Abstract
Hu antigen C (HuC) has three RNA-binding domains (RBDs). The N-terminal two, RBD1 and RBD2, are linked in tandem and bind to the AU-rich elements (AREs) in the 3′-untranslated region of particular mRNAs. The solution structures of HuC RBD1 and RBD2 were determined by NMR methods. The HuC RBD1 and RBD2 structures are quite similar to those of Sxl RBD1 and RBD2, respectively. The individual RBDs of HuC, RBD1 and RBD2 in isolation can interact rather weakly with the minimal ARE motif, AUUUA, while the didomain fragment, RBD1–RBD2, of HuC binds more tightly to a longer ARE RNA, UAUUUAUUUU. Chemical shift perturbations by the longer RNA on HuC RBD1–RBD2 were mapped on and around the two β-sheets and on the C-terminal region of RBD1. The HuC RBD1–RBD2 residues that exhibited significant chemical shift perturbations coincide with those conserved in Sxl RBD1–RBD2. These data indicate that the RNA-binding characteristics of the HuC and Sxl didomain fragments are similar, even though the target RNAs and the biological functions of the proteins are different.
INTRODUCTION
The AU-rich element (ARE) is a sequence consisting mostly of many uridines and some adenosines in the 3′-untranslated region of mRNA and was first identified as a cis-acting degradation signal of the mRNAs of certain lymphokines, cytokines and proto-oncogenes (1,2). AREs have been found in a number of mRNAs and are divided into three classes with respect to the copy number of the AUUUA motif: several copies are dispersed in class I and are clustered in class II, while class III lacks this motif (3). Furthermore, many proteins that can bind to AREs have been reported: hnRNP A1 and hnRNP C (4), AUF1 (5), AUH (6), Hel-N1/HuB (hereafter called HuB), HuC/Ple21 (hereafter called HuC), HuD and HuA/HuR (hereafter called HuR) (7–11). All of these proteins, except AUH, have one to three copies of the RNA-recognition motif (RRM), which is a type of RNA-binding domain (RBD) consisting of about 90 amino acid residues (12).
HuB, HuC, HuD and HuR from vertebrates are RBD-containing ARE-binding proteins and belong to the ELAV/Hu protein family. In addition to these four vertebrate members, ELAV and RBP9 from Drosophila and ELR-1 from Caenorhabditis elegans are included in the ELAV/Hu family (reviewed in 13; 14,15). The vertebrate Hu proteins bind tightly to ARE-containing mRNAs in vitro (7–11,16). The ELAV/Hu proteins have three copies of the RBD (RBD1, RBD2 and RBD3), the N-terminal two of which are nearly directly linked in tandem (RBD1–RBD2), while RBD2 and RBD3 are separated by another region. The RBD1–RBD2 fragments of HuC, HuD and HuR bind tightly to the ARE (9,11,17). On the other hand, the RBD3 fragments of HuC and HuD bind to poly(A) (9,11). The RBD1–RBD2 and RBD3 segments are likely to be responsible for the ARE- and poly(A)-binding activities, respectively, in all of the ELAV/Hu proteins, because the amino acid sequences of the corresponding RBDs are highly conserved within this family.
HuR stabilizes ARE-containing mRNAs (18–20). Thus, HuR has been suggested to protect the ARE-containing mRNA from an ARE-associated nuclease(s), although the nuclease itself has not yet been identified. HuR is expressed ubiquitously in various tissues (11,21), while the other ELAV/Hu proteins are expressed predominantly in neuronal cells and are thought to participate in neuronal differentiation (reviewed in 13; 15). One of the neuronal ELAV/Hu proteins, HuB, enhances the expression level of ARE-containing mRNAs (22,23). HuR and HuD have also been shown to shuttle between the nucleus and cytoplasm and were suggested to contribute to the export of ARE-containing mRNAs (19,24). Therefore, ARE binding by RBD1–RBD2 is important for the biological functions of the ELAV/Hu proteins. On the other hand, ELAV in Drosophila has been suggested to be involved in the regulation of alternative splicing (25). Interestingly, the amino acid sequences of the ELAV/Hu RBD1–RBD2 segments are significantly homologous (53% identity) to that of Drosophila Sex-lethal (Sxl), an alternative splicing regulator, which binds specifically to pre-mRNA polyuridine tracts (26).
As in the case of the ELAV/Hu RBD1–RBD2 and ARE, many multiple-RBD proteins bind to single-stranded RNA in a sequence-specific manner. In general, the RNA-binding sites in the individual RBDs are thought to be in the β-sheet (27,28). Recently, the structures of three complexes, hnRNP A1 RBD1–RBD2 (29), Sxl RBD1–RBD2 (30) and poly(A)-binding protein RBD1–RBD2 (31), have been determined by X-ray crystallography. Among them, the case of hnRNP A1 is exceptional, because it forms a dimer consisting of four RBDs that recognizes two copies of single-stranded telomeric DNA (29). In contrast, Sxl RBD1–RBD2 forms a V-shaped cleft recognizing the UGUUUUUUU region of the target pre-mRNA in a tight and specific manner (30). The two RBDs of poly(A)-binding protein form a continuous eight-stranded β-sheet platform which recognizes poly(A) (AAAAAAAA) (31). These three structures suggest that the mechanisms of single-stranded RNA recognition are diverse, even though they have similar didomain architectures, RBD1–RBD2, in common.
In the present study, we selected one of the ELAV/Hu proteins, mouse HuC, and analyzed the interaction of the ARE with RBD1–RBD2 by NMR spectroscopy. First, the solution structures of two fragments, RBD1 and RBD2, were determined separately. Next, it was shown that the two RBDs arranged in tandem cooperate in binding to an ARE RNA, UAUUUAUUUU. Then, the chemical shift perturbations of the ARE RNA on HuC RBD1–RBD2 were analyzed on the basis of the solution structures of RBD1 and RBD2. These results indicate that the RNA-binding mechanism of HuC RBD1–RBD2 is similar to that of Sxl RBD1–RBD2 (30,32).
MATERIALS AND METHODS
Sample preparation
The HuC fragments, RBD1, RBD2 and RBD1–RBD2, correspond to Asp36–Arg123, Asp124–Gln208 and Asp36–Gln208, respectively, of mouse HuC protein (9). The proteins were overexpressed in Escherichia coli strain BL21(DE3) (33) using the expression vector pK7 (34). Non-labeled HuC fragments were expressed in LB medium, while the 15N- and 13C/15N-labeled HuC fragments were expressed in modified minimal medium (35) containing 1 g/l 15NH4Cl and/or 2 g/l [13C]d-glucose. After 24 h cultivation, the cells were harvested and sonicated. The supernatants were applied to a CM-Toyopearl cation exchange column (Tosoh, Japan) and the HuC fragments were eluted with a concentration gradient of ammonium formate buffer (pH 6.0). Finally, the HuC fragments were purified by a Mono-S cation exchange column (Pharmacia).
The RNAs were synthesized by the phosphoramidite method, followed by a deprotection procedure using methylamine (MA) and anhydrous triethylamine/hydrogen fluoride in N-methylpyrrolidinone (TEA·HF/NMP) (36). The RNAs were purified on a Mono-Q anion exchange column (Pharmacia) and desalted.
NMR sampling
The HuC monodomain fragments, RBD1 and RBD2, were prepared in 20 mM potassium oxalate buffer (pH 3.6) containing 10% 2H2O. For structure determination, RBD1 and RBD2 were concentrated to 1–2 mM. For the chemical shift perturbation studies, the concentration of the monodomain fragment was adjusted to 300 µM and an RNA was added to achieve the molecular ratio as indicated in Figure 2. The didomain fragment, RBD1–RBD2, was prepared in 20 mM potassium phosphate buffer (pH 6.0) containing 10% 2H2O and was concentrated to 1 mM. For measurement of 13C/15N-labeled RBD1–RBD2 complexed with RNA, ~100 U/ml of the RNase inhibitor SIN101 (Toyobo, Japan) and 1 mM deuterated dithiothreitol were added to prevent RNA degradation.
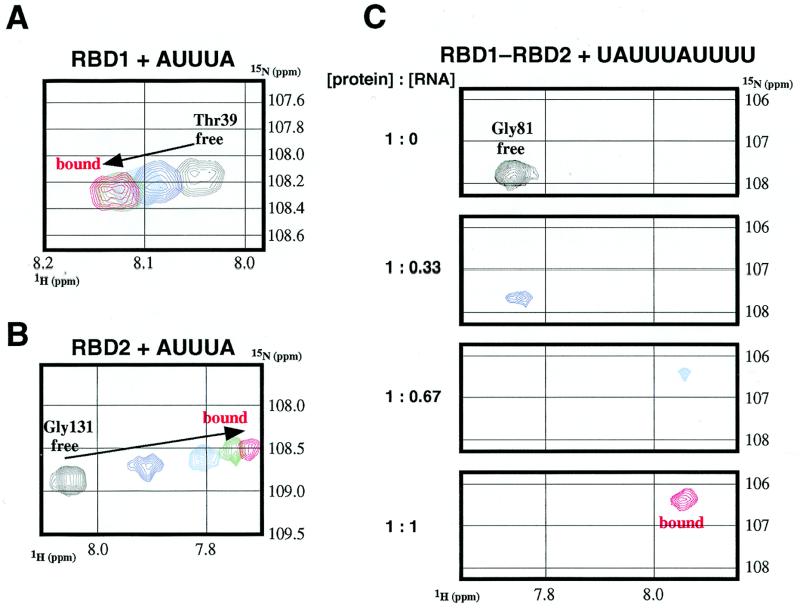
Figure 2.
Changes in the 2D 1H-15N HSQC crosspeaks of the HuC monodomain (RBD1 and RBD2) and didomain (RBD1–RBD2) fragments upon addition of AUUUA and UAUUUAUUUU, respectively. (A) The Thr39 crosspeaks of HuC RBD1 at [protein]:[AUUUA] ratios of 1:0 (black), 1:0.33 (blue), 1:0.67 (cyan), 1:1 (green) and 1:1.33 (red). (B) The Gly131 crosspeaks of HuC RBD2 at [protein]:[AUUUA] ratios of 1:0 (black), 1:0.33 (blue), 1:0.67 (cyan), 1:1 (green) and 1:1.33 (red). (C) The Gly81 crosspeaks of HuC RBD1–RBD2 at [protein]:[UAUUUAUUUU] ratios of 1:0 (black), 1:0.33 (blue), 1:0.67 (cyan) and 1:1 (red).
NMR spectroscopy
NMR spectra were measured at a probe temperature of 298 K with Bruker DMX500 and DRX600 spectrometers. The water suppression was performed by selective pre-irradiation or a jump–return pulse. The DQF-COSY, TOCSY, NOESY and ROESY spectra (reviewed in 37) were measured to determine the tertiary structures of RBD1 and RBD2. The 2D 1H-15N HSQC spectra (38) were measured to detect the chemical shift perturbation. The 3D 15N-edited TOCSY-HSQC and 3D 15N-edited NOESY-HSQC spectra were measured to confirm the proton resonance assignments of RBD1 and RBD2. The HMQC-J (39) and HNHA (40) spectra were measured to obtain the φ angle constraints. For 13C/15N-labeled RBD1–RBD2, the 3D 15N-edited NOESY-HSQC and HNCA spectra were measured to obtain the sequence-specific assignments of the RNA-free form, while the 3D 15N-edited NOESY-HSQC, 3D 15N-edited TOCSY-HSQC, HNCA, HN(CO)CA, CBCB(CO)NH, HNCO (reviewed in 41) and HNCACB (42) spectra were measured to establish the sequence-specific assignments of the complex.
Structure calculations
The tertiary structures of HuC RBD1 and RBD2 were calculated by the distance geometry/simulated annealing protocol using XPLOR 3.1 (43). The procedure for tertiary structure determination is the same as that for Sxl RBD1 described previously (35). Hydrogen bond constraints were applied to amide protons for which the proton–deuterium exchange rates were slow and the acceptor oxygens could be unambiguously determined during initial rounds of structure calculation. In the case of HuC RBD1, 607 middle/long-range NOE constraints, 88 dihedral angle constraints and 29 hydrogen bond constraints were finally used. For HuC RBD2, 588 middle/long-range NOE constraints, 88 dihedral angle constraints and 31 hydrogen bond constraints were finally used. All of the 100 structure calculations were converged for both RBD1 and RBD2 and the 20 with the lowest energy were used for the average structure calculations. After the energy minimization step, the qualities of the average structures were checked by PROCHECK-NMR (44). The coordinates have been deposited in the Protein Data Bank (accession nos 1D8Z and 1D9A for RBD1 and RBD2, respectively). The figures of the tertiary structures were prepared with MOLMOL (45).
Kd calculation
The Kd value between HuC RBD1 and AUUUA was estimated from the chemical shift changes of the amide protons of Leu41 and Arg116. The chemical shift change δ was fitted to δ([R]) = δmax{Kd + [P] + [R] – [(Kd + [P] + [R])2 – 4[P][R]]1/2}/(2[P]), where [P] and [R] are the concentrations of RBD1 and AUUUA, respectively. Kd and δmax were treated as fitting parameters during the curve fitting. The Kd values were converged to 40 µM for both Leu41 and Arg116, within the standard deviations.
Model building
The model of the HuC RBD1–RBD2·UAUUUAUUUU complex was built from the crystal structure of the Sxl RBD1–RBD2·UGUUUUUUUU complex (30). The RBD1 (Ser123–Ala201) and RBD2 (Asp210–Ala289) moieties of Sxl were replaced by the solution structures of HuC RBD1 (Ser37–Ala115) and RBD2 (Asp124–Ala203), respectively. The interdomain linker of HuC RBD1–RBD2 (Arg116–Arg123) and the backbone of UAUUUAUUU were constructed from the backbone coordinates of the Sxl RBD1–RBD2·UGUUUUUUUU complex. The energy of the model structure was minimized using XPLOR 3.1 (43)
RESULTS AND DISCUSSION
The solution structures of HuC RBD1 and RBD2
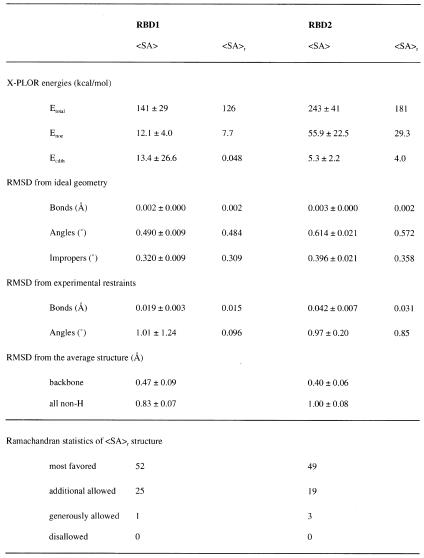
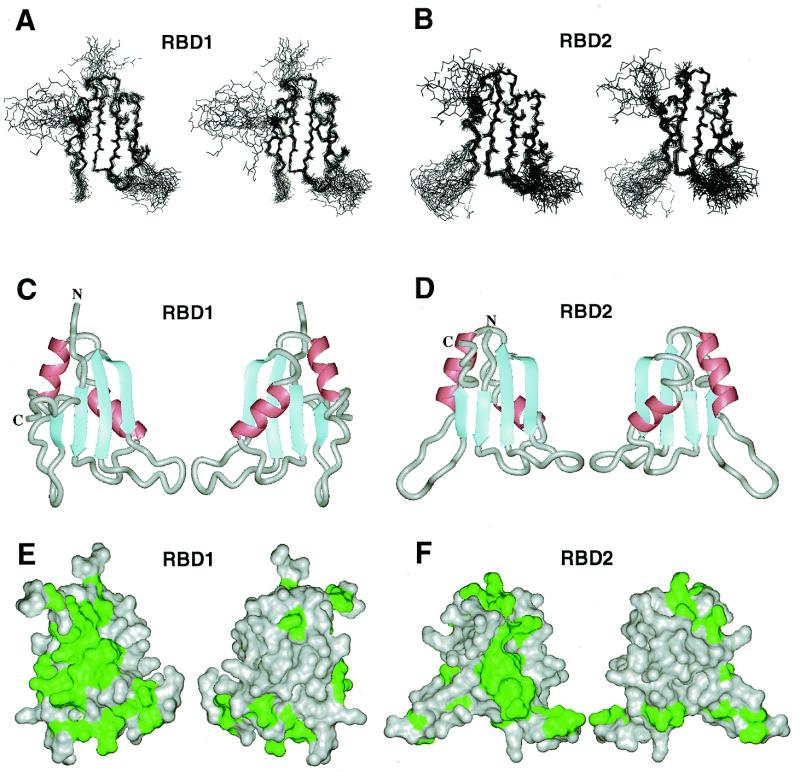
The solution structures of the two fragments of HuC, RBD1 and RBD2 were determined by 1H and 15N NMR spectroscopy and the distance geometry/simulated annealing protocol. The statistics of the 20 converged structures are summarized in Table 1. The tertiary structures of RBD1 and RBD2 have a similar βαββαβ topology (Fig. 1A–D), consisting of an antiparallel β-sheet supported by two α-helices. This topology is also common within other RBDs whose tertiary structures have been solved. HuC RBD1 and RBD2 exhibit the highest similarities to Sxl RBD1 (35) and RBD2 (46,47), respectively. Figure 1E and F shows the solvent-accessible surfaces of HuC RBD1 and RBD2, in which the residues that are identical between HuC and Sxl are colored green. Residues participating in formation of the hydrophobic core are buried in the globules and are therefore invisible in Figure 1E and F. Most of the exposed conserved residues appear either on the surfaces of the β-sheet or in the loops connecting the strands (Fig. 1E and F). On the other hand, the residues around the α-helices on the opposite sides are not conserved. In addition, the characteristic aromatic residues in the β2/β3 loop of Sxl RBD1 (35) are not conserved in HuC RBD1. Therefore, the regions on and around the β-sheets may participate in the common function between HuC and Sxl, i.e. RNA binding. On the other hand, the non-conserved residues on the α-helix sides of the RBDs and in the β2/β3 loop of RBD1 might play distinct roles in HuC and Sxl.
Table 1. Statistics of the 20 lowest energy structures of HuC RBD1 and RBD2.
<SA> is the ensemble of 20 structures. <SA>r is the restrained energy-minimized average structure. The RMSD value from the average structure was calculated for residues 40–63, 65–71, 82–101 and 111–114 in RBD1 and for residues 126–149, 151–157, 170–189 and 199–202 in RBD2. Ramachandran statistics were calculated by PROCHECK-NMR (44).
Figure 1.
The solution structures of HuC RBD1 and RBD2. Superimposition of the 20 lowest energy backbone structures of HuC (A) RBD1 and (B) RBD2 in stereoview. The mean structures of HuC (C) RBD1 and (D) RBD2 (left, the β-sheet side; right, the α-helix side) in ribbon representation. The identical amino acid residues between HuC and Sxl are indicated in green on the solvent-accessible surfaces of HuC (E) RBD1 and (F) RBD2 (left, the β-sheet side; right, the α-helix side).
The 2D 1H-15N HSQC and 3D 15N-edited NOESY-HSQC spectra of RBD1–RBD2 (the longer fragment consisting of both RBD1 and RBD2) were measured and found to be quite similar to the sum of those of the individual domains, RBD1 and RBD2 (data not shown). The C-terminal flanking region (Arg81–Arg88) of the monodomain RBD1 fragment serves as a linker between the two typical RBD folds in the didomain RBD1–RBD2 fragment and is therefore designated hereafter as the interdomain linker. The residues of the interdomain linker of RBD1–RBD2 showed no middle/long-range NOEs and only sharp sequential NOEs; no interactions between the RBD1 and RBD2 moieties were detected (data not shown). Therefore, this indicates that the orientation between the RBD1 and RBD2 moieties of RBD1–RBD2 is not fixed. Correspondingly, this mobility is also observed for Sxl RBD1–RBD2 (32,48).
ARE binding of HuC RBD1 and RBD2
First, we tested whether the individual RBD fragments, RBD1 and RBD2, can bind to ARE. We chose the pentamer AUUUA, which is known as an ARE minimal sequence (2,3), as a ligand. The 2D 1H-15N HSQC spectra of 15N-labeled RBD1 and RBD2 were measured with stepwise addition of AUUUA. Both RBD1 and RBD2 showed continuous chemical shift changes upon addition of AUUUA (Fig. 2A and B). These changes indicate that both RBD1 and RBD2 interact weakly with AUUUA, in fast exchange on the NMR time scale. From the profile of the chemical shift changes, the Kd value between RBD1 and AUUUA was estimated to be ~40 µM. The Kd value for RBD2 was not quantitatively estimated, as the interaction was too weak.
We also performed similar experiments with longer ARE RNAs, UAUUUAUUUU and AUUUAUUUUA. Although the chemical shift changes were somewhat larger, the exchange between the free and bound states remained fast on the NMR time scale (data not shown). Thus, we conclude that RBD1 and RBD2 in isolation interact primarily with the pentamer AUUUA, while surrounding uridine residues slightly increase the affinity.
ARE binding of HuC RBD1–RBD2
Next, we examined the interaction between the didomain fragment, RBD1–RBD2, and AUUUA by NMR. However, many HSQC crosspeaks disappeared upon addition of the RNA (data not shown). This indicates that RBD1–RBD2 interacts with AUUUA with an intermediate exchange rate on the NMR time scale. As each of the RBD1 and RBD2 domains can interact with AUUUA, it is possible that two molecules of AUUUA interact with RBD1–RBD2. Then, we measured the imino proton resonances of seven longer ARE RNAs (AUUUAUUUA, UAUUUAUUUU, AUUUUAUUUA, AUUUUAUUUU, UAUUUUAUUUU, UAUUUUUUUU and UUUUUUUUUU) complexed with RBD1–RBD2 (data not shown) and found that UAUUUAUUUU can form the most stable complex with RBD1–RBD2, with respect to exchange of the imino protons with the solvent water. The HSQC crosspeaks of the RNA-free and RNA-bound states of RBD1–RBD2 were observed separately (Fig. 2C), which is the typical pattern of slow exchange on the NMR time scale. Therefore, the complex of HuC RBD1–RBD2 with UAUUUAUUUU is much more stable than that with AUUUA.
In summary, the N-terminal tandem RBDs in HuC cooperate to achieve stronger and more specific binding to the decamer UAUUUAUUUU, while the individual RBDs can bind only weakly to the pentamer AUUUA. The strong affinity of RBD1–RBD2 for UAUUUAUUUU seems to be suitable for HuC to accomplish its putative functions, such as protection and/or transport of ARE-containing mRNAs. These results are consistent with the conclusion from a previous UV crosslinking study that the RBD1–RBD2 region of HuC is required for tight and specific RNA binding (9). Correspondingly, a similar cooperation between the tandem RBDs of Sxl has been indicated by NMR spectroscopy: fast exchange was observed for Sxl RBD2 (47,49) (no data are available for Sxl RBD1), while slow exchange was observed for Sxl RBD1–RBD2 (49).
While the pentamer AUUUA has been suggested to be the minimal element of the ARE (2,3), our results show that HuC RBD1–RBD2 requires a longer RNA, such as UAUUUAUUUU, to achieve tight binding. The UAUUUAUUUU sequence agrees with the motif RU2–5R1–2U2–5R (R = A or G) deduced from an in vitro selection study of HuC (9). Thus, HuC may not target all of the AUUUA motifs, but only certain AREs containing a longer continuous motif, such as UAUUUAUUUU. For example, the class II ARE, which contains a cluster of AUUUA motifs, may be a target that HuC would bind tightly.
Mapping of RNA chemical shift perturbations on HuC RBD1, RBD2 and RBD1–RBD2
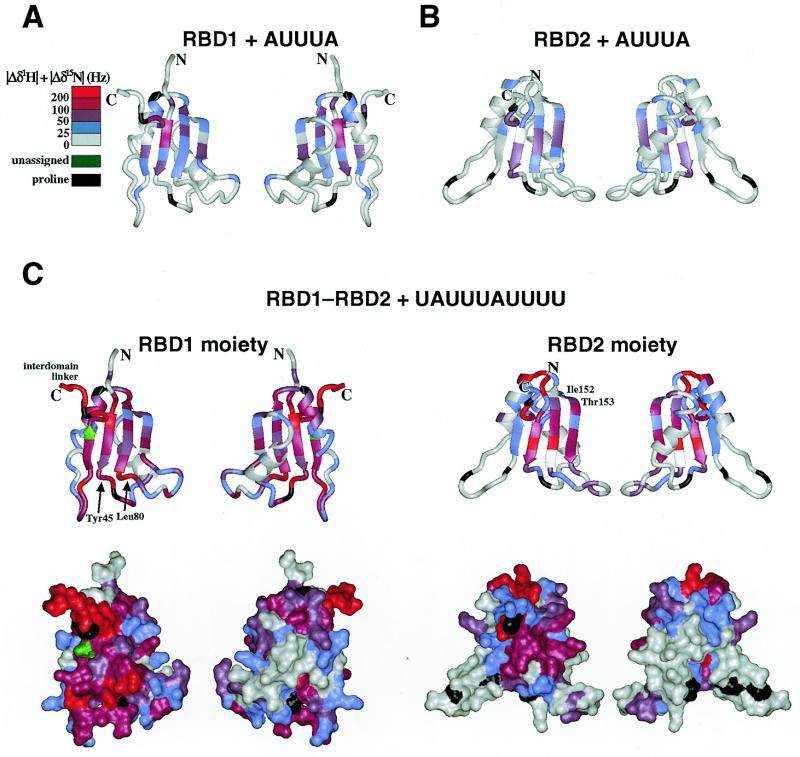
In the cases of the monodomain fragments, RBD1 and RBD2, with AUUUA, the backbone 1H-15N chemical shift differences between the free and RNA-bound states in fast exchange were easily obtained by tracing the stepwise shifts of the crosspeaks from one to the other. Significant chemical shift perturbations are clearly mapped on the β-sheets of RBD1 and RBD2 (Fig. 3A and B), indicating that their RNA-binding sites are located on the β-sheet surfaces. The β-sheet surface of either RBD1 or RBD2 is involved in the aforementioned binding of AUUUA with the RBD1–RBD2 didomain fragment, probably in a similar manner to those of the RBD1 and RBD2 monodomain fragments.
Figure 3.
The backbone 1H and 15N chemical shift changes of HuC (A) RBD1 and (B) RBD2 upon addition of AUUUA at a molar ratio of 1.33 and those of HuC (C) RBD1–RBD2 upon addition of UAUUUAUUUU at a molar ratio of 1.0. The chemical shift changes [│Δδ1H│ + │Δδ15N│] are color coded at the five levels indicated on the two sides (left, the β-sheet side; right, the α-helix side) of the ribbon models [A, B and C (top)] and the solvent-accessible surfaces [C (bottom)] of the HuC RBD1 and RBD2 solution structures. The C-terminal region of RBD1 corresponds to the interdomain linker. For the Ser113 residue, shown in green, the backbone 1H–15N crosspeak was not assigned. The Pro residues, which lack the backbone 1H–15N crosspeak, are indicated in black.
On the other hand, because of the slow exchange between the free and bound states, the resonance assignment of RBD1–RBD2 complexed with UAUUUAUUUU was performed independently from that of the free form; all of the backbone 1H-15N HSQC crosspeaks, except for that of Ser113, were assigned. Figure 3C shows the RBD1–RBD2·UAUUUAUUUU chemical shift perturbations mapped on the solution structures of RBD1 and RBD2. The residues with large chemical shift perturbations were mapped not only on the β-sheet surfaces but also on the surrounding loops and the interdomain linker (Fig. 3C). In contrast, no appreciable chemical shift perturbations were observed for these regions of the individual domains in isolation (Fig. 3A and B). Therefore, UAUUUAUUUU interacts with both of the two β-sheet surfaces together with the surrounding amino acid residues of RBD1–RBD2, while AUUUA interacts primarily with the β-sheet surface of either RBD. In more detail, these additional interactions with the RNA, characteristic of the tandem arrangement of RBD1–RBD2, involve three regions around Tyr45, Leu80 and Ile152–Thr153 and the entire interdomain linker (the C-terminal flanking region in the RBD1 structure) (Fig. 3C). Furthermore, even for the residues on the β-sheet surfaces, the extents of the chemical shift changes are larger and the interactions are therefore more intensive for RBD1–RBD2·UAUUUAUUUU than for RBD1·AUUUA and RBD2·AUUUA. Consequently, the two RBDs of HuC RBD1–RBD2 cooperate with the interdomain linker to bind the longer RNA tightly, whereas the interdomain linker is very flexible and therefore does not fix the two RBDs to each other in the RNA-free state (described above).
Similarity between HuC RBD1–RBD2 and Sxl RBD1–RBD2
The HuC RBD1–RBD2 residues that exhibited significant chemical shift perturbations (Fig. 3C) coincide surprisingly well with those conserved in Sxl RBD1–RBD2 (Fig. 1E and F). This suggests that the HuC and Sxl RBD1–RBD2 fragments bind with RNA in quite similar manners. In this context, a chemical shift perturbation analysis has been done for Sxl RBD1–RBD2 complexed with GUUUUUUUUC (32). However, we cannot directly compare the chemical shift changes between HuC and Sxl because ~15% of the backbone crosspeaks, most of which are from the RNA-interacting amino acid residues, have not been assigned for the Sxl RBD1–RBD2·GUUUUUUUUC complex (32). Nevertheless, as far as the assigned crosspeaks of Sxl are concerned, the RNA chemical shift perturbation patterns are similar between the HuC and Sxl RBD1–RBD2 fragments. First, the chemical shift perturbations were mapped mostly on the β-sheets of both HuC and Sxl (32). Second, some residues around the β-sheets in Sxl exhibited large chemical shift perturbations (32), including the regions corresponding to those around Tyr45 and Ile152–Thr153 in HuC. Third, the interdomain linker between RBD1 and RBD2 exhibits large chemical shift perturbations in both HuC and Sxl (32), which may be caused by rearrangement of the domains and/or direct contact with the RNA.
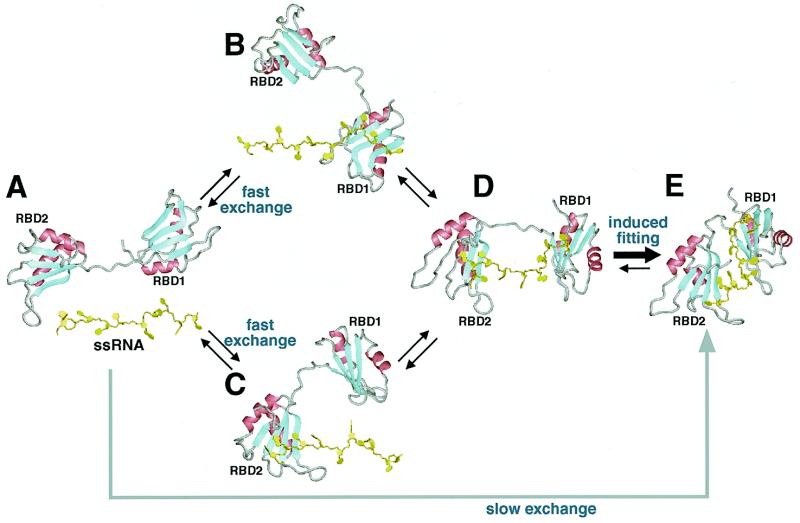
Thus, we built a structural model of the HuC RBD1–RBD2·UAUUUAUUUU complex (Fig. 4B) on the basis of the crystal structure of the Sxl RBD1–RBD2·UGUUUUUUUU complex (Fig. 4A; 30). In the model, the RNA directly contacts the β-sheets, the regions around Tyr45, Leu80 and Ile152–Thr153 and the interdomain linker, all of which exhibit significant chemical shift perturbations in the HuC RBD1–RBD2·UAUUUAUUUU complex (Fig. 4B). We therefore concluded that the HuC and Sxl RBD1–RBD2 proteins have remarkably similar RNA-binding modes. The deep RNA-binding cleft is likely to be formed by reorientation of the two tandem RBDs and a conformational change in the flexible interdomain linker, as suggested for Sxl RBD1–RBD2 (30,48,50). Moreover, we propose here that tight complex formation involving the entire RNA and RBD1–linker–RBD2 molecules may be preceded by weaker interactions involving either of the two RBDs and part of the binding sequence of the RNA (Fig. 5). Actually, we showed in this study that both the HuC RBD1 and RBD2 monodomain fragments and the RBD1–RBD2 didomain fragment can bind to AUUUA, a part of the longer binding sequence (UAUUUAUUUU). Similarly, the Sxl RBD2 monodomain fragment can bind specifically to the target RNAs, although the binding is much weaker than that of the didomain fragment (47).
Figure 4.
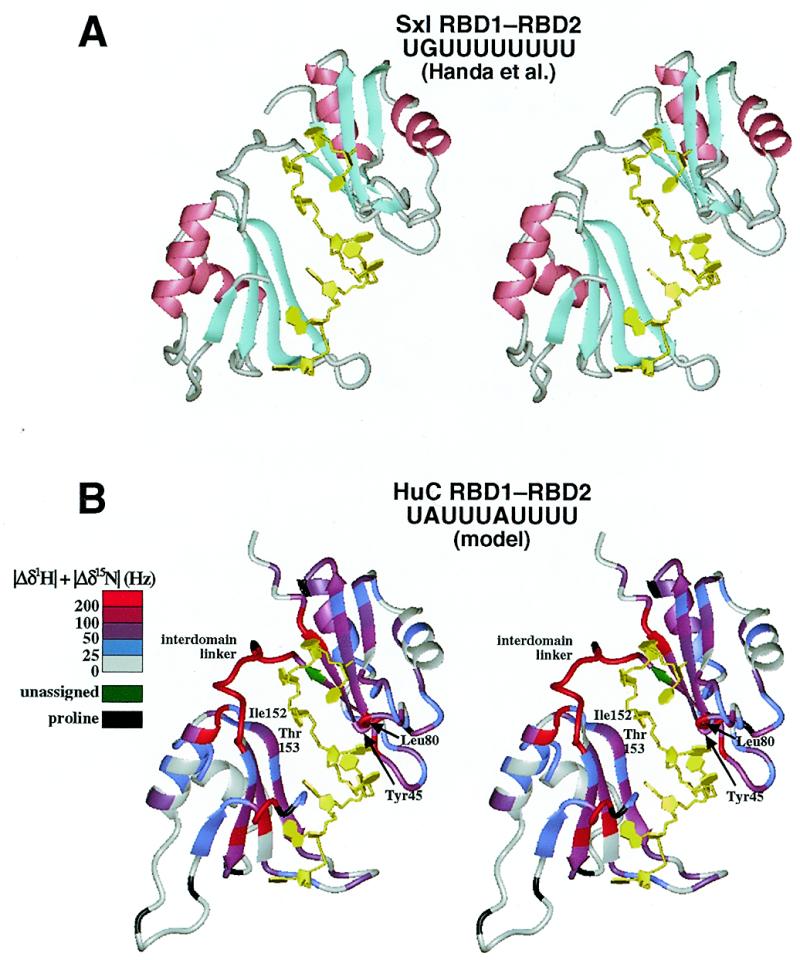
Modeling of the HuC RBD1–RBD2·UAUUUAUUUU structure. (A) The crystal structure of Sxl RBD1–RBD2 complexed with UGUUUUUUUU (30) used as the template for modeling (stereoview). (B) A model structure of the HuC RBD1–RBD2·UAUUUAUUUU complex (stereoview), with color coding of the chemical shift changes as in Figure 3.
Figure 5.
A possible mechanism of UAUUUAUUUU recognition by HuC RBD1–RBD2. The didomain fragment and the single-stranded RNA (A) associate with each other, first at either the RBD1 moiety (B) or the RBD2 moiety (C) (fast exchange) and then at both of the RBD1 and RBD2 moieties (D) to form, through induced fitting, a tight complex (E) (slow exchange).
Phylogeny of HuC and Sxl
The HuC and Sxl RBD1–RBD2 proteins are similar not only in RNA-binding mode but also in primary and tertiary structure, as described above. Thus, HuC and Sxl must be derived from a common ancestor that had a didomain architecture. The HuC homologs, i.e. the ELAV/Hu proteins, are found in both vertebrates and invertebrates (13), whereas Sxl and its homologs are found only in insects (51–55). The ELAV/Hu proteins consist not only of the N-terminal RBD1–RBD2 region but also of a third C-terminal RBD connected through a long linker region. On the other hand, the Sxl family proteins mostly consist of the RBD1–RBD2 region. These facts strongly suggest that Sxl protein was derived from the RBD1–RBD2 region of an ELAV/Hu-type ancestor during insect evolution. Presumably, the ancestor of Sxl changed its specificity from AU-rich elements to the present target sequences with a much longer stretch of U, which was probably coupled with the acquisition of different biological functions. In fact, the Drosophila (fruitfly) Sxl protein is known to regulate sex-specific alternative splicing of its own and transformer mRNA (56–58), whereas the Sxl proteins from Chrysomya rufifacies (blowfly), Musca domestica (housefly) and Ceratitis capitata (medfly) seem to have some other, unknown function(s) (53–55).
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas to S.Y. from the Ministry of Education, Science, Culture and Sports of Japan. M.I. was supported by the JPSP Fellowships for Japanese Junior Scientists.
PDB accession nos 1D8Z, 1D9A
REFERENCES
- 1.Caput D., Beutler,B., Hartog,K., Thayer,R., Brown-Shimer,S. and Cerami,A. (1986) Proc. Natl Acad. Sci. USA, 83, 1670–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw G. and Kamen,R. (1986) Cell, 46, 659–667. [DOI] [PubMed] [Google Scholar]
- 3.Xu N., Chen,C.Y. and Shyu,A.B. (1997) Mol. Cell. Biol., 17, 4611–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton B.J., Nagy,E., Malter,J.S., Arrick,B.A. and Rigby,W.F. (1993) J. Biol. Chem., 268, 8881–8887. [PubMed] [Google Scholar]
- 5.Zhang W., Wagner,B.J., Ehrenman,K., Schaefer,A.W., DeMaria,C.T., Crater,D., DeHaven,K., Long,L. and Brewer,G. (1993) Mol. Cell. Biol., 13, 7652–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagawa J., Waldner,H., Meyer-Monard,S., Hofsteenge,J., Jeno,P. and Moroni,C. (1995) Proc. Natl Acad. Sci. USA, 92, 2051–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine T.D., Gao,F., King,P.H., Andrews,L.G. and Keene,J.D. (1993) Mol. Cell. Biol., 13, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J., Dalmau,J., Szabo,A., Rosenfeld,M., Huber,J. and Furneaux,H. (1995) Neurology, 45, 544–550. [DOI] [PubMed] [Google Scholar]
- 9.Abe R., Sakashita,E., Yamamoto,K. and Sakamoto,H. (1996) Nucleic Acids Res., 24, 4895–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung S., Jiang,L., Cheng,S. and Furneaux,H. (1996) J. Biol. Chem., 271, 11518–11524. [DOI] [PubMed] [Google Scholar]
- 11.Ma W.J., Cheng,S., Campbell,C., Wright,A. and Furneaux,H. (1996) J. Biol. Chem., 271, 8144–8151. [DOI] [PubMed] [Google Scholar]
- 12.Burd C.G. and Dreyfuss,G. (1994) Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 13.Antic D. and Keene,J.D. (1997) Am. J. Hum. Genet., 61, 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakamatsu Y. and Weston,J.A. (1997) Development, 124, 3449–3460. [DOI] [PubMed] [Google Scholar]
- 15.Fujita M., Kawano,T., Ohta,A. and Sakamoto,H. (1999) Biochem. Biophys. Res. Commun., 260, 646–652. [DOI] [PubMed] [Google Scholar]
- 16.Abe R., Yamamoto,K. and Sakamoto,H. (1996) Nucleic Acids Res., 24, 2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung S., Eckrich,M., Perrone-Bizzozero,N., Kohn,D.T. and Furneaux,H. (1997) J. Biol. Chem., 272, 6593–6598. [DOI] [PubMed] [Google Scholar]
- 18.Levy N.S., Chung,S., Furneaux,H. and Levy,A.P. (1998) J. Biol. Chem., 273, 6417–6423. [DOI] [PubMed] [Google Scholar]
- 19.Fan X.C. and Steitz,J.A. (1998) EMBO J., 17, 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng S.S., Chen,C.Y., Xu,N. and Shyu,A.B. (1998) EMBO J., 17, 3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Good P.J. (1995) Proc. Natl Acad. Sci. USA, 92, 4557–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain R.G., Andrews,L.G., McGowan,K.M., Pekala,P.H. and Keene,J.D. (1997) Mol. Cell. Biol., 17, 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antic D., Lu,N. and Keene,J.D. (1999) Genes Dev., 13, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasashima K., Terashima,K., Yamamoto,K., Sakashita,E. and Sakamoto,H. (1999) Genes Cells, 4, 667–683. [DOI] [PubMed] [Google Scholar]
- 25.Koushika S.P., Lisbin,M.J. and White,K. (1996) Curr. Biol., 6, 1634–1641. [DOI] [PubMed] [Google Scholar]
- 26.Szabo A., Dalmau,J., Manley,G., Rosenfeld,M., Wong,E., Henson,J., Posner,J.B. and Furneaux,H.M. (1991) Cell, 67, 325–333. [DOI] [PubMed] [Google Scholar]
- 27.Kenan D.J., Query,C.C. and Keene,J.D. (1991) Trends Biochem. Sci., 16, 214–220. [DOI] [PubMed] [Google Scholar]
- 28.Oubridge C., Ito,N., Evans,P.R., Teo,C.H. and Nagai,K. (1994) Nature, 372, 432–438. [DOI] [PubMed] [Google Scholar]
- 29.Ding J., Hayashi,M.K., Zhang,Y., Manche,L., Krainer,A.R. and Xu,R.M. (1999) Genes Dev., 13, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- 31.Deo C.D., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- 32.Lee A.L., Volkman,B.F., Robertson,S.A., Rudner,D.Z., Barbash,D.A., Cline,T.W., Kanaar,R., Rio,D.C. and Wemmer,D.E. (1997) Biochemistry, 36, 14306–14317. [DOI] [PubMed] [Google Scholar]
- 33.Studier F.W. and Moffatt,B.A. (1986) J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- 34.Kigawa T., Muto,Y. and Yokoyama,S. (1995) J. Biomol. NMR, 6, 129–134. [DOI] [PubMed] [Google Scholar]
- 35.Inoue M., Muto,Y., Sakamoto,H., Kigawa,T., Takio,K., Shimura,Y. and Yokoyama,S. (1997) J. Mol. Biol., 272, 82–94. [DOI] [PubMed] [Google Scholar]
- 36.Wincott F., DiRenzo,A., Shaffer,C., Grimm,S., Tracz,D., Workman,C., Sweedler,D., Gonzalez,C., Scaringe,S. and Usman,N. (1995) Nucleic Acids Res., 23, 2677–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markley J.L. (1989) Methods Enzymol., 176, 12–64. [DOI] [PubMed] [Google Scholar]
- 38.Bodenhausen G. and Ruben,D.J. (1980) Chem. Phys. Lett., 69, 185–189. [Google Scholar]
- 39.Kay L.E. and Bax,A. (1990) J. Magn. Reson., 86, 110–126. [Google Scholar]
- 40.Vuister G.W. and Bax,A. (1993) J. Am. Chem. Soc., 115, 7772–7777. [Google Scholar]
- 41.Clore G.M. and Gronenborn,A.M. (1994) Methods Enzymol., 239, 349–363. [DOI] [PubMed] [Google Scholar]
- 42.Wittekind M. and Mueller,L. (1993) J. Magn. Reson. B, 101, 201–205. [Google Scholar]
- 43.Brünger A.T. (1993) X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- 44.Laskowski R.A., Rullmannn,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- 45.Koradi R., Billeter,M. and Wüthrich,K. (1996) J. Mol. Graphics, 14, 51–55. [DOI] [PubMed] [Google Scholar]
- 46.Lee A.L., Kanaar,R., Rio,D.C. and Wemmer,D.E. (1994) Biochemistry, 33, 13775–13786. [DOI] [PubMed] [Google Scholar]
- 47.Chi S.W., Muto,Y., Inoue,M., Kim,I., Sakamoto,H., Shimura,Y., Yokoyama,S., Choi,B.S. and Kim,H. (1999) Eur. J. Biochem., 260, 649–660. [DOI] [PubMed] [Google Scholar]
- 48.Crowder S.M., Kanaar,R., Rio,D.C. and Alber,T. (1999) Proc. Natl Acad. Sci. USA, 96, 4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanaar R., Lee,A.L., Rudner,D.Z., Wemmer,D.E. and Rio,D.C. (1995) EMBO J., 14, 4530–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hughson F.M. and Schedl,P. (1999) Nature Struct. Biol., 6, 499–502. [DOI] [PubMed] [Google Scholar]
- 51.Bell L.R., Maine,E.M., Schedl,P. and Cline,T.W. (1988) Cell, 55, 1037–1046. [DOI] [PubMed] [Google Scholar]
- 52.O’Neil M.T. and Belote,J.M. (1992) Genetics, 131, 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller-Holtkamp F. (1995) J. Mol. Evol., 41, 467–477. [DOI] [PubMed] [Google Scholar]
- 54.Saccone G., Peluso,I., Artiaco,D., Giordano,E., Bopp,D. and Polito,L.C. (1998) Development, 125, 1495–1500. [DOI] [PubMed] [Google Scholar]
- 55.Meise M., Hilfiker-Kleiner,D., Dubendorfer,A., Brunner,C., Nothiger,R. and Bopp,D. (1998) Development, 125, 1487–1494. [DOI] [PubMed] [Google Scholar]
- 56.Sosnowski B.A., Belote,J.M. and McKeown,M. (1989) Cell, 58, 449–459. [DOI] [PubMed] [Google Scholar]
- 57.Bell L.R., Horabin,J.I., Schedl,P. and Cline,T.W. (1991) Cell, 65, 229–239. [DOI] [PubMed] [Google Scholar]
- 58.Sakamoto H., Inoue,K., Higuchi,I., Ono,Y. and Shimura,Y. (1992) Nucleic Acids Res., 20, 5533–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.