ABSTRACT
The lytic bacteriophage EO1 has been newly isolated. This phage infects Escherichia coli O157:H7 and has a broad antibacterial spectrum, including against Shigella. The complete genome sequence of phage EO1 was determined; its full length is 166,941 bp, and it has a G+C content of 35.46%.
ANNOUNCEMENT
Escherichia coli O157:H7 is a Gram-negative pathogen that can be a major cause of various clinical diseases, such as hemolytic uremic syndrome, hemorrhagic colitis, and thrombocytopenia (1, 2). Therefore, bacteriophages have been reported as promising biological control agents to target pathogenic bacteria without destroying human, plant, and animal cells (3).
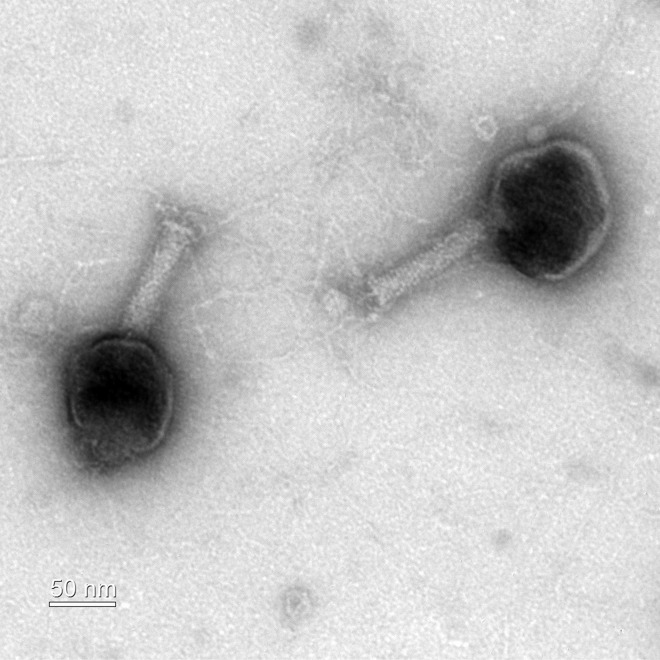
Phage EO1, which forms clear plaques, was isolated in April 2021 from a sewage sample (Tancheon Sewage Treatment Plant, Seoul, South Korea), using E. coli O157:H7 ATCC 43890 as the host bacterium. The sample was grown in Luria-Bertani broth (LB) at 37°C, centrifuged at 15,000 × g for 5 min, and filtered through a 0.45-μm-diameter pore syringe filter. Phage single plaques were isolated three times using the spotting assay (4) and double agar overlay method (5). Phage EO1 showed a broad host range, infecting 15 out of 21 different strains tested, including E. coli and Shigella spp. (Table 1). Transmission electron microscopy (TEM) images showed the EO1 phage with an icosahedral head of 93 ± 6 nm and a tail of 104 ± 2 nm (n = 5) (Fig. 1).
TABLE 1.
Host range determination of phage EO1a
| Bacterial strain | Sensitivity to lysis by EO1b | Source or reference |
|---|---|---|
| Gram-negative bacteria | ||
| E. coli O157:H7 ATCC 35150 | + | KCTC |
| E. coli O157:H7 ATCC 43888 | + | IVI |
| E. coli O157:H7 ATCC 43890 | ++ | KCTC |
| E. coli O157:H7 ATCC 43894 | ++ | NCCP |
| E. coli O157:H7 ATCC 43895 | + | KCTC |
| E. coli ATCC 15597 | + | IVI |
| E. coli ATCC 23724 | ++ | ATCC |
| E. coli K-12 MG1655 | ++ | ATCC |
| E. coli DH5α | ++ | ATCC |
| Shigella flexneri KCTC 2993 | + | ATCC |
| Shigella. flexneri 2a 2457T | + | ATCC |
| Shigella flexneri KCTC 2517 | + | ATCC |
| Shigella flexneri NCCP 10852 | + | ATCC |
| Shigella sonnei KCTC 22530 | + | 6 |
| Shigella boydii IB 2474 | + | ATCC |
| Vibrio parahaemolyticus KCTC 2471 | − | KCTC |
| Salmonella enterica serovar Typhimurium ATCC 14028 | − | ATCC |
| Cronobacter sakazakii ATCC 29544 | − | ATCC |
| Gram-positive bacteria | ||
| Listeria monocytogenes ATCC 15313 | − | ATCC |
| Bacillus cereus ATCC 27348 | − | ATCC |
| Staphylococcus aureus ATCC 29213 | − | ATCC |
The spot test (4) was used to determine the host range against 21 strains. Phage lysate (1 × 109 PFU) was spotted onto the bacterial lawns and incubated overnight at 37°C. The efficiency of plating (EOP) was measured as the ratio between the number of plaques on the test bacteria and the number of plaques of E. coli O157:H7 ATCC 43890.
++, EOP of 0.1 to 1; +, EOP of less than 0.1; −, no susceptibility to phage EO1.
FIG 1.
Transmission electron microscopy (TEM) image of E. coli O157:H7 phage EO1. The phage suspension was applied onto a copper grid (200 mesh) and negatively stained with 2% (vol/vol) uranyl acetate (pH 4.5). Phage morphology was observed using an energy-filtering Libra 120 transmission electron microscope (Carl Zeiss, Oberkochen, Germany).
The SDS-proteinase K method and the phenol-chloroform method were used for phage suspension lysis and DNA purification from lysate after spinning down the cell, respectively (7). A TruSeq Nano DNA library prep kit was used for preparation of the DNA library, and sequencing was carried out on an Illumina MiSeq instrument in 2 × 300-bp paired-end format (8). All tools were run with default parameters unless otherwise specified. Quality trimming was performed using Trimmomatic, and the resulting contigs were assembled by performing de novo assembly (SPAdes v.3.13.0). A total of 2,011,220 reads (578,421,935 bp) were trimmed and assembled, resulting in a complete genome sequence with a length of 166,941 bp, a G+C content of 35.46%, and an average coverage of 513.93×. Using RAST (9), GeneMarkS (10), and FGENESV, 139 open reading frames (ORFs) were annotated. The physical ends of the genome were identified by comparing the coverage values of the length of the phage EO1 to closely related phages in the NCBI database, Escherichia phage PEC04 (GenBank accession number KR233165.1; 97.03% identity with 95% query coverage) and Escherichia phage HY01 (NC-027349.1; 97.03% identity with 95% query coverage), using BLAST (11) and ViroBLAST (12).
Based on blastp (13) and InterProScan (14), protein-coding genes predicted to be involved in various functions were classified: structure and packaging (coil, tail fiber protein, tail protein, outer membrane lipoprotein, head chaperone, baseplate hub, major head protein, head scaffolding protein, terminase family protein, neck protein, and large terminase subunit), host lysis (peptidase and holin), DNA metabolism (DNA polymerase, DNA primase, recombinase protein, DNA topoisomerase, DNA ligase, endonuclease, thymidylate synthase, dCTP pyrophosphatase, RNA polymerase, exonuclease, deoxynucleoside monophosphate kinase, and RNA ligase), and additional functions (thioredoxin and naphthalene 1,2-dioxygenase).
Since EO1 forms clear plaques and there are no lysogen-forming genes (e.g., Cl protein or integrase) in the genome, it can be assumed that it is a strictly lytic phage that has great potential for use as a biocontrol agent.
Data availability.
The complete genome sequence of Escherichia phage EO1 was deposited at GenBank under the accession number OQ223305.1. The associated BioProject, BioSample, and SRA accession numbers are PRJNA942357, SAMN33687158, and SRR24296758, respectively.
ACKNOWLEDGMENTS
This work was carried out with the support of the Cooperative Research Program for Agriculture Science and Technology Development (PJ01608201; Rural Development Administration, Republic of Korea). This work was also supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (number 2022M3H4A1A03085361).
Contributor Information
Yoonjee Chang, Email: ychang@kookmin.a.kr.
Simon Roux, DOE Joint Genome Institute.
REFERENCES
- 1.Necel A, Bloch S, Nejman-Faleńczyk B, Grabski M, Topka G, Dydecka A, Kosznik-Kwaśnicka K, Grabowski Ł, Jurczak-Kurek A, Wołkowicz T, Węgrzyn G, Węgrzyn A. 2020. Characterization of a bacteriophage, vB_Eco4M-7, that effectively infects many Escherichia coli O157 strains. Sci Rep 10:3743. doi: 10.1038/s41598-020-60568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan X, Zhang S, Wang J, Li C, Li N, Yu S, Kong L, Zeng H, Yang G, Huang Y, Li H, Zhang J, Wu Q, Ding Y. 2021. Isolation and characterization of a novel Escherichia coli Kayfunavirus phage DY1. Virus Res 293:198274. doi: 10.1016/j.virusres.2020.198274. [DOI] [PubMed] [Google Scholar]
- 3.Yazdi M, Bouzari M, Ghaemi EA, Shahin K. 2020. Isolation, characterization and genomic analysis of a novel bacteriophage VB_EcoS-Golestan infecting multidrug-resistant Escherichia coli isolated from urinary tract infection. Sci Rep 10:7690. doi: 10.1038/s41598-020-63048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S, Chang Y. 2022. Anti-Salmonella polyvinyl alcohol coating containing a virulent phage PBSA191 and its application on chicken eggshell. Food Res Int 162:111971. doi: 10.1016/j.foodres.2022.111971. [DOI] [PubMed] [Google Scholar]
- 5.Manohar P, Tamhankar AJ, Lundborg CS, Nachimuthu R. 2019. Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter species. Front Microbiol 10:574. doi: 10.3389/fmicb.2019.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim M, Ryu S. 2011. Characterization of a T5-like coliphage, SP35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl Environ Microbiol 77:2042–2050. doi: 10.1128/AEM.02504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikalova L, Bosak J, Hříbková H, Dědičová D, Benada O, Šmarda J, Šmajs D. 2017. Novel temperate phages of Salmonella enterica subsp. salamae and subsp. diarizonae and their activity against pathogenic S. enterica subsp. enterica isolates. PLoS One 12:e0170734. doi: 10.1371/journal.pone.0170734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mount DW. 2007. Using the basic local alignment search tool (BLAST). CSH Protoc 2007:pdb.top17. doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- 12.Deng W, Nickle DC, Learn GH, Maust B, Mullins JI. 2007. ViroBLAST: a stand-alone BLAST Web server for flexible queries of multiple databases and user’s datasets. Bioinformatics 23:2334–2336. doi: 10.1093/bioinformatics/btm331. [DOI] [PubMed] [Google Scholar]
- 13.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of Escherichia phage EO1 was deposited at GenBank under the accession number OQ223305.1. The associated BioProject, BioSample, and SRA accession numbers are PRJNA942357, SAMN33687158, and SRR24296758, respectively.