ABSTRACT
The Bacillus cereus phage BSG01 has a siphovirus morphology that can belong to the order Caudovirales. It consists of 81,366 bp, with a GC content of 34.6%, and contains 70 predicted open reading frames. BSG01 includes lysogeny-related genes (tyrosine recombinase and antirepressor protein), indicating that it is a temperate phage.
ANNOUNCEMENT
Bacillus cereus is a Gram-positive, spore-forming pathogen that can cause diarrhea and emetic syndrome (1, 2). Antibiotics have been used to control the pathogen but are generally not recommended because they induce antibiotic resistance problems (3). Therefore, the use of bacteriophages (phages) is emerging as an alternative approach for the control of this pathogen (4). In this study, we report the complete genome sequence of B. cereus phage BSG01, which was isolated from a topsoil sample from Naksan Park (Seoul, South Korea). The phage was isolated by spotting an enriched soil sample on the lawns of a Bacillus strain (5). Phage purification and propagation were performed using B. cereus ATCC 27348 as the host bacterium. Briefly, a single plaque was picked and suspended in phosphate-buffered saline (PBS) for phage purification using the phage titration method (6), and this step was repeated three times. Then, the bacterial culture (early exponential phase) was treated with the phage at a multiplicity of infection (MOI) of 1 and incubated at 37°C for 4 h for phage amplification. Phage morphology was confirmed by transmission electron microscopy (TEM) analysis after negative staining with 2% (vol/vol) uranyl acetate (pH 4.5). Furthermore, DNA extraction and purification of phage lysate were performed by phenol-chloroform extraction and ethanol precipitation methods, as described previously (7). The DNA library was constructed using the TruSeq Nano DNA library preparation kit and sequenced with an Illumina MiSeq sequencer (2 × 300-bp paired-end reads). A total of 2,502,910 reads (587,879,675 bp) were trimmed using Trimmomatic v.0.39 (8), and the resulting contigs were de novo assembled using SPAdes v.3.13.0 (9) (mean coverage, 855.66×). The open reading frames (ORFs) of the phage genome were predicted using RAST (10), GeneMarkS (11), and FGENESV (5). ORF annotation, phage genome completeness, and phage genome similarity were confirmed using NCBI BLAST (12). The physical ends of the genome were recognized by comparing the sequence identity of the length of the phage.
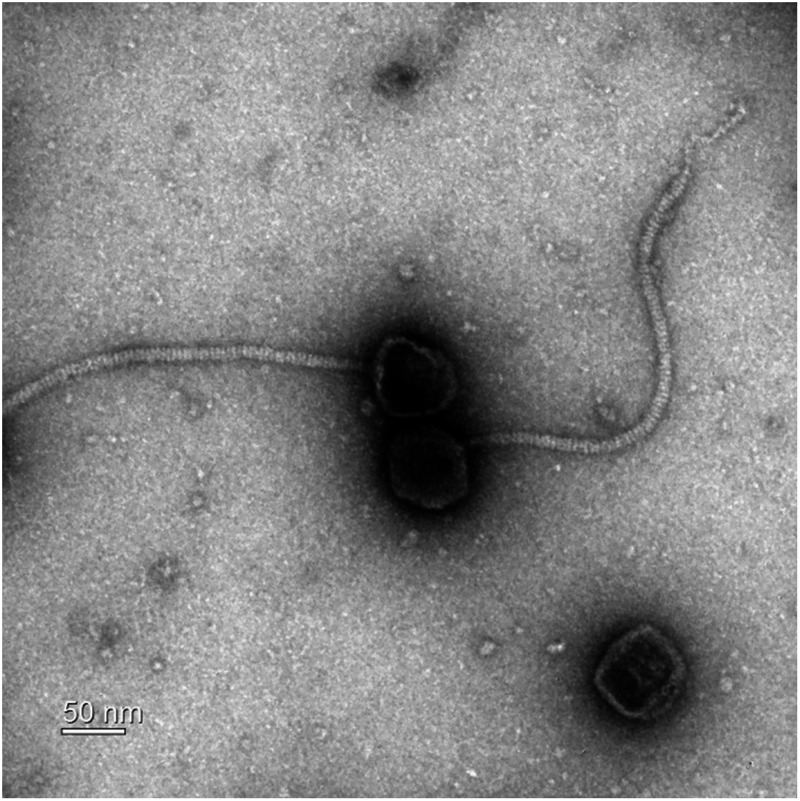
BSG01 has an icosahedral head and a noncontractile flexible tail (Fig. 1). The head diameter was 70.5 ± 0.7 nm (n = 5). The tail length and width were 371.0 ± 8.8 nm (n = 5) and 10.6 ± 0.2 nm (n = 5), respectively. BSG01 has a siphovirus morphology that can belong to the order Caudovirales (13).
FIG 1.
TEM image of phage BSG01 negatively stained with 2% (vol/vol) uranyl acetate. The sample was examined using a Libra 120 transmission electron microscope (Carl Zeiss, Oberkochen, Germany) with an accelerating voltage of 120 kV. Scale bar, 50 nm.
The complete genome of phage BSG01 consists of 81,366 bp, with a GC content of 34.6%, and contains 70 predicted ORFs. Predicted ORFs were annotated in five groups, including structure and packaging (including terminase-like protein and tail fiber protein), transcription regulation (transcriptional regulator and RNA polymerase sigma factor), DNA replication/modification (including DNA helicase and DNA primase), host lysis (endolysin), and additional functions (including putative peptidase inhibitor and putative GTP-binding protein). Furthermore, the genome was predicted to contain lysogeny-related genes (tyrosine recombinase and antirepressor protein), suggesting that BSG01 is a temperate phage having both lytic and lysogenic life cycles. BSG01 is closely related to Bacillus phages PBC4 (GenBank accession number NC_070843), BCP6 (GenBank accession number MW392801), and BCPST (GenBank accession number MW392802), with genome sequence identity of 94% and with percent identity of 96 to 98%.
Data availability.
The complete genome sequence of phage BSG01 was deposited in GenBank under the accession number OQ436521. The associated BioProject, BioSample, and SRA accession numbers are PRJNA956347, SAMN34211098, and SRR24186942, respectively.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MIST) (No. 2021R1F1A1058773). Also, this work was carried out with the support of the “R&D Program for Forest Science Technology (Project No. 2021375A00-2223-BD02)” provided by Korea Forest Service (Korea Forestry Promotion Institute).
Contributor Information
Yoonjee Chang, Email: ychang@kookmin.ac.kr.
Kenneth M. Stedman, Portland State University
REFERENCES
- 1.Kong M, Ryu S. 2015. Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl Environ Microbiol 81:2274–2283. doi: 10.1128/AEM.03485-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandara N, Jo J, Ryu S, Kim K-P. 2012. Bacteriophages BCP1-1 and BCP8-2 require divalent cations for efficient control of Bacillus cereus in fermented foods. Food Microbiol 31:9–16. doi: 10.1016/j.fm.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Shin H, Lee J-H, Park J, Heu S, Ryu S. 2014. Characterization and genome analysis of the Bacillus cereus-infecting bacteriophages BPS10C and BPS13. Arch Virol 159:2171–2175. doi: 10.1007/s00705-014-2030-6. [DOI] [PubMed] [Google Scholar]
- 4.Alomari M, Dec M, Urban-Chmiel R. 2021. Bacteriophages as an alternative method for control of zoonotic and foodborne pathogens. Viruses 13:2348. doi: 10.3390/v13122348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S, Chang Y. 2022. Anti-Salmonella polyvinyl alcohol coating containing a virulent phage PBSE191 and its application on chicken eggshell. Food Res Int 162:111971. doi: 10.1016/j.foodres.2022.111971. [DOI] [PubMed] [Google Scholar]
- 6.Duc HM, Son HM, Yi HPS, Sato J, Ngan PH, Masuda Y, Honjoh K-i, Miyamoto T. 2020. Isolation, characterization and application of a polyvalent phage capable of controlling Salmonella and Escherichia coli O157: H7 in different food matrices. Food Res Int 131:108977. doi: 10.1016/j.foodres.2020.108977. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q, Xing S, Sun Q, Pei G, Cheng S, Liu Y, An X, Zhang X, Qu Y, Tong Y. 2017. Characterization and complete genome sequence analysis of a novel virulent Siphoviridae phage against Staphylococcus aureus isolated from bovine mastitis in Xinjiang, China. Virus Genes 53:464–476. doi: 10.1007/s11262-017-1445-z. [DOI] [PubMed] [Google Scholar]
- 8.Bolger A, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes: implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Zheng K, Liu B, Liang Y, You S, Zhang W, Zhang X, Jie Y, Shao H, Jiang Y, Guo C, He H, Wang H, Sung YY, Mok WJ, Wong LL, McMinn A, Wang M. 2021. Characterization and genomic analysis of Marinobacter phage vb_mals-ps3, representing a new lambda-like temperate siphoviral genus infecting algae-associated bacteria. Front Microbiol 12:726074. doi: 10.3389/fmicb.2021.726074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of phage BSG01 was deposited in GenBank under the accession number OQ436521. The associated BioProject, BioSample, and SRA accession numbers are PRJNA956347, SAMN34211098, and SRR24186942, respectively.