Abstract
We recently described a drug-based selectable and counterselectable genetic platform for the animal model system Drosophila melanogaster, consisting of four resistance and two sensitivity markers that allow direct selection for, or counterselection against, a desired genotype. This platform eliminates the need to identify modified progeny by traditional laborious screening using dominant eye and body color markers, white+ and yellow+, respectively. The four resistance markers permit selection of animals using G418 sulfate, Puromycin HCl, Blasticidin S, or Hygromycin B, while the two sensitivity markers allow counterselection of animals against Ganciclovir or Acyclovir, and 5-Fluorocytosine. The six markers can be used alone or in combination to perform co-selection, combination selection and counterselection, as well as co-counterselection. To make this novel selection and counterselection genetics platform easily accessible to and rapidly implementable by the scientific community, we used a synthetic assembly DNA cloning platform, GoldenBraid 2.0 (GB2.0). GB2.0 relies on two Type IIs restriction enzymes that are alternatingly used during successive cloning steps to make increasingly complex genetic constructs. Here we describe how to perform synthetic assembly DNA cloning using GB2.0 to build such complex plasmids, using the assembly of both components of the binary LexA/LexA-Op overexpression system, a G418 sulfate-selectable LexA transactivator plasmid, and a Blasticidin S-selectable LexA-Op responder plasmid, as an example. We demonstrate the functionality of these plasmids by including the expression pattern obtained after co-injection, followed by co-selection using G418 sulfate an Blasticidin S, resulting in co-transgenesis of both plasmids. Protocols are provided on how to obtain, adapt, and clone DNA parts for synthetic assembly cloning after de novo DNA synthesis or PCR amplification of desired DNA parts, how to assemble those DNA parts into multipartite transcription units, followed by how to further assemble multiple transcription units into genetic constructs of increasing complexity to perform multiplexed transgenic selection and counterselection, or any other, genetic strategies using Drosophila melanogaster. The protocols we present can be easily adapted to incorporate any of the six selectable and counterselectable, or any other, markers to generate plasmids of unmatched complexity for various genetic applications. A protocol on how to generate transgenic animals using these synthetically assembled plasmids is described in an accompanying Current Protocols article (Venken,Matinyan, Gonzalez, & Dierick, 2023).
Basic Protocol 1:
Obtaining and cloning a de novo synthesized DNA part for synthetic assembly DNA cloning.
Basic Protocol 2:
Obtaining and cloning a DNA part amplified by PCR from existing DNA resources for synthetic assembly DNA cloning.
Alternate Protocol 2:
Obtaining, adapting, and cloning a DNA part amplified by PCR from existing DNA resources for synthetic assembly DNA cloning.
Basic Protocol 3:
Synthetic assembly DNA cloning of individual DNA parts into a multipartite transcription unit.
Basic Protocol 4:
Synthetic assembly DNA cloning of multiple transcription units into genetic constructs of increasing complexity.
Keywords: Synthetic assembly cloning, GoldenBraid 2.0, Selection, Counterselection, Multiplexed, Transgenesis, Drosophila melanogaster, Gene expression analysis
INTRODUCTION
We recently developed a drug-based platform for multiplexed selection and counterselection genetic strategies using Drosophila melanogaster (Matinyan et al., 2021a, 2021b). We demonstrated that four selectable markers, encoding Neomycin phosphotransferase II, Puromycin HCl N-acetyltransferase, Blasticidin S-resistance, and Hygromycin B phosphotransferase, provide animal resistance against the drugs G418 sulfate, Puromycin HCl, Blasticidin S, and Hygromycin B, respectively (Matinyan et al., 2021b). In addition, two counterselectable markers that encode a mutant version of thymidine kinase called sr39TK, and the chimeric FCY1/FUR1 fusion protein called FCU1, make animals sensitive to the drugs Ganciclovir or Acyclovir, and 5-Fluorocytosine, respectively (Matinyan et al., 2021b). We further demonstrated that marker associated drug resistance or sensitivity is specific to the corresponding drug, allowing to combine multiple markers in multiplexed genetic strategies to perform co-selection, combination selection and counterselection, and co-counterselection to obtain animals with desired genotypes (Matinyan et al., 2021b). We then applied this selection/counterselection platform to generate dual transgenic animals in a single step allowing immediate expression analysis using binary overexpression systems (e.g., GAL4/UAS and LexA/LexA-Op) without having to make individual transgenic animals for each of the components of the binary overexpression system (i.e., GAL4 and UAS, or LexA and LexA-Op) followed by crossing both components together (Matinyan et al., 2021b). We also generated selectable and counterselectable balancer chromosomes that allow selection for, or against, the presence of this modified chromosome during cross schemes, and make dual modified transgenes, i.e., P[acman] Bacterial Artificial Chromosomes (BACs) that contain a resistance marker for selectable transgenesis purposes and a fluorescent marker for gene expression analysis (Matinyan et al., 2021b).
To make this novel genetic platform easy to implement by the scientific community, we integrated plasmid construction needed for multiplexed selection and counterselection with synthetic assembly DNA cloning (Matinyan et al., 2021b). While traditional Type II restriction enzyme cloning is difficult to use to build complex DNA constructs, synthetic assembly DNA cloning strategies are designed to generate complex end-products by stitching together different DNA parts in a standardized hierarchical manner (Figure 1 and Figure 2) (Casini et al., 2015; Ellis et al., 2011; Xie and Fussenegger, 2018; Blasi et al., 2021). We used the previously developed GoldenBraid 2.0 (GB2.0) platform for this purpose (Matinyan et al., 2021b; Sarrion-Perdigones et al., 2013) (Figure 2). GB2.0 depends on two Type IIs restriction enzymes (Esp3I or its isoschizomer BsmBI, and BsaI) that are alternatingly used in successive cloning steps to build multipartite genetic constructs (Sarrion-Perdigones et al., 2013, 2011) (Figure 2A). DNA parts are first accommodated, now commonly referred to as “domesticated” to the GB2.0 workflow by cloning them in a Universal Part Domesticator vector backbone using Esp3I. This step ensures that domesticated parts “behave” during downstream procedures by not having any (or as few as possible) recognition sites for any of the Type IIs restriction enzymes used during assembly cloning (i.e., Esp3I and BsaI). Domestication can then be followed by assembly of multiple domesticated DNA parts into a first tier of assembly vectors, called Alpha level destination vectors, using BsaI, resulting in a plasmid containing a transcription unit (Figure 2A). Alpha level transcription units can then further be assembled, typically as pairs, into a second tier of assembly vectors, called Omega level destination vectors using Esp3I, resulting in a plasmid containing two transcription units (Figure 2A). Omega assemblies can be further pairwise assembled back into Alpha level destination vectors, and so on (Figure 2A).
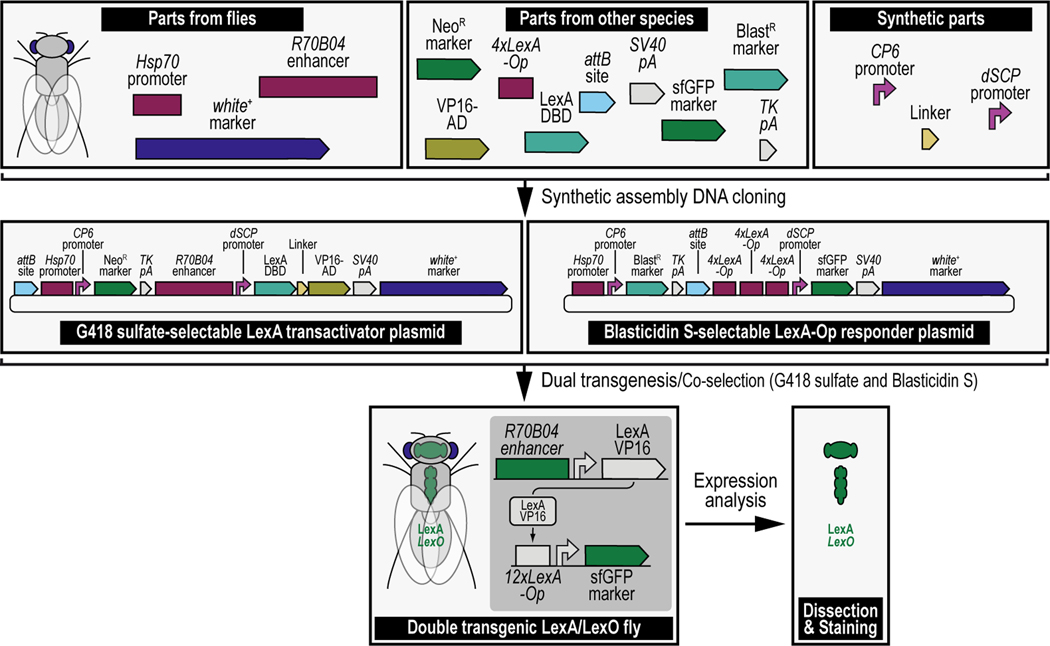
Figure 1. Simplified schematic of synthetic assembly DNA cloning for Drosophila melanogaster.
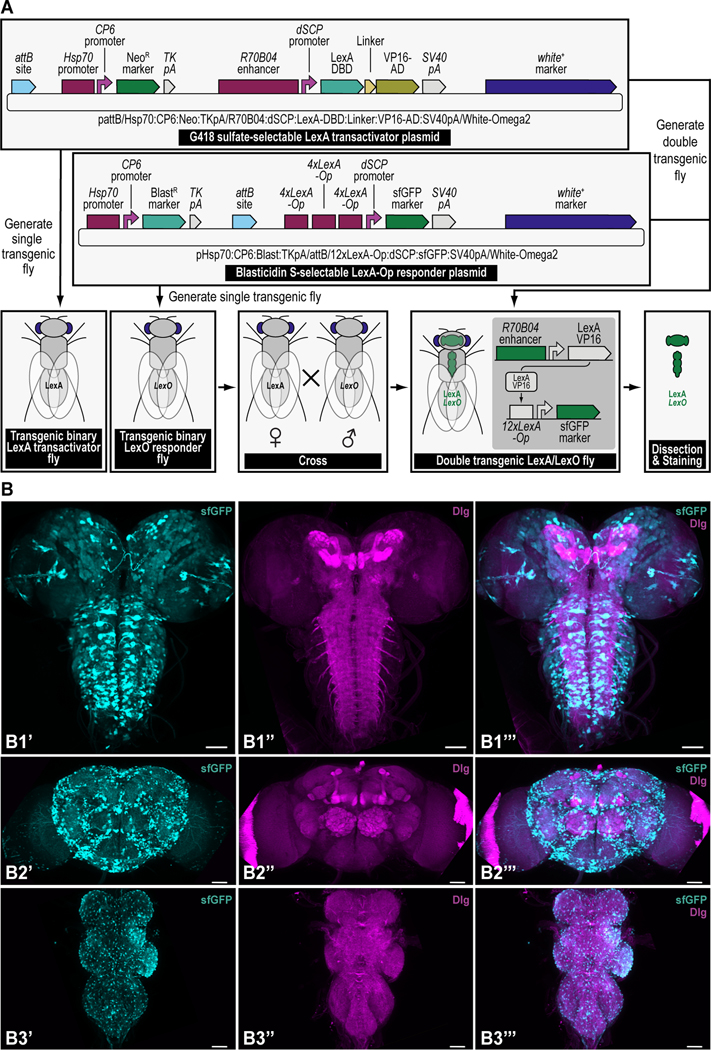
Different DNA parts, obtained from flies directly (Top left), other species (Top middle), or through synthetic means (Top right), are put together using synthetic assembly DNA cloning to generate continuously growing plasmids of increasing complexity. The resulting end products can be two transgenes (used as an example throughout this protocol), a first transgene providing G418 sulfate selection (provided by the NeoR marker) and encoding the binary LexA transactivator driven by the R70B04 enhancer (i.e., the G418 sulfate-selectable LexA transactivator plasmid) (Middle left), and a second transgene providing Blasticidin S selection (provided by the BlastR marker) and encoding the binary LexA-Op responder, reporting green fluorescent protein (sfGFP) reporter expression driven by the LexA binary transactivator, whose expression is regulated by the R70B04 enhancer (i.e., the Blastidicin S-selectable LexA-Op responder plasmid) (Middle right). A double transgenic fly can then be obtained directly by injecting both transgenes (i.e., dual transgenesis) followed by co-selection using both G418 sulfate and Blasticidin S (shown, Bottom) (see Venken et al., 2023), or indirectly by injecting each transgene separately, followed by selection using G418 sulfate for one transgene (LexA transactivator), and Blasticidin S for the other transgene (LexA-Op responder), subsequently followed by crossing both transgenes together and coselect using both G418 sulfate and Blasticidin S (not shown). The double transgenic fly can then be analyzed for expression patterns (see Figure 15).
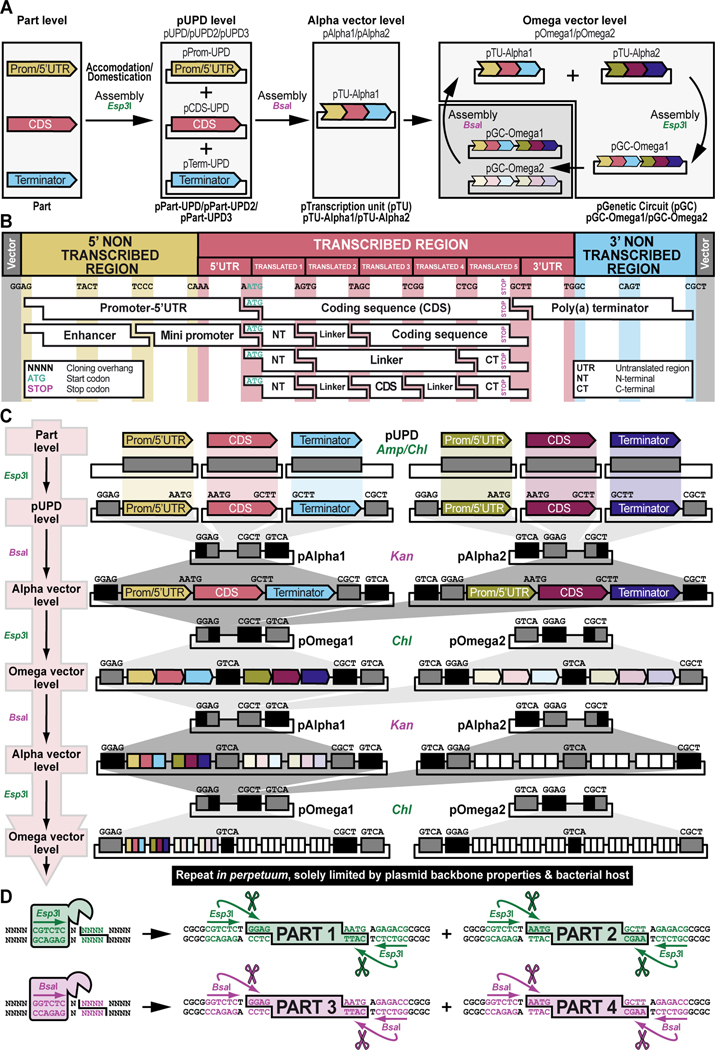
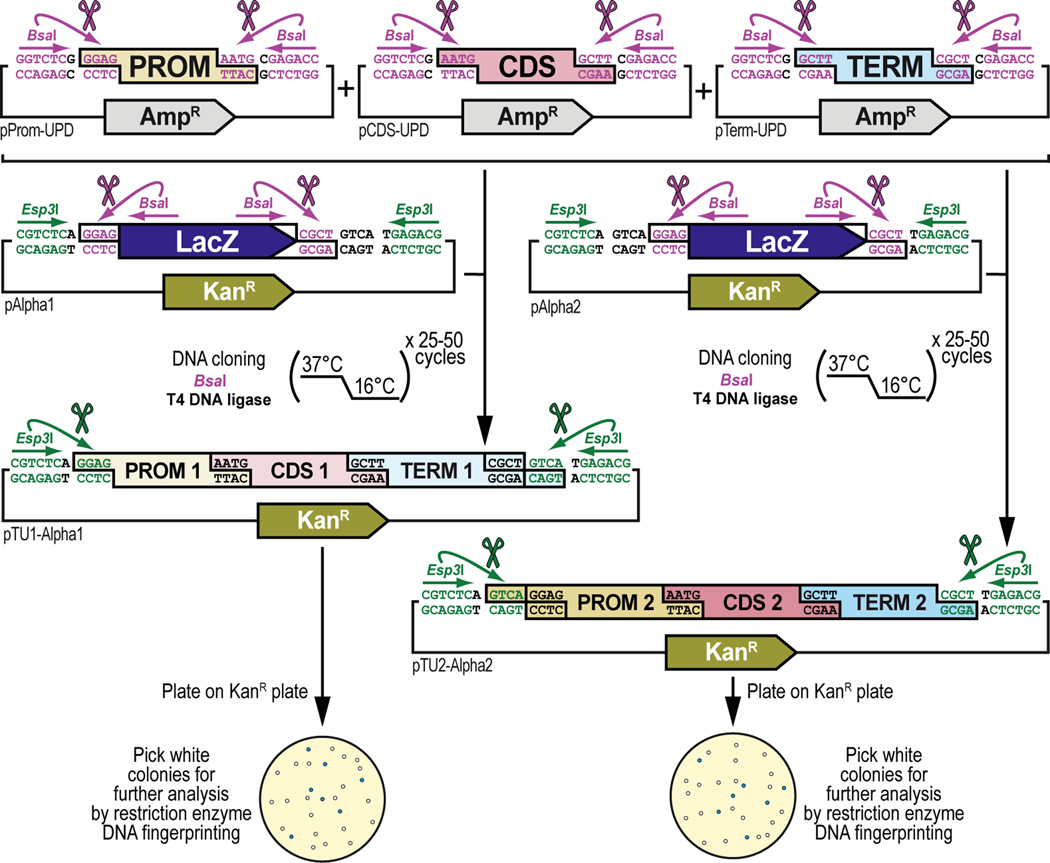
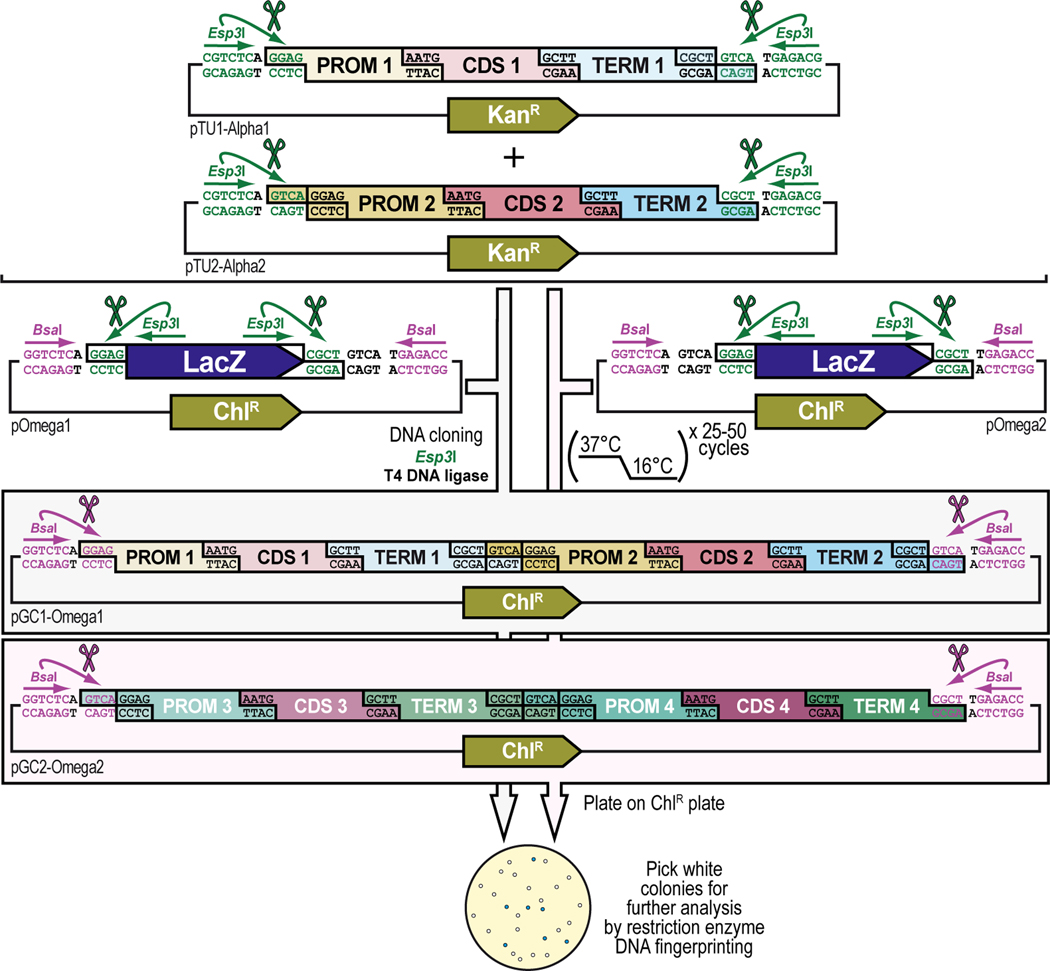
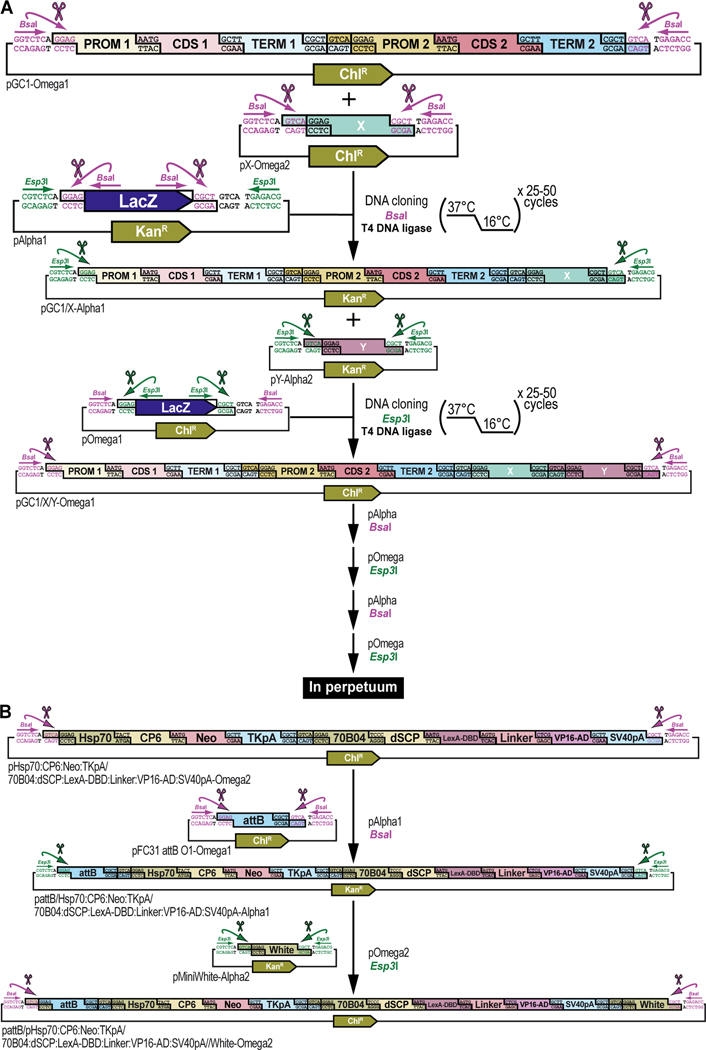
Figure 2. Schematic overview of GoldenBraid 2.0 synthetic assembly DNA cloning to build plasmids for multiplexed selection, counterselection or other genetic strategies using Drosophila melanogaster.
(A) Simplified schematic of the synthetic assembly DNA cloning workflow in GoldenBraid 2.0. DNA parts, including promoter (Prom), coding DNA sequence (CDS), and terminator (Term) (Part level), are first accommodated (also known as domesticated) to the GoldenBraid 2.0 (GB2.0) workflow via synthetic assembly DNA cloning, with the Type IIs restriction enzyme Esp3I, into a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3). This produces a library of DNA part plasmids (pPart-UPD), i.e., pProm-UPD, pCDS-UPD, or pTerm-UPD, respectively (pUPD level), with defined overhangs (see B), allowing ordered assembly in the next assembly level (see C). These parts are further assembled into an Alpha level destination vector (pAlpha1 or pAlpha2) using the Type IIs restriction enzyme BsaI, resulting in a plasmid containing a transcription unit, pTranscription unit (pTU), defined by at least a promoter, coding sequence and terminator (pTU-Alpha1 or pTU-Alpha2), or another assembly of various complexity (Alpha vector level). Finally, Alpha level vectors can be further assembled into an Omega level destination vector (pOmega1 or pOmega2) using again the Type IIs restriction enzyme Esp3I, resulting in a plasmid containing a genetic circuit, pGenetic circuit (pGC), defined by two transcription units or other DNA assemblies of various complexity (pGC-Omega1 or pGC-Omega2) (Omega vector level). Conveniently, those Omega assemblies can be further pairwise assembled into an Alpha level destination vector, and the resulting product can serve as a reagent for further assembly reactions into an Omega level destination vector, and so on. (B) Underlying cloning grammar defined by orthogonal restriction enzyme overhangs to guide ordered synthetic assembly DNA cloning of DNA parts in a multipartite fashion by GoldenBraid 2.0 assembly. GB2.0 cloning, like Golden Gate assembly from which it is derived, features a predefined system of 4-nucleotide cloning overhangs used to assemble linearized DNA parts together in a defined fashion. These overhangs form a “cloning grammar” which governs part identity and assembly order such that a promoter type part always assembles 5’ of a coding sequence which assembles 5’ of a terminator sequence and so on. Several 5’ – 3’ cloning overhangs and their associated part identities are shown to illustrate how different parts can be assembled in meaningful ways. (C) Detailed cloning workflow defined by orthogonal restriction enzyme overhangs to guide parts and intermediate assemblies through the GoldenBraid 2.0 assembly pipeline to reach final status. DNA parts are cloned in Universal Part Domesticator plasmids (pUPD, pUPD2 or pUPD3) using the Type IIs restriction enzyme Esp3I and selected for with ampicillin or chloramphenicol. Cloned parts are combined in a meaningful manner into an Alpha level destination vector (pAlpha1 or pAlpha2) using the Type IIs restriction enzyme BsaI and selected for with kanamycin. Alpha level assemblies can be further pairwise combined into an Omega level destination vector (pOmega1 or pOmega2) using again the Type IIs restriction enzyme Esp3I and selected for with chloramphenicol. Those Omega assemblies can be further pairwise combined into an Alpha level destination vector, and the resulting Alpha assembly can be further pairwise combined into an Omega level destination vector, and so on. This infinitely iterative process always involves a pair of vectors of the same level but different identity (e.g., pAlpha1 and pAlpha2 is a valid assembly into pOmega1 or pOmega2, but pAlpha1 and pAlpha1 is not) governed by their own cloning grammars. This is true for both Alpha and Omega assemblies: pAlpha1 always combines with pAlpha2 in a defined order into any of the two Omega level plasmids, and pOmega1 and pOmega2 always combine in a defined order into any of the two Alpha level plasmids. The ordered assembly of pAlpha1 and pAlpha2 into Omega level vectors is defined by the 5’ “GGAG” and 3’ “GTCA” grammar of pAlpha1 and the 5’ “GTCA” and 3’ “CGCT” grammar of pAlpha2, combined by switching antibiotic selection from kanamycin (for Alpha vectors) to chloramphenicol (for Omega vectors) (see Figure 9 and Figure 10). The same overhangs define the ordered assembly of pOmega1 and pOmega2 into Alpha level vectors, except that antibiotic selection is switched from chloramphenicol (for Omega vectors) to kanamycin (for Alpha vectors) (see Figure 11 and Figure 12). Not all Alpha or Omega assemblies have to be multipart assemblies, as long as the 5’ end has “GGAG” grammar and the 3’ end has “CGCT” grammar. This allows flexibility in the assemblies when a less complex part, e.g., just an attB attachment site, is needed to expand an already complex assembly (see Figure 3 and Figure 13). (D) Simplified schematic of the two Type IIs restriction enzymes used by GoldenBraid 2.0 assembly. Esp3I (Top), or its isoschizomer BsmBI, is used during accommodating/domesticating assemblies going from Part level to pUPD level (see A and C), as well as during assemblies going from Alpha vector level to Omega vector level (see A and C). BsaI (Bottom) is used during assemblies going from pUPD level to Alpha vector level (see A and C), as well as during assemblies going from Omega vector level back to Alpha vector level (see A and C). Both enzymes bind to a unique 6-bp recognition site and create sticky ends one base pair away from their binding site, leaving a 5’ sticky 4-nucleotide sequence behind that can be used for annealing purposes during the ligation step of assembly cloning. Since this 5’ sticky 4-nucleotide sequence is nonspecific (“NNNN”), it can be made user-specific, i.e., programmable, as needed (see B), allowing ordered assemblies to happen using those overhangs, as illustrated for two parts for each enzyme, part 1 and 2 can ligate together using “AATG” sticky ends after cutting with Esp3I, while part 3 and 4 can ligate together using “AATG” sticky ends after cutting with BsaI.
To ensure correct assembly of DNA parts in a meaningful manner, GB2.0 uses 4-nucleotide cloning overhangs that are specific to each part (Sarrion-Perdigones et al., 2013, 2011) (Figure 2B). These overhangs form a “cloning grammar” which directs DNA part identity and assembly order such that a “promoter” DNA part always assembles 5’ of a “coding DNA sequence” DNA part which assembles 5’ of a “terminator” DNA part and so on, like the “grammar” of a language sentence (Figure 2B). Subsequent pairwise assemblies of two Alpha vectors into an Omega vector, or two Omega vectors back into an Alpha vector, are directed by similar “cloning grammars” (Figure 2C). These “cloning grammar” designs are possible because the Type IIs restriction enzymes used in GB2.0, Esp3I (or its isoschizomer, BsmBI) and BsaI, bind to a unique 6-bp recognition site but cut one base pair away from their binding site, leaving a 5’ sticky 4-nucleotide sequence behind that can be used for annealing purposes during assembly cloning (Figure 2D). Since this 5’ sticky 4-nucleotide sequence is nonspecific (“NNNN)”, the user can program it to direct assembly order as needed (Figure 2B and Figure 2C). The “programmed” overhang grammar can then be used to determine the order in which multiple DNA parts assemble during a multipartite cloning reaction (e.g., the 3’ end of a 5’UTR always links to the 5’ end of a coding DNA sequence using the four-nucleotide overhang “AATG”) (Figure 2B). Continued assemblies are only limited by plasmid backbone properties and bacterial host properties. High-copy number plasmid backbones can hold up to about 20 kb of insert, while a low-copy number plasmid backbones can maintain several hundreds of kb of insert. Some bacterial hosts cannot maintain large insert plasmids while others can. Going from one vector to another in the “braiding” process in GB2.0 also depends on different antibiotic selection markers in each of the vectors, ampicillin or chloramphenicol for Universal Part Domesticator, kanamycin for Alpha, and chloramphenicol for Omega vector backbones, so that only the products generated in the destination vector produce viable colonies (Figure 2C).
In our previous work, we developed a GB2.0 compatible DNA part library and vector toolkit to build plasmids for drug-selectable and/or counterselectable genetic strategies in Drosophila melanogaster (Matinyan et al., 2021b). This library of more than 100 DNA parts and vector backbones is publicly available through the DNA repository Addgene (www.addgene.org) so that users can make their own transgenic constructs for selection and counterselection, or any other, genetic strategies. The basic cloning vector backbones in the library allow anyone to integrate new DNA parts into the GB2.0 platform to further grow this resource for the community (Matinyan et al., 2021b).
In this work, we describe how to implement GB2.0 to build plasmids for selection and counterselection, or other genetic strategies in the fruit fly Drosophila melanogaster using the dual transgenesis application for generating transgenic fly lines containing both components of LexA/LexA-Op binary overexpression (Yagi et al., 2010; Pfeiffer et al., 2010; Lai and Lee, 2006). Each component of the binary LexA “transactivator”/LexA-Op “responder” overexpression system is coupled to a different selection marker. The LexA “transactivator” component is coupled to the G418 sulfate-resistant marker (G418R), while the LexA-Op “responder” component is coupled to the Blasticidin S-resistant marker (BlastR) (Figure 1). Importantly, the principles described here are not limited to the generation of plasmids for binary overexpression but can also be utilized to build plasmids for other genetic strategies that incorporate one or more of the selection and/or counterselection markers depending on the user’s needs, or other plasmids designed for a genetic strategy not requiring selection and/or counterselection.
We provide step-by-step protocols to build genetic constructs of increasing complexity through synthetic assembly DNA cloning, using the building of G418 sulfate-selectable LexA transactivator (Figure 3) and Blasticidin S-selectable LexA-Op responder (Figure 4) plasmids for tissue-specific overexpression as an example. We begin by describing how to obtain and adapt DNA parts for synthetic assembly DNA cloning by de novo DNA synthesis (Basic Protocol 1) or PCR amplification (Basic Protocol 2 and Alternate Protocol 2) followed by cloning these DNA parts in Universal Part Domestication plasmids. We then go on to demonstrate how to assemble those DNA parts into multipartite transcription units (Basic Protocol 3), followed by how to further assemble multiple transcription units into genetic constructs of increasing complexity to perform multiplexed transgenic selection and counterselection (Basic Protocol 4). These protocols can be adapted to incorporate any of the six selectable and counterselectable, or other markers to generate plasmids of unmatched complexity for various genetic applications. By the end of this protocol, the user should be able to clone using the GB2.0 synthetic assembly methodology starting from basic DNA parts to complex genetic circuits.
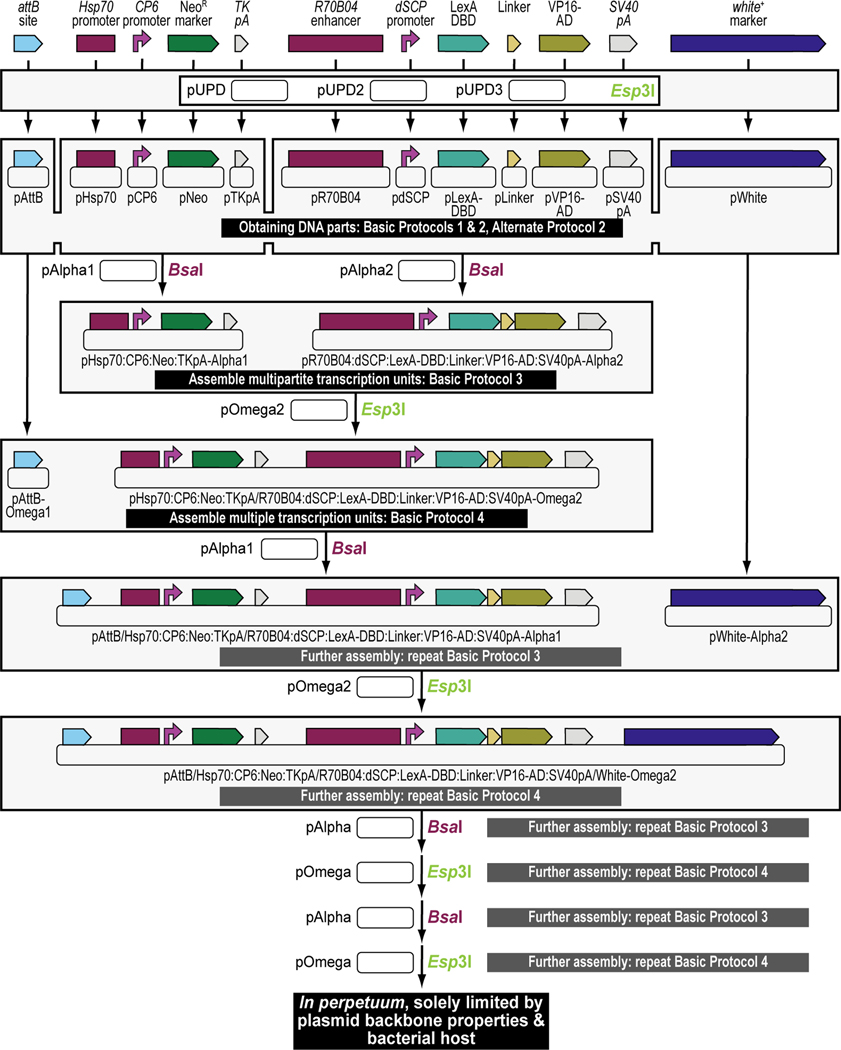
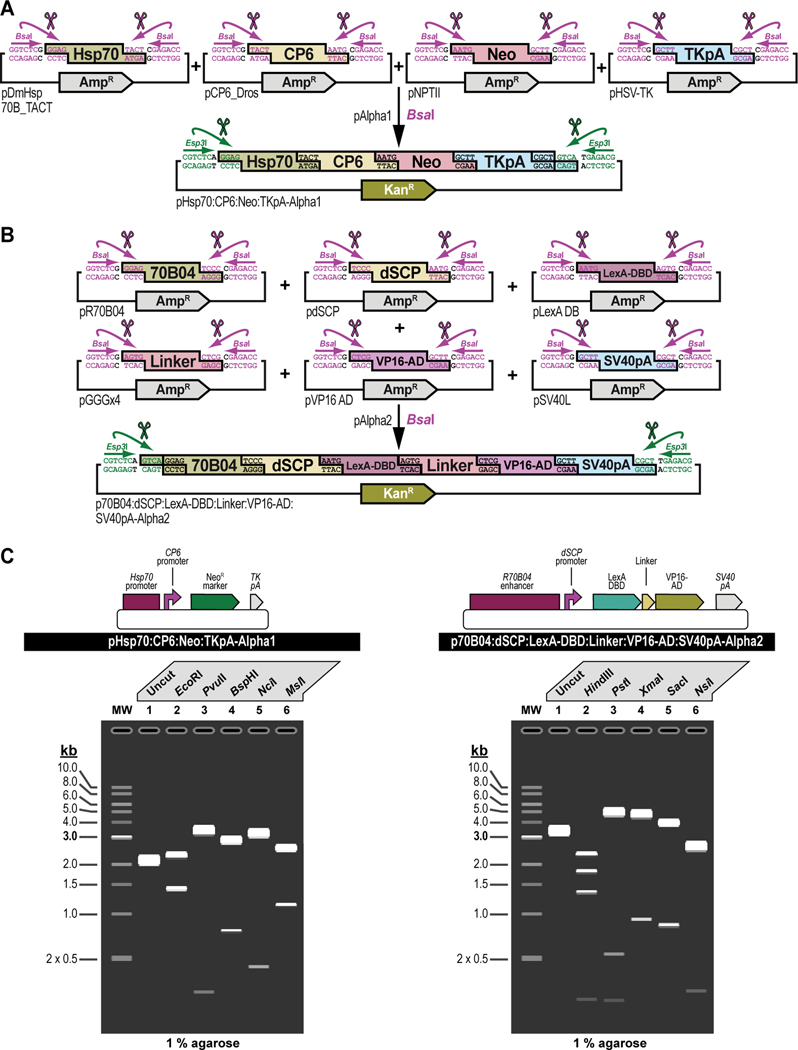
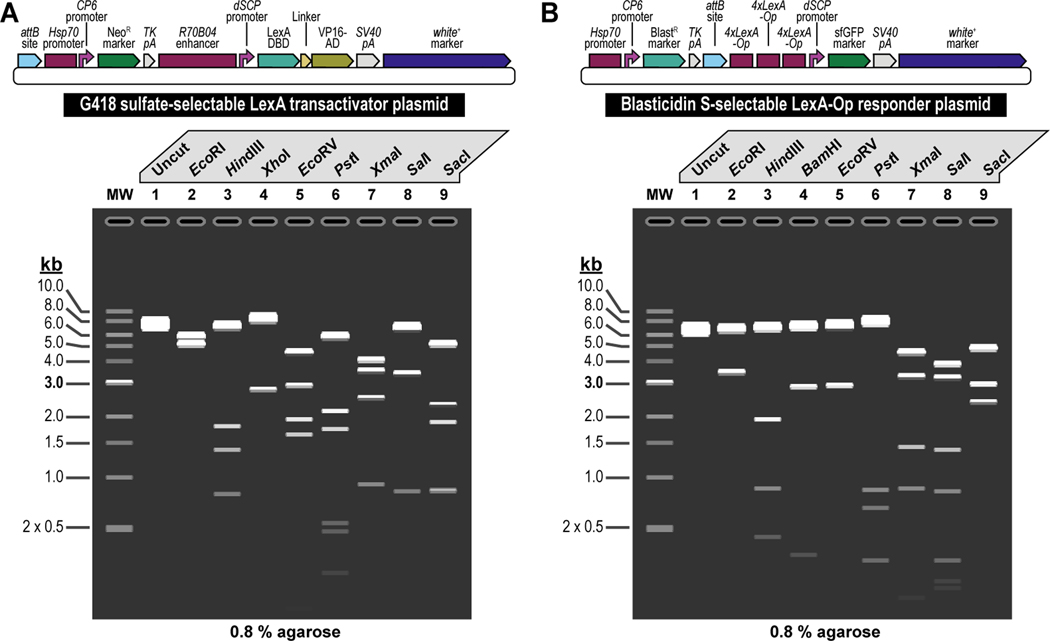
Figure 3. Experimental steps during a typical synthetic assembly DNA cloning workflow to build a genetic construct of continuously increasing complexity: building a G418 sulfate-selectable LexA transactivator plasmid for tissue-specific overexpression as an example.
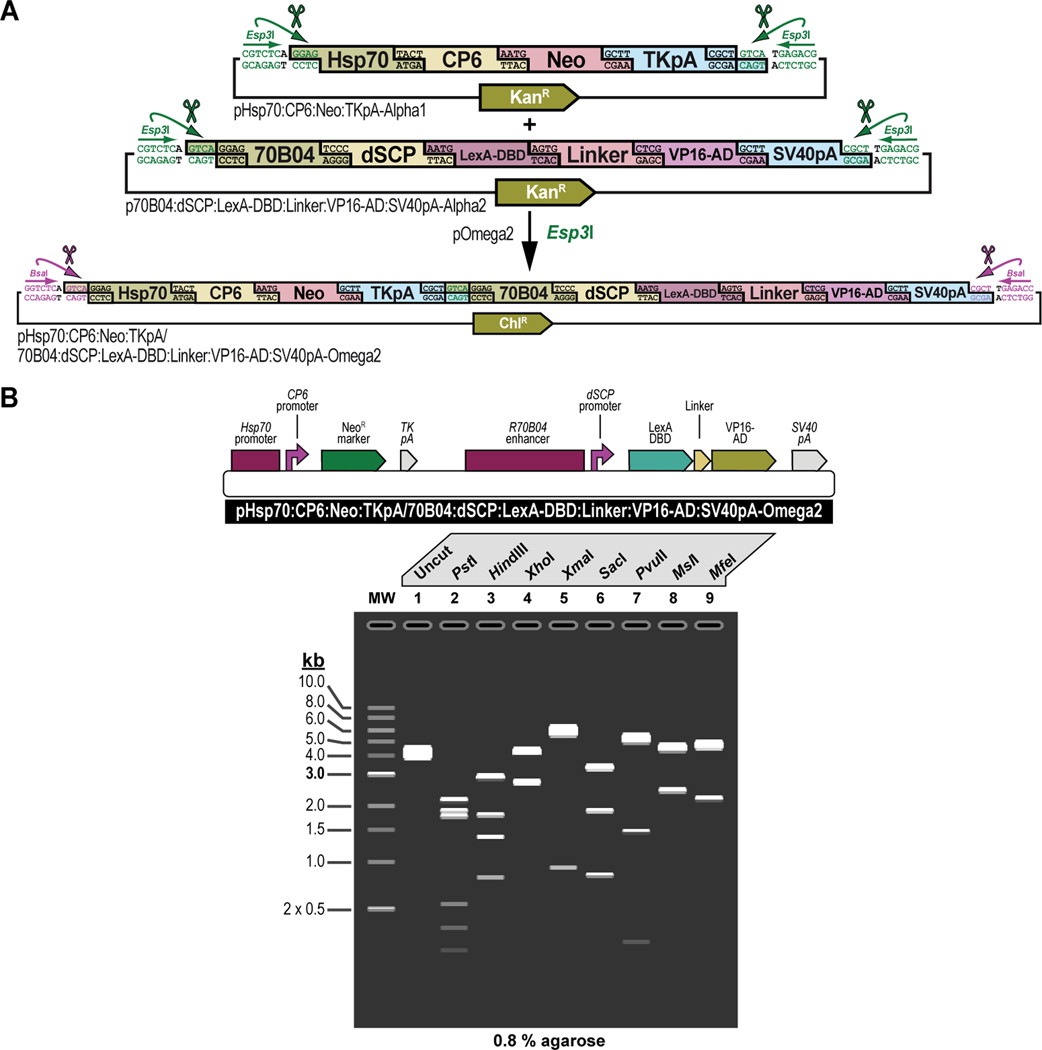
First, all parts needed for assembly are accommodated or “domesticated” into a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3) using Esp3I, as described in Basic Protocol 1, Basic Protocol 2, and Alternate Protocol 2. Parts include a ФC31 bacteriophage attB attachment site for site-specific transgenesis (attB site), the Hsp70 promoter from Drosophila melanogaster (Hsp70 promoter), the synthetic Escherichia coli CP6 promoter (CP6 promoter), the Neomycin phosphotransferase II of transposon Tn5 (NeoR marker), the minimal polyadenylation signal of the thymidine kinase gene from the herpes simplex virus (TK pA), the R70B04 enhancer from Drosophila melanogaster (R70B04 enhancer), the Drosophila melanogaster synthetic core promoter (dSCP promoter), the DNA binding domain of the LexA repressor from Escherichia coli (LexA DBD), a (GlyGlyGlySer)4 peptide linker (Linker), the transcription factor activation domain of VP16 from the herpes simplex virus (VP16-AD), the late polyadenylation signal from simian vacuolating virus 40 (SV40 pA), and the dominant “eye” screening marker called “mini-white” from Drosophila melanogaster (white+ marker). Next, several of these parts are assembled in Alpha level vector backbones to form transcription units using BsaI, as described in Basic Protocol 3. The Hsp70 promoter, the CP6 promoter, the NeoR marker, and the TK pA are assembled in pAlpha1 resulting in the plasmid pHsp70:CP6:Neo:TKpA-Alpha1, while the R70B04 enhancer, the dSCP promoter, the LexA DBD, Linker, the VP16-AD, and the SV40 pA are assembled in pAlpha2 resulting in the plasmid pR70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2. Alpha assemblies can be further combined in Omega level vector backbones using Esp3I, to form genetic circuits, defined by two transcription units or other DNA assemblies of various complexity, as described in Basic Protocol 4. Alpha assemblies, pHsp70:CP6:Neo:TKpA-Alpha1 and pR70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2 are combined in pOmega2 using Esp3I, resulting in plasmid pHsp70:CP6:Neo:TKpA/R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Omega2. This assembly, together with the attB site located in a pOmega1 plasmid are further combined in pAlpha1 using BsaI, resulting in plasmid pAttB/Hsp70:CP6:Neo:TKpA/R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha1 (repeat Basic Protocol 3), which then, together with the white+ marker located in a pAlpha2 plasmid, is additionally expanded in a pOmega2 plasmid using Esp3I to form the final plasmid, pAttB/Hsp70:CP6:Neo:TKpA/R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA/White-Omega2, also known as the G418 sulfate-selectable LexA transactivator plasmid for tissue-specific overexpression within the R70B04 expression domain (repeat Basic Protocol 4). If needed, this plasmid can be further expanded using additional rounds of synthetic assembly DNA cloning using BsaI (repeat Basic Protocol 3) or Esp3I (repeat Basic Protocol 4), as indicated. Essentially, assemblies could occur in perpetuum, solely limited by plasmid backbone properties (a high-copy number plasmid backbone can maintain up to about 20 kilobases of insert, while a low-copy number plasmid backbone could maintain several hundreds of kilobases of insert) and the large plasmid maintenance capabilities of the bacterial host (some bacterial hosts cannot maintain large insert plasmids while others can). Combined with an appropriate LexA-Op responder plasmid, e.g., the Blasticidin S-selectable LexA-Op responder plasmid (see Figure 4), both plasmids can then be used to obtain a double transgenic fly through multiplexed dual selection transgenesis (see Venken et al., 2023) that can be used to determine gene expression patterns (See Figure 15).
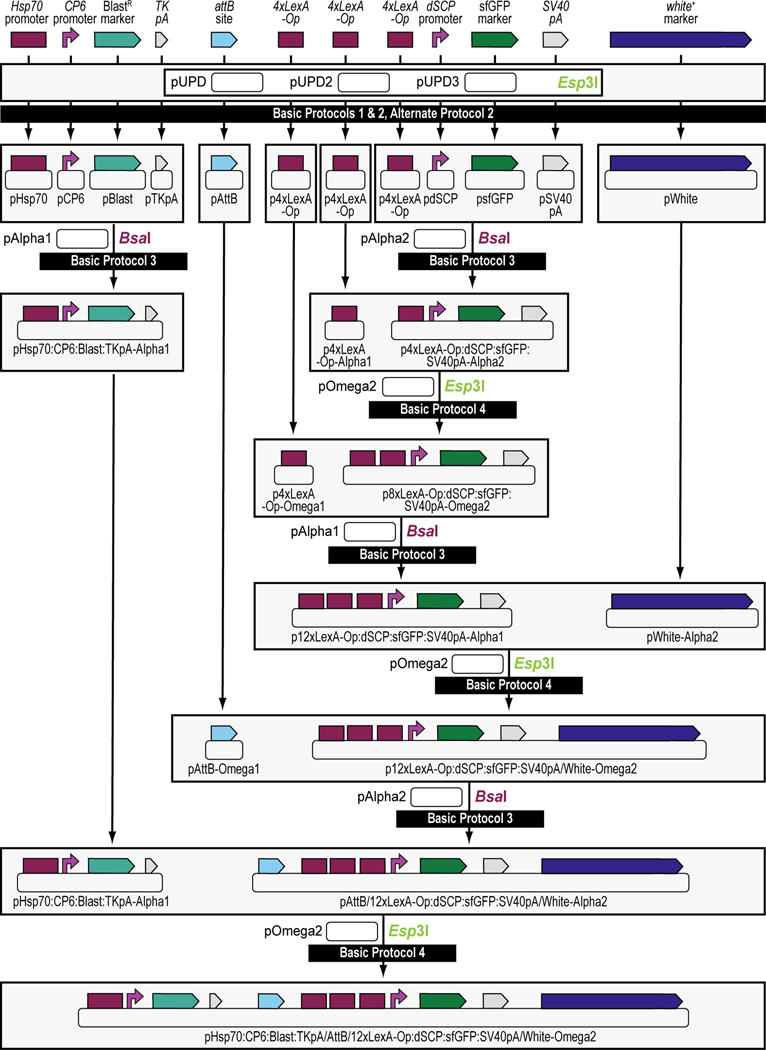
Figure 4. Schematic overview of the experimental steps during the synthetic assembly cloning workflow to build a Blasticidin S-selectable LexA-Op responder plasmid for tissue-specific overexpression in Drosophila melanogaster.
First, all parts needed for assembly are domesticated into a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3) using Esp3I (Basic Protocol 1, Basic Protocol 2, or Alternate Protocol 2). Parts are: the Hsp70 promoter from Drosophila melanogaster (Hsp70 promoter), the synthetic Escherichia coli CP6 promoter (CP6 promoter), the Blasticidin S resistance deaminase gene (BlastR marker), the minimal polyadenylation signal of the thymidine kinase gene from the herpes simplex virus (TK pA), a ФC31 bacteriophage attB attachment site for site-specific transgenesis (attB site), three times 4 copies of the binding site for the LexA DNA binding domain (4xLexA-Op), the Drosophila melanogaster synthetic core promoter (dSCP promoter), the green fluorescent protein reporter sfGFP (sfGFP marker), the late polyadenylation signal from simian vacuolating virus 40 (SV40 pA), and the dominant “eye” screening marker called “mini-white” from Drosophila melanogaster (white+ marker). Next several of these parts are assembled in Alpha level vector backbones to form transcription units using BsaI (see Basic Protocol 3). The Hsp70 promoter, the CP6 promoter, the BlastR marker, and the TK pA are assembled in pAlpha1, resulting in plasmid pHsp70:CP6:Blast:TKpA-Alpha1, while one copy of 4xLexA-Op, the dSCP promoter, the sfGFP marker, and the SV40 pA are assembled together in pAlpha2, resulting in plasmid p4xLexA-Op:dSCP:sfGFP:SV40pA-Alpha2. The latter assembly, together with the second copy of 4xLexA-Op located in pAlpha1 are combined in pOmega2 using Esp3I, resulting in plasmid p8xLexA-Op:dSCP:sfGFP:SV40pA-Alpha2 (see Basic Protocol 4), which together with the third and final copy of 4xLexA-Op located in pOmega1 are combined in pAlpha1 using BsaI, resulting in plasmid p12xLexA-Op:dSCP:sfGFP:SV40pA-Alpha2 (see Basic Protocol 3). This assembly, together with the white+ marker located in a pAlpha2 plasmid, is further combined in a pOmega2 plasmid using Esp3I resulting in plasmid p12xLexA-Op:dSCP:sfGFP:SV40pA/White-Omega2 (see Basic Protocol 4), to which the attB site located in a pOmega1 plasmid is added in pAlpha2 using BsaI, resulting in plasmid pAttB/12xLexA-Op:dSCP:sfGFP:SV40pA/White-Alpha2 (see Basic Protocol 3). During a final step, the Hsp70:CP6:Blast:TKpA-Alpha1 transcription unit located in pAlpha1 and the previous assembly in pAlpha2 are combined together in pOmega2 using Esp3I to form the final plasmid pHsp70:CP6:Blast:TKpA/AttB/12xLexA-Op:dSCP:sfGFP:SV40pA/White-Omega2, also known as the Blasticidin S-selectable LexA-Op responder plasmid (see Basic Protocol 4). Combined with an appropriate LexA transactivator plasmid, e.g., the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3), both plasmids can then be used to obtain a double transgenic fly through multiplexed dual selection transgenesis (see Venken et al., 2023) that can be used to determine gene expression patterns (See Figure 15).
BASIC PROTOCOL 1
Obtaining and cloning a de novo synthesized DNA part for synthetic assembly DNA cloning.
Introductory paragraph
This protocol will demonstrate how to obtain a DNA part and integrate it into, or domesticate it for, the GoldenBraid 2.0 (GB2.0) workflow starting from a de novo synthesized DNA fragment (Figure 5). We will explain the general principles of this protocol (Figure 5A and Figure 5B) and apply those principles to the domestication of the antibiotic resistance gene encoding the Neomycin phosphotransferase II protein (Figure 5C and Figure 5D), obtained from transposon Tn5, used as one of the DNA parts to build the G418 sulfate-selectable LexA transactivator plasmid (Figure 3). By the end of this protocol the user should be able to analyze the DNA sequence for a desired DNA part, modify the sequence to be both codon-optimized for optimal expression in Drosophila melanogaster and compatible with GB2.0 synthetic DNA assembly cloning, as well as order the DNA part by de novo DNA synthesis (Figure 5A and Figure 5C). They should also be able to clone the modified sequence into a Universal Part Domestication plasmid vector backbone and verify the accuracy of the resulting clones using restriction enzyme DNA fingerprinting and Sanger DNA sequencing (Figure 5B and Figure 5D). For existing sequences that originate from a plasmid or genomic DNA and do not require codon optimization for optimal expression in Drosophila melanogaster, see Basic Protocol 2 and Alternate Protocol 2 for the corresponding domestication protocol. The DNA parts generated in this protocol can be combined into a multipartite transcription unit or any other genetically encoded assembly with a specific function (see Basic Protocol 3).
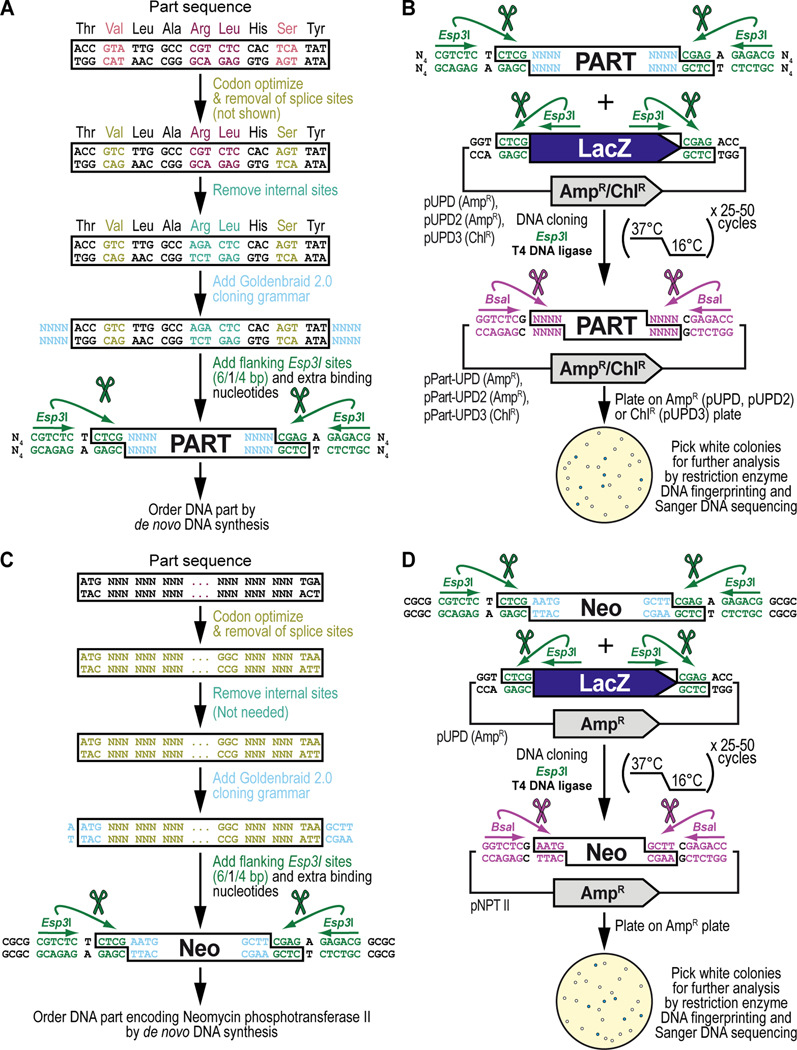
Figure 5. Obtaining and cloning a de novo synthesized DNA part for synthetic assembly cloning. (A) General principles of obtaining a DNA part by de novo DNA synthesis.
Designing the sequence for a de novo synthesized DNA part begins with codon optimization for expression in Drosophila melanogaster (shown in light green) including removal of Drosophila splice acceptor and donor sites (not shown in this example of too short of a sequence, see Text), followed by identification and manual removal of internal binding sites for the Type IIs restriction enzymes Esp3I (“CGTCTC”) (shown in cyan) and BsaI (“GGTCTC”) (not shown). Next, on either side of the DNA part, the desired 4-nucleotide Goldenbraid 2.0 cloning grammar (see Figure 2B) is added (shown in light blue), as well as a pair of inverted Esp3I sites (shown in dark green) that will generate “CTCG” overhangs required for domestication (see B). For improved restriction enzyme binding, 4 extra nucleotides, “NNNN” abbreviated to “N4” (we typically use “CGCG”) (see C), are added to the free ends of the DNA fragment as well (shown in black). An application of the principles described here are illustrated below for obtaining the DNA sequence encoding the selectable marker Neomycin phosphotransferase II (see C), used as one of the parts to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3). (B) General principles of cloning a de novo synthesized DNA part into a pUPD vector backbone. Once synthesized, the DNA fragment is combined with a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3), the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates (ampicillin for pUPD and pUPD2, or chloramphenicol for pUPD3), assembled plasmids are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting and Sanger DNA sequencing (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment. An application of the principles described here is illustrated below for cloning of the DNA part encoding the selectable marker Neomycin phosphotransferase II (see D), used as one of the parts to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3). (C) DNA synthesis of the DNA part encoding Neomycin phosphotransferase II (Neo). Designing the DNA part encoding Neomycin phosphotransferase II (Neo) begins with codon optimization for expression in Drosophila melanogaster (shown in light green) including removal of Drosophila splice acceptor and donor sites (see Text), followed by identification and manual removal of internal binding sites for the Type IIs restriction enzymes Esp3I (CGTCTC) and BsaI (GGTCTC) (both not needed in this example). Next, on either side of the DNA part, the 5’ “AATG” and 3’ “GCTT” Goldenbraid 2.0 cloning grammars are integrated/added (shown in light blue) to couple it to a 5’UTR and 3’UTR, respectively (see Figure 2B), as well as a pair of inverted Esp3I sites (shown in dark green) that will generate overhangs (CTCG) required for domestication (see D). For improved restriction enzyme binding, the tetranucleotide “CGCG” is added last (shown in black). (D) Cloning of the DNA part encoding Neomycin phosphotransferase II (Neo) into pUDP. Once synthesized, the “Neo” fragment is combined with the Universal Part Domesticator plasmid pUPD, the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using 10x T4 DNA ligase buffer). After overnight selection on bacterial plates supplemented with ampicillin for pUPD, assembled plasmids are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting and Sanger DNA sequencing (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment.
Materials
Reagents, solutions, and starting samples or test organisms/cells
De novo synthesized DNA part (commercially obtained)
EB buffer (10 mM Tris-Cl, pH 8.5) from QIAprep spin miniprep kit (see below)
- At least one of the following Universal Part Domestication plasmids (pUPD):
- pUPD (Sarrion-Perdigones et al., 2011) (Table 1)
- pUPD2, provided by the public plasmid repository, Addgene (https://www.addgene.org/) (Addgene, cat. no. 165856) (Matinyan et al., 2021b) (Table 1)
- pUPD3, provided by the public plasmid repository, Addgene (https://www.addgene.org/) (Addgene, cat. no. 118043) (Matinyan et al., 2021b) (Table 1)
1xLB agar (see Reagents and Solutions section for recipe)
Ampicillin powder (VWR, cat. no. IC19014605) to make a 1,000x stock solution in 50% EtOH diluted with MilliQ H2O, followed by filter sterilization (100 mg/ml)
MilliQ H2O, sterilized by autoclaving
Chloramphenicol powder (VWR, cat. no. 45000–618) to make a 1,000x stock solution in 100% EtOH, sterilization not required (12.5 mg/ml)
2xLB-0.5 medium (see Reagents and Solutions section for recipe)
Glycerol (Fisher Scientific, cat. no. BP229–1) to make a 40% glycerol solution in MilliQ H2O, sterilized by autoclaving
QIAprep spin miniprep kit (QIAGEN, cat. no. 27106) for plasmid purification
EcoRI-HF restriction enzyme for restriction enzyme DNA fingerprinting (New England Biolabs, cat. no. R3101L)
10x rCutSmart Buffer for restriction enzyme digestions (NEB B6004S)
Universal DNA sequencing primers for Sanger DNA sequencing of pUPD vectors: T7 (TAATACGACTCACTATAGGG), SP6 (ATTTAGGTGACACTATAGA), M13 Forward (GTAAAACGACGGCCAG) and M13 Reverse (CAGGAAACAGCTATGAC)
Esp3I restriction enzyme for synthetic assembly DNA cloning (New England Biolabs, cat. no. R0734L)
T4 DNA ligase and 10x T4 DNA ligase buffer (Promega, cat. no. M1804)
Home-made chemocompetent E. coli cells (Sarrion‐Perdigones et al., 2020), using the DH10B-T1R strain (ThermoFisher Scientific, cat. no., 12331013)
X-Gal (5-bromo-4-chloro-3-indolyl-ß-D-galactopyranoside) powder (VWR, cat. no. 97061–648) to make a 1,000x stock solution (2%) in dimethyl sulfoxide (DMSO), sterilization not required
Table 1. Summary of vectors described in this work.
Plasmid name, brief description, bacterial antibiotic resistance, Addgene stock number, and bibliographic reference are indicated for all plasmids mentioned in this work. Addgene, public plasmid repository (https://www.addgene.org/)
| Plasmid name | Description | Resistance | Addgene | Reference |
|---|---|---|---|---|
| pUPD | Universal Part Domesticator plasmid: “Domestication” level plasmid to adapt parts to Goldenbraid 2.0 | AmpicillinR | NA | (Sarrion-Perdigones et al., 2011) |
| pUPD2 | Universal Part Domesticator plasmid 2: “Domestication” level plasmid to adapt parts to Goldenbraid 2.0 | AmpicillinR | 165856 | (Matinyan et al., 2021b) |
| pUPD3 | Universal Part Domesticator plasmid: 3”Domestication” level plasmid to adapt parts to Goldenbraid 2.0 | ChloramphenicolR | 118043 | (Sarrion-Perdigones et al., 2019) |
| pAlpha1 | Alpha level destination plasmid 1: “Alpha” level Goldenbraid 2.0 plasmid | KanamycinR | 118044 | (Sarrion-Perdigones et al., 2019) |
| pAlpha2 | Alpha level destination plasmid 2: “Alpha” level Goldenbraid 2.0 plasmid | KanamycinR | 118045 | (Sarrion-Perdigones et al., 2019) |
| pOmega1 | Omega level destination plasmid 1: “Omega” level Goldenbraid 2.0 plasmid | ChloramphenicolR | 118046 | (Sarrion-Perdigones et al., 2019) |
| pOmega2 | Omega level destination plasmid 2: “Omega” level Goldenbraid 2.0 plasmid | ChloramphenicolR | 118047 | (Sarrion-Perdigones et al., 2019) |
| pDmHsp70B_TACT | “Domesticated part” plasmid encoding the Drosophila melanogaster Hsp70 promoter | AmpicillinR | 165780 | (Matinyan et al., 2021b) |
| pCP6_Dros | “Domesticated part” plasmid encoding the synthetic Escherichia coli CP6 promoter containing a Drosophila melanogaster Kozak/Cavener translational initiation sequence | AmpicillinR | 165794 | (Matinyan et al., 2021b) |
| pNPTII | “Domesticated part” plasmid encoding a Drosophilized Neomycin phosphotransferase II of transposon Tn5 | AmpicillinR | 165836 | (Matinyan et al., 2021b) |
| pHSV-TK | “Domesticated part” plasmid encoding a minimal polyadenylation signal of the thymidine kinase gene from the herpes simplex virus | AmpicillinR | 165801 | (Matinyan et al., 2021b) |
| pR70B04 | “Domesticated part” plasmid encoding the R70B04 enhancer from Drosophila melanogaster | AmpicillinR | 165792 | (Matinyan et al., 2021b) |
| pdSCP | “Domesticated part” plasmid encoding the Drosophila melanogaster synthetic core promoter | AmpicillinR | 165776 | (Matinyan et al., 2021b) |
| pLexA BD | “Domesticated part” plasmid encoding the Drosphilized DNA binding domain of the LexA repressor from Escherichia coli | AmpicillinR | 165821 | (Matinyan et al., 2021b) |
| pGGGSx4 | “Domesticated part” plasmid encoding the Drosphilized peptide linker (GlyGlyGlySer)4 | AmpicillinR | 165807 | (Matinyan et al., 2021b) |
| pVP16 AD | “Domesticated part” plasmid encoding the Drosphilized transcription factor activation domain of VP16 from the herpes simplex virus | AmpicillinR | 165822 | (Matinyan et al., 2021b) |
| pSV40L | “Domesticated part” level plasmid encoding the late polyadenylation signal from simian vacuolating virus 40 | AmpicillinR | 165798 | (Matinyan et al., 2021b) |
| pFC31 attB O1 | “Omega” level plasmid containing the attB site from the bacteriophage phiC31 cloned in Omega level destination plasmid 1 | ChloramphenicolR | 165843 | (Matinyan et al., 2021b) |
| pMiniWhiteA2 | “Alpha” level plasmid encoding the dominant “eye” screening marker called “mini-white” from Drosophila melanogaster cloned in Alpha level destination plasmid 2 | KanamycinR | 165873 | (Matinyan et al., 2021b) |
Hardware and instruments
Disposable inoculating loops (VWR, cat. no. 12000–806)
Bacterial plates (VWR, cat. no. 25384–092)
37°C incubator (VWR, cat. no. 89409–216)
14-ml disposable culture tubes (VWR, cat. no. 60818–689)
32°C incubator-shaker (Amerex, cat. no. 747/747R)
1.7-ml microcentrifuge tubes (VWR, cat. no. 20170–038)
Refrigerated tabletop centrifuge that can accommodate 14-ml tubes (Fisher Scientific, cat. no. 75230115)
Tabletop microcentrifuge that can accommodate 1.7-ml tubes (Fisher Scientific, cat. no. 75002435)
Spectrophotometer (DeNovix, cat. no. DS-11+)
Reagents and equipment for agarose gel electrophoresis (Voytas, 2001)
Gel documentation system
0.2-ml PCR strip tubes with individually attached caps (VWR, cat. no. 53509–304)
PCR machine (Applied Biosystems, cat. no. 4375786)
Ice bucket
Dry bead bath (Lab Armor, cat. no. 74309–706), set at 42°C for bacterial transformation
Glass spreading beads (VWR, cat. no. 26396–508)
Protocol steps
Obtaining a desired DNA part for synthetic assembly DNA cloning by de novo DNA synthesis
-
1
Choose the DNA part you wish to integrate into, or domesticate for, the GB2.0 workflow. This DNA part can be a promoter, coding region, polyA terminator or any other DNA sequence. Here, we will demonstrate domestication of the coding DNA sequence for the antibiotic resistance gene encoding the Neomycin phosphotransferase II (NPT II) protein, obtained from transposon Tn5, used as one of the DNA parts to build the G418 sulfate-selectable LexA transactivator plasmid (Figure 3). The starting DNA sequence, with “ATG” start and “TGA” stop codons highlighted in bold black, is:
ATGATTGAACAAGATGGATTGCACGCAGGTTCTCCGGCCGCTTGGGTGGAGAGGCTATTCGGCTATGACTGGGCACAACAGACAATCGGCTGCTCTGATGCCGCCGTGTTCCGGCTGTCAGCGCAGGGGCGCCCGGTTCTTTTTGTCAAGACCGACCTGTCCGGTGCCCTGAATGAACTGCAAGACGAGGCAGCGCGGCTATCGTGGCTGGCCACGACGGGCGTTCCTTGCGCAGCTGTGCTCGACGTTGTCACTGAAGCGGGAAGGGACTGGCTGCTATTGGGCGAAGTGCCGGGGCAGGATCTCCTGTCATCTCACCTTGCTCCTGCCGAGAAAGTATCCATCATGGCTGATGCAATGCGGCGGCTGCATACGCTTGATCCGGCTACCTGCCCATTCGACCACCAAGCGAAACATCGCATCGAGCGAGCACGTACTCGGATGGAAGCCGGTCTTGTCGATCAGGATGATCTGGACGAAGAGCATCAGGGGCTCGCGCCAGCCGAACTGTTCGCCAGGCTCAAGGCGAGCATGCCCGACGGCGAGGATCTCGTCGTGACCCATGGCGATGCCTGCTTGCCGAATATCATGGTGGAAAATGGCCGCTTTTCTGGATTCATCGACTGTGGCCGGCTGGGTGTGGCGGACCGCTATCAGGACATAGCGTTGGCTACCCGTGATATTGCTGAAGAGCTTGGCGGCGAATGGGCTGACCGCTTCCTCGTGCTTTACGGTATCGCCGCTCCCGATTCGCAGCGCATCGCCTTCTATCGCCTTCTTGACGAGTTCTTCTGA
-
2
Using an online tool such as the Codon Optimization Tool from Integrated DNA Technologies (IDT) (https://www.idtdna.com/CodonOpt), optimize the coding DNA sequence for expression in Drosophila melanogaster (Figure 5A and Figure 5C). If implementing this method to another species not listed within this tool, use the online codon optimization tool available from GENEius (https://geneius.de) which allows the import of custom codon usage tables for many species, available from the Codon Usage Database (http://www.kazusa.or.jp/codon/) (Nakamura et al., 2000). If integrating a non-coding DNA part into the GB2.0 workflow, skip to Step 4. The resulting DNA sequence of this DNA part, from IDT, with “ATG” start and “TAA” stop codons highlighted in bold black, is:
ATGATTGAGCAGGATGGACTGCACGCTGGCTCGCCAGCTGCCTGGGTGGAGCGTCTGTTCGGCTACGATTGGGCCCAGCAGACCATCGGCTGCTCCGATGCTGCCGTGTTCCGTCTGTCGGCCCAGGGACGCCCCGTGCTGTTCGTCAAGACCGACCTGAGTGGAGCCCTGAACGAGCTCCAGGATGAGGCAGCTCGTCTGAGCTGGCTGGCCACAACAGGAGTGCCGTGTGCTGCCGTGCTGGATGTGGTGACCGAAGCTGGACGCGATTGGCTGCTGCTGGGCGAGGTGCCAGGCCAGGACCTGCTGTCCAGCCATCTGGCCCCAGCCGAGAAGGTGTCCATTATGGCCGATGCTATGCGTCGCCTGCACACCCTGGACCCAGCCACCTGCCCCTTTGACCACCAGGCCAAGCACCGTATTGAGCGTGCCCGAACCCGTATGGAGGCTGGACTGGTGGACCAGGATGACCTGGATGAGGAGCACCAGGGCCTGGCTCCAGCCGAGCTGTTTGCCCGTCTGAAGGCCCGTATGCCCGATGGCGAGGACCTGGTGGTGACCCACGGCGACGCCTGCCTGCCCAACATTATGGTGGAGAACGGACGCTTCAGTGGCTTCATTGATTGTGGACGCCTGGGCGTGGCCGACCGTTACCAGGATATCGCCCTGGCCACCCGAGATATCGCCGAGGAGCTGGGAGGTGAGTGGGCCGACCGATTCCTGGTGCTCTACGGCATTGCAGCCCCAGATAGCCAGCGTATTGCCTTCTACCGTCTGCTGGACGAGTTCTTCTAA
-
3
Once you have obtained the codon-optimized sequence for the DNA part, analyze the sequence for putative splice donor and acceptor sites used in Drosophila melanogaster using the online splice site prediction tool from the Berkley Drosophila Genome Project (https://www.fruitfly.org/seq_tools/splice.html) (Reese et al., 1997). Select the option for detecting both 5’ and 3’ splice sites in the sequence of the DNA part for the organism Drosophila. You do not need to detect 5’ and 3’ splice sites in the reverse direction. The default threshold of 0.4 for prediction score does not need to be changed (i.e., any score lower than 0.4 can be considered a false positive). Once analyzed, manually change all detected sites (Figure 5A and Figure 5C) taking care that the coding sequence does not change. Afterwards, reanalyze the sequence of the DNA part for any additional sites that may have been overlooked or introduced and manually remove those as well. The example sequence only had a single detected splice donor site (highlighted in bold dark blue in the sequence above) that was adjusted, highlighted below in bold light green, changing the GGT codon (underlined in the sequence above) to GGC, which both code for Glycine:
ATGATTGAGCAGGATGGACTGCACGCTGGCTCGCCAGCTGCCTGGGTGGAGCGTCTGTTCGGCTACGATTGGGCCCAGCAGACCATCGGCTGCTCCGATGCTGCCGTGTTCCGTCTGTCGGCCCAGGGACGCCCCGTGCTGTTCGTCAAGACCGACCTGAGTGGAGCCCTGAACGAGCTCCAGGATGAGGCAGCTCGTCTGAGCTGGCTGGCCACAACAGGAGTGCCGTGTGCTGCCGTGCTGGATGTGGTGACCGAAGCTGGACGCGATTGGCTGCTGCTGGGCGAGGTGCCAGGCCAGGACCTGCTGTCCAGCCATCTGGCCCCAGCCGAGAAGGTGTCCATTATGGCCGATGCTATGCGTCGCCTGCACACCCTGGACCCAGCCACCTGCCCCTTTGACCACCAGGCCAAGCACCGTATTGAGCGTGCCCGAACCCGTATGGAGGCTGGACTGGTGGACCAGGATGACCTGGATGAGGAGCACCAGGGCCTGGCTCCAGCCGAGCTGTTTGCCCGTCTGAAGGCCCGTATGCCCGATGGCGAGGACCTGGTGGTGACCCACGGCGACGCCTGCCTGCCCAACATTATGGTGGAGAACGGACGCTTCAGTGGCTTCATTGATTGTGGACGCCTGGGCGTGGCCGACCGTTACCAGGATATCGCCCTGGCCACCCGAGATATCGCCGAGGAGCTGGGAGGCGAGTGGGCCGACCGATTCCTGGTGCTCTACGGCATTGCAGCCCCAGATAGCCAGCGTATTGCCTTCTACCGTCTGCTGGACGAGTTCTTCTAA
-
4
Analyze the resulting sequence for Esp3I and BsaI Type IIs restriction enzyme sites. Since both enzymes are used during the entire GB2.0 workflow, they should be removed from the DNA sequence via synonymous codon switching to prevent unwanted cutting within the sequence (Figure 5A and Figure 5C). If you are domesticating a non-coding DNA part, such as a promoter or terminator, it may not be possible to remove the Type IIs restriction enzyme sites without potentially compromising the functionality of the DNA part, although this can be experimentally tested during downstream functional analysis. The GB2.0 workflow can tolerate up to one recognition site for each Type IIs restriction enzyme (Esp3I and BsaI) within a DNA part and still assemble the DNA part as two pieces with moderate efficiency. Although the DNA part is cut, both pieces can still reconstitute back to the original DNA part during multipartite assembly, as long as the overhangs generated by the cut are not complementary to any of the overhangs used by the “cloning “grammars” (Figure 2B and Figure 2C). DNA parts with more than one recognition site for one of the Type IIs restriction enzymes (Esp3I or BsaI) will become increasingly more difficult to assemble. In our example sequence, no Esp3I or BsaI sites are present.
-
5
Next, add the four-nucleotide cloning overhangs corresponding to the grammar of the DNA part to the 5’ and 3’ ends of the sequence (Figure 5A and Figure 5C). For a standard coding DNA sequence, as in our example, the 5’ overhang is “AATG”, while the 3’ overhang is “GCTT”, to couple it to a 5’UTR and 3’UTR, respectively (see Figure 2B). Note that the 5’ overhang includes the start codon sequence “ATG” plus an additional 5’ “A” nucleotide. Other types of DNA parts (e.g., promoter, linker, terminator, and so on) have their own predefined cloning overhangs (Figure 2B). The resulting sequence, with the four-nucleotide “grammar” cloning overhangs highlighted in bold light blue, is:
AATGATTGAGCAGGATGGACTGCACGCTGGCTCGCCAGCTGCCTGGGTGGAGCGTCTGTTCGGCTACGATTGGGCCCAGCAGACCATCGGCTGCTCCGATGCTGCCGTGTTCCGTCTGTCGGCCCAGGGACGCCCCGTGCTGTTCGTCAAGACCGACCTGAGTGGAGCCCTGAACGAGCTCCAGGATGAGGCAGCTCGTCTGAGCTGGCTGGCCACAACAGGAGTGCCGTGTGCTGCCGTGCTGGATGTGGTGACCGAAGCTGGACGCGATTGGCTGCTGCTGGGCGAGGTGCCAGGCCAGGACCTGCTGTCCAGCCATCTGGCCCCAGCCGAGAAGGTGTCCATTATGGCCGATGCTATGCGTCGCCTGCACACCCTGGACCCAGCCACCTGCCCCTTTGACCACCAGGCCAAGCACCGTATTGAGCGTGCCCGAACCCGTATGGAGGCTGGACTGGTGGACCAGGATGACCTGGATGAGGAGCACCAGGGCCTGGCTCCAGCCGAGCTGTTTGCCCGTCTGAAGGCCCGTATGCCCGATGGCGAGGACCTGGTGGTGACCCACGGCGACGCCTGCCTGCCCAACATTATGGTGGAGAACGGACGCTTCAGTGGCTTCATTGATTGTGGACGCCTGGGCGTGGCCGACCGTTACCAGGATATCGCCCTGGCCACCCGAGATATCGCCGAGGAGCTGGGAGGCGAGTGGGCCGACCGATTCCTGGTGCTCTACGGCATTGCAGCCCCAGATAGCCAGCGTATTGCCTTCTACCGTCTGCTGGACGAGTTCTTCTAAGCTT
-
6
Typically, new DNA parts that are integrated into the GB2.0 workflow are cloned into a plasmid vector backbone for domestication purposes known as the Universal Part Domesticator plasmid (pUPD) (Figure 5B and Figure 5D). Three Universal Part Domesticator plasmids exist: pUPD, pUPD2 and pUPD3 (Table 1). The ampicillin-resistant pUPD is the vector developed in the original GoldenBraid method (Sarrion-Perdigones et al., 2011). An upgraded ampicillin-resistant version, called pUPD2, in which the entry site for the DNA part is flanked on either side by pairs of inverted bacterial transcription terminator DNA parts was developed to increase plasmid stability (Matinyan et al., 2021b). The pUPD3 vector is chloramphenicol-resistant (Matinyan et al., 2021b) and is useful to re-domesticate any DNA part obtained by PCR amplification from a pre-existing ampicillin-resistant plasmid to eliminate potential Ampicillin-based cross-contamination issues during bacterial selection (see Basic Protocol 2 and Alternate Protocol 2). Re-domestication can be necessary to change the grammar of a DNA part to use it in a different position in an assembly. To clone the DNA part into one of these pUPD vectors, a “domesticator” cloning overhang “CTCG” has to be added to both ends to the DNA part that can anneal to “CTCG” overhangs present in the pUPD vectors (Figure 5A and Figure 5C). These overhangs are the same for both the 5’ and 3’ ends and identical for all pUPD backbones, since cloning of DNA parts in pUPD vectors using Esp3I will reconstitute BsaI sites on either end of the DNA part that will be needed to release different DNA parts from pUPD vectors to combine them into Alpha level destination vectors during the next step in the GB2.0 workflow (see Basic Protocol 3). Note that the 3’ overhang (“CGAG”) is the reverse complement of the 5’ overhang sequence (“CTCG”). The resulting sequence, with the four-nucleotide “domesticator” cloning overhangs highlighted in bold dark green, is:
CTCGAATGATTGAGCAGGATGGACTGCACGCTGGCTCGCCAGCTGCCTGGGTGGAGCGTCTGTTCGGCTACGATTGGGCCCAGCAGACCATCGGCTGCTCCGATGCTGCCGTGTTCCGTCTGTCGGCCCAGGGACGCCCCGTGCTGTTCGTCAAGACCGACCTGAGTGGAGCCCTGAACGAGCTCCAGGATGAGGCAGCTCGTCTGAGCTGGCTGGCCACAACAGGAGTGCCGTGTGCTGCCGTGCTGGATGTGGTGACCGAAGCTGGACGCGATTGGCTGCTGCTGGGCGAGGTGCCAGGCCAGGACCTGCTGTCCAGCCATCTGGCCCCAGCCGAGAAGGTGTCCATTATGGCCGATGCTATGCGTCGCCTGCACACCCTGGACCCAGCCACCTGCCCCTTTGACCACCAGGCCAAGCACCGTATTGAGCGTGCCCGAACCCGTATGGAGGCTGGACTGGTGGACCAGGATGACCTGGATGAGGAGCACCAGGGCCTGGCTCCAGCCGAGCTGTTTGCCCGTCTGAAGGCCCGTATGCCCGATGGCGAGGACCTGGTGGTGACCCACGGCGACGCCTGCCTGCCCAACATTATGGTGGAGAACGGACGCTTCAGTGGCTTCATTGATTGTGGACGCCTGGGCGTGGCCGACCGTTACCAGGATATCGCCCTGGCCACCCGAGATATCGCCGAGGAGCTGGGAGGCGAGTGGGCCGACCGATTCCTGGTGCTCTACGGCATTGCAGCCCCAGATAGCCAGCGTATTGCCTTCTACCGTCTGCTGGACGAGTTCTTCTAAGCTTCGAG
-
7
As mentioned above, GB2.0 synthetic assembly DNA cloning uses an alternating pair of Type IIs restriction enzymes, Esp3I and BsaI, during its cloning workflow. Esp3I is used during cloning of new DNA parts into any of the pUPD vectors. Thus, it is necessary to add the enzyme’s DNA binding site to both ends of the DNA part’s sequence (Figure 5A and Figure 5C). Unlike typical Type II restriction enzymes, Esp3I and BsaI are Type IIs restriction enzymes that do not cut directly at their DNA binding sites, but rather one “spacer” nucleotide away from their binding site in a directional manner (Figure 2D). The restriction site for Esp3I is CGTCTCNNNNN, in which the dark green hexanucleotide “CGTCTC” sequence represents the binding site, the black N represents the spacer, and the dark green random (“NNNN”) tetranucleotide sequence represents the cutting overhang sequence. Complete the Esp3I restriction site at both ends, by adding “binding” (“CGTCTC”) and “spacer” (“T”) sequence, to both the 5’ and 3’ ends of your DNA part sequence. The four-nucleotide domesticator cloning overhangs (“CTCG”) added during the previous step act as the cutting overhang sequence. Note that the 3’ end requires reverse complementing the “spacer” (“A”) and “binding” site (“GAGACG”) sequences. The resulting sequence, with the binder and spacer sequences highlighted in bold dark green and bold black, respectively, is:
CGTCTCTCTCGAATGATTGAGCAGGATGGACTGCACGCTGGCTCGCCAGCTGCCTGGGTGGAGCGTCTGTTCGGCTACGATTGGGCCCAGCAGACCATCGGCTGCTCCGATGCTGCCGTGTTCCGTCTGTCGGCCCAGGGACGCCCCGTGCTGTTCGTCAAGACCGACCTGAGTGGAGCCCTGAACGAGCTCCAGGATGAGGCAGCTCGTCTGAGCTGGCTGGCCACAACAGGAGTGCCGTGTGCTGCCGTGCTGGATGTGGTGACCGAAGCTGGACGCGATTGGCTGCTGCTGGGCGAGGTGCCAGGCCAGGACCTGCTGTCCAGCCATCTGGCCCCAGCCGAGAAGGTGTCCATTATGGCCGATGCTATGCGTCGCCTGCACACCCTGGACCCAGCCACCTGCCCCTTTGACCACCAGGCCAAGCACCGTATTGAGCGTGCCCGAACCCGTATGGAGGCTGGACTGGTGGACCAGGATGACCTGGATGAGGAGCACCAGGGCCTGGCTCCAGCCGAGCTGTTTGCCCGTCTGAAGGCCCGTATGCCCGATGGCGAGGACCTGGTGGTGACCCACGGCGACGCCTGCCTGCCCAACATTATGGTGGAGAACGGACGCTTCAGTGGCTTCATTGATTGTGGACGCCTGGGCGTGGCCGACCGTTACCAGGATATCGCCCTGGCCACCCGAGATATCGCCGAGGAGCTGGGAGGCGAGTGGGCCGACCGATTCCTGGTGCTCTACGGCATTGCAGCCCCAGATAGCCAGCGTATTGCCTTCTACCGTCTGCTGGACGAGTTCTTCTAAGCTTCGAGAGAGACG
-
8
Finally, add at least four extra base pairs to either end of the sequence of the DNA part as Type IIs restriction enzymes do not efficiently bind free DNA ends (Figure 5A and Figure 5C). The exact sequence of these extra bases does not matter but we typically add “CGCG” to either end (3’ reverse complement being “GCGC”) as shown below (with the additional “CGCG” sequence highlighted in bold black):
CGCGCGTCTCTCTCGAATGATTGAGCAGGATGGACTGCACGCTGGCTCGCCAGCTGCCTGGGTGGAGCGTCTGTTCGGCTACGATTGGGCCCAGCAGACCATCGGCTGCTCCGATGCTGCCGTGTTCCGTCTGTCGGCCCAGGGACGCCCCGTGCTGTTCGTCAAGACCGACCTGAGTGGAGCCCTGAACGAGCTCCAGGATGAGGCAGCTCGTCTGAGCTGGCTGGCCACAACAGGAGTGCCGTGTGCTGCCGTGCTGGATGTGGTGACCGAAGCTGGACGCGATTGGCTGCTGCTGGGCGAGGTGCCAGGCCAGGACCTGCTGTCCAGCCATCTGGCCCCAGCCGAGAAGGTGTCCATTATGGCCGATGCTATGCGTCGCCTGCACACCCTGGACCCAGCCACCTGCCCCTTTGACCACCAGGCCAAGCACCGTATTGAGCGTGCCCGAACCCGTATGGAGGCTGGACTGGTGGACCAGGATGACCTGGATGAGGAGCACCAGGGCCTGGCTCCAGCCGAGCTGTTTGCCCGTCTGAAGGCCCGTATGCCCGATGGCGAGGACCTGGTGGTGACCCACGGCGACGCCTGCCTGCCCAACATTATGGTGGAGAACGGACGCTTCAGTGGCTTCATTGATTGTGGACGCCTGGGCGTGGCCGACCGTTACCAGGATATCGCCCTGGCCACCCGAGATATCGCCGAGGAGCTGGGAGGCGAGTGGGCCGACCGATTCCTGGTGCTCTACGGCATTGCAGCCCCAGATAGCCAGCGTATTGCCTTCTACCGTCTGCTGGACGAGTTCTTCTAAGCTTCGAGAGAGACGGCGC
-
9
The sequence for your DNA part (Neo) is now ready for DNA synthesis. Order the sequence as a double-stranded DNA fragment from a commercial source. We typically order the DNA parts as a gBlock from IDT (https://www.idtdna.com/) or as a gene strand from Eurofins (https://eurofinsgenomics.com/), although several other vendors provide similar services. Depending on the size of your DNA part, your fragment will arrive within 2 to 4 business days as either lyophilized DNA or as a solution of resuspended DNA in buffer. DNA parts that are larger than 1 kilobase can be divided into smaller sub-DNA parts designed in such a way they can reconstitute during synthetic assembly DNA cloning using Esp3I and 4-bp overhangs present at each division point of the to-be-reconstituted DNA part.
-
10
Upon receiving your fragment encoding the “Neo” DNA part, spin down the contents of the tube and, if DNA is lyophilized, resuspend in EB buffer at a concentration of 20 ng/μl. Vortex and spin down as necessary.
Obtaining and preparing a Universal Part Domestication plasmid for synthetic assembly DNA cloning a de novo synthesized DNA part
-
11Order the desired Universal Part Domestication plasmid vector backbone(s), pUPD, pUPD2 and/or pUPD3, from Addgene (www.addgene.org). Plasmids ordered from Addgene will arrive as agar stabs.
- If the desired Universal Part Domestication plasmid is already present in the lab, identify the glycerol stock.
-
12Using a disposable inoculating loop, streak out each agar stab (or glycerol stock) to single colonies on bacterial plates containing 1xLB agar supplemented with:
- 100 μg/ml ampicillin (pUPD and/or pUPD2)
- 12.5 μg/ml chloramphenicol (pUPD3)
-
13
Incubate plates overnight in a 37°C incubator.
-
14For each plasmid, pick a single colony using a disposable inoculation loop and inoculate into a 14-ml disposable culture tube containing 5 ml of 2xLB-0.5 medium supplemented with appropriate antibiotic:
- 100 μg/ml ampicillin (pUPD and/or pUPD2)
- 12.5 μg/ml chloramphenicol (pUPD3)
-
15
Grow the culture(s) overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
16
For each plasmid for which you don’t have a glycerol stock yet, separate 0.25 ml of the overnight grown culture in a 1.7-ml microcentrifuge tube and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
17Spin down remaining portion of each culture for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
18
Isolate plasmid DNA using the QIAprep spin miniprep kit according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
19
Elute DNA in 50 μl of EB buffer. Measure the concentration using a spectrophotometer.
-
20Verify the identity of Universal Part Domestication plasmids via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Digest 400 ng of the resulting plasmids with the restriction enzyme EcoRI-HF resulting in two distinct bands, one band corresponding to the plasmid backbone and the other band to the LacZ stuffer containing the colorimetric LacZ α-fragment (Figure 5B and Figure 5D):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI-HF
- MilliQ H2O up to 25 μl
- Alternatively, Universal Part Domestication plasmids can be verified using a commercial Sanger DNA sequencing service and the universal DNA sequencing primers, T7 and SP6 (pUPD), or M13 Forward and M13 Reverse (pUPD2 or pUPD3).
-
21
Once you have verified the identity of the Universal Part Domestication plasmids, dilute each to a final concentration of 75 ng/μl using EB buffer.
Synthetic assembly DNA cloning a de novo synthesized DNA part in a Universal Part Domestication plasmid
-
22In a 0.2-ml PCR strip tube with individually attached cap, add synthesized DNA part (Neo), Universal Part Domesticator vector (pUPD) and enzymes according to the following recipe:
- 40 ng DNA part (Neo)
- 75 ng Universal Part Domesticator vector (pUPD)
- 1 μl Esp3I
- 1 μl of T4 DNA ligase
- 2 μl of 10x T4 DNA ligase buffer
- MilliQ H2O up to 20 μl
- While Esp3I’s isoschizomer, BsmBI, can also be used during synthetic assembly DNA cloning, Esp3I is preferred over BsmBI, since Esp3I cuts optimally at 37°C, while BsmBI cuts optimally at 55°C, a temperature not recommended for T4 DNA ligase function.
-
23Briefly vortex and spin down tube contents and place into a PCR machine. Use the following GB2.0 cloning protocol to drive the synthetic DNA assembly cloning reaction:
- 25 to 50 cycles, each consisting of Esp3I-mediated cutting at 37°C for 2 minutes followed by T4 DNA ligase-mediated ligation at 16°C for 5 minutes
- Heat inactivation of enzymes at 85°C for 20 minutes
- Hold at 16°C until ready for further processing
-
24
Keep sample cool at 16°C in the PCR machine or move to refrigerator (4°C) until ready to transform. Transform sample within 24 to 48 hours of assembly reaction. Do not freeze.
-
25On ice, thaw a microcentrifuge tube containing 50 μl aliquot of home-made chemocompetent DH10B-T1R E. coli cells (Sarrion‐Perdigones et al., 2020).
- Although other bacterial strains can be used for chemical transformation as well (e.g., DH5a and similar), DH10B is recommended for assemblies of increasing size.
-
26
Add 2 μl of synthetic DNA assembly reaction and incubate on ice for 10 to 15 minutes.
-
27Heat shock cells by transferring the tube into a dry bead bath, set at 42°C for bacterial transformation, for 45 seconds.
- A water bath, set at 42°C can be used instead of a dry bead bath.
-
28
Immediately after heat shocking the cells, transfer them to ice and incubate for an additional 2 minutes.
-
29
Add 450 μl of 2xLB-0.5 liquid media (without antibiotics) to the microcentrifuge tube containing transformed cells and transfer to the 32°C shaking incubator, by taping microcentrifuge tube to the shaker at an angle to ensure maximal aeration of culture.
-
30
Allow cells to recover shaking at 32°C for 45 minutes.
-
31Plate 50 μl of recovered cells onto bacterial plates containing 1xLB agar supplemented with ampicillin (100 μg/ml) and X-Gal, using glass spreading beads.
- For pUPD3, use bacterial plates containing 1xLB agar supplemented with chloramphenicol (12.5 μg/ml) and X-gal.
-
32
Remove glass spreading beads and place plates into a 37°C incubator and incubate overnight. This should yield plenty of colonies. Depending on the efficiency of the cloning reaction, most colonies should be white (Figure 5B and Figure 5D). Presence of lots of blue colonies represent a lower cloning efficiency.
-
33Pick two white colonies using a fresh disposable inoculation loop for each colony and inoculate into a 14-ml disposable culture tubes containing 5 ml of 2xLB-0.5 medium supplemented with ampicillin (100 μg/ml).
- For pUPD3, use 2xLB-0.5 medium supplemented with chloramphenicol (12.5 μg/ml).
-
34
Grow the cultures overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
35
Separate 0.25 ml of the overnight grown cultures in a 1.7-ml microcentrifuge tube each and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
36Spin down remaining portion of the cultures for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
37
Isolate plasmid DNA using the QIAprep spin miniprep kit (QIAGEN) according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
38
Elute DNA in 50 μl of EB buffer. Measure the concentration using a spectrophotometer.
Verification of synthetic assembly DNA cloning a de novo synthesized DNA part in a Universal Part Domestication plasmid
-
39Verify the identity of assembled plasmid via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis. Digest 400 ng of the resulting plasmids with the restriction enzyme EcoRI-HF resulting in 2 distinct bands corresponding to the plasmid backbone and cloned insert (DNA part):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI
- MilliQ H2O up to 25 μl
Perform uncut digestion in parallel:- 400 ng of plasmid
- MilliQ H2O up to 25 μl
If EcoRI sites are present within the DNA part, one extra band for each EcoRI site will be observed for the cloned insert (DNA part) during restriction enzyme DNA fingerprinting.
We recommend using a DNA manipulation software package to simulate the enzyme digest before performing the experimental digestions. We use the SnapGene software (SnapGene, https://www.snapgene.com/) to help us plan our digests, though several alternatives are available.
-
40
Digest plasmids at 37°C for at least one hour.
-
41
After digestion, briefly vortex and spin down samples.
-
42
Add DNA loading buffer.
-
43
Run your samples on a 1% agarose gel and after visualization using a gel documentation system, confirm by comparing the actual enzyme digest to the one in silico predicted by Snapgene.
-
44
Using a commercial service, confirm plasmids by Sanger DNA sequencing using the universal sequencing primers, T7 and SP6 (pUPD), or M13 Forward and M13 Reverse (pUPD2 and pUPD3), to ensure that no errors are present that were potentially introduced during commercial DNA synthesis.
-
45
Separate DNA samples and glycerol stocks of confirmed synthetically assembled DNA constructs for long term storage at −20°C and −80°C, respectively.
-
46
This is plasmid pNPTII encoding DNA part “Neo” (Addgene, cat. no. 165836) (Table 1) (Matinyan et al., 2021b)
BASIC PROTOCOL 2
Obtaining and cloning a DNA part amplified by PCR from existing DNA resources for synthetic assembly DNA cloning.
Introductory paragraph
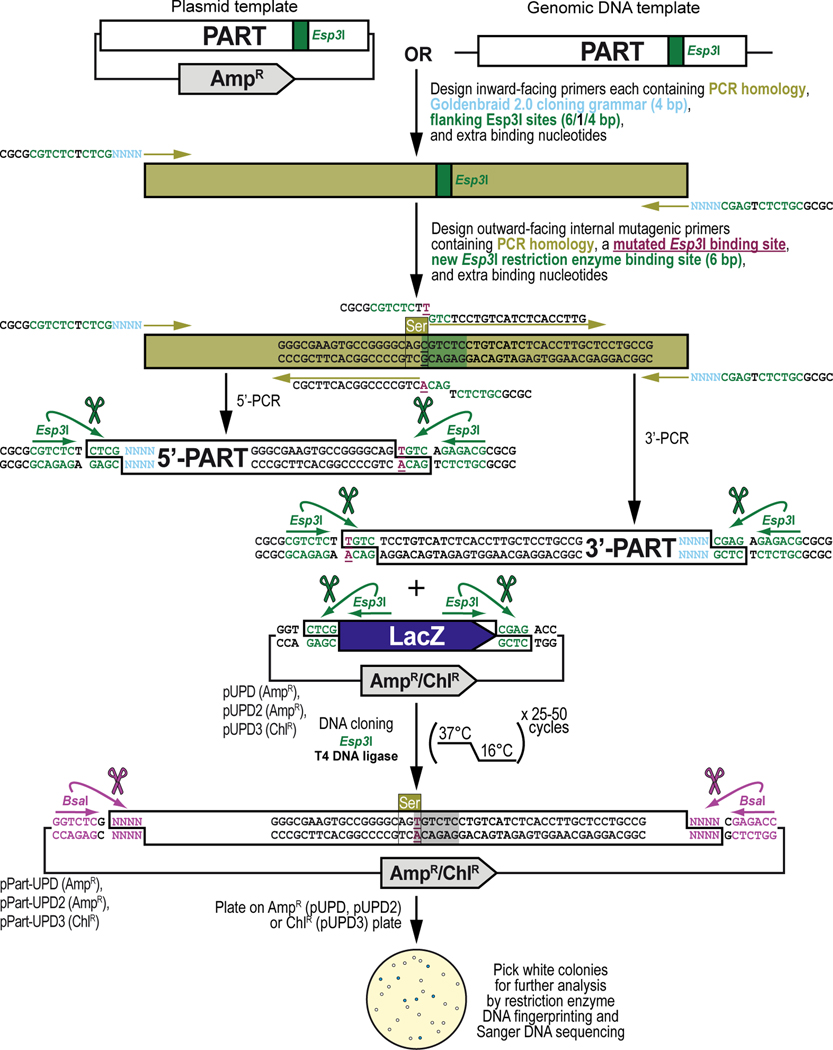
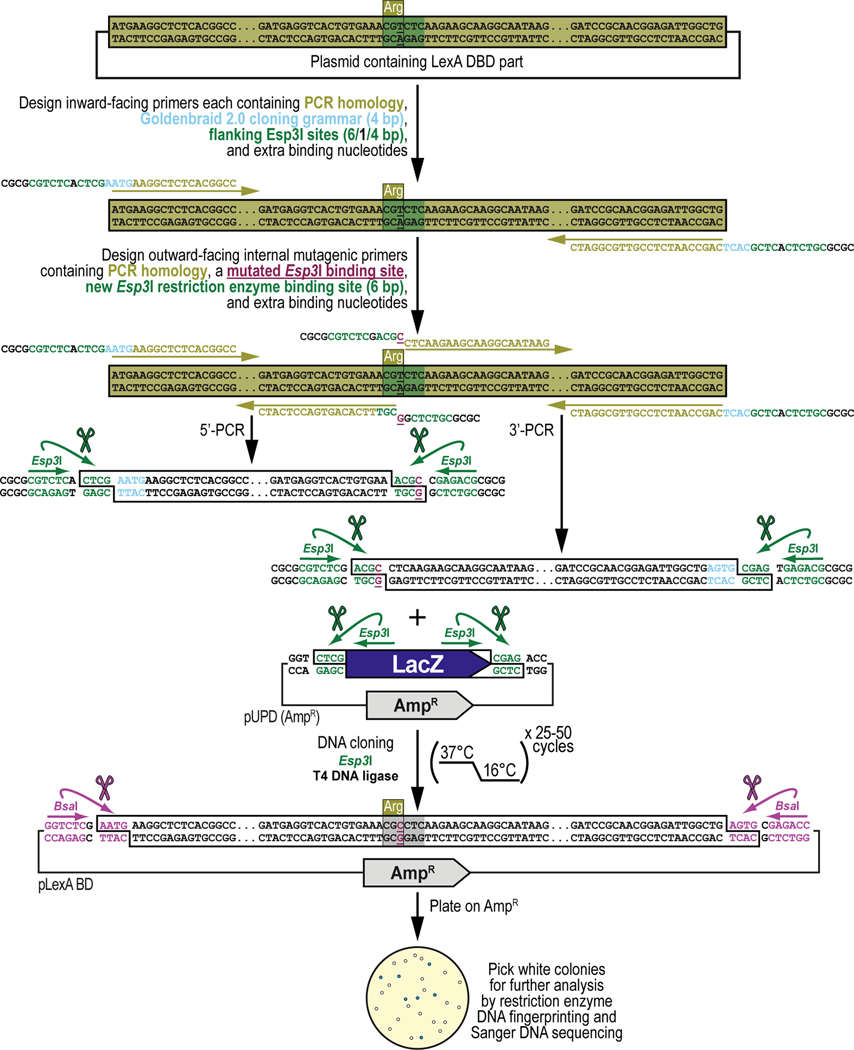
This protocol will demonstrate how to obtain a DNA part by PCR amplification and integrate it into, or domesticate it for, the GoldenBraid 2.0 (GB2.0) workflow starting from a sequence already existing in a plasmid or sample containing genomic DNA template present within the lab, or available as a living sample or published material from another lab or public repository (Figure 6). The sequence can already be, but is not necessarily, codon-optimized for optimal expression in Drosophila melanogaster. We will explain the general principles of this protocol (Figure 6A) and apply those principles to the PCR amplification and cloning (without codon optimization) of the antibiotic resistance gene encoding the Neomycin phosphotransferase II protein (Figure 6B), obtained from transposon Tn5. By the end of this protocol, the user should be able to obtain a DNA part by PCR amplification from an existing plasmid source and clone the amplified sequence into a Universal Part Domestication plasmid vector backbone and verify the accuracy of the resulting clones using restriction enzyme DNA fingerprinting and Sanger DNA sequencing (Figure 6A and Figure 6B). If binding sites for the Type IIs restriction enzymes Esp3I and/or BsaI are present and require removal, see Alternate Protocol 2 for the corresponding PCR amplification and cloning protocol. The DNA parts cloned in Universal Part Domestication plasmid vector backbones using this protocol can then be combined into a multipartite transcription unit or any other genetically encoded assembly with any specific other function (see Basic Protocol 3).
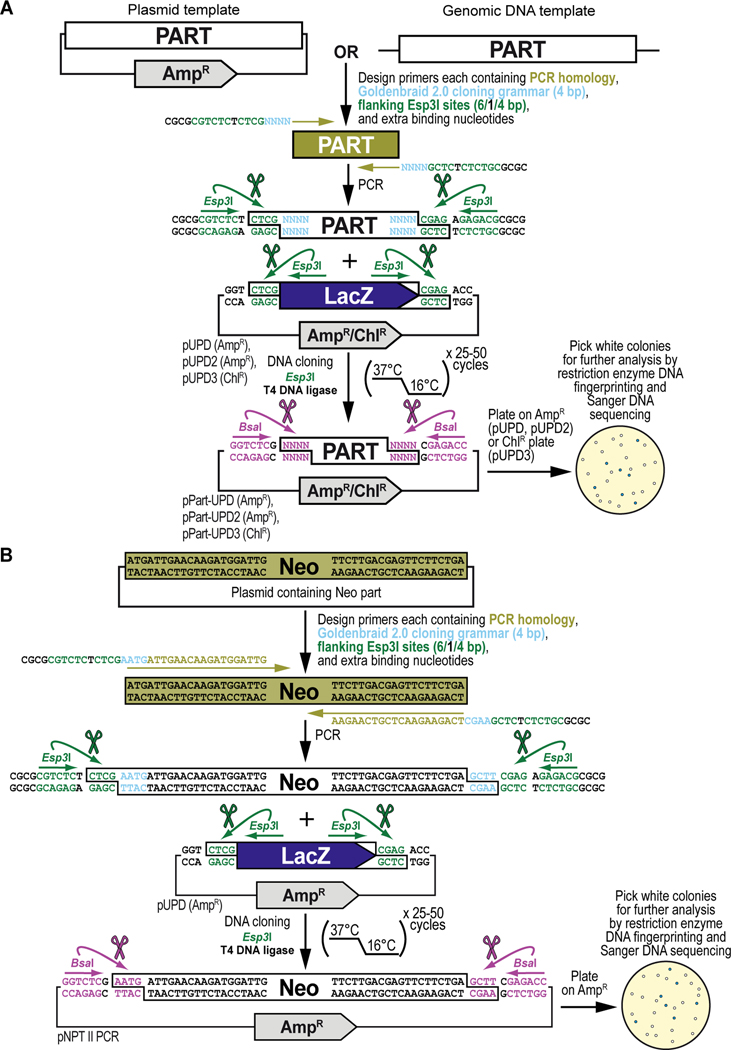
Figure 6. Obtaining and cloning a DNA part by PCR from existing DNA resources for synthetic assembly cloning. (A) General principles of obtaining and cloning a DNA part by PCR into a pUPD vector backbone.
DNA sequences from existing plasmids or genomic DNA can be integrated into the GoldenBraid2.0 synthetic DNA workflow using PCR amplification. Primers are designed that consist of 20–25 bases homologous to the DNA sequence to be amplified (PCR homology, shown in light green), and 19-nucleotide overhangs which add the desired 4-nucleotide GB2.0 cloning grammar (“NNNN”, shown in light blue) (see Figure 2B), as well as a pair of inverted Esp3I sites (shown in dark green) that will generate “CTCG” overhangs required for domestication (see below), and 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in black). Once PCR-amplified, the DNA fragment is combined with a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3), the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates (ampicillin for pUPD and pUPD2, or chloramphenicol for pUPD3), assembled plasmids are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting and Sanger DNA sequencing, while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment. An application of the principles described here are illustrated below for the DNA sequence encoding the selectable marker Neomycin phosphotransferase II (see B). (B) Obtaining and cloning of the DNA part encoding Neomycin phosphotransferase II (Neo) into pUPD. Primers are designed, to amplify Neomycin phosphotransferase II (Neo), consisting of 20–21 bases homologous to the DNA sequence to be amplified (PCR homology, shown in light green), and 19-nucleotide overhangs which will add the desired 4-nucleotide GB2.0 cloning grammar, “AATG” to the 5’ end and “GCTT” to the 3’ end to couple it to a 5’UTR and 3’UTR, respectively (see Figure 2B), as well as a pair of inverted Esp3I sites (shown in dark green) that will generate “CTCG” overhangs required for domestication (see below), and 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in black). Once PCR-amplified, the “Neo” DNA fragment is combined with the Universal Part Domesticator plasmid pUPD, the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates supplemented with ampicillin for pUPD, assembled plasmids are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting and Sanger DNA sequencing (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment.
Materials
Reagents, solutions, and starting samples or test organisms/cells
Template plasmid or genomic DNA template
Custom primers for PCR amplification and synthetic assembly DNA cloning, see below (Sigma-Aldrich)
dNTP nucleotide mix (Thermo Scientific, R1121)
Phusion high-fidelity DNA polymerase and 10x Buffer HF (NEB M0350S or M0350L)
MilliQ H2O, sterilized by autoclaving
DNA Clean and Concentrator kit for purification of PCR-amplified DNA products (Zymogen D4033 or D4034)
EB buffer (10 mM Tris-Cl, pH 8.5) from QIAprep spin miniprep kit (see below)
- At least one of the following Universal Part Domestication plasmids (pUPD):
- pUPD (Sarrion-Perdigones et al., 2011) (Table 1)
- pUPD2, provided by the public plasmid repository, Addgene (https://www.addgene.org/) (Addgene, cat. no. 165856) (Matinyan et al., 2021b) (Table 1)
- pUPD3, provided by the public plasmid repository, Addgene (https://www.addgene.org/) (Addgene, cat. no. 118043) (Matinyan et al., 2021b) (Table 1)
1xLB agar (see Reagents and Solutions section for recipe)
Ampicillin powder (VWR, cat. no. IC19014605) to make a 1,000x stock solution in 50% EtOH diluted with MilliQ H2O, followed by filter sterilization (100 mg/ml)
Chloramphenicol powder (VWR, cat. no. 45000–618) to make a 1,000x stock solution in 100% EtOH, sterilization not required (12.5 mg/ml)
2xLB-0.5 medium (see Reagents and Solutions section for recipe)
Glycerol (Fisher Scientific, cat. no. BP229–1) to make a 40% glycerol solution in MilliQ H2O, sterilized by autoclaving
QIAprep spin miniprep kit (QIAGEN, cat. no. 27106) for plasmid purification
EcoRI-HF restriction enzyme for restriction enzyme DNA fingerprinting (New England Biolabs, cat. no. R3101L)
10x rCutSmart Buffer for restriction enzyme digestions (NEB B6004S)
Universal DNA sequencing primers for Sanger DNA sequencing of pUPD vectors: T7 (TAATACGACTCACTATAGGG), SP6 (ATTTAGGTGACACTATAGA), M13 Forward (GTAAAACGACGGCCAG) and M13 Reverse (CAGGAAACAGCTATGAC)
Esp3I restriction enzyme for synthetic assembly DNA cloning (New England Biolabs, cat. no. R0734L)
T4 DNA ligase and 10x T4 DNA ligase buffer (Promega, cat. no. M1804)
Home-made chemocompetent E. coli cells (Sarrion‐Perdigones et al., 2020), using the DH10B-T1R strain (ThermoFisher Scientific, cat. no., 12331013)
X-Gal (5-bromo-4-chloro-3-indolyl-ß-D-galactopyranoside) powder (VWR, cat. no. 97061–648) to make a 1,000x stock solution in dimethyl sulfoxide (DMSO), sterilization not required (2%)
Hardware and instruments
0.2-ml PCR strip tubes with individually attached caps (VWR, cat. no. 53509–304)
PCR machine (Applied Biosystems, cat. no. 4375786)
Spectrophotometer (DeNovix, cat. no. DS-11+)
Reagents and equipment for agarose gel electrophoresis (Voytas, 2001)
Gel documentation system
Disposable inoculating loops (VWR, cat. no. 12000–806)
Bacterial plates (VWR, cat. no. 25384–092)
37°C incubator (VWR, cat. no. 89409–216)
14-ml disposable culture tubes (VWR, cat. no. 60818–689)
32°C incubator-shaker (Amerex, cat. no. 747/747R)
1.7-ml microcentrifuge tubes (VWR, cat. no. 20170–038)
Refrigerated tabletop centrifuge that can accommodate 14-ml tubes (Fisher Scientific, cat. no. 75230115)
Tabletop microcentrifuge that can accommodate 1.7-ml tubes (Fisher Scientific, cat. no. 75002435)
Ice bucket
Dry bead bath (Lab Armor, cat. no. 74309–706), set at 42°C for bacterial transformation
Glass spreading beads (VWR, cat. no. 26396–508)
Protocol steps
Obtaining a desired DNA part for synthetic assembly DNA cloning by PCR amplification
-
1Begin by designing a primer pair to amplify your DNA fragment (Figure 6A). Design primers to include 20 to 25 nucleotides homologous to the DNA fragment for PCR amplification (PCR homology, highlighted in bold light green) as well as 19-nucleotide overhangs. These overhangs will add the desired 4-nucleotide GB2.0 cloning grammar (“NNNN”, shown in bold light blue) (see Figure 2B), a pair of inverted Esp3I sites (shown in bold dark green) that include the “binding” site for Esp3I (“CGTCTC”), “spacer” nucleotide (“T”), and “domesticator” cloning overhang (“CTCG”) required for domestication into a pUPD backbone, as well as 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in bold black). Generic primer sequences are provided below:
- Forward Primer: 5’-CGCGCGTCTCTCTCGNNNN(N20–25)-3’
- Reverse Primer: 5’-CGCGCGTCTCTCTCGNNNN(N20–25)-3’
-
2
As an example, we will demonstrate PCR amplification-based domestication of coding DNA sequence for the antibiotic resistance gene encoding the Neomycin phosphotransferase II (NPT II) protein, obtained from transposon Tn5 (no Esp3I or BsaI sites are present within this sequence) (Figure 6B). The starting DNA sequence, with “ATG” start and “TGA” stop codons highlighted in bold black, is:
ATGATTGAACAAGATGGATTGCACGCAGGTTCTCCGGCCGCTTGGGTGGAGAGGCTATTCGGCTATGACTGGGCACAACAGACAATCGGCTGCTCTGATGCCGCCGTGTTCCGGCTGTCAGCGCAGGGGCGCCCGGTTCTTTTTGTCAAGACCGACCTGTCCGGTGCCCTGAATGAACTGCAAGACGAGGCAGCGCGGCTATCGTGGCTGGCCACGACGGGCGTTCCTTGCGCAGCTGTGCTCGACGTTGTCACTGAAGCGGGAAGGGACTGGCTGCTATTGGGCGAAGTGCCGGGGCAGGATCTCCTGTCATCTCACCTTGCTCCTGCCGAGAAAGTATCCATCATGGCTGATGCAATGCGGCGGCTGCATACGCTTGATCCGGCTACCTGCCCATTCGACCACCAAGCGAAACATCGCATCGAGCGAGCACGTACTCGGATGGAAGCCGGTCTTGTCGATCAGGATGATCTGGACGAAGAGCATCAGGGGCTCGCGCCAGCCGAACTGTTCGCCAGGCTCAAGGCGAGCATGCCCGACGGCGAGGATCTCGTCGTGACCCATGGCGATGCCTGCTTGCCGAATATCATGGTGGAAAATGGCCGCTTTTCTGGATTCATCGACTGTGGCCGGCTGGGTGTGGCGGACCGCTATCAGGACATAGCGTTGGCTACCCGTGATATTGCTGAAGAGCTTGGCGGCGAATGGGCTGACCGCTTCCTCGTGCTTTACGGTATCGCCGCTCCCGATTCGCAGCGCATCGCCTTCTATCGCCTTCTTGACGAGTTCTTCTGA
-
3Design primers to include 20 to 25 nucleotides homologous to the DNA fragment for PCR amplification (PCR homology, highlighted in bold light green) as well as 19-nucleotide overhangs. These overhangs will add the desired 4-nucleotide GB2.0 cloning grammar, which for a standard coding DNA sequence, as in our example, will be “AATG” for the 5’ end and “GCTT” for the 3’ end (shown in bold light blue), to couple it to a 5’UTR and 3’UTR, respectively (see Figure 2B). Note that the 5’ overhang includes the start codon sequence “ATG” plus an additional “A” nucleotide. Other types of DNA parts (e.g., promoter, linker, terminator, and so on) have their own predefined cloning overhangs (Figure 2B). In addition, these overhangs will add a pair of inverted Esp3I sites (shown in bold dark green) that include the “binding” site for Esp3I (“CGTCTC”), “spacer” nucleotide (“T”), and “domesticator” cloning overhang (“CTCG”) required for domestication into a pUPD backbone, as well as 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in bold black). The primer pair for amplification of our example sequence are shown below (Note that the 3’ overhang is the reverse complement of the sequence GCTT):
- Forward primer: CGCGCGTCTCTCTCGAATGATTGAACAAGATGGATTG
- Reverse primer: CGCGCGTCTCTCTCGAAGCTCAGAAGAACTCGTCAAGAA
Sequence of the resulting PCR amplicon (Neo PCR) is:
CGCGCGTCTCTCTCGAATGATTGAACAAGATGGATTGCACGCAGGTTCTCCGGCCGCTTGGGTGGAGAGGCTATTCGGCTATGACTGGGCACAACAGACAATCGGCTGCTCTGATGCCGCCGTGTTCCGGCTGTCAGCGCAGGGGCGCCCGGTTCTTTTTGTCAAGACCGACCTGTCCGGTGCCCTGAATGAACTGCAAGACGAGGCAGCGCGGCTATCGTGGCTGGCCACGACGGGCGTTCCTTGCGCAGCTGTGCTCGACGTTGTCACTGAAGCGGGAAGGGACTGGCTGCTATTGGGCGAAGTGCCGGGGCAGGATCTCCTGTCATCTCACCTTGCTCCTGCCGAGAAAGTATCCATCATGGCTGATGCAATGCGGCGGCTGCATACGCTTGATCCGGCTACCTGCCCATTCGACCACCAAGCGAAACATCGCATCGAGCGAGCACGTACTCGGATGGAAGCCGGTCTTGTCGATCAGGATGATCTGGACGAAGAGCATCAGGGGCTCGCGCCAGCCGAACTGTTCGCCAGGCTCAAGGCGAGCATGCCCGACGGCGAGGATCTCGTCGTGACCCATGGCGATGCCTGCTTGCCGAATATCATGGTGGAAAATGGCCGCTTTTCTGGATTCATCGACTGTGGCCGGCTGGGTGTGGCGGACCGCTATCAGGACATAGCGTTGGCTACCCGTGATATTGCTGAAGAGCTTGGCGGCGAATGGGCTGACCGCTTCCTCGTGCTTTACGGTATCGCCGCTCCCGATTCGCAGCGCATCGCCTTCTATCGCCTTCTTGACGAGTTCTTCTGAGCTTCGAGAGAGACGCGCG
-
4
Order both primers from your preferred vendor.
-
5
Once primers are received lyophilized, prepare a master and working stock of 100 μM (100 pmol/μl) and 10 μM (10 pmol/μl), respectively, for each, using MilliQ H2O.
-
6Amplify the desired DNA part (Neo PCR) by PCR (Figure 6A and Figure 6B), using the Phusion high-fidelity DNA polymerase by aliquoting the following components in a 0.2-ml PCR strip tube with individually attached cap:
- 1 ng diluted plasmid template DNA
- Forward primer (10 pmol/μl): 2.5 μl
- Reverse primer (10 pmol/μl): 2.5 μl
- dNTP nucleotide mix: 1 μl
- Phusion high-fidelity DNA polymerase: 0.5 μl
- 10x Buffer HF: 5 μl
- MilliQ H2O: up to 50 μl
-
7Briefly spin tubes and place into PCR machine. Amplify the DNA part (Neo PCR) using the following cycling conditions:
- Initial denaturation at 98°C for 30 seconds
- 30 amplification cycles consisting of a denaturation step at 98°C for 10 seconds, primer annealing at 64°C for 30 seconds (calculate annealing temperature when preferred), and a polymerization step at 72° for 30 seconds/kilobase
- Final extension at 72°C for 10 minutes
- Hold at 16°C until ready for further processing
-
8
Remove sample from PCR machine. Gently flick tubes followed by briefly spinning down samples.
-
9
Do a post-PCR amplification DNA clean-up using the DNA Clean and Concentrator kit according to manufacturer’s instruction and elute sample in 20 μl of EB Buffer.
-
10
Measure the DNA concentration using a spectrophotometer. Check 2 μl of purified PCR-amplified product(s) using agarose gel electrophoresis and gel documentation to verify fragment integrity and size.
Obtaining and preparing a Universal Part Domestication plasmid for cloning a PCR-amplified DNA part
-
11Order the desired Universal Part Domestication plasmid vector backbones, pUPD, pUPD2 and/or pUPD3, from Addgene (www.addgene.org). Plasmids ordered from Addgene will arrive as agar stabs.
- If the desired Universal Part Domestication plasmid is already present in the lab, identify the glycerol stock.
-
12Using a disposable inoculating loop, streak out each agar stab (or glycerol stock) to single colonies on bacterial plates containing 1xLB agar supplemented with:
- 100 μg/ml ampicillin (pUPD or pUPD2)
- 12.5 μg/ml chloramphenicol (pUPD3)
-
13
Incubate plates overnight in a 37°C incubator.
-
14For each plasmid, pick a single colony using a disposable inoculation loop and inoculate into a 14-ml disposable culture tubes containing 5 ml of 2xLB-0.5 medium supplemented with appropriate antibiotic:
- 100 μg/ml ampicillin (pUPD and/or pUPD2)
- 12.5 μg/ml chloramphenicol (pUPD3)
-
15
Grow the cultures overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
16
For each plasmid for which you don’t have a glycerol stock yet, separate 0.25 ml of the overnight grown culture in a 1.7-ml microcentrifuge tube and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
17Spin down remaining portion of each culture for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
18
Isolate plasmid DNA using the QIAprep spin miniprep kit according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
19
Elute DNA in 50 μl of EB buffer. Measure the concentration using a spectrophotometer.
-
20Verify the identity of Universal Part Domestication plasmids via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Digest 400 ng of the resulting plasmids with the restriction enzyme EcoRI-HF resulting in two distinct bands, one band corresponding to the plasmid backbone and the other band to the LacZ stuffer containing the colorimetric LacZ α-fragment (Figure 6A and Figure 6B):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI-HF
- MilliQ H2O up to 25 μl
- Alternatively, Universal Part Domestication plasmids can be verified using a commercial Sanger DNA sequencing service and the universal DNA sequencing primers, T7 and SP6 (pUPD), or M13 Forward and M13 Reverse (pUPD2 and pUPD3).
-
21
Once you have verified the identity of the Universal Part Domestication plasmids, dilute each to a final concentration of 75 ng/μl as using EB buffer.
Synthetic assembly DNA cloning a PCR-amplified DNA part in a Universal Part Domestication plasmid
-
22In a 0.2-ml PCR strip tube with individually attached cap, add PCR amplified DNA part (Neo PCR), Universal Part Domesticator vector (pUPD) and enzymes according to the following recipe:
- 40 ng PCR-amplified DNA part
- 75 ng Universal Part Domesticator vector (pUPD, pUPD2 or pUPD3)
- 1 μl Esp3I
- 1 μl of T4 DNA ligase
- 2 μl of 10x T4 DNA ligase buffer
- MilliQ H2O up to 20 μl
-
23Briefly vortex and spin down tube contents and place into a PCR machine. Use the following GB2.0 cloning protocol to drive the synthetic DNA assembly cloning reaction. We recommend at least 25 cycles for efficient cloning (Figure 6A and Figure 6B). More complex assemblies, involving multiple DNA parts or DNA fragments of highly variable sizes, require additional cycles:
- 25 to 50 cycles, each consisting of Esp3I-mediated cutting at 37°C for 2 minutes followed by T4 DNA ligase-mediated ligation at 16°C for 5 minutes
- Heat inactivation of enzymes at 85°C for 20 minutes
- Hold at 16°C until ready for further processing
-
24
Keep sample cool at 16°C in the PCR machine or move to refrigerator (4°C) until ready to transform. Transform sample within 24 to 48 hours of assembly reaction. Do not freeze.
-
25On ice, thaw a microcentrifuge tube containing 50 μl aliquot of home-made chemocompetent DH10B-T1R E. coli cells (Sarrion‐Perdigones et al., 2020).
- Although other bacterial strains can be used for chemical transformation as well (e.g., DH5a and similar), DH10B is recommended for assemblies of increasing size.
-
26
Add 2 μl of synthetic DNA assembly reaction and incubate on ice for 10 to 15 minutes.
-
27Heat shock cells by transferring the tube into a dry bead bath, set at 42°C for bacterial transformation, for 45 seconds.
- A water bath, set at 42°C can be used instead of a dry bead bath.
-
28
Immediately after heat shocking the cells, transfer them to ice and incubate for an additional 2 minutes.
-
29
Add 450 μl of 2xLB-0.5 liquid media (without antibiotics) to the microcentrifuge tube containing transformed cells and transfer to the 32°C shaking incubator, by taping microcentrifuge tube to the shaker at an angle to ensure maximal aeration of culture.
-
30
Allow cells to recover shaking at 32°C for 45 minutes.
-
31Plate 50 μl of recovered cells onto bacterial plates containing 1xLB agar supplemented with ampicillin (100 μg/ml) and X-Gal using glass spreading beads.
- For pUPD3, use bacterial plates containing 1xLB agar supplemented with chloramphenicol (12.5 μg/ml) and X-gal
-
32
Remove glass spreading beads and place plates into a 37°C incubator and let grow overnight. This should yield plenty of colonies. Depending on the efficiency of the cloning reaction, most colonies should be white (Figure 6A and Figure 6B). Presence of lots of blue colonies represent a lower cloning efficiency.
-
33Pick two white colonies using a fresh disposable inoculation loop for each colony and inoculate into a 14-ml disposable culture tubes containing 5 mL of 2xLB-0.5 medium supplemented with ampicillin (100 μg/ml).
- For pUPD3, use 2xLB-0.5 medium supplemented with chloramphenicol (12.5 μg/ml)
-
34
Grow the cultures overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
35
Separate 0.25 ml of the overnight grown culture(s) in a 1.7-ml microcentrifuge tube each and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
36Spin down remaining portion of the culture(s) for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
37
Isolate plasmid DNA using the QIAprep spin miniprep kit according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
38
Elute DNA in 50 μl of EB buffer. Measure resultant concentration using a spectrophotometer.
Verification of synthetic assembly DNA cloning a PCR amplified DNA part in a Universal Part Domestication plasmid
-
39Verify the identity of assembled plasmid via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Digest 400 ng of the resulting plasmids with the restriction enzyme EcoRI-HF resulting in 2 distinct bands corresponding to the plasmid backbone and cloned insert (DNA part):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI
- MilliQ H2O up to 25 μl
Perform uncut digestion in parallel:- 400 ng of plasmid
- MilliQ H2O up to 25 μl
If EcoRI sites are present within the DNA part, one extra band for each EcoRI site will be observed for the cloned insert (DNA part) during restriction enzyme DNA fingerprinting.
We recommend using a DNA manipulation software package to simulate the enzyme digest before performing the experimental digestions. We use the SnapGene software (SnapGene, https://www.snapgene.com/) to help us plan our digests, though several alternatives are available.
-
40
Digest plasmids at 37°C for at least one hour.
-
41
After digestion, briefly vortex and spin down samples.
-
42
Add DNA loading buffer.
-
43
Run your samples on a 1% agarose gel and after visualization using a gel documentation system, confirm by comparing the actual enzyme digest to the one in silico predicted by Snapgene.
-
44
Using a commercial service, confirm plasmids by Sanger DNA sequencing using the universal DNA sequencing primers, T7 and SP6 (pUPD), or M13 Forward and M13 Reverse (pUPD2 and pUPD3), to ensure that no errors are present that got potentially introduced during PCR amplification.
-
45
Separate DNA samples and glycerol stocks of confirmed synthetically assembled DNA constructs for long term storage at −20°C and −80°C, respectively.
-
46
This is plasmid pNPTII-PCR encoding DNA part “Neo-PCR” (Unpublished).
ALTERNATE PROTOCOL 2
Obtaining, adapting, and cloning a DNA part amplified by PCR from existing DNA resources for synthetic assembly DNA cloning.
Introductory paragraph
This protocol will demonstrate how to obtain and adapt a DNA part by PCR amplification and integrate it into, or domesticate it for, the GoldenBraid 2.0 (GB2.0) workflow starting from a sequence already existing in a plasmid or sample containing genomic DNA template present within the lab, or available as a living sample or published material from another lab or public repository, but also has binding sites for the Type IIs restriction enzymes Esp3I and/or BsaI present that require removal before cloning (Figure 7 and Figure 8). The sequence can already be, but is not necessarily, codon-optimized for optimal expression in Drosophila melanogaster. We will explain the general principles of this protocol (Figure 7) and apply those principles to the PCR amplification and cloning (without codon optimization) of the DNA binding domain of the LexA repressor from Escherichia coli (LexA DBD) (Figure 8), used as one of the DNA parts to build the G418 sulfate-selectable LexA transactivator plasmid (Figure 3). By the end of this protocol, the user should be able to obtain and adapt the DNA by PCR amplification from an existing plasmid source and clone the amplified sequence into a Universal Part Domestication plasmid vector backbone and verify the accuracy of the resulting clones using restriction enzyme DNA fingerprinting and Sanger DNA sequencing (Figure 7 and Figure 8). If binding sites for the Type IIs restriction enzymes Esp3I and/or BsaI are not present, see Basic Protocol 2 for the corresponding PCR amplification and cloning protocol. This resulting DNA parts cloned in Universal Part Domestication plasmid vector backbones using this protocol can then be combined into a multipartite transcription unit or any other genetically encoded assembly with some other function (see Basic Protocol 3).
Figure 7. General principles of obtaining, adapting, and cloning a DNA part by PCR into a pUPD vector backbone.
Some DNA sequence available from existing DNA resources may contain internal binding sites for one or both Type IIs restriction enzymes (Esp3I and/or BsaI) that are used during the Goldenbraid 2.0 workflow and, while not mandatory (see Text), ideally should be removed since they can interfere with the synthetic DNA assembly process resulting in lower efficiencies or even failure of assembly reactions. These sites are removed via PCR amplification using mutagenic primers that change one (or more) of the bases within the binding site of Esp3I (shown) or BsaI (not shown). This is straightforward in a coding sequence, as sites can be removed without changing the amino acid sequence using synonymous codon switching, but care needs to be taken when sites are removed in non-coding DNA sequence ensuring not to affect any sequence critical for the function of the DNA part that needs to be integrated within the GB2.0 assembly workflow. To make the change, one additional outward-facing primer pair is designed that overlaps with the internal binding site, in addition to the first pair of inward-facing primer pair that is designed as described in Basic Protocol 2 (see Figure 6). Besides a mutation designed to remove the internal binding site for the Type IIs restriction enzyme (shown in dark purple), the outward-facing primers also contain 20–25 bases homologous to the DNA sequence to be amplified (PCR homology, shown in light green), and overhangs which add a pair of additional inverted binding sites for Esp3I (“CGTCTC”, shown in dark green), and 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in black). Thus, a DNA part with a single internal binding site for a Type IIs restriction enzyme will require 2 separate PCRs resulting in a 5’ and 3’ section of the DNA part which then seamlessly reconstitute the DNA part but without the binding site for the Type IIs restriction enzyme during a GB2.0 assembly reaction in a domesticator plasmid. Once PCR-amplified, both DNA fragments (encoding the 5’ and 3’ sections of the DNA part) are combined with a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3), the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates (ampicillin for pUPD and pUPD2, or chloramphenicol for pUPD3), assembled plasmids are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting and Sanger DNA sequencing (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment. An application of the principles described here is illustrated below for the DNA binding domain of the LexA repressor from Escherichia coli (LexA DBD) (see Figure 8), used as one of the parts to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3).
Figure 8. Obtaining, adapting and cloning of the DNA part encoding the DNA binding domain of the LexA repressor from Escherichia coli (LexA DBD) into pUPD.
A first pair of PCR primers, inward-facing, are designed, to amplify the DNA binding domain of the LexA repressor from Escherichia coli (LexA DBD), used as one of the parts to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3). Both primers consist of 18–22 bases homologous to the DNA sequence to be amplified (PCR homology, shown in light green), and 19-nucleotide overhangs which will add the desired 4-nucleotide GB2.0 cloning grammar (shown in light blue), “AATG” to the 5’ end and “AGTG” to the 3’ end to couple it to a 5’UTR and peptide linker, respectively (see Figure 2B), as well as a pair of inverted Esp3I sites (shown in dark green) that will generate “CTCG” overhangs required for domestication (see below), and 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in black). A second pair of PCR primers, outward facing, are designed that overlap with the internal binding site for Esp3I. Besides a mutation designed to remove the internal binding site for the Type IIs restriction enzyme Esp3I (shown in dark purple), those primers also have 20–21 bases that are homologous to the DNA sequence to be amplified (PCR homology, shown in light green), and overhangs which add a pair of additional inverted binding sites for Esp3I (“CGTCTC”, shown in dark green), and 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in black). Both primers will then be used to amplify two sections of the DNA part encoding the DNA binding domain of the LexA repressor, a 5’ and 3’ section that then, during a GB2.0 assembly reaction in a domesticator plasmid, can seamlessly reconstitute the DNA part but without the binding site for the Type IIs restriction enzyme. Once PCR-amplified, both DNA fragments, the 5’ and 3’ sections of the DNA binding domain of the LexA repressor, are combined with the Universal Part Domesticator plasmid, pUPD, the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates (ampicillin for pUPD and pUPD2, or chloramphenicol for pUPD3), assembled plasmids are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting and Sanger DNA sequencing (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment.
Materials
Reagents, solutions, and starting samples or test organisms/cells
Template plasmid or genomic DNA template
Custom primers for PCR amplification and synthetic assembly DNA cloning, see below (Sigma-Aldrich)
dNTP nucleotide mix (ThermoScientific, R1121)
Phusion high-fidelity DNA polymerase and 10x Buffer HF (NEB M0350S or M0350L)
MilliQ H2O, sterilized by autoclaving
DNA Clean and Concentrator kit for purification of PCR-amplified DNA products (Zymogen D4033 or D4034)
EB buffer (10 mM Tris-Cl, pH 8.5) from QIAprep spin miniprep kit (see below)
- At least one of the following Universal Part Domestication plasmids (pUPD):
- pUPD (Sarrion-Perdigones et al., 2011) (Table 1)
- pUPD2, provided by the public plasmid repository, Addgene (https://www.addgene.org/) (Addgene, cat. no. 165856) (Matinyan et al., 2021b) (Table 1)
- pUPD3, provided by the public plasmid repository, Addgene (https://www.addgene.org/) (Addgene, cat. no. 118043) (Matinyan et al., 2021b) (Table 1)
1xLB agar (see Reagents and Solutions section for recipe)
Ampicillin powder (VWR, cat. no. IC19014605) to make a 1,000x stock solution in 50% EtOH diluted with MilliQ H2O, followed by filter sterilization (100 mg/ml)
Chloramphenicol powder (VWR, cat. no. 45000–618) to make a 1,000x stock solution in 100% EtOH, sterilization not required (12.5 mg/ml)
2xLB-0.5 medium (see Reagents and Solutions section for recipe)
Glycerol (Fisher Scientific, cat. no. BP229–1) to make a 40% glycerol solution in MilliQ H2O, sterilized by autoclaving
QIAprep spin miniprep kit (QIAGEN, cat. no. 27106) for plasmid purification
EcoRI-HF restriction enzyme for restriction enzyme DNA fingerprinting (New England Biolabs, cat. no. R3101L)
10x rCutSmart Buffer for restriction enzyme digestions (NEB B6004S)
Universal DNA sequencing primers for Sanger DNA sequencing of pUPD vectors: T7 (TAATACGACTCACTATAGGG), SP6 (ATTTAGGTGACACTATAGA), M13 Forward (GTAAAACGACGGCCAG) and M13 Reverse (CAGGAAACAGCTATGAC)
Esp3I restriction enzyme for synthetic assembly DNA cloning (New England Biolabs, cat. no. R0734L)
T4 DNA ligase and 10x T4 DNA ligase buffer (Promega, cat. no. M1804)
Home-made chemocompetent E. coli cells (Sarrion‐Perdigones et al., 2020), using the DH10B-T1R strain (ThermoFisher Scientific, cat. no., 12331013)
X-Gal (5-bromo-4-chloro-3-indolyl-ß-D-galactopyranoside) powder (VWR, cat. no. 97061–648) to make a 1,000x stock solution in dimethyl sulfoxide (DMSO), sterilization not required (2%)
Hardware and instruments
0.2-ml PCR strip tubes with individually attached caps (VWR, cat. no. 53509–304)
PCR machine (Applied Biosystems, cat. no. 4375786)
Spectrophotometer (DeNovix, cat. no. DS-11+)
Reagents and equipment for agarose gel electrophoresis (Voytas, 2001)
Gel documentation system
Disposable inoculating loops (VWR, cat. no. 12000–806)
Bacterial plates (VWR, cat. no. 25384–092)
37°C incubator (VWR, cat. no. 89409–216)
14-ml disposable culture tubes (VWR, cat. no. 60818–689)
32°C incubator-shaker (Amerex, cat. no. 747/747R)
1.7-ml microcentrifuge tubes (VWR, cat. no. 20170–038)
Refrigerated tabletop centrifuge that can accommodate 14-ml tubes (Fisher Scientific, cat. no. 75230115)
Tabletop microcentrifuge that can accommodate 1.7-ml tubes (Fisher Scientific, cat. no. 75002435)
Ice bucket
Dry bead bath (Lab Armor, cat. no. 74309–706), set at 42°C for bacterial transformation
Glass spreading beads (VWR, cat. no. 26396–508)
Protocol steps
Obtaining and adapting a desired DNA part for synthetic assembly DNA cloning by PCR amplification
-
1
If amplifying a DNA part from a plasmid, make note of the antibiotic resistance marker present on the backbone as it will be important later when choosing which Universal Part Domesticator (pUPD) backbone to insert the PCR product into. Otherwise, if amplifying from genomic DNA, make sure your sample is of sufficient “high-molecular weight” quality to ensure efficient PCR amplification of all DNA parts needed.
-
2Begin by designing a first, inward-facing, primer pair to amplify your DNA fragment (Figure 7). Design primers to include 18 to 25 nucleotides homologous to the DNA fragment for PCR amplification (PCR homology, highlighted in bold light green) as well as 19-nucleotide overhangs. These overhangs will add the desired 4-nucleotide GB2.0 cloning grammar (“NNNN”, shown in bold light blue) (see Figure 2B), a pair of inverted Esp3I sites (shown in bold dark green) that include the “binding” site for Esp3I (“CGTCTC”), “spacer” nucleotide (“T”), and “domesticator” cloning overhang (“CTCG”) required for domestication into a pUPD backbone, as well as 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in bold black). Generic primer sequences are provided below:
- Forward Primer: 5’-CGCGCGTCTCTCTCGNNNN(N18–25)-3’
- Reverse Primer: 5’-CGCGCGTCTCTCTCGNNNN(N18–25)-3’
-
3
Removal of internal Esp3I and/or BsaI cloning sites is done via synonymous codon switching using a second, outward-facing primer pair, overlapping the binding site (Figure 7). Mutating Type IIs binding sites requires one separate primer pair and one additional PCR amplification reaction each. To remove one internal cloning site for one of the two Type IIs restriction enzymes will require two primer pairs (i.e., one inward-facing and one outward-facing primer pair) and two PCR amplification reactions. To remove two internal cloning sites for one of the two Type IIs restriction enzymes will require three primer pairs (one inward-facing and two outward-facing primer pairs) and three PCR reactions amplification, and so on. The design of these primers will be like those described above, however, instead of the desired 4-nucleotide GB2.0 cloning grammar (“NNNN”, shown in bold light blue above) these overhangs will be a customized 4-nucleotide sequence within the binding site to be removed including the nucleotide to be mutated via PCR amplification (Figure 7). Shown below is an example DNA sequence containing an internal Esp3I binding site (Highlighted in bold dark green), with the location of the to-be synonymous mutation, eliminating the Esp3I binding site, underlined:
GGGCGAAGTGCCGGGGCAGCGTCTCCTGTCATCTCACCTTGCTCCTGCCG- Note that another base within the 6 bp binding site can be mutated if it is matched on the other strand of the corresponding outward facing primer. The first base cannot be changed to a G because that would generate a BsaI site. Similarly, for an existing BsaI site, the first base cannot be mutated to a C because that would generate an Esp3I site.
The primer pair design will feature 18 to 25 nucleotides homologous to the sequence (PCR homology, highlighted in bold light green), including the portion of the binding site to be mutated (Highlighted in bold underlined dark purple), and an 11-base pair overhang comprised of a novel Esp3I binding site (Highlighted in bold dark green) including accompanying spacer (7 nucleotides) as well as the 4 extra bases for improved enzyme binding (Highlighted in bold black). Generic primer templates are provided below:- Forward Primer: 5’-CGCGCGTCTCTTGTCTCCTGTCATCTCACCTTG-3’
- Reverse Primer: 5’-CGCGCGTCTCTGACACTGCCCCGGCACTTCGC-3’
Combining these two primer pairs to remove the Type IIs binding site for Esp3I results in one primer pair for amplifying the N-terminal fragment:- Forward Primer: 5’-CGCGCGTCTCTCTCGNNNN(N20–25)-3’
- Reverse Primer: 5’-CGCGCGTCTCTGACACTGCCCCGGCACTTCGC-3’
And a second primer pair for amplifying the C-terminal fragment:- Forward Primer: 5’-CGCGCGTCTCTTGTCTCCTGTCATCTCACCTTG-3’
- Reverse Primer: 5’-CGCGCGTCTCTCTCGNNNN(N20–25)-3’
If you are domesticating a non-coding DNA part, such as a promoter or terminator, it may not be possible to remove the Type IIs restriction enzyme sites without potentially compromising the functionality of the DNA part, although this can be experimentally tested during downstream functional analysis. The GB2.0 workflow can tolerate up to one recognition site for each Type IIs restriction enzyme (Esp3I and BsaI) within a DNA part and still assemble the DNA part as two pieces with moderate efficiency. Although the DNA part is cut, both pieces can still reconstitute back to the original DNA part during multipartite assembly, if the overhangs generated by the cut are not complementary to any of the overhangs used by the cloning “grammar” (Figure 2B). DNA parts with more than one recognition site for one of the Type IIs restriction enzymes (Esp3I or BsaI) will become increasingly more difficult to assemble.
-
4
As an example, we will demonstrate PCR amplification-based domestication of a DNA fragment encoding for DNA binding domain of the LexA repressor from Escherichia coli (LexA DBD) (Figure 8), used as one of the DNA parts to build the G418 sulfate-selectable LexA transactivator plasmid (Figure 3). The starting DNA sequence, with “ATG” start codon highlighted in bold black, and binding site for Esp3I highlighted in bold dark green, with the location of the to-be synonymous mutation, eliminating the Esp3I binding site, underlined, is:
ATGAAGGCTCTCACGGCCCGACAACAGGAAGTTTTTGATTTGATACGGGATCATATATCCCAAACGGGTATGCCTCCGACCCGCGCAGAGATAGCACAGCGACTGGGCTTTCGATCGCCTAACGCCGCGGAGGAGCACTTGAAGGCACTGGCCCGCAAGGGTGTCATTGAAATCGTGTCCGGTGCGAGCCGCGGAATCCGGCTGTTGCAGGAGGAGGAAGAGGGCCTGCCACTGGTGGGACGCGTGGCCGCTGGCGAGCCGCTGCTGGCCCAGCAACACATAGAGGGACACTATCAGGTGGACCCCTCCTTGTTTAAGCCAAATGCTGATTTCCTGTTGCGGGTGTCGGGAATGTCCATGAAGGACATCGGTATTATGGATGGTGACCTCCTCGCCGTCCATAAGACACAGGATGTCAGGAACGGCCAGGTAGTCGTTGCCAGGATAGACGATGAGGTCACTGTGAAACGTCTCAAGAAGCAAGGCAATAAGGTCGAGCTGCTGCCGGAGAATAGCGAGTTCAAGCCGATCGTGGTGGATCTGCGACAGCAGTCCTTTACTATCGAGGGCTTGGCCGTGGGTGTGATCCGCAACGGAGATTGGCTG
-
5Begin by designing a first, inward-facing, primer pair to amplify your DNA fragment (Figure 8). Design primers to include 18 to 25 nucleotides homologous to the DNA fragment for PCR amplification (PCR homology, highlighted in bold light green) as well as 19-nucleotide overhangs. These overhangs will add the desired 4-nucleotide GB2.0 cloning grammar, which for an N-terminal coding sequence, as in our example, will be “AATG” for the 5’ end, and “AGTG” for the 3’ end (shown in bold light blue), to couple it to a 5’UTR and peptide linker, respectively (see Figure 2B). Note that the 5’ overhang includes the start codon sequence “ATG” plus an additional “A” nucleotide. Other types of DNA parts (e.g., promoter, linker, terminator, and so on) have their own predefined cloning overhangs (Figure 2B). In addition, these overhangs will add a pair of inverted Esp3I sites (shown in bold dark green) that include the “binding” site for Esp3I (“CGTCTC”), “spacer” nucleotide (“T”), and “domesticator” cloning overhang (“CTCG”) required for domestication into a pUPD backbone, as well as 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in bold black). The primer pair is:
- Forward primer: 5’-CGCGCGTCTCTCTCGAATGAAGGCTCTCACGGCC-3’
- The “ATG” is part of the PCR homology
- Reverse Primer: 5’-CGCGCGTCTCTCTCGCACTCAGCCAATCTCCGTTGCGGATC-3’
-
6Design a second, outward-facing primer pair, overlapping the binding site. This primer pair design will feature 18 to 25 nucleotides homologous to the sequence (PCR homology, highlighted in bold light green), including the portion of the binding site to be mutated (Highlighted in bold underlined dark purple), and an 11-base pair overhang comprised of a novel Esp3I binding site (Highlighted in bold dark green) including accompanying spacer (7 nucleotides) as well as the 4 extra bases for improved enzyme binding (Highlighted in bold black). The primer pair is:
- Forward Primer: 5’-CGCGCGTCTCGACGCCTCAAGAAGCAAGGCAATAAG-3’
- Reverse Primer: 5’-CGCGCGTCTCGGCGTTTCACAGTGACCTCATC-3’
Combining these two primer pairs to remove the Type IIs binding site for Esp3I results in one primer pair for amplifying the N-terminal fragment:- Forward primer: 5’-CGCGCGTCTCACTCGAATGAAGGCTCTCACGGCC-3’
- Reverse Primer: 5’-CGCGCGTCTCGGCGTTTCACAGTGACCTCATC-3’
And a second primer pair for amplifying the C-terminal fragment:- Forward Primer: 5’-CGCGCGTCTCGACGCCTCAAGAAGCAAGGCAATAAG-3’
- Reverse Primer: 5’-CGCGCGTCTCACTCGCACTCAGCCAATCTCCGTTGCGGATC-3’
-
7
Sequence of the resulting N-terminal PCR amplicon is (N-terminal LexA DBD):
CGCGCGTCTCACTCGAATGAAGGCTCTCACGGCCCGACAACAGGAAGTTTTTGATTTGATACGGGATCATATATCCCAAACGGGTATGCCTCCGACCCGCGCAGAGATAGCACAGCGACTGGGCTTTCGATCGCCTAACGCCGCGGAGGAGCACTTGAAGGCACTGGCCCGCAAGGGTGTCATTGAAATCGTGTCCGGTGCGAGCCGCGGAATCCGGCTGTTGCAGGAGGAGGAAGAGGGCCTGCCACTGGTGGGACGCGTGGCCGCTGGCGAGCCGCTGCTGGCCCAGCAACACATAGAGGGACACTATCAGGTGGACCCCTCCTTGTTTAAGCCAAATGCTGATTTCCTGTTGCGGGTGTCGGGAATGTCCATGAAGGACATCGGTATTATGGATGGTGACCTCCTCGCCGTCCATAAGACACAGGATGTCAGGAACGGCCAGGTAGTCGTTGCCAGGATAGACGATGAGGTCACTGTGAAACGCCGAGACGCGCG
-
8
Sequence of the resulting C-terminal PCR amplicon is (C-terminal LexA DBD):
CGCGCGTCTCGACGCCTCAAGAAGCAAGGCAATAAGGTCGAGCTGCTGCCGGAGAATAGCGAGTTCAAGCCGATCGTGGTGGATCTGCGACAGCAGTCCTTTACTATCGAGGGCTTGGCCGTGGGTGTGATCCGCAACGGAGATTGGCTGAGTGCGAGTGAGACGCGCG
-
9
Order both primer pairs from your preferred vendor.
-
10
Once primers are received lyophilized, prepare a master and working stock of 100 μM (100 pmol/μl) and 10 μM (10 pmol/μl), respectively, for each, using MilliQ H2O.
-
11Amplify both sections, 5’ and 3’, of the DNA part (LexA DBD) using the Phusion high-fidelity DNA polymerase by aliquoting the following components in a 0.2-ml PCR strip tube with individually attached cap:
- 1 ng diluted plasmid template DNA
- Forward primer (10 pmol/μl): 2.5 μl
- Reverse primer (10 pmol/μl): 2.5 μl
- dNTP nucleotide mix: 1 μl
- Phusion high-fidelity DNA polymerase: 0.5 μl
- 10x Buffer HF: 5 μl
- MilliQ H2O: up to 50 μl
-
12Briefly spin tubes and place into PCR machine. Amplify the 5’ and 3’ sections of the DNA part using the following cycling conditions:
- Initial denaturation at 98°C for 30 seconds
- 30 amplification cycles consisting of a denaturation step at 98°C for 10 seconds, primer annealing at 64°C for 30 seconds (calculate annealing temperature when preferred), and a polymerization step at 72° for 30 seconds
- Final extension at 72°C for 10 minutes
- Hold at 16°C until ready for further processing
-
13
Remove sample from PCR machine. Gently flick tubes followed by briefly spinning down samples.
-
14
Do a post-PCR amplification DNA clean-up using the DNA Clean and Concentrator kit according to manufacturer’s instruction and elute sample in 20 μl of EB Buffer.
-
15
Measure the DNA concentration using a spectrophotometer. Check 2 μl of both purified PCR-amplified products using agarose gel electrophoresis and gel documentation to verify fragment integrity and size.
Obtaining and preparing a Universal Part Domestication plasmid for synthetic assembly DNA cloning a PCR-amplified adapted DNA part
-
16Order the desired Universal Part Domestication plasmid vector backbones, pUPD, pUPD2 and/or pUPD3, from Addgene (www.addgene.org). Plasmids ordered from Addgene will arrive as agar stabs.
- If the desired Universal Part Domestication plasmid is already present in the lab, identify the glycerol stock.
-
17Using a disposable inoculating loop, streak out each agar stab (or glycerol stock) to single colonies on bacterial plates containing 1xLB agar supplemented with:
- 100 μg/ml ampicillin (pUPD and/or pUPD2)
- 12.5 μg/ml chloramphenicol (pUPD3)
-
18
Incubate plates overnight in a 37°C incubator.
-
19For each plasmid, pick a single colony using a disposable inoculation loop and inoculate into a 14-ml disposable culture tubes containing 5 ml of 2xLB-0.5 medium supplemented with appropriate antibiotic:
- 100 μg/ml ampicillin (pUPD and/or pUPD2)
- 12.5 μg/ml chloramphenicol (pUPD3)
-
20
Grow the cultures overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
21
For each plasmid for which you don’t have a glycerol stock yet, separate 0.25 ml of the overnight grown culture in a 1.7-ml microcentrifuge tube and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
22Spin down remaining portion of each culture for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
23
Isolate plasmid DNA using the QIAprep spin miniprep kit according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
24
Elute DNA in 50 μl of EB buffer. Measure resultant concentration using a spectrophotometer.
-
25Verify the identity of Universal Part Domestication plasmids via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Digest 400 ng of the resulting plasmids with the restriction enzyme EcoRI-HF resulting in two distinct bands, one band corresponding to the plasmid backbone and the other band to the LacZ stuffer containing the colorimetric LacZ α-fragment (Figure 7 and Figure 8):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI-HF
- MilliQ H2O up to 25 μl
- Alternatively, Universal Part Domestication plasmids can be verified using a commercial Sanger DNA sequencing service and the universal DNA sequencing primers, T7 and SP6 (pUPD), or M13 Forward and M13 Reverse (pUPD2 and pUPD3).
-
26
Once you have verified the identity of the Universal Part Domestication plasmids, dilute each to a final concentration of 75 ng/μl as using EB buffer.
Synthetic assembly DNA cloning a PCR-amplified adapted DNA part in a Universal Part Domestication plasmid
-
27In a 0.2-ml PCR strip tube with individually attached cap, add each PCR-amplified fragment (5’ and 3’) of the DNA part (LexA DBD), Universal Part Domesticator vector (pUPD) and enzymes according to the following recipe:
- 40 ng of each PCR-amplified fragment (5’ and 3’) of the DNA part (LexA DBD)
- 75 ng Universal Part Domesticator vector (pUPD)
- 1 μl Esp3I
- 1 μl of T4 DNA ligase
- 2 μl of 10x T4 DNA ligase buffer
- MilliQ H2O up to 20 μl
-
28Briefly vortex and spin down tube contents and place into a PCR machine. Use the following GB2.0 cloning protocol to drive the synthetic DNA assembly cloning reaction. We recommend at least 25 cycles for efficient cloning of both sections for the DNA part (Figure 7 and Figure 8). More complex assemblies, involving more DNA parts or DNA fragments of highly variable sizes, require additional cycles:
- 25 to 50 cycles, each consisting of Esp3I-mediated cutting at 37°C for 2 minutes followed by T4 DNA ligase-mediated ligation at 16°C for 5 minutes
- Heat inactivation of enzymes at 85°C for 20 minutes
- Hold at 16°C until ready for further processing
-
29
Keep sample cool at 16°C in the PCR machine or move to refrigerator (4°C) until ready to transform. Transform sample within 24 to 48 hours of assembly reaction. Do not freeze.
-
30On ice, thaw a microcentrifuge tube containing 50 μl aliquot of home-made chemocompetent DH10B-T1R E. coli cells (Sarrion‐Perdigones et al., 2020).
- Although other bacterial strains can be used for chemical transformation as well (e.g., DH5a and similar), DH10B is recommended for assemblies of increasing size.
-
31
Add 2 μl of synthetic DNA assembly reaction and incubate on ice for 10 to 15 minutes.
-
32Heat shock cells by transferring the tube into a dry bead bath, set at 42°C for bacterial transformation, for 45 seconds.
- A water bath, set at 42°C can be used instead of a dry bead bath.
-
33
Immediately after heat shocking the cells, transfer them to ice and incubate for an additional 2 minutes.
-
34
After 2 minutes, add 450 μl of 2xLB-0.5 liquid media (without antibiotics) to the microcentrifuge tube containing transformed cells and transfer to the 32°C shaking incubator, by taping microcentrifuge tube to the shaker at an angle to ensure maximal aeration of culture.
-
35
Allow cells to recover shaking at 32°C for 45 minutes.
-
36Plate 50 μl of recovered cells onto bacterial plates containing 1xLB agar supplemented with ampicillin (100 μg/ml) and X-Gal using glass spreading beads.
- For pUPD3, use bacterial plates containing 1xLB agar supplemented with chloramphenicol (12.5 μg/ml) and X-gal
-
37
Remove glass spreading beads and place plates into a 37°C incubator and let grow overnight. This should yield plenty of colonies. Depending on the efficiency of the cloning reaction, most colonies should be white (Figure 7 and Figure 8). Presence of lots of blue colonies represent a lower cloning efficiency.
-
38Pick two white colonies using a fresh disposable inoculation loop for each colony and inoculate into a 14-ml disposable culture tubes containing 5 mL of 2xLB-0.5 medium supplemented with ampicillin (100 μg/ml).
- For pUPD3, use 2xLB-0.5 medium supplemented with chloramphenicol (12.5 μg/ml)
-
39
Grow the cultures overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
40
Separate 0.25 ml of the overnight grown culture(s) in a 1.7-ml microcentrifuge tube each and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
41Spin down remaining portion of the culture(s) for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
42
Isolate plasmid DNA using the QIAprep spin miniprep kit according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
43
Elute DNA in 50 μl of EB buffer. Measure the concentration using a spectrophotometer.
Verification of synthetic assembly DNA cloning a PCR-amplified adapted DNA part in a Universal Part Domestication plasmid
-
Verify the identity of assembled plasmid via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis. Digest 400 ng of the resulting plasmids with the restriction enzyme EcoRI-HF resulting in 2 distinct bands corresponding to the plasmid backbone and cloned insert (DNA part):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI
- MilliQ H2O up to 25 μl
Perform uncut digestion in parallel:- 400 ng of plasmid
- MilliQ H2O up to 25 μl
If EcoRI sites are present within the DNA part, one extra band for each EcoRI site will be observed for the cloned insert (DNA part) during restriction enzyme DNA fingerprinting.
We recommend using a DNA manipulation software package to simulate the enzyme digest before performing the experimental digestions. We use the SnapGene software (SnapGene, https://www.snapgene.com/) to help us plan our digests, though several alternatives are available.
Digest plasmids at 37°C for at least one hour.
After digestion, briefly vortex and spin down samples.
Add DNA loading buffer.
Run your samples on a 1% agarose gel and after visualization using a gel documentation system, confirm by comparing the actual enzyme digest to the one in silico predicted by Snapgene.
Using a commercial service, confirm plasmids by Sanger DNA sequencing, using the universal DNA sequencing primers, T7 and SP6 (pUPD), or M13 Forward and M13 Reverse (pUPD2 and pUPD3), to ensure that no errors are present that got potentially introduced during PCR amplification.
Separate DNA samples and glycerol stocks of confirmed synthetically assembled DNA constructs for long term storage at −20°C and −80°C, respectively.
This is plasmid pLexA BD encoding DNA part “LexA DBD”, provided by the public plasmid repository, Addgene (https://www.addgene.org/) (Addgene, cat. no. 165821) (Table 1) (Matinyan et al., 2021b)
BASIC PROTOCOL 3
Synthetic assembly DNA cloning of individual DNA parts into a multipartite transcription unit.
Introductory paragraph
This protocol will demonstrate how to combine individual DNA parts previously obtained by de novo DNA synthesis (see Basic Protocol 1) or PCR amplification (see Basic Protocol 2 and Alternate Protocol 2) into a multipartite transcription unit or any other genetically encoded assembly with some other specific function using the synthetic assembly DNA cloning workflow GoldenBraid 2.0 (GB2.0) (Figure 9). The most basic GoldenBraid2.0 (GB2.0) transcription unit consists of three DNA parts: a promoter, a coding DNA sequence, and a polyA terminator (Figure 9). Each DNA part will have been cloned into a Universal Part Domesticator vector backbone (pUPD, pUPD2 or pUPD3) and verified using both restriction enzyme DNA fingerprinting and Sanger sequencing (see Figure 5, Figure 6, Figure 7, and Figure 8). More complex assemblies feature a larger number of DNA parts, each with their own preset assembly grammar (Figure 2B), resulting in cloning reactions involving up to eight to ten different DNA parts. We will explain the general principles of this protocol using three custom DNA parts encoding a promoter (PROM), coding DNA sequence (CDS), and a polyA terminator (TERM) that will be combined into a transcription unit in Alpha level destination vectors (pAlpha1 and pAlpha2) (Figure 9). We will then apply those principles to the synthetic assembly DNA cloning of two individual transcription units used to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3). Synthetic assembly cloning of four DNA parts, the Hsp70 promoter from Drosophila melanogaster (Hsp70), the synthetic Escherichia coli CP6 promoter (CP6), the Neomycin phosphotransferase II of transposon Tn5 (Neo), and the minimal polyadenylation signal of the thymidine kinase gene from the herpes simplex virus (Tk pA) (see Figure 3), in pAlpha1 dictated by their grammar identities (see Figure 2B) results in the transcription unit Hsp70:CP6:Neo:TkpA (Figure 10A). Synthetic assembly cloning of six DNA parts, the R70B04 enhancer from Drosophila melanogaster (70B04), the Drosophila melanogaster synthetic core promoter (dSCP), the DNA binding domain of the LexA repressor from Escherichia coli (LexA-DBD), the synthetic (GlyGlyGlySer)4 peptide linker (Linker), the transcription factor activation domain of VP16 from the herpes simplex virus (VP16-AD), and the late polyadenylation signal from simian vacuolating virus 40 (SV40pA) (see Figure 3), in pAlpha2 dictated by their grammar identities (see Figure 2B) results in the transcription unit R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA (Figure 10B). By the end of this protocol, the user should be able to perform a multipartite GB2.0 synthetic assembly reaction in Alpha level destination vectors using prebuilt DNA parts that are publicly available, or custom-built DNA parts, to generate a multipartite transcription unit or any other genetically encoded assembly with some other specific function. This resulting transcription units cloned in Alpha level destination vectors using this protocol can then be combined into genetic constructs of increasing complexity to perform multiplexed transgenic selection and counterselection genetic strategies using Drosophila melanogaster (see Basic Protocol 4).
Figure 9. General principles of synthetic assembly cloning of multiple DNA parts into a multipartite transcription unit.
Transcription units are the basic functional unit of GoldenBraid2.0 synthetic DNA assembly, capable of expressing an encoded protein or with some other specific function. The simplest transcription unit (TU) consists of a promoter (PROM), coding DNA sequence (CDS), and a polyA terminator (TERM), which all exist as DNA parts cloned in a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3). These DNA parts are assembled in a single, one-pot cloning step using the BsaI enzyme into an Alpha destination vector. The three plasmids needed, each encoding a DNA part (pProm-UPD encoding PROM, pCDS-UPD encoding CDS, and pTerm-UPD encoding TERM), are combined with an Alpha destination vector (pAlpha1 or pAlpha2), the Type IIs restriction enzyme BsaI, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using BsaI) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates (kanamycin for Alpha vectors), assembled plasmids (pTU1-Alpha1 or pTU2-Alpha2) are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment. The resulting product will consist of three DNA parts (shown), less or more (both not shown), assembled in the order dictated by their grammar identities (see Figure 2B). Different choices for promoters (PROM 1 versus PROM 2), coding DNA sequences (CDS 1 versus CDS 2), and polyA terminator (TERM 1 versus TERM 2) allows for assemblies of different transcription units (TU1 consisting of PROM 1, CDS 1 and TERM 1, versus TU2 consisting of PROM 2, CDS 2 and TERM 2) in different Alpha destination vectors (pAlpha1 versus pAlpha2, respectively) that can be brought together during further assembly steps (see Figure 11). Note the difference in the position of the internal BsaI sites in the pAlpha1 versus pAlpha2 vectors relative to the Esp3I sites. This accommodates using the same entry grammar from pUPD or Omega level vectors, while creating different exit grammars that are compatible for Omega level entry (see Figure 2C). Application of the principles described here are illustrated below (see Figure 10) for the synthetic assembly cloning of two individual transcription units used to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3): Hsp70:CP6:Neo:TKpA (see Figure 10A and Figure 10C, left) and R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA (see Figure 10B and Figure 10C, right).
Figure 10. Practical examples of synthetic assembly cloning of multiple DNA parts into multipartite transcription units. (A) Synthetic assembly cloning of the transcription unit Hsp70:CP6:Neo:TKpA into pAlpha1.
Synthetic assembly cloning of four parts, the Hsp70 promoter from Drosophila melanogaster (Hsp70), the synthetic Escherichia coli CP6 promoter (CP6), the Neomycin phosphotransferase II of transposon Tn5 (Neo), and the minimal polyadenylation signal of the thymidine kinase gene from the herpes simplex virus (Tk pA) (see Figure 3), in the destination vector pAlpha1 dictated by their grammar identities (see Figure 2B) results in the transcription unit Hsp70:CP6:Neo:TkpA, one of the units used to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3). (B) Synthetic assembly cloning of the transcription unit R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA into pAlpha2. Synthetic assembly cloning of six parts, the R70B04 enhancer from Drosophila melanogaster (70B04), the Drosophila melanogaster synthetic core promoter (dSCP), the DNA binding domain of the LexA repressor from Escherichia coli (LexA-DBD), a (GlyGlyGlySer)4 peptide linker (Linker), the transcription factor activation domain of VP16 from the herpes simplex virus (VP16-AD), and the late polyadenylation signal from simian vacuolating virus 40 (SV40pA) (see Figure 3), in the destination vector pAlpha2 dictated by their grammar identities (see Figure 2B) results in the transcription unit R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA, a second unit used to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3). (C) Verification of synthetic assembly cloning of transcription units. Simulated gels showing restriction enzyme DNA fingerprinting for the transcription units assembled in pAlpha destination vectors: transcription unit Hsp70:CP6:Neo:TkpA assembled in pAlpha1 (pHsp70:CP6:Neo:TKpA-Alpha1) (Left, Top), and transcription unit R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA assembled in pAlpha2 (p70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2) (Right, Top). Appropriate assembly for pHsp70:CP6:Neo:TKpA-Alpha1 is confirmed after restriction enzyme DNA fingerprinting using no enzyme to show uncut (Lane 1), and digestions using enzymes EcoRI-HF (Lane 2), PvuII-HF (Lane 3), BspHI (Lane 4), NciI (Lane 5), and MslI (Lane 6) (Left, Bottom). Appropriate assembly for p70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2 is confirmed after restriction enzyme DNA fingerprinting using no enzyme to show uncut (Lane 1), and digestions using enzymes HindIII-HF (Lane 2), PstI-HF (Lane 3), XmaI (Lane 4), SacI-HF (Lane 5), and NsiI-HF (Lane 6) (Right, Bottom). Digestions are visualized on a 1% agarose gel.
Materials
Reagents, solutions, and starting samples or test organisms/cells
- 1x of each standard GB2.0 Alpha level cloning vector backbone, provided by the public plasmid repository, Addgene (https://www.addgene.org/):
- pAlpha1 (Addgene, cat. no. 118044) (Sarrion-Perdigones et al., 2019) (Table 1)
- pAlpha2 (Addgene, cat. no. 118045) (Sarrion-Perdigones et al., 2019) (Table 1)
1x of each DNA part plasmid, custom made, encoding a promoter (PROM), coding DNA sequence (CDS), and a polyA terminator (TERM) (see Basic Protocol 1, Basic Protocol 2, and Alternate Protocol 2)
- 1x of each DNA part plasmid, provided by the public plasmid repository, Addgene (https://www.addgene.org/):
- pDmHsp70B_TACT encoding the Hsp70 promoter from Drosophila melanogaster (Addgene, cat. no. 165780) (Matinyan et al., 2021b) (Table 1)
- pCP6_Dros encoding the synthetic Escherichia coli CP6 promoter (Addgene, cat. no. 165794) (Matinyan et al., 2021b) (Table 1)
- pNPTII, encoding the Neomycin phosphotransferase II of transposon Tn5 (Addgene, cat. no. 165836) (Matinyan et al., 2021b) (Table 1) (see Basic Protocol 1)
- pHSV-TK, encoding the minimal polyadenylation signal of the thymidine kinase gene from the herpes simplex virus (Addgene, cat. no. 165801) (Matinyan et al., 2021b) (Table 1)
- pR70B04, encoding the R70B04 enhancer from Drosophila melanogaster (Addgene, cat. no. 165792) (Matinyan et al., 2021b) (Table 1)
- pdSCP, encoding the Drosophila melanogaster synthetic core promoter (Addgene, cat. no. 165776) (Matinyan et al., 2021b) (Table 1)
- pLexA BD, encoding the DNA binding domain of the LexA repressor from Escherichia coli (Addgene, cat. no. 165821) (Matinyan et al., 2021b) (Table 1) (see Alternate Protocol 2)
- pGGGSx4, encoding the synthetic (GlyGlyGlySer)4 peptide linker (Addgene, cat. no. 165807) (Matinyan et al., 2021b) (Table 1)
- pVP16 AD, encoding the transcription factor activation domain of VP16 from the herpes simplex virus (Addgene, cat. no. 165822) (Matinyan et al., 2021b) (Table 1)
- pSV40L, encoding the late polyadenylation signal from simian vacuolating virus 40 (Addgene, cat. no. 165798) (Matinyan et al., 2021b) (Table 1)
1xLB agar (see Reagents and Solutions section for recipe)
Ampicillin powder (VWR, cat. no. IC19014605) to make a 1,000x stock solution in 50% EtOH diluted with MilliQ H2O, followed by filter sterilization (100 mg/ml)
MilliQ H2O, sterilized by autoclaving
Chloramphenicol powder (VWR, cat. no. 45000–618) to make a 1,000x stock solution in 100% EtOH, sterilization not required (12.5 mg/ml)
Kanamycin sulfate powder (VWR, cat. no. 45000–640) to make a 1,000x stock solution in MilliQ H2O, followed by filter sterilization (30 mg/ml)
2xLB-0.5 medium (see Reagents and Solutions section for recipe)
Glycerol (Fisher Scientific, cat. no. BP229–1) to make a 40% glycerol solution in MilliQ H2O, sterilized by autoclaving
QIAprep spin miniprep kit (QIAGEN, cat. no. 27106) for plasmid purification
10x rCutSmart Buffer for restriction enzyme digestions (NEB B6004S)
Universal DNA sequencing primers for Sanger DNA sequencing of pUPD vectors: T7 (TAATACGACTCACTATAGGG), SP6 (ATTTAGGTGACACTATAGA), M13 Forward (GTAAAACGACGGCCAG) and M13 Reverse (CAGGAAACAGCTATGAC)
Additional DNA sequencing primers for Sanger DNA sequencing of assemblies cloned in standard GB2.0 Alpha level cloning vector backbones: SeqFOR (CTTTTTACGGTTCCTGGCCTTTTG) and SeqREV (AATGAGCTGATTTAACAAAAATTTAACGCG)
BsaI-HFv2 restriction enzyme for synthetic assembly DNA cloning (New England Biolabs, cat. no. R3733L)
T4 DNA ligase and 10x T4 DNA ligase buffer (Promega, cat. no. M1804)
Home-made chemocompetent E. coli cells (Sarrion‐Perdigones et al., 2020), using the DH10B-T1R strain (ThermoFisher Scientific, cat. no., 12331013)
X-Gal (5-bromo-4-chloro-3-indolyl-ß-D-galactopyranoside) powder (VWR, cat. no. 97061–648) to make a 1,000x stock solution in dimethyl sulfoxide (DMSO) (2%)
Restriction enzymes for restriction enzyme DNA fingerprinting: BspHI (New England Biolabs, cat. no. R0517L), EcoRI-HF (New England Biolabs, cat. no. R3101L), HindIII-HF (New England Biolabs, cat. no. R3104L), MslI (New England Biolabs, cat. no. R0571L), NciI (New England Biolabs, cat. no. R0196L), NsiI-HF (New England Biolabs, cat. no. R3127L) PstI-HF (New England Biolabs, cat. no. R3140L), PvuII-HF (New England Biolabs, cat. no. R3151L), SacI-HF (New England Biolabs, cat. no. R3156L), and XmaI (New England Biolabs, cat. no. R0180L).
Hardware and instruments
Disposable inoculating loops (VWR, cat. no. 12000–806)
Bacterial plates (VWR, cat. no. 25384–092)
37°C incubator (VWR, cat. no. 89409–216)
14-ml disposable culture tubes (VWR, cat. no. 60818–689)
32°C incubator-shaker (Amerex, cat. no. 747/747R)
1.7-ml microcentrifuge tubes (VWR, cat. no. 20170–038)
Refrigerated tabletop centrifuge that can accommodate 14-ml tubes (Fisher Scientific, cat. no. 75230115)
Tabletop microcentrifuge that can accommodate 1.7-ml tubes (Fisher Scientific, cat. no. 75002435)
Spectrophotometer (DeNovix, cat. no. DS-11+)
Reagents and equipment for agarose gel electrophoresis (Voytas, 2001)
Gel documentation system
0.2-ml PCR strip tubes with individually attached caps (VWR, cat. no. 53509–304)
PCR machine (Applied Biosystems, cat. no. 4375786)
Ice bucket
Dry bead bath (Lab Armor, cat. no. 74309–706), set at 42°C for bacterial transformation
Glass spreading beads (VWR, cat. no. 26396–508)
Protocol steps
Obtaining and preparing standard cloning and DNA part vectors for synthetic assembly DNA cloning of a multipartite transcription unit
-
1
Order the necessary standard GB2.0 cloning vector backbones, pAlpha1 and pAlpha2 (Table 1), from Addgene (www.addgene.org). Plasmids ordered from Addgene will arrive as agar stabs.
-
2
Identify glycerol stocks for plasmids containing the necessary custom DNA parts encoding promoter (PROM), coding DNA sequence (CDS), and a polyA terminator (TERM), to build a custom transcription unit (Figure 9). Alternatively, order the necessary publicly available DNA part plasmids from Addgene (www.addgene.org): pDmHsp70B_TACT, pCP6_Dros, pNPTII and pHSV-TK to build transcription unit Hsp70:CP6:Neo:TkpA (Figure 10A), and pR70B04, pdSCP, pLexA BD, pGGGSx4, pVP16 AD, and pSV40L to build transcription unit R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA (Figure 10B). Plasmids ordered from Addgene will arrive as agar stabs.
-
3Using a disposable inoculating loop, streak out each glycerol stock or agar stab to single colonies on bacterial plates containing 1xLB agar and the appropriate antibiotic:
- Standard GB2.0 cloning vector backbones, pAlpha1 and pAlpha2: 30 μg/ml kanamycin
- DNA part plasmids, custom-built or publicly available: 100 μg/ml ampicillin (DNA parts cloned in pUPD or pUPD2) or 12.5 μg/ml chloramphenicol (DNA parts cloned in pUPD3)
-
4
Incubate plates overnight in a 37°C incubator.
-
5For each plasmid, pick a single colony using a disposable inoculation loop and inoculate into a 14-ml disposable culture tube containing 5 ml of 2xLB-0.5 medium supplemented with appropriate antibiotic:
- Standard GB2.0 cloning vector backbones, pAlpha1 and pAlpha2: 30 μg/ml kanamycin
- DNA part plasmids, custom or publicly available: 100 μg/ml ampicillin (DNA parts cloned in pUPD or pUPD2) or 12.5 μg/ml chloramphenicol (DNA parts cloned in pUPD3)
-
6
Grow the cultures overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
7
For each plasmid for which you don’t have a glycerol stock yet, separate 0.25 ml of the overnight grown culture in a 1.7-ml microcentrifuge tube and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
8Spin down remaining portion of each culture for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
9
Isolate plasmid DNA using the QIAprep spin miniprep kit according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
10
Elute DNA in 50 μl of EB buffer. Measure the concentration using a spectrophotometer.
-
11Verify the identity of isolated standard GB2.0 cloning vector backbones, pAlpha1 and pAlpha2, via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Digest 400 ng of the plasmids with the restriction enzyme EcoRI-HF (pAlpha1) or HindIII (pAlpha2) resulting in 2 distinct bands, one band corresponding to the plasmid backbone and the other band to the LacZ stuffer containing the colorimetric LacZ α-fragment (Figure 9):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI-HF (pAlpha1) or HindIII (pAlpha2)
- MilliQ H2O up to 25 μl
Alternatively, the two standard GB2.0 cloning vectors can be verified using Sanger DNA sequencing and DNA sequencing primers SeqFOR and SeqREV.
-
12Verify the identity of isolated DNA part plasmids via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Digest 400 ng of the plasmids with the restriction enzyme EcoRI-HF resulting in two distinct bands, one band corresponding to the plasmid backbone and the other band to the cloned insert (DNA part):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI-HF
- MilliQ H2O up to 25 μl
If EcoRI sites are present within the DNA part, one extra band for each EcoRI site will be observed for the cloned insert (DNA part) during restriction enzyme DNA fingerprinting.
Alternatively, DNA part plasmids can be verified using a Sanger DNA sequencing and the universal DNA sequencing primers, T7 and SP6 (pUPD), or M13 Forward and M13 Reverse (pUPD2 and pUPD3).
-
13
Once you have verified the identity of all plasmids, dilute each ordered or custom-built DNA part plasmid with MilliQ H2O to a final concentration of 40 ng/μl. Similarly, dilute the standard GB2.0 cloning vectors (pAlpha1 and pAlpha2) to a final concentration of 75 ng/μl as above.
Synthetic assembly DNA cloning of individual DNA parts into a multipartite transcription unit
-
14In a 0.2-ml PCR strip tube with individually attached cap, assemble all DNA parts needed for a certain transcription unit into the appropriate pAlpha destination vector according to the following recipe:
- 40 ng of each DNA part plasmid
- 75 ng of appropriate pAlpha destination vector
- 1 μl of BsaI-HFv2 enzyme
- 1 μl of T4 DNA ligase
- 2 μl of 10x T4 DNA ligase buffer
- MilliQ H2O up to 20 μl
For synthetic assembly cloning of a generic transcription unit in pAlpha1 or in pAlpha2, use three custom DNA parts encoding promoter (PROM), coding DNA sequence (CDS), and polyA terminator (TERM) (Figure 9). For synthetic assembly cloning of the transcription unit Hsp70:CP6:Neo:TkpA in pAlpha1, use four DNA parts encoding the Hsp70 promoter from Drosophila melanogaster (Hsp70), the synthetic Escherichia coli CP6 promoter (CP6), the Neomycin phosphotransferase II of transposon Tn5 (Neo), and the minimal polyadenylation signal of the thymidine kinase gene from the herpes simplex virus (Tk pA) (Figure 10A). For synthetic assembly cloning of the transcription unit R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pAlpha2, use six DNA parts encoding the R70B04 enhancer from Drosophila melanogaster (70B04), the Drosophila melanogaster synthetic core promoter (dSCP), the DNA binding domain of the LexA repressor from Escherichia coli (LexA-DBD), a (GlyGlyGlySer)4 peptide linker (Linker), the transcription factor activation domain of VP16 from the herpes simplex virus (VP16-AD), and the late polyadenylation signal from simian vacuolating virus 40 (SV40pA) (Figure 10B).
-
15Briefly vortex and spin down tube contents and place into a PCR machine. Use the following GB2.0 cloning protocol to drive the synthetic DNA assembly cloning reaction. We recommend at least 25 cycles for efficient cloning of all DNA parts (Figure 9):
- 25 to 50 cycles, each consisting of BsaI-HFv2-mediated cutting at 37°C for 2 minutes followed by T4 DNA ligase-mediated ligation at 16°C for 5 minutes
- Heat inactivation of enzymes at 85°C for 20 minutes
- Hold at 16°C until ready for further processing
-
16
Keep sample cool at 16°C in the PCR machine or move to refrigerator (4°C) until ready to transform. Transform sample within 24 to 48 hours of assembly reaction. Do not freeze.
-
17On ice, thaw a microcentrifuge tube containing 50 μl aliquot of home-made chemocompetent DH10B-T1R E. coli cells (Sarrion‐Perdigones et al., 2020).
- Although other bacterial strains can be used for chemical transformation as well (e.g., DH5a and similar), DH10B is recommended for assemblies of increasing size.
-
18
Add 2 μl of synthetic DNA assembly reaction and incubate on ice for 10 to 15 minutes.
-
19Heat shock cells by transferring the tube into a dry bead bath, set at 42°C for bacterial transformation, for 45 seconds.
- A water bath, set at 42°C can be used instead of a dry bead bath.
-
20
Immediately after heat shocking the cells, transfer them to ice and incubate for an additional 2 minutes.
-
21
After 2 minutes, add 450 μl of 2xLB-0.5 liquid media (without antibiotics) to the microcentrifuge tube containing transformed cells and transfer to the 32°C shaking incubator, by taping microcentrifuge tube to the shaker at an angle to ensure maximal aeration of culture.
-
22
Allow cells to recover shaking at 32°C for 45 minutes.
-
23
Plate 50 μl of recovered cells onto bacterial plates containing 1xLB agar supplemented with kanamycin (30 μg/ml) and X-Gal using glass spreading beads.
-
24
Remove glass spreading beads and place plates into a 37°C incubator and let grow overnight. This should yield plenty of colonies. Depending on the efficiency of the cloning reaction, most colonies should be white (Figure 9). Presence of lots of blue colonies represent a lower cloning efficiency.
-
25
For each assembly reaction, pick two white colonies using a fresh disposable inoculation loop for each colony and inoculate into a 14-ml disposable culture tubes containing 5 mL of 2xLB-0.5 medium supplemented with kanamycin (30 μg/ml).
-
26
Grow the cultures overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
27
Separate 0.25 ml of the overnight grown culture(s) in a 1.7-ml microcentrifuge tube each and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
28Spin down remaining portion of the cultures for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
29
Isolate plasmid DNA using the QIAprep spin miniprep kit according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
30
Elute DNA in 50 μl of EB buffer. Measure the concentration using a spectrophotometer.
Verification of synthetic assembly DNA cloning individual DNA parts into a multipartite transcription unit
-
31Verify the identity of assembled plasmids via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Design enzyme digestions that will result in a series of easy to distinguish DNA bands. Digest 400 ng of the resulting plasmids as indicated below:
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of restriction enzyme (see below)
- MilliQ H2O up to 25 μl
For synthetic assembly cloning of a generic transcription unit (PROM:CDS:TERM) in pAlpha1 or in pAlpha2 (Figure 9), determine appropriate restriction enzymes as desired. For synthetic assembly cloning of the transcription unit Hsp70:CP6:Neo:TkpA in pAlpha1, perform restriction enzyme DNA fingerprinting using no enzyme (uncut), and digestions using enzymes EcoRI-HF, PvuII-HF, BspHI, NciI, and MslI (Figure 10C, Left). For synthetic assembly cloning of the transcription unit R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pAlpha2, perform restriction enzyme DNA fingerprinting using no enzyme (uncut), and digestions using enzymes HindIII-HF, PstI-HF, XmaI, SacI-HF, and NsiI-HF (Figure 10C, right).
We recommend using a DNA manipulation software package to simulate the enzyme digest before performing the experimental digestions. We use the SnapGene software (SnapGene, https://www.snapgene.com/) to help us plan our digests, though several alternatives are available.
-
32
Digest plasmids at 37°C for at least one hour.
-
33
After digestion, briefly vortex and spin down samples.
-
34
Add DNA loading buffer.
-
35
Run your samples on a 1% agarose gel and after visualization using a gel documentation system, confirm by comparing the actual enzyme digest to the one in silico predicted by Snapgene.
-
36
As each individual DNA part was previously sequenced, and GB2.0 cloning does not involve PCR amplification, further verification via Sanger DNA sequencing is not necessary. Sanger DNA sequencing may be required if there is ambiguity in the DNA digest banding pattern, though we recommend further analysis using a different set of restriction enzymes prior to Sanger DNA sequencing. If Sanger DNA sequencing is preferred, do so with DNA sequencing primers SeqFOR and SeqREV.
-
37
Separate DNA samples and glycerol stocks of confirmed synthetically assembled DNA constructs for long term storage at −20°C and −80°C, respectively.
-
38
Resulting plasmids are pTU1-Alpha1 (Figure 9), pTU2-Alpha2 (Figure 9), pHsp70:CP6:Neo:TKpA-Alpha1 (Figure 10A), and p70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2 (Figure 10B).
-
39
Dilute the DNA prep of each transcription unit, pTU1-Alpha1, pTU2-Alpha2, pHsp70:CP6:Neo:TKpA-Alpha1, and p70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2, to a final concentration of 40 ng/μl using MilliQ H2O. You will use these diluted samples as a reagent in the next protocol to assemble a basic genetic circuit (see Basic Protocol 4).
BASIC PROTOCOL 4
Synthetic assembly DNA cloning of multiple transcription units into genetic constructs of increasing complexity.
Introductory paragraph
This protocol will demonstrate how to combine multiple transcription units previously cloned in Alpha level destination vectors (see Basic Protocol 3) into genetic circuits, defined by two transcription units or other DNA assemblies of various complexity, using the synthetic assembly DNA cloning workflow GoldenBraid 2.0 (GB2.0) (Figure 11 and Figure 12). We will explain the general principles of this protocol using two custom transcription units (TUs), TU1 cloned in pAlpha1 (pTU1-Alpha1) and TU2 cloned in pAlpha2 (pTU2-Alpha2) that will be combined in Omega level destination vectors (pOmega1 and pOmega2) (Figure 11). We will then apply those principles to the synthetic assembly DNA cloning of a genetic circuit consisting of two transcription unit used to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3). Synthetic assembly cloning of two previously obtained transcription units (see Basic Protocol 3), Hsp70:CP6:Neo:TKpA located in pAlpha1 and R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA located in pAlpha2, results in the genetic circuit consisting of two transcription units, called Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA, in pOmega2 dictated by their Alpha level grammar identities (Figure 12). By the end of this protocol, the user should be able to perform a multipartite GB2.0 synthetic assembly reaction in Omega level destination vectors using prebuilt transcription units that are publicly available, or custom-built transcription units as described above (see Basic Protocol 3), to generate genetic constructs comprised of two transcription units capable of expressing multiple protein products or other DNA assemblies of various complexity affecting a variety of other cellular functions.
Figure 11. General principles of synthetic assembly cloning of multiple transcription units into genetic constructs of increasing complexity.
Transcription unit vector products (see Figure 9 see Figure 10) can be further pairwise assembled into an Omega level destination vector using the Type IIs restriction enzyme Esp3I. The resulting vector is defined as a Genetic Circuit (GC), comprised of two transcription units or other DNA assemblies of various complexity and capable of expressing multiple protein products or affecting a variety of cellular functions, respectively. GoldenBraid2.0 assembly is iterative, always bringing together a pair of vectors of the same level, but not the same cloning identity (e.g., assembling Alpha1 and Alpha2 together is valid, while assembling Alpha1 and Alpha1 or Alpha2 and Alpha2 together is not) using a similar grammar system to ensure ordered assembly of vector inserts (see Figure 2C). Thus, an insert in Alpha1 always assembles 5’ of the insert in the accompanying vector, Alpha2. Both plasmid assemblies, each encoding a transcription unit, are combined with an Omega destination vector (pOmega1 or pOmega2), the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates (chloramphenicol for Omega vectors), assembled plasmids (pGC1-Omega1 and pGC2-Omega2) are identified as white colonies that are characterized further by restriction enzyme fingerprinting (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment. The resulting product will consist of two transcription units assembled in the order dictated by their Alpha level cloning grammar identities. Different choices for transcription units (TU1, TU2, TU3, and TU4) previously assembled in different Alpha destination vectors (TU1 or TU3 in pAlpha1 versus TU2 or TU4 in pAlpha2) allows for assemblies of two transcription units (TU1/TU2 versus TU3/TU4) in different Omega destination vectors (pOmega1 versus pOmega2, respectively) that can be brought together during further assembly steps (see Figure 13). Note the difference in the position of the internal Esp3I sites in the pOmega1 versus pOmega2 vectors relative to the BsaI sites. This accommodates entry grammar from Alpha level vectors, while creating different exit grammars that are compatible for Alpha level entry (see Figure 2C). Application of the principles described here are illustrated below for the synthetic assembly cloning of a genetic circuit consisting of two transcription units used to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3): Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA (see Figure 12).
Figure 12. A practical example of synthetic assembly cloning of multiple transcription units into genetic constructs of increasing complexity.
(A) Synthetic assembly cloning of the dual transcription unit Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA into pOmega2. Synthetic assembly cloning of two transcription units, Hsp70:CP6:Neo:TKpA located in pAlpha1 and R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA located in pAlpha2, results in the dual transcription unit Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pOmega2, used to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3). (B) Verification of synthetic assembly cloning of a dual transcription unit. Simulated gel showing restriction enzyme DNA fingerprinting for the assembled dual transcription unit in the pOmega2 destination vector: pHsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA (Top). Appropriate assembly of pHsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA is confirmed after restriction enzyme DNA fingerprinting using no enzyme to show uncut (Lane 1), and digestions using enzymes PstI-HF (Lane 2), HindIII-HF (Lane 3), XhoI (Lane 4), XmaI (Lane 5), SacI-HF (Lane 6), PvuII-HF (Lane 7), MslI (Lane 8), and MfeI-HF (Lane 9) (Bottom). Digestions are visualized on a 0.8% agarose gel.
Since GB2.0 synthetic assembly is iterative, the Omega level assembly products can be further expanded pairwise into an Alpha level destination vector (see Basic Protocol 3). The resulting Alpha level assembly product can then be further expanded in a pairwise fashion back into an Omega level destination vector (see Basic Protocol 4), and so on. We will explain the general principles of this approach by expanding the two transcription units previously assembled in an Omega level vector (see above) with more previously assembled material back into an Alpha level destination vector. The resulting assembly product can be further expanded with more previously assembled material back into an Omega level destination vector (Figure 13A). Such iterative assemblies are only limited by plasmid backbone properties and bacterial host properties. A high-copy number plasmid backbone can maintain up to about 20 kb of insert, while a low-copy number plasmid backbone can maintain several hundreds of kb of insert. Some bacterial hosts cannot maintain large insert plasmids while others can. We will then apply those principles to the synthetic assembly DNA cloning of the final G418 sulfate-selectable LexA transactivator plasmid (Figure 3), by adding a phiC31 bacteriophage attB attachment site to the dual transcription unit Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA. The resulting assembly product will then be expanded with the dominant “eye” screening marker called “mini-white” from Drosophila melanogaster to finalize building the G418 sulfate-selectable LexA transactivator plasmid (Figure 13B). Altogether this protocol describes synthetic assembly DNA cloning using the iterative GB2.0 workflow to build sophisticated plasmids for multiplexed selection, counterselection and any other genetic strategies using Drosophila melanogaster.
Figure 13. Further assembly steps to expand genetic constructs towards final complexity for a multiplexed selection and counterselection genetic strategy using Drosophila melanogaster.
(A) General principles of further assembly steps using pAlpha and pOmega level vector backbones to expand genetic constructs towards final complexity. Omega level assembly products (see Figure 11 and Figure 12) can be further pairwise expanded into an Alpha level destination vector again using the BsaI cloning enzyme. GoldenBraid2.0 assembly is iterative, always bringing together a pair of vectors of the same level, but not the same cloning identity (e.g., assembling Omega1 and Omega2 together is valid, while assembling Omega1 and Omega1 or Omega2 and Omega2 together is not) using a similar grammar system to ensure ordered assembly of vector inserts (see Figure 2C). Thus, an insert in Omega1 always assembles 5’ of the insert in the accompanying vector, Omega2. Hence, the two transcription units assembled in pOmega1 can be expanded with any assembly product (“X”) located in pOmega2 into one of the Alpha destination vectors. Both plasmid assemblies, each encoding an assembly of various complexity, are combined with an Alpha destination vector (pAlpha1 or pAlpha2), the Type IIs restriction enzyme BsaI, and T4 DNA ligase (together with 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using BsaI) and 16°C (favoring ligation using T4 DNA ligase). Assembled plasmids are identified and characterized similarly as described before (see Figure 9 and Figure 10). The resulting product will consist of two transcription units expanded with assembly “X” in the order dictated by their Omega level cloning grammar identities. This assembly product itself can be further pairwise expanded into an Omega level destination vector again using the Esp3I cloning enzyme. Hence, two transcription units expanded with assembly “X” in pAlpha1, can be expanded with any assembly product (“Y”) located in pAlpha2 using one of the Omega destination vectors. Both plasmid assemblies, each encoding an assembly of various complexity, are combined with an Omega destination vector (pOmega1 or pOmega2), the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (together with 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting) and 16°C (favoring ligation). Assembled plasmids are identified and characterized similarly as described before (see Figure 11 and Figure 12). The resulting product will consist of two transcription units and assembly “X”, expanded with assembly “Y” in the order dictated by their Alpha level cloning grammar identities. Application of the principles described here are illustrated below (see B) for finalizing the G418 sulfate-selectable LexA transactivator plasmid pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA/White-Omega2 (see Figure 3). (B) Further assembly steps to finalize the G418 sulfate-selectable LexA transactivator plasmid pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA/White-Omega2. Additional assembly steps required to build the G418 sulfate-selectable LexA transactivator plasmid for tissue-specific overexpression within the R70B04 expression domain. The dual transcription unit Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pOmega2 is further expanded with a phiC31 bacteriophage attB attachment site for site-specific transgenesis (attB) in pOmega1 to form pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pAlpha1, which itself is further expanded with a dominant “eye” screening marker called “mini-white” from Drosophila melanogaster (White) in pAlpha2 to form pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA/White in pOmega2 (abbreviated to G418 sulfate-selectable LexA transactivator plasmid) (see Figure 3).
Materials
Reagents, solutions, and starting samples or test organisms/cells
- 1x of each standard GB2.0 Omega level cloning vector backbone, provided by the public plasmid repository, Addgene (https://www.addgene.org/):
- pOmega1 (Addgene, cat. no. 118046) (Sarrion-Perdigones et al., 2019) (Table 1)
- pOmega2 (Addgene, cat. no. 118047) (Sarrion-Perdigones et al., 2019) (Table 1)
- 1x of each standard GB2.0 Alpha level cloning vector backbone, provided by the public plasmid repository, Addgene (https://www.addgene.org/):
- pAlpha1 (Addgene, cat. no. 118044) (Sarrion-Perdigones et al., 2019) (Table 1)
- pAlpha2 (Addgene, cat. no. 118045) (Sarrion-Perdigones et al., 2019) (Table 1)
- 1x of each transcription unit plasmid, custom made (see Basic Protocol 3):
- pTU1-Alpha1, encoding transcription unit 1 (TU1) cloned in pAlpha1
- pTU2-Alpha2, encoding transcription unit 2 (TU2) cloned in pAlpha2
- 1x of each transcription unit plasmid, previously built (see Basic Protocol 3):
- pHsp70:CP6:Neo:TKpA-Alpha1, encoding transcription unit Hsp70:CP6:Neo:TKpA cloned in pAlpha1
- pR70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2, encoding transcription unit R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA cloned in pAlpha2
- 1x of other previously built plasmids to finalize building of the G418 sulfate-selectable LexA transactivator plasmid, provided by the public plasmid repository, Addgene (https://www.addgene.org/):
- pFC31 attB O1, encoding the attB attachment site from the bacteriophage ФC31, cloned in pOmega1 (Addgene, cat. no. 165843) (Matinyan et al., 2021b) (Table 1)
- pMiniWhiteA2, encoding the dominant “eye” screening marker called “mini-white” from Drosophila melanogaster, cloned in pAlpha2 (Addgene, cat. no. 165873) (Matinyan et al., 2021b) (Table 1)
Chloramphenicol powder (VWR, cat. no. 45000–618) to make a 1,000x stock solution in 100% EtOH, sterilization not required (12.5 mg/ml)
1xLB agar (see Reagents and Solutions section for recipe)
Kanamycin sulfate powder (VWR, cat. no. 45000–640) to make a 1,000x stock solution in MilliQ H2O, followed by filter sterilization (30 mg/ml)
MilliQ H2O, sterilized by autoclaving
2xLB-0.5 medium (see Reagents and Solutions section for recipe)
Glycerol (Fisher Scientific, cat. no. BP229–1) to make a 40% glycerol solution in MilliQ H2O, sterilized by autoclaving
QIAprep spin miniprep kit (QIAGEN, cat. no. 27106) for plasmid purification
10x rCutSmart Buffer for restriction enzyme digestions (NEB B6004S)
DNA sequencing primers for Sanger DNA sequencing of assemblies cloned in standard GB2.0 Alpha and Omega level cloning vector backbones: SeqFOR (CTTTTTACGGTTCCTGGCCTTTTG) and SeqREV (AATGAGCTGATTTAACAAAAATTTAACGCG)
Restriction enzymes for synthetic assembly DNA cloning: Esp3I (New England Biolabs, cat. no. R0734L) and BsaI-HFv2 (New England Biolabs, cat. no. R3733L)
T4 DNA ligase and 10x T4 DNA ligase buffer (Promega, cat. no. M1804)
Home-made chemocompetent E. coli cells (Sarrion‐Perdigones et al., 2020), using the DH10B-T1R strain (ThermoFisher Scientific, cat. no., 12331013)
X-Gal (5-bromo-4-chloro-3-indolyl-ß-D-galactopyranoside) powder (VWR, cat. no. 97061–648) to make a 1,000x stock solution in dimethyl sulfoxide (DMSO) (2%)
Restriction enzymes for restriction enzyme DNA fingerprinting: BamHI-HF (New England Biolabs, cat. no. R3136L), EcoRI-HF (New England Biolabs, cat. no. R3101L), EcoRV-HF (New England Biolabs, cat. no. R3195L), HindIII-HF (New England Biolabs, cat. no. R3104L), MfeI-HF (New England Biolabs, cat. no. R3589L), MslI (New England Biolabs, cat. no. R0571L), PstI-HF (New England Biolabs, cat. no. R3140L), PvuI-HF (New England Biolabs, cat. no. R3151L), SacI-HF (New England Biolabs, cat. no. R3156L), SalI-HF (New England Biolabs, cat. no. R3138L), XhoI (New England Biolabs, cat. no. R0146L), and XmaI (New England Biolabs, cat. no. R0180L).
Hardware and instruments
Disposable inoculating loops (VWR, cat. no. 12000–806)
Bacterial plates (VWR, cat. no. 25384–092)
37°C incubator (VWR, cat. no. 89409–216)
14-ml disposable culture tubes (VWR, cat. no. 60818–689)
32°C incubator-shaker (Amerex, cat. no. 747/747R)
1.7-ml microcentrifuge tubes (VWR, cat. no. 20170–038)
Refrigerated tabletop centrifuge that can accommodate 14-ml tubes (Fisher Scientific, cat. no. 75230115)
Tabletop microcentrifuge that can accommodate 1.7-ml tubes (Fisher Scientific, cat. no. 75002435)
Spectrophotometer (DeNovix, cat. no. DS-11+)
Reagents and equipment for agarose gel electrophoresis (Voytas, 2001)
Gel documentation system
0.2-ml PCR strip tubes with individually attached caps (VWR, cat. no. 53509–304)
PCR machine (Applied Biosystems, cat. no. 4375786)
Ice bucket
Dry bead bath (Lab Armor, cat. no. 74309–706), set at 42°C for bacterial transformation
Glass spreading beads (VWR, cat. no. 26396–508)
Protocol steps
Obtaining and preparing “standard cloning”, “transcription unit” and “other previously built” vectors for synthetic assembly DNA cloning of a genetic construct of increasing complexity to perform multiplexed transgenic selection, counterselection and other genetic strategies using Drosophila melanogaster
-
1
Order the necessary standard GB2.0 cloning vector backbones, pOmega1, pOmega2, pAlpha1, and pAlpha2 (Table 1), from Addgene (www.addgene.org). Plasmids ordered from Addgene will arrive as agar stabs.
-
2
Identify glycerol stocks for plasmids containing the necessary custom transcription units, pTU1-Alpha1 and pTU2-Alpha2 to build a custom genetic circuit consisting of two transcription units (Figure 11), as well as the previously built transcription units, pHsp70:CP6:Neo:TKpA-Alpha1 and pR70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2 to build the genetic circuit consisting of two transcription units, called Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pOmega2 (Figure 12).
-
3
In addition, order the other necessary publicly available DNA part plasmids from Addgene (www.addgene.org) to finalize building of the G418 sulfate-selectable LexA transactivator plasmid (Figure 3 and Figure 13): pFC31 attB O1, and pMiniWhiteA2 (Table 1). Plasmids ordered from Addgene will arrive as agar stabs.
-
4Using a disposable inoculating loop, streak out each glycerol stock or agar stab to single colonies on bacterial plates containing 1xLB agar and the appropriate antibiotic:
- Standard GB2.0 Omega level cloning vector backbones, pOmega1 and pOmega2: 12.5 μg/ml chloramphenicol
- Standard GB2.0 Alpha level cloning vector backbones, pAlpha1 and pAlpha2: 30 μg/ml kanamycin
- Alpha level transcription unit plasmids (custom-built, previously built, or publicly available): 30 μg/ml kanamycin
- Other assemblies in Omega level destination plasmids (previously built, or publicly available): 12.5 μg/ml chloramphenicol
- Other assemblies in Alpha level destination plasmids (previously built, or publicly available): 30 μg/ml kanamycin
-
5
Incubate plates overnight in a 37°C incubator.
-
6
For each plasmid, pick a single colony using a disposable inoculation loop and inoculate into a 14-ml disposable culture tube containing 5 ml of 2xLB-0.5 medium supplemented with appropriate antibiotic (see Step 4)
-
7
Grow the cultures overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
8
For each plasmid for which you don’t have a glycerol stock yet, separate 0.25 ml of the overnight grown culture in a 1.7-ml microcentrifuge tube and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
9Spin down remaining part of each culture for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
10
Isolate plasmid DNA using the QIAprep spin miniprep kit according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
11
Elute DNA in 50 μl of EB buffer. Measure the concentration using a spectrophotometer.
-
12Verify the identity of the standard GB2.0 cloning vector backbones, pOmega1, pOmega2, pAlpha1, and pAlpha2, via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Digest 400 ng of the plasmids with the restriction enzyme BamHI-HF (pOmega1), PstI-HF (pOmega2), EcoRI-HF (pAlpha1), or HindIII-HF (pAlpha2), resulting in 2 distinct bands, one band corresponding to the plasmid backbone and the other band to the LacZ stuffer containing the colorimetric LacZ α-fragment (Figure 9 and Figure 11):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of BamHI-HF (pOmega1), PstI-HF (pOmega2), EcoRI-HF (pAlpha1), or HindIII-HF (pAlpha2)
- MilliQ H2O up to 25 μl
Alternatively, the four standard GB2.0 cloning vectors, pOmega1, pOmega2, pAlpha1, and pAlpha2 can be verified using Sanger DNA sequencing and DNA sequencing primers SeqFOR and SeqREV.
-
13Verify the identity of isolated transcription unit plasmids via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Digest 400 ng of the plasmids with the restriction enzyme EcoRI-HF (for a transcription unit cloned in pAlpha1) or HindIII-HF (for a transcription unit cloned in pAlpha2) resulting in two distinct bands, one band corresponding to the plasmid backbone and the other band to the cloned insert (i.e., transcription unit):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI-HF (for a transcription unit cloned in pAlpha1) or HindIII-HF (for a transcription unit cloned in pAlpha2)
- MilliQ H2O up to 25 μl
If EcoRI or HindIII sites are present within the transcription unit, one extra band for each EcoRI or HindIII site will be observed for the cloned insert (i.e., transcription unit) during restriction enzyme DNA fingerprinting.
Alternatively, transcription unit plasmids can be verified using Sanger DNA sequencing and DNA sequencing primers SeqFOR and SeqREV.
-
14Verify the identity of other assemblies in Alpha and Omega level destination plasmids via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Digest 400 ng of the plasmids with the restriction enzyme EcoRI-HF (for an assembly previously cloned in pAlpha1), HindIII-HF (for an assembly previously cloned in pAlpha2), BamHI-HF (for an assembly previously cloned in pOmega1), PstI-HF (for an assembly previously cloned in pOmega2), resulting in two distinct bands, one band corresponding to the plasmid backbone and the other band to the cloned insert (i.e., other assembly):
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of EcoRI-HF (for an assembly previously cloned in pAlpha1), HindIII-HF (for an assembly previously cloned in pAlpha2), BamHI-HF (for an assembly previously cloned in pOmega1), PstI-HF (for an assembly previously cloned in pOmega2)
- MilliQ H2O up to 25 μl
If EcoRI, HindIII, BamHI, or PstI sites are present within the insert, one extra band for each additional restriction site will be observed for the cloned insert (i.e., other assembly) during restriction enzyme DNA fingerprinting.
Alternatively, plasmids containing other assemblies can be verified using Sanger DNA sequencing and DNA sequencing primers SeqFOR and SeqREV.
-
15
Once you have verified the identity of all plasmids, dilute each standard GB2.0 cloning vectors (pOmega1, pOmega2, pAlpha1, and pAlpha2) with MilliQ H2O to a final concentration of 75 ng/μl. Similarly, dilute each “transcription unit” and “other assembly” plasmid with MilliQ H2O to a final concentration of 40 ng/μl.
Synthetic assembly DNA cloning of two transcription units into a genetic circuit consisting of two transcription units
-
16In a 0.2-ml PCR strip tube with individually attached cap, assemble both transcription units needed for a certain genetic circuit into the appropriate pOmega destination vector according to the following recipe:
- 40 ng of each transcription unit plasmid
- 75 ng of appropriate pOmega destination vector
- 1 μl of Esp3I enzyme
- 1 μl of T4 DNA ligase
- 2 μl of 10x T4 DNA ligase buffer
- MilliQ H2O up to 20 μl
For synthetic assembly cloning of a generic genetic circuit consisting of two generic transcription units in pOmega1 or in pOmega2, use transcription unit 1 (TU1), and transcription unit 2 (TU2) (Figure 11). For synthetic assembly cloning of the genetic circuit consisting of two transcription units, called Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA, in pOmega2, use the transcription units Hsp70:CP6:Neo:TKpA and R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA (Figure 12).
-
17Briefly vortex and spin down tube contents and place into a PCR machine. Use the following GB2.0 cloning protocol to drive the synthetic DNA assembly cloning reaction. We recommend at least 25 cycles for efficient cloning of all DNA parts (Figure 11):
- 25 to 50 cycles, each consisting of Esp3I-mediated cutting at 37°C for 2 minutes followed by T4 DNA ligase-mediated ligation at 16°C for 5 minutes
- Heat inactivation of enzymes at 85°C for 20 minutes
- Hold at 16°C until ready for further processing
-
18
Keep sample cool at 16°C in the PCR machine or move to refrigerator (4°C) until ready to transform. Transform sample within 24 to 48 hours of assembly reaction. Do not freeze.
-
19On ice, thaw a microcentrifuge tube containing 50 μl aliquot of home-made chemocompetent DH10B-T1R E. coli cells (Sarrion‐Perdigones et al., 2020).
- Although other bacterial strains can be used for chemical transformation as well (e.g., DH5a and similar), DH10B is recommended for assemblies of increasing size.
-
20
Add 2 μl of synthetic DNA assembly reaction and incubate on ice for 10 to 15 minutes.
-
21Heat shock cells by transferring the tube into a dry bead bath, set at 42°C for bacterial transformation, for 45 seconds.
- A water bath, set at 42°C can be used instead of a dry bead bath.
-
22
Immediately after heat shocking the cells, transfer them to ice and incubate for an additional 2 minutes.
-
23
After 2 minutes, add 450 μl of 2xLB-0.5 liquid media (without antibiotics) to the microcentrifuge tube containing transformed cells and transfer to the 32°C shaking incubator, by taping microcentrifuge tube to the shaker at an angle to ensure maximal aeration of culture.
-
24
Allow cells to recover shaking at 32°C for 45 minutes.
-
25
Plate 50 μl of recovered cells onto bacterial plates containing 1xLB agar supplemented with chloramphenicol (12.5 μg/ml) and X-Gal using glass spreading beads.
-
26
Remove glass spreading beads and place plates into a 37°C incubator and let grow overnight. This should yield plenty of colonies. Depending on the efficiency of the cloning reaction, majority colonies should be white (Figure 11). Presence of lots of blue colonies represent a lower cloning efficiency.
-
27
For each assembly reaction, pick two white colonies using a fresh disposable inoculation loop for each colony and inoculate into a 14-ml disposable culture tubes containing 5 ml of 2xLB-0.5 medium supplemented with chloramphenicol (12.5 μg/ml).
-
28
Grow the cultures overnight in a shaking incubator at 150 rpm and 32°C. Angle tubes for maximal aeration.
-
29
Separate 0.25 ml of the overnight grown culture(s) in a 1.7-ml microcentrifuge tube each and add 0.25 ml of 40% glycerol to make a glycerol stock. Move glycerol stock to ultrafreezer at −80°C.
-
30Spin down remaining part of the cultures for 5 minutes at 4,000 RPM using a refrigerated tabletop centrifuge that can accommodate 14-ml tubes. Decant the supernatant.
- OPTIONAL: Freeze pellet at −20°C or −80°C for at least 30 minutes prior to proceeding to the next step. Freezing the pellet will simplify resuspension of bacterial pellet for DNA isolation purposes.
-
31
Isolate plasmid DNA using the QIAprep spin miniprep kit according to manufacturer’s instruction, and a tabletop microcentrifuge that can accommodate 1.7-ml tubes.
-
32
Elute DNA in 50 μl of EB buffer. Measure the concentration using a spectrophotometer.
Verification of synthetic assembly DNA cloning of two individual transcription units into a genetic circuit
-
33Verify the identity of assembled plasmids via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Design enzyme digestions that will result in a series of easy to distinguish DNA bands. Digest 400 ng of the resulting plasmids as indicated below:
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of restriction enzyme (see below)
- MilliQ H2O up to 25 μl
For synthetic assembly cloning of a generic genetic circuit consisting of two transcription units (GC) in pOmega1 or in pOmega2 (Figure 11), determine appropriate restriction enzymes as desired. For synthetic assembly cloning of the genetic circuit consisting of two transcription units, called Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pOmega2, perform restriction enzyme DNA fingerprinting using no enzyme (uncut), and digestions using enzymes PstI-HF, HindIII-HF, XhoI, XmaI, SacI-HF, PvuI-HF, MslI, and MfeI-HF (Figure 12).
We recommend using a DNA manipulation software package to simulate the enzyme digest before performing the experimental digestions. We use the SnapGene software (SnapGene, https://www.snapgene.com/) to help us plan our digests, though several alternatives are available.
-
34
Digest plasmids at 37°C for at least one hour.
-
35
After digestion, briefly vortex and spin down samples.
-
36
Add DNA loading buffer.
-
37
Run your samples on a 0.8% agarose gel and after visualization using a gel documentation system, confirm by comparing the actual enzyme digest to the one in silico predicted by Snapgene.
-
38
As each individual DNA part was previously sequenced, and GB2.0 cloning does not involve PCR amplification, further verification via Sanger DNA sequencing is not necessary. Sanger DNA sequencing may be required if there is ambiguity in the resulting DNA digest banding pattern, though we recommend further analysis using a different set of restriction enzymes prior to Sanger DNA sequencing. If Sanger DNA sequencing is preferred, do so with DNA sequencing primers SeqFOR and SeqREV.
-
39
Separate DNA samples and glycerol stocks of confirmed synthetically assembled DNA constructs for long term storage at −20°C and −80°C, respectively.
-
40
Resulting plasmids are pGC1-Omega1 (Figure 11), pGC2-Omega2 (Figure 11), and pHsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Omega2 (Figure 12).
-
41
Dilute the DNA prep of the resultant gene circuits, pGC1-Omega1, pGC2-Omega2, and pHsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Omega2, to a final concentration of 40 ng/μl using MilliQ H2O. You will use these diluted samples for further assembly.
Further assembly steps to expand genetic constructs towards final complexity
-
42
The Omega level assembly products obtained through this protocol (Figure 11 and Figure 12) can be further pairwise expanded back into an Alpha level destination vector using the BsaI cloning enzyme (repeat Basic Protocol 3). The two transcription units assembled in pOmega1 (pGC1-Omega1) can be expanded with any assembly product (e.g., “X”) located in pOmega2 using one of the Alpha destination vectors (Figure 13A). The resulting product (pGC1/X-Alpha1) will consist of two transcription units expanded with assembly “X” in the order dictated by their Omega level cloning grammar identities. This assembly product itself can be further pairwise expanded again into an Omega level destination vector using the Esp3I cloning enzyme (repeat Basic Protocol 4). Hence, two transcription units expanded with assembly “X” in pAlpha1 (pGC1/X-Alpha1), can be expanded with any assembly product (e.g., “Y”) located in pAlpha2 using one of the Omega destination vectors. The resulting product (pGC1/X/Y-Omega1) will consist of two transcription units and assembly “X”, expanded with assembly “Y” in the order dictated by their Alpha level cloning grammar identities. Applying those principles to the synthetic assembly DNA cloning of the final G418 sulfate-selectable LexA transactivator plasmid (Figure 3), further expands the dual transcription unit Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pOmega2 with a phiC31 bacteriophage attB attachment site for site-specific transgenesis (attB) in pOmega1 to form pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pAlpha1 (repeat Basic Protocol 3) (Figure 13B). This assembly product is then further expanded with a dominant “eye” screening marker called “mini-white” from Drosophila melanogaster (White) in pAlpha2 to form pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA/White in pOmega2 (repeat Basic Protocol 4) (Figure 13B), abbreviated as the G418 sulfate-selectable LexA transactivator plasmid.
Verification of the final synthetically assembled products
-
43Verify the identity of the final assembled plasmids via restriction enzyme DNA fingerprinting, followed by agarose gel electrophoresis and gel documentation. Design enzyme digestions that will result in a series of easy to distinguishable DNA bands. Digest 400 ng of the resulting plasmids as indicated below:
- 400 ng of plasmid
- 2.5 μl of rCutSmart buffer
- 0.5 μl of restriction enzyme (see below)
- MilliQ H2O up to 25 μl
For the verification of the final synthetically assembled product encoding the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3), perform restriction enzyme DNA fingerprinting using no enzyme (uncut), and digestions using enzymes EcoRI-HF, HindIII-HF, XhoI, EcoRV-HF, PstI-HF, XmaI, SalI-HF and SacI-HF (Figure 14A). For the verification of the final synthetically assembled product encoding the Blasticidin S-selectable LexA-Op responder plasmid whose assembly workflow was only schematically illustrated (see Figure 4), perform restriction enzyme DNA fingerprinting using no enzyme (uncut), and digestions using enzymes EcoRI-HF, HindIII-HF, BamHI-HF, EcoRV-HF, PstI-HF, XmaI, SalI-HF and SacI-HF (Figure 14A).
We recommend using a DNA manipulation software package to simulate the resulting enzyme digest before performing the experimental digestions. We use the SnapGene software (SnapGene, https://www.snapgene.com/) to help us plan our digests, though several alternatives are available. [*Figure 14 near here]
-
44
Digest plasmids at 37°C for at least one hour.
-
45
After digestion, briefly vortex and spin down samples.
-
46
Add DNA loading buffer.
-
47
Run your samples on a 0.8% agarose gel and after visualization using a gel documentation system, confirm by comparing the actual enzyme digest to the one in silico predicted by Snapgene.
-
48
As each individual DNA part was previously sequenced, and GB2.0 cloning does not involve PCR amplification, further verification via Sanger DNA sequencing is not necessary. Sanger DNA sequencing may be required if there is ambiguity in the resulting DNA digest banding pattern, though we recommend further analysis using a different set of restriction enzymes prior to Sanger DNA sequencing. If Sanger DNA sequencing is preferred, do so with DNA sequencing primers SeqFOR and SeqREV.
-
49
Separate DNA samples and glycerol stocks of confirmed synthetically assembled DNA constructs for long term storage at −20°C and −80°C, respectively.
-
50
Resulting plasmids are the G418 sulfate-selectable LexA transactivator plasmid (Figure 3 and Figure 14A), and the Blasticidin S-selectable LexA-Op responder plasmid (Figure 4 and Figure 14B).
-
51
Perform multiplexed dual selection transgenesis using both synthetically assembled plasmids (Figure 15A), as presented in an accompanying Current Protocols article (Venken et al., 2023).
-
52
Determine gene expression patterns (Figure 15B).
Figure 14. Verification of the final synthetic genetic DNA assemblies, the G418 sulfate-selectable LexA transactivator and the Blasticidin S-selectable LexA-Op responder plasmids, for tissue-specific overexpression in Drosophila melanogaster.
(A) Simplified schematic of the synthetically assembled transgene providing G418 sulfate selection (NeoR) and encoding the binary LexA transactivator driven by the R70B04 enhancer (see Figure 3): the G418 sulfate-selectable LexA transactivator plasmid (Top). Simulated gel showing restriction enzyme DNA digestions for the final assembled plasmid. To confirm appropriate assembly, several restriction enzymes are being tested for DNA fingerprinting that once digested, were visualized on a 0.8% agarose gel: EcoRI-HF (Lane 2), HindIII-HF (Lane 3), XhoI (Lane 4), EcoRV-HF (Lane 5), PstI-HF (Lane 6), XmaI (Lane 7), SalI-HF (Lane 8), and SacI-HF (Lane 9). In addition to the digested plasmid, a lane showing uncut clone is shown as well (Lane 1) (Bottom). (B) Simplified schematic of the synthetically assembled transgene providing Blasticidin S selection (BlastR) and encoding the binary LexA-Op responder reporter, providing green fluorescent protein (sfGFP) reporter expression driven by the LexA binary transactivator, whose expression is regulated by the R70B04 enhancer (see Figure 4): the Blasticidin S-selectable LexA-Op responder plasmid (Top). Simulated gel showing restriction enzyme DNA digestions for the final assembled plasmid. To confirm appropriate assembly, several restriction enzymes are being tested for DNA fingerprinting that once digested, were visualized on a 0.8% agarose gel: EcoRI-HF (Lane 2), HindIII-HF (Lane 3), BamHI-HF (Lane 4), EcoRV-HF (Lane 5), PstI-HF (Lane 6), XmaI (Lane 7), SalI-HF (Lane 8), and SacI-HF (Lane 9). In addition to the digested plasmid, a lane showing uncut clone is shown as well (Lane 1) (Bottom). Simulated agarose gels were produced using the SnapGene cloning software (v6.1, www.snapgene.com).
Figure 15. Example of a gene expression pattern obtained after multiplexed dual selection transgenesis in Drosophila melanogaster, using a G418 sulfate-selectable LexA transactivator plasmid together with a Blasticidin S-selectable LexA-Op responder plasmid, both obtained by synthetic assembly DNA cloning.
(A) Simplified experimental workflow of obtaining expression patterns from a double transgenic fly after multiplexed dual selection transgenesis of synthetically assembled plasmids for gene overexpression analysis. Two plasmids, 1) a synthetically assembled transgene providing G418 sulfate selection (NeoR) and encoding the binary LexA transactivator driven by the R70B04 enhancer (see Figure 3), and, 2) a synthetically assembled transgene providing Blasticidin S selection (BlastR) and encoding the binary LexA-Op responder, reporting green fluorescent protein (sfGFP) reporter expression driven by the LexA binary transactivator present within the R70B04 expression domain (see Figure 4), are used for multiplexed dual selection transgenesis. A double transgenic fly can be obtained directly by injecting both transgenes followed by co-selection using both G418 sulfate and Blasticidin S (see Venken et al., 2023), or indirectly by injecting each transgene separately, followed by selection using G418 sulfate for one transgene (LexA transactivator), and Blasticidin S for the other transgene (LexA-Op responder), subsequently followed by crossing both transgenes together and coselect using both G418 sulfate and Blasticidin S. The resulting double transgenic fly is used for gene expression analysis. (B) Gene expression patterns obtained after multiplexed dual selection transgenesis of synthetically assembled plasmids for gene overexpression analysis. Expression patterns obtained from the double transgenic fly containing both the G418 sulfate-selectable LexA transactivator transgene (see Figure 3) and the Blasticidin S-selectable LexA-Op responder transgene (see Figure 4), shows green fluorescent protein (sfGFP) reporter expression in the larval central nervous system (CNS) (B1’-B1‴), as well as the adult brain (B2’-B2‴) and ventral nerve cord (VNC) (B3’-B3‴). Panels on the left represent staining against the green fluorescent protein reporter sfGFP (B1’, B2’, and B3’), panels in the middle represent staining against the neuropil marker Discs Large (Dlg) (B1’’, B2’’, and B3’’), and panels on the right represent the merge of both staining patterns (B1‴, B2‴, and B3‴). Larval CNS staining shows scattered cell bodies and projections particularly prominent in the VNC, with sparse brain lobe staining. Detailed patterns of the adult CNS are described elsewhere (Matinyan et al., 2021b). Scale bars represent 50 μm. Primary antibodies used are polyclonal rabbit anti-GFP (Invitrogen A-11122, 1/500), and mouse monoclonal anti-Dlg (Developmental Studies Hybridoma Bank, 4F3, 1/100). Secondary antibodies are chicken anti-rabbit AlexaFluor 488 (Invitrogen A-21441, 1/500) and goat anti-mouse AlexaFluor568 (Invitrogen A-11004, 1/500). The images represent maximum intensity projection stacks obtained on a Zeiss Axio Imager M2 with an ApoTome2 and processed using Zen software Blue Version 2.3 pro HWL. Detailed staining protocols are described elsewhere (Matinyan et al., 2021b; Gnerer et al., 2015).
REAGENTS AND SOLUTIONS
1xLB bacterial agar solution for bacterial plates (per liter):
10 g tryptone (VWR, cat. no. 97063–390)
5 g yeast extract (Alfa Aesar, cat. no. H26769–22)
5 g NaCl (Alfa Aesar, cat. no. A12313–0B)
10 g agar (Fisher Scientific, cat. no. BP1423–2)
Deionized water up to 1 liter
Sterilize by autoclaving
2xLB-0.5 bacterial growth medium (per liter):
20 g tryptone (VWR, cat. no. 97063–390)
10 g yeast extract (Alfa Aesar, cat. no. H26769–22)
5 g NaCl (Alfa Aesar, cat. no. A12313–0B)
Deionized water up to 1 liter
Sterilize by autoclaving
COMMENTARY
Background information
While traditional cut-and-paste cloning, using a Type II restriction endonuclease cutting and T4 DNA ligase steps, works well enough to make DNA constructs with limited complexity, assembly of plasmids with dozens of DNA parts requires more advanced cloning methods (Casini et al., 2015; Ellis et al., 2011; Xie and Fussenegger, 2018; Blasi et al., 2021). Modern synthetic assembly DNA cloning methods allow for assembly of complex, multipartite genetic circuits in a single assembly step within a single tube, therefore often called “one-pot” assemblies (Cheng and Lu, 2012; Smanski et al., 2016; Casini et al., 2015). Several one-pot cloning methodologies now exist using various approaches ranging from site-specific recombination using integrases and excisionases commonly known as Gateway cloning (Hartley et al., 2000), isothermal chew back and annealing cloning commonly known as Gibson cloning (Gibson et al., 2009), cutting and ligation using Type IIs restriction endonucleases and T4 DNA ligase, such as Golden Gate (Engler et al., 2008) and Goldenbraid cloning (Sarrion-Perdigones et al., 2011), and even CRISPR/Cas9 systems, such as C-Brick cloning (Li et al., 2016).
Of particular interest are cloning systems that use Type IIs restriction endonucleases for rapid, PCR-free assembly of complex transgenes, pioneered by the introduction of Golden Gate cloning (Engler et al., 2008). Unlike typical Type II “sticky” restriction endonucleases that cut at their specific binding site that always leave the same “sticky” and palindromic overhangs behind (e.g., the commonly used Type II restriction enzyme, EcoRI, always leaves a 5’-sticky “AATT” sequence behind for annealing purposes), Type IIs restriction enzymes rather cut away from their binding sites directionally, independent of the DNA sequence at the cut site (e.g., the commonly used Type IIs restriction enzyme, BsaI, always cuts one nucleotide away from its binding site, leaving a 5’-sticky “NNNN” sequence behind for annealing purposes) (Figure 2D) (Szybalski et al., 1991). This feature makes it possible to design DNA sequences, either via PCR amplification or commercial synthesis, such that a Type IIs restriction enzyme will cut at predetermined user-specified, also called “programmable”, sites (i.e., define “NNNN” to be “CAAA”, “GCTT”, and so on) (Figure 2B, Figure 2C and Figure 2D) (Engler et al., 2008). The resulting 4 nucleotide overhangs produced by different cuts can be standardized to create a cloning “grammar” (Figure 2B, Figure 2C and Figure 2D) (Engler et al., 2008; Weber et al., 2011; Sarrion-Perdigones et al., 2011). The “programmed” overhang grammar can then be used to determine the order in which multiple DNA parts assemble during a multipartite cloning reaction (e.g., the 3’ end of a 5’UTR always links to the 5’ end of a coding DNA sequence using the four-nucleotide overhang “AATG”), forming the foundation of the Golden Gate cloning methodology, as well as derivatives of it (Figure 2B, Figure 2C and Figure 2D) (Engler et al., 2008). The main disadvantage of the original Golden Gate cloning method is its reliance on a single Type IIs restriction enzyme, BsaI, such that once DNA parts are assembled into a construct, the resulting transgene becomes “fixed” and can no longer be modified further using the method (Engler et al., 2008; Sarrion-Perdigones et al., 2011).
GoldenBraid cloning, a derivative of Golden Gate cloning, uses a second Type IIs restriction enzyme, BsmBI (or its isoschizomer Esp3I), in addition to BsaI (Sarrion-Perdigones et al., 2011). Moreover, GoldenBraid uses a multilevel cloning system consisting of two kinds of vectors, called Alpha and Omega level destination vectors, with a pair of vector backbones in each level (Figure 2B and Figure 2C) (Sarrion-Perdigones et al., 2011). Multiple DNA parts are assembled via BsaI digestion in a single, one-pot cloning step into one of two Alpha level destination vectors (Figure 2A and Figure 2C). Different assemblies performed in two Alpha level vectors can then be further assembled during a second cloning step via BsmBI (or Esp3I) digestion into one of two Omega level destination vector (Figure 2A and Figure 2C) (Sarrion-Perdigones et al., 2011). Different assemblies performed in two Omega level vectors can then be further assembled back into an Alpha level destination vector during a second BsaI-mediated cloning step, the product of which can then be even further assembled back into an Omega level backbone, and so on (Figure 2A and Figure 2C) (Sarrion-Perdigones et al., 2011). GoldenBraid assembly is theoretically infinitely iterative as the product of one reaction can always be used as a reagent for further assembly, only limited by plasmid backbone properties (a high-copy number plasmid backbone can maintain up to about 20 kb of insert, while a low-copy number plasmid backbone can maintain several hundreds of kb of insert) and the capabilities of large plasmid maintenance by the bacterial host (some bacterial hosts cannot maintain large insert plasmids while others can) (Figure 2A, Figure 2C and Figure 3).
A subsequent update to GoldenBraid known as Goldenbraid2.0 (GB2.0) introduced the Universal Part Domesticator plasmid, pUPD (Sarrion-Perdigones et al., 2013). The pUPD vector allows generation of compatible DNA part vector libraries by cloning all parts needed for final synthetic DNA assembly in it (Figure 2A and Figure 2C) (Sarrion-Perdigones et al., 2013). Entry of each DNA part, obtained through de novo DNA synthesis or PCR, into pUPD is mediated via BsmBI (or Esp3I) assembly while exit is mediated via BsaI-mediated cutting to move different parts together in one of the two Alpha level vectors (Figure 2A and Figure 2C). Compatible DNA part libraries exist for a number of model systems including the plant Arabidopsis thaliana (Sarrion-Perdigones et al., 2013; Vazquez-Vilar et al., 2017), the fungus (Hernanz-Koers et al., 2018), the social amoebae Dictyostelium discoideum (Kundert et al., 2020), the yeast Saccharomyces cerevisiae (Agmon et al., 2015; Pérez-González et al., 2017), human cell lines (Sarrion-Perdigones et al., 2019, 2022; Sarrion‐Perdigones et al., 2020), and most recently the fruit fly, Drosophila melanogaster (Matinyan et al., 2021b), providing numerous opportunities to generate plasmids of sophisticated complexity for various genetic applications in each of these species.
Critical parameters
Good-quality purified plasmid DNA is necessary for efficient synthetic assembly DNA cloning reactions. Purify plasmid DNA use the original “master” aliquot to make a “working” aliquot by diluting using MilliQ H2O the standard cloning vectors to 75 ng/μl and all other vectors to 40 ng/μl for easier pipetting into tubes or plates used for the synthetic assembly DNA cloning reactions. Always store both “master” and “working” aliquot of each plasmid for synthetic assembly DNA cloning reactions at −20°C.
Pick just two or three colonies for each synthetic assembly DNA cloning reaction, and you will likely find a correctly assembled clone after restriction enzyme DNA fingerprinting (and Sanger DNA sequencing when cloning in Universal Plasmid Domestication vectors). Synthetic assembly DNA cloning reactions using Type IIs restriction enzymes are very efficient, whether 2, 3, 4, or 5 parts are put together in one of the Alpha or Omega destination vectors.
Find efficiently cutting and cheap Type II restriction enzymes to verify synthetic assembly DNA cloning reactions by restriction enzyme DNA fingerprinting. This is key for verification of correct assembly of all fragments into the destination vector. You should do at least one digestion for synthetic assembly DNA cloning in one of the Universal Part Domestication plasmids, and at least two parallel digestions for synthetic assembly DNA cloning in one of the Alpha or Omega destination vectors that cut your assembly product into very distinctive DNA fragments. Assembly reactions that become increasingly larger or complex should be confirmed by increasingly more parallel digestions to confirm correct assembly.
Run uncut DNA while you run the different plasmid restriction digestions during agarose gel electrophoresis. Uncut plasmid produces two bands on a gel, representing the open-circular (or nicked) and supercoiled DNA (truly uncut). At the same overall size, supercoiled DNA runs faster than open circular DNA. A relaxed plasmid configuration (cut) is also possible, generating a third band running between supercoiled and open circular DNA, but this band is a minor one and is often not seen for small plasmids. However, for assemblies increasing in size, this band will become more obvious. Moreover, assemblies resulting in plasmid sizes larger than about 30 kilobase, this band will start running below the supercoiled DNA, and always at the same height since regular agarose gel electrophoreses can’t distinguish between bands that are larger than that, i.e., a linearized 30 kilobase plasmid will essentially run at the same height of a linearized 40 kilobase plasmid, and so on. Uncut DNA will inform you of unwanted plasmid dimerization, which will produce bands of a larger size than expected.
Troubleshooting
Fragment design and de novo DNA synthesis issues
When designing a DNA part for de novo DNA synthesis, you may encounter issues such as the inability to remove a putative splicing acceptor/donor site from within a coding sequence without affecting the amino acid sequence. You should aim to reduce the predicted strength of the site by changing bases adjacent to the putative splicing site until the score predicted by the splice prediction tool from the Berkley Drosophila Genome Project (https://www.fruitfly.org/seq_tools/splice.html) (Reese et al., 1997) is as low as possible. Weakened sites are unlikely to significantly affect expression of your transgene. You may also find that the codon-optimized sequence contains homopolymers or other difficult to synthesize sequences which you will have to manually adjust. When domesticating non-coding DNA parts, removal of internal binding sites for the Type IIs restriction enzymes may affect the functionality of the resulting DNA part, although this can be experimentally tested during downstream functional analysis. The GB2.0 workflow can tolerate up to one recognition site for each Type IIs restriction enzyme (Esp3I and BsaI) within a DNA part and still assemble the part as two pieces with reasonable efficiency. Although the part is cut, both pieces can still reconstitute back to the original part during multipartite assembly, as long as the overhangs generated by the cut are not complementary to any of the overhangs used by the cloning “grammar” (Figure 2B and Figure 2C). DNA parts with more than one recognition site for one of the Type IIs restriction enzymes (Esp3I or BsaI) will become increasingly more difficult to assemble.
Assembly design and cloning issues
Before beginning synthetic assembly DNA cloning using GB2.0, we recommend drafting a design of the desired final construct including all DNA parts that will be required to test its functionality. It is helpful to draft a schema of the cloning steps, alternating between Alpha and Omega level destination vectors, needed to generate the final assembly product. To do so, we recommend using a modern cloning software tool during the design process, such as SnapGene (Snapgene, www.snapgene.com) or MacVector (www.macvector.com). Several alternatives are also available. These software packages help to visualize in silico the construct design and overall cloning strategy, allowing the user to make and correct mistakes digitally before attempting them experimentally. Another notable feature of these programs is their ability to simulate restriction enzyme digestions and allowing the user to decide which restriction enzymes are preferred for plasmid verification using restriction enzyme DNA fingerprinting, which will result in visually distinct bands during DNA agarose gel electrophoresis.
There are several possible reasons why you may see few to no white colonies when you plate cells transformed with the DNA assembly reaction mix even after successfully designing a cloning step in silico. If you see no colonies at all, make sure that you are plating the chemically transformed cells onto plates supplemented with the appropriate antibiotic, assemblies in pUPD or pUPD2 with ampicillin, assemblies in pAlpha1/2 with kanamycin, and assemblies in pUPD3 or pOmega1/2 with chloramphenicol plates, and that your cells have a decent competency. If you see that most if not all resulting colonies are blue (i.e., empty destination vector that never got cut, or religated), this means that the assembly either failed or was only poorly efficient. This may be due to several reasons such as adding incorrect DNA samples to the mix, using the wrong vector backbone, using the wrong Type IIs restriction enzyme, using a defective aliquot of Type IIs restriction enzyme, or running the synthetic DNA assembly reaction for too few cycles. If you see only a few colonies on a plate that are white and/or blue, this likely means that the assembly was complex, with few completely assembled plasmids in the assembly reaction mix. For complex assemblies of more than four DNA parts we recommend that instead of plating only 50 μl of the recovered cells, that you also plate the remaining 450 μl of transformed culture on a second plate of appropriate antibiotic to improve your chances of recovering a white colony containing the correct assembly DNA product.
Understanding results
Successful GB2.0 assembly DNA cloning should yield white colonies containing the desired assembly product as verified by Type II restriction enzyme DNA fingerprinting and/or Sanger DNA sequencing. You should aim to design Type II restriction enzyme digestions that yield clear, easy to interpret DNA banding patterns while providing enough information to tell if a small DNA part has been excluded during the assembly cloning process. Depending on the complexity and size of the synthetic DNA assembly, we typically perform 2 to 4 different restriction enzyme digestions of the plasmid DNA for small assembly end products (up to 10 kilobase), depending on the size of the assembly product. Assembly reactions that become increasingly larger and/or complex should be confirmed by increasingly more digestions run in parallel to confirm correct assembly. We also highly encourage the user to run uncut plasmid in an additional lane for each of their constructs to verify that there are no gross defects with the DNA prep. While rare, it is possible to see a correct DNA banding pattern after digestion but have the uncut lane show the vector to be the incorrect size. This happens when more than one assembled insert is cloned into a single vector backbone. Running uncut plasmid will help identify and discard these rare but anomalous outcomes.
Time considerations
Each GB2.0 cloning steps takes about three to four days depending on the efficiency of the user. On the first day, the user will set up the cloning reaction in the morning and transform 2 μl of the resulting assembly mix in the afternoon, spread transformed cells on bacterial plates supplemented with appropriate antibiotic selection, and allow transformed cells to grow overnight. This timeline should allow for about 30 – 35 assembly cycles during the cloning reaction which is sufficient for efficient cloning of most constructs. On the second day, the user will inoculate at least two cultures (supplemented with appropriate antibiotic selection) containing individually picked white bacterial colonies for each construct and set the cultures shaking overnight. On the third day, the user will then spin down the saturated cultures and isolate plasmid DNA using a commercial miniprep kit (or similar) in the morning, followed by Type II restriction enzyme digestion and visualization of the resulting DNA banding pattern on a DNA agarose gel in the afternoon. If this is not the final construct, the user can then continue assembling using the verified DNA prep as a reagent for the next cloning reaction the following day. The user will repeat each of these steps until the final product is generated which may take several weeks depending on the design and complexity of the final assembly product.
INTERNET RESOURCES
| https://www.addgene.org/ | Public plasmid repository, Addgene |
| https://www.idtdna.com/CodonOpt | Codon Optimization Tool from Integrated DNA Technologies (IDT) |
| https://geneius.de | Online codon optimization tool available from GENEius |
| http://www.kazusa.or.jp/codon/ | Codon Usage Database |
| https://www.fruitfly.org/seq_tools/splice.html | Online splice site prediction tool from the Berkley Drosophila Genome Project |
| https://www.idtdna.com/ | DNA part provider as gBlock |
| https://eurofinsgenomics.com/ | DNA part provider as gene strands |
| https://www.snapgene.com/ | DNA manipulation software, SnapGene |
ACKNOWLEDGMENTS
This research was supported by external grants from the Foundation For Angelman Syndrome Therapeutics grant FT2016-002 (K.J.T.V.), the Cancer Prevention and Research Institute of Texas grant R1313 (K.J.T.V.), and the National Institutes of Health grants T32GM008231 (N.M.) R21HG006726 (K.J.T.V.), R21GM110190 (K.J.T.V.), R21OD022981 (K.J.T.V.), and R01MH107474, (H.A.D.).
This work was additionally supported by internal start-up and seed funds kindly provided by Baylor College of Medicine (K.J.T.V.), the Albert and Margaret Alkek Foundation (K.J.T.V.), and the McNair Medical Institute at The Robert and Janice McNair Foundation (K.J.T.V.).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
LITERATURE CITED
- Agmon N, Mitchell LA, Cai Y, Ikushima S, Chuang J, Zheng A, Choi W-J, Martin JA, Caravelli K, Stracquadanio G, et al. 2015. Yeast Golden Gate (yGG) for the Efficient Assembly of S. cerevisiae Transcription Units. ACS Synthetic Biology 4:853–859. [DOI] [PubMed] [Google Scholar]
- Blasi RD, Zouein A, Ellis T, and Ceroni F. 2021. Genetic Toolkits to Design and Build Mammalian Synthetic Systems. Trends in Biotechnology 39:1004–1018. [DOI] [PubMed] [Google Scholar]
- Casini A, Storch M, Baldwin GS, and Ellis T. 2015. Bricks and blueprints: methods and standards for DNA assembly. Nature Reviews Molecular Cell Biology 16:568–576. [DOI] [PubMed] [Google Scholar]
- Cheng AA, and Lu TK 2012. Synthetic Biology: An Emerging Engineering Discipline. Annual Review of Biomedical Engineering 14:155–178. [DOI] [PubMed] [Google Scholar]
- Ellis T, Adie T, and Baldwin GS 2011. DNA assembly for synthetic biology: from parts to pathways and beyond. Integrative Biology 3:109–118. [DOI] [PubMed] [Google Scholar]
- Engler C, Kandzia R, and Marillonnet S. 2008. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS ONE 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, and Smith HO 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6:343–345. [DOI] [PubMed] [Google Scholar]
- Gnerer JP, Venken KJT, and Dierick HA 2015. Gene-specific cell labeling using MiMIC transposons. Nucleic Acids Research 43:e56–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, and Brasch MA 2000. DNA Cloning Using In Vitro Site-Specific Recombination. Genome Research 10:1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernanz-Koers M, Gandía M, Garrigues S, Manzanares P, Yenush L, Orzaez D, and Marcos JF 2018. FungalBraid: A GoldenBraid-based modular cloning platform for the assembly and exchange of DNA elements tailored to fungal synthetic biology. Fungal Genetics and Biology 116:51–61. [DOI] [PubMed] [Google Scholar]
- Kundert P, Sarrion-Perdigones A, Gonzalez Y, Katoh-Kurasawa M, Hirose S, Lehmann P, Venken KJT, and Shaulsky G. 2020. A GoldenBraid cloning system for synthetic biology in social amoebae. Nucleic Acids Research 48:4139–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S-L, and Lee T. 2006. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nature Neuroscience 9:703–709. [DOI] [PubMed] [Google Scholar]
- Li S-Y, Zhao G-P, and Wang J. 2016. C-Brick: A New Standard for Assembly of Biological Parts Using Cpf1. ACS Synthetic Biology 5:1383–1388. [DOI] [PubMed] [Google Scholar]
- Matinyan N, Gonzalez Y, Dierick HA, and Venken KJT 2021a. Determining effective drug concentrations for selection and counterselection genetics in Drosophila melanogaster. STAR Protocols 2:100783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matinyan N, Karkhanis MS, Gonzalez Y, Jain A, Saltzman A, Malovannaya A, Sarrion-Perdigones A, Dierick HA, and Venken KJT 2021b. Multiplexed drug-based selection and counterselection genetic manipulations in Drosophila. Cell Reports 36:109700–109700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, and Ikemura T. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Research 28:292–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-González A, Kniewel R, Veldhuizen M, Verma HK, Navarro-Rodríguez M, Rubio LM, and Caro E. 2017. Adaptation of the GoldenBraid modular cloning system and creation of a toolkit for the expression of heterologous proteins in yeast mitochondria. BMC Biotechnology 17:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo T-TB, Hibbard KL, Murphy C, Jenett A, Truman JW, and Rubin GM 2010. Refinement of Tools for Targeted Gene Expression in Drosophila. Genetics 186:735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, and Haussler D. 1997. Improved Splice Site Detection in Genie. Journal of Computational Biology 4:311–323. [DOI] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Chang L, Gonzalez Y, Gallego-Flores T, Young DW, and Venken KJT 2019. Examining multiple cellular pathways at once using multiplex hextuple luciferase assaying. Nature Communications 10:5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juárez P, Fernández-del-Carmen A, Granell A, and Orzaez D. 2011. GoldenBraid: An Iterative Cloning System for Standardized Assembly of Reusable Genetic Modules. PLoS ONE 6:e21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion‐Perdigones A, Gonzalez Y, and Venken KJT 2020. Rapid and Efficient Synthetic Assembly of Multiplex Luciferase Reporter Plasmids for the Simultaneous Monitoring of Up to Six Cellular Signaling Pathways. Current Protocols in Molecular Biology 131:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Gonzalez Y, and Venken KJT 2022. Synthetic Assembly DNA Cloning of Multiplex Hextuple Luciferase Reporter Plasmids. Methods in Molecular Biology 2524:409–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Vazquez-Vilar M, Palací J, Castelijns B, Forment J, Ziarsolo P, Blanca J, Granell A, and Orzaez D. 2013. GoldenBraid 2.0: A Comprehensive DNA Assembly Framework for Plant Synthetic Biology. Plant Physiology 162:1618–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA, and Voigt CA 2016. Synthetic biology to access and expand nature’s chemical diversity. Nature Reviews Microbiology 14:135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szybalski W, Kim SC, Hasan N, and Podhajska AJ 1991. Class-IIS restriction enzymes — a review. Gene 100:13–26. [DOI] [PubMed] [Google Scholar]
- Vazquez-Vilar M, Quijano-Rubio A, Fernandez-del-Carmen A, Sarrion-Perdigones A, Ochoa-Fernandez R, Ziarsolo P, Blanca J, Granell A, and Orzaez D. 2017. GB3.0: a platform for plant bio-design that connects functional DNA elements with associated biological data. Nucleic Acids Research 45:2196–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas D. 2001. Agarose gel electrophoresis. Current Protocols in Molecular Biology Chapter 2:2.5A.1–2.5A.9. [DOI] [PubMed]
- Weber E, Engler C, Gruetzner R, Werner S, and Marillonnet S. 2011. A Modular Cloning System for Standardized Assembly of Multigene Constructs. PLoS ONE 6:e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, and Fussenegger M. 2018. Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nature Reviews Molecular Cell Biology 19:507–525. [DOI] [PubMed] [Google Scholar]
- Yagi R, Mayer F, and Basler K. 2010. Refined LexA transactivators and their use in combination with the Drosophila Gal4 system. Proceedings of the National Academy of Sciences 107:16166–16171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.