Abstract
Lipid conjugation supports delivery of small interfering RNAs (siRNAs) to extrahepatic tissues, expanding the therapeutic potential of siRNAs beyond liver indications. However, siRNA silencing efficacy in extrahepatic tissues remains inferior to that routinely achieved in liver, partially due to the low rate of endosomal escape following siRNA internalization. Improving siRNA endosomal release into cytoplasm is crucial to improving efficacy of lipid-conjugated siRNAs. Given the ability of ionizable lipids to enhance endosomal escape in a context of lipid nanoparticles (LNP), here, we provide the first report on the effect of an ionizable lipid conjugate on siRNA endosomal escape, tissue distribution, efficacy, and toxicity in vivo. After developing a synthetic route to covalently attach the ionizable lipid, DLin-MC3-DMA, to siRNAs, we demonstrate that DLin-MC3-DMA enhances endosomal escape in cell culture without compromising siRNA efficacy. In mice, DLin-MC3-DMA conjugated siRNAs exhibit a similar overall tissue distribution profile to the similarly hydrophobic cholesterol-conjugated siRNA. However, only DLin-MC3-DMA conjugated siRNAs accumulated in vascular compartments, suggesting an effect of conjugate structure on intratissue distribution. Interestingly, we observed non-specific modulation of gene expression in tissues with high accumulation of DLin-MC3-DMA siRNAs (>20 pmol/mg of tissue) while limited non-specific gene modulation has been observed in tissues with lower siRNA accumulation. These findings suggest modulating the nature of the conjugate is a promising strategy to alter siRNA intratissue and intracellular trafficking. Fine-tuning the nature of the conjugate to optimize endosomal escape while minimizing toxicity will be critical for the progression of therapeutic siRNA applications beyond the liver.
Keywords: RNA interference, therapeutic oligonucleotides, siRNA extrahepatic delivery, lipid conjugation, endosomal escape, ionizable lipid
Introduction
Small interfering RNA (siRNA) therapeutics function in the cytoplasm of cells, where they engage the RNA-induced silencing complex (RISC) to degrade disease-causing messenger RNA (mRNA) in a sequence-specific manner. siRNAs are revolutionizing medicine due to their ability to potently silence previously “undruggable” genes with unprecedented duration of effect. Efficient delivery to disease tissues is crucial for clinical success of siRNAs [1]; yet, the vast majority of siRNAs – even when fully chemically modified – are rapidly eliminated from the body after systemic administration [2]. Covalent conjugation of chemical moieties to siRNAs overcomes this issue by promoting siRNA uptake into tissues and reducing clearance from the blood. Conjugating N-acetylgalactosamine (GalNAc) [3, 4] to fully chemically stabilized siRNA enables highly selective, sustainable, and functional delivery to hepatocytes with minimal adverse effects [4-8]. GalNAc conjugation is the basis of multiple FDA-approved siRNA drugs for liver diseases [7-10], and represents a major breakthrough in the therapeutic siRNA field. Yet, functional delivery of siRNA to tissues beyond liver remains challenging.
Lipid conjugation has emerged as an extrahepatic delivery platform for siRNAs following systemic administration [11], with cholesterol being intensively studied. Cholesterol conjugation supports siRNA accumulation and productive silencing in kidney [12], muscle [13], and placenta [14], but the vast majority of cholesterol-siRNAs accumulate in the liver (up to 80%) [15, 16]. Moreover, cholesterol conjugates are highly toxic at high concentrations, limiting its clinical potential [17].
The impact of other classes of lipid conjugates – fatty acids, steroids, and vitamins – on extrahepatic siRNA delivery has also been evaluated [2, 18-21]. These analyses revealed a correlation between degree of siRNA hydrophobicity and its tissue distribution profile [16, 22]. Less hydrophobic compounds bind high density lipoprotein in serum and mostly accumulate in kidneys, whereas more hydrophobic siRNAs primarily bind low density lipoprotein and mostly distribute to liver [2, 22, 23]. In addition to enhance liver and/or kidney tissue accumulation, lipid conjugation enables functional siRNA delivery to extrahepatic and extrarenal tissues including fat, adrenal gland, lung, heart and muscle [2]. Indeed, we found that systemic administration in mice of docosanoic acid conjugated siRNA leads to durable and safe siRNA-mediated silencing in both skeletal and cardiac muscles [21]. We also find that chemical structure of the conjugate impacts intra-organ spatial distribution and cell-type internalization [2, 20]. These reports demonstrate that lipid conjugation is a promising approach for improving siRNA extrahepatic functional delivery. However, the level of extrahepatic silencing by lipid-siRNAs (30–60% gene knockdown) remains lower than that routinely achieved in liver (80–90% knockdown) due, in part, to a low rate of siRNA endosomal escape after cell internalization. Only 1-2% of intracellular siRNA is released from endosome into cytoplasm, limiting RISC activity [24, 25]. Enhancing siRNA ability to cross the endosomal lipid bilayer into cytoplasm, regardless of endocytosis type (e.g., clathrin, caveolae, phagocytosis, macropinocytosis) [26], is key to expanding the utility of siRNA therapies.
Numerous lipids have been explored in the context of LNP to enhance endosomal escape of siRNAs [27]. The ionizable amino lipid, dilinoleylmethyl-4-dimethylaminobutyrate (DLin-MC3-DMA), is a gold standard cationic lipid for delivery of LNP siRNA systems to, and subsequent gene silencing in, hepatocytes [28]. DLin-MC3-DMA is composed of two unsaturated acyl chains and an amino function exhibiting a pKa between 6.2 and 6.4 and, therefore, is positively charged inside the endosome (pH ~5). These characteristics result in association of DLin-MC3-DMA from the LNP with anionic lipids from the endosomal membrane, leading to endosomal membrane disruption (from bilayer structure to hexagonal structure) and endosomal siRNA release into cytoplasm [29]. While Dlin-MC3-DMA lipid has been used in a context of LNP to enhance siRNA endosomal release, to our knowledge, direct conjugation of such a lipid to siRNAs and its impact on siRNA endosomal escape have never been investigated.
Here, we directly conjugate an analog of DLin-MC3-DMA to siRNAs. The DLin-MC3-DMA lipid analog retained key properties that enhance endosomal escape (Figure 1) while being compatible with standard oligonucleotide synthesis methods. We describe a straightforward scheme for synthesis of a DLin-MC3-DMA analog functionalized solid support (CPG) to obtain DLin-MC3-DMA conjugated siRNAs then evaluate the impact on siRNA tissue distribution, endosomal escape, and silencing efficacy. We demonstrate that DLin-MC3-DMA promotes endosomal escape of siRNAs in a cell reporter system without compromising RISC activity. DLin-MC3-DMA conjugated siRNAs show a similar tissue distribution profile in vivo as cholesterol-conjugated siRNAs due to a similar degree of compound hydrophobicity. However, DLin-MC3-DMA conjugated siRNA showed significantly higher accumulation in the vascular compartment within tissues. In tissues with high DLin-MC3-DMA siRNA accumulation (>20 pmol/mg of tissue), significant non-specific modulation of gene expression was observed. Our findings demonstrate that engineering lipid conjugates is a promising strategy to improve siRNA intracellular trafficking. However, fine-tuning the nature of the conjugate is needed to balance improved endosomal escape with minimal toxicity.
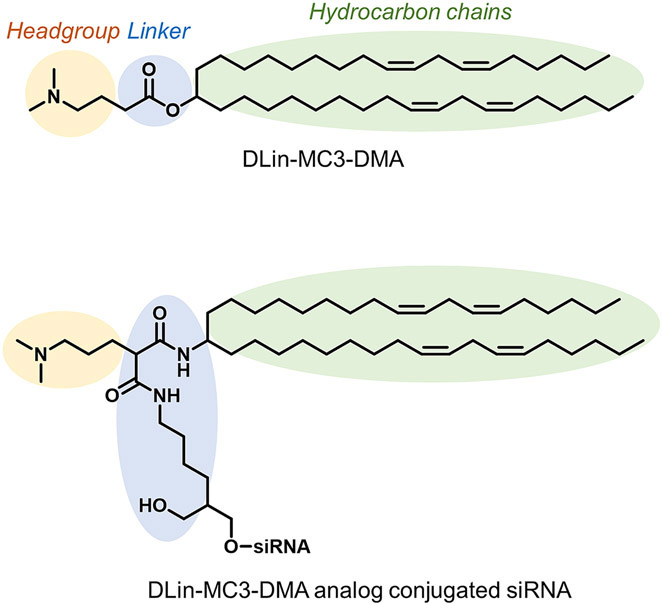
Figure 1:
Structure of DLin-MC3-DMA and Dlin-MC3-DMA analog conjugated siRNAs
Materials and Methods
Preparation of DLin-MC3-DMA functionalized CPG
All synthetic schemes, protocols, and NMR spectra are presented in supplemental information (Schemes S1-S9 and Figures S1-S16)
Oligonucleotide synthesis, deprotection, and purification
Oligonucleotides were synthesized on a Mermaid 12 synthesizer following standard protocols. DLin-MC3-DMA conjugated sense strands were synthesized at 10-μmole scale on a custom synthesized DLin-MC3-DMA functionalized CPG support (Compound 8, Figure 2), and were cleaved and deprotected using 40% aq. methylamine at 45°C for 1h. Antisense strands were synthesized at 10 μmole scale on CPG functionalized with Unylinker® (ChemGenes, Wilmington, MA). They were first deprotected with a solution of bromotrimethylsilane:pyridine (3:2, v/v) in dichloromethane for (E)-vinylphosphonate deprotection, then cleaved and deprotected with 40% aq. methylamine at 45°C for 1h. Oligonucleotides were purified using an Agilent Prostar System (Agilent, Santa Clara, CA) over a C18 column for DLin-MC3-DMA conjugated sense strands and over an ion-exchange column for antisense strands. Purified oligonucleotides were desalted by size-exclusion chromatography and characterized by Liquid Chromatography-Mass Spectrometry (LC-MS) analysis on an Agilent 6530 accurate-mass Q-TOF LC/MS (Agilent technologies, Santa Clara, CA) (Figures S17-S19).
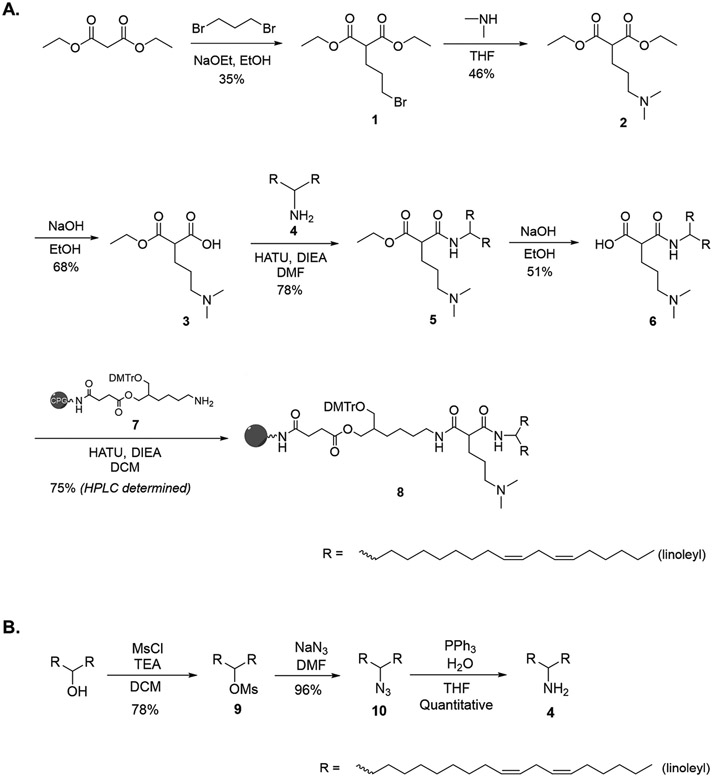
Figure 2:
Synthetic route of (A.) DLin-MC3-DMA analog functionalized CPG to prepare DLin-MC3-DMA conjugated RNA (B.) Dimeric linoleyl compound 4
DLin-MC3-DMA siRNA physicochemical characterization
Relative hydrophobicity of DLin-MC3-DMA and cholesterol conjugated siRNAs were determine by measuring the retention time on an Agilent Prostar System equipped with a Water HxSil C18 column (75 × 4.6) in a gradient of 100% buffer A (0.1 M trimethylamine acetate in water) to 100% buffer B (0.1 M trimethylamine acetate in acetonitrile) at a flow rate of 1 ml/min for 15 min at 60°C. Hydrodynamic diameters of siRNA were determined by dynamic light scattering (DLS) using a Zetasizer ZEN3600 (Malvern Instruments, UK). siRNAs solutions (10 nmol in 1 mL PBS) were analyzed at 25°C in triplicate. All scattered photons were collected at a 173°-scattering angle. Scattering intensity data were processed using instrumental software to obtain the hydrodynamic diameter and the size distribution of each sample. For transmission electron microscopy, freshly prepared carbon support films (either Copper or Gold 400 mesh grids) were used and the entire procedure was carried out in an enclosed chamber with a relative humidity of 60%. Solution of siRNAs (33 μM) were spread on the freshly prepared grids and excess sample droplets were wicked away with a wedge of filter paper (Whatman #1). The grids were negatively stained with 1% uranyl acetate in water. The excess stain were wicked off and the grid were air-dried. The samples were examined using a FEI (Thermo-Fisher) Tecnai 12, transmission electron microscope with an accelerating voltage of 120KV. The images were recorded using Digital Micrograph software running a Gatan, Rio9 digital camera system.
Preparation and treatment of Galectin-8 cells for siRNA efficacy
Gal8-YFP-MDA-MB-231 cells were kindly provided by the Duvall lab at Vanderbilt University [30]. Cells were cultured in DMEM media (Invitrogen) supplemented with 100 U/ml penicillin streptomycin (Invitrogen), and 10% fetal bovine serum (Atlanta Biologicals) at 37°C and 5% CO2. To measure silencing activity, a 7-point dose response was done using RNAiMAX (Thermo Fisher) using the manufacturer’s standard protocol for reverse transfection. Briefly, siRNA was diluted to 4x final concentration in 25 μl serum-free OptiMEM and mixed with 25 μl diluted RNAiMAX (0.3 μl/well in Optimem). After 10 min incubation, 50 μl of the resulting mixture was added to each well in a 96-well plate. During this incubation, cells were harvested by trypsinization and resuspended at 5000 cells/50 μl in DMEM containing 6% FBS and no antibiotics. 50 μl of the diluted cell suspension was added to each well already containing the RNAiMAX-siRNA complex, for a final volume of 100 μl/well, and the plates were gently rocked back and forth to mix. 72 hours after treatment, the cells were lysed and processed according to the manufacturer’s recommended protocol using the Quantigene bDNA Assay 2.0 (Thermo Fisher QS0011) and were normalized to the housekeeping gene, PPIB, as previously described [31]. Briefly, cells were lysed in 250 μl diluted lysis mixture (1:2 lysis mixture:water) with 0.167 μg/μl proteinase K (Thermo Fisher QS0103) for 30 minutes at 55°C. Cell lysates were mixed thoroughly and 20-80 μl of lysate were added to a bDNA capture plate, along with 0-60 μl additional diluted lysis mixture without proteinase K to obtain 80 μl total volume. Probe sets were diluted as specified in Thermo Fisher protocol and 20 μl of either human HTT (Thermo Fisher SA-50339) or PPIB (Thermo Fisher SA-10003) were added to each well of the capture plate to a final volume of 100 μl. Luminescence was detected on a Tecan M 1000.
Preparation and treatment of Galectin-8 cells for endosomal escape analysis
Gal8-YFP-MDA-MB-231 cells were seeded at a density of 100,000 cells/well in 35-mm glass-bottom dishes (MatTek P35G-1.5-14-C) in DMEM containing 3% FBS and grown overnight. The next day, the cells were transfected with siRNA (10 nM final concentration) using RNAiMAX (Thermo Fisher, 7.5 μl/well in 1 ml of media). After 24 hours, the cells were washed twice with FluoroBrite DMEM (Thermo Fisher) and imaged immediately on a Leica DMi8 widefield fluorescence microscope. After image acquisition, images were exported in 16-bit TIFF format from the Leica LAS X software and processed in Fiji v1.53c to quantify the percentage of cells containing Gal8-YFP large foci, and the mean Gal8-YFP fluorescence intensity per cell [32, 33].
Injection of DLin-MC3-DMA conjugated siRNAs into mice
Animal experiments were performed in accordance with animal care ethics approval and guidelines of the University of Massachusetts Chan Medical School Institutional Animal Care and Use Committee (IACUC, protocol number A-2411). In all experiments, 7-week-old female FVB/NJ mice (n=6 per group) were injected subcutaneously with phosphate buffered saline (PBS), or a 20 mg/kg dose of non-targeting control siRNA (Ntc), DLin-MC3-DMA conjugated siRNA, cholesterol siRNA, or unconjugated siRNA.
Peptide nucleic acid (PNA) hybridization assay
Quantification of antisense strands in tissues was performed using a PNA hybridization assay as described [34, 35]. Briefly, tissues (10 mg) were lysed in 200 μl MasterPure tissue lysis solution (EpiCentre) containing 0.2 mg/ml proteinase K (Invitrogen). Sodium dodecyl sulphate (SDS) was precipitated from lysates by adding 30 μL 3 M potassium chloride and pelleted centrifugation at 5000 × g for 15 minutes. siRNAs in cleared supernatant were hybridized to a Cy3-labeled PNA probe fully complementary to the antisense strand (PNABio, Thousand Oaks, CA, USA). Samples were analyzed by HPLC (Agilent, Santa Clara, CA) over a DNAPac PA100 anion-exchange column (Thermo Fisher Scientific). Cy3 fluorescence was monitored and peaks integrated. Final concentrations were ascertained using calibration curves.
Fluorescence microscopy
All fluorescence images were acquired with Leica DMi8 inverted tiling miscroscope (Leica Microsystems) and images were processed using the Leica software suite (LAS X and LAS X Lite). To prepare the slides, mice were euthanized and perfused with PBS at 48h post-injection. Tissues were collected and immersed in 10% formalin solution overnight at 4°C. Tissues were embedded in paraffin and sliced into 4-μm sections that were mounted on glass slides. Tissue sections on glass slides were deparaffinized in xylene, rehydrated in ethanol and stained with DAPI (Molecular Probes). Sections were imaged at 5 × and 40 × magnification.
In vivo mRNA silencing experiments
At 1-week post-injection, tissues were collected. mRNA was quantified using the QuantiGene 2.0 Assay (Affymetrix). Tissue punches were lysed in 300 μl Homogenizing Buffer (Affymetrix) containing 0.2 mg/ml proteinase K (Invitrogen). Diluted lysates and probe sets (mouse Htt, mouse Ppib, or mouse Hprt) were added to the bDNA capture plate and signal was amplified and detected as described by Coles et al. [36]. Luminescence was detected on a Tecan M1000 (Tecan, Morrisville, NC, USA).
Statistical analysis
Data were analyzed using GraphPad Prism 8 software (GraphPad Software, Inc., San Diego, CA). Concentration-dependent IC50 curves for in vitro experiments were fitted using a log(inhibitor) versus response—variable slope (four parameter). In vivo data were analyzed using non-parametric one-way ANOVA with Dunnett test for multiple comparisons.
Results
Synthesis of DLin-MC3-DMA analog conjugated siRNAs
To conjugate ionizable lipid to siRNA, we synthesized a solid support (CPG) functionalized with a DLin-MC3-DMA analog conjugate (Figure 2, Schemes S1-S9). The synthetic route of DLin-MC3-DMA CPG was carefully built so that each chemical element was compatible with oligonucleotide synthesis conditions while maintaining key lipid characteristics for endosomal membrane disruption – i.e., two unsaturated acyl chains (linoleyl) and one amino group with pka 6.2 to 6.4.
To prepare DLin-MC3-DMA CPG, diethyl malonate was reacted with dibromopropane to yield compound 1 (Figure 2A, Scheme S1 and Figures S1-S2). During this step, a mixture of compound 1 and a dimer product (from a reaction between compound 1 and diethyl malonate) was obtained. Clean isolation of compound 1 was achieved by column chromatography (35% yield). To incorporate the amine group essential for endosomal membrane disruption, compound 1 was reacted with dimethylamine to obtain compound 2 (Figure 2A, Scheme S2, Figures S3-S4). Controlled deprotection of compound 2 using stoichiometric amounts of sodium hydroxide was performed to achieve 68% yield of compound 3 (Figure 2A, Scheme S3, Figures S5-S6). Controlled deprotection leading to an unprotected carboxylic acid group of compound 3 enabled conjugation of dimeric linoleyl (compound 4) through an amide linkage to obtain compound 5 at 78% yield (Figure 2A, Scheme S4, Figures S7-S8). Dimeric linoleyl 4 was generated from mesylation, azidation, and amination of DLin-MeOH, as described by Jayaraman et al. [37] (Figure 2B, Schemes S6-S8, Figures S11-S16).
Deprotecting the second carboxylic acid group of compound 5 yielded compound 6 (Figure 2A, Scheme S5, Figures S9-S10), allowing attachment of a functionalized CPG 7, prepared as described by Nikan et al. [38]. The final DLin-MC3-DMA functionalized CPG (compound 8) was obtained with a coupling efficiency of 75% (Figure 2A, Scheme S9).
Using DLin-MC3-DMA functionalized CPG 8, we synthesized DLin-MC3-DMA conjugated siRNA sense strands in good purity following standard solid-phase synthesis and deprotection protocols (See Materials and Methods section). Oligonucleotides were purified by High Performance Liquid Chromatography (HPLC) and characterized by mass spectrometry (Figures S17-S19). As controls, unconjugated and cholesterol conjugated siRNAs were synthesized. Cholesterol and DLin-MC3-DMA lipids were covalently attached to the 3′ end of the siRNA sense strand, which tolerates a range of covalent modifications [15]. siRNAs were fully chemically stabilized for maximal stability and retention, and minimal innate immune activation using validated chemical modification patterns [39-46]. RNA sequences and chemical modification patterns used in this manuscript are reported in Table S1.
DLin-MC3-DMA conjugate impacts siRNA hydrophobicity and aggregation without interfering with RISC activity
To evaluate the impact of DLin-MC3-DMA conjugate structure on siRNA physicochemical properties, we measured overall hydrophobicity and aggregation of unconjugated, DLin-MC3-DMA, or cholesterol conjugated siRNAs (Figure 3). To determine hydrophobicity, we subjected compounds to reversed-phase HPLC and monitored retention time, which increases with increasing hydrophobicity (Figure 3A) [47]. Consistent with previous reports, lipid conjugation (either DLin-MC3-DMA or cholesterol) increased siRNA hydrophobicity when compared to unconjugated siRNA (retention time 11.8 vs 5.7 min, respectively) [2, 22]. Although DLin-MC3-DMA conjugated siRNAs exhibited a similar degree of hydrophobicity to that of cholesterol-conjugated siRNAs, DLin-MC3-DMA compounds eluted in a broader peak (~3.5 min) compared to cholesterol compounds, which showed a sharp elution profile (~1 min). The broad elution profile of DLin-MC3-DMA conjugated siRNA is likely due to the formation of aggregates.
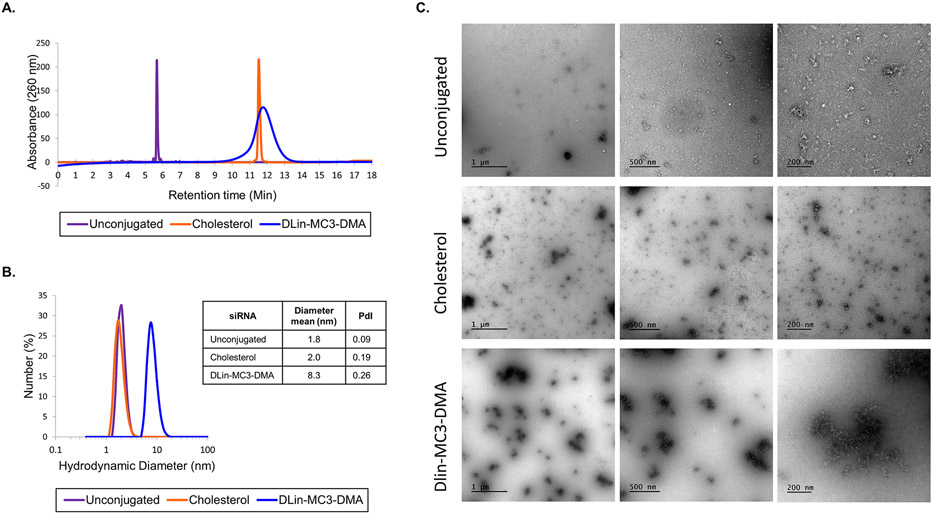
Figure 3:
DLin-MC3-DMA conjugated siRNAs have distinct physicochemical properties (A.) High Performance Liquid Chromatography spectra at pH 6.4 of unconjugated, cholesterol, and DLin-MC3-DMA-conjugated siRNA sense strands show variation in retention time (B.) Hydrodynamic diameter (nm) of unconjugated, cholesterol, and DLin-Mc3-DMA-conjugated siRNA determined by Dynamic Light Scattering. PdI = Polydispersity Index. (C.) Transmission electron microscopy for unconjugated, cholesterol and DLin-MC3-DMA siRNAs at 33 μM. Images are taken with different magnifications: X87 (scale = 200 nm), X43 (scale = 500 nm) and X26.5 (scale = 1 μm) magnifications.
To determine the effect of the nature of the conjugate on aggregation, we measured hydrodynamic diameters of unconjugated, cholesterol, and DLin-MC3-DMA conjugated siRNAs using Dynamic Light Scattering (DLS) (Figure 3B). The mean diameter of cholesterol-conjugated siRNAs and unconjugated siRNAs was ~2.0 nm, indicating that they did not aggregate. By contrast, the mean diameter of DLin-MC3-DMA conjugated siRNAs was 8.3 nm, indicative of self-assembly into small aggregates and micelles. Interestingly, the average particle size of DLin-MC3-DMA siRNAs was approximately double the average diameter of other previously-reported dimeric compounds [20], suggesting that the chemical structure of the conjugate, not just valency, significantly impacts siRNA physicochemical properties. Formation of aggregates was confirmed by Transmission Electron Microscopy (TEM) (Figure 3C), where DLin-MC3-DMA conjugate enables self-assembly of the siRNAs. In contrast, limited aggregations was observed with unconjugated and cholesterol siRNAs, demonstrating that DLin-MC3-DMA conjugate is responsible for siRNA aggregation.
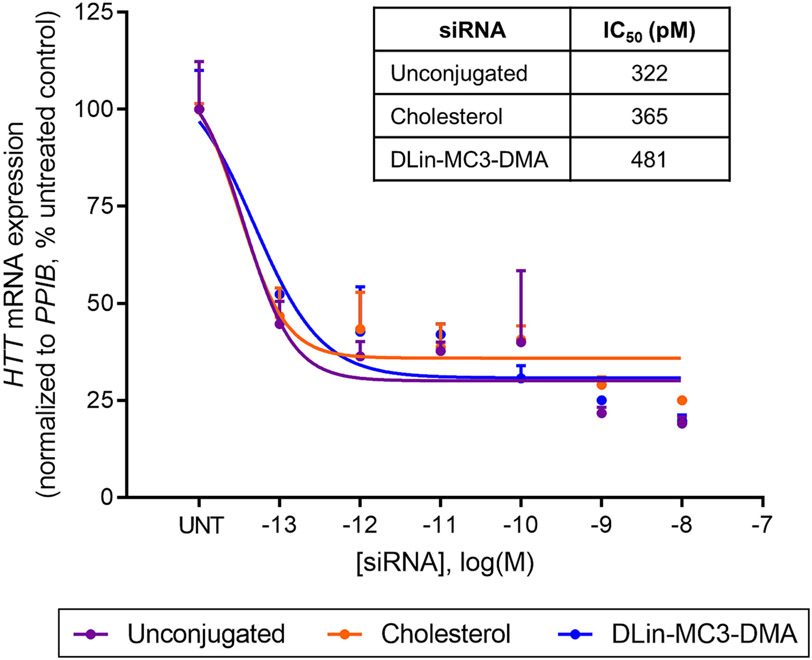
To evaluate the effect of DLin-MC3-DMA conjugation on siRNA silencing efficacy in vitro, we transfected unconjugated, cholesterol, or DLin-MC3-DMA conjugated siRNAs into MDA-MB-231 cells stably expressing Galectin8 fused to yellow fluorescent protein (Gal8-YFP) [30]. This reporter line enables subsequent analysis of endosomal escape (see DLin-MC3-DMA conjugate enhances siRNA endosomal escape section). siRNAs were designed to target a well-validated gene, huntingtin (HTT) [48]. Similar potency was observed for all three compounds, evidenced by similar half-maximal inhibitory concentrations (IC50): 322 pM (unconjugated), 365 pM (cholesterol), and 481 pM (DLin-MC3-DMA). No change in the level of housekeeping gene expression has been observed except at high concentrations (>1nM) (Figure S20), concentration corresponding to an order of magnitude higher than IC50 values. These data show that DLin-MC3-DMA conjugated siRNAs affect the targeting gene and housekeeping gene expression levels in a similar manner to unconjugated and cholesterol conjugated siRNAs, suggesting that DLin-MC3-DMA conjugate does not lead to significant toxicity in vitro. Thus, we demonstrated that Dlin-MC3-DMA conjugation does not substantially affect RISC activity and RNAi potency and cell viability (Figure 4).
Figure 4:
DLin-MC3-DMA conjugate does not prevent RISC activity. HTT mRNA levels measured in MDA-MB-231 cells expressing Gal8-YFP using QuantiGene® (Affymetrix) and presented as percent of untreated control (n=3, mean ± SD). Cells were transfected with siRNAs using RNAiMAX (72h). log IC50 values of unconjugated, cholesterol, and DLin-MC3-DMA siRNAs from dose response curves.
Our experiments are the first to demonstrate successful synthesis of ionizable lipid conjugated siRNAs with distinct physicochemical properties that retain gene silencing activity in vitro.
DLin-MC3-DMA conjugate enhances siRNA endosomal escape
To evaluate the impact of DLin-MC3-DMA conjugate on endosomal escape, Cy3-labeled siRNAs conjugated to either cholesterol or DLin-MC3-DMA were incubated with MDA-MB-231-Gal8-YFP cells (Figure 5) [30]. In this experiment, RNAiMAX was used as transfecting agent to insure that cholesterol and DLin-MC3-DMA conjugated siRNAs get internalized inside the cells to a similar extent. Galectin proteins are normally dispersed homogenously throughout the cytoplasm. When endosomes are disrupted, galectins redistribute by binding to glycosylation moieties located on the inner face of endosomal membranes [49]. Thus, we can use fluorescence microscopy to visualize redistribution of Gal8-YFP and assess endosomal integrity by measuring fluorescence intensity in cells, where an increase in fluorescence is indicative of an increase in endosomal membrane disruption [50-52].
Figure 5:
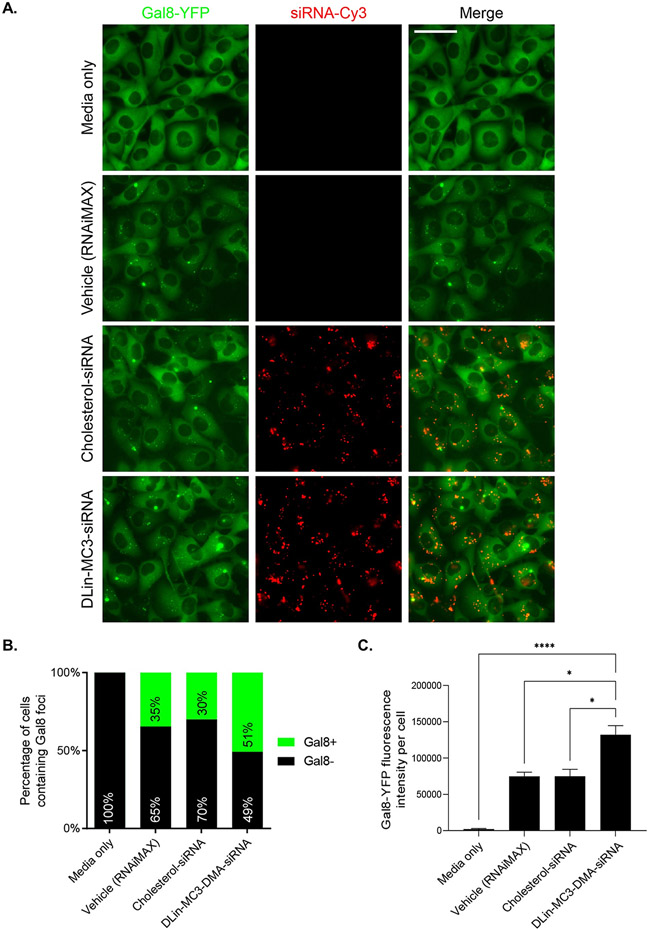
DLin-MC3-DMA conjugate enhances endosomal escape in cell culture. (A) Fluorescence microscopy of Gal8-YFP reporter cells after 24h incubation of media only, RNAiMAX only, or conjugated siRNAs. (B.) Counted number of large foci Gal8+ and Gal8− cells for each treatment group (n>300 counted per group) (C.) Quantification of average Gal8-YFP fluorescence intensity per cell. Error bars represent SEM (n = >300 cells counted per group). Data analysis: Multiple comparisons = One-way ANOVA, Dunnett test (****P<0.0001, *P<0.1).
After a 24 h incubation of vehicle, cholesterol, or DLin-MC3-DMA conjugated siRNAs, we observed a significant amount of Gal8 foci (small and large) in cells, demonstrating endosomal disruption (Figure 5A). The effect of vehicle on endosomal integrity is consistent with previous reports using LNP delivery [50]. Cholesterol siRNAs did not increase endosome disruption compared to vehicle, with a similar number of large foci Gal8+ cells (30% vs 35%, respectively) (Figure 5B, Figure S21) and a similar Gal8-YFP fluorescence intensity per cell observed (~70,000 FLU) (Figure 5C). By contrast, DLin-MC3-DMA conjugated siRNA enhanced endosomal disruption, evidenced by a significant increase in large foci Gal8+ cells (51%) (Figure 5B, Figure S21) and a ~2-fold increase in Gal8-YFP fluorescence intensity per cell (130,000 FLU) compared to vehicle (Figure 5C).
These data demonstrate that the structure of DLin-MC3-DMA analog conjugated to siRNAs maintains the key lipid characteristics for endosomal membrane disruption – i.e., two unsaturated acyl chains (linoleyl) and one amino group with pka 6.2 to 6.4. Conjugation of ionizable lipid to siRNAs significantly improves endosomal disruption and is a promising strategy to enhance siRNA endosomal escape.
DLin-MC3-DMA conjugate enhances tissue retention and promotes broad distribution
Next, we investigated the effect of the DLin-MC3-DMA conjugate on siRNA tissue retention, distribution, and efficacy in mice. Mice were subcutaneously (SC) injected with Cy3-labeled unconjugated, cholesterol, or DLin-MC3-DMA siRNAs targeting Htt at a dose of 20 mg/kg. We have previously shown that the presence of the Cy3 label attached to the 5′ end of the siRNA sense strand has minimal impact on overall tissue distribution in the context of lipophilic siRNAs [2]. 48h post-injection, antisense strand accumulation was measured in tissues using the PNA hybridization assay (Figure 6) [35] and siRNA spatial-distribution in tissue was evaluated by Cy3 fluorescence microscopy (Figure 7, Figures S22-S23).
Figure 6:
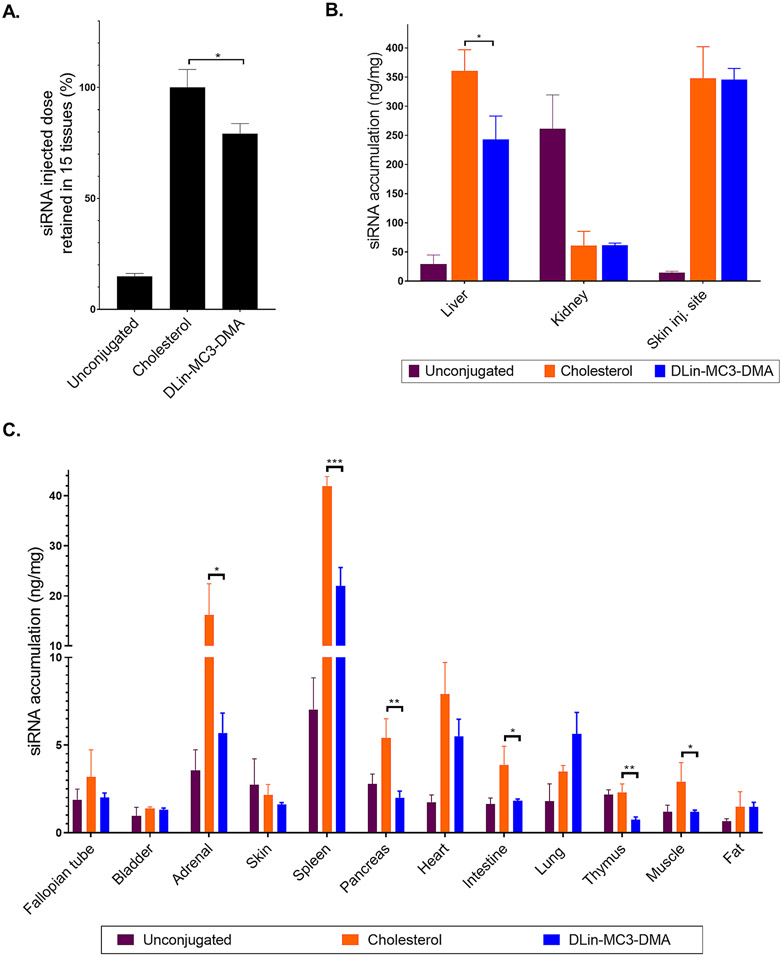
DLin-MC3-DMA conjugate allows quantitative siRNA tissue retention and broad tissue distribution (A.) siRNA injected dose retained in tissues at 48h after SC injection of unconjugated (unconj), cholesterol, and DLin-MC3-DMA conjugated siRNAs (20 mg/kg, n=3, mean ± SD) (B.) Bar graph showing siRNA quantification in liver, kidney (cortex), and skin (injection site) measured by PNA hybridization assay (SC, 20 mg/kg, n=3, mean ± SD) (C.) Bar graph showing siRNA quantification in tissues measured by PNA hybridization assay (SC, 20 mg/kg, n=3, mean ± SD). Data analysis: Multiple comparisons = One-way ANOVA, Dunnett test (***P<0.001, **P<0.01, *P<0.1).
Figure 7:
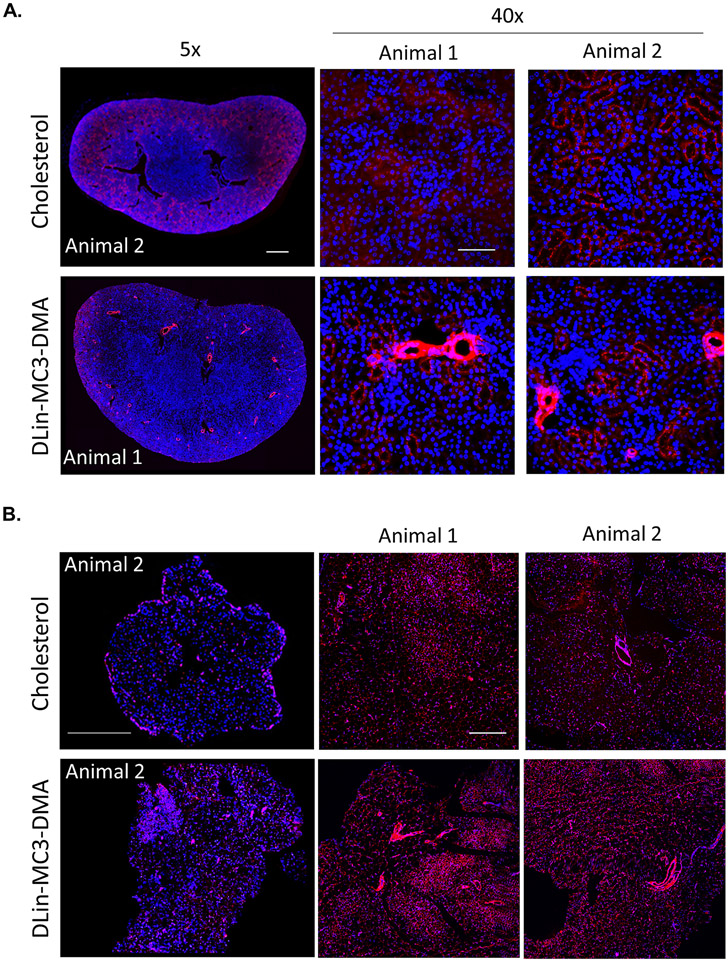
DLin-MC3-DMA siRNAs accumulate to vascular compartment. Representative fluorescence images of (A.) kidney (cortex) and (B.) fat from mice injected subcutaneously (n=2, 20 mg/kg) with cholesterol and DLin-MC3-DMA conjugated Cy3-labeled siRNAs (red). Nuclei stained with DAPI (blue). Tissues were collected 48h after injection. Images taken at 5× or 40× magnifications and collected at the same laser intensity and acquisition time. Scale, 1 mm for 5× and 50 μm for 40×
To estimate the fraction of injected siRNA dose retained after 48h, we determined antisense strand accumulation in fifteen tissues that comprise most of the mouse body (Figure 6A). The relative mass of each tissue was experimentally defined or approximated based on published mouse organ weights [53-55]. Within minutes of injection, unconjugated siRNAs were cleared rapidly from the body, with only 15% being retained (mostly in kidneys) (Figure 6A). The addition of the cholesterol conjugate significantly enhanced overall retention to almost 100%, with minimal loss due to kidney filtration. DLin-MC3-DMA conjugation also significantly enhanced overall tissue retention relative to unconjugated siRNAs, but to a lesser extent than cholesterol siRNAs (80%, P<0.1).
DLin-MC3-DMA siRNAs distributed to liver, spleen, lung, heart, fat significantly more than unconjugated siRNAs, indicating that DLin-MC3-DMA conjugation alters compound tissue distribution and potentially enhance tissue delivery. Although cholesterol and DLin-MC3-DMA compounds have a similar degree of hydrophobicity, DLin-MC3-DMA siRNAs showed lower accumulation in liver, adrenal glands, spleen, pancreas, intestine, thymus, and muscle (Figure 6B-C). These findings confirm that hydrophobicity is not the sole determinant of overall siRNA retention and accumulation in tissues.
Conjugate structure not only affected siRNA tissue accumulation, but also spatial distribution within tissues (Figure 7, Figures S22-S23). In kidney and fat, based on anatomical structure DLin-MC3-DMA conjugated siRNAs seem to accumulate significantly in the vascular compartment, whereas cholesterol conjugated siRNAs did not. These data demonstrate that the DLin-MC3-DMA conjugate not only alters in vitro siRNA localization (Figure 5) but also in vivo cell-type distribution.
DLin-MC3-DMA conjugate induces non-specific modulation of gene expression in several tissues
To evaluate if DLin-MC3-DMA conjugate leads to an improvement in productive silencing in tissues, mice were SC injected with a single dose (n = 6 per group, 20 mg/kg) of DLin-MC3-DMA siRNAs targeting either Htt [48] or Cyclophilin B (Ppib) [56]. Both targets have validated siRNA sequences and are expressed in a wide range of tissues at different levels (Htt low; Ppib high). To maximize siRNA efficacy, the DLin-MC3-DMA conjugate was attached via a dTdT cleavable linker to siRNAs [57]. Control mice were treated with PBS or DLin-MC3-DMA siRNAs with a non-targeting control (NTC) sequence. At 1-week post-injection, Htt, Ppib, and Hprt (hypoxanthine-guanine phosphoribosyl transferase, a housekeeping gene) mRNA levels were measured in clearance tissues (i.e., liver, kidney, spleen) as well as heart, fat, and muscle (Figures 8-9).
Figure 8:
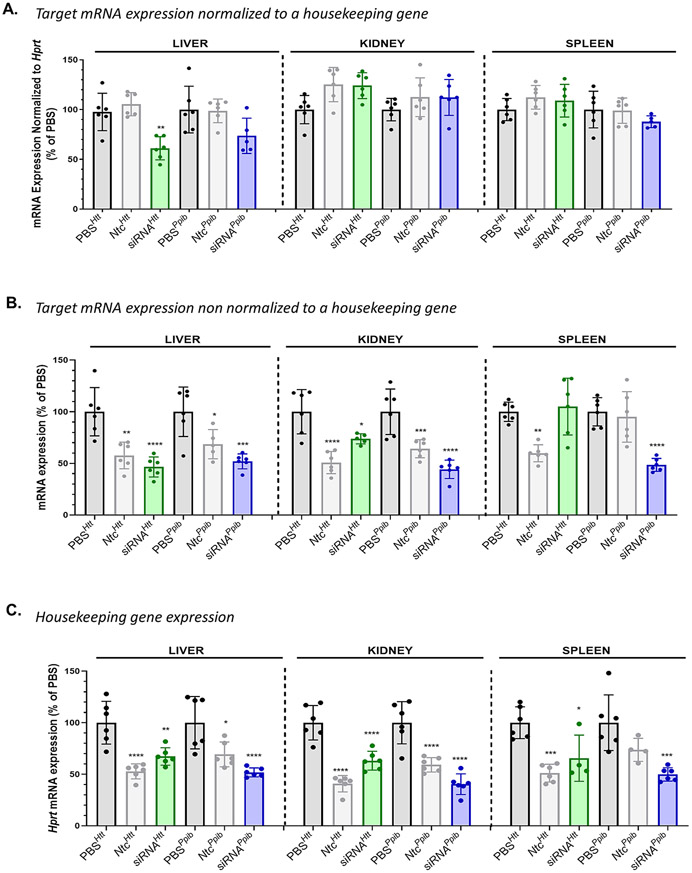
DLin-MC3-DMA conjugate induced non-specific gene modulation in tissues accumulating >20 pmol siRNA/mg tissue. Observed Htt and Ppib mRNA silencing (A.) normalized to Hprt (an housekeeping gene) (B.) non normalized to Hprt in liver, kidney, and spleen after SC injection of DLin-MC3-DMA siRNAs (1 week, 20 mg/kg, n=5-6, mean ± SD) (C.) Bar graph showing Hprt mRNA silencing in liver, kidney, and spleen after SC injection of DLin-MC3-DMA siRNAs (1 week, 20 mg/kg, n=5-6, mean ± SD). Data analysis: Multiple comparisons = One-way ANOVA, Dunnett test (****P<0.0001, ***P<0.001, **P<0.01, *P<0.1).
Figure 9:
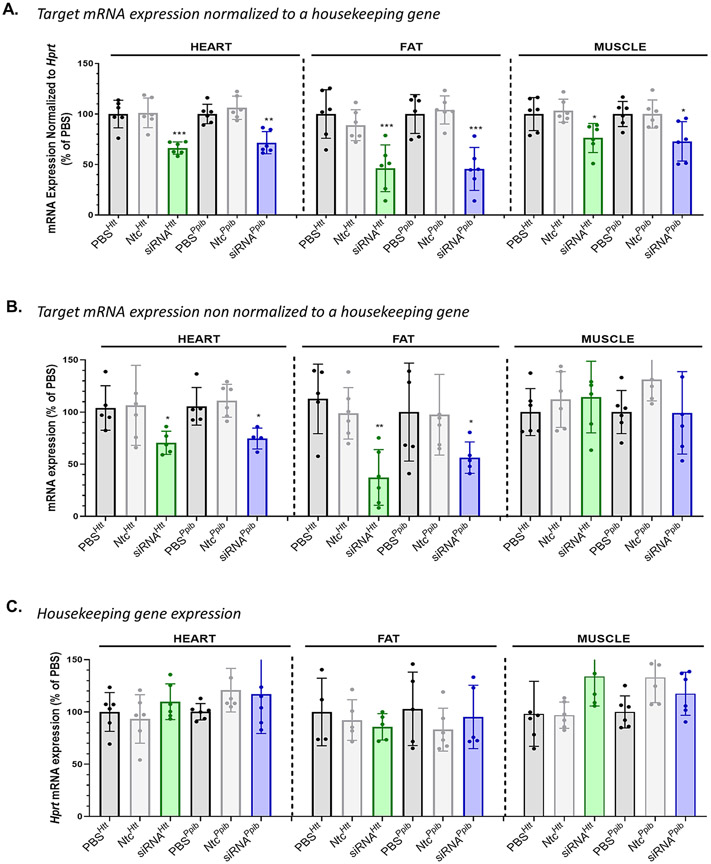
DLin-MC3-DMA conjugate does not induce non-specific gene modulation in tissues accumulating less than 20 pmol of siRNA/mg of tissue. Observed Htt and Ppib mRNA silencing (A.) normalized to Hprt (an housekeeping gene) (B.) non normalized to Hprt in heart, fat, and muscle after SC injection of DLin-MC3-DMA siRNAs (1 week, 20 mg/kg, n=5-6, mean ± SD) (C.) Bar graph showing Hprt mRNA silencing in heart, fat, and muscle after SC injection of DLin-MC3-DMA siRNAs (1 week, 20 mg/kg, n=5-6, mean ± SD). Data analysis: Multiple comparisons = One-way ANOVA, Dunnett test (**P<0.01, *P<0.1).
Among clearance tissues, Htt silencing (normalized to Hprt) by DLin-MC3-DMA siRNA was significant only in liver (40% silencing, P<0.01, Figure 8A). These results were unexpected, as siRNA accumulation in those tissues (>20 pmol siRNA/mg tissue) far exceeds levels required for productive silencing (Figure 6) [2, 22]. To understand this observation, we analyzed Htt and Ppib expression non-normalized to Hprt (Figure 8B) and found that levels of silencing induced by DLin-MC3-DMA siRNAs were significant (up to 60% silencing) for both targets, with the exception of Htt silencing in spleen. The low expression of Htt gene in the spleen tissue (11.6 nTPM) could explain the variability of the data, showing no observed Htt silencing when using Htt-targeting DLin-MC3-DMA siRNAs. DLin-MC3-DMA Ntc compounds also modulated Htt and Ppib expression, which was surprising given that we determined in a previous study that the NTC sequences and chemical modified pattern used did not impact Htt or Ppib mRNA expression [2, 46]. Moreover, in the current study, we did not observe downregulation of Htt or Ppib following unconjugated or cholesterol NTC siRNA delivery (Figure S24). These findings suggest that the downregulation observed with DLin-MC3-DMA NTC siRNAs is not due to the sequence or to the chemical pattern of the siRNAs but due to the nature of the conjugate. We also observed significant downregulation of Hprt by all DLin-MC3-DMA conjugated siRNAs in these tissues (Figure 8C). These data demonstrate that the DLin-MC3-DMA conjugate induces non-specific modulation of gene expression in liver, kidney, and spleen, resulting in non-specific modulation of gene expression.
A different scenario occurred in heart, fat, and muscle, where DLin-MC3-DMA conjugated siRNAs accumulated to a lower extent (<20 pmol siRNA/mg tissue). Significant Htt and Ppib silencing was observed in these tissues (For both targets: ~35% silencing in heart, 60% silencing in fat and ~30% silencing in muscle) when normalized to Hprt (Figure 9A). No downregulation of targets was observed for NTC compounds (Figure 9B) and Hprt mRNA expression level was not affected by any siRNAs (Figure 9C). Moreover, although DLin-MC3-DMA conjugated siRNAs accumulated to a less extent in heart and muscle compared to cholesterol siRNAs (Figure 6C), Dlin-MC3-DMA siRNAs induced better silencing (37±6% vs 10±7% in heart and 30±14% vs 17±13% in muscle) (Figure 9A, Figure S25).
Our collective findings suggest accumulation of DLin-MC3-DMA-siRNA in tissues beyond 20 pmol/mg results in non-specific modulation of gene expression, likely related to toxicity driven by enhanced endosomal escape. Interestingly, an increase of gene silencing in tissues with lower siRNA accumulation was observed compared to cholesterol siRNAs which accumulate to a better extent in those tissues. These data indicate that enhancement of silencing may be a result of an improvement of endosomal escape induced by DLin-MC3-DMA conjugate.
Discussion
The clinical success of GalNAc-conjugated siRNAs to treat liver diseases demonstrates the critical importance of conjugation for efficient delivery of chemically stabilized siRNAs to tissues [39]. Lipid conjugation of siRNAs supports broad tissue distribution beyond the liver, and modulating lipid structures profoundly impacts siRNA pharmacokinetics/pharmacodynamics, tissue distribution, and efficacy [2, 11, 16, 20, 22]. This paper demonstrates, for the first time, that direct conjugation of ionizable lipid to siRNA promotes endosomal escape, alters spatial distribution, and impacts tissue accumulation and efficacy. In tissues where DLin-MC3-DMA siRNAs accumulate to a high extent (>20 pmol/mg of tissue), significant non-specific modulation of gene expression is observed. However, in tissues with lower accumulation, an improvement of silencing is observed, likely due to an increase of siRNA endosomal escape engendered by DLin-MC3-DMA conjugate. These findings demonstrate that engineering lipid conjugates is essential to improving siRNA activity and that rational fine-tuning is needed to develop safe siRNAs.
While there are extensive reports on the design of lipids for efficient delivery in the context of LNPs [37], exploration of diverse ionizable lipid conjugates for direct conjugation to siRNAs has never been described. To conjugate DLin-MC3-DMA to siRNAs, we developed a straightforward route to synthesize a DLin-MC3-DMA functionalized CPG that was compatible with oligonucleotide synthesis conditions. The advantage of the synthetic methodology described herein is that it can be used for the conjugation of a broad range of ionizable lipids. Thus, modulation of the amino group, acyl chains and linker can be rapidly achieved to expand the chemical diversity of ionizable lipid conjugates with the goal of reducing toxicity while improving siRNA endosomal escape and activity.
We observed both increased numbers of, and more intense, Gal8 foci following DLin-MC3-DMA conjugated siRNA delivery compared to cholesterol siRNAs. These findings indicate that the DLin-MC3-DMA conjugate increases endosomal escape and impacts intracellular trafficking of siRNAs. However, no difference in efficacy in vitro was observed with DLin-MC3-DMA and cholesterol conjugated siRNAs when using a transfecting agent. This is likely because the in vitro system is saturated. The observed improvement in siRNA endosomal escape when using DLin-MC3-DMA suggests that the conjugate interacts with and disrupts the endosomal membrane, which is consistent with previous reports on LNPs containing DLin-MC3-DMA lipids [27]. This effect is likely attributed to the ionizable properties of the DLin-MC3-DMA conjugate. Furthermore, no internalization of DLin-MC3-DMA conjugated siRNAs and no gene silencing was observed in vitro without a transfecting agent (data not shown), limiting the use of in vitro systems to evaluate the impact of the conjugate on activity and trafficking in a carrier-free setting. The micelle formation of DLin-MC3-DMA conjugated siRNAs is by structure unlikely contributing to the enhancement of endosomal release, as micelle structures with other lipid conjugated siRNAs do not impact endosomal release [20], but may play a role in defining the apparent pKa of the amino function (pKa between 6.2 and 6.4) required for endosomal membrane disruption.
In vivo, unconjugated siRNAs showed less than ~15% body retention, but lipid conjugates significantly improved siRNA overall body retention (80-100%) and extrahepatic/extrarenal tissue exposure [2]. The higher overall retention and tissue distribution of lipid conjugated siRNAs is primarily explained by the capability of lipophilic siRNAs to bind spontaneously with serum lipoproteins [16, 22, 23]. DLin-MC3-DMA and cholesterol conjugated siRNAs have similar hydrophobicity and thus present similar tissue distribution profiles. However, DLin-MC3-DMA conjugated siRNAs show less tissue retention and accumulation in primary clearance tissues (liver, spleen, adrenal glands). The mechanism underlying decreased DLin-MC3-DMA siRNA tissue accumulation is unknown but may be due to the effect of DLin-MC3-DMA valency and charge on membrane fluidity and receptor interactions, which likely impact the degree of siRNA accumulation in tissues and clearance kinetics. Lower tissue accumulation of DLin-MC3-DMA conjugated siRNAs might also be due to its observed entrapment in endothelial cells, which form a barrier between vessels and tissue and control the flow of substances and fluid into and out of a tissue. The observed preferential endothelial cell delivery is very promising as vascular endothelium represents an important therapeutic target in inflammation, ischemia, edema, thrombosis, diabetes, oxidative stress, atherosclerosis and hypertension, and metabolic and oncological diseases. The use of non-toxic ionizable lipids could open a possibility to deliver siRNA to endothelial cells and therefore treat endothelium-related diseases.
In tissues with high siRNA accumuation (>20 pmol/mg of tissue), DLin-MC3-DMA conjugate likely causes toxicity by inducing endosomal stress thereby creating an imbalance in the endosomal trafficking pathway and impacting cell viability. Interestingly, the impact of DLin-MC3-DMA conjugate on endocytic pathways is tolerated in vitro but not in vivo. These findings demonstrate a lack of direct correlation between in vitro and in vivo models, supporting the need for evaluation in both systems to better design lipid conjugates.
Lipid conjugation is an emerging approach to further enhance siRNA distribution and efficacy in tissues after systemic administration. This study reports the first example of ionizable lipid conjugation of siRNAs and demonstrate that the chemical nature of the conjugate is a strong determinant of siRNA pharmacokinetics, distribution profiles, and toxicity in vivo. While the ionizable conjugate presented in this study has limited potential due to non-specific gene modulation, the study proves that rational engineering of lipid conjugates is the path to alter both intratissue and intracellular siRNA distribution. A careful design of the conjugate is imperative to enhance siRNA performance without inducing toxicity. Future expansion of the chemical space of ionizable lipids will establish a path toward enhancing siRNA delivery and potency to extrahepatic tissues.
Supplementary Material
Acknowledgements
We thank all the Khvorova lab members for insightful discussions and support, and Emily Haberlin for helping with the manuscript writing and editing. We thank the Electron Microscopy Core facility for performing the TEM experiment supported by Award Number S10 OD025113-01 from the National Center for Research Resources.
Funding
This work was supported by National Institutes of Health grants [RO1GM10880304, S10 OD020012].
Footnotes
Supplementary Material
This article includes supplementary material.
Declaration of interests
A.K. discloses ownership of stock in RXi Pharmaceuticals and Advirna; is a founder of Atalanta Therapeutics.
References
- 1.Khvorova A and Watts JK, The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol, 2017. 35(3): p. 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biscans A., et al. , Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Research, 2018. 47(3): p. 1082–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair JK, et al. , Multivalent N-Acetylgalactosamine-Conjugated siRNA Localizes in Hepatocytes and Elicits Robust RNAi-Mediated Gene Silencing. Journal of the American Chemical Society, 2014. 136(49): p. 16958–16961. [DOI] [PubMed] [Google Scholar]
- 4.Rajeev KG, et al. , Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. Chembiochem, 2015. 16(6): p. 903–8. [DOI] [PubMed] [Google Scholar]
- 5.Sardh E., et al. , Phase 1 Trial of an RNA Interference Therapy for Acute Intermittent Porphyria. N. Engl. J. Med, 2019. 380(6): p. 549–558. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y., Preclinical and Clinical Advances of GalNAc-Decorated Nucleic Acid Therapeutics. Mol Ther Nucleic Acids, 2017. 6: p. 116–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raal FJ, et al. , Inclisiran for the Treatment of Heterozygous Familial Hypercholesterolemia. N. Engl. J. Med, 2020. 382(16): p. 1520–1530. [DOI] [PubMed] [Google Scholar]
- 8.Balwani M., et al. , Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria. N. Engl. J. Med, 2020. 382(24): p. 2289–2301. [DOI] [PubMed] [Google Scholar]
- 9.Garrelfs S., et al. , LB002ILLUMINATE-A, A PHASE 3 STUDY OF LUMASIRAN, AN INVESTIGATIONAL RNAI THERAPEUTIC, IN CHILDREN AND ADULTS WITH PRIMARY HYPEROXALURIA TYPE 1 (PH1). Nephrology Dialysis Transplantation, 2020. 35(Supplement_3). [Google Scholar]
- 10.Adams D., et al. , Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med, 2018. 379(1): p. 11–21. [DOI] [PubMed] [Google Scholar]
- 11.Osborn MF and Khvorova A, Improving siRNA Delivery In Vivo Through Lipid Conjugation. Nucleic Acid Ther, 2018. 28(3): p. 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan H., et al. , Effects of cholesterol-tagged small interfering RNAs targeting 12/15-lipoxygenase on parameters of diabetic nephropathy in a mouse model of type 1 diabetes. American Journal of Physiology-Renal Physiology, 2008. 295(2): p. F605–F617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan T., et al. , Silencing Myostatin Using Cholesterol-conjugated siRNAs Induces Muscle Growth. Mol Ther Nucleic Acids, 2016. 5(8): p. e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turanov AA, et al. , RNAi modulation of placental sFLT1 for the treatment of preeclampsia. Nat Biotechnol, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soutschek J., et al. , Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature, 2004. 432(7014): p. 173–8. [DOI] [PubMed] [Google Scholar]
- 16.Wolfrum C., et al. , Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol, 2007. 25(10): p. 1149–57. [DOI] [PubMed] [Google Scholar]
- 17.Østergaard ME, et al. , Conjugation of hydrophobic moieties enhances potency of antisense oligonucleotides in the muscle of rodents and non-human primates. Nucleic Acids Research, 2019. 47(12): p. 6045–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubo T., et al. , Sixteen Different Types of Lipid-Conjugated siRNAs Containing Saturated and Unsaturated Fatty Acids and Exhibiting Enhanced RNAi Potency. ACS Chemical Biology, 2021. 16(1): p. 150–164. [DOI] [PubMed] [Google Scholar]
- 19.Nishina K., et al. , Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol Ther, 2008. 16(4): p. 734–40. [DOI] [PubMed] [Google Scholar]
- 20.Biscans A., et al. , The valency of fatty acid conjugates impacts siRNA pharmacokinetics, distribution, and efficacy in vivo. Journal of controlled release, 2019. 302: p. 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biscans A., et al. , Docosanoic acid conjugation to siRNA enables functional and safe delivery to skeletal and cardiac muscles. Molecular Therapy, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osborn MF, et al. , Hydrophobicity drives the systemic distribution of lipid-conjugated siRNAs via lipid transport pathways. Nucleic Acids Research, 2018. 47(3): p. 1070–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarett SM, et al. , Lipophilic siRNA targets albumin in situ and promotes bioavailability, tumor penetration, and carrier-free gene silencing. Proc Natl Acad Sci U S A, 2017. 114(32): p. E6490–e6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilleron J., et al. , Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol, 2013. 31(7): p. 638–46. [DOI] [PubMed] [Google Scholar]
- 25.Stalder L., et al. , The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. Embo j, 2013. 32(8): p. 1115–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doherty GJ and McMahon HT, Mechanisms of endocytosis. Annu Rev Biochem, 2009. 78: p. 857–902. [DOI] [PubMed] [Google Scholar]
- 27.Cullis PR and Hope MJ, Lipid Nanoparticle Systems for Enabling Gene Therapies. Molecular Therapy, 2017. 25(7): p. 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam YYC, Chen S, and Cullis PR, Advances in Lipid Nanoparticles for siRNA Delivery. Pharmaceutics, 2013. 5(3): p. 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semple SC, et al. , Rational design of cationic lipids for siRNA delivery. Nature Biotechnology, 2010. 28(2): p. 172–176. [DOI] [PubMed] [Google Scholar]
- 30.Kilchrist KV, et al. , Gal8 Visualization of Endosome Disruption Predicts Carrier-Mediated Biologic Drug Intracellular Bioavailability. ACS Nano, 2019. 13(2): p. 1136–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ly S., et al. , Single-Stranded Phosphorothioated Regions Enhance Cellular Uptake of Cholesterol-Conjugated siRNA but Not Silencing Efficacy. Mol Ther Nucleic Acids, 2020. 21: p. 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J., et al. , Fiji: an open-source platform for biological-image analysis. Nat Methods, 2012. 9(7): p. 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, and Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat Methods, 2012. 9(7): p. 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Godinho B., et al. , Pharmacokinetic Profiling of Conjugated Therapeutic Oligonucleotides: A High-Throughput Method Based Upon Serial Blood Microsampling Coupled to Peptide Nucleic Acid Hybridization Assay. Nucleic Acid Ther, 2017. 27(6): p. 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roehl I, Schuster M, and Seiffert S, Oligonucleotide detection method. US Patent US20110201006A1, 2011: p. 1–9.
- 36.Coles AH, et al. , A High-Throughput Method for Direct Detection of Therapeutic Oligonucleotide-Induced Gene Silencing In Vivo. Nucleic Acid Ther, 2016. 26(2): p. 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayaraman M., et al. , Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. Int. Ed. Engl, 2012. 51(34): p. 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikan M., et al. , Docosahexaenoic Acid Conjugation Enhances Distribution and Safety of siRNA upon Local Administration in Mouse Brain. Mol Ther Nucleic Acids, 2016. 5(8): p. e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster DJ, et al. , Advanced siRNA Designs Further Improve In Vivo Performance of GalNAc-siRNA Conjugates. Mol Ther, 2018. 26(3): p. 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allerson CR, et al. , Fully 2‘-Modified Oligonucleotide Duplexes with Improved in Vitro Potency and Stability Compared to Unmodified Small Interfering RNA. Journal of Medicinal Chemistry, 2005. 48(4): p. 901–904. [DOI] [PubMed] [Google Scholar]
- 41.Nallagatla SR and Bevilacqua PC, Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA, 2008. 14(6): p. 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson AL, et al. , Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA, 2006. 12(7): p. 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geary RS, et al. , Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev, 2015. 87: p. 46–51. [DOI] [PubMed] [Google Scholar]
- 44.Haraszti RA, et al. , 5΄-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res, 2017. 45(13): p. 7581–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parmar R., et al. , 5'-(E)-Vinylphosphonate: A Stable Phosphate Mimic Can Improve the RNAi Activity of siRNA-GalNAc Conjugates. Chembiochem, 2016. 17(11): p. 985–9. [DOI] [PubMed] [Google Scholar]
- 46.Hassler MR, et al. , Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res, 2018. 46(5): p. 2185–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith M and Jungalwala FB, Reversed-phase high performance liquid chromatography of phosphatidylcholine: a simple method for determining relative hydrophobic interaction of various molecular species. J Lipid Res, 1981. 22(4): p. 697–704. [PubMed] [Google Scholar]
- 48.Alterman JF, et al. , Hydrophobically Modified siRNAs Silence Huntingtin mRNA in Primary Neurons and Mouse Brain. Mol Ther Nucleic Acids, 2015. 4(12): p. e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thurston TL, et al. , Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature, 2012. 482(7385): p. 414–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wittrup A., et al. , Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nature biotechnology, 2015. 33(8): p. 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du Rietz H., et al. , Imaging small molecule-induced endosomal escape of siRNA. Nature Communications, 2020. 11(1): p. 1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munson MJ, et al. , A high-throughput Galectin-9 imaging assay for quantifying nanoparticle uptake, endosomal escape and functional RNA delivery. Communications biology, 2021. 4(1): p. 211–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reed DR, Bachmanov AA, and Tordoff MG, Forty mouse strain survey of body composition. Physiol Behav, 2007. 91(5): p. 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wanke R., et al. , Overgrowth of Skin in Growth Hormone Transgenic Mice Depends on the Presence of Male Gonads. Journal of Investigative Dermatology, 1999. 113(6): p. 967–971. [DOI] [PubMed] [Google Scholar]
- 55.Taniguchi T., et al. , Plasmodium berghei ANKA causes intestinal malaria associated with dysbiosis. Scientific Reports, 2015. 5(1): p. 15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reynolds A., et al. , Rational siRNA design for RNA interference. Nat Biotechnol, 2004. 22(3): p. 326–30. [DOI] [PubMed] [Google Scholar]
- 57.Biscans A., et al. , The chemical structure and phosphorothioate content of hydrophobically modified siRNAs impact extrahepatic distribution and efficacy. Nucleic Acids Research, 2020. 48(14): p. 7665–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.