Abstract
Using a combination of several approaches we estimated and characterized a total of 314 regulatory DNA-binding proteins in Escherichia coli, which might represent its minimal set of transcription factors. The collection is comprised of 35% activators, 43% repressors and 22% dual regulators. Within many regulatory protein families, the members are homogeneous in their regulatory roles, physiology of regulated genes, regulatory function, length and genome position, showing that these families have evolved homogeneously in prokaryotes, particularly in E.coli. This work describes a full characterization of the repertoire of regulatory interactions in a whole living cell. This repertoire should contribute to the interpretation of global gene expression profiles in both prokaryotes and eukaryotes.
INTRODUCTION
The large number of sequenced bacterial genomes with their relative simplicity will play an important role in understanding the prospects and limits in the interpretation and analysis of the human genome. Escherichia coli has a particular place within the bacterial genomes, given the legacy of experimental knowledge in molecular biology that it encompasses (1,2). This large amount of information has to be gathered and organized in accessible ways for its analysis. Our laboratory has been engaged in gathering information on transcriptional regulation and operon organization in E.coli and organizing it into a database, RegulonDB (3,4). This database supports global studies of transcriptional regulation, such as the prediction of promoters, regulatory sites, and operon organization in the complete genome, as well as preliminary observations on the architecture and connectivity of the regulatory network in E.coli (5,6). It has been shown that in addition to sequence comparisons, other sources of information are relevant for the understanding of the regulation of gene expression, such as the relative position of motifs in the upstream DNA regions of transcriptional regulation (7,8) as well as the relative position of helix–turn–helix (HTH) motifs within the protein sequence (9).
In order to characterize and define the set of regulatory DNA-binding proteins in the cell, we applied two informative sources to provide an estimate of the complete set of transcription regulators in E.coli. First, we exhaustively collected information from the literature and, second, we exhaustively searched the E.coli genome by computational methods and completed the putative set. Finally, we analyzed this set of regulatory proteins in terms of their structural and functional properties. The collection we present here contributes to the organized knowledge available on E.coli and should represent a relevant reference collection for the analysis of global gene expression profiles.
MATERIALS AND METHODS
Detection and prediction of transcription factors in E.coli
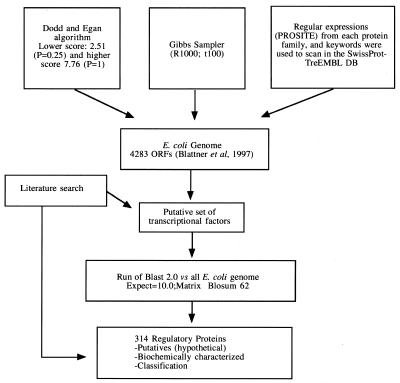
Prediction of protein function using computational tools are becoming more important as the gap between the increasing number of sequences and experimental characterization of the respective proteins widens (10). One problem with the current predictive methods is that they usually fail to detect all members of a protein/gene family that carry out a given function. For this reason, the general strategy to detect all transcriptional factors schematized in Figure 1 involved several methods, including the use of regular expressions and profile-based algorithms. The sequential order of the complete search was as follows.
Figure 1.
Prediction of regulatory proteins in the E.coli genome. The method is divided into two stages. In the first stage, a set of putative transcription factors is generated. This results from searches in the SwissProt database (keywords and regular expressions were used) and from scanning all 4283 ORFs of the E.coli genome using Dodd and Egan, PROSITE patterns and Gibbs sampler algorithms. In addition, a literature search was performed for evidence concerning new transcription factors. We filtered proteins that are not transcription regulators (a transcription factor has a DNA-binding functional motif). In the second part, a sequence comparison was performed using Blast 2.0 to detect additional proteins. Additionally, annotations of the E.coli genome were used together with the collection in RegulonDB (3,32). The DNA-binding protein structures used in the Gibbs sampler calibration were: ArgR (58), BirA (59,60), Crp (61), Fis (62), FruR (63), LacI (64), LexA (65), NarL (66), OmpR (67), PurR (68), TetR (69,70) and TrpR (71).
• Regulatory proteins were searched for in the weekly updated SwissProt Data Base v.34.0 (11,12) and in the E.coli genome (5). The search was done using several keywords, as well as patterns derived from PROSITE (6). We removed 12 proteins that are architecturally similar to regulators but lack a DNA-binding domain (DBD), as previously reported (14). This leaves 159 proteins, out of which 154 are either part of the known set or are recognized by a previous method, leaving only five new candidates. Literature information generated in this search was used to find additional members and also to complete different properties of previously gathered regulators.
• It is known that most of the prokaryotic regulatory proteins recognize DNA operator sequences using a HTH motif (15). The weight matrix designed for HTH recognition by Dodd and Egan (16,17) was used to scan the whole E.coli genome. This produced 276 proteins, out of which we manually excluded 88 for several reasons (transposons, insertion sequences and β-galactosidase), finally keeping 188 proteins. Of these, 159 were detected by another method, whereas 29 are new putative regulatory proteins. One must be aware that this method fails to detect an HTH motif in typical regulators such as the TrpR repressor, due to the presence of an Ile at position 12. In parallel, we ran a Gibbs sampler search (18) using a weight matrix built with known HTH proteins. In order to calibrate the method, several tests were made with a set of 15 known 3D structures from E.coli and phage proteins. The best result with this training set was obtained running 1000 independent searches 100 times, with no gaps allowed, corresponding to the HTH motif. This pattern reflects the arrangement of the secondary structure as computed by the DSSP program (19). With this method 131 proteins were detected, 123 of which are known, generating only eight new candidates.
• From the 244 regulators annotated in the E.coli genome (5), 57 proteins were excluded since they either lack a DBD, are enzymes or the literature indicates that they are not DNA-binding regulatory proteins. An example is the d-xylose-binding periplasmic protein XylF, which is known not to be a transcription regulator (20).
• We re-analyzed the initial blast scan (using the default parameters and Blosum 62 matrix) (21) and added a few more proteins showing a sequence identity >25% with any known regulator. Identity values <25% in fragments of at least 100 amino acid residues were not considered significant. This empirical calibration is widely used, since sequence similarity above this threshold implies structural and functional similarity (22). In this way, a few more proteins with a DBD different from the HTH were added. Examples include the Cold Shock Domain (CSD), β-strand, HLH and zinc finger domain. This also helped to eliminate proteins with an architecture similar to regulators (i.e. sugar transporters such as MgbL, XylF and RbsB).
• During all of this process we continually searched the literature and completed a total of 159 transcription regulators supported by either 3D structure, mutation in the regulatory gene, or mutation in the DNA-binding site.
Briefly, the criteria to accept a regulatory protein were:
(i) the presence of a known DBD, preferably HTH;
(ii) proteins for which there is experimental evidence as transcription regulators and that are recognized by any of the methods used;
(iii) proteins detected by any two methods out of PROSITE, Blattner annotation, Dodd and Egan weight matrix or Gibbs sampler;
(iv) proteins detected by only one method that share sequence similarity to a known regulator;
(v) proteins with at least 25% identity to a known transcription regulator, especially when the matching segment includes the HTH region.
The final set after searching, filtering and selecting by these automated methods and by manual inspection groups 314 proteins, of which 159 have experimental support and 155 are predicted regulatory proteins. Based on the set of known regulators we estimated 65–67% true positives and 9–15% false positives with the different methods used (see Table 1).
Table 1. Evaluation of the dataset.
| Method | True (%) | False positive (%) |
|---|---|---|
| Dodd and Egan | 65.0 | 15.0 |
| Gibbs | 50.7 | 30.0 |
| PROSITE | 67.9 | 9.3 |
| Blattner annotations | 97.6 | 21.0 |
We used the dataset of 159 experimentally supported proteins to evaluate the performance of the methods. The evaluations of the Dodd and Egan and Gibbs methods were done by comparing with the set of 128 proteins that have a reported HTH motif.
A fold recognition (23) was performed for all DNA-binding transcription factors as an independent method to evaluate these predictions, showing that ~83% of the known regulators and 77% of the predicted regulators show a DBD fold.
All DNA-binding transcription factors were grouped into families based on information available in the literature, as well as on sequence comparisons (ClustalW, using default parameters; 25). Several families have been proposed previously, such as the enhancer binding protein (EBP), the LuxR/UhpA and the OmpR families (25,26) (many of them are involved in two component systems), as well as the GalR/LacI (20), the LysR (27,28), AraC/XylS (29), the ArsR (30) and the CRP (31) families. The criterion to define a family is based on sequence comparison. If a protein shares at least 25% identity in its complete sequence or within the DBD with any member of a family, then it is considered part of that family.
RESULTS
We will first present an overview of the transcription factors, emphasizing their structural and functional properties, such as their DNA-binding motif and their regulatory roles. Then we will describe the regulators distributed amongst their evolutionary families and discuss some functional correlations.
The repertoire of DNA-binding transcription regulators
To fully understand gene regulation it is necessary to study it in the context of cellular processes such as cell division, differentiation and responses to several environmental changes. The purpose of this work was to analyze the organization of 314 (known and predicted) E.coli transcription regulators in terms of both structural and physiological properties. It should be clear that all computational predictions are preliminary, awaiting experimental confirmation. This repertoire has been incorporated in a database on transcriptional regulation and operon organization, RegulonDB v.3.0 (4), accessible through the web at http://www.cifn.unam.mx/Computational_Biology/regulondb/ . The database indicates whether the proteins are known transcription factors or are predicted as such and describes for each protein the relative position of the HTH, its function in regulation, sequence, references and family membership.
An important question requiring an answer is whether the data set of 314 regulators contains the total number of DNA-binding regulatory proteins in E.coli. Based on the estimated percentage of false positives of the methods used, the minimum would be of the order of 100 predicted proteins (65% of 155), adding to the 159 already characterized. This eliminates ~20% of the estimated 314 proteins. On the other hand, we know that several methods fail to detect all true positives. In the case of profile-based algorithms, 30% can be missed (based on a comparison with a pattern search or sequence comparisons). This gives an upper limit of around 350 regulatory proteins, adding 8% to the set (see Table 1).
Previous rough estimates pointed to around 400 regulatory genes in the E.coli genome (6), assuming a 1:10 ratio of regulatory to regulated genes, and 10% constitutively expressed genes. Current information describes 933 genes grouped in 361 transcriptional units (32) and subject to regulation by 78 different transcription factors. These numbers give a 1:12 ratio of regulatory to regulated genes. Assuming that this set corresponds to 25% of all regulatory genes, since they regulate 25% of the total set of genes in E.coli, surprisingly we obtain the number of 78 × 4 = 312 DNA-binding transcription regulators in the E.coli genome, corresponding very closely to the proposed set of 314 regulators based on sequence analysis. The precise matching of these numbers may be mere coincidence. Overall, based on this information and on the sensitivity of the methods, we consider that the universe of DNA-binding regulatory proteins in E.coli contains of the order of 300–350 proteins.
The characterized regulatory proteins have a diversity of functions. Some proteins regulate the bacterial housekeeping σ70 promoters, the σ54 promoters (some EBP proteins) or both types of promoters (NtrC, a dual protein). Several regulators affect a particular pathway, like l-cysteine biosynthesis (CysB regulator). Many regulatory proteins control operons with one or more promoters (8), while others are involved in catabolic regulons (Crp regulator) or have structural and regulatory roles (e.g. ArgR and Fis). In a few cases regulatory proteins directly affect expression of other regulators, and when this happens negative autoregulation is by far the most common type of interaction (6).
Regulatory proteins can be grouped into evolutionary families based on their sequence similarity. The complete set of E.coli K-12 chromosomal regulators falls into 25 families. The HTH DNA-binding motif is detected in 248 known and predicted transcription regulators. The remaining predictions are based on homology to known transcription regulators. Additional DNA-binding motifs such as zinc fingers (33), antiparallel β-sheets (34), RNA-binding like motifs (35) and HLH (36) have been described in regulatory proteins of E.coli, although they contribute a small fraction compared with the regulators with an HTH motif.
Regulatory activity and relative HTH position
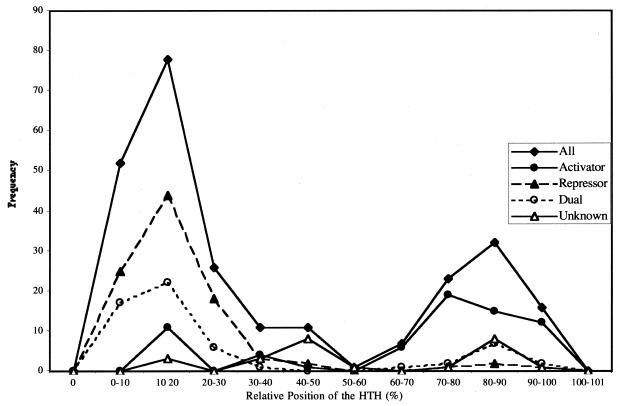
We have previously observed an interesting correlation between the relative position of the HTH motif in the protein sequence and its role as a negative or positive activator. Repressor proteins usually have the HTH motif in the N-terminus, whereas activator proteins tend to have the HTH close to the C-terminus end. Furthermore, this position is conserved across different evolutionary families of regulatory proteins (9). A preferred position was also observed within the HLH family of eukaryotic transcription factors. It is interesting to re-estimate whether this distribution is conserved in the current larger set of proposed regulators in E.coli. Figure 2 shows the distribution of all regulators with an HTH and the separate distributions of activators, repressors and dual proteins. Repressor proteins have a strong tendency for the HTH to be located in the N-terminus (96%), with only 4% having it in the C-terminus, whereas activator proteins have a strong preference for the HTH being in the C-terminus (78%), with only 22% having it in the N-terminus. There are few proteins with the HTH in a central position, mostly proteins with sizes <100 amino acid residues.
Figure 2.
HTH distribution in the complete set of E.coli proteins. The distribution of the relative HTH location is shown for different subsets of the collection. On the x-axis, 0% represents the N-terminus and 100% the C-terminus end of the protein. The y-axis shows the frequency of regulators. A total of 234 transcriptional factors bearing an HTH motif were taken into account for this analysis. Closed triangles, repressors; closed circles, activators; open circles, dual proteins (activator and repressor); open triangles, those with unknown function.
This distribution of positions was used to predict whether a putative transcription regulator is expected to be a repressor or an activator. If a protein has an N-terminal HTH, it will be predicted to be a repressor protein, unless it belongs to the LysR family of dual regulators. As discussed in an earlier work, members of the LysR family dominate the peak of dual proteins at the N-terminus (open circles in Fig. 2) (9). A protein with the HTH in the C-terminus is assumed to be an activator. Using these simple rules, we correctly predicted the function of ~70% of cases in the known dataset, with 15% false positives.
The regulatory function of regulators is based on experimental evidence and the prediction rule explained above. We predict as dual all proteins whose sequence is similar to any member of the LysR family. Dual proteins are either activators of several genes and repressors of their own expression (proteins of the LysR family) (28) or activators and repressors of different sets of genes, such as Crp and FruR (37). Around 50 proteins in the set do not have a described function, because there is not enough information available.
The set of known and predicted proteins is formed by around 92 activators, 113 repressors and 59 dual proteins, corresponding to 34.8, 42.8 and 22.3%, respectively. A previous evaluation with a much smaller database some years ago gave 10, 55 and 38%, respectively (8). The current numbers show a more even distribution of repressors, activators and dual proteins. Although dual proteins are not limited to the LysR family, it is quite remarkable that this family alone accounts for almost 25% of all dual regulatory proteins in E.coli. The LysR family might have been selected in evolution with a large number of members in E.coli for positive control of the regulation of several pathways in amino acid biosynthesis (38) and negative control of their own expression.
Dual proteins exert an activator effect on some promoters and a negative effect on others. They show a similar behavior to repressor proteins. This structural observation supports the unexpected suggestion that dual proteins might have initially been repressor proteins that acquired the ability to activate. As discussed before, given the mechanistic requirements for repression, we conceived that the opposite situation where activators became repressors by simple displacement of their DNA-binding site in relation to promoter initiation (8) was most likely.
As already mentioned, negative autoregulation is predominant in transcription factors of E.coli (6). The updated number in RegulonDB shows that only six regulators are positively autoregulated (PhoB, GutM, TdcA, CadC, RhaS and RhaR), while three regulators exert both positive and negative regulation of their own expression (Ada, CRP and NtrC) (39). LysR is a unique family given the extensive negative autoregulation affecting its members. This family accounts for 25% of all negatively autoregulated proteins.
Organization of regulators in families
The 314 known and putative regulatory proteins can be clustered into subsets based on their sequence similarity. This generates 20 families of evolutionarily related proteins, with a similarity of at least 25% within members of the same family. In addition, these groups match the 20 families that have been described when studying regulatory proteins across all prokaryotes. In other words, we found that all evolutionary families of transcription regulators found in bacteria have representatives in E.coli. These groups vary considerably in their number of members, as summarized in Table 2. The average number of members per family is of the order of 10, ranging from LysR, the largest family in E.coli with 45 members, to families with one or two members, such as the ArgR, CRP and Fur families. Families with very few members have homologs in different bacteria, providing evidence that they form separate identifiable groups.
Table 2. Regulatory protein families: E.coli versus prokaryotes.
| Family | Escherichia coli | Total | Prokaryotes | |||
|---|---|---|---|---|---|---|
| Known | Predicted | No. | Percent | No. | Percent | |
| CRP | 2 | 0 | 2 | 0.6 | 28 | 7.1 |
| AsnC | 2 | 1 | 3 | 0.9 | 16 | 18.75 |
| IclR | 2 | 6 | 8 | 2.5 | 12 | 66.6 |
| Cold | 8 | 1 | 9 | 2.8 | 50 | 18 |
| TetR/AcrR | 4 | 5 | 9 | 2.8 | 22 | 40.9 |
| DeoR | 7 | 7 | 14 | 4.4 | 24 | 58.3 |
| EBP | 9 | 5 | 14 | 4.4 | 56 | 25 |
| GalR/LacI | 12 | 2 | 14 | 4.4 | 49 | 28.5 |
| OmpR | 13 | 4 | 17 | 5.1 | 61 | 26.2 |
| Luxr/UhpA | 11 | 6 | 17 | 5.4 | 59 | 28.8 |
| GntR | 8 | 12 | 20 | 6.4 | 38 | 52.6 |
| AraC/XylS | 14 | 13 | 27 | 8.6 | 80 | 33.75 |
| LysR | 18 | 27 | 45 | 14.3 | 173 | 26 |
| Others | 49 | 66 | 115 | 36.7 | 132 | 293.4 |
| Total | 159 | 155 | 314 | 100 | 701 | 44.79 |
All regulatory protein families in prokaryotes (E.coli included) were obtained by an exhaustive search in the SwissProt database and in the E.coli genome. The first column is the family name, the second column describes the number of known and the third column of predicted regulatory proteins. The fourth and fifth columns are the total number of members. The last two columns describe the number of members per family currently known within all eubacteria and the percentage that the E.coli members represent in all eubacteria per family, respectively.
It certainly makes sense for E.coli to have a large diversity of regulatory proteins given its ability to grow and to respond to different environmental conditions and given its capacity to live in the mammalian gut as well as as a free-living organism. Escherichia coli must survive under very diverse environments, including wide ranges of temperature (8–50°C), osmolarity and pH (40). For instance, E.coli provides 30% of all prokaryotic proteins within the GalR/LacI family (Table 2), which may also reflect the fact that many other bacteria do not require sugar metabolism because all carbon compounds are imported from the host (41). Note however, that this fraction is going to diminish as more bacterial genomes are sequenced. For instance, the DeoR family has been predicted to be formed by 14, 4 and 6 members in the genomes of E.coli (5), Haemophilus influenzae (42) and Bacillus subtilis (43), respectively. Currently, ~30 members of the DeoR family have been found in species ranging from Gram-positive organisms such as Lactococcus lactis (44) and Streptococcus mutans (45) to Gram-negative bacteria such as Agrobacterium tumefaciens (46) and Pseudomonas aeruginosa (47).
The LysR family is most abundant in Proteobacteria (purple bacteria), in the α and γ subgroups. Few proteins of the LysR family have been found in the β subgroup and none within the δ subgroup, whereas members of the family have been described in Gram-positive bacteria (28). The large genetic distances between prokaryotes with members of this family and vast differences in G+C content suggest a LysR progenitor that arose early in prokaryotic evolution (28). In contrast, members of the AraC/XylS family are widely distributed in diverse prokaryote genera. Most of the members of this family occur in the γ subdivision of the Proteobacteria (purple bacteria). A few have been found in low and high G+C Gram-positive bacteria and in cyanobacteria (48). The AraC/XylS and LysR families have few members in archaeabacteria or in eukaryotes. Members of the GntR family have been described in E.coli, B.subtilis, Pseudomonas putida and Klebsiella aerogenes (49). An interesting observation related to a still earlier evolutionary scenario is that some regulatory protein families, such as GalR/LacI and GntR, are only present within eubacteria, while some other families, like the AsnC family, show a wider distribution in both eubacteria and archeobacterial organisms (50).
Our knowledge of the distribution of regulatory families in different bacteria will change as more genomes are finished. However, the fraction of regulatory proteins within genomes should not vary that much in the future. For instance, Helicobacter pylori is an example of a bacterium living in a quite stable environment and its annotated genome indicates 13 regulatory proteins, accounting for 0.82% of the total 1590 genes it contains (41). It may be that some regulatory proteins are not required in microorganisms that live in relatively stable environments. Nonetheless, the cell has alternative coccoid and bacillus forms, which should involve regulatory interactions for their differentiation. Such interactions may involve DNA-binding motifs other than the HTH or σ factors, as used by B.subtilis for differentiation (51).
Families of transcription regulators share a type of DNA-binding motif, as well as inducer binding or oligomerization motifs. These similarities define a signature sequence shared by members of the family. In most of the cases the region that characterizes a regulatory family contains the HTH motif. The HTH is not only the general main signature in the DNA-binding transcription regulators of prokaryotes, but it also provides the internal distinction among different families. Remember that one of the first helix motifs considered to provide for a non-specific binding ability is common to many regulatory proteins, whereas the second helix motif confers the DNA-binding specificity. The domain organization has been experimentally identified in most transcription regulatory proteins. For instance, the DBD from the GalR/LacI family is around 59 amino acids long (20). The family contains a second domain of around 270 residues implicated in multimerization and induction. The second domain has also been found in proteins that bind sugars, such as RbsB (ribose transport). The DBD of AraC/XylS has around 60 residues, while the LysR family shares a highly conserved N-terminus DBD (consisting of a HTH motif and flanking sequences). In most of the LysR members the less conserved C-terminus domain has a sensory function (52). In the case of CRP protein, the C-terminus domain contains a HTH DNA-binding motif, while the N-terminus domain is a large structure of around 170 residues containing a nucleotide-binding site that shows homology with cAMP-dependent protein kinases (37). The main feature in the set of transcription factors is the presence of the HTH. In fact, this main feature characterizes almost all DNA-binding regulatory proteins in E.coli. In most protein families the domain covalently linked to the DBD is less well conserved and is involved in several responses (for instance, in AraC the N-terminus domain is involved in allosteric regulation and dimerization by the co-inducer) (52). The IclR and EBP families are the only families with a signature that involves other domains in addition to the DBD. The DBD of IclR is located in the N-terminus, while the most conserved domain is located in the C-terminus. In the EBP family the ATPase and the σ-interaction sites are more conserved than the DBD.
Conservation of protein sizes within families
Given the high degree of similarity of proteins within a family, it is not surprising that their size in amino acid residues is also rather conserved. Observing the size distribution within families, it is natural to group them into two classes. Families with members of rather homogeneous size, and families showing a more heterogeneous size distribution.
Homogeneous families can be defined by having at least 70% of their members of conserved size, as shown by an overall standard deviation (SD) smaller than 20 amino acids. Table 3 shows the mean size and standard deviation for all families. This analysis shows homogeneous groups with a small SD, such as the IclR, DeoR, GntR, LuxR/UhpA, Cold Shock, OmpR, TetR/AcrR and GalR/LacI families. The IclR family has one protein with 315 residues (MhpR) and seven proteins with a mean length of 272.4 ± 10 residues. DeoR is a family of conserved size except for the two subunits of GatR.
Table 3. Functional conservation of transcription factor families.
| Family | Function | Mean (± SD) | Physiological function | Parallel | Antiparallel | Percent | Members |
|---|---|---|---|---|---|---|---|
| Cold | Activator | 70.0 ± 1.8 | Low temperatures: Cold Shock | 50.0 | 50.0 | 90 | 9 |
| AsnC | Dual | 156 ± 6.9 | Amino acid biosynthesis | a | a | 66 | 3 |
| TetR/AcrR | Repressor | 210.0 ±17.1 | Tetracycline resistance | 42.8 | 57.1 | 66 | 9 |
| LuxR/UhpA | Activator | 219.2 ± 13.2 | Biosynthesis and glycerol metabolism | 52.9 | 47.1 | 40 | 17 |
| CRP | Dual | 230 ± 20 | Global responses | a | a | 100 | 2 |
| OmpR | Activator | 230.6 ± 7.3 | Adaptive response | 40.0 | 60.0 | 45 | 16 |
| GntR | Repressor | 246.0 ± 1.4 | Carbon metabolism | 54.5 | 45.4 | 55 | 20 |
| DeoR | Repressor | 256.8 ± 12.7 | Sugar metabolism | 31.2 | 68.7 | 50 | 14 |
| IclR | Repressor | 272.4 ± 19.5 | Carbon source uptake | 14.3 | 85.7 | 60 | 8 |
| GalR/LacI | Repressor | 333.7 ± 12.3 | Carbon source uptake | 35.7 | 64.3 | 90 | 14 |
| AraC/XylS | Activator | Heterogeneous | Virulence, transposition, sugar metabolism | 40.0 | 60.0 | 22 | 27 |
| EBP | Activator | Heterogeneous | Nitrogen assimilation, aromatic amino acid synthesis and several functions | 53.8 | 46.1 | 70 | 14 |
| LysR | Dual | Heterogeneous | Amino acid biosynthesis | 29.5 | 70.4 | 50 | 45 |
| Others | Several | 233.9 ± 258.6 | Several functions | 50.0 | 50.0 | a | 116 |
The most conserved families in terms of length, with at least eight members in E.coli, are shown. For each family (name in the first column), the main regulatory function is indicated followed by the mean of the sequence size for each family and the standard deviation. The fourth column indicates the most common function of the regulated genes. The number of genes with a parallel direction of transcription and replication and antiparallel organization are indicated in the fifth and sixth columns. Column seven describes the percentage of members associated with one physiological function and the last column indicates the number of members in E.coli per family. Although the CRP and AsnC families have too few members in E.coli to assign a main physiological function to the family, in most prokaryotes their function is conserved (data not shown).
aNot calculated.
The GntR family has two small proteins (b3694 and DgoR) of 98 and 177 residues and two much larger ones (b1439 and YjiR) of 468 and 470 residues. Otherwise the remaining 16 proteins have a mean length of 246.0 ± 1.4 residues, showing a highly homologous group. A sequence comparison between the biggest proteins in the family (b1439 and YijR) shows an identity of 35%. This result might reflect a genetic duplication inside families in E.coli and the less frequent success of shuffling of motifs to generate larger functional regulatory proteins.
Similarly, 70% of the LuxR/UhpA members fall within a mean size of 219.1 ± 13.2 residues. The maltose activator, MalT, is included in this group. It represents, with 901 residues, one of the biggest proteins in the family and in the collection (26). Some proteins of the LuxR/UhpA family belong to receiver-response regulators. MalT is a protein that lacks receiver modules and is not a response regulator, but possesses homology in the DBD to several members of the family. In general, the organization of MalT shows three domains: the DBD located in the C-terminus where the HTH is found and two domains of ~400 and 200 residues in the N-terminus, which are not shared with the other members of the family.
The OmpR-like proteins belong to a very homogeneous family in size; 93% of the members fall within a mean size of 230.6 ± 7.3 residues. Smaller proteins tend to have a higher percentage identity than bigger proteins. This is a consequence of the definition of families, mostly in terms of the DBD, which is the most conserved region. Smaller proteins have only the DBD, while bigger proteins have additional domains, probably as a result of acquisition of novel domains in the ancestral protein of the family. Given this pattern of size and distribution in these families, the most plausible events in evolution seem, first, the emergence of the ancestral protein of the family as a result of joining the HTH motif and a second larger motif, followed by divergence by gene duplication, with a few successful cases of additional shuffling of motifs generating a few larger members (see Fig. 3).
Figure 3.
Hypothetical alternative evolutionary pathways for the emergence of current transcription regulatory families. A DBD with a length of ~60–100 amino acid residues has at least two pathways along which to evolve into protein families. In the first group, a second or third domain (with length relatively conserved) was added to the DBD, to its N- or C-terminus. This fusion produced as a consequence the divergence of several members of the family as a whole unit. In the second heterogeneous group, several domains were added to the N- or C-terminus of the DBD. As a consequence, several sub-groups depending on the size, physiological function and regulatory role should be identified in the same family.
As mentioned before, heterogeneous families in terms of the distribution in size of their protein members form a second group. For instance, the LysR family shows a bimodal distribution with one subset of 15 proteins with a size between 215 and 299 amino acids (mean 284.8 ± 24.6). We detected a second subset of 29 transcription factors with a length from 300 to 354 amino acids (mean 312.9 ± 11.6). Similarly, the NagR/XylS family shows two subsets, one with a mean of 304 ± 3.3 and a second with a mean of 403.6 ± 4.03. These subsets might reflect two different evolutionary events in ancestral members of the family, each one generating later similar members by gene duplication.
AraC/XylS, an activator protein family that shows heterogeneity in terms of the location of the HTH motif (9), also shows a high variability in the size of its members. The family can be divided into three subsets: (i) a subset of two small proteins of size 107 and 129 with their HTH in the N-terminus; (ii) a larger set of 16 proteins within the range 239–292 residues (mean 267.8 ± 18.1) with the HTH in the last third of the sequence; (iii) a group of nine larger proteins with a size between 300 and 400 residues (mean 332.8 ± 35.3), with their HTH in a central position. Similarly, the EBP family shows a high variability with proteins ranging from 98 to 668 residues. The larger subsets are four proteins with a mean of 454 ± 13.2 residues, a subset of five proteins with a mean of 538 ± 30.6 residues and three larger proteins with around 668 ± 25 residues.
In general, within a family proteins of similar length show a higher degree of sequence similarity. In some cases the high sequence similarity and length conservation suggest that the members of these families are derived from a common ancestor through pathways in evolution involving early duplications and subsequent acquisition of additional motifs, followed by gene duplication, individual amino acid substitutions and small insertions/deletions that could lead to incremental changes in size (53,54). Such might be the case with the Cold Shock, GntR, IclR and GalR/LacI families and the two AraC/XylS members of 107 and 129 residues. Size conservation and presence of the HTH motif in a conserved relative position, as well as homogeneity in the motif that defines the family, all contribute to the notion of a homogeneous family of regulatory proteins. On the other hand, there are families with a more heterogeneous behavior, suggesting a more diverse evolutionary history involving multi-domain shuffling and acquisition of novel domains, such as the EBP and AraC/XylS families (see Fig. 3).
Regulatory families and the physiology of the genes they regulate
Regulatory proteins within a family share structural properties as discussed before. They also tend to affect genes and transcription units involved in related metabolic functions. In Table 3 we describe the most common physiological classes of genes regulated by the different regulatory families. We calculated the percentage of members for each family dedicated to the regulation of one physiological function. Some families regulate genes that can clearly be associated with a particular physiological class. For instance, of all regulators associated with carbon uptake, 20% belong to the GalR/LacI family, while members of the LysR family control 40% of the biosynthesis of amino acids pathways. On the other hand, members of different families, such as the Cold Shock, ArsR (arsenical resistance) and TetR/AcrR (antibiotic resistance) families tend to control resistance responses. Because E.coli can grow on many different carbon sources (galactose, melibiose, lactose, rhamnose, etc.), at least 50 transcription factors are devoted to regulating degradation of carbon compounds, which involve five different regulatory families.
A regulatory family is not always involved in regulating the same metabolic response. This is the case, for instance, for the growth phase-dependent expression of CspD protein, a member of the CspA family. All other regulatory proteins of this family are induced by cold shock (55), however, expression of cspD is induced by stationary phase growth in a way that does not involve the stationary phase σ factor σS.
We did not observe a clear correspondence between size homogeneity and conservation or dominance in the type of physiological function. For instance, the LuxR/UhpA and LysR families regulate around half of the known genes involved in glycerol metabolism and amino acid biosynthesis, respectively. This raises the possibility that regulation of some metabolic pathways may involve a larger number or chemical diversity of signal molecules (co-inducers or co-repressors) and therefore a more structurally diverse regulatory family as opposed to other families (i.e. GalR/LacI family) where a more homogeneous set of regulators is sufficient.
DISCUSSION
Based on different sequence similarity search strategies we have defined a set of 314 DNA-binding transcriptional regulators in E.coli K12. The definition of this set was facilitated by the predominant occurrence of the HTH DNA-binding motif in regulatory proteins in E.coli, and in fact also in the prokaryotic and eukaryotic kingdoms. Other DNA-binding motifs are also present in E.coli, but in only a few regulatory proteins. Based on the specificity of recognition we estimate around 300–350 transcription regulators in E.coli.
Regulatory proteins share a significant amount of protein similarity, enabling their clustering into families of plausible common evolutionary origin. The diagnostic region shared within a family imposes an identity of ~25% on members within one family. We find in this way that all 20 families of HTH regulatory proteins of the bacterial kingdom have representatives in E.coli. Most of these families appear to be quite homogeneous groups whose members share several properties. These are families with proteins of rather similar length and with their HTH domain localized in the same relative position either in the N- or C-terminus. Regulators within a family tend to be mostly repressors or mostly activators, with the dual regulators concentrated in the most abundant family, LysR, with 45 members in E.coli. These families group regulators that tend to affect genes involved in related biological functions. All these common structural and functional properties support the notion of a family even if some of them have only one member in E.coli. The additional correlation between the HTH in the N-terminus for repressors and the HTH positioning for known activators in the C-terminus was used, in combination with family membership, to generate a predicted functional role for most of the 314 regulatory proteins in E.coli. This produces a picture of a quite even distribution of 34.8, 42.8 and 22.3% of activators, repressors and dual regulators, respectively.
Evolution is, however, more flexible, as illustrated by multi-domain regulatory proteins that are more difficult to group. Thus, some families group a less homogeneous set of proteins. The existence of multiple domains makes their clustering more difficult to achieve, grouping proteins with a more elaborate evolutionary history with some motifs appearing only in some members of the family. In some cases, multiple domains reflect the existence of several evolutionary events, such as shuffling in prokaryotic proteins of the two component systems.
An interesting question related to these observations is that of the different size or abundance of regulatory proteins within the different families. Huynen and van Nimwegen (56) have shown that genes within one family have similar functions, but as the requirements of this function vary over time so does the presence of the gene family in the genome. Activation of transcription by different types of metabolites (e.g. ions, amino acid derivatives, nucleic acids, sugars, etc.) suggests an ancient divergence of signal recognition within regulatory families. The evolutionary flexibility within regulatory families can be appreciated when observing the structural diversity of the different co-inducers that stimulate various transcription factors that belong to the same family, as opposed to the highly conserved DBD domain. An additional source of diversity is the presence of self-transmissible plasmids, which probably move freely throughout the prokaryotic community and may have promoted a more recent and rapid dissemination and evolution (28).
A good number of documented regulatory families show a tendency for members of similar size. We hypothesize that the largest proteins (such as the monomeric maltose activator, MalT) could have other functions in addition to transcription regulation (like the proline dehydrogenase of PutA), while smaller or medium sized proteins, usually acting as multimers, could rarely have additional functions.
The amount of information on known binding sites is limited to around 1/6 (50 out of 300) of all potential transcription regulators of E.coli. It is interesting to address the question of whether the grouping of proteins into families would limit or structure their DNA-binding available space. It is possible to imagine one scenario where proteins of the same family could recognize similar DNA-binding sites. Whether a mechanism of co-evolution between one family and one set of DNA-binding sites exists, and its functional implications, should be investigated in the future.
We analyzed as another potential trait that could help to characterize the different regulatory families their gene location in operons or as transcription-isolated genes, as well as their neighboring genes, especially if these are also regulatory proteins. One salient feature is that 30% of all transcription regulators occur as isolated transcription units. This organization may be relevant for their independent transcription regulation, uncoupled from the set of genes they regulate.
We considered it equally interesting to study the orientation of transcription and replication in this set of regulatory proteins, given the rational hypothesis presented years ago about such distributions (57). The predicted tendency for an antiparallel orientation of replication and transcription is in fact confirmed, although not very strongly. It will be interesting to test with global studies if this set of genes is expressed in low amounts as assumed. Transcription regulators have a pattern of position and orientation in the genome that does not differ particularly from the complete set of genes of E.coli. This same conclusion seems justified when studying their organization into operons or into single transcription units.
In brief, we have learned that E.coli transcription regulators are grouped into families reflecting their common evolutionary origin. These families share conserved functional and structural properties. Less than 10% of all genes in E.coli participate as transcription regulators. Other proteins certainly participate in regulation of transcription, without necessarily binding to the DNA, increasing the fraction of regulator genes. These global properties of the repertoire of transcription regulators in E.coli provide a reference to compare in the future among other bacterial and eukaryotic genomes.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Guillermo Dávila, Jay Gralla, Boris Magasanik and Shoshana Wodak for valuable comments on this work, as well as Heladia Salgado for computational support. E.P.R. was supported by a doctoral fellowship from CONACYT and DGEP-UNAM. Part of this work was supported by grants from CONACYT and DGAPA to J.C.V.
REFERENCES
- 1.Neidhardt F.C., Ingraham,J.L, Low,K.B., Magasanik,B., Schaechter,M. and Umbarger,H.E. (eds) (1987) Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. American Society for Microbiology, Washington, DC.
- 2.Neidhardt F.C., Curtiss,R., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley.M., Schaerchter,M. and Umbarger,H.E. (eds) (1996) Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. American Society for Microbiology, Washington, DC.
- 3.Huerta A.M., Salgado,H., Thieffry,D. and Collado-Vides,J. (1998) Nucleic Acids Res., 26, 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salgado H., Santos-Zavaleta,A., Gama-Castro,S., Millán-Zárate,D., Blattner,F.R. and Collado-Vides,J. (2000) Nucleic Acids Res., 28, 65–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner F.R., Plunkett,G., Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) Science, 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- 6.Thieffry D., Huerta,A., Pérez-Rueda,E. and Collado-Vides,J. (1998) Bioessays, 20, 433–440. [DOI] [PubMed] [Google Scholar]
- 7.Collado-Vides J., Magasanik,B. and Gralla,J.D. (1991) Microbiol. Rev., 55, 371–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gralla J.D. and Collado-Vides,J. (1996) In Neidhart,F.C., Curtiss,R., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 1232–1246.
- 9.Pérez-Rueda E., Gralla,J. and Collado-Vides,J. (1998) J. Mol. Biol., 275, 165–170. [DOI] [PubMed] [Google Scholar]
- 10.Bork P. and Koonin,E.V. (1998) Nature Genet., 18, 313–318. [DOI] [PubMed] [Google Scholar]
- 11.Bairoch A. and Apweiler.R. (1996) Nucleic Acids Res., 24, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bairoch A. and Boeckmann,B. (1992) Nucleic Acids Res., 20, 2019–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bairoch A., Bucher,P. and Hoffman,K. (1997) Nucleic Acids Res., 25, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brazma A., Jonassen,I., Eidhammer,I. and Gilbert,D. (1998) J. Comput. Biol., 5, 279–305. [DOI] [PubMed] [Google Scholar]
- 15.Harrison S.C. (1991) Nature, 353, 715–719. [DOI] [PubMed] [Google Scholar]
- 16.Dodd I.B. and Egan,J.B. (1987) J. Mol. Biol., 194, 557–564. [DOI] [PubMed] [Google Scholar]
- 17.Yudkin M.D. (1987) Protein Eng., 1, 371–372. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wooton,J.C. (1993) Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 19.Kabsch W. and Sander,C. (1983) Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 20.Weickert M.J. and Adhya,S. (1992) J. Biol. Chem., 267, 15869–15874. [PubMed] [Google Scholar]
- 21.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L. (1998) Curr. Opin. Struct. Biol., 8, 372–379. [DOI] [PubMed] [Google Scholar]
- 23.Jones D.T. (1999) J. Mol. Biol., 287, 797–815. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henikoff S., Wallace,J.C. and Brown,J.P. (1990) Methods Enzymol., 183, 111–132. [DOI] [PubMed] [Google Scholar]
- 26.Pao G.M. and Saier,M.H.,Jr (1995) J. Mol. Evol., 40, 136–154. [DOI] [PubMed] [Google Scholar]
- 27.Henikoff S., Haughn,G.W., Calvo,J.M. and Wallace,J.C. (1988) Proc. Natl Acad. Sci. USA, 85, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schell M.A. (1993) Annu. Rev. Microbiol., 47, 597–626. [DOI] [PubMed] [Google Scholar]
- 29.Gallegos M.-T., Schelif,R., Bairoch,A., Hoffman,K. and Ramos,J.L. (1997) Microbiol. Mol. Biol. Rev., 61, 393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bairoch A. (1993) Nucleic Acids Res., 21, 2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spiro S. (1994) Antonie van Leewenhoek, 66, 23–36. [DOI] [PubMed] [Google Scholar]
- 32.Salgado H., Santos,A., Garza-Ramos,U., Van Helden,J., Diaz,E. and Collado-Vides,J. (1999) Nucleic Acids Res., 27, 59–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rey L., Murillo,J., Hernando,Y., Hidalgo,E., Cabrera,E., Imperial,J. and Ruiz-Argueso,T. (1993) Mol. Microbiol., 8, 471–481. [DOI] [PubMed] [Google Scholar]
- 34.Somers W.S. and Phillips,S.E. (1992) Nature, 359, 387–393. [DOI] [PubMed] [Google Scholar]
- 35.Graumman P. and Marahiel,M.A. (1996) Bioessays, 18, 309–315. [DOI] [PubMed] [Google Scholar]
- 36.Roth A. and Messer,W. (1995) EMBO J., 14, 2106–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolb A., Busby,S., Buc,H., Garges,S. and Adhya,S. (1993) Annu. Rev. Biochem., 62, 749–795. [DOI] [PubMed] [Google Scholar]
- 38.Newman E.B., Lin,R.T. and D’Ari,R. (1996) In Neidhardt,F.C., Curtiss,R., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaerchter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 1513–1526.
- 39.Magasanik B. and Neidhardt,F.C. (1987) In Neidhart,F.C., Ingraham,J.L., Low,K.B., Magasanik,B., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 1318–1325.
- 40.Death A. and Ferenci,T. (1994) J. Bacteriol., 176, 5101–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomb J.F., White,O., Kerlavage,A.R., Clayton,R.A., Sutton,G.G., Fleischmann,R.D., Ketchum,K.A., Klenk,H.P., Gill,S., Dougherty,B.A. et al. (1997) Nature, 388, 539–547. [DOI] [PubMed] [Google Scholar]
- 42.Fleischmann R.D., Adam,M.D., White,O., Clayton,R.A., Kirkness,E.F., Kerlavage,A.R., Bult,C.J., Tomb,J.F., Dougherty,B.A., Merrick,J.M. et al. (1995) Science, 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 43.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessieres,P., Bolotin,A., Borchert,S. et al. (1997) Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 44.Van Rooijen R.T.J., Dechering,K.J., Niek,C., Wilmink,J. and de Vos,W.M. (1993) Protein Eng., 6, 201–206. [DOI] [PubMed] [Google Scholar]
- 45.Rosey E.L. and Stewart,G.C. (1992) J. Bacteriol., 174, 6159–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H. and Farrand,S.K. (1997) J. Bacteriol., 179, 7559–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweizer H.P. and Po,C. (1996) J. Bacteriol., 178, 5215–5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olsen G.J., Woese,C.R. and Overbeek,R. (1994) J. Bacteriol., 176, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haydon D. and Guest,J. (1991) FEMS Microbiol. Lett., 79, 291–296. [DOI] [PubMed] [Google Scholar]
- 50.Kyrpides N.C. and Ouzounis,C.A. (1995) Trends Biochem. Sci., 20, 140–141. [DOI] [PubMed] [Google Scholar]
- 51.Haldenwang W.G. (1995) Microbiol. Rev., 59, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ninfa A.J. (1996) In Neidhardt,F.C., Curtiss,R., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaerchter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 1246–1262.
- 53.Nguyen C.C. and Saier,M.H.,Jr (1995) FEBS Lett., 377, 98–102. [DOI] [PubMed] [Google Scholar]
- 54.Savageau M. (1986) Proc. Natl Acad. Sci. USA, 83, 1198–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamanaka K., Fang,L. and Inouye.M. (1998) Mol. Microbiol., 27, 247–255. [DOI] [PubMed] [Google Scholar]
- 56.Huynen M.A. and van Nimwegen,E. (1998) Mol. Biol. Evol., 15, 583–589. [DOI] [PubMed] [Google Scholar]
- 57.Brewer B.J. (1988) Cell, 53, 679–686. [DOI] [PubMed] [Google Scholar]
- 58.Sunnerhagen M., Nilges,M., Otting,G. and Carey,J. (1997) Nature Struct. Biol., 4, 819–825. [DOI] [PubMed] [Google Scholar]
- 59.Streaker E.D. and Beckett,D. (1998) J. Mol. Biol., 278, 787–800. [DOI] [PubMed] [Google Scholar]
- 60.Wilson K.P., Shewchuk,L.M., Brennan,R.G., Otsuka,A.J. and Matthews,B.W. (1992) Proc. Natl Acad. Sci. USA, 89, 9257–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber I.T. and Steitz,T.A. (1987) J. Mol. Biol., 198, 311–326. [DOI] [PubMed] [Google Scholar]
- 62.Kostrewa D., Granzin,J., Stock,D., Choe,H.-W., Labahn,J. and Saenger,W. (1992) J. Mol. Biol., 226, 209–226. [DOI] [PubMed] [Google Scholar]
- 63.Penin F., Georjon,C., Montserret,R., Bockmann,A., Lesage,A., Yang,Y., Bonod-Bidaud,C., Cortay,J.C., Negre,D., Cozzone,A.J. and Deleage,G. (1997) J. Mol. Biol., 270, 496–510. [DOI] [PubMed] [Google Scholar]
- 64.Lewis M., Chang,G., Horton,N.C., Kercher,M.A., Pace,H.C., Schumacher,M.A., Brennan,R.R. and Lu,P. (1996) Science, 271, 1247–1254. [DOI] [PubMed] [Google Scholar]
- 65.Fogh,R.H., Ottleben,G., Rueterjans,H., Schnarr,M., Boelens,R. and Kaptein,R. (1994) EMBO J., 13, 3936–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baikalov I., Schroeder,I., Kaczor-Grzeskowiak,M., Cascio,D., Gunsalus,R.P. and Dickerson,R.E. (1998) Biochemistry, 37, 3665–3676. [DOI] [PubMed] [Google Scholar]
- 67.Martinez-Hackert E. and Stock,M. (1997) J. Mol. Biol., 269, 301–312. [DOI] [PubMed] [Google Scholar]
- 68.Schumacher M.A., Glasfeld,A., Zalkin,H. and Brennan,R.G. (1997) J. Biol. Chem., 272, 22648–22653. [DOI] [PubMed] [Google Scholar]
- 69.Hinrichs W., Kisker,C., Duevel,C., Mueller,A., Tovar,K., Hillen,W. and Saenger,W. (1994) Science, 264, 418–420. [DOI] [PubMed] [Google Scholar]
- 70.Kisker C., Hinrichs,W., Tovar,K., Hillen,W. and Saenger,W. (1995) J. Mol. Biol., 247, 260–280. [DOI] [PubMed] [Google Scholar]
- 71.Zhang R.-G., Joachimiak,A., Lawson,C.L., Schevitz,R.W., Otwinowski,Z. and Sigler,P.B. (1987) Nature, 327, 591–597. [DOI] [PubMed] [Google Scholar]