5 (41.7%) | 0 (0%) | 0 (0%) | 4 (33.3%) | 0.040 | 5 (41.7%) | 0 (0%) | 0 (0%) |
|
E. coli (n = 8) |
0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (12.5%) | 3 (37.5%) | 0 (0%) | 0 (0%) | 4 (50%) | 0.040 | 7 (87.5%) | 1 (12.5%) | 0 (0%) |
*Chi square test; P-value was set to 0.05
R0 = Isolates are sensitive to all antimicrobials tested
Key: R1-R8 = Number of antimicrobial classes in which a given isolate was resistant
MDR = Multi-drug resistance; number of isolates resistant to three or more of antibacterial classes
XDR = Extensive drug resistance; number of isolates resistant to all classes of antibiotics except one class of antibiotic
PDR = Pan drug resistance; number of isolates resistant to all classes of antibiotics
% of MDR, XDR and PDR were calculated out of number of isolates belonging each bacteria
Discussion
This survey assessed the prevalence of various bacterial species isolated from different wound infections among a group of patients in Upper Egypt. Of the 146 population subjects included in this survey, 120 patients elucidated positive bacterial growth with a high isolation rate of 82.2%. This was approximately similar to the finding of Mohammed et al. who reported an isolation rate of 83.9% for wound infections from inpatients and outpatients with pus and/or wound discharge in Northwest Ethiopia [11]. Our overall isolation rate was relatively higher than that of previous studies conducted in Ethiopia [12, 13]. The difference could be attributed to the etiology of bacterial infection and the source of wound infections from which samples were obtained. Another reason that may explain this variation is the adopted protocol of infection control and antibiotic prophylaxis that may play a crucial role in bacterial growth. Also, the fastidious nature of some bacteria may be responsible for their inability to grow[14].
The prevalence of poly-bacterial species (55%) was higher than that of mono-bacterial species (45%). This was different from the findings of Hassan et al. [15] who reported a higher prevalence of mono-bacterial isolates at 60% and 40% of mixed bacterial species. Likewise, Ares et al. [16] and Bessa et al. [2] reported a predominance of mono-microbial infections over poly-microbial infections with rates of 88.6% and 72.8%, respectively. The presence of monotype or poly-microbial communities of bacteria has a multifactorial nature. For instance, the wound state, microbial density, previous treatment of the wound, dermal moisture, and nutrient availability [17] could explain the difference.
Our findings revealed almost similar proportions of Gram-positive bacteria (51.8%) and Gram-negative strains (48.2%). This was in line with Ares et al. [16] and Bessa et al. [2]. On the other hand, some prior studies showed a predominance of Gram-negative isolates over Gram-positive strains [15, 17, 18]. The diversity of results could be related to the variations in participants’ demographic characteristics. Moreover, nosocomial infection increases the prevalence of MDR bacteria [15].
Our findings revealed that S. aureus and P. aeruginosa were the dominant pathogens. This was consistent with the findings of Puca et al. [19]. S. aureus represented 62.5% of all isolates and 85.2% of isolated Gram-positive strains that have been isolated from all samples of different wound infections. This was comparable to the findings of Ahmed et al. [20] in a study conducted in Upper Egypt. They stated that S. aureus was identified in 61% of specimens, surgical site infections, abscesses, and burn infections were the most common sites of S. aureus isolates representing 59%, 56%, and 52%, respectively. This was to some extent in agreement with the present results. The prevalence of S. aureus was 65%, which coincided with the prevalence reported by Mulu et al. [21] among 151 wound swabs. In like manner, different studies confirmed the predominance of S. aureus isolated from wound infections [14, 22, 23]. This is not surprising because S. aureus is the common skin commensal. Also, exogenous or endogenous infections could be the source of the S. aureus infection.
CoNS accounted for 10.8% of all isolates which was comparable to a retrospective study conducted in intensive care unit of Ain Shams University Hospitals in Egypt, which reported a rate of 12.5% from different infection sites [3]. Another study conducted in Ethiopia showed CNS prevalence of 14.5%, which is relatively higher than our findings [24].
Among all isolates, P. aeruginosa was the second pathogen that was isolated from wound infections, with a prevalence of 40%. This prevalence was compatible with that recorded by Manikandan and Amsath [25]. Another Gram-negative species prevalence was for Klebsiella spp. (11.6%), Proteus spp. (10%), and E. coli (6.7%). A previous study in Egypt revealed a predominance of Klebsiella spp. followed by E. coli [3]. Another study demonstrated the dominance of E. coli among all identified Gram-negative species, followed by Proteus spp., with the least prevalence of P. aeruginosa [24]. The diversity among different studies could be attributed to the number of participants, environmental factors, providing health services, and individual health care conditions [26].
Beta-lactam ring-based antibiotics had the highest resistance profile to Gram-positive strains, with a prevalence ranging from 84.6 to 98.7%. Our findings revealed a remarkable increase in the prevalence of MRSA compared to a previous survey that was conducted by Ahmed et al. [27] who reported a 24% prevalence of MRSA in the same hospital eight years ago. This raises the alarm about the escalating and noticeable increase in the prevalence of MRSA in Egypt. Another study carried out in Ghana [28] confirmed absolute resistance to oxacillin. S. aureus was sensitive to vancomycin with a rate of 85.2%, while vancomycin-resistant S. aureus (VRSA) was detected in 14.8% of cases. This was relatively lower than that concluded in Egypt by Ghoniem et al. [29] who reported that the ratio of VRSA was 20.7%. On the other hand, another study has been held in Egypt by Amr et al. [30] demonstrated a lower prevalence of VRSA (8.8%). These discrepancies could be related to the variations in the bacterial culture method and the different geographical settings of these studies.
Regarding Gram-negative sensitivity, All Gram-negative isolates tested positive for absolute resistance to the cefazolin antibiotic in our study. P. aeruginosa was the most commonly isolated Gram-negative species, with resistance rates ranging from 62.5 to 100% to the ß-lactam antibiotic. Similarly, P. aeruginosa had a high prevalence of levofloxacin resistance (83.3%). These findings were quite similar to Nikokar et al. [31] results of the resistance profile rates of P. aeruginosa isolated from burn infections and post-surgical sites and wound infection against the cephalosporin antibiotics cefazolin (83.7%) and ceftazidime (68.8%), gentamicin (37.2%), and imipenem (23.3%). While they confirmed a high resistance rate against ciprofloxacin (66.3%), our findings supported the lowest resistance being against ciprofloxacin (16.7%).
Klebsiella spp. exhibited high resistance to ß-lactam antibiotics. This was in agreement with previous literature conducted in Bangladesh [7]. This was consistent with the conclusion of a review conducted in Asia [32] that highlighted the rising danger of MDR of Klebsiella spp. Antimicrobial resistance as a result of antibiotic misuse is a life threatening issue affecting both males and females of all ages without regard to certain genders or age groups [33]. The current study confirmed that idea, as no difference was detected regarding associating patients’ gender and age groups with the resistance profile of the tested antimicrobials against Gram-positive and Gram-negative isolates.
The prevalence of the MDR profile was slightly higher among the Gram-negative isolates compared to Gram-positive species. The MDR of Gram-negative rate was 54.9%, which is close to the MDR rates reported in two studies conducted in Ethiopia. The first study reported an MDR prevalence of 51% among bacteria isolated from open fracture wounds [34]. The second study found that 59.3% of bacteria from wound infections had an MDR profile [35]. Relatively similar results were published in Egypt, which recorded 73.9% and 70% MDRs of E. coli and Pseudomonas spp., respectively, against the majority of tested antibiotics. Regarding Gram-positive isolates, the overall MDR was 50%, with a higher MDR profile for S. aureus (56%) followed by CoNS (15.4%). This was less than the findings of Melake et al. who found that the prevalence of MDR of S. aureus and CoNS isolated from burn wounds in Egypt were 73.5% and 47%, respectively.
The main limitations of the current study can be summarized as follows; (1) the use of a disk-diffusion approach to detect the bacterial susceptibility, which has relative reliability, (2) the generalization of the study findings has to be interpreted with caution as possible due to heterogeneity of subjects and settings in different geographic areas; and (3) the availability of anaerobic bacteria was not accounted for in the current study due to the shortage in the culture conditions.
Conclusions
Within the limitations of the current findings, it can be concluded that there was a dominance of polymicrobial wound samples. S. aureus and P. aeruginosa were the most commonly isolated pathogens. A high rate of MRSA was revealed among S. aureus isolates. Vancomycin and linezolid were the most effective drugs against S. aureus and ciprofloxacin was the most effective one against P. aeruginosa. Several isolates elucidated MDR profile for all tested classes of antibiotics, which indicates a serious exacerbation of bacterial resistance and a difficulty finding treatment options for all infections.
Acknowledgements
The authors are thankful to the technical staff in the hospital laboratories for helping in performing the investigations.
Author’ contributions
E.A. was responsible for data analysis, and drafting manuscripts; manuscript critical revising gave final approval and agrees to be accountable for all aspects of work ensuring integrity and accuracy. A.R. was responsible for study conception and design, data analysis and interpretation, and drafting a manuscript. A.D. and G.G. were responsible for the manuscript’s critical revising gave final approval and agrees to be accountable for all aspects of the work ensuring integrity and accuracy.
Funding
Research is self-funded.
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Informed consent was obtained from all subjects and their legal guardian. All experimental protocols were approved by the ethics committee/Institutional Review Board of Faculty of Pharmacy, Deraya University (Approval number: 1/2019). The study has been performed in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eman Farouk Ahmed, Email: eman.farouk@pharm.sohag.edu.eg.
Asia Helmi Rasmi, Email: asia.helmy@deraya.edu.eg.
Abdou M. A. Darwish, Email: abdoudarwish@hotmail.com
Gamal Fadl Mahmoud Gad, Email: gmal.fadl@mu.edu.eg.
References
- 1.Dai T, Huang YY, Sharma SK, Hashmi JT, Kurup DB, Hamblin MR. Topical antimicrobials for burn wound infections. Recent Pat Antiinfect Drug Discov. 2010;5(2):124–51. doi: 10.2174/157489110791233522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessa LJ, Fazii P, Di Giulio M, Cellini L. Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: some remarks about wound infection. Int Wound J. 2015;12(1):47–52. doi: 10.1111/iwj.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fahim NAE. Prevalence and antimicrobial susceptibility profile of multidrug-resistant bacteria among intensive care units patients at Ain Shams University Hospitals in Egypt-a retrospective study. J Egypt Public Health Assoc. 2021;96(1):7. doi: 10.1186/s42506-020-00065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadri SS. Key takeaways from the US CDC’s 2019 antibiotic resistance threats report for frontline providers. Crit Care Med 2020. [DOI] [PMC free article] [PubMed]
- 5.Bagdonas R, Tamelis A, Rimdeika R. Staphylococcus aureus infection in the surgery of burns. Medicina. 2003;39(11):1078–81. [PubMed] [Google Scholar]
- 6.Kesah C, Ben Redjeb S, Odugbemi T, Boye CB, Dosso M, Ndinya Achola J, Koulla-Shiro S, Benbachir M, Rahal K, Borg M. Prevalence of methicillin‐resistant Staphylococcus aureus in eight african hospitals and Malta. Clin Microbiol Infect. 2003;9(2):153–6. doi: 10.1046/j.1469-0691.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- 7.Alam MM, Islam MN, Hawlader MDH, Ahmed S, Wahab A, Islam M, Uddin KR, Hossain A. Prevalence of multidrug resistance bacterial isolates from infected wound patients in Dhaka, Bangladesh: a cross-sectional study. Int J Surg Open. 2021;28:56–62. doi: 10.1016/j.ijso.2020.12.010. [DOI] [Google Scholar]
- 8.Sakeena MHF, Bennett AA, McLachlan AJ. Non-prescription sales of antimicrobial agents at community pharmacies in developing countries: a systematic review. Int J Antimicrob Agents. 2018;52(6):771–82. doi: 10.1016/j.ijantimicag.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 9.Taati Moghadam M, Khoshbayan A, Chegini Z, Farahani I, Shariati A. Bacteriophages, a New Therapeutic Solution for Inhibiting Multidrug-Resistant Bacteria causing wound infection: lesson from animal models and clinical trials. Drug Des Devel Ther. 2020;14:1867–83. doi: 10.2147/DDDT.S251171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collee JG, Miles R, Watt B. Tests for identification of bacteria. Mackie and McCartney practical medical microbiology. 1996;14:131–49. [Google Scholar]
- 11.Mohammed A, Seid ME, Gebrecherkos T, Tiruneh M, Moges F. Bacterial isolates and their antimicrobial susceptibility patterns of wound infections among inpatients and outpatients attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol 2017, 2017. [DOI] [PMC free article] [PubMed]
- 12.Mengesha RE, Kasa BG, Saravanan M, Berhe DF, Wasihun AG. Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder Teaching and Referral Hospital, Mekelle, Ethiopia. BMC Res Notes. 2014;7:575. doi: 10.1186/1756-0500-7-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dessalegn L, Shimelis T, Tadesse E, Gebre-selassie S. Aerobic bacterial isolates from post-surgical wound and their antimicrobial susceptibility pattern: a hospital based cross-sectional study. J Med Res. 2014;3(2):18–23. [Google Scholar]
- 14.Mulu W, Kibru G, Beyene G, Damtie M. Postoperative nosocomial infections and antimicrobial resistance pattern of Bacteria isolates among patients admitted at Felege Hiwot Referral Hospital, Bahirdar, Ethiopia. Ethiop J Health Sci. 2012;22(1):7–18. [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan MA, Abd El-Aziz S, Elbadry HM, Samy A, Tamer TM. Prevalence, antimicrobial resistance profile, and characterization of multi-drug resistant bacteria from various infected wounds in North Egypt. Saudi J Biol Sci 2022. [DOI] [PMC free article] [PubMed]
- 16.Asres G, Legese M, Woldearegay G. Prevalence of multidrug resistant Bacteria in postoperative wound infections at Tikur Anbessa specialized hospital, Addis Ababa, Ethiopia. Arch Med. 2017;9(4):12. [Google Scholar]
- 17.Upreti N, Rayamajhee B, Sherchan SP, Choudhari MK, Banjara MR. Prevalence of methicillin resistant Staphylococcus aureus, multidrug resistant and extended spectrum beta-lactamase producing gram negative bacilli causing wound infections at a tertiary care hospital of Nepal. Antimicrob Resist Infect Control. 2018;7:121. doi: 10.1186/s13756-018-0408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moges F, Eshetie S, Abebe W, Mekonnen F, Dagnew M, Endale A, Amare A, Feleke T, Gizachew M, Tiruneh M. High prevalence of extended-spectrum beta-lactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS ONE. 2019;14(4):e0215177. doi: 10.1371/journal.pone.0215177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puca V, Marulli RZ, Grande R, Vitale I, Niro A, Molinaro G, Prezioso S, Muraro R, Di Giovanni P. Microbial Species isolated from infected Wounds and Antimicrobial Resistance Analysis: data emerging from a three-years retrospective study. Antibiot (Basel) 2021, 10(10). [DOI] [PMC free article] [PubMed]
- 20.Ahmed EF, Gad GF, Abdalla AM, Hasaneen AM, Abdelwahab SF. Prevalence of methicillin resistant Staphylococcus aureus among egyptian patients after surgical interventions. Surg Infect (Larchmt) 2014;15(4):404–11. doi: 10.1089/sur.2013.212. [DOI] [PubMed] [Google Scholar]
- 21.Mulu A, Moges F, Tessema B, Kassu A. Pattern and multiple drug resistance of bacterial pathogens isolated from wound infection at University of Gondar Teaching Hospital, Northwest Ethiopia. Ethiop Med J. 2006;44(2):125–31. [PubMed] [Google Scholar]
- 22.Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13(1):1–10. doi: 10.1186/1476-0711-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emele FE, Izomoh MI, Alufohai E. Microorganisms associated with wound infection in Ekpoma, Nigeria. West Afr J Med. 1999;18(2):97–100. [PubMed] [Google Scholar]
- 24.Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13:14. doi: 10.1186/1476-0711-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manikandan C, Amsath A. Antibiotic susceptibility of bacterial strains isolated from wound infection patients in Pattukkottai, Tamilnadu, India. Int J Curr Microbiol App Sci. 2013;2(6):195–203. [Google Scholar]
- 26.Shebl RI, Mosaad YO. Frequency and antimicrobial resistance pattern among bacterial clinical isolates recovered from different specimens in Egypt. Cent Afr J Public Heal. 2019;5(1):36–45. doi: 10.11648/j.cajph.20190501.16. [DOI] [Google Scholar]
- 27.Ahmed EF, Gad GF, Abdalla AM, Hasaneen AM, Abdelwahab SF. Prevalence of methicillin resistant Staphylococcus aureus among egyptian patients after surgical interventions. Surg Infect. 2014;15(4):404–11. doi: 10.1089/sur.2013.212. [DOI] [PubMed] [Google Scholar]
- 28.Saba CKS, Amenyona JK, Kpordze SW. Prevalence and pattern of antibiotic resistance of Staphylococcus aureus isolated from door handles and other points of contact in public hospitals in Ghana. Antimicrob Resist Infect Control. 2017;6:44. doi: 10.1186/s13756-017-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghoniem EM, El Hendawy GR, Moteleb TMA, Hassan HA, Khalil HAER. Characterization of vancomycin-resistant Staphylococcus aureus in the National Liver Institute. Menoufia Med J. 2014;27(4):825. doi: 10.4103/1110-2098.149802. [DOI] [Google Scholar]
- 30.Amr GE, Al Gammal S. Emergence of vancomycin resistant Staphylococcus aureus isolated from patients in ICUs of Zagazig University Hospitals. Egypt J Med Microbiol (EJMM) 2017, 26(2).
- 31.Nikokar I, Tishayar A, Flakiyan Z, Alijani K, Rehana-Banisaeed S, Hossinpour M, Amir-Alvaei S, Araghian A. Antibiotic resistance and frequency of class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran J Microbiol. 2013;5(1):36–41. [PMC free article] [PubMed] [Google Scholar]
- 32.Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19(1):1. doi: 10.1186/s12941-019-0343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodin MW. Antimicrobial Resistance: a major threat to the Promise of healthy aging. Health Affairs Forefront; 2023.
- 34.Yishak A, Biruk L. Microbial susceptibility of bacteria isolated from open fracture wounds presenting to the black-lion hospital, Addis Ababa University. Afr J Microbiol Res. 2009;3:939–51. [Google Scholar]
- 35.Melake NA, Eissa NA, Keshk TF, Sleem AS. Prevalence of multidrug-resistant bacteria isolated from patients with burn infection. Menoufia Med J. 2015;28(3):677. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.