ABSTRACT
Hepatitis B virus (HBV) is endemic in many parts of sub-Saharan Africa. Here, we present 5 full-length HBV recombinant genomes from blood donors in Beira, Mozambique. The genomes are recombinants between genotypes E and A and are distantly related to one another, based on their genetic distances.
ANNOUNCEMENT
Hepatitis B virus (HBV) (family Hepadnaviridae, genus Orthohepadnavirus) is a viral infection of the liver that can lead to cirrhosis and hepatocellular carcinoma. HBV infection is endemic in many sub-Saharan African countries. The virus contains a relaxed circular, partially double-stranded DNA genome that is ~3,200 bases in length. The genome includes four open reading frames (ORFs), coding for the polymerase (P), surface (S), core (C), and X proteins (1). HBV can be classified into 10 genotypes (genotypes A through J), and intergenotypic recombination has been reported (2–7). As in other sub-Saharan African countries, HBV is endemic in Mozambique (8, 9); however, few studies have examined HBV genotypes or recombination in Mozambique. Here, we present 5 full-length HBV genotype E/A recombinant viruses from blood donors in Beira, Mozambique.
In a previous study approved by the National Bioethics Committee in Mozambique (approval number 263/CNBS/2014), we evaluated partial HBV genomes from 57 HBV DNA-positive blood donor samples that were collected between November 2015 and April 2016. The majority of the viruses were nonrecombinant genotype A viruses, while 6 were putative recombinant viruses (10). In the current study, full-length genomes of these putative recombinant viruses were amplified and analyzed further. Viral DNA was extracted from blood donor plasma using the QIAmp UltraSens virus kit. The extracted DNA underwent a completion/ligation (C/L) step using the C/L reagents CutSmart buffer, bovine serum albumin (BSA), deoxynucleoside triphosphates (dNTPs), ATP, T4 DNA polymerase, and T4 DNA ligase, followed by a rolling circle amplification (RCA) step using the TempliPhi kit from Cytiva Life Sciences. Full-length HBV PCR was conducted using PicoMaxx high-fidelity PCR master mix (Agilent), HBV forward primer 5′-CCG GAA AGC TTG AGC TCT TCT TTT TCA CCT CTG CCT AAT CA-3′, and HBV reverse primer 5′-CCG GAA AGC TTG AGC TCT TCA AAA AGT TGC ATG GTG CTG G-3′. Amplified PCR products were run on a 1% agarose gel, and 3.2-kb products were cut and extracted using the QIAEX II gel extraction kit.
PCR products were submitted to the University of Cincinnati College of Medicine Genomics, Epigenomics, and Sequencing Core for next-generation sequencing (NGS). The New England BioLabs NEBNext Ultra II FS DNA library preparation kit was used to prepare the library, and sequencing was performed on an Illumina HiSeq 1000 sequencer with 1 × 51-bp single reads. A consensus sequence was generated by mapping the raw reads to the reference genome (GenBank accession number AY233282) from South Africa using UGENE v44.0 (11). Of note, a small portion of the full-length genome sequence corresponds to the primers utilized to amplify the virus and was not determined de novo. Consensus sequences were aligned to reference genomes from the Hepatitis Virus Diversity Research (HVDR) database using Clustal X v2.1 (12, 13). The alignment was visualized in FigTree v1.4.4. Consensus sequences were evaluated for recombination using the jumping profile Hidden Markov Model (jpHMM) program (14). Genetic distances were calculated in MEGA11 with the Kimura two-parameter model (15). All tools were run with default parameters unless otherwise specified. Details about raw reads and consensus sequences are available in Table 1.
TABLE 1.
HBV recombinant genome characteristics
| Patient identification no. | GenBank accession no. | No. of raw reads | Avg read depth (×) | Consensus length (bp) | Consensus GC content (%) |
|---|---|---|---|---|---|
| BSB0632 | OP514931 | 376,456 | 6,495 | 3,002 | 49.1 |
| BSB0670 | OP514932 | 599,795 | 9,635 | 3,175 | 48.4 |
| BSB1014 | OP514933 | 318,498 | 5,079 | 3,198 | 48.3 |
| BSB1230 | OP514934 | 388,153 | 6,161 | 3,213 | 48.4 |
| BSB1364 | OP514935 | 226,661 | 3,762 | 3,073 | 48.2 |
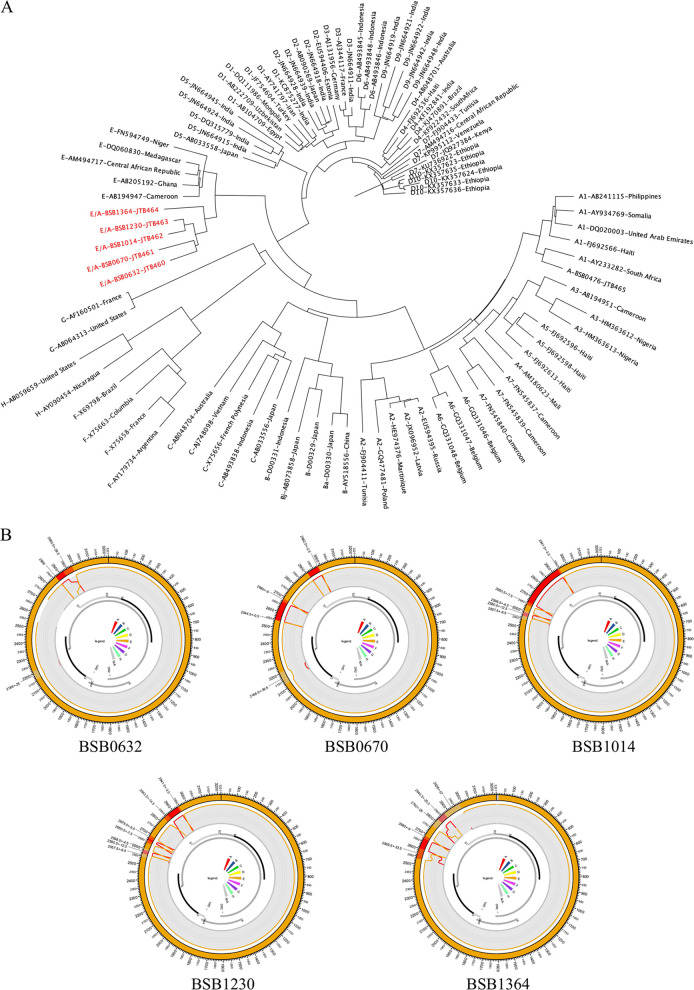
Of the 6 samples that were previously identified as putative recombinants, 1 sample could not be amplified by full-length PCR. The 5 putative recombinant viruses from Mozambique clustered separately from all genotype E references, as shown in Fig. 1A. The recombination prediction profiles confirmed that these samples were genotype E/A recombinants, with much of their genomes belonging to genotype E (Fig. 1B). The average genetic distance for these 5 samples was 2.58% (range, 1.17 to 3.49%), compared to 1.83% (range, 0.23 to 3.43%) for 14 nonrecombinant genotype E samples from Ghana (16, 17). Recombinant E/A isolates have been reported previously in West and Central African countries (4–7, 18–22), although the current study highlights the broader range of these unique variants.
FIG 1.
(A) Phylogenetic tree of full-length recombinant sequences from Mozambique. Each sequence is labeled with its GenBank accession number, country of origin, and genotype. Full-length recombinant sequences from Mozambique are colored in red. (B) Recombination prediction profiles. Profiles were evaluated using the jpHMM program, which employs a probabilistic approach to compare a nucleic acid or protein sequence S to a given multiple alignment A of a sequence family for which a classification into subtypes is available. jpHMM does not align and compare a database sequence S to the multiple alignment A as a whole; rather, it aligns local segments of S to those segments of individual sequences from A that are most similar to them. Recombination analysis of circular genomes is performed with artificially linearized sequences of the circular genomes using linear models. Samples are identified by their patient identification number.
Data availability.
The raw sequence data are available under BioProject PRJNA885128, with SRA accession numbers SRR21743134 through SRR21743138. The consensus HBV genomes are available in GenBank under accession numbers OP514931 through OP514935.
ACKNOWLEDGMENTS
Thanks go to the blood donors who voluntarily participated in this study, to the blood bank at Maputo Central Hospital, and to the laboratory at the Instituto Nacional de Saúde (Maputo, Mozambique).
This study was supported by the National Research Fund of Mozambique (FNI). NGS was conducted by the Genomics, Epigenomics, and Sequencing Core in the Department of Environmental Health, University of Cincinnati College of Medicine, and was supported in part by NIEHS grant P30 ES006096.
Contributor Information
Jason T. Blackard, Email: jason.blackard@uc.edu.
Simon Roux, DOE Joint Genome Institute.
REFERENCES
- 1.Araujo NM. 2015. Hepatitis B virus intergenotypic recombinants worldwide: an overview. Infect Genet Evol 36:500–510. doi: 10.1016/j.meegid.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 2.Glebe D, Goldmann N, Lauber C, Seitz S. 2021. HBV evolution and genetic variability: impact on prevention, treatment and development of antivirals. Antiviral Res 186:104973. doi: 10.1016/j.antiviral.2020.104973. [DOI] [PubMed] [Google Scholar]
- 3.Pourkarim M, Amini-Bavil-Olyaee S, Kurbanov F, Ranst M, Tacke F. 2014. Molecular identification of hepatitis B virus genotypes/subgenotypes: revised classification hurdles and updated resolutions. World J Gastroenterol 20:7152–7168. doi: 10.3748/wjg.v20.i23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurbanov F, Tanaka Y, Fujiwara K, Sugauchi F, Mbanya D, Zekeng L, Ndembi N, Ngansop C, Kaptue L, Miura T, Ido E, Hayami M, Ichimura H, Mizokami M. 2005. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J Gen Virol 86:2047–2056. doi: 10.1099/vir.0.80922-0. [DOI] [PubMed] [Google Scholar]
- 5.Garmiri P, Loua A, Haba N, Candotti D, Allain J-P. 2009. Deletions and recombinations in the core region of hepatitis B virus genotype E strains from asymptomatic blood donors in Guinea, West Africa. J Gen Virol 90:2442–2451. doi: 10.1099/vir.0.012013-0. [DOI] [PubMed] [Google Scholar]
- 6.Boyce CL, Ganova-Raeva L, Archampong TNA, Lartey M, Sagoe KW, Obo-Akwa A, Kenu E, Kwara A, Blackard JT. 2017. Identification and comparative analysis of hepatitis B virus genotype D/E recombinants in Africa. Virus Genes 53:538–547. doi: 10.1007/s11262-017-1469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyce CL, Willis S, Archampong TNA, Lartey M, Sagoe KW, Obo-Akwa A, Kenu E, Kwara A, Blackard JT. 2019. Identification of hepatitis B virus genotype A/E recombinants in Ghana. Virus Genes 55:707–712. doi: 10.1007/s11262-019-01690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viegas EO, Tembe N, Macovela E, Gonçalves E, Augusto O, Ismael N, Sitoe N, De Schacht C, Bhatt N, Meggi B, Araujo C, Sandström E, Biberfeld G, Nilsson C, Andersson S, Jani I, Osman N. 2015. Incidence of HIV and the prevalence of HIV, hepatitis B and syphilis among youths in Maputo, Mozambique: a cohort study. PLoS One 10:e0121452. doi: 10.1371/journal.pone.0121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokx J, Gillet P, De Weggheleire A, Casas EC, Maendaenda R, Beulane AJ, Jani IV, Kidane S, Mosse CD, Jacobs J, Bottieau E. 2011. Seroprevalence of transfusion-transmissible infections and evaluation of the pre-donation screening performance at the Provincial Hospital of Tete, Mozambique. BMC Infect Dis 11:141. doi: 10.1186/1471-2334-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew A, Ismael N, Meeds H, Vubil A, Zicai AF, Mabunda N, Blackard JT. 2023. Hepatitis B virus genotypes and drug resistance mutations circulating in blood donors in Beira, Mozambique. PLoS One 18:e0281855. doi: 10.1371/journal.pone.0281855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okonechnikov K, Golosova O, Fursov M, UGENE Team . 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 12.Bell TG, Yousif M, Kramvis A. 2016. Bioinformatic curation and alignment of genotyped hepatitis B virus (HBV) sequence data from the GenBank public database. Springerplus 5:1896. doi: 10.1186/s40064-016-3312-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 14.Schultz A-K, Bulla I, Abdou-Chekaraou M, Gordien E, Morgenstern B, Zoaulim F, Dény P, Stanke M. 2012. jpHMM: recombination analysis in viruses with circular genomes such as the hepatitis B virus. Nucleic Acids Res 40:W193–W198. doi: 10.1093/nar/gks414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Stecher G, Kumar S. 2021. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol Biol Evol 38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huy TTT, Ishikawa K, Ampofo W, Izumi T, Nakajima A, Ansah J, Tetteh JO, Nii-Trebi N, Aidoo S, Ofori-Adjei D, Sata T, Ushijima H, Abe K. 2006. Characteristics of hepatitis B virus in Ghana: full length genome sequences indicate the endemicity of genotype E in West Africa. J Med Virol 78:178–184. doi: 10.1002/jmv.20525. [DOI] [PubMed] [Google Scholar]
- 17.Zahn A, Li C, Danso K, Candotti D, Owusu-Ofori S, Temple J, Allain J-P. 2008. Molecular characterization of occult hepatitis B virus in genotype E-infected subjects. J Gen Virol 89:409–418. doi: 10.1099/vir.0.83347-0. [DOI] [PubMed] [Google Scholar]
- 18.Lingani M, Akita T, Ouoba S, Nagashima S, Boua PR, Takahashi K, Kam B, Sugiyama A, Nikiema T, Yamamoto C, Somé A, Derra K, Ko K, Sorgho H, Tarnagda Z, Tinto H, Tanaka J. 2020. The changing epidemiology of hepatitis B and C infections in Nanoro, rural Burkina Faso: a random sampling survey. BMC Infect Dis 20:46. doi: 10.1186/s12879-019-4731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodgers MA, Vallari AS, Harris B, Yamaguchi J, Holzmayer V, Forberg K, Berg MG, Kenmenge J, Ngansop C, Awazi B, Mbanya D, Kaptue L, Brennan C, Cloherty G, Ndembi N. 2017. Identification of rare HIV-1 group N, HBV AE, and HTLV-3 strains in rural South Cameroon. Virology 504:141–151. doi: 10.1016/j.virol.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Bekondi C, Olinger CM, Boua N, Talarmin A, Muller CP, Le Faou A, Venard V. 2007. Central African Republic is part of the West-African hepatitis B virus genotype E crescent. J Clin Virol 40:31–37. doi: 10.1016/j.jcv.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Olinger CM, Venard V, Njayou M, Oyefolu AOB, Maïga I, Kemp AJ, Omilabu SA, Le Faou A, Muller CP. 2006. Phylogenetic analysis of the precore/core gene of hepatitis B virus genotypes E and A in West Africa: new subtypes, mixed infections and recombinations. J Gen Virol 87:1163–1173. doi: 10.1099/vir.0.81614-0. [DOI] [PubMed] [Google Scholar]
- 22.Abdou Chekaraou M, Brichler S, Mansour W, Le Gal F, Garba A, Dény P, Gordien E. 2010. A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J Gen Virol 91:1609–1620. doi: 10.1099/vir.0.018127-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequence data are available under BioProject PRJNA885128, with SRA accession numbers SRR21743134 through SRR21743138. The consensus HBV genomes are available in GenBank under accession numbers OP514931 through OP514935.