ABSTRACT
Across species, ovulation is a process induced by a myriad of signaling cascades that ultimately leads to the release of encapsulated oocytes from follicles. Follicles first need to mature and gain ovulatory competency before ovulation; however, the signaling pathways regulating follicle maturation are incompletely understood in Drosophila and other species. Our previous work has shown that the bHLH-PAS transcription factor Single-minded (Sim) plays important roles in follicle maturation downstream of the nuclear receptor Ftz-f1 in Drosophila. Here, we demonstrate that Tango (Tgo), another bHLH-PAS protein, acts as a co-factor of Sim to promote follicle cell differentiation from stages 10 to 12. In addition, we discover that re-upregulation of Sim in stage-14 follicle cells is also essential to promote ovulatory competency by upregulating octopamine receptor in mushroom body (OAMB), matrix metalloproteinase 2 (Mmp2) and NADPH oxidase (NOX), either independently of or in conjunction with the zinc-finger protein Hindsight (Hnt). All these factors are crucial for successful ovulation. Together, our work indicates that the transcriptional complex Sim:Tgo plays multiple roles in late-stage follicle cells to promote follicle maturation and ovulation.
Keywords: Follicle maturation, Ovulatory competency, Ovulation, BHLH-PAS transcription factors, Sim, Tgo
Summary: Sim plays two roles throughout oogenesis: in stage 10B-12 follicles, it functions with Tgo to promote follicle cell maturation; in mature follicles, it actively regulates genes required for follicle rupture.
INTRODUCTION
Reproductive health is a vital component of every society. In the USA alone, 10% of human females of reproductive age suffer from infertility (Chandra et al., 2014). Some of the leading causes of infertility are ovulation disorders, which affect one in four couples who have difficulty conceiving [Health (UK), 2013]. Ovulation is the process of releasing a fertilizable oocyte from a mature ovarian follicle. In mammals, this process is induced by a luteinizing hormone (LH) surge that activates multiple pathways, including progesterone, EGFR-Ras-ERK1/2 and prostaglandin signaling (Richards and Ascoli, 2018; Robker et al., 2018; Duffy et al., 2019; Tokmakov et al., 2020). Despite many years of investigation, we still do not know the detailed mechanisms that precisely regulate follicle rupture – the actual step of releasing the oocyte.
Drosophila melanogaster offers a wide array of genetic tools and displays significant conservation of ovulation pathways with mammals, making it a valuable model to decipher the genetic pathways for follicle rupture. In Drosophila, the adrenergic system is responsible for inducing multiple pathways for follicle rupture and ovulation (Monastirioti et al., 1996; Cole et al., 2005; Middleton et al., 2006; Deady and Sun, 2015). Octopamine (OA), the Drosophila counterpart of norepinephrine, binds to its receptor octopamine receptor in mushroom body (OAMB) in mature follicle cells and increases intracellular Ca2+ (Deady and Sun, 2015). In posterior follicle cells, the increase of intracellular Ca2+ induces the activation of Matrix metalloproteinase 2 (Mmp2), which breaks down the posterior follicle wall to allow oocyte release (Deady et al., 2015). In mainbody follicle cells, the Ca2+ activates NADPH oxidase (NOX) and superoxide dismutase 3 (SOD3), which increase ROS production to facilitate follicle rupture (Li et al., 2018). Although we are gaining a better understanding of the rupture process, it is largely unknown how these ovulatory factors are precisely regulated. In other words, how do follicles become fully mature and competent to ovulate?
The Drosophila ovary is composed of ∼16 ovarioles, where follicles develop through 14 stages from anterior to posterior (Spradling, 1993). To reach maturity, somatic follicle cells surrounding the germline cells must undergo eight or nine rounds of mitosis from stages 1-6, three rounds of endocycles from stages 7-10A, and synchronized gene amplification and chorion synthesis from stages 10B-14 (Klusza and Deng, 2011). Our previous work has shown that ecdysteroid signaling induces the transition from the endocycle to synchronized chorion gene amplification by upregulating Ttk69, while it induces the expression of NR5A nuclear receptor Fushi tarazu-factor 1 (Ftz-f1) at stage 10B to regulate follicle cell differentiation, maturation and competency to ovulatory stimuli (Sun et al., 2008; Knapp et al., 2020). Specifically, Ftz-f1 directly activates another transcription factor, Single-minded (Sim), at stages 10B-12. This mediates final differentiation of follicle cells, which is manifested by the downregulation of Ttk69, Cut, Broad-Complex (Br-C), Ecdysone receptor isoform A (EcR.A) and EcR.B1, and by upregulation of the zinc-finger transcription factor Hindsight (Hnt) (Knapp et al., 2019, 2020). Hnt subsequently activates OAMB expression in all follicle cells and Mmp2 expression in posterior follicle cells at stage 14 (Deady et al., 2017). Previous work found that Sim is downregulated at stage 13 and re-upregulated at stage 14 after its expression from stage 10B to 12 (Knapp et al., 2020). However, it is unknown why Sim has such a dynamic pattern in later stages and how Sim precisely regulates follicle cell differentiation and final maturation.
Sim is well studied during embryonic development, where it functions as a master regulator of CNS midline cell transcription and development (Nambu et al., 1990). It belongs to the basic-helix-loop-helix-PAS (bHLH-PAS) protein family and forms a transcriptional activator with another bHLH-PAS protein, Tango (Tgo; Nambu et al., 1991; Ohshiro and Saigo, 1997; Sonnenfeld et al., 1997; Estes et al., 2001; Jiang and Crews, 2003). The role of Sim is not only limited to CNS midline development but is also crucial for left-right asymmetry of embryonic gut, axon guidance in larval brain, lamina-retina neuron association during optic lobe development and genital disc development (Pielage et al., 2002; Umetsu et al., 2006; Maeda et al., 2007; Freer et al., 2011). Tgo can not only heterodimerize with Sim but also with other bHLH-PAS proteins, including Trachealess and Spineless (Ohshiro and Saigo, 1997; Emmons et al., 1999). The process of dimerization is crucial for Tgo nuclear localization (Ward et al., 1998), while the specific PAS domain confers the transcriptional specificity (Zelzer et al., 1997). Despite these studies, little is known about the roles of Sim and Tgo in adult physiology and reproduction.
In this article, we demonstrate that Tgo is co-expressed with Sim throughout late oogenesis and that its nuclear translocation depends on Sim, which is consistent with the previous report. Moreover, disruption of Tgo and Sim produces similar defects in follicle cell differentiation, dorsal appendage formation and egg morphology, suggesting that Sim and Tgo form a heterodimer complex to regulate follicle cell differentiation. Most importantly, we characterize a novel role for Sim in stage-14 follicles that is distinct from its role in follicle cell differentiation from stages 10B to 12. At stage 14, Sim promotes ovulatory competency by upregulating the expression of key proteins required for follicle rupture, including OAMB, NOX and Mmp2. In addition, the role of Sim in promoting ovulatory competency is either independent of or in conjunction with Hnt. Our results suggest that Sim acts as a master regulator to promote ovulatory competency of the follicles. We also demonstrated for the first time that the tunable CRISPR-Cas9 system (Port et al., 2020) can effectively disrupt gene expression in a tissue-specific manner in follicle cells with 16 copies of the genome. Altogether, our work demonstrates that the Sim:Tgo complex plays essential roles in promoting follicle differentiation and maturation in late oogenesis. Considering the conserved nature of this family of transcription factors in neurogenesis (Crews and Fan, 1999), the role of this complex in follicle development may be conserved in other species.
RESULTS
Tgo is dynamically co-expressed with Sim in follicle cells during late oogenesis
Sim and Tgo form a heterodimer complex that regulates CNS midline cell development (Sonnenfeld et al., 1997). To test whether Tgo is also a heterodimerization partner for Sim in late oogenesis, we first examined the Tgo expression in Drosophila ovaries using an anti-Tgo antibody. Tgo begins to be detected faintly in the cytoplasm of follicle cells in mid-stage egg chambers (stages 7-9) and is upregulated at late stage 10A (Fig. 1A and Fig. S1A,B). Tgo is then enriched in follicle cell nuclei from stages 10B to 12 but becomes downregulated at stage 13 with low levels of cytoplasmic signal still detected (Fig. 1B-D,F and Fig. S1F,G). At stage 14, Tgo is re-upregulated and enriched in follicle cell nuclei (Fig. 1E). The specificity of the Tgo staining is confirmed by the observation that Tgo expression in follicle cells is significantly reduced upon tgo knockdown via RNA interference (RNAi; Fig. S1A-H).
Fig. 1.
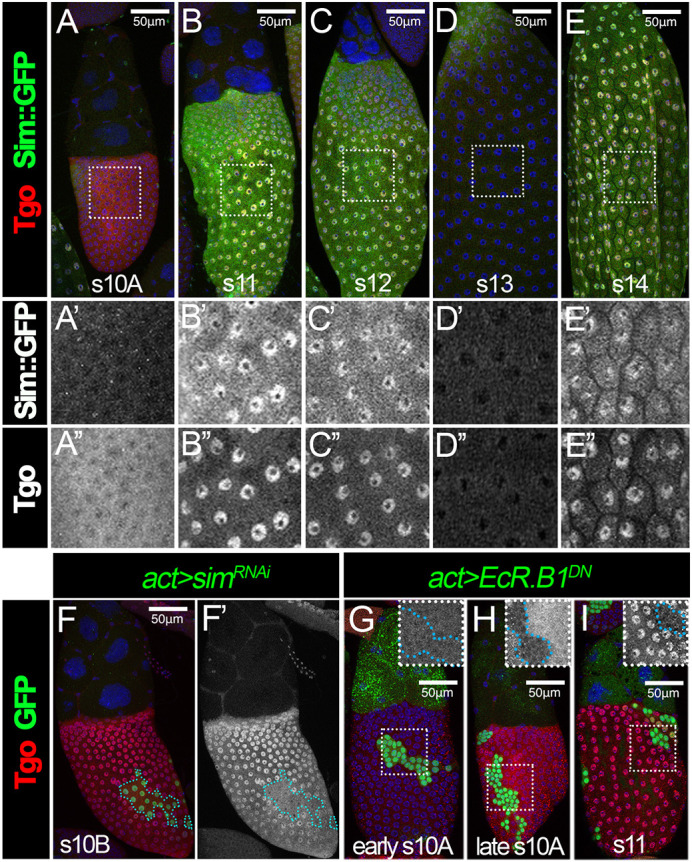
Tgo is dynamically expressed in follicle cells during late oogenesis. (A-E) Expression of Sim::GFP and Tgo in stages 10-14. Sim::GFP expression is detected using anti-GFP antibody (green) and Tgo is detected using anti-Tgo antibody (red) in stage 10A (A), stage 11 (B), stage 12 (C), stage 13 (D) and stage 14 (E) follicles. The higher magnifications of the areas outlined are shown in A′-E′ (Sim::GFP) and A″-E″ (Tgo). All images from A-E were acquired using the same microscope settings. (F) Tgo expression (red) in stage 10B follicles with flip-out Gal4 clones (green) overexpressing simRNAi (act>simRNAi). (F′) Tgo expression (white) in wild-type versus clone populations (outlined in blue). (G-I) Tgo expression (red) in stage 10A (G), stage 10B (H) and stage 11 (I) follicles with flip-out Gal4 clones (green) overexpressing EcR.B1DN (act>EcR.B1DN). Insets show higher magnifications of Tgo expression in the areas outlined. The clone boundary is outlined with a cyan dashed line. Nuclei are marked by DAPI in blue. Scale bars: 50 μm.
The nuclear localization of Tgo coincides with the expression of Sim at stages 10B-12 and stage 14 (Fig. 1B-E), which is consistent with our previous report (Knapp et al., 2020). Previous work showed that Tgo nuclear localization depends on Sim in CNS midline precursor cells (Ward et al., 1998). To test whether Sim is also required for Tgo nuclear localization in follicle cells, we examined the Tgo expression in sim-knockdown follicle cells. Tgo did not show nuclear enrichment but was instead maintained in the cytoplasm of sim-knockdown follicle cells (Fig. 1F). This result confirms that Tgo nuclear localization depends on Sim in follicle cells and suggests that Tgo is likely the heterodimerization partner of Sim for its functions in late oogenesis.
We noticed an increase in Tgo expression at late stage 10A in comparison with earlier stages, which coincides with the endocycle-to-gene-amplification transition initiated by ecdysone signaling (Fig. 1A; Sun et al., 2008). To test whether ecdysone signaling is required for the upregulation of Tgo expression, we examined the Tgo expression in follicle cells that overexpress a dominant-negative (DN) form of EcR (EcRDN). In earlier stages, EcRDN-overexpressing follicle cells showed the same level of Tgo expression as adjacent wild-type follicle cells (Fig. 1G). However, we did not observe Tgo upregulation in EcRDN-overexpression follicle cells at late stage 10A, when the adjacent wild-type follicle cells had a significant increase of Tgo expression (Fig. 1H). At stage 11, we also did not observe Tgo nuclear localization in EcRDN-overexpressing follicle cells (Fig. 1I). This is likely because EcRDN prevents Ftz-F1 expression, which is crucial for Sim expression (Knapp et al., 2020). This suggests that ecdysone signaling is required for the upregulation of Tgo expression at the stage 10A/10B transition.
Tgo acts as a heterodimer partner of Sim for follicle cell differentiation
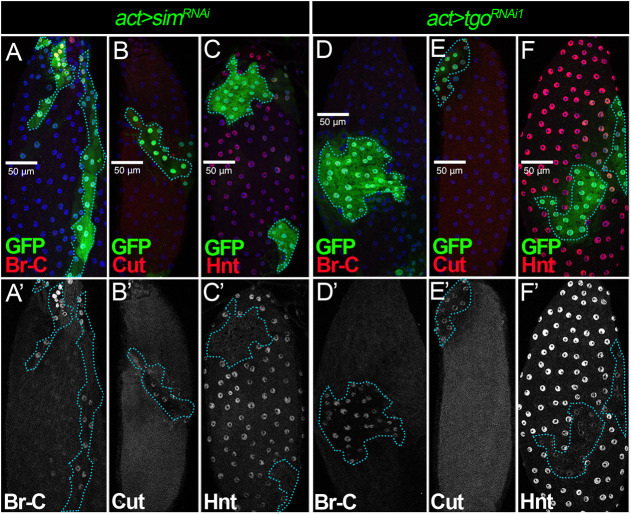
Disrupting the expression of Sim (as well as its upstream regulator Ftz-f1) prevents follicle cells from differentiating into a mature state at stage 14, which is characterized by the upregulation of Hnt and the downregulation of Cut, Ttk69, Br-C, EcR.A and EcR.B1 (Knapp et al., 2019). To determine whether Tgo is indeed the partner for Sim in regulating follicle cell differentiation, we tested whether Tgo depletion affects the expression of some of these factors (Br-C, Cut and Hnt) in stage 14 follicle cells. Consistent with previous findings, sim-knockdown follicle cells had extended expression of Br-C and Cut, and reduced expression of Hnt at stage 14 (Fig. 2A-C; Knapp et al., 2020). Similarly, tgo-knockdown follicle cells failed to downregulate Br-C and Cut (Fig. 2D,E), and upregulate Hnt (Fig. 2F) at stage 14. These results indicate that Tgo is also required to promote follicle cell differentiation and is likely the partner of Sim.
Fig. 2.
Tgo is a heterodimer partner of Sim for follicle cell differentiation. (A-C′) Br-C (A,A′), Cut (B,B′) and Hnt (C,C′) expression (red in A-C and white in A′-C′) in stage 14 follicles with flip-out clones (marked by GFP) overexpressing simRNAi (act>simRNAi). (D-F′) Br-C (D,D′), Cut (E,E′) and Hnt (F,F′) expression (red in D-F and white in D′-F′) in stage 14 follicles with flip-out clones (marked by GFP) overexpressing tgoRNAi1 (act>tgoRNAi1). The clone boundary is outlined by a cyan dashed line. Nuclei are marked by DAPI in blue. Scale bars: 50 μm.
The Sim:Tgo complex is required for ovulation
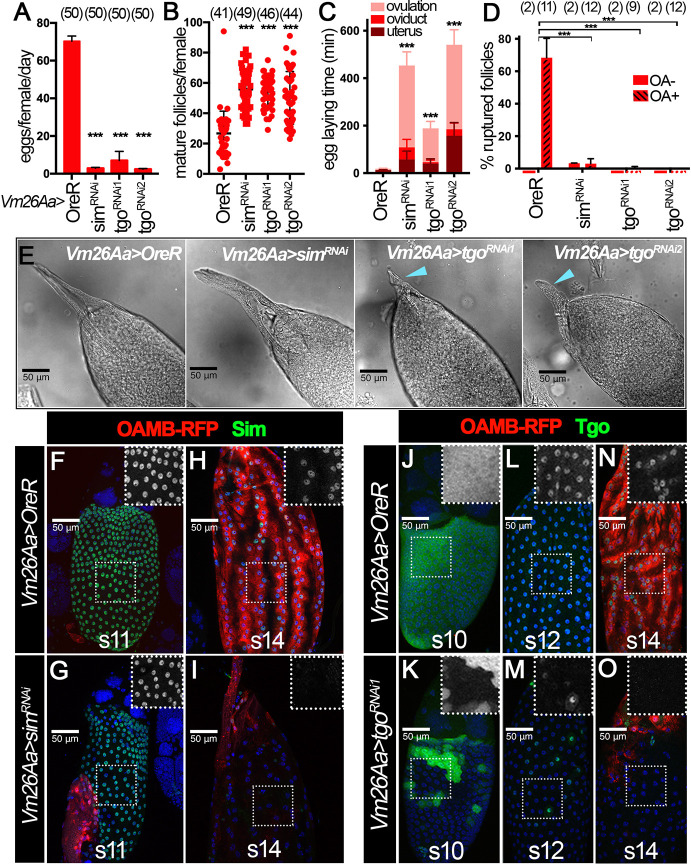
To further support that Sim and Tgo expression at stages 10-12 promotes follicle cell differentiation into maturation, we attempted to test whether knockdown of sim or tgo in follicle cells led to an ovulation defect similar to that observed upon knockdown of ftz-f1 (Knapp et al., 2020). To do so, we used Vm26Aa-Gal4, a Gal4 driver expressed in follicle cells starting at stage 10 and expressed in no other developmental context or tissues besides the female ovary (Peters et al., 2013). Knockdown of sim or tgo using Vm26Aa-Gal4 resulted in almost complete inhibition of egg laying (Fig. 3A). In addition, we observed an increased number of stage-14 egg chambers inside the ovary, indicating an egg-retention phenotype and ovulation defect (Fig. 3B). To determine whether the reduction in egg laying is indeed due to an ovulation defect, we estimated the time spent in ovulation using an assay described in previous reports (Deady and Sun, 2015; Sun and Spradling, 2013). sim- and tgo-knockdown females showed a dramatic increase in ovulation time. Control females usually take ∼5 min to ovulate an egg (Fig. 3C and Table S1). In contrast, sim- and tgo-knockdown females take more than 2 h to ovulate an egg (Fig. 3C and Table S1).
Fig. 3.
The Sim:Tgo complex is required for ovulation. (A-C) Quantification of egg laying (A), mature follicle numbers after egg laying (B) and egg laying time (C) in control, simRNAi, tgoRNAi1 or tgoRNAi2 females with Vm26Aa-Gal4 and Oamb-RFP. The number of females is indicated above each bar. (D) Quantification of OA-induced follicle rupture using mature follicles isolated from control, simRNAi, tgoRNAi1 or tgoRNAi2 females with Vm26Aa-Gal4 and Oamb-RFP. The number of wells analyzed (containing 25-35 follicles) is noted above each bar. (E) Representative DIC images showing dorsal appendage morphology in control, simRNAi, tgoRNAi1 or tgoRNAi2 stage 14 follicles with Vm26Aa-Gal4 and Oamb-RFP. Blue arrowheads indicate stunted dorsal appendages. (F-I) Sim expression (green) in stage 11 (F,G) and stage 14 (H,I) follicles from control (F,H) and simRNAi (G,I) females with Vm26Aa-Gal4 and Oamb-RFP. The insets are higher magnifications of Sim expression (white) in outlined areas. (J-O) Tgo expression (green) in stage 10 (J,K), stage 12 (L,M) and stage 14 (N,O) follicles from control (J,L,N) and tgoRNAi1 (K,M,O) females with Vm26Aa-Gal4 and Oamb-RFP. The insets are higher magnifications of Tgo expression (white) in outlined areas. Nuclei are marked by DAPI in blue. ***P<0.001 [unpaired (two-sample equal variance) Student's t-test]. Data are mean±s.d. (B,D) and mean±s.e.m. (A,C). Scale bars: 50 μm.
To further probe whether this ovulation defect is due to the incompetency of a follicle to undergo OA-induced follicle rupture, we attempted to isolate mature follicles and carry out the ex vivo OA-induced follicle rupture assay described previously (Deady and Sun, 2015; Knapp et al., 2018; Beard et al., 2023). Upon OA stimulation, follicle cells of a mature follicle will recede toward the dorsal appendage region in order to release the encapsulated oocyte. The Oamb-RFP reporter was used as in previous experiments to identify the mature follicles with an intact follicle-cell layer. However, we immediately noticed that sim-knockdown follicles did not express OAMB-RFP in most of the follicle cells, except anterior follicle cells (see description below). Therefore, we used anterior OAMB-RFP expression to identify the mature follicles and used the bright-field signal to ensure these follicles had an intact follicle-cell layer (see MM). After a 3 h incubation with OA, we examined the follicle rupture by staining the tissue with DAPI. We found that all mature follicles isolated in this way had an intact follicle-cell layer before culture, and about 60% of follicles from control females ruptured after 3 h (Fig. 3D and Fig. S2A). In contrast, follicles isolated from sim- and tgo-knockdown females showed less than 5% rupture rate after 3 h (Fig. 3D and Fig. S2B-D).
During follicle isolation, we noticed that tgo-knockdown mature follicles were rounder with shortened dorsal appendages, reminiscent of ftz-f1-knockdown follicles (Knapp et al., 2020), while sim-knockdown follicles were morphologically normal (Fig. 3E and Fig. S2E). This inconsistency prompted us to examine the knockdown efficiency in these genotypes. To our surprise, simRNAi driven by Vm26Aa-Gal4 showed no obvious effect on Sim protein expression from stages 10B to 12 (Fig. 3F,G), although both tgoRNAi lines showed drastic reduction of Tgo protein from stages 10B to 12 (Fig. 3J-M and Fig. S2F-H). This inefficient knockdown of Sim protein via simRNAi is further supported by the observation that, in these knockdown follicles, Tgo is properly localized to the follicle cell nuclei (Fig. S2F,J-K) and that Sim::GFP fusion protein is still detected in these follicle cells (Fig. S2M-P) from stages 10B to 12. At stage 14, both simRNAi and tgoRNAi lines were able to knock down Sim and Tgo protein, respectively (Fig. 3H,I,N,O and Fig. S2I,L,Q,R). In addition, both led to a severe reduction of OAMB-RFP expression.
To test whether the inefficient knockdown of Sim protein in simRNAi line is the cause for the morphological difference between simRNAi and tgoRNAi follicles, we attempted to knock down sim using C204-Gal4, the expression of which starts in mainbody and posterior follicle cells at stage 8 (Manseau et al., 1997; Sun et al., 2008). Owing to the limited availability of the Sim antibody, we used Tgo nuclear localization to assess the sim knockdown efficiency. We found that Tgo nuclear localization was significantly reduced in stages 10B-12 and 14 follicles with C204-Gal4 driving simRNAi expression (Fig. S3A-F), indicating that Sim protein is efficiently knocked down with this manipulation. Follicles with this manipulation showed shorter dorsal appendages and rounder morphology (reflected by a reduction in the AP/DV ratio of the follicles; Fig. S3G-H), as seen in tgo- and ftz-f1-knockdown follicles (Fig. 3E). In addition, follicle cell differentiation and maturation was disrupted, manifested by the extended expression of Br-C and Cut, and by the prevention of Hnt upregulation at stage 14 (Fig. S3I-N). All these data support the conclusion that Sim and Tgo form a complex and function downstream of Ftz-f1 in promoting follicle cell differentiation and maturation, which is crucial for successful ovulation.
Re-upregulation of Sim at stage 14 is essential for ovulation
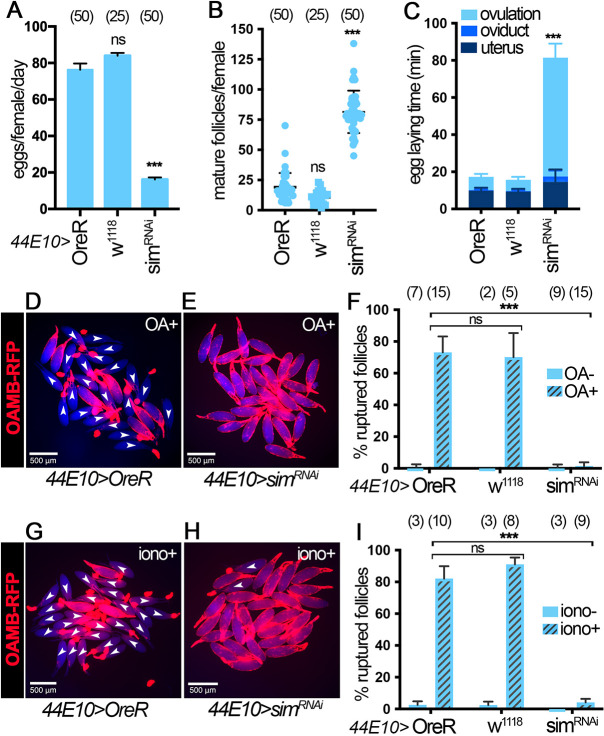
The fact that females with Vm26Aa-Gal4 driving simRNAi expression showed a severe ovulation defect without affecting Sim expression at stages 10B-12 prompted us to propose that re-upregulation of Sim at stage 14 plays an active role in mediating follicle rupture and ovulation. To test this hypothesis explicitly, we used three stage 14 follicle cell Gal4 drivers to deplete Sim specifically in stage 14 follicle cells: 47A04-Gal4 (Deady and Sun, 2015), 44E10-Gal4 (Deady and Sun, 2015) and CG13083-Gal4 (Knapp et al., 2019). All three Gal4 drivers start expression in stage 14 follicle cells with CG13083-Gal4 starting at the earliest stage (possibly in late stage 13) and 47A04-Gal4 starting at the latest stage. We first examined the sim knockdown efficiency using these three Gal4 lines. Consistent with the Gal4 expression pattern, Sim protein is not reduced from stages 10B-12 in all three conditions (Fig. S4A,B,D,E,G,H). In addition, Sim protein is not efficiently depleted in early stage 14 follicles (including stage 14A and 14B, as defined by Deady et al., 2017), except in the case of CG13083-Gal4, which showed some reduction at this stage (Fig. S4A-I). In contrast, Sim protein is clearly depleted in late stage 14 follicles with all three Gal4 drivers (Fig. S4A-I). Next, we examined whether this manipulation caused an ovulation defect. Regardless of the Gal4 driver used, sim depletion with either of these Gal4 lines led to a significant decrease in egg laying (Fig. 4A and Fig. S5A,J) without defects in stage 14 follicle formation (Fig. 4B and Fig. S5B,K). In fact, both 44E10-Gal4 and CG13083-Gal4 led to mature follicle retention (Fig. 4B and Fig. S5K), a strong indication of ovulation defect. In addition, sim knockdown with all the Gal4 lines led to an increase in ovulation time (Fig. 4C, Fig. S5C,L and Table S1). Together, these data suggest that Sim in stage 14 follicle cells is essential for ovulation in vivo.
Fig. 4.
Sim knockdown with the 44E10-Gal4 driver leads to ovulation defects. (A-C) Quantification of egg laying (A), mature follicle numbers after egg laying (B) and egg laying time (C) in OreR control, w1118 control and simRNAi females with 44E10-Gal4 and Oamb-RFP. The number of females is indicated above each bar. (D,E) Representative images show mature follicles from control (D) and simRNAi (E) females with 44E10-Gal4 and Oamb-RFP after 3 h in culture with 20 µM OA (OA+). Oamb-RFP is shown in red and bright-field signal is shown in blue. Ruptured follicles are labeled with white arrowheads. (F) Quantification of OA-induced follicle rupture. The number of wells analyzed (containing 25-35 follicles) is indicated above each bar. (G,H) Representative images show mature follicles from control (G) and simRNAi (H) females with 44E10-Gal4 and Oamb-RFP after 3 h in culture with 2 µM ionomycin (iono+). Oamb-RFP is shown in red and bright-field signal is shown in blue. Ruptured follicles are labeled with white arrowheads. (I) Quantification of ionomycin-induced follicle rupture. The number of wells analyzed (containing 25-35 follicles) is indicated above each bar. ***P<0.001 [unpaired (two-sample equal variance) Student's t-test]. Data are mean±s.d. (B,F,I) and mean±s.e.m. (A,C). Scale bars: 500 μm.
Next, we aimed to determine whether sim is required for OA-induced follicle rupture using our ex vivo follicle rupture assay. Mature follicles from control females ruptured normally in response to OA treatment, whereas mature follicles from simRNAi females using 44E10-Gal4 or 47A04-Gal4 drivers showed a severe defect in follicle rupture (Fig. 4D-F and Fig. S5D-F). Owing to the difficulty in isolating mature follicles with CG13083-Gal4-mediated knockdown of sim, which prevents OAMB-RFP expression, we did not perform the follicle rupture assay with this manipulation. In addition, we tested whether sim-depleted follicles can rupture in response to ionomycin, a Ca2+ ionophore, which can bypass OAMB to increase intracellular Ca2+ (Deady and Sun, 2015). Interestingly, sim-knockdown follicles exhibited significant follicle rupture defects in response to ionomycin stimulation (Fig. 4G-I and Fig. S5G-I). All these data suggest that Sim in stage 14 follicle cells is required for follicle rupture and likely regulates components downstream of Ca2+, in addition to OAMB.
Depletion of Sim using tunable CRISPR-Cas9 system results in similar defects to those observed in simRNAi females
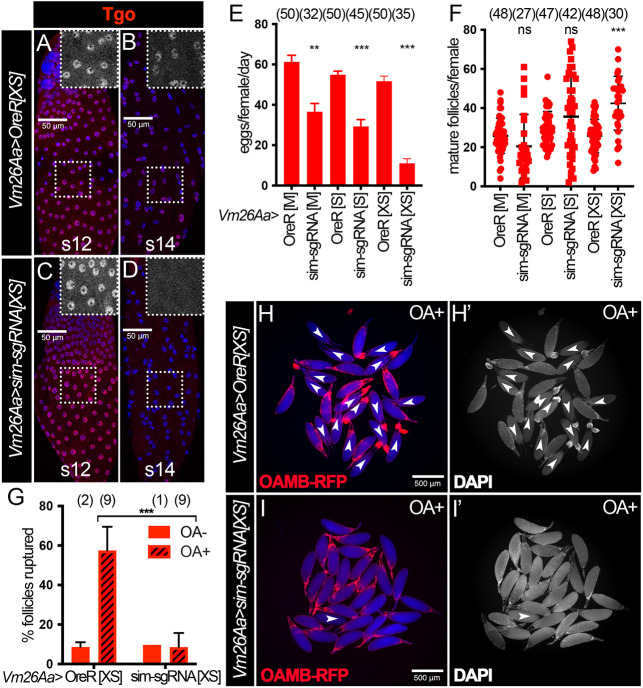
The simRNAi used in this study has no predictable off targets. To further eliminate the off-target concern, we used a tunable CRISPR-Cas9 system (Port et al., 2020) to disrupt Sim expression. The ability of the CRISPR-Cas9 system to efficiently knockout genes in follicle cells in late oogenesis has never been tested, as follicle cells undergo three rounds of endoreplication and contain 16 copies of the genome. To this end, we used follicle cell-specific Vm26Aa-Gal4 driving two guide RNAs targeting the first and second common coding exons of the sim gene, as well as different levels of Cas9 expression in follicle cells. UAS-Cas9XS expresses Cas9 at the highest levels, UAS-Cas9S expresses Cas9 at medium levels and UAS-Cas9M expresses Cas9 at the lowest levels (Port et al., 2020). This genetic manipulation disrupts Tgo nuclear localization (inferring Sim expression) in stage 14 follicle cells but not before stage 14 (Fig. 5A-D). In addition, these females showed reduced egg laying capacity and a trend of increased number of mature follicles in the ovaries (Fig. 5E,F), reminiscent of sim-knockdown females. Most importantly, mature follicles from sim-knockout females with the CRISPR-Cas9 system also showed defective OA-induced follicle rupture and reduced OAMB-RFP expression (Fig. 5G-I). These data demonstrate that tissue-specific CRISPR-Cas9 system is sufficient to knockout genes in follicle cells, and that Sim in stage-14 follicle cells play an active role in mediating follicle rupture and ovulation.
Fig. 5.
Depletion of Sim using the tunable CRISPR-Cas9 system results in similar defects to those in simRNAi females. (A-D) Tgo expression (red) in stage 12 (A,C) and stage 14 (B,D) follicles from control (A,B) and simsgRNA (C,D) females with Vm26Aa-Gal4 and UAS-Cas9XS. The insets are higher magnifications of Tgo expression (white) in outlined areas. All images from A-D were acquired using the same microscope settings. Nuclei are marked by DAPI in blue. (E,F) Quantification of egg laying (E) and mature follicle retention (F) in control and simsgRNA females with Vm26Aa-Gal4 and three different UAS-Cas9 constructs: M, S and XS. The number of females is indicated above each bar. (G) Quantification of OA-induced follicle rupture assessed by DAPI staining. The number of wells analyzed is indicated above each bar. (H-I′) Representative images show mature follicles from control (H,H′) and simsgRNA (I,I′) females with Vm26Aa-Gal4, UAS-Cas9XS and Oamb-RFP after 3 h culture with 20 µM OA (OA+). Oamb-RFP is shown in red; bright-field signal is shown in blue; DAPI signal is shown in white in H′-I′. Ruptured follicles are labeled with white arrowheads and usually have corpus luteum accumulation at the posterior end by the dorsal appendage. ***P<0.001, **P<0.01 [unpaired (two-sample equal variance) Student's t-test]. Data are mean±s.d. (F,G) and mean±s.e.m. (E). Scale bars: 50 μm (A-D); 500 μm (H-I′).
Sim at stage 14 upregulates Oamb expression in mature follicles in conjunction with Hnt
To explore the mechanism of Sim in ovulation, we investigated whether Sim regulates known ovulatory genes. As mentioned previously, mature follicles with Vm26Aa-Gal4 driving simRNAi expression showed almost no detectable Oamb-RFP in most of the follicle cells (Fig. 3I). We hypothesized that Sim is required for Oamb transcription in mature follicle cells. Consistent with this hypothesis, mature follicles with CG13083-Gal4 driving simRNAi expression also showed no detectable Oamb-RFP in most follicle cells, except in the anterior region (Fig. 6A,B,F and Fig. S6A-D). In contrast, knockdown of sim with 44E10-Gal4 or 47A04-Gal4 had much weaker (if any) effect on Oamb-RFP expression (Fig. S6E-L) but still showed significant reduction of Oamb mRNA levels (Fig. S6M-N). All these data indicate that Sim in stage 14 follicles is required for Oamb transcription.
Fig. 6.
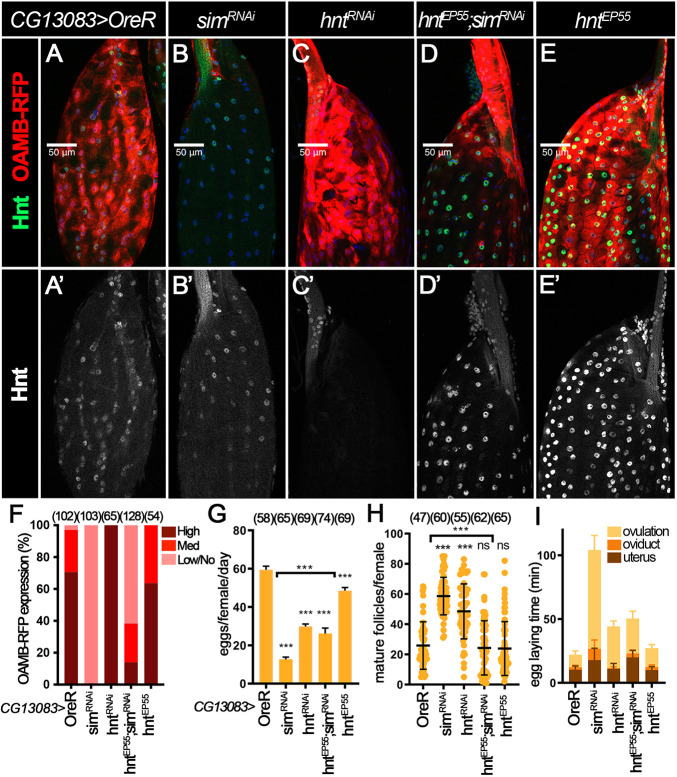
Sim at stage 14 upregulates Oamb expression in mature follicles in conjunction with Hnt. (A-E′) Hnt expression (green in A-E and white in A′-E′) and OAMB-RFP expression (red in A-E; detected by RFP antibody) in control (A), simRNAi (B), hntRNAi (C), hntEP55;simRNAi (D) and hntEP55 (E) stage 14 follicles with CG13083-Gal4 and Oamb-RFP. Nuclei are marked by DAPI in blue. (F) Quantification of main body OAMB-RFP expression levels in late stage 14 follicles (characterized by absence of nurse cell nuclei, follicle morphology and high RFP expression in the dorsal appendages region). The number of mature follicles quantified is indicated above each bar. (G-I) Quantification of egg laying (G), mature follicle retention (H) and egg laying time (I) in control, simRNAi, hntRNAi, hntEP55;simRNAi and hntEP55 females with CG13083-Gal4 and Oamb-RFP. The number of females is indicated above each bar. ***P<0.001 [unpaired (two-sample equal variance) Student's t-test]. Data are mean±s.d. (H) and mean±s.e.m. (G,I). Scale bars: 50 μm.
Our previous work showed that Hnt upregulation in stage 14 follicle cells is required for Oamb transcription (Deady et al., 2017). We therefore examined Hnt expression in sim-knockdown follicles with CG13083-Gal4 driver. We observed that, upon sim knockdown, Hnt could still be upregulated in early and middle stage 14 follicle cells (Fig. S6A and S6C) but showed an obvious reduction in late stage 14 follicle cells (Fig. 6A,B and Fig. S6B,D). A similar reduction of Hnt expression in late stage 14 follicle cells was also observed when simRNAi was driven by 44E10-Gal4 (Fig. S6F and H) but this reduction was not obvious with 47A04-Gal4 (Fig. S6J,L). Consistent with the protein expression, hnt mRNA is also drastically reduced in follicle cells with 44E10-Gal4 but not 47A04-Gal4 driving simRNAi expression (Fig. S6M-N). Together, these data indicate that Sim in stage 14 follicles also regulates Hnt expression.
To test whether Sim activates Oamb expression via Hnt, we examined whether hnt overexpression (with UAS-hntEP55) in sim-knockdown follicles could restore Oamb-RFP expression. Using this genetic manipulation, Hnt protein is restored in mature follicle cells (Fig. 6D). Surprisingly, the defective Oamb-RFP expression associated with sim-knockdown could not be fully rescued by hnt overexpression (Fig. 6D-F). In addition, Oamb mRNA levels were also not rescued by hnt overexpression in sim-knockdown follicles with 47A04-Gal4 (Fig. S6N). These results indicate that hnt is not the only target downstream of Sim to regulate Oamb transcription. Consistent with this idea, hnt knockdown using CG13083-Gal4 did not affect Oamb-RFP expression, unlike sim-knockdown follicles (Fig. 6C). In addition, hnt overexpression showed only a partial rescue of egg laying and ovulation defects in sim-knockdown follicles (Fig. 6G-I, Table S1). Therefore, these results lead us to conclude that Sim regulates OAMB expression in mature follicle cells independently of and in conjunction with Hnt.
Sim at stage 14 upregulates Mmp2 in mature follicles in conjunction with Hnt
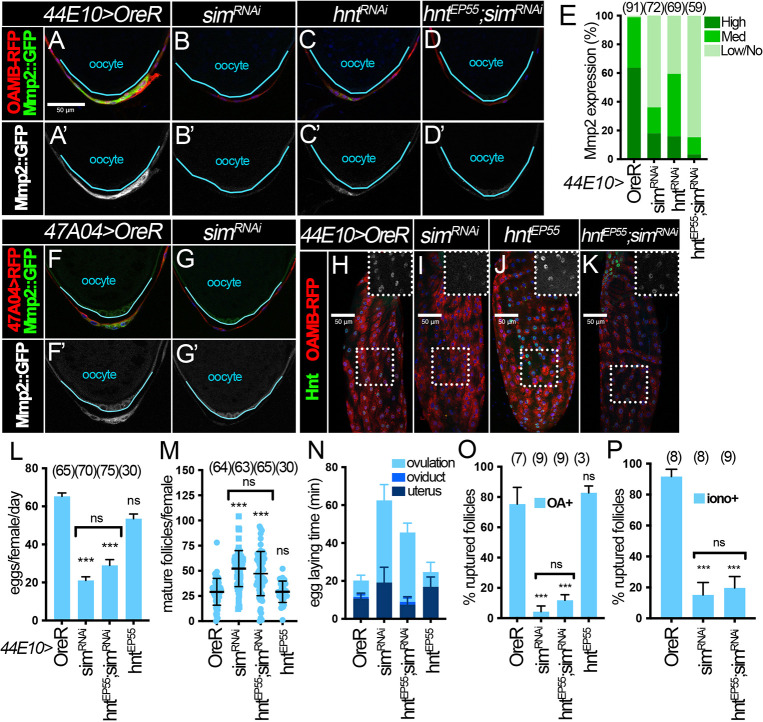
The fact that ionomycin failed to induce follicle rupture in sim-knockdown follicles (Fig. 4G-I and Fig. S5G-I) suggests that sim could regulate components downstream of Ca2+, such as Mmp2 and ROS signaling components. To test whether late Sim regulates Mmp2 expression in posterior follicle cells, we examined Mmp2::GFP reporter expression in sim-knockdown follicles. Owing to technical difficulties in recombining CG13083-Gal4 and Mmp2::GFP, we were unable to examine Mmp2::GFP expression in sim-knockdown follicles with CG13083-Gal4. Instead, we examined Mmp2::GFP expression in sim-knockdown follicles with 44E10-Gal4 or 47A04-Gal4. We found that Mmp2::GFP is significantly reduced in posterior follicle cells of late stage 14 follicles with sim knockdown relative to controls (Fig. 7A,B,E-G). Moreover, Mmp2 mRNA levels were also significantly reduced in sim-knockdown follicles (Fig. S6M,N). All these data indicate that Sim is required for Mmp2 expression.
Fig. 7.
Sim at stage 14 upregulates Mmp2 in mature follicles in conjunction with Hnt. (A-D′) Representative images show Mmp2::GFP expression (green in A-D and white in A′-D′) in posterior follicle cells of control (A), simRNAi (B), hntRNAi (C) and hntEP55;simRNAi (D) mature follicles with 44E10-Gal4 and OAMB-RFP. Oocytes are outlined in cyan. All images in A-D were acquired using the same microscope settings. Nuclei are marked by DAPI in blue in all panels. (E) Quantification of Mmp2::GFP expression levels in posterior follicle cells of unruptured late stage 14 follicles (characterized by the highest RFP expression, long dorsal appendages and absence of nurse cell nuclei). Posterior polar cell expression is not considered when quantifying Mmp2::GFP expression intensity. The number of mature follicles quantified is indicated above each bar. (F,G) Representative images show Mmp2::GFP expression (green in F,G and white in F′,G′) in control (F,F′) and simRNAi (G,G′) late stage 14 follicles marked by 47A04-Gal4>UAS-RFP (red). Oocytes are outlined in blue. All images from F and G were acquired using the same microscope settings. (H-K) Hnt expression (green) in late stage 14 follicles from control (H), simRNAi (I), hntEP55 (J), and hntEP55;simRNAi (K) females with 44E10-Gal4 and Oamb-RFP. Late stage 14 follicle cells are marked by the highest level of OAMB-RFP expression (red). The insets are higher magnifications of Hnt expression (white) in the outlined areas. All images in H-K were acquired using the same microscope settings. (L-N) Quantification of egg laying (L), mature follicle retention (M) and egg laying time (N) in control, simRNAi, hntEP55;simRNAi and hntEP55 females with 44E10-Gal4 and Oamb-RFP. The number of females is indicated above each bar. (O,P) Quantification of OA-induced follicle rupture (O) and ionomycin-induced follicle rupture (P) in control, simRNAi and hntEP55;simRNAi (O,P) and hntEP55 (P) females with 44E10-Gal4 and Oamb-RFP. The number of wells analyzed (containing 25-35 follicles) is indicated above each bar. ***P<0.001; ns, not significant [unpaired (two-sample equal variance) Student's t-test].Data are mean±s.d. (M,O,P) and mean±s.e.m. (L,N). Scale bars: 50 μm.
We also noticed that sim-knockdown follicles showed a more-severe Mmp2 reduction than hnt-knockdown follicles (Fig. 7C ,E), which prompted us to investigate whether Sim regulates Mmp2 expression via Hnt. We evaluated Mmp2::GFP expression in follicle cells with sim knockdown and hnt overexpression simultaneously, and found that these follicles showed similar (if not worse) Mmp2::GFP reduction to sim-knockdown follicles (Fig. 7D,E,H-K). In addition, hnt overexpression did not rescue the ovulation and follicle rupture defect caused by sim knockdown (Fig. 7L-P and Table S1), similar to the results with CG13083-Gal4 (Fig. 6G-I). Altogether, our results suggest that Sim regulates Mmp2 expression independently of and in conjunction with Hnt.
Sim is required for NOX expression independent of Hnt
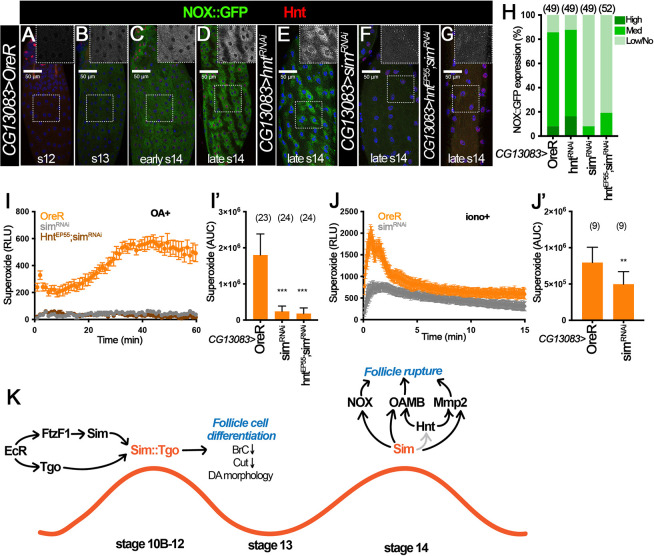
Next, we examined whether Sim regulates NOX expression in stage 14 follicles. Our previous work indicates that NOX is present in stage 14 follicle cells to produce ROS required for follicle rupture (Li et al., 2018). However, NOX expression throughout oogenesis and its regulation are completely unknown. To address this issue, we generated Nox::GFP knock-in allele by replacing NoxCRIMIC.TG4.0 allele with the Double Header GFP/T2A-Gal4 coding cassette (Li-Kroeger et al., 2018). NOX::GFP was not detected in follicle cells before stage 13 (Fig. 8A), and it was faintly detected in a few follicle cells at late stage 13 (Fig. 8B and Fig. S7A). Its expression started to upregulate at early stage 14 and peaked at late stage 14 (Fig. 8C,D and Fig. S7A). This upregulation of NOX::GFP is earlier than the upregulation of Hnt at stage 14 (Fig. 8C), suggesting that Hnt may not regulate NOX expression. Consistent with this idea, hnt knockdown with CG13083-Gal4 did not disrupt NOX::GFP expression in mature follicle cells (Fig. 8E,H). In contrast, NOX::GFP was drastically reduced in sim-knockdown follicles with CG13083-Gal4 (Fig. 8F,H). Consistent with the NOX::GFP protein expression defect, OA- and ionomycin-induced superoxide production were significantly reduced in sim-knockdown follicles (Fig. 8I,J). It is interesting that sim knockdown with 44E10-Gal4 or 47A04-Gal4 did not show a clear reduction of NOX::GFP expression (Fig. S7B-E) but showed the reduction of Nox mRNA (Fig. S6M,N). In addition, hnt overexpression did not restore NOX::GFP expression in sim-knockdown follicles with CG13083-Gal4 (Fig. 8G,H), nor did it restore Nox mRNA expression in sim-knockdown follicles with 47A04-Gal4 (Fig. S6N). All these data suggest that Sim upregulates NOX expression in stage 14 follicle cells independently of Hnt. In conclusion, we demonstrated that Sim functions like a master regulator in late follicles to regulate ovulatory competency by upregulating all known ovulatory genes, including OAMB, Mmp2 and NOX (Fig. 8K).
Fig. 8.
Sim is required for NOX expression independently of Hnt. (A-D) NOX::GFP (green, detected by anti-GFP antibody) and Hnt (red) in stage 12 (A), stage 13 (B), early stage 14 (C) and late stage 14 (D) follicles from NOX::GFP control females with CG13083-Gal4. The insets are higher magnifications of NOX::GFP expression (white) in the outlined areas. (E-G) NOX::GFP (green) and Hnt expression (red) in hntRNAi (E), simRNAi (F) and hntEP55;simRNAi (G) late stage 14 follicles with CG13083-Gal4. All images in A-G were acquired using the same microscope settings. Nuclei are marked by DAPI in blue. (H) Quantification of NOX::GFP expression levels in late stage 14 follicles (characterized by long dorsal appendages, lack of nurse cell nuclei and Hnt expression). The number of mature follicles quantified is indicated above each bar. (I,J) Measurement of OA-induced (I) and ionomycin-induced (J) superoxide production in stage 14 follicles from control (orange), simRNAi (gray) and hntEP55;simRNAi (brown) females with CG13083-Gal4 and Oamb-RFP. The superoxide is detected by L-012 luminescent signal (relative luminescence). (I′,J′) Quantification of the area under the curve representing superoxide production for each genotype and condition. The number of wells analyzed is indicated above each bar. **P<0.01 ***P<0.001 [unpaired (two-sample equal variance) Student's t-test]. Data are mean±s.d. (I′,J′) and mean±s.e.m. (I,J). (K) The role of Sim and Tgo in follicle differentiation and ovulation in late oogenesis in Drosophila. Downstream of ecdysteroid signaling, Sim and Tgo form a complex to promote follicle differentiation by downregulating Br-C and Cut, and regulating follicle morphology at stage 10B-12. Sim is re-upregulated at stage 14 and plays an active role in regulating the expression of Hnt, Mmp2, NOX and OAMB proteins required for OA-induced follicle rupture. Scale bars: 50 μm.
DISCUSSION
Development of a mature follicle is a long and complex process conserved across metazoans, and requires the orchestration of many signaling pathways between somatic follicle cells and germ cells to ensure their proper differentiation and maturation (Merkle et al., 2020; Doherty et al., 2022). A large body of work has focused on the formation and development of follicles during early stages (Duhart et al., 2017; Spradling et al., 2022), whereas less work has focused on the late follicle development and the maturation process that leads to their competency for ovulatory stimuli. Our previous work in Drosophila identified the nuclear receptor Ftz-f1, which plays a conserved role in follicle differentiation and maturation during late oogenesis downstream of ecdysteroid signaling (Knapp et al., 2020). Ftz-f1 directly activates the bHLH-PAS transcription factor Sim. However, the mechanism of Sim in follicle differentiation and maturation is unknown. This article contributes to filling this knowledge gap by uncovering how the bHLH-PAS protein complex Sim:Tgo functions within the Drosophila ovulatory network to regulate key genes required for the follicle maturation and follicle rupture pathways, which are conserved in pre-ovulatory mammalian follicles to ensure reproductive success.
We discovered that both Sim and Tgo have dynamic expression in follicle cells of late oogenesis. Ecdysteroid signaling at stage 10A/10B transition not only induces Ftz-f1 expression, which upregulates Sim (Knapp et al., 2020), but also upregulates Tgo expression (Fig. 8K), the heterodimeric partner of Sim (Ohshiro and Saigo, 1997; Sonnenfeld et al., 1997). The dimerization of Sim and Tgo leads to their nuclear localization, which is similar to the findings in other developmental systems. It is believed that the protein level of Tgo is typically static during embryogenesis, while the change of Sim expression determines their nuclear localization and transcriptional activity. In contrast, we clearly demonstrate that Tgo protein level is dynamic in follicle cells, which is an observation that has not been reported previously. Not only is Tgo upregulated at stage 10A, but it is also downregulated at stage 13, and appears to be re-upregulated and translocated into the nucleus at stage 14, which is concurrent with the re-upregulation of Sim (Fig. 1). It is unclear what signals induce this re-upregulation. Our previous work showed that ecdysone signaling is also activated at stage 14 and essential for follicle rupture and ovulation (Knapp and Sun, 2017). With the role of ecdysone signaling at stage 10A for Sim and Tgo upregulation, we tested whether the ecdysone signaling is also responsible for Sim or Tgo upregulation at stage 14. Unfortunately, knockdown of shade, the enzyme converting ecdysone to active 20-hydroyecdysone, in mature follicle cells with 44E10-Gal4 did not prevent either Tgo or Sim upregulation and nuclear localization at stage 14 (Fig. S8). More work is required to identify the signal for the re-upregulation of Sim and Tgo at stage 14.
Similar to many developmental systems, Sim and Tgo likely form a complex, translocate into the nucleus and promote follicle cell differentiation, including downregulating Br-C and Cut. Sim:Tgo represses gene expression by indirectly activating the repressive factors in CNS development (Estes et al., 2001). Therefore, we suspect that downregulation of Br-C and Cut by Sim:Tgo may also be indirect. This is also consistent with the fact that downregulation of Br-C and Cut is observed at stage 13, when Sim and Tgo are also downregulated (Fig. 1D). Future work will focus on identifying the molecular mechanism of this regulation.
The role of Sim:Tgo in follicle maturation does not seem to be limited to stages 10B to 12. With multiple stage 14 Gal4 drivers, we were able to disrupt late Sim expression (stage 14) without altering its early expression (stages 10B-12). Our data strongly suggests that late Sim acts as a master regulator of all known genes involved in follicle rupture and ovulation, including hnt, Nox, Oamb and Mmp2 (Fig. 8K). Unfortunately, we were unable to efficiently disrupt late Tgo expression using stage 14 Gal4 drivers (data not shown). Therefore, we were unable to provide conclusive evidence that Tgo also partners with Sim to regulate ovulatory gene expression in stage 14 follicle cells. However, the nuclear localization of Tgo in stage 14 follicle cells also depends on Sim (Fig. S2J), and Sim and Tgo form a complex to regulate gene expression in every studied tissue or cell type to date (Sonnenfeld et al., 1997; Estes et al., 2001). Consequently, we suspect that Tgo is still the partner of Sim in stage 14 follicle cells. It takes less than 10 h for follicles to develop from stages 10B to 14 (Spradling, 1993); therefore, we find it intriguing to see multiple genes turned on and off in this short period of time, which suggests that the regulation of follicle maturation is highly complex and dynamic. A comprehensive understanding of this process in flies and other model systems may allow us to identify genetic causes for ovulation-related infertility, including polycystic ovarian syndrome, and lead to better treatment development for these health problems.
We also identified, for the first time, a factor that regulates NOX expression in follicle cells. Our work showed that late Sim is crucial for upregulating Nox expression in follicle cells (Fig. 8A-H). It is unclear whether Nox is a direct target of Sim. Previous work in CNS midline development identified the Sim-binding motif as ACGTG (Wharton et al., 1994). Further work by Hong and his group also found that Sim:Tgo could bind to a slight variable motif DDRCGTG (Hong et al., 2013; Shin and Hong, 2015, 2016). Using the latter motif to search the Nox gene region, we found six DDRCGTG motifs (only one ACGTG motif) within 3 kb upstream of the Nox-RC isoform, which is the isoform expressed in follicle cells according to our RNAseq experiments (Knapp et al., 2020). In addition, there are 14 additional DDRCGTG motifs (only six ACGTG motifs) in the Nox gene region (Table S2). Therefore, Sim could potentially regulate Nox expression through direct binding to its enhancer. In addition to Nox, Sim may directly regulate Mmp2 and Oamb expression. There are three DDRCGTG motifs (one ACGTG motif) clustered within 350 bp in the Mmp2 enhancer that directs Mmp2 expression in follicle cells. There are two DDRCGTG motifs (none ACGTG motifs) clustered together in the Oamb-RFP enhancer. Our data show that Hnt does not seem to regulate the Oamb-RFP enhancer (Fig. 6C), despite the fact that it can regulate Oamb gene expression (Deady et al., 2017). This information suggests that Hnt and Sim likely bind to different enhancer regions of Oamb if both Hnt and Sim regulate Oamb expression directly. Future work will be required to decipher whether Sim directly regulates the expression of these genes. Moreover, it would be interesting to identify additional differentially up- or downregulated genes in sim-knockdown follicles using RNAseq, with the hope of finding other previously unreported genes that could play crucial roles in follicle maturation and ovulation that are unknown to date. These discoveries will aid in the process of understanding the precise transcription factor network that is spatiotemporally controlling follicle maturation and ovulation – key processes for reproductive success.
The role of Sim:Tgo in follicle maturation and ovulation may be conserved across metazoans. Both Ftz-f1 and its mammalian homolog SF-1 play an important role in promoting follicle development and maturation (Knapp et al., 2020; Pelusi et al., 2008). In addition, many of the ovulatory genes and signaling pathways downstream of Sim also play conserved roles in Drosophila and mammalian ovulation, including matrix metalloproteinases, the adrenergic system and ROS (Curry and Smith, 2006; Deady and Sun, 2015; Deady et al., 2015; Kannisto et al., 1985; Li et al., 2018; Shkolnik et al., 2011).
There are two mammalian sim homologs (sim1 and sim2) (Chrast et al., 1997; Ema et al., 1996; Kewley et al., 2004). These mammalian Sim proteins also require heterodimerization with their partner protein ARNT, which is homologous to Tgo (Ema et al., 1996; Michaud et al., 2000). Interestingly, both Drosophila and mammalian Sim (murine, specifically) have been shown to play important roles in CNS development (Michaud et al., 1998, 2000). Published RNAseq data evaluating human granulosa cells (Yerushalmi et al., 2014) have demonstrated that hSIM2 is induced after ovulation, whereas hSIM1 is not detected. Data reported in the Human Protein Atlas (Uhlén et al., 2015) also report that hSIM2 is expressed at medium levels in follicle cells, whereas hSIM1 is not expressed in the human ovary. Together, this information suggests that it is highly likely that Sim and its mammalian partner ARNT play a role in mammalian follicle maturation.
MATERIALS AND METHODS
Drosophila genetics
Drosophila melanogaster flies were reared on standard cornmeal-molasses food at 25°C unless otherwise noted. All RNAi-mediated depletion experiments included the UAS-dcr2 and were carried out at 29°C upon animal eclosion to increase knockdown efficiency. The following Gal4 drivers were used: 47A04-Gal4 and 44E10-Gal4 from the Janelia Gal4 collection (Pfeiffer et al., 2008), CG13083-Gal4 (Knapp et al., 2019), Vm26Aa-Gal4 (Peters et al., 2013) and C204-Gal4 (Manseau et al., 1997; Sun et al., 2008). The C204-Gal4 was the only Gal4 line that also contained tub-Gal80ts to avoid early developmental defects. To knock down or overexpress genes under the control of the above-mentioned Gal4 drivers, the following transgenic lines were used: UAS-simRNAi [Vienna Drosophila Resource Center (VDRC), stock 26888], UAS-tgoRNAi1 [Bloomington Drosophila Stock Center (BDSC), stock 53351], UAS-tgoRNAi2 (BDSC, stock 26740), UAS-tgo (BDSC, stock 9583), UAS-hntRNAi (VDRC, stock 3788), UAS-hntEP55 (BDSC, stock 5358), UAS-shdRNAi1 (VDRC, stock 108911), UAS-shdRNAi2 (VDRC, stock 17203) and UAS-EcRDN (BDSC, stock 6872). Isolation and identification of stage 14 follicles for follicle rupture assay were performed using Oamb-RFP (Knapp et al., 2019) or 47A04-Gal4 driving UAS-RFP (47A04>RFP) or UAS-RG6 (47A04>RG6; Jiang et al., 2021). The following protein trap lines were used: Mmp2::GFP (Deady et al., 2015), Sim::GFP (VDRC, stock 318096; (Sarov et al., 2016) and NOX::GFP. The Nox::GFP reporter is generated by replacing NoxCRIMIC.TG4.0 allele with the Double Header GFP/T2A-Gal4-coding cassette following the exact cross scheme of Li-Kroeger et al. (2018). For the CRISPR-Cas9 mediated disruption, Vm26Aa-Gal4 was used to drive tunable UAS-Cas9 constructs (XS, S and M; VDRC stocks 340000, 340001 and 340002, respectively; Port et al., 2020) and UAS-simsgRNA (VDRC, stock 341711). Control flies for all experiments were prepared by crossing Gal4 driver to Oregon-R or w1118 flies.
Clone induction and generation of mosaics
For generation of flip-out clones, the hsFLP;; act<CD2<Gal4, UAS-GFP/TM3 stock was crossed to the indicated transgenes of interest. For clone induction, adult female progeny with correct genotypes were heat shocked for 45 min at 37°C to induce FLP/FRT-mediated recombination and incubated at 25°C for 2-4 days before dissection. During the time post-heat shock, flies were mated and provided with wet yeast paste accordingly to induce the production of the desired follicle stages required for the experiment (Jia et al., 2016).
Ovulation and follicle rupture assays
The egg laying and mature follicle retention assays were conducted as described in previous literature, along with the egg laying time analysis (Deady and Sun, 2015; Knapp and Sun, 2017; Beard et al., 2023). In short, 5- to 6-day-old virgin females fed with wet yeast for 1 day were used to access the number of eggs laid over a 2-day period after mating with Oregon-R males, or to examine the location of eggs in the reproductive tract after 6-h mating with Oregon-R males. These data were then used to calculate the average number of eggs laid/female/day and the average time of ovulating an egg.
The OA- and ionomycin-induced follicle rupture assays were performed as previously described (Knapp et al., 2018) except for the following modification when using Vm26Aa-Gal4 driver. Owing to the loss of the Oamb-RFP reporter in sim-knockdown follicle cells, we were unable to use Oamb-RFP to isolate mature follicles and examine follicle rupture, as in previous work. Instead, we relied on high levels of OAMB-RFP in anterior follicle cells to identify the mature follicles and used the bright-field microscopy to ensure these follicles had an intact follicle-cell layer at the posterior (a transparent layer of follicle cells over the white yolk of the oocyte at 80× magnification). A couple of groups of 25-35 follicles isolated using this method were fixed and stained using DAPI in a master fixation buffer (4% EM-grade paraformaldehyde+0.1 µg/ml DAPI) to determine that these follicles indeed had an intact layer of follicle cells. The rest of the mature follicles in groups of 25-35 were incubated for 3 h with OA medium (Grace's Medium+10% FBS+100 U/ml penicillin-streptomycin+20 µM OA) (Knapp et al., 2018). After incubation, follicles were fixed and stained with DAPI in the master fixation buffer for 15 min. In all instances, washing was not performed after fixation and DAPI staining to avoid washing away the corpus luteum and disturbing the follicles. The follicles were immediately visualized and imaged using a Leica MZ10F fluorescence stereoscope, and the number of ruptured follicles was quantified according to DAPI signal (marking follicle cell nuclei). We counted the follicle as ruptured if more than 80% of the follicle lacked DAPI signals (indicating follicle cells were absent) and if the follicle had an accumulation of follicle cells expressing DAPI in the anterior region (indicating corpus luteum formation), similar to observations previously described (Knapp et al., 2018). Each data point represents the percentage (mean percentage±s.d.) of ruptured follicles per group of ∼25-35 follicles.
qRT-PCR
For quantitative RT-PCR, total RNA was extracted from 60 stage 14 follicles using TRIzol (Invitrogen) and the Direct-zol RNA MicroPrep Kit (Zymo Research). Fifteen 5- to 6-day-old virgin flies were fed wet yeast 3 days before dissection. Mature follicles were isolated using the Oamb-RFP reporter or the 47A04>RFP. cDNA synthesis and real-time PCR amplification with three technical repeats were performed as previously described (Knapp and Sun, 2017; Li et al., 2018). Primers for oamb.K3, mmp2 and nox have been described previously (Knapp and Sun, 2017; Li et al., 2018). The hnt primer pair used was 5′-ACATCCGGTGCCACAATTAC-3′ and 5′-GTGAACGTCAGGTGGCAGTAG-3′. The Rps17 primer was used as an internal control. The data are presented as mean±s.d. from at least two biological replicates, to ensure reproducibility.
Superoxide detection
Measurement of superoxide production was performed as previously described (Li et al., 2018) with slight modifications. Five- to 6-day-old virgin flies were fed wet yeast 3 days before dissection. Five intact mature follicles were isolated using OAMB-RFP and bright-field microscopy and placed in each well of a 96-well plate injected with 100 µl of Grace's insect medium containing 200 µM of L-012 (Wako Chemicals) and either 20 µM OA or 2 µM ionomycin. For OA-induced ROS detection, plates were placed in a CLARIOstar microplate reader (BMG Labtech) for luminescence reading for 60 min (60 cycles, 63 s each, measurement interval time of 0.5 s). For ionomycin-induced ROS detection, luminescence reading lasted 15 min (300 cycles, 3 s each, measurement interval time of 0.5 s, which was enough to catch the fast initial peak. A minimum of three wells (technical repeats) were used in each experiment per genotype, and the mean±s.e.m. of the technical repeats was calculated. Each experiment was performed at least twice.
Immunostaining and microscopy
Adult female ovaries were dissected in cold Grace's medium and fixed in 4% EM-grade paraformaldehyde in PBT (1×PBS+0.2% Triton X-100) for 13-15 min at room temperature. The tissue was washed vigorously with PBT, blocked in antibody buffer (PBT+0.5% BSA+2% normal goat serum) and stained using primary antibodies overnight at 4°C. After incubation, tissue was washed with PBT and incubated with secondary antibodies for 2 h at room temperature. After this incubation, tissue was washed with PBT and stained with DAPI (final concentration 0.5 μg/ml) for 15 min. The following primary antibodies were used: mouse anti-Hnt (1G9, 1:75), mouse anti-Tgo (1:15), mouse anti-Cut (2B10, 1:15) and mouse anti-Br-C (25E9.D7, 1:15) from Developmental Study Hybridoma bank; rabbit anti-GFP (1:4000; Invitrogen, #A11122), rabbit anti-RFP (1:2000, MBL International, #PM005 lot 040) and guinea pig anti-Sim (1:1000; a gift from Dr Stephen Crews, University of North Carolina at Chapel Hill School of Medicine, NC, USA). Alexa Fluor 488 and Alexa Fluor 568 goat secondary antibodies (1:1000; Invitrogen, Alexa Fluor 568 rabbit #A11011, Alexa Fluor 568 mouse #A11004, Alexa Fluor 568 guinea pig #A11075, Alexa Fluor 488 rabbit #A11008, Alexa Fluor 488 mouse #A11001, Alexa Fluor 488 guinea pig #A11073) were used. Fluorescent and DIC images were acquired using a Leica TCS SP8 confocal microscope and assembled using Adobe Photoshop software and ImageJ/Fiji software (can be downloaded from https://fiji.sc/).
Mature follicles with egg morphology defects were imaged using Leica DMi8 microscope to quantify anterior-posterior and dorso-ventral axis length (microns) using ImageJ/Fiji software. The average AP/DV ratio for each genotype was calculated using Excel by dividing the AP length by the DV length of each follicle and subsequently calculating the mean±s.d. of all individual AP/DV values.
For Tgo expression quantification at stages 10-12, the same microscopic settings were used to image four to six follicles at each stage and analysis was performed using Fiji/ImageJ software. For each follicle at a particular stage, the total number of mainbody follicle cells was quantified using the DAPI channel. Areas where follicle cells were overlapping (such as in the oocyte-nurse cell junction) and follicle edges were avoided during the quantification to avoid introducing error. The total number of follicle cells expressing Tgo (in the green channel) were then quantified, making sure we were assessing the same main body follicle region. The percentage of follicle cells expressing Tgo was calculated per follicle at each stage for each genotype and subsequently the mean±s.d. of all individual values were calculated.
For the NOX::GFP expression quantification, the same microscopic settings were used, and analysis was performed using Fiji/ImageJ software. After subtraction of background using a rolling ball radius of 25 pixels for an 8-bit image, each individual follicle cell was contoured and the NOX::GFP intensity was measured. Subsequently, the mean signal intensity of each follicle cell was used to calculate the mean±s.d. at each stage.
Statistical analysis
Statistical tests were carried out using Excel primarily and in some instances Prism 7 (GraphPad). Quantification results are displayed as mean±s.d. or mean±s.e.m., as indicated in figure legends. Statistical analysis between two groups was conducted using a two-tailed Student’s t-test. For the egg distribution assay used to calculate egg-laying time, a Chi-square analysis was performed to assess significance.
Supplementary Material
Acknowledgements
We thank Drs Stephen Crews, Wu-Min Deng, Allan Spradling, Celeste Berg, Steven Henikoff, Gerald Rubin, Shinya Yamamoto, Takano-Shimizu, Ken Wan and Liria Masuda-Nakagawa for sharing fly lines and reagents; Bloomington Drosophila Stock Center and Vienna Drosophila Resource Center for fly stocks; and Developmental Studies Hybridoma Bank for antibodies. We thank Drs Wei Li and Yuping Huang in Dr Sun's laboratory for technical support. We appreciate constructive comments from anonymous reviewers. The Leica SP8 confocal microscope is supported by a National Institutes of Health Award (S10OD016435) to Akiko Nishiyama.
Footnotes
Author contributions
Conceptualization: J.S.; Methodology: R.O., E.M.K., B.Z., J.S.; Formal analysis: R.O., J.S.; Investigation: R.O., E.M.K., B.Z., J.S.; Data curation: R.O., E.M.K., B.Z.; Writing - original draft: R.O., J.S.; Writing - review & editing: R.O., E.M.K., B.Z., J.S.; Visualization: R.O., J.S.; Supervision: J.S.; Project administration: J.S.; Funding acquisition: J.S.
Funding
J.S. is supported by a University of Connecticut Start-Up Fund, and by National Institutes of Health/National Institute of Child Health and Human Development grants (R01-HD086175 and R01-HD097206). Deposited in PMC for release after 12 months.
Data availability
All relevant data can be found within the article and its supplementary information.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.201566.reviewer-comments.pdf.
References
- Beard, A., Oramas, R. and Sun, J. (2023). Assessing ovulation in Drosophila melanogaster. In Drosophila Oogenesis: Methods and Protocols (ed. Giedt M. S. and Tootle T. L.), pp. 253-276. New York, NY: Springer US. [DOI] [PubMed] [Google Scholar]
- Chandra, A., Copen, C. E. and Stephen, E. H. (2014). Infertility Service Use in the United States: Data from the National Survey of Family Growth, 1982-2010. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics. [PubMed] [Google Scholar]
- Chrast, R., Scott, H. S., Chen, H., Kudoh, J., Rossier, C., Minoshima, S., Wang, Y., Shimizu, N. and Antonarakis, S. E. (1997). Cloning of two human homologs of the Drosophila single-minded Gene SIM1 on Chromosome 6q and SIM2 on 21q within the Down syndrome chromosomal region. Genome Res. 7, 615-624. 10.1101/gr.7.6.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S. H., Carney, G. E., Mcclung, C. A., Willard, S. S., Taylor, B. J. and Hirsh, J. (2005). Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. J. Biol. Chem. 280, 14948-14955. 10.1074/jbc.M414197200 [DOI] [PubMed] [Google Scholar]
- Crews, S. T. and Fan, C.-M. (1999). Remembrance of things PAS: regulation of development by bHLH–PAS proteins. Curr. Opin. Genet. Dev. 9, 580-587. 10.1016/S0959-437X(99)00003-9 [DOI] [PubMed] [Google Scholar]
- Curry, T. E. and Smith, M. F. (2006). Impact of extracellular matrix remodeling on ovulation and the folliculo-luteal transition. Semin. Reprod. Med. 24, 228-241. 10.1055/s-2006-948552 [DOI] [PubMed] [Google Scholar]
- Deady, L. D. and Sun, J. (2015). A follicle rupture assay reveals an essential role for follicular adrenergic signaling in Drosophila ovulation. PLoS Genet. 11, e1005604. 10.1371/journal.pgen.1005604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deady, L. D., Shen, W., Mosure, S. A., Spradling, A. C. and Sun, J. (2015). Matrix metalloproteinase 2 is required for ovulation and corpus luteum formation in Drosophila. PLoS Genet. 11, e1004989. 10.1371/journal.pgen.1004989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deady, L. D., Li, W. and Sun, J. (2017). The zinc-finger transcription factor Hindsight regulates ovulation competency of Drosophila follicles. eLife 6, e29887. 10.7554/eLife.29887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, C. A., Amargant, F., Shvartsman, S. Y., Duncan, F. E. and Gavis, E. R. (2022). Bidirectional communication in oogenesis: a dynamic conversation in mice and Drosophila. Trends Cell Biol. 32, 311-323. 10.1016/j.tcb.2021.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, D. M., Ko, C. M., Jo, M., Brannstrom, M. and Curry, T. E. (2019). Ovulation: parallels with inflammatory processes. Endocr. Rev. 40, 369-416. 10.1210/er.2018-00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhart, J. C., Parsons, T. T. and Raftery, L. A. (2017). The repertoire of epithelial morphogenesis on display: Progressive elaboration of Drosophila egg structure. Mech. Dev. 148, 18-39. 10.1016/j.mod.2017.04.002 [DOI] [PubMed] [Google Scholar]
- Ema, M., Morita, M., Ikawa, S., Tanaka, M., Matsuda, Y., Gotoh, O., Saijoh, Y., Fujii, H., Hamada, H., Kikuchi, Y.et al. (1996). Two new members of the murine Sim gene family are transcriptional repressors and show different expression patterns during mouse embryogenesis. Mol. Cell. Biol. 16, 5865-5875. 10.1128/MCB.16.10.5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons, R. B., Duncan, D., Estes, P. A., Kiefel, P., Mosher, J. T., Sonnenfeld, M., Ward, M. P., Duncan, I. and Crews, S. T. (1999). The spineless-aristapedia and tango bHLH-PAS proteins interact to control antennal and tarsal development in Drosophila. Development 126, 3937-3945. 10.1242/dev.126.17.3937 [DOI] [PubMed] [Google Scholar]
- Estes, P., Mosher, J. and Crews, S. T. (2001). Drosophila single-minded represses gene transcription by activating the expression of repressive factors. Dev. Biol. 232, 157-175. 10.1006/dbio.2001.0174 [DOI] [PubMed] [Google Scholar]
- Freer, S. M., Lau, D. C., Pearson, J. C., Talsky, K. B. and Crews, S. T. (2011). Molecular and functional analysis of drosophila single-minded larval central brain expression. Gene Expr. Patterns 11, 533-546. 10.1016/j.gep.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health (Uk), N. C. C. for W. and C. (2013). Investigation of Fertility Problems and Management Strategies. Royal College of Obstetricians & Gynaecologists. [Google Scholar]
- Hong, J.-W., Park, K. W. and Levine, M. S. (2013). Temporal regulation of single-minded target genes in the ventral midline of the Drosophila central nervous system. Dev. Biol. 380, 335-343. 10.1016/j.ydbio.2013.05.015 [DOI] [PubMed] [Google Scholar]
- Jia, D., Xu, Q., Xie, Q., Mio, W. and Deng, W.-M. (2016). Automatic stage identification of Drosophila egg chamber based on DAPI images. Sci. Rep. 6, 18850. 10.1038/srep18850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. and Crews, S. T. (2003). The Drosophila dysfusion Basic Helix-Loop-Helix (bHLH)-PAS Gene Controls Tracheal Fusion and Levels of the Trachealess bHLH-PAS Protein. Mol. Cell. Biol. 23, 5625-5637. 10.1128/MCB.23.16.5625-5637.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, K., Zhang, J., Huang, Y., Wang, Y., Xiao, S., Hadden, M. K., Woodruff, T. K. and Sun, J. (2021). A platform utilizing Drosophila ovulation for nonhormonal contraceptive screening. Proc. Natl. Acad. Sci. USA 118, e2026403118. 10.1073/pnas.2026403118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannisto, P., Owman, C. and Walles, B. (1985). Involvement of local adrenergic receptors in the process of ovulation in gonadotrophin-primed immature rats. J. Reprod. Fertil. 75, 357-362. 10.1530/jrf.0.0750357 [DOI] [PubMed] [Google Scholar]
- Kewley, R. J., Whitelaw, M. L. and Chapman-Smith, A. (2004). The mammalian basic helix–loop–helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 36, 189-204. 10.1016/S1357-2725(03)00211-5 [DOI] [PubMed] [Google Scholar]
- Klusza, S. and Deng, W.-M. (2011). At the crossroads of differentiation and proliferation: precise control of cell-cycle changes by multiple signaling pathways in Drosophila follicle cells. BioEssays News Rev. Mol. Cell. Dev. Biol. 33, 124-134. 10.1002/bies.201000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, E. and Sun, J. (2017). Steroid signaling in mature follicles is important for Drosophila ovulation. Proc. Natl. Acad. Sci. USA 114, 699-704. 10.1073/pnas.1614383114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, E. M., Deady, L. D. and Sun, J. (2018). Ex vivo follicle rupture and in situ zymography in Drosophila. Bio-Protoc. 8, e2846. 10.21769/BioProtoc.2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, E. M., Li, W. and Sun, J. (2019). Downregulation of homeodomain protein Cut is essential for Drosophila follicle maturation and ovulation. Development 146, dev179002. 10.1242/dev.179002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, E. M., Li, W., Singh, V. and Sun, J. (2020). Nuclear receptor Ftz-f1 promotes follicle maturation and ovulation partly via bHLH/PAS transcription factor Sim. eLife 9, e54568. 10.7554/eLife.54568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Young, J. F. and Sun, J. (2018). NADPH oxidase-generated reactive oxygen species in mature follicles are essential for Drosophila ovulation. Proc. Natl. Acad. Sci. USA 115, 7765-7770. 10.1073/pnas.1800115115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Kroeger, D., Kanca, O., Lee, P.-T., Cowan, S., Lee, M. T., Jaiswal, M., Salazar, J. L., He, Y., Zuo, Z. and Bellen, H. J. (2018). An expanded toolkit for gene tagging based on MiMIC and scarless CRISPR tagging in Drosophila. eLife 7, e38709. 10.7554/eLife.38709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, R., Hozumi, S., Taniguchi, K., Sasamura, T., Murakami, R. and Matsuno, K. (2007). Roles of single-minded in the left–right asymmetric development of the Drosophila embryonic gut. Mech. Dev. 124, 204-217. 10.1016/j.mod.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Manseau, L., Baradaran, A., Brower, D., Budhu, A., Elefant, F., Phan, H., Philp, A. V., Yang, M., Glover, D., Kaiser, K.et al. (1997). GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of drosophila. Dev. Dyn. 209, 310-322. [DOI] [PubMed] [Google Scholar]
- Merkle, J. A., Wittes, J. and Schüpbach, T. (2020). Signaling between somatic follicle cells and the germline patterns the egg and embryo of Drosophila. Curr. Top. Dev. Biol. 140, 55-86. 10.1016/bs.ctdb.2019.10.004 [DOI] [PubMed] [Google Scholar]
- Michaud, J. L., Rosenquist, T., May, N. R. and Fan, C.-M. (1998). Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 12, 3264-3275. 10.1101/gad.12.20.3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud, J. L., Derossi, C., May, N. R., Holdener, B. C. and Fan, C.-M. (2000). ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech. Dev. 90, 253-261. 10.1016/S0925-4773(99)00328-7 [DOI] [PubMed] [Google Scholar]
- Middleton, C. A., Nongthomba, U., Parry, K., Sweeney, S. T., Sparrow, J. C. and Elliott, C. J. H. (2006). Neuromuscular organization and aminergic modulation of contractions in the Drosophila ovary. BMC Biol. 4, 17. 10.1186/1741-7007-4-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastirioti, M., Charles, E., Linn, C. E., Jr and White, K. (1996). Characterization of Drosophila Tyramine β-hydroxylasegene and isolation of mutant flies lacking octopamine. J. Neurosci. 16, 3900-3911. 10.1523/JNEUROSCI.16-12-03900.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu, J. R., Franks, R. G., Hu, S. and Crews, S. T. (1990). The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell 63, 63-75. 10.1016/0092-8674(90)90288-P [DOI] [PubMed] [Google Scholar]
- Nambu, J. R., Lewis, J. O., Wharton, K. A., Jr and Crews, S. T. (1991). The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67, 1157-1167. 10.1016/0092-8674(91)90292-7 [DOI] [PubMed] [Google Scholar]
- Ohshiro, T. and Saigo, K. (1997). Transcriptional regulation of breathless FGF receptor gene by binding of TRACHEALESS/dARNT heterodimers to three central midline elements in Drosophila developing trachea. Development 124, 3975-3986. 10.1242/dev.124.20.3975 [DOI] [PubMed] [Google Scholar]
- Peters, N. C., Thayer, N. H., Kerr, S. A., Tompa, M. and Berg, C. A. (2013). Following the “tracks”: Tramtrack69 regulates epithelial tube expansion in the Drosophila ovary through Paxillin, Dynamin, and the homeobox protein Mirror. Dev. Biol. 378, 154-169. 10.1016/j.ydbio.2013.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelusi, C., Ikeda, Y., Zubair, M. and Parker, K. L. (2008). Impaired follicle development and infertility in female mice lacking steroidogenic factor 1 in ovarian granulosa cells. Biol. Reprod. 79, 1074-1083. 10.1095/biolreprod.108.069435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, B. D., Jenett, A., Hammonds, A. S., Ngo, T.-T. B., Misra, S., Murphy, C., Scully, A., Carlson, J. W., Wan, K. H., Laverty, T. R.et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715-9720. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage, J., Steffes, G., Lau, D. C., Parente, B. A., Crews, S. T., Strauss, R. and Klämbt, C. (2002). Novel behavioral and developmental defects associated with Drosophila single-minded. Dev. Biol. 249, 283-299. 10.1006/dbio.2002.0770 [DOI] [PubMed] [Google Scholar]
- Port, F., Strein, C., Stricker, M., Rauscher, B., Heigwer, F., Zhou, J., Beyersdörffer, C., Frei, J., Hess, A., Kern, K.et al. (2020). A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila. eLife 9, e53865. 10.7554/eLife.53865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, J. A. S. and Ascoli, M. (2018). Endocrine, paracrine, and autocrine signaling pathways that regulate ovulation. Trends Endocrinol. Metab. 29, 313-325. 10.1016/j.tem.2018.02.012 [DOI] [PubMed] [Google Scholar]
- Robker, R. L., Hennebold, J. D. and Russell, D. L. (2018). Coordination of ovulation and oocyte maturation: a good egg at the right time. Endocrinology 159, 3209-3218. 10.1210/en.2018-00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov, M., Barz, C., Jambor, H., Hein, M. Y., Schmied, C., Suchold, D., Stender, B., Janosch, S., Kj, V. V., Krishnan, R. T.et al. (2016). A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 5, e12068. 10.7554/eLife.12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, D.-H. and Hong, J.-W. (2015). Midline enhancer activity of the short gastrulation shadow enhancer is characterized by three unusual features for cis-regulatory DNA. BMB Rep. 48, 589-594. 10.5483/BMBRep.2015.48.10.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, D.-H. and Hong, J.-W. (2016). Transcriptional activity of the short gastrulation primary enhancer in the ventral midline requires its early activity in the presumptive neurogenic ectoderm. BMB Rep. 49, 572-577. 10.5483/BMBRep.2016.49.10.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik, K., Tadmor, A., Ben-Dor, S., Nevo, N., Galiani, D. and Dekel, N. (2011). Reactive oxygen species are indispensable in ovulation. Proc. Natl. Acad. Sci. USA 108, 1462-1467. 10.1073/pnas.1017213108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenfeld, M., Ward, M., Nystrom, G., Mosher, J., Stahl, S. and Crews, S. (1997). The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development 124, 4571-4582. 10.1242/dev.124.22.4571 [DOI] [PubMed] [Google Scholar]
- Spradling, A. C. (1993). Developmental Genetics of Oogenesis. Drosophila Melanogaster (ed. Bate M. and Martinez-Arias A.). Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Spradling, A. C., Niu, W., Yin, Q., Pathak, M. and Maurya, B. (2022). Conservation of oocyte development in germline cysts from Drosophila to mouse. eLife 11, e83230. 10.7554/eLife.83230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. and Spradling, A. C. (2013). Ovulation in Drosophila is controlled by secretory cells of the female reproductive tract. eLife 2, e00415. 10.7554/eLife.00415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., Smith, L., Armento, A. and Deng, W.-M. (2008). Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J. Cell Biol. 182, 885-896. 10.1083/jcb.200802084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokmakov, A. A., Stefanov, V. E. and Sato, K.-I. (2020). Dissection of the ovulatory process using ex vivo approaches. Front. Cell Dev. Biol. 8, 605379. 10.3389/fcell.2020.605379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., Sivertsson, Å., Kampf, C., Sjöstedt, E., Asplund, A.et al. (2015). Tissue-based map of the human proteome. Science 347, 1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Umetsu, D., Murakami, S., Sato, M. and Tabata, T. (2006). The highly ordered assembly of retinal axons and their synaptic partners is regulated by Hedgehog/Single-minded in the Drosophila visual system. Development 133, 791-800. 10.1242/dev.02253 [DOI] [PubMed] [Google Scholar]
- Ward, M. P., Mosher, J. T. and Crews, S. T. (1998). Regulation of bHLH-PAS protein subcellular localization during Drosophila embryogenesis. Development 125, 1599-1608. 10.1242/dev.125.9.1599 [DOI] [PubMed] [Google Scholar]
- Wharton, K. A., Franks, R. G., Kasai, Y. and Crews, S. T. (1994). Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development 120, 3563-3569. 10.1242/dev.120.12.3563 [DOI] [PubMed] [Google Scholar]
- Yerushalmi, G. M., Salmon-Divon, M., Yung, Y., Maman, E., Kedem, A., Ophir, L., Elemento, O., Coticchio, G., Dal Canto, M., Mignini Renzinu, M.et al. (2014). Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol. Hum. Reprod. 20, 719-735. 10.1093/molehr/gau031 [DOI] [PubMed] [Google Scholar]
- Zelzer, E., Wappner, P. and Shilo, B.-Z. (1997). The PAS domain confers target gene specificity of Drosophila bHLH/PAS proteins. Genes Dev. 11, 2079-2089. 10.1101/gad.11.16.2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.