Abstract
Background and Objectives
Charcot-Marie-Tooth disease (CMT) is a syndrome of a hereditary neurodegenerative condition affecting the peripheral nervous system and is a single gene disorder. Deep phenotyping coupled with advanced genetic techniques is critical in discovering new genetic defects of rare genetic disorders such as CMT.
Methods
We applied multidisciplinary investigations to examine the neurophysiology and nerve pathology in a family that fulfilled the diagnosis of CMT2. When phenotype-guided first-tier genetic tests and whole-exome sequencing did not yield a molecular diagnosis, we conducted full genome analysis by examining phased whole-genome sequencing and whole-genome optical mapping data to search for the causal variation. We then performed a systematic review to compare the reported patients with interstitial microdeletion in the short arm of chromosome 4.
Results
In this family with CMT2, we reported the discovery of a heterozygous 85-kb microdeletion in the short arm of chromosome 4 (4p16.3)[NC_000004.12:g.1733926_1819031del] spanning 3 genes [TACC3 (intron 6-exon 16), FGFR3 (total deletion), and LETM1 (intron 10-exon14)] that cosegregated with disease phenotypes in family members. The clinical features of peripheral nerve degeneration in our family are distinct from the well-known 4p microdeletion syndrome of Wolf-Hirschhorn syndrome, in which brain involvement is the major phenotype.
Discussion
In summary, we used the full genome analysis approach to discover a new microdeletion in a family with CMT2. The deleted segment contains 3 genes (TACC3, FGFR3, and LETM1) that likely play a role in the pathogenesis of nerve degeneration.
Charcot-Marie-Tooth disease (CMT) is a syndrome of inherited neuropathies affecting motor and sensory nerves caused by single-gene pathogenic variants.1-8 The defective proteins derived from pathogenic variants impair the structural integrity or functions of peripheral nerves, leading to nerve degeneration.1,2 Traditionally, CMT is classified according to the nerve conduction velocity (NCV) and inheritance pattern. The autosomal dominant CMT is classified further as CMT1 (NCV <35 m/s), CMT2 (NCV >45 m/s), or dominant intermediate CMT (NCV 35–45 m/s). The inheritance is autosomal recessive in CMT4 and X-linked in CMTX.1,2
Most CMT conditions are found to be single-gene disorders.1,4,9 The clinical applications of next-generation sequencing such as panel-based target sequencing and whole-exome sequencing (WES) have facilitated the identification of disease-causing variants in neuromuscular disorders. However, in contrast to the high diagnostic yield of WES for myopathy (35%–76%) and congenital myasthenic syndrome (50%–70%), the yield for CMT is only 20%–45%.1,10 A possible reason for the lower diagnostic yield is the inability for WES to detect large structural variation, mitochondrial DNA defect, and gene contraction or expansion.10 Full genome analysis (FGA) is an approach that scans the whole genome comprehensively with 2 complementary techniques: whole-genome optical mapping and long-read whole-genome sequencing (WGS) that circumvents the shortcomings of the short-read sequence analysis.11 FGA detects all classes of genetic variations, including single nucleotide variants, small insertion/deletions, and large structural variants (deletions, duplication, inversion, and translocation), and provides a way to identify new genetic defects for rare genetic disorders such as CMT.1,4,9,10
In this study, we report the discovery of an interstitial microdeletion of 85 kb in the region of 4p16.3 by FGA in a CMT family with an autosomal dominant inheritance pattern. This may represent as new gene locus and new disease mechanism for CMT.
Methods
Standard Protocol Approvals, Registrations, and Patient Consents
The Ethics Committee of the National Taiwan University Hospital, Taipei, Taiwan (2009093RINB, 201305034RIND, and 201612036RIND), and the Ethics Committee of the Academia Sinica (AS-IRB01-19041), Taipei, Taiwan, approved this study. All participants signed the informed consent following the standard protocol of the Ethics Committee of the National Taiwan University Hospital and the Ethics Committee of the Academia Sinica before joining the study. The index patient signed the Neurology Journals' Consent-to-Disclose form with the Traditional Chinese translation.
Patient Recruitment and Clinical Evaluations
All participants received detailed evaluation including history taking, neurologic examinations, and additional tests as indicated according to our established protocols: physiology (nerve conduction studies, autonomic function test, and needle EMG), psychophysics (quantitative sensory testing), and pathology examinations (nerve biopsy and skin biopsy).12-14
Genetic Analyses
To discover the genotype of this family, we took a 3-tier genetic analysis approach.1,4,10 In the first-tier approach, we screened the patient and family members for the most common genetic variants for hereditary neuropathies: peripheral myelin protein 22 (PMP22) duplication and transthyretin (TTR) pathogenic variants. We performed WES as a second-tier approach to search for disease-causing variants when the first-tier testing results were negative. When WES returned no candidate variants for the condition, we conducted full genome analysis on the patient and key members of the family with 2 complementary techniques: (1) linked-read whole-genome sequencing and (2) whole-genomic optic mapping. Breakpoint junction assay was applied to confirm the genotype discovered in FGA in the family members.
Genetic Tests of PMP22 Duplication
The diagnostic method was based on PCR. The target sequence is a 3.6-kb region that includes the 1.7-kb hotspot repetition in CMT type 1A. We used the published sequence (GenBank accession numbers: U41165-distal and U41166-proximal) to design the primers. Primers were forward (5′-AGGTTGTTTACTCCTTCTTC-3′) and reverse (5′-AGATGGAATAGTAGAGCTCAC-3′). The Expand Long Template PCR system (Roche Applied Science, Mannheim, Germany) was used to amplify the targeted sequence in the volume of 50 μL including 0.4 μM of the above primers, 1.5 mM MgCl2, 100 ng genomic DNA, 80 μM dNTPs, and 2.6 U Taq DNA polymerase. The steps of amplification using a multiblock system (MBS) thermocycler (Thermo Hybaid, Ashford, UK) comprised (1) denaturation: 94°C for 3 minutes; (2) 10 cycles: 30 seconds at 94°C, 30 seconds at 54°C, and 3 minutes at 68°C; (3) 25 cycles: 30 seconds at 94°C, 30 seconds at 54°C, and 3 minutes at 72°C; and (4) extension: 7 minutes at 68°C, including a 20-second autoextend function. Then, we used the EcoRI (New England Biolabs, Beverly, MA) at 37°C for 1 hour to digest the amplicons, which electrophoresed at 80 V on 0.8% agarose gels.
Genetic Tests of TTR
We extracted the genomic DNA from the peripheral venous blood and amplified the 4 exons and the neighboring intron regions of the human TTR gene by PCR. We used the Gel/PCR DNA Fragments Extraction Kit (Geneaid, Taipei, Taiwan) to purify the amplicons, which were then subjected to direct sequencing. The targeted exons were sequenced with the ABI3730 automatic DNA sequencer (Applied Biosystems, Foster City, CA).
Whole-Exome Sequencing
WES was performed using an exome capture kit probe (Agilent V6) and a NovaSeq 6000 sequencer (Illumina). The genes of protein-coding exons and flanking intronic sequences (50 bp) were sequenced with a 300-bp paired-end run and an average of 108-fold coverage.
We performed the sequence alignment to the human reference genome (GRCh38) with a Burrows-Wheeler Aligner (BWA), and the variant calling was performed with the Genome Analysis Tool Kit (GATK V3.5, Broad Institute).15 First, we annotated variants by using ANNOVAR16,17 and by using the information of the inheritance pattern from OMIM,18 variant pathogenicity from ClinVar,19 and the allele frequency from the Taiwan Biobank.20 The filtering criteria for single nucleotide variants (SNVs) and small indels comprised variant severity (splicing, nonsense, insertion, deletion, splicing, or nonsynonymous variant) and a maximal minor allele frequency of <0.01. With the guidance of American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology guidelines,21-25 the variants were classified. Furthermore, for copy number analysis (CNV), with SAM/BAM file as input, the GATK germline CNV (gCNV) caller can generate a VCF file containing the CNVs found during the variant calling process.26
Full Genome Analysis
We collected the fresh peripheral blood samples from the patient (S305) and his 2 biological parents (S206 and S207). After RBC removal by RBC lysis buffer (Qiagen), high-molecular-weight genomic DNAs (HMW gDNAs) were extracted with the Bionano Prep™ kit (Bionano Genomics). HMW gDNA samples were further transferred to (1) 10x Genomics (10xG) Linked‐Read WGS on the NovaSeq 6000 Sequencing System (Illumina) with 40–60X read depth and (2) Bionano DLE optical genome mapping (OGM) on the Bionano Saphyr system (Bionano Genomics) with 60X coverage. We processed 10xG WGS FASTQ files with human reference genome (GRCh38, hg38) from the Genome Reference Consortium. The Long Ranger BASIC and ALIGN Pipelines were used for 10x Genomics Linked-Read Alignment, Structural Variant Calling, Phasing, and Variant Calling. We used the optical images of labeled molecules to generate de novo assembled genome maps with the default setting in the Bionano Solve pipeline. With the Variant Annotation Pipeline of Bionano Solve, all structural variations (SVs) including deletions, insertions, inversions, and translocations were annotated to hg38. The de novo phased genome assemblies from 10xG WGS were compared to identify SNVs and (SVs from 10xG WGS and OGM through the well-established FGA pipeline.11,27 Briefly, the suspected SNVs and SVs were derived from the FGA pipeline, and common SNVs/SVs and synonymous SNVs were based on information from the 1000 Genomes Project, Genome Aggregation Database (gnomAD28), Exome Aggregation Consortium, benign variants in ClinVar, and ACMG recommendations InterVar.29,30 Then, we evaluate each gene affected by SNVs, SVs, or combination of SNVs and SVs in detail. We did Sanger sequencing of PCR products from the individuals to confirm all candidate variants.
Breakpoint Junction Assay
For confirming the genotype discovered in full genome analysis in the family members, we conducted breakpoint junction assay. Breakpoint (BP) assay was performed by PCR and Sanger sequencing on the forward and reverse sides of 2 breakpoints. The primers were as follows: d85k-F1: TGAGACTAACCTGGGCAACA, d85k-R1: CAGGCTGAGGCATCTCTAGTTT, d85k-F2: AAGCCACTTTCACGTAATC, and d85k-R3: TGCAGGAGCAGAAGAAGTCA. The wild-type allele was amplified with BP left-side primer set (d85k-F1 and d85k-R1) and with BP right-side primer set (d85k-F2 and d85k-R3) from gDNAs (20 ng). Only deleted allele could be amplified by the primer set with d85k-F1 and d85k-R3. After PCR amplification, agarose electrophoresis and Sanger sequencing were performed.
Systematic Review of Patients With Interstitial Deletion in the Short Arm of Chromosome 4
To compare the phenotype and the involved genes among the reported patients and our family, we searched the “Wolf-Hirschhorn syndrome” or “4p microdeletion syndrome” in PubMed, and the articles in and after 2000 were included. The inclusion criteria were that the gene defect was interstitial deletion smaller than 1 Mb. The articles using the genetic test with inadequate resolution to identify the involved genes would be excluded (e.g., only karyotype study). In summary, the phenotype studies with fluorescence in situ hybridization, array comparative genomic hybridization, and WES were included. The search was conducted on April 25, 2022. After the removal of the duplicate items, the titles and abstracts were reviewed by H.W. Hsueh, and obviously, irrelevant results were removed. Finally, a full-text review in the final reference list was conducted to retrieve the associated information.
Availability of Data and Material
The data sets generated during analysis and/or during the current study are available from the corresponding author on reasonable request.
Results
Clinical Profiles of the Family
The proband (S204) was a 53-year-old man with a length-dependent neuropathy, i.e., slowly progressive weakness, numbness, and neuralgia arising from the foot since the age of 20 years. Despite these symptoms, he could walk and execute daily activities independently until age 50 years. The numbness and hypoesthesia ascended to the thigh and to the elbow with accentuation at the distal parts of toes and fingers. Morning erection disappeared at the same time. The gait became unsteady and wide based. There was no orthostatic dizziness, urinary problem, or gastrointestinal symptoms.
The neurologic examination revealed muscle wasting over first dorsal interosseous muscles and foot muscles with characteristic pes cavus and high arched foot (Figure 1A).The muscle strength was mildly decreased in hands and feet with generalized areflexia. The sensory examination revealed a symmetric decrease in pinprick and vibration senses in lower limbs and below the elbow. The cerebellar system and the extrapyramidal system were normal. The nerve conduction studies showed markedly reduced amplitudes of compound muscle action potentials and sensory nerve action potentials, indicating a pattern of axonal degeneration. The clinical and neurophysiologic manifestations were consistent with the diagnosis of CMT2.
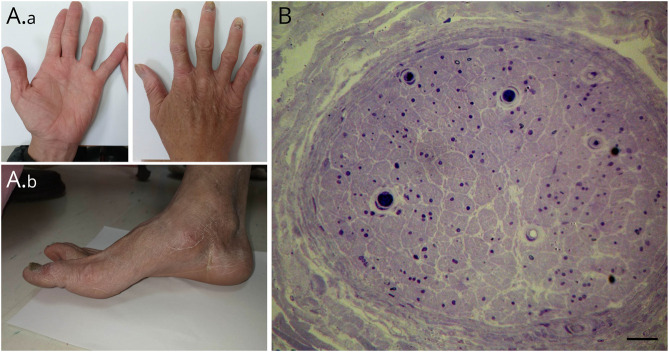
Figure 1. Clinical Signs and Histopathology of the Sural Nerve in the Index Patient (S204).
(A) The atrophic intrinsic muscle of the bilateral hands (A.a) and pes cavus (A.b) was noted in the index patient. (B) The histopathology of the sural nerve from the index patient revealed a marked depletion of myelinated nerve fibers in the sural nerve. Scale bar = 20 μm.
A workup for the causes of peripheral neuropathy on common etiologies revealed no specific abnormalities, except a well-controlled diabetes mellitus with minimal medication in the past 3 years. Pathologic examinations of the nerve biopsy showed a marked reduction of myelinated nerve fibers in the left sural nerve (Figure 1B). The skin biopsy on the distal leg revealed complete denervation in the epidermis. The innervation of sweat glands was depleted as well, providing the evidence of autonomic nerve degeneration (eFigure 1, links.lww.com/NXG/A611).
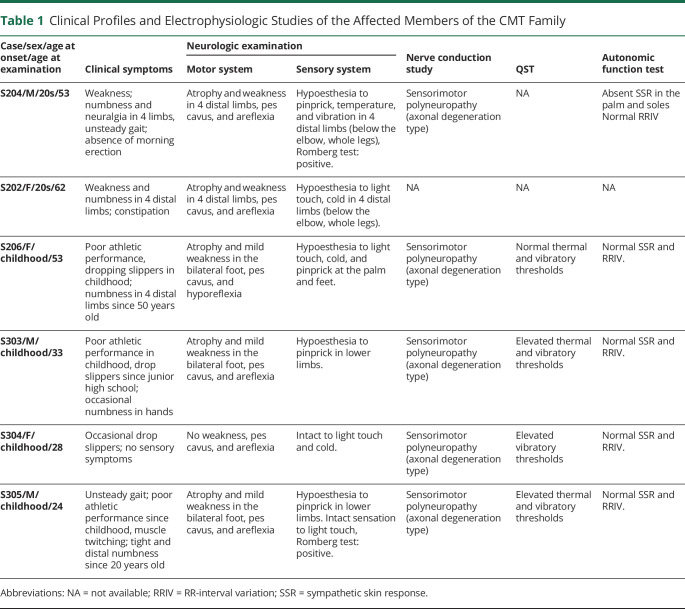
He was born of nonconsanguineous parents, with similarly affected family members (siblings and nephew) showing a pattern of autosomal dominant inheritance (Figure 2A). The clinical symptoms of S102, S208, and S307 were reported by the family members who were evaluated in the clinic, but we did not examine them in person because S102 had died, whereas S208 and S307 declined the invitation to join the study. The phenotype of S208 was uncertain because he refused any evaluations, but S307 had been evaluated at an outside hospital and was given a diagnosis of polyneuropathy. Common etiologies and risk factors in the family members were excluded by laboratory tests (metabolic, endocrine, and autoimmune factors). The clinical profiles and electrodiagnostic studies of the affected and unaffected members are summarized in Tables 1, 2 and eTable1, links.lww.com/NXG/A611.
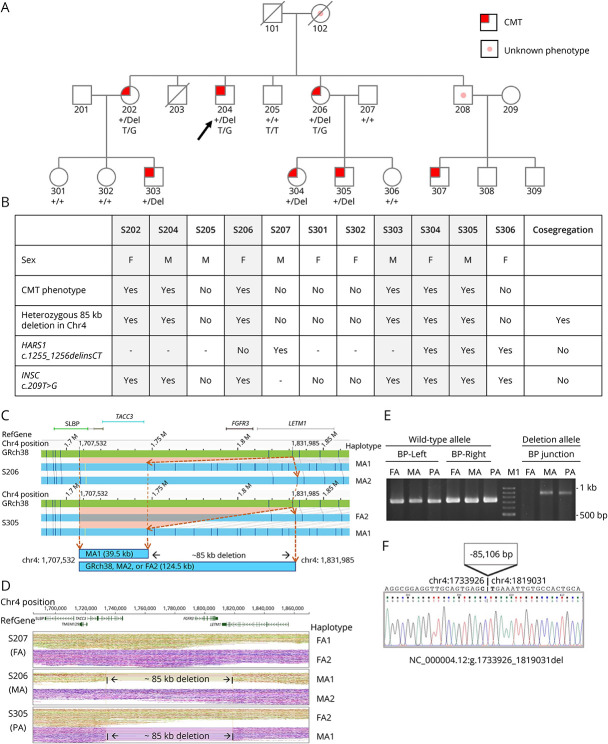
Figure 2. Pedigree of the Family With Charcot-Marie-Tooth Disease and the Heterozygous 85-kb Deletion Disrupting TACC3-FGFR3-LETM1 Genes Identified by Full Genome Analysis and Breakpoint Assay.
The pedigree (A) of the family and the segregation analysis (B) of each family member. (C) De novo assembly in optical genome mapping (OGM) presented a missing 85-kb fragment in affected patients. The DLE labeling sites (dark blue lines) in GRch38 reference (green contig) and 2 haplotypes (blue contigs) in S206 and S305 showed that they shared a deletion MA1 haplotype. The dashed orange lines and arrowheads denoted the position in chr4:1,707,532 and chr4:1,831,985 in their wild-type and deletion haplotypes. The orange rectangle area represents the deletion referred to the GRch38 map. (D) Linked-read WGS with phased haplotypes in the S305 trio was illustrated. RefGene showed the location of reference genes. After phased analysis, the trio haplotypes of S207 (FA, S305's father), S206 (MA, S305's mother), and S305 (PA) were shown based on variant sequencing. S206 and S305 shared the same haplotype MA1 with an 85-kb deletion, which spanned 3 genes TACC3-FGFR3-LETM1. (E) Breakpoint (BP) PCR and Sanger sequencing were performed in the S305 trio. Primers sets were designed nearby the breakpoints of the upstream (BP-left site with d85k-F1 and d85k-R1 primers) and the downstream (BP-right site with d85k-F2 and d85k-R3 primers) for wild-type allele and for deletion allele (BP junction set with d85k-F1 and d85k-R3 primers). Only the person who carried the deletion allele could be amplified by the BP-specific primer set. The results showed that S305 (PA) and S206 (MA) carried the deletion. (F) The panel showed the precise breakpoint sequencing from S305 by Sanger sequencing on the BP junction PCR product. The nucleotides in bold were the breaking sites, which resulted in the loss of an 85,106 bp fragment from chr 4:1,733,926 to chr 4:1,819,031. FA, S207; MA, S206; PA, S305; M1: 100 bp DNA ladder marker (Zymeset).
Table 1.
Clinical Profiles and Electrophysiologic Studies of the Affected Members of the CMT Family
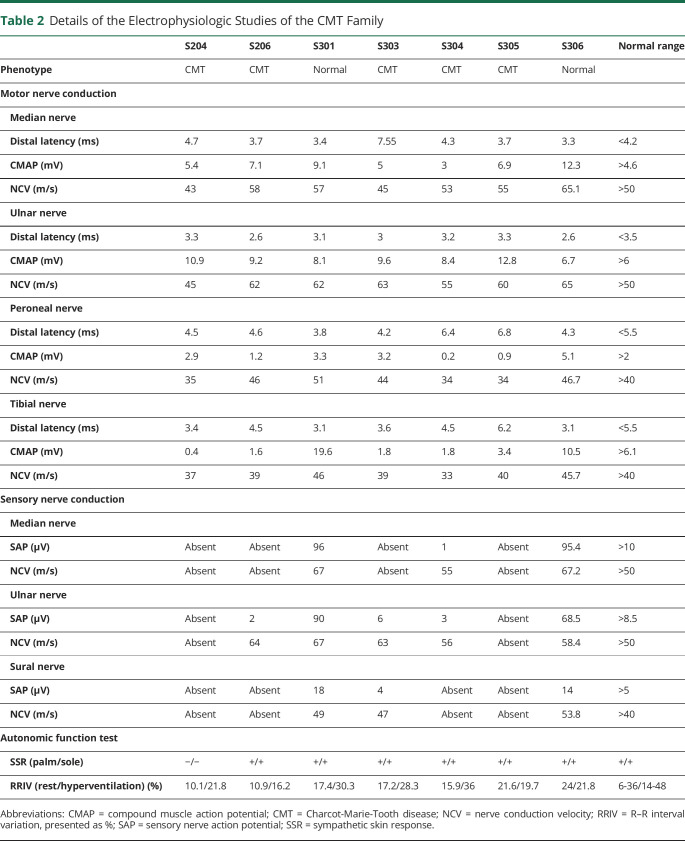
Table 2.
Details of the Electrophysiologic Studies of the CMT Family
First-Tier and Second-Tier Genetic Test
Considering the polyneuropathy affecting motor and sensory nerves with an autosomal dominant inheritance, we examined the most common genetic variants for this phenotype in Taiwan as the first-tier genetic test, i.e., PMP22 duplication for CMT disease and TTR gene variants for hereditary transthyretin amyloidosis.31 However, both tests failed to identify pathogenic variants. We then conducted WES for single nucleotide variants and small insertion/deletion initially on the proband's affected nephew (S305). Two heterozygous variants (c.1255_1256delinsCT in HARS1; c.209T>G in INSC) were identified. However, the segregation analysis with deep phenotyping excluded the possibility of pathogenicity in these 2 variants (Figure 2B). In addition, the blood mitochondrial sequencing also resulted in negative findings.
Identification of a Large Deletion Affecting the TACC3-FGFR3-LETM1 Genes by Full Genome Analysis
All the above tests resulted in negative findings. We thus performed the FGA on the S305 core trio with S206 (affected mother) and S207 (unaffected father). The FGA analysis pipeline identified a heterozygous deletion in chromosome 4p16.3 in S305 and his mother (S206) based on strong evidence in both 10xG linked-read WGS and Bionano optical genome mapping (Figure 2, C and D). This heterozygous microdeletion spanned 3 genes, TACC3 (intron 6-exon 16), FGFR3 (whole gene), and LETM1 (intron 10-exon14). After validation with breakpoint junction PCR and Sanger sequencing, the precise breakpoints were identified [NC_000004.12:g.1733926_1819031del (−85,106bp)] (Figure 2, E and F).
The interstitial deletion is not found in the Bionano or gnomAD (SVs v2.1) databases, and only a small 122bp deletion (g.1770422_1770543) is found in the NCBI Curated Common Structural Variants (nssv17665245) (eTable 2, links.lww.com/NXG/A611). This copy number variant was also identified by WES after reanalyzing the data. All recruited family members were then subjected to breakpoint junction PCR analysis and showed that the deletion segregated with the phenotype, confirming this deletion as a pathogenic allele (Figure 2B).
Comparison of the Clinical Presentations of the Family With 4p Microdeletion Syndrome in the Literature
The deletion in the short arm of chromosome 4 was related to Wolf-Hirschhorn syndrome (WHS), whose phenotype was distinct from our family. We did a systematic review to recruit patients with interstitial microdeletion smaller than 1 Mb in the 4p. In total, 488 articles were collected, and their abstract was reviewed to retrieve the original articles with phenotype study, case series, and case reports.
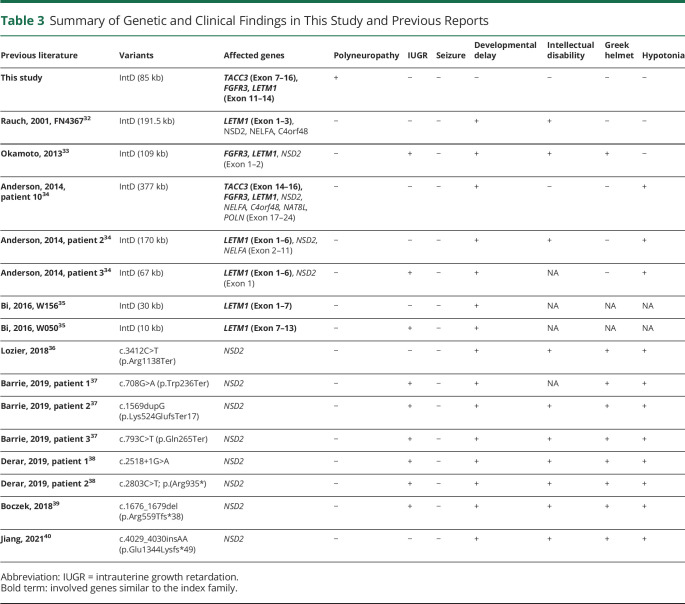
Finally, 5 articles (8 patients) were included. In addition, we included another 5 articles (8 patients) with pathogenic variants in the NSD2 gene, which is next to the LETM1 gene and is critical for Wolf-Hirschhorn syndrome. The summary of the phenotype and gene defect was summarized in Table 3 and Figure 3. In reported patients with interstitial deletion small than 1 Mb, the most frequent symptoms were developmental delay, and the intrauterine growth retardation, intellectual disability, and the craniofacial type “Greek warrior helmet” were also reported. Notably, seizure was not noted in these patients. The patient from the study by Okamoto et al.33 and patient 3 from the study by Anderson et al.34 had the interstitial deletion close to our family. Two patients (W156 and W050) from the study by W. Bi et al.35 had a small interstitial deletion, only involving the LETM1 gene. Although many reported patients had hypotonia, but none of them reported clear neuropathy symptoms or signs like our family. In the patients with pathogenic variants in NSD2, they had intrauterine growth retardation, developmental delay, intellectual disability, typical craniofacial type of WHS, and hypotonia. Notably, none of them had seizure or peripheral neuropathy. As a result, our CMT family had a unique phenotype among the patients with small interstitial deletion in the short arm of chromosome 4.
Table 3.
Summary of Genetic and Clinical Findings in This Study and Previous Reports
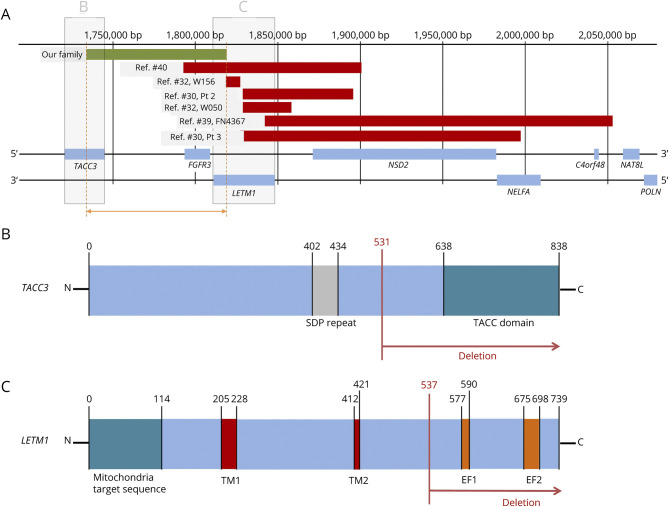
Figure 3. Gene Map of the Affected Genes in This Study and Previous Reports.
(A) Gene map of the affected genes in our patients and previous reports with the size of interstitial deletion smaller than 200 kb (green line: our case, red line: interstitial deletions). The diagram of deleted domains of TACC3 (B) and LETM1 (C) in our family. The FGFR3 was totally deleted in our patients. TM1, TM2 = transmembrane domains 1 and 2; EF1, EF2 = EF hand domains 1 and 2.
Discussion
This study reports the discovery of an 85-kb deletion in the short arm of chromosome 4 consisting of 3 genes [TACC3 (intron 6-exon 16), FGFR3 (total deletion), and LETM1 (intron 10-exon14)], which may serve as a new genetic mechanism for autosomal dominant CMT. Such a phenotype of peripheral nerve degeneration is distinct from the well-known microdeletion syndrome of the Wolf-Hirschhorn syndrome (WHS), which affects brain development.41 Furthermore, we also illustrate a diagnostic approach for hereditary neuropathy by applying long-read sequencing and optical mapping techniques for full genome analysis.
This study demonstrates a practical approach to identify the underlying etiology of hereditary neuropathy by deep phenotyping and integrating multidiscipline examinations to guide the genetic test and to confirm the pathogenicity of candidate variants.42 Current approach in the field is that if the first-tier–targeted genetic tests failed to identify specific variants, then WES is usually performed, with a yield rate of 30%–40% in hereditary neuropathy. There are several possibilities for negative results in the initial WES: rare secondary causes, mitochondrial DNA defect, and nuclear genetic defect.10 In our case, the strong family history, affected family member not living in the same area, and extensive workup on different cases (S204 and S305) excluded the possibility of secondary causes. Mitochondrial DNA sequencing did not find plausible gene defects. Heteroplasmy is present in mitochondrial DNA, but mitochondrial DNA is mainly inherited from mother to children. As a result, mitochondrial DNA defect is less likely in this family given the inheritance of the condition from father (S208) to son (S307). However, the phenotype of S208 was unknown, and the possibility that S307 may have other secondary etiologies or different genetic mechanism for his polyneuropathy cannot be excluded.
Short-read WGS detects variants in noncoding sequences and structural variants more readily than WES, but, with a read length of 300 bp, WGS typically misses the large structural variants (e.g., microsatellite expansion/contraction and large insertion/deletion). By contrast, long-read sequencing generates sequencing reads from 10 kb to 100 kb (and, on rare occasions, up to 2 Mb) that, when combined with OGM, can be assembled into contiguous sequences 50 Mb and can detect large structural variants unambiguously. As a result, the FGA approach that incorporates the linked-read sequencing and OGM has the advantage of detecting intermediate to large structural variants easily, establishing the rearrangement breakpoints precisely, and determining the phasing of variants (cis or trans) in autosomal recessive disease.11 In a recent study, FGA identified structural variants and small variants in the clinical setting of WES-negative cases and achieved a diagnostic rate of 35%.11 We conducted FGA for the proband and key members of the family and identified the interstitial microdeletion with precise breakpoints in the short arm of chromosome 4, showing the utility of the FGA on WES-negative cases. In summary, FGA solves the inherent limitation of short-read sequencing-based analysis. Furthermore, this study demonstrates a practical diagnostic algorithm to identify the genetic defect in a CMT family with negative results on the target-genetic test and WES.
The most well-known deletion in the short arm of chromosome 4 is WHS, which includes heterogenous symptoms: specific craniofacial type “Greek warrior helmet,” growth delay, intellectual disability, seizure, and congenital heart defects.41 The variations in the phenotype are correlated with the size of deletion.43,44 WHS is considered as a spectrum of disease based on the diagnosis by karyotyping.41 Genetic analyses of multiple cases with various techniques (e.g., fluorescent in situ hybridization, array-based comparative genomic hybridization, and next-generation sequencing) showed that the disease-causing genetic defect was restricted to the region of 4p16.3, with several genes (e.g., LETM1, NSD2, and NELFA) critical in the pathogenesis of WHS.34,41 Although the affected family members in our study have a microdeletion in 4p16.3, the clinical phenotype of our family is distinct from WHS. Two possibilities likely account for the discrepancy. First, the size of deletion (85 kb) in our family is much smaller than the average deletion size in WHS (5–18 Mb), and the severity of WHS was related to the length of deletion.43 Furthermore, 2 critical genes (NSD2 and NELFA) of WHS are spared in our family and limit the clinical manifestations to those affecting the peripheral nerves. We surveyed the phenotypes of WHS with (1) interstitial deletion smaller than 1 Mb or (2) pathogenic variants in the NSD2 gene, which is next to the LETM1 gene and is critical for WHS. The patients with interstitial deletion including NSD2 or NELFA gene or variants in NSD2 gene all had features of WHS. Although some of these patients had hypotonia, but none of them reported definite neuropathy symptoms or signs as seen in our family. In summary, a small deletion sparing of the critical regions of WHS explains the late-onset symptoms and a lack of CNS involvement.
The microdeletion found in our patient family affects 3 genes: partial deletion of LETM1 (intron 10 to exon 14) and TACC3 (intron 6 to exon 16) and complete deletion of FGFR3. All 3 genes exert the effect on the integrity of structure and corresponding functions on peripheral nerves. The LETM1 encodes leucine zipper and EF-hand containing transmembrane protein 1 located in the inner membrane of the mitochondria,45 and its deletion leads to WHS.34,41,45 The onset age of the 2 patients (W156 and W050) in the reference study35 was younger than our family, and the central nerve system was involved (Table 3). The deleted segment in these 2 cases shared exon 7 (exon 1–7 deletion in W156 and exon 7–13 deletion in W050), which was responsible for the second transmembrane domain of the LETM1 protein.46 By contrast, the deleted segment (exon 11–14 deletion) of our family only affects the EF hands.46 The involvement of different domains likely explains the distinct phenotype between ours and the previously reported cases.35 TACC3 encodes the transforming acidic coiled-coil (TACC)-containing protein 3 and is important for the microtubule plus end dynamics.47 TACC3 is expressed at the growth cones of embryonic neurons and promotes axon growth. The length of axons was reduced if TACC3 was silenced.48 The TACC domain, deleted in our patients, binds to and interacts with the microtubule.48 An absence of the TACC domain likely impairs the functions of microtubules and causes the defects of axon growth. The FGFR3 gene encodes fibroblast growth factor receptor 3 and plays a role in axonal maturation, i.e., small diameter in axons of FGFR3-deficient mice.49 The recent identification of anti-FGFR3 antibody in patients with polyneuropathy provides evidence that the perturbation of FGFR3 may have detrimental effects on peripheral nerves.50,51 Future studies are required to dissect the effects of individual genes or their combinations in this family.
This study has several limitations. First, this finding is derived from a single family, and some family members refused genetic tests. Identification of a similar deletion in another family and further molecular studies will be needed to confirm the microdeletion as the pathogenic variant in this family. Second, the youngest unaffected member (S305) was examined at the age of 18 years. Although her athletic performance was perfect at the time of physical examination, as compared to the poor athletic performance of typical patients with CMT, the possibility that she may develop CMT symptom later in life cannot be excluded. Serial follow-ups of the clinical symptoms and nerve conduction studies are warranted. Third, the individual effect of the 3 genes on peripheral nerves cannot be resolved at this moment. The mechanisms require further investigations on the effects of the individual genes and their interactions. Finally, the involvement of the peripheral nerve system in WHS is not clearly defined in the literature,44 as the possibility of neuropathy in newborns with WHS cannot be excluded. An evaluation of peripheral nerves in WHS probably will expand the phenotype of WHS.
We performed FGA to identify an interstitial microdeletion (85 kb) in the short arm of chromosome 4 in a family with autosomal dominant CMT neuropathy. The deleted segment contains 3 genes (TACC3, FGFR3, and LETM1) that likely play a role in the pathogenesis of nerve degeneration.
Acknowledgment
The authors confirm that they have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines. The authors thank the patients' family for their participation in this study and the National Center for Genome Medicine and Hsiao-Huei Chen for technical support.
Glossary
- CMT
Charcot-Marie-Tooth disease
- CNV
copy number analysis
- FGA
full genome analysis
- MBS
multiblock system
- NCV
nerve conduction velocity
- OGM
optical genome mapping
- SNVs
single nucleotide variants
- WES
whole-exome sequencing
- WGS
whole-genome sequencing
- WHS
Wolf-Hirschhorn syndrome
Appendix. Authors
Study Funding
Ministry of Science and Technology, Taiwan, to S.-T.H. (110-2320-B-002-018), National Taiwan University Hospital to S.-T.H. (UN110-014) and H.-W H. (UN110-N4820), National Science and Technology Council to S.-T.H. (111-2634-F-002-017), and Academia Sinica to P.-Y. K. (AS-GMM-111-01).
Disclosure
H.W. Hsueh, H.J. Kao, C.C. Chao, S.J. Hsueh, Y.N. Huang, W.J. Lin, J.P. Su, H.T. Shy, T.Y. Yeh, C.C. Lin, P.Y. Kwok, N.C. Lee, and S.T. Hsieh report no disclosures relevant to the manuscript. Go to Neurology.org/NG for full disclosures.
References
- 1.Pipis M, Rossor AM, Laura M, Reilly MM. Next-generation sequencing in Charcot-Marie-Tooth disease: opportunities and challenges. Nat Rev Neurol. 2019;15(11):644-656. [DOI] [PubMed] [Google Scholar]
- 2.Klein CJ. Charcot-Marie-Tooth disease and other hereditary neuropathies. Continuum (Minneap Minn). 2020;26(5):1224-1256. [DOI] [PubMed] [Google Scholar]
- 3.Cortese A, Zhu Y, Rebelo AP, et al. Biallelic mutations in SORD cause a common and potentially treatable hereditary neuropathy with implications for diabetes. Nat Genet. 2020;52(5):473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortese A, Wilcox JE, Polke JM, et al. Targeted next-generation sequencing panels in the diagnosis of Charcot-Marie-Tooth disease. Neurology. 2020;94(1):e51-e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juneja M, Azmi A, Baets J, et al. PFN2 and GAMT as common molecular determinants of axonal Charcot-Marie-Tooth disease. J Neurol Neurosurg Psychiatry. 2018;89(8):870. [DOI] [PubMed] [Google Scholar]
- 6.Juneja M, Burns J, Saporta MA, Timmerman V. Challenges in modelling the Charcot-Marie-Tooth neuropathies for therapy development. J Neurol Neurosurg Psychiatry. 2019;90(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein CJ, Wu Y, Vogel P, et al. Ubiquitin ligase defect by DCAF8 mutation causes HMSN2 with giant axons. Neurology. 2014;82(10):873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebelo AP, Saade D, Pereira CV, et al. SCO2 mutations cause early-onset axonal Charcot-Marie-Tooth disease associated with cellular copper deficiency. Brain. 2018;141(3):662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanhangad M, Cornett K, Brewer MH, et al. Unique clinical and neurophysiologic profile of a cohort of children with CMTX3. Neurology. 2018;90(19):e1706-e1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson R, Spendiff S, Roos A, et al. Advances in the diagnosis of inherited neuromuscular diseases and implications for therapy development. Lancet Neurol. 2020;19(6):522-532. [DOI] [PubMed] [Google Scholar]
- 11.Shieh JT, Penon-Portmann M, Wong KHY, et al. Application of full-genome analysis to diagnose rare monogenic disorders. NPJ Genom Med. 2021;6(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan CL, Tseng TJ, Lin YH, Chiang MC, Lin WM, Hsieh ST. Cutaneous innervation in Guillain-Barré syndrome: pathology and clinical correlations. Brain. 2003;126(Pt 2):386-397. [DOI] [PubMed] [Google Scholar]
- 13.Chiang HY, Chien HF, Shen HH, et al. Reinnervation of muscular targets by nerve regeneration through guidance conduits. J Neuropathol Exp Neurol. 2005;64(7):576-587. [DOI] [PubMed] [Google Scholar]
- 14.Lin YH, Hsieh SC, Chao CC, Chang YC, Hsieh ST. Influence of aging on thermal and vibratory thresholds of quantitative sensory testing. J Peripher Nerv Syst. 2005;10(3):269-281. [DOI] [PubMed] [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.wANNOVAR. Accessed March 4, 2021. wannovar.wglab.org/
- 18.OMIM. Accessed March 4, 2021. omim.org/
- 19.National Library of Medicine. ClinVar. ncbi.nlm.nih.gov/clinvar/ [DOI] [PubMed]
- 20.Taiwan Biobank. Accessed March 4, 2021. taiwanview.twbiobank.org.tw/
- 21.Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18(7):696-704. [DOI] [PubMed] [Google Scholar]
- 22.Abou Tayoun AN, Pesaran T, DiStefano MT, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018;39(11):1517-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biesecker LG, Harrison SM. The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet Med. 2018;20(12):1687-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh R, Harrison SM, Rehm HL, Plon SE, Biesecker LG. Updated recommendation for the benign stand-alone ACMG/AMP criterion. Hum Mutat. 2018;39(11):1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical Genome Resource. Sequence Variant Interpretation. Accessed October 6, 2021. clinicalgenome.org/working-groups/sequence-variant-interpretation/ [Google Scholar]
- 26.Mehrtash B, Jack MF, Samuel KL, Andrey NS, Laura DG, Mark W, et al. ATK-gCNV: A Rare Copy Number Variant Discovery Algorithm and Its Application to Exome Sequencing in the UK Biobank. bioRxiv 2022: 2022.08.25.504851. [Google Scholar]
- 27.Kao HJ, Chiang HL, Chen HH, et al. De novo mutation and skewed X-inactivation in girl with BCAP31-related syndrome. Hum Mutat. 2020;41(10):1775-1782. [DOI] [PubMed] [Google Scholar]
- 28.gnomAD. Accessed March 1, 2023. gnomad.broadinstitute.org/
- 29.Li Q, Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100(2):267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.wInterVar. Accessed March 1, 2023. wintervar.wglab.org/
- 31.Yang NC, Lee MJ, Chao CC, et al. Clinical presentations and skin denervation in amyloid neuropathy due to transthyretin Ala97Ser. Neurology. 2010;75(6):532-538. [DOI] [PubMed] [Google Scholar]
- 32.Rauch A, Schellmoser S, Kraus C, et al. First known microdeletion within the Wolf-Hirschhorn syndrome critical region refines genotype-phenotype correlation. Am J Med Genet. 2001;99(4):338-342. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto N, Ohmachi K, Shimada S, Shimojima K, Yamamoto T. 109 kb deletion of chromosome 4p16.3 in a patient with mild phenotype of Wolf-Hirschhorn syndrome. Am J Med Genet A. 2013;161(6):1465-1469. [DOI] [PubMed] [Google Scholar]
- 34.Andersen EF, Carey JC, Earl DL, et al. Deletions involving genes WHSC1 and LETM1 may be necessary, but are not sufficient to cause Wolf-Hirschhorn Syndrome. Eur J Hum Genet. 2014;22(4):464-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi W, Cheung SW, Breman AM, Bacino CA. 4p16.3 microdeletions and microduplications detected by chromosomal microarray analysis: new insights into mechanisms and critical regions. Am J Med Genet A. 2016;170(10):2540-2550. [DOI] [PubMed] [Google Scholar]
- 36.Lozier ER, Konovalov FA, Kanivets IV, et al. De novo nonsense mutation in WHSC1 (NSD2) in patient with intellectual disability and dysmorphic features. J Hum Genet. 2018;63(8):919-922. [DOI] [PubMed] [Google Scholar]
- 37.Barrie ES, Alfaro MP, Pfau RB, et al. De novo loss-of-function variants in NSD2 (WHSC1) associate with a subset of Wolf-Hirschhorn syndrome. Cold Spring Harb Mol Case Stud. 2019;5(4):a004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derar N, Al-Hassnan ZN, Al-Owain M, et al. De novo truncating variants in WHSC1 recapitulate the Wolf-Hirschhorn (4p16.3 microdeletion) syndrome phenotype. Genet Med. 2019;21(1):185-188. [DOI] [PubMed] [Google Scholar]
- 39.Boczek NJ, Lahner CA, Nguyen TM, et al. Developmental delay and failure to thrive associated with a loss-of-function variant in WHSC1 (NSD2). Am J Med Genet A. 2018;176(12):2798-2802. [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y, Sun H, Lin Q, et al. De novo truncating variant in NSD2gene leading to atypical Wolf-Hirschhorn syndrome phenotype. BMC Med Genet. 2019;20(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battaglia A, Carey JC. The delineation of the Wolf-Hirschhorn syndrome over six decades: illustration of the ongoing advances in phenotype analysis and cytogenomic technology. Am J Med Genet A. 2021;185(9):2748-2755. [DOI] [PubMed] [Google Scholar]
- 42.Delude CM. Deep phenotyping: the details of disease. Nature. 2015;527(7576):S14–S15. [DOI] [PubMed] [Google Scholar]
- 43.Sukarova-Angelovska E, Kocova M, Sabolich V, Palcevska S, Angelkova N. Phenotypic variations in Wolf-Hirschhorn syndrome. Balkan J Med Genet. 2014;17(1):23-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Battaglia A, Carey JC, South ST. Wolf-Hirschhorn syndrome: a review and update. Am J Med Genet C Semin Med Genet. 2015;169(3):216-223. [DOI] [PubMed] [Google Scholar]
- 45.Durigon R, Mitchell AL, Jones AW, et al. LETM1 couples mitochondrial DNA metabolism and nutrient preference. EMBO Mol Med. 2018;10(9):e8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura S, Matsui A, Akabane S, et al. The mitochondrial inner membrane protein LETM1 modulates cristae organization through its LETM domain. Commun Biol. 2020;3(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding ZM, Huang CJ, Jiao XF, Wu D, Huo LJ. The role of TACC3 in mitotic spindle organization. Cytoskeleton (Hoboken). 2017;74(10):369-378. [DOI] [PubMed] [Google Scholar]
- 48.Nwagbara BU, Faris AE, Bearce EA, et al. TACC3 is a microtubule plus end-tracking protein that promotes axon elongation and also regulates microtubule plus end dynamics in multiple embryonic cell types. Mol Biol Cell. 2014;25(21):3350-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jungnickel J, Gransalke K, Timmer M, Grothe C. Fibroblast growth factor receptor 3 signaling regulates injury-related effects in the peripheral nervous system. Mol Cell Neurosci. 2004;25(1):21-29. [DOI] [PubMed] [Google Scholar]
- 50.Kovvuru S, Cardenas YC, Huttner A, Nowak RJ, Roy B. Clinical characteristics of fibroblast growth factor receptor 3 antibody-related polyneuropathy: a retrospective study. Eur J Neurol. 2020;27(7):1310-1318. [DOI] [PubMed] [Google Scholar]
- 51.Tholance Y, Moritz CP, Rosier C, et al. Clinical characterisation of sensory neuropathy with anti-FGFR3 autoantibodies. J Neurol Neurosurg Psychiatry. 2020;91(1):49-57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated during analysis and/or during the current study are available from the corresponding author on reasonable request.