Summary
Phytophthora species are the most destructive plant pathogens worldwide and the main threat to agricultural and natural ecosystems; however, their pathogenic mechanism remains largely unknown. Here, we show that Avh113 effector is required for the virulence of Phytophthora sojae and is important for development of Phytophthora root and stem rot (PRSR) in soybean (Glycine max). Ectopic expression of PsAvh113 enhanced viral and Phytophthora infection in Nicotiana benthamiana. PsAvh113 directly associated with the soybean transcription factor GmDPB, inducing its degradation by the 26S proteasome. The internal repeat 2 (IR2) motif of PsAvh113 was important for its virulence and interaction with GmDPB, while silencing and overexpression of GmDPB in soybean hairy roots altered the resistance to P. sojae. Upon binding to GmDPB, PsAvh113 decreased the transcription of the downstream gene GmCAT1, which acts as a positive regulator of plant immunity. Furthermore, we revealed that PsAvh113 suppressed the GmCAT1‐induced cell death by associating with GmDPB, thereby enhancing plant susceptibility to Phytophthora. Together, our findings reveal a vital role of PsAvh113 in inducing PRSR in soybean and offer a novel insight into the interplay between defence and counter‐defence during the P. sojae infection of soybean.
Keywords: Effector, host‐pathogen interaction, Phytophthora root and stem rot, P. sojae, soybean, transcription factor
Introduction
Microbial pathogens and their plant hosts are involved in a perpetual arms race, which is well demonstrated by the classical zig‐zag model (Jones and Dangl, 2006). A fierce battle of attack, counter‐attack and counter‐counter‐attack arises in the cytoplasmic space, which frequently governs the outcome of pathogen effector–plant interactions (Jones and Dangl, 2006; Wang et al., 2019). Phytophthora, a filamentous eukaryotic pathogen, is responsible for many devastating plant diseases (Kamoun et al., 2015). Phytophthora root and stem rot (PRSR) caused by Phytophthora sojae is one of the most devastating diseases of soybean (Glycine max), resulting in annual losses of $1–2 billion globally (Tyler et al., 2006). Despite extensive studies, the mechanism of its hemibiotrophic pathogenesis is still inadequately understood. Accumulating evidence demonstrates that P. sojae secretes an arsenal of host cytoplasmic effectors, some of which interfere with plant innate immunity by modifying host target functions and disrupting the immune signalling network (Ai et al., 2021; Hou et al., 2019; Jing et al., 2016; Kong et al., 2017; Lin et al., 2021; Lu et al., 2023; Qiao et al., 2013; Song et al., 2015; Wang and Wang, 2018). However, the host targets and their molecular mechanisms regulated by the majority of P. sojae effectors remain unexplored.
DRFT1‐polypeptide1 (DP1; also known as Dimerization Partner 1) belongs to a family of transcription factors that heterodimerize with E2 PROMOTER BINDING FACTOR (E2F) to bind to target DNA (Girling et al., 1993). E2F, a nuclear factor in HeLa cells infected with adenovirus, is able to stimulate the early expression of the E2 virus gene, named E2F, which is responsible for gene transcription (Kovesdi et al., 1987). Numerous studies have shown that DP proteins can heterodimerize with E2F to form a complex, which exhibits greater DNA‐binding ability and enhances the transcription of E2F target genes by binding to the joint E2 recognition site (Helin et al., 1993). The E2F/DP family of transcription factors includes typical E2Fs and atypical E2Fs (DP/E2F‐like [DEL]) as well as their related DP proteins, and plays a pivotal role in controlling the activity of cyclin‐dependent kinase and the progression of cell cycle in plants and animals (Kent and Leone, 2019; Lammens et al., 2009).
To date, a number of E2F family members have been identified in the animal kingdom; for example, seven E2F and two DP proteins in mammals, two E2F proteins in Drosophila, and one DP protein in Caenorhabditis elegans (Dimova and Dyson, 2005). E2F family members have also been identified in multiple plant species, including Arabidopsis, Nicotiana benthamiana, carrot and rice (Kosugi and Ohashi, 2002a). Among these plant species, Arabidopsis is the most well‐characterized for E2F/DP proteins. Studies show that the Arabidopsis genome encodes three typical E2Fs (AtE2FA, AtE2FB and AtE2FC), three atypical E2Fs (AtDEL1, AtDEL2 and AtDEL3) and two DPs (AtDPA and AtDPB) (Mariconti et al., 2002). For instance, AtE2FA and AtE2FB activate the expression of genes involved in the initiation and progression of the S‐phase, whereas AtE2FC and AtDEL act as repressors and induce the G2 or G2/M cell cycle arrest, which potentially negatively regulates some E2F target genes involved in the G1/S‐phase (Vlieghe et al., 2005). However, several studies show that the E2F/DP family proteins are also involved in multiple biological processes, such as mitosis, response to DNA damage, DNA repair and cell differentiation, apoptosis and plant development (DeGregori and Johnson, 2006). The Arabidopsis e2fabc triple mutant is highly susceptible to the bacterial pathogen Pseudomonas syringae pv. maculicola (Psm) strain ES4326, and does not express some of the immune response‐related genes (Wang et al., 2014). AtDEL1 participates in the immune response by regulating the expression of the EDS5 gene, which encodes a salicylic acid (SA) transporter (Chandran et al., 2014). However, the above‐mentioned studies focused on the mechanism of E2F proteins in plants and animals, and little is known about the role of DP proteins in plants.
The catalase (CAT) family consists of highly conserved enzymes that convert hydrogen peroxide (H2O2) into water and oxygen, and plays a critical role in the response to abiotic and biotic stresses (Petrov and Van Breusegem, 2012). Phylogenetic analysis suggests that most animals contain a single CAT gene, whereas angiosperms such as N. benthamiana, Arabidopsis, maize, pumpkin and rice contain three CAT genes (Mhamdi et al., 2010). In Arabidopsis, AtCAT1 is an essential gene that responds to numerous abiotic stresses by scavenging H2O2. Moreover, AtCAT2 and AtCAT3 are involved in eliminating H2O2 in both light and dark conditions, leading to the homeostasis of reactive oxygen species (ROS) (Mhamdi et al., 2010). The mutation of AtCAT2 results in enhanced resistance to bacterial pathogens, mainly because the reduced CAT activity increases H2O2 accumulation. Recent studies show that the accumulated SA binds to AtCAT2 to inhibit its enzymatic activity upon exposure to biotrophic pathogens (Yuan et al., 2017). Although CAT enzymes play an important role in the association with several viral effectors through distinct mechanisms (Inaba et al., 2011; Jiao et al., 2021; Mathioudakis et al., 2013; Roshan et al., 2018; Yang et al., 2020), two CRN effectors of P. sojae ectopically regulate programmed cell death (PCD) by disrupting CAT activity and perturbing H2O2 homeostasis in N. benthamiana (Zhang et al., 2015). Nevertheless, how P. sojae modulates soybean CATs to overcome host defence remains largely unexplored.
Here, we report the identification of a virulence effector of P. sojae, PsAvh113, using the potato virus X (PVX)‐based virus‐induced virulence effector (VIVE) assay, and its role in facilitating P. sojae infection. We show that PsAvh113 is important for Phytophthora infection in both N. benthamiana and soybean. PsAvh113 associates with the soybean transcription factor GmDPB and causes its degradation by the 26S proteasome. Furthermore, GmDPB binds directly to the promoter of GmCAT1 and induces its expression. PsAvh113 suppressed the GmCAT1‐induced cell death by associating with GmDPB. Taken together, these results indicate that PsAvh113 inhibits the expression of GmCAT1 by associating with transcription factor GmDPB, thus enhancing the susceptibility to P. sojae in soybean.
Results
PsAvh113 contributes to Phytophthora infection in plants
To identify the key virulence effectors of P. sojae, we performed the VIVE rapid functional screening assay in N. benthamiana (Shi et al., 2020). Compared with the mock treatment, N. benthamiana plants infected with PVX carrying PsAvh113 displayed strong viral symptoms and high‐level viral RNA accumulation in leaves, whereas those inoculated with the empty vector displayed mild viral symptoms and lower viral RNA abundance (Figures 1a,b and S1a). These results suggest that PsAvh113 encodes a potential virulence effector that promotes viral infection. PsAvh113 is predicted to a typical P. sojae RxLR effector, and its amino acid sequence was highly conserved among various strains of P. sojae (Figure S1b). The expression of PsAvh113 was significantly induced at an early stage of infection, reaching a peak at 12 h post‐inoculation, which was approximately 50‐fold higher than its expression at mycelia (Figure S1c).
Figure 1.
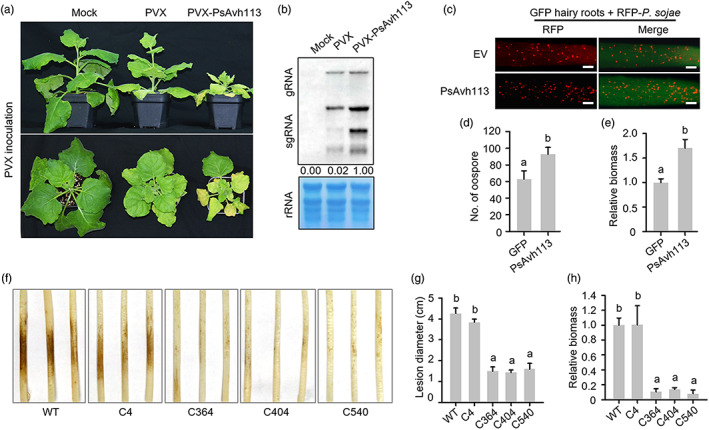
PsAvh113 is an important virulence factor of Phytophthora sojae. (a) Phenotypic comparison of Nicotiana benthamiana plants infiltrated with PVX‐PsAvh113, PVX, or no construct (mock treatment; negative control) at 21 days post‐infiltration (dpi). PsAvh113 expression retarded plant growth and caused leaf yellowing and atrophy. (b) RNA blot showing the accumulation of PVX genomic and subgenomic RNAs at 14 dpi. (c) Oospore production in soybean hairy roots expressing PsAvh113 or empty vector (EV) and inoculated with RFP‐labelled P. sojae (RFP‐P6497). Scale bars, 0.25 mm. (d) Analysis of the number of P. sojae oospores in soybean hairy roots at 48 hpi using a confocal microscope. (e) Quantification of P. sojae biomass in soybean hairy roots by qPCR. (f) Comparison of disease symptoms in soybean hypocotyls inoculated with the PsAvh113‐edited P. sojae generated using the CRISPR/Cas9 technology. Pathogen virulence was greatly impaired in soybean hypocotyls. Disease symptoms in etiolated hypocotyls were observed and photographed at 3 dpi. The C4 transformant without PsAvh113‐editing events was used as a control. (g) Lesion length on soybean hypocotyls upon inoculation with different PsAvh113 knockout transformants. (h) Relative biomass of P. sojae as determined by qRT‐PCR. Data represent mean ± standard error (SE). Different letters in (d), (e), (g), and (h) indicate statistically significant differences (P < 0.01; Duncan's multiple range test). The experiment was performed in triplicate with similar results.
To confirm the contribution of PsAvh113 to virulence in the host plant, the PsAvh113 gene was transiently expressed in HC6 soybean hairy roots (Figure S1d), which were then inoculated with the RFP‐labelled wild‐type (WT) P. sojae strain P6497 (P6497‐RFP). Compared with hairy roots expressing the empty vector (EV) control, those expressing PsAvh113 produced more oospores (Figure 1c,d) and accumulated greater P. sojae biomass (Figure 1e). To further investigate the importance of PsAvh113 in plant infection, we knocked out PsAvh113 in the WT P. sojae strain P6497 using the CRISPR/Cas9 system. Protoplasts of P. sojae were co‐transformed with the two unique sgRNAs (Figure S1e) and the hSpCas9 expression plasmid. Subsequently, many transformants were generated and identified by genomic DNA (gDNA)‐based PCR, and three homozygous PsAvh113 knockout mutants (C364, C404 and C540) were inoculated in soybean hypocotyls (Figure S1f). Compared with the WT strain, the three PsAvh113 knockout mutants showed no difference in growth (Figure S1g) but had a significantly reduced ability to cause disease (Figure 1f). Furthermore, all PsAvh113 knockout mutants of P. sojae caused smaller lesions on soybean seedlings than in the WT strain (Figure 1g). Consistently, P. sojae biomass was markedly lower in soybean seedlings infected with PsAvh113 knockout mutants than in seedlings infected with the WT strain (Figure 1h).
The ability of PsAvh113 to promote disease development prompted us to examine whether its expression in plants can facilitate infection. We generated two independent Arabidopsis transgenic lines expressing PsAvh113 and identified by Western blotting, which exhibited dwarfed and moderately retarded development and slightly delayed flowering compared with WT plants (Figure S2a–d). Upon inoculation with P. parasitica, both transgenic lines showed significantly more susceptibility, higher pathogen biomass and larger lesions than WT plants (Figure S2e–g). Together, these experiments indicate that PsAvh113 is required for full virulence of pathogen.
Internal repeat 2 (IR2) motif is required for the virulence activity of PsAvh113
Domain and motif searches using the Simple Modular Architecture Research Tool (SMART) database identified two IRs (IR1 and IR2) in the C‐terminal region of PsAvh113, which differed by only one amino acid (Figure 2a). To determine the virulence role of IR motifs in PsAvh113, we generated five PsAvh113 mutant proteins, including PsAvh113▵IR1 (IR1 region was substituted with alanine residues), PsAvh113▵IR2 (IR2 region was substituted with alanine residues) and three residue substitution mutants of PsAvh113 which substituted Ser108 with Met (S108M), Met122 with Ser (M122S) or Ser108 with Met and Met122 with Ser (S108M/M122S) (Figure 2a), and then determined subcellular localization of PsAvh113, PsAvh113▵IR1, PsAvh113▵IR2, PsAvh113S108M and PsAvh113M122S in N. benthamiana leaves by confocal microscopy (Figure S3a–f). The results showed that, like YFP‐PsAvh113 (WT protein), PsAvh113▵IR1 was uniformly distributed in the cytoplasm and the nucleus; however, the fluorescence intensity of YFP‐PsAvh113▵IR2 in the nucleus was significantly lower than that of PsAvh113 (Figure S3a–c), suggesting that the IR2 region is associated with the nuclear localization of PsAvh113. Similar to PsAvh113, PsAvh113S108M was uniformly distributed in the cytoplasm and the nucleus, while weak fluorescence was observed in the nucleus of the epidermic cells that expressed PsAvh113M122S (Figure S3d–f), indicating that Met122 residue of PsAvh113 plays a key role in the nuclear localization. Intriguingly, similar to PsAvh113, PsAvh113▵IR1 induced strong disease symptoms and viral RNA accumulation, whereas PsAvh113▵IR2 caused slight disease symptoms and lower viral RNA accumulation in the VIVE assay (Figure 2b,c), suggesting that the IR2 region of PsAvh113 is required for PVX infection. Similar results were obtained when PsAvh113, PsAvh113 ▵IR1 and PsAvh113 ▵IR2 were transiently expressed in soybean hairy roots, and the transformed roots were subsequently inoculated with P. sojae (Figure 2d–f). To further explore whether the virulence function of PsAvh113 is due to differenence in two amino acid residues, we analysed three residue substitution mutants PsAvh113S108M, PsAvh113M122S and PsAvh113S108M/M122S using a transient expression system in N. benthamiana leaves. Similar to PsAvh113, PsAvh113S108M induced severe disease symptoms and higher viral RNA accumulation, whereas PsAvh113M122S and PsAvh113S108M/M122S caused mild disease symptoms and lower viral RNA accumulation, indicating that both mutants did not completely lose their virulence activity (Figure 2g,h). These results show that the IR2 motif of PsAvh113 is required for its virulence activity and the methionine (M122) of the IR2 region is an important residue for PVX infection.
Figure 2.
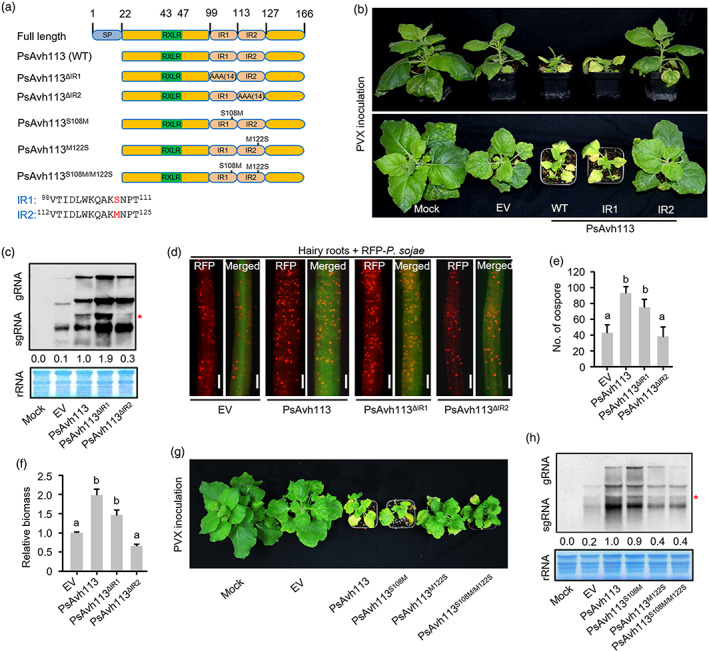
IR2 motif of PsAvh113 plays a crucial role in Phytophthora infection. (a) Schematic diagram of full‐length (wild‐type; [WT]) and mutant versions of PsAvh113. PsAvh113 contains two internal repeats (IR1 and IR2). All residues in the IR region were substituted with alanine residues in the mutant proteins PsAvh113▵IR1 and PsAvh113▵IR2. The residues in two IR regions showed one amino acid difference, and three mutual substitution mutants of the residues between Ser118 and Met122 were shown. SP, signal peptide. The number above each structure indicates the position of the amino acid. (b) Phenotypic comparison of N. benthamiana plants infiltrated with PVX, PVX‐PsAvh113, PVX‐PsAvh113 ▵IR1 , or PVX‐PsAvh113 ▵IR2 . Plants infected with PVX or PVX‐PsAvh113▵ IR2 showed mild disease symptoms. Photographs were taken at 21 dpi. (c) RNA blot showing the accumulation of PVX genomic and subgenomic RNA at 14 dpi. (d) Oospore production in soybean hairy roots expressing PsAvh113, PsAvh113 ▵IR1 , PsAvh113 ▵IR2 or EV. Scale bars, 0.25 mm. (e) Oospore count determined at 48 hpi under a confocal microscope. (f) Quantification of P. sojae biomass in soybean hairy roots by qPCR. (g) Phenotypic comparison of N. benthamiana plants infiltrated with PVX, PVX‐PsAvh113, PVX‐PsAvh113 S108M , PVX‐PsAvh113 M122S or PVX‐PsAvh113 S108M/M122S . Plants infected with PVX‐PsAvh113 M122S or PVX‐PsAvh113 S108M/M122S showed mild disease symptoms. Photographs were taken at 21 dpi. (h) RNA blot showing the accumulation of PVX genomic and subgenomic RNA at 14 dpi. The red asterisk in (c) and (h) represents sgRNA expression that might be caused by PsAvh113 and its mutants. Data represent mean ± SE. Different letters in (e) and (f) indicate statistically significant differences (P < 0.01; Duncan's multiple range test). The experiment was performed in triplicate with similar results.
To evaluate whether the nuclear localization of PsAvh113 is required for virulence, we fused a nuclear export sequence (NES) or nuclear localization sequence (NLS) to the N terminus of PsAvh113, respectively. Confocal imaging showed that NLS‐PsAvh113 was exclusively present in nuclei, whereas NES‐PsAvh113 was almost completely excluded from the nuclei (Figure S4a). Compared with the YFP control, N. benthamiana leaves expressing NLS‐PsAvh113 showed significantly reduced P. parasitica resistance, larger lesions and higher pathogen biomass, similar to those expressing PsAvh113, whereas leaves expressing NES‐PsAvh113 showed no increase in the severity of disease symptoms (Figure S4b,c). Expression of PsAvh113, NLS‐PsAvh113 and NES‐PsAvh113 was confirmed by western blot (Figure S4d). Similar disease symptoms were also observed in the VIVE assay when PsAvh113, NLS‐PsAvh113 and NES‐PsAvh113 were expressed in N. benthamiana leaves (Figure S4e,f). Collectively, these results suggest that the virulence activity of PsAvh113 is dependent on its nuclear localization in planta.
PsAvh113 interacts with GmDPB in soybean
To identify the potential host targets of PsAvh113, we performed yeast two‐hybrid (Y2H) screening using PsAvh113 as a bait against a soybean cDNA library prepared from P. sojae‐infected plants. Among the PsAvh113‐associated cDNA fragments, yeast cells co‐transformed with PsAvh113 and Glyma.01G228100 formed colonies on selection media (Figure 3a). The Glyma.01G228100 gene, which was repeatedly identified in four independent screens, encodes a putative DPB transcription factor in soybean (Figure S5a). Interestingly, we found that mutation in the nucleotide sequence of IR2, but not in that of IR1, abolished the interaction between PsAvh113 and GmDPB in Y2H assays (Figure 3a). To further examine the direct association between PsAvh113 and GmDPB, we conducted the bimolecular fluorescence complementation (BiFC) assay. PsAvh113, PsAvh113 mutants and GmDPB were fused to the N‐ and C‐terminal halves of the yellow fluorescent protein (YFP; YN and YC), respectively, and the resulting constructs were transformed into N. benthamiana leaves in various combination. After 48 h, strong YFP signal was detected in leaf cells co‐expressing YC‐GmDPB and YN‐PsAvh113 or YN‐PsAvh113 ▵IR1 but not in cells expressing YC‐GmDPB + YN‐PsAvh113 ▵IR2 , YC‐GmDPB + YN or YC + YN‐PsAvh113 (Figures 3b and S5b). To confirm the PsAvh113–GmDPB interaction, we performed a co‐immunoprecipitation (co‐IP) assay by co‐expressing PsAvh113, PsAvh113 ▵IR1 or PsAvh113 ▵IR2 with GmDPB in N. benthamiana leaves. Total proteins were extracted from the co‐infiltrated leaves, and incubated with anti‐MYC resin. Immunoblotting with anti‐HA antibody revealed that the abundance of PsAvh113‐YFP‐3*HA and PsAvh113type="InGeometric_Shapes">▵IR1‐YFP‐3*HA in the precipitate was high, whereas that of PsAvh113type="InGeometric_Shapes">▵IR2‐YFP‐3*HA was low or non‐existent (Figure 3c). These results imply that the IR2 region of PsAvh113 mediates its association with GmDPB, and is required for pathogenesis. Furthermore, we generated a mutant variant of GmDPB (GmDPB‐M) lacking 86 aa, which is required for its heterodimerization with E2F. In both Y2H and co‐IP assays, GmDPB‐M was unable to interact with PsAvh113, PsAvh113▵IR1 and PsAvh113▵IR2 (Figure S5c–e), indicating that region of forming heterodimerization is important for the PsAvh113–GmDPB interacation.
Figure 3.
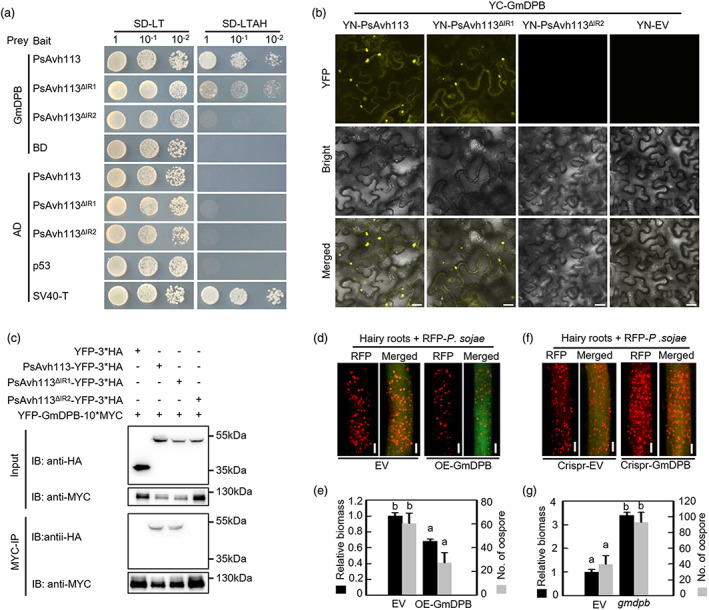
PsAvh113 physically interacts with GmDPB, which acts as a positive regulator of resistance to P. sojae. (a) Y2H assay showing that the IR2 domain of PsAvh113 mediates its interaction with GmDPB. The yeast strain AH109 was transformed with the bait plasmid pGBKT7 (BD) carrying PsAvh113, PsAvh113 ▵IR1 or PsAvh113 ▵IR2 together with the prey plasmid pGADT7 (AD) carrying GmDPB. Transformants were selected on minimal medium. SV40‐T and p53 were used as a positive control combination. ‐TL and ‐TLHA indicate SD/‐Trp‐Leu and SD/‐Trp‐Leu‐His‐Ade plates, respectively. Colony formation on ‐TLHA plates indicates an interaction between the two proteins. (b) BiFC assay showing the interaction between PsAvh113 or PsAvh113 mutants and GmDPB. (c) Co‐IP assays showing that PsAvh113 and PsAvh113 ▵IR1 interact with GmDPB, whereas PsAvh113 ▵IR2 does not. Total proteins were extracted from N. benthamiana leaves expressing YFP‐GmDPB‐10*MYC and PsAvh113‐YFP‐3*HA or YFP‐3*HA‐tagged PsAvh113 ▵IR1 or PsAvh113 ▵IR2 . Immune complexes were pulled down using anti‐MYC agarose gel. Proteins that co‐precipitated with GmDPB were detected by western blotting. (d–g) Confocal microscopy analysis (d, f) and quantification (e, g) of oospores and pathogen biomass in soybean hairy roots overexpressing WT, GmDPB (d, e) or edited GmDPB (f, g) along with the EV control. Scale bars, 0.25 mm. Confocal microscopy analysis was conducted at 48 hpi. In (e) and (g), black columns represent pathogen biomass, and grey columns represent oospore count. Data represent mean ± SE. Different letters in (e) and (g) indicate statistically significant differences (P < 0.01; Duncan's multiple range test). The experiment was performed in triplicate with similar results.
GmDPB enhances plant resistance against Phytophthora infection
To investigate the potential role of GmDPB in plant immunity, we firstly overexpressed the GmDPB gene in soybean hairy roots and then confirmed GmDPB expression by western blot analysis (Figures 3d and S6a). Soybean root tissues transformed with Agrobacterium carrying GmDPB or EV were inoculated with RFP‐labelled P. sojae strain P6497. At 48 hpi, the oospore count in roots overexpressing GmDPB was lower than that in roots transformed with the EV control (Figure 3d). Additionally, qPCR analysis showed that the accumulation of P. sojae biomass was reduced in the GmDPB‐overexpressing roots compared with the EV control (Figure 3e). Next, we knocked down GmDPB in soybean hairy roots using the CRISPR/Cas9 system. The hairy root lines harbouring nucleotide deletions in the GmDPB coding sequence, as determined by Sanger sequencing, were challenged with P6497‐RFP (Figure S6b). The GmDPB‐edited hairy roots displayed increased susceptibility to P. sojae infection compared with the control hairy roots (Figure 3f). The oospore count and P. sojae biomass were higher in GmDPB‐edited hairy roots than in hairy roots carrying EV (Figure 3g). These results indicate that GmDPB promotes disease resistance against P. sojae in soybean.
To further support the role of DPB in plant defence, we performed inoculation assays in N. benthamiana and Arabidopsis. Consistent with our observations in soybean, N. benthamiana leaves overexpressing GmDPB were more resistant to P. parasitica relative to leaves habouring YFP gene (Figure S6c–e), whereas the AtDPB knockout mutant displayed significantly decreased resistance to P. parasitica compared with wild type leaves (Figure S6f–h). Taken together, these results suggest that DPB genes promotes disease resistance in Arabidopsis, N. benthamiana and soybean.
PsAvh113 induces the degradation of GmDPB in planta
To gain insights into the effect of PsAvh113 on GmDPB function, fusions of PsAvh113‐Flag and its mutated derivatives with YFP were co‐expressed with YFP‐GmDPB‐M YC in N. benthamiana leaves via agroinfiltration. The results exhibited that a lower amount of GmDPB accumulated in the presence of PsAvh113 than in the YFP control, and the degradation of GmDPB was increased with an increasing amount of PsAvh113 (Figure 4a). Immunoblot analysis suggested that PsAvh113▵IR1, but not PsAvh113▵IR2 affected the reduction of GmDPB protein in co‐infiltrated N. benthamiana leaves (Figure 4b). Furthermore, the reduced protein of GmDPB can be rescued in N. benthamiana leaves coinfiltrated PsAvh113 with GmDPB upon treatment with the proteasome inhibitor MG132 (Figure 4c). Taken together, PsAvh113 causes the degradation of GmDPB by 26S proteasome. These data prompted us to examine where the PsAvh113 degrades GmDPB by 26S proteasome, fusions of PsAvh113 and its mutated derivatives with CFP were co‐expressed with ‐GmDPB‐YFP in N. benthamiana leaves via agroinfiltration, respectively. Confocal microscopy analysis showed that GmDPB‐YFP was primarily appeared in the nucleus and cytoplasm in the absence of PsAvh113 (Figure S6i,j). However, GmDPB‐YFP signals were observed in cytoplasm in the presence of PsAvh113‐CFP or PsAvh113type="InGeometric_Shapes">▵IR1‐CFP (Figure 4d). Furthermore, similar to that of the EV control, GmDPB‐YFP appeared in the nucleus and cytoplasm in the presence of PsAvh113▵IR2 (Figure 4d). The percentage of cells expressing GmDPB with EV or together with PsAvh113▵IR2 that exhibited clear and strong fluorescence in the nucleus was about 80%, whereas when GmDPB was co‐expressed together with PsAvh113 or PsAvh113▵IR1, this percentage was decreased to approximately 30% (Figure 4e). Furthermore, western blot analysis was performed using the cytoplasmic and nuclear extracts fractionated from N. benthamiana leaves co‐expressing PsAvh113‐CFP or PsAvh113 mutants ‐CFP and GmDPB‐YFP. Compared with the co‐expressing of GmDPB‐YFP and CFP in N. benthamiana (control), the GmDPB protein in the nuclear fraction and cytoplasm were markedly reduced in presence of CFP‐PsAvh113 and CFP‐PsAvh113▵IR1 (Figure 4f). To further confirm the degradation of GmDPB fusions of NES‐PsAvh113, NLS‐PsAvh113 or YFP‐HA were co‐expressed with GmDPB‐YFP‐HA in N. benthamiana leaves via agroinfiltration (Figure S7). Immunoblot analysis suggested that both NES‐PsAvh113 and NLS‐PsAvh113 affected the reduction of GmDPB protein in co‐infiltrated N. benthamiana leaves and the reduced protein of GmDPB can be rescued upon treatment with the proteasome inhibitor MG132 (Figure S7), indicating PsAvh113‐induced the degradation of GmDPB by 26S proteasome occurs in both the cytoplasm and nucleus. These results indicate that PsAvh113 remarkably inhibits the accumulation of GmDPB protein in nucleus and cytoplasm.
Figure 4.
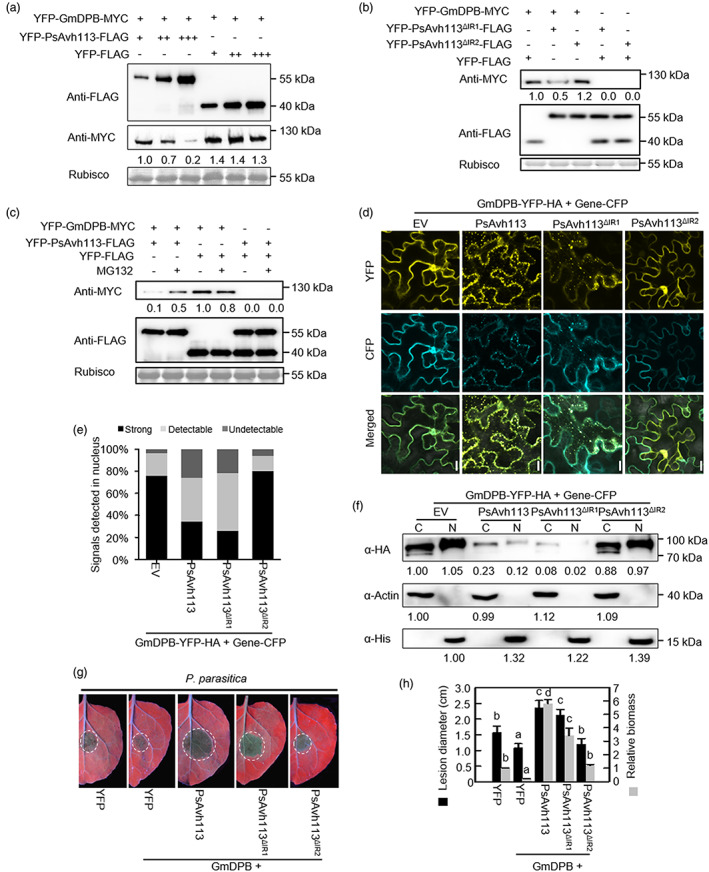
PsAvh113 affects the stability of GmDPB protein. (a) Western blot showing that co‐expression of GmDPB with PsAvh113 reduces GmDPB abundance, and PsAvh113 affects the stability of GmDPB in a dose‐dependent manner. (b) Western blot showing that co‐infiltration of GmDPB with PsAvh113 ▵IR1 , but not PsAvh113 ▵IR2 , reduces GmDPB abundance, and IR2 motif of PsAvh113 associtaes with affects the stability of GmDPB in N. benthamiana. (c) Western blot displaying that the degradation of GmDPB protein was prevented by application of proteasome inhibitor MG132 in N. benthamiana leaves co‐expressed GmDPB with PsAvh113. (d) Subcellular localization of CFP‐tagged WT and mutant PsAvh113 proteins and YFP‐HA‐labelled GmDPB in N. benthamiana leaves upon agroinfiltration. Fluorescence was detected by confocal microscopy at 48 hpi. Scale bars, 40 μm. (e) A total of 50 epidermal cells of N. benthamiana expressing GmDPB‐YFP‐HA alone, GmDPB‐YFP‐HA together with PsAvh113 or PsAvh113 mutants, respectively, were screened for the distribution of fluorescence. The strong means a strong signal in the nucleus, the detectable means a partial signal in nucleus and the undetectable means no signal in the nucleus. (f) Western blot analysis of GmDPB‐YFP‐HA in cytoplasmic and nuclear extracts from N. benthamiana leaves. The cytoplasmic and nuclear fractions were analysed of GmDPB‐YFP‐HA with PsAvh113 or PsAvh113 mutants by western blot with anti‐HA antibody, and GmDPB‐YFP‐HA with CFP was used as control. Actin and histone H3 were detected as fractionation markers for the cytoplasm and the nucleus, respectively. (g) Disease symptoms on N. benthamiana leaves co‐expressing the indicated constructs. Leaves were detached from the plants and inoculated with P. parasitica. Disease symptoms were monitored and photographed under ultraviolet (UV) light at 2 dpi. (h) qRT‐PCR analysis of relative Phytophthora biomass (grey columns) and P. parasitica lesion size (black columns) of 15 infected leaves were counted at 48 hpi. Data represent mean ± SE. Different letters indicate statistically significant differences (P < 0.01; Duncan's multiple range test). The experiment was performed in triplicate with similar results.
To elucidate whether PsAvh113 affects GmDPB‐induced resistant to Phytophthora, we co‐expressed GmDPB with PsAvh113, PsAvh113 ▵IR1 or PsAvh113 ▵IR2 in N. benthamiana leaves prior to pathogen challenge. Co‐expression of PsAvh113 or PsAvh113 ▵IR1 with GmDPB resulted in greater susceptibility to P. parasitica than the resistance induced by the expression of GmDPB alone (Figure 4g,h). This inhibiting effect was not observed when PsAvh113 ▵IR2 was used in place of PsAvh113 for co‐expression with GmDPB (Figure 4g,h). Taken together, the results suggest that PsAvh113 decreases the stability of GmDPB, leading to increased Phytophthora infection.
GmDPB functions as a transcriptional activator of GmCAT
A previous study showed that DPB binds to the GCGGGAA motif in the promoter of target genes in human (Cheneby et al., 2018). Interestingly, we detected two cis‐elements within ~1.5 kb region upstream of the coding sequence of GmCAT1 (Glyma.14G223500) using the PLACE program (Higo et al., 1999) (Figure S8a,b). There are four CAT genes in soybean, but the GCGGGAA motif was not detected in promoter region of other three CAT genes (Figure S8c). The GmCAT1 protein is localized to the peroxisomes (Figure S8d). To determine whether GmDPB binds to the promoter of GmCAT1, we performed yeast one‐hybrid (Y1H) assays. Our results showed that GmDPB fused to the GAL4 transcriptional activation domain (AD) activated the expression of the AbA reporter gene driven by the GmCAT1 promoter (Figure 5a). Moreover, we analysed the promoter regions of one allele, GmCAT2, and found that the GmCAT2 promoter has no GCGGGAA motif, supporting the notion that GmDPB was unable to bind to GmCAT2 promoter in the Y1H assay (Figure 5a). Next, we performed a transactivation assay in N. benthamiana leaves using the LUC reporter construct, which contained four tandem copies of the cis‐elements of the GmCAT1 promoter, and the GmDPB effector construct, and showed that GmDPB acts as a transcriptional activator (Figure 5b). Subsequently, we performed an EMSA using the purified MYC‐tagged GmDPB protein along with the Cy3‐labelled double‐stranded oligonucleotide probe (Table S1). In the presence of GmDPB, the probe showed a shift in its position and migrated more slowly than the free probe; however, this shift was not observed in the presence of EV (Figure 5c), suggesting that GmDPB binds to the GCGGGAA motif. Consistent with these results, the transcript level of GmCAT1 was significantly up‐regulated in GmDPB‐overexpressing hairy roots but markedly down‐regulated in GmDPB‐edited lines compared with EV (Figure 5d,e).
Figure 5.
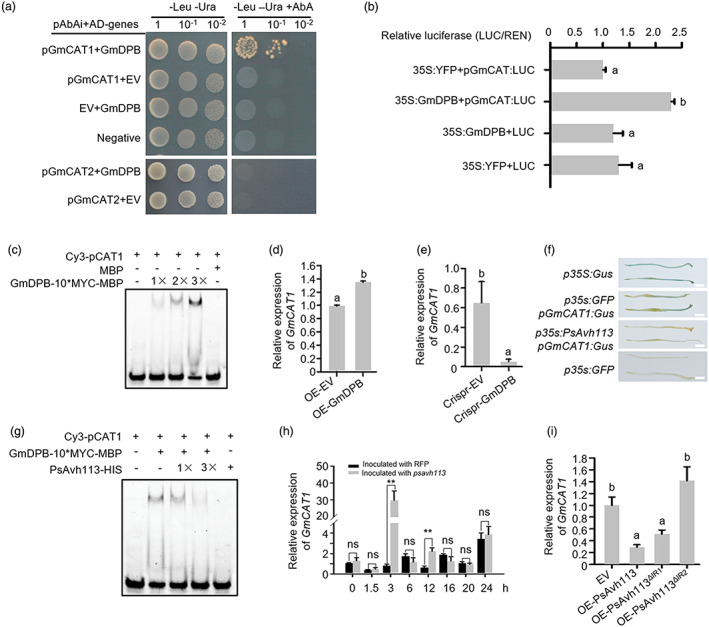
GmDPB specifically binds to the promoter of GmCAT1 to activate gene expression. (a) Y1H assay showing that GmDPB binds to the GmCAT1 promoter, but not to the GmCAT2 promoter. The yeast strain Y1H Gold was co‐transformed with the pGmCAT‐containing pAbAi bait plasmid and AD‐GmDPB. Yeast transformants were selected on minimal medium containing 150 ng/mL AbA. (b) Dual luciferase assay showing that GmDPB induces GmCAT1 expression in N. benthamiana leaves. (c) Verification of the binding of GmDPB to the GmCAT1 promoter via EMSA. The DNA‐binding ability of GmDPB increased with its increasing amount. (d) Expression analysis of GmCAT1 in GmDPB‐overexpressing soybean hairy roots by qRT‐PCR. (e) GmCAT1 expression is inhibited in GmDPB‐edited soybean hairy roots, as examined by qRT‐PCR. (f) PsAvh113 represses the GmCAT1 promoter activity in 20‐day‐old soybean hairy roots treated with P. sojae oospores for 48 h. PsAvh113 and pGmCAT were transiently co‐expressed in soybean hairy roots, which were then subjected to the GUS staining assay. Scale bars, 4 mm. (g) Verification of the association between PsAvh113 and pGmCAT1 by EMSA. The binding capacity decreased with the increase in PsAvh113 concentration. (h) Expression profiles of GmCAT1 in soybean hairy roots inoculated with the P6497 (WT) strain and PsAvh113 knockout line of P. sojae. Asterisks indicate significant differences according to Student's t‐test (**P < 0.01, ns, no significant difference). (i) Analysis of GmCAT1 transcript levels in soybean hairy roots overexpressing PsAvh113, PsAvh113 ▵IR1 or PsAvh113 ▵IR2 . Data represent mean ± SE. Different letters in (b), (d), (e) and (i) indicate statistically significant differences (P < 0.01; Duncan's multiple range test). Experiments were repeated twice with similar results.
PsAvh113 represses the DNA‐binding activity of GmDPB both in vitro and in vivo
To examine the regulatory effect of GmDPB on the expression of its target genes, we performed transient expression assays in soybean hairy roots by expressing the GUS reporter gene under the control of a 1.8 kb fragment of the GmCAT1 promoter. When the reporter construct (pGmCAT1:GUS) and control construct (p35S:GUS or p35S:GFP) were co‐transformed into soybean hairy roots, we detected GUS activity driven by the GmCAT1 promoter or by 35S promoter (p35S:GUS) as the positive control (Figure 5f). In addition, GUS staining was weaker at the tip of hairy roots expressing the GmCAT1 promoter together with PsAvh113 than that together with GFP (Figure 5f). To determine whether PsAvh113 affects the GmDPB‐induced expression of GmCAT1, we examined the DNA‐binding ability of GmDPB in the presence of PsAvh113 by performing an EMSA assay. Interestingly, we found that the DNA‐binding ability of GmDPB was reduced by PsAvh113 in a dose‐dependent manner, but not by PsAvh113 ▵IR2 (Figures 5g and S8e), indicating that PsAvh113 inhibits the DNA‐binding activity of GmDPB through its IR2 motif in vitro.
Next, we analysed the expression of GmCAT1 during P. sojae infection in soybean. The transcript level of GmCAT1 was significantly induced in soybean plants inoculated with the PsAvh113 knockout mutant compared with those infected with the WT strain P6497 (Figure 5h). However, the expression of GmCAT2 showed no change or slight reduction in soybean when inoculated with the PsAvh113 knockout mutant compared with those infected with the WT strain P6497 (Figure S8f). Meanwhile, the transcript levels of GmCAT1 were down‐regulated in hairy roots expressing PsAvh113 or PsAvh113 ▵IR1 compared with roots carrying EV or PsAvh113 ▵IR2 (Figure 5i). Finally, we tested the expression of AtCAT in Arabidopsis transgenic lines expressing PsAvh113, and the results obtained were similar to those in soybean (Figure S8g). Taken together, these results indicate that PsAvh113 affects the stability of GmDPB and the DNA‐binding activity of the encoded protein, thereby disrupting GmDPB‐regulated functions of the downstream gene GmCAT1.
GmCAT1 contributes to resistance against Phytophthora pathogens
Our finding that GmCAT1 is the potential target of GmDPB led us to further investigate its biological roles in plant defence. To understand the potential role of GmCAT1 in soybean, we expressed the GmCAT1 gene in soybean hairy roots and found that GmCAT1 was strongly increased at the transcriptional level (Figures 6a and S9a). Upon P6497‐RFP inoculation, fewer oospores were produced in GmCAT1‐overexpressing roots compared with hairy roots expressing EV (Figure 6b). Compared with that expressing empty vector control, the results of qPCR analysis showed decreased accumulation of P. sojae biomass in GmCAT1‐overexpressing hairy roots (Figure 6b). Furthermore, to edit the GmCAT1 gene, we designed two gRNAs including gRNA1, which targeted the GmDPB‐binding site in the GmCAT1 promoter (pGmCAT1), and gRNA2, which targeted the coding sequence of GmCAT1, and performed genetic transformation of HC6 hairy roots (Figure S9b). Compared with the EV control, the GmCAT1 or pGmCAT1‐edited hairy roots exhibited stronger disease symptoms upon P. sojae infection, as indicated by the increased oospore count and biomass (Figure 6c,d), suggesting that GmCAT suppresses the progression of P. sojae infection in soybean. The expression of GmCAT1 was reduced in pGmCAT1‐edited and GmCAT1‐edited hairy roots (Figure 6e).
Figure 6.
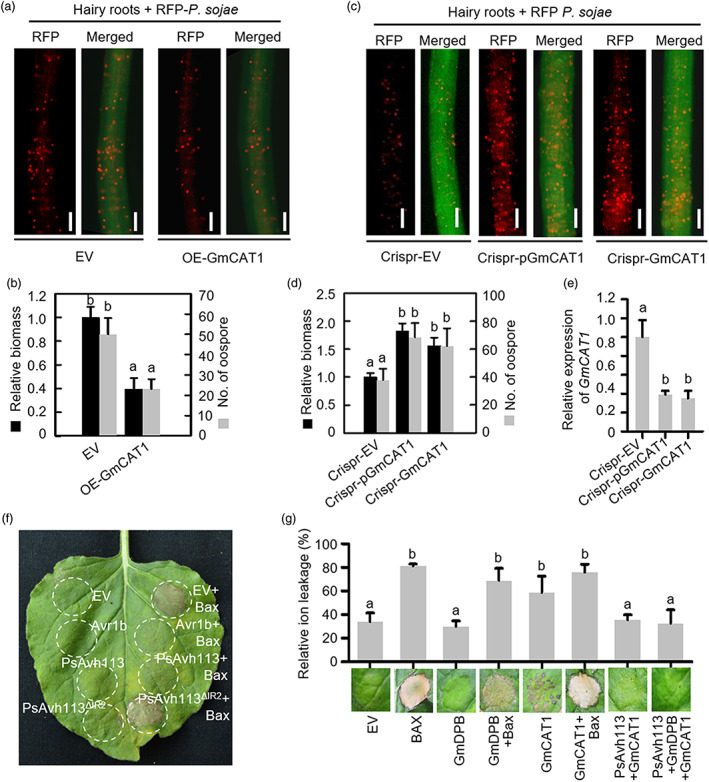
PsAvh113 inhibits the positive immune regulator GmCAT1‐induced cell death. (a–d) Confocal microscopy analysis (a, c) and quantification (d) of oospores and pathogen biomass in soybean hairy roots overexpressing WT GmCAT1 (a, b) or edited GmCAT1 (c, d) along with the EV control. Scale bar, 0.25 mm. Confocal microscopy analysis was conducted at 48 hpi. In (b) and (d), black columns represent biomass, and grey columns represent oospore count. (e) The transcript levels of GmCAT1 in GmCAT1‐edited hairy roots. In (b–e), data represent mean ± SE, and different letters indicate statistically significant differences (P < 0.01; Duncan's multiple range test). (f) Overexpression of PsAvh113 in N. benthamiana suppresses Bax‐triggered cell death. N. benthamiana leaves were infiltrated with A. tumefaciens containing PVX‐PsAvh113, PVX‐PsAvh113▵IR2, PVX, or PVX‐Avr1b, either alone or with A. tumefaciens cells carrying PVX‐Bax, which were infiltrated 24 h later. EV, empty vector. (g) GmCAT1 induces cell death in N. benthamiana leaves, whereas overexpression of PsAvh113 in N. benthamiana suppresses PCD triggered by GmCAT1. Pictures were taken at 5 dpi. Experiments were repeated three times with similar results.
These findings prompted us to test whether GmCAT1 is involved in programmed cell death in plants. We firstly assessed the PsAvh113 using a transient expression assay in N. benthamiana and found that both PsAvh113 and PsAvh113▵IR2 did not trigger cell death (Figures 6f and S9c,d). Similar to Avr1b, PsAvh113 suppresses Bax‐induced cell death in N. benthamiana leaves, while PsAvh113▵IR2 fails to inhibit cell death which is comparable to EV (Figure 6f). Furthermore, GmDPB is not able to induce cell death and also cannot inhibit Bax‐induced cell death (Figure 6g). Interestingly, GmCAT1 induces slightly PCD, whereas the GmCAT1‐induced cell death is blocked by PsAvh113 alone or PsAvh113 together with GmDPB (Figure 6g). Overall, our data showed that GmCAT1 positively regulates plant resistance to Phytophthora pathogens, which is likely due to GmCAT1–triggered cell death. Subsequently, this enhanced resistance of GmCAT1 is inhibited by PsAvh113 during P. sojae infection, thereby increasing soybean susceptibility to Phytophthora.
Discussion
Many efforts have been made to identify virulence factors required for pathogen colonization and disease development, and to elucidate the molecular mechanisms of microbial pathogenicity. However, most virulence effectors have not been characterized in Phytophthora pathogens. In this study, we exploited a newly developed screening assay, VIVE (Shi et al., 2020), and successfully identified PsAvh113 as a potential virulence effector in P. sojae, which was responsible for growth stunting and severe viral symptoms in N. benthamiana plants. Subsequently, using PsAvh113 as a molecular bait, we identified the GmDPB gene as its target, thereby providing mechanistic insights into the virulence function of PsAvh113 and the development of PRSR.
During evolution, IR regions may appear within a protein, which play an important role in protein stability and function as well as genome evolution (Jorda and Kajava, 2010). For example, the genome of Ustilago maydis encodes 15 effector proteins containing IRs (Mueller et al., 2008); Mycosphaerella graminicola genome harbours 23 genes predicted to encode surface‐associated proteins possessing tandem IR regions (Rudd et al., 2010); and Aspergillus fumigatus genome contains 292 genes with IRs (Levdansky et al., 2007). These repeat motifs are usually highly variable in size, mainly because of homologous recombination or slippage during replication (O'Dushlaine et al., 2005). Variations in repeat lengths and numbers increase functional diversity and allow adaptation to environmental changes or escape from host immunity (Ma et al., 2018). Our data revealed that the PsAvh113 protein harbours two IR regions (IR1 and IR2), both of which differ by only a single amino acid residue. Our results show that IR2, but not IR1, is required for the virulence activity of PsAvh113 and for its interaction with GmDPB. And the methionine (M122) of IR2 region is an important residue for PVX infection. This is largely different from another oomycete effector PsAvh23, in which contains two IRs; however, unlike the IRs in PsAvh113, both IRs in PsAvh23 are responsible for its virulence activity and interaction with the ADA2 subunit of the histone acetyltransferase complex (Kong et al., 2017), indicating that oomycete pathogens have also evolved effectors harbouring different IRs to suppress host immunity, the biological function of IR1 in PsAvh113 needs further investigations by protein crystal structure in the future.
The E2F/DP family of transcription factors is widely believed to play a pivotal role in regulating cell cycle progression, DNA replication, DNA damage repair and stress response (Perrot‐Rechenmann, 2010; Wang et al., 2014). In animals and plants, the E2F/DP family includes typical E2Fs and their related dimerizing partners, called DP proteins, both of which heterodimerize to bind to the E2F site in the promoters of target genes (Kosugi and Ohashi, 2002b). Because DP proteins function as essential DNA‐binding partners of E2Fs, most of the previous studies focused on how DP proteins enhance the transcriptional activation or repression of E2F target genes, and the self‐regulatory roles of DP protein have not been fully explored. In the present study, we showed that the GmDPB positively regulates the host immune response to Phytophthora pathogens, and acts as the direct host target of PsAvh113. Moreover, upon binding, nuclear signals of GmDPB were tough to detect in epidermal cells of N. benthamiana leaves. In addition, there was a modest increase in the decay rate of GmDPB in the presence of PsAvh113, indicating that the PsAvh113‐induced punctate localization in the cytoplasm is detrimental for the stability of GmDPB. Our findings are in agreement with previous results showing that the tumour suppressor protein ARF binds to and induces the nucleolar relocalization of DP1, leading to the inhibition of E2F‐activated genes (Datta et al., 2005). More interestingly, we observed that GmDPB‐M, which lacked the region important for heterodimerization with E2Fs, also failed to associate with PsAvh113. This prompted us to examine whether GmDPB associates with PsAvh113 either through GmE2Fs, or PsAvh113 and GmE2F independently bind GmDPB through the heterodimerization domain in the future. The latter would suggest the possibility that GmE2Fs and PsAvh113 compete with each other to bind to GmDPB.
Our results showed that GmDPB functions as a transcription activator of GmCAT1 by binding to its promoter. Furthermore, PsAvh113‐mediated inhibition of GmDPB activity reduced the expression of GmCAT1. Plant CAT, as a type of peroxisomal enzyme, plays crucial roles in maintaining H2O2 homeostasis and regulating PCD (Mhamdi et al., 2010; Zhang et al., 2015). In the present study, GmCAT1 could slightly induce cell death in N. benthamiana and positively regulated disease development by P. sojae. Analogous to from RxLR effector, analysis of CRN (crinkling‐ and necrosis‐inducing proteins) effectors showed that the PsCRN63 effector induces cell death, while PsCRN115 suppresses cell death in N. benthamiana leaves (Liu et al., 2011). Subsequently, PsCRN63/115 effectors regulate plant PCD through directly interfering with CATs, and perturbing H2O2 homeostasis, thereby, overcoming plant immune response in N. benthamiana (Zhang et al., 2015). These data suggested that P. sojae have evolved two different types of effector proteins and distinct mechanism to assault the similar immune‐related protein, and ensured P. sojae infection and PRSR disease development in soybean. However, the relationship among these effectors remains unknown, which might be a key point for further investigation. Moreover, similar to PsAvh113 of P. sojae, an effector PstGSRE1 from Puccinia striiformis f. sp. tritici has been shown to interact with and disrupt the nuclear localization of transcription factor TaLOL2 and suppress ROS‐mediated cell death induced by TaLOL2, thus promoting stripe rust of wheat (Qi et al., 2019). Compared to three CAT genes in arabidopsis, there are four CAT genes in soybean, all of which form one clade and are close to AtCAT2. Intriguingly, GmCAT1 and AtCAT2 showed differential plant defence responses to pathogens, this is probably due to infection with biotrophic and necrotrophic pathogens (Cheng et al., 2018; Dat et al., 2003; Mur et al., 2006; Yuan et al., 2017; Zhang et al., 2015). We also do not rule out that soybean catalases have distinct functions in catalysing H2O2, the precise mechanism of GmCAT1 remains elusive and needs further investigation. Taken together, it is conceivable that a virulence effector of P. sojae associates with a host factor and alters its stability, subsequently, inhitits the GmCAT1‐induced cell death and leads to disease symptoms during infection (Figure 7). In conclusion, our results indicate that the PsAvh113–GmDPB–GmCAT1 interaction serves as a potential defence mechanism, probably in favour of P. sojae infection.
Figure 7.
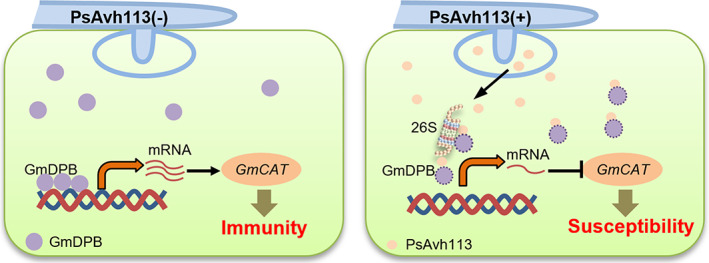
Proposed model demonstrating the role of PsAvh113 in regulating plant immunity. The PsAvh113 virulence effector of P. sojae interacts with GmDPB to manipulate GmCAT1 expression, which eventually leads to PRSR. During P. sojae infection, PsAvh113 interacts with the GmDPB transcription factor, affecting its stability. The degradation of GmDPB reduced the ability of binding to the promoter GmCAT1, thereby directly inhibiting its expression and activity. Subsequently, the reduced expression of GmCAT1 decreases PCD triggered by itself and enhances the susceptibility to P. sojae in soybean.
Experimental procedures
Plants and microbe growth conditions
Arabidopsis and N. benthamiana plants were grown in a greenhouse (25 °C, 67% relative humidity, 16 h light photoperiod). pEG101‐PsAvh113 was introduced into Arabidopsis ecotype Col‐0 to generate transgenic Arabidopsis plants using dipping method. The hairy roots are generated from soybean cotyledons by Agrobacterium rhizogenes‐mediated transformation. The P. parasitica isolate PBS042 and the P. sojae isolate P6497 were grown on 10% (v/v) V8 juice agar in the dark at 25 °C. Agrobacterium tumefaciens strains GV3101 and A. rhizogenes strains K599 were cultured on LB medium supplemented with appropriate antibiotics at 28 °C. Escherichia coli strains Rosetta and DH5α were cultured on LB medium at 37 °C supplemented with appropriate antibiotics. Given that Arabidopsis CAT mRNA abundance is controlled by the circadian clock. In order to avoid the effect of circadian rhythm on expression of soybean CAT genes, the sampling time is set to 2 : 00–4 : 00 PM for those plants that are grown in a greenhouse.
Yeast one/two‐hybrid assays
The yeast one‐hybrid assay was performed to examine DNA‐Protein binding using the Gold yeast one‐hybrid system (Liu et al., 2019). A 300 bp promoter fragment of GmCAT1 containing GCGGGAA box (as pGmCAT1) was cloned into the pAbAi vector. The CDS of GmDPB was inserted into the pGADT7 vector (AD‐GmDPB). The resultant constructs and empty vector pGADT7 were co‐transformed into the pGmCAT reporter strain by PEG/LiAc yeast transformation method, respectively. The transformed colonies were plated on SD/−Leu/‐Ura medium containing 1.5 mg/mL Aureobasidin A (AbA) to observe yeast growth. An empty reporter vector was used as a negative control. Y2H assay was carried out as previously described (Gui et al., 2022; Zhang et al., 2019).
Recombinant protein expression and purification
The plasmid pMAL‐GmDPB that contains the His‐tag was transformed into the E. coli strain Ro setta. To obtain the fusion protein, Ro setta cells harbouring the pMAL‐GmDPB construct were cultured in LB liquid medium supplemented with Ampicillin at 37 °C for 2 h and then induced by 0.5 mM IPTG at 16 °C for 18 h. The induced Ro setta cells were sonicated until the solution became clear. The supernatant was collected by centrifugation, and the recombinant proteins were purified using an Ni+‐NTA beads.
Electrophoretic mobility shift assay (EMSA)
The His‐tagged recombinant constructs (His‐GmDPB, His‐PsAvh113) were introduced in E. coli Rosetta and purified as described above (Cheng et al., 2018; Del Pozo et al., 2007). DNA fragments were end‐labelled with Cy3. The fluorescence‐labelled DNA (2 pM) was incubated with different amount of purified protein in 30 μL binding system (10 mm Tris–HCl, pH 8.8, 100 mm KCl, 1 mm EDTA, 0.1 mm DTT, 1.5 mm BSA and 5% glycerol) at 25 °C for 30 min. Then 10 × DNA loading buffer was added to the reaction. For DNA competition assays, 10‐fold non‐labelled competitor DNA was also added to the reaction. For protein competition assays, 10‐fold His‐PsAvh113 was added in the reaction. The reaction mixture was electrophoresed at 4 °C on an 8% native polyacrylamide gel in 0.5 × TBE (45 mm Tris, 45 mm Borate, 1 mm EDTA) solution for 100 min at 100 V in the dark. Fluorescence‐labelled DNA on the gel was directly detected with Amersham Typhoon.
DAB staining assays
Hydrogen peroxide (H2O2) in soybean hairy roots was detected by staining with 3,3′‐diaminobenzidine (DAB) tetrahydrochloride hydrate (Cheng et al., 2018). The hairy roots infected by A. tumefaciens carrying the construct of overexpression or silencing of targeting gene were harvested at 0 and 48 h after inoculation. The samples were immersed in DAB solution (1 mg/mL, pH 3.8) overnight at room temperature and then were treated using transparent solution (100% Trichloroacetaldehyde) for observation.
Agrobacterium‐mediated transformation in soybean hairy roots and N. benthamiana
Gene overexpression and knockdown in soybean hairy roots were performed as previously described (Zhang et al., 2019). Briefly, soybean cotyledons were collected and transformed with the overexpression vector (pFGC5941, pBI121 or pBI101) and CRISPR vector (pGES201) that contained interesting gene or fragment using A. rhizogenes K599. The treated cotyledons were sealed and placed in a greenhouse at 25 °C. After 3–4 weeks, transformed hairy roots were used to conduct the relative experiments. For transient expression analysis, N. benthamiana leaves were transformed by A. tumefaciens GV3101 as described previously with minor modification (Qiao et al., 2013). For stabilization assays, protesome inhibitor MG132 was infiltrated into plant leaves that coexpressed PsAvh113 and GmDPB after the agroinfiltration for 24 h.
Histochemical GUS assay
The transcriptional activity was performed as previously described (Mao et al., 2020). The hairy roots were co‐transformed with A. rhizogenes containing pFGC5941‐GmDPB and pBI101‐pCAT1 with or without PsAvh113. Hairy roots with pBI121‐35S::GUS or p35s:GFP were transformed separately. The hairy roots were collected and soaked in GUS Buffer (20 mg/mL X‐Gluc, 10 mm Na2EDTA, 100 mm Na2H2PO4, 0.5 mm K3[Fe(CN)6], 0.5 mm K4[Fe(CN)6]·3H2O, 0.1% Triton‐X‐100, pH 7.0). Following staining for 2 days at 25 °C, the samples were washed in 95% (v/v) ethanol and then photographed.
Dual‐luciferase reporter assay
LUC and REN luciferase activity was detected using a dual‐luciferase reporter assay system (Promega, USA) on Glomax machine (Promega, USA) as previously described (Iglesias‐Ara et al., 2018). Briefly, the effector construct pEG101‐GmDPB and reporter construct pGreen‐pGmCAT1 were introduced into Agrobacterium strain GV3101 separately and then expressed in tobacco leaves by A. tumefaciens‐mediated transient transformation. Leaf samples were collected and ground to powder at 4 °C separately. Luciferase Assay Substrate was resuspended in Luciferase Assay BufferII to generate LARII and 50 × Stop Glo substrate was added into Stop Glo Buffer to generate Stop Glo Reagent. After measuring firely luciferase activity, 20 μL Stop Glo Reagent was added into the mix to Renilla luciferase activity. The firefly LUC activity was normalized to the REN activity. The experiment was replicated three times independently.
Real‐time quantitative PCR
Leaf discs were sampled from the infected leaves of N. benthamiana, Arabidopsis or soybean 2 dpi upon P. parasitica or P. sojae inoculations. An equal amount of samples were used to extract genomic DNA using a genomic DNA isolation kit (TIANGEN BIOTECH, Beijing). Phytophthora biomass in inoculated leaves was determined by quantitative PCR (qPCR) using Phytophthora‐specific primers (Table S1).
For analysis of gene expression, RNA was extracted using TRIzol Reagent (Thermo) according to the manufacturer's instructions. A 1‐μg aliquot of total RNA was reverse‐transcribed by priming with oligo (dT18) in a 20‐μL reaction volume using the HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme Biotech). PCR amplification was conducted using gene‐specific primers as previously described (Gui et al., 2022). qRT‐PCRs were performed on Lightcycler 480II (Roche) using a ChamQ Universal SYBR qPCR Master Mix Kit (Vazyme, Nanjing, China).
Phytophthora infection assays
Arabidopsis was infected with P. parasitica as described before (Zhu et al., 2020). In brief, leaves of 4‐weeks‐old plants (Col‐0, PsAvh113‐L1/L2 lines and atdpb mutant) were inoculated with P. parasitica PBS042 by adding a droplet containing 2000 zoospores. Approximately 30 inoculated leaves were incubated in plastic plates padded with wet filter paper in a greenhouse at 25 °C for 2 days. Subsequently, disease development was evaluated by measuring and photographing the lesion diameter. In tobacco, expressing GmDPB, GmCAT1 and PsAvh113 were individually expressed in N. benthamiana leaves and followed by P. parasitica PBS042 inoculation. In soybean, target genes were transiently transformed in soybean hairy roots mediated by A. rhizogenes. Subsequently, hairy roots expressing target gene were inoculated with P. sojae RFP‐P6497. The number of oospores was counted and the relative biomass of P. sojae was assayed. These experiments were repeated three times with similar results.
Microscopy
To detect protein interactions in vivo, the full‐length coding sequences of the GmDPB and PsAvh113 genes were individually cloned into the YC and YN vectors as previously described (Lu et al., 2010). Combinations of the plasmid constructs (YN‐PsAvh113/PsAvh113 ▵IR1 /PsAvh113 ▵IR2 + YC‐GmDPB, YN‐EV + YC‐GmDPB, YN‐PsAvh113/PsAvh113 ▵IR1 /PsAvh113 ▵IR2 + YC‐EV) were coinfitrated into N. benthamiana, respectively. To analyse GmDPB nuclear reposition regulated by PsAvh113 in the epidermal leaves of N. benthamiana, p35S:GmDPB‐YFP‐HA or p35S:PsAvh113/PsAvh113 ▵IR1 /PsAvh113 ▵IR2 ‐CFP constructs were transformed into Agrobacterium strain GV3101, respectively. The OD600 of A. tumefaciens was brought to an optical density of 1.2, and equal volumes of each culture were mixed for infiltrating the epidermal N. benthamiana leaves. YFP fluorescence was excited with the 514‐nm line ray of the argon laser and detected in the range between 520 and 540 nm.CFP fluorescence was excited with the 458 nm ray line of the argon laser and recorded in one of the confocal channels in the 465–520 nm emission range. The CFP/YFP signals were imaged 2–3 days after infiltration using an Olympus Fluoview FV3000 confocal laser microscope (Olympus, Tokyo, Japan).
Potato virus X (PVX) infection assays and northern blotting
PCR products of PsAvh113, PsAvh113 ▵IR1 and PsAvh113 ▵IR2 (without signal peptides) were ligated into pGR106 vector containing the entire PVX genome (Shi et al., 2020). About 2‐weeks‐old N. benthamiana plants were infiltrated with the A. tumefaciens (OD = 0.6) harbouring the plasmids pGR106‐EV, pGR106‐PsAvh113 and pGR106‐PsAvh113‐muants. After 2–3 weeks, the infiltrated plants were photographed and total RNA of those plants were extracted using Trizol Reagent. Viral RNAs were detected by probes corresponding to the PVX coat protein‐encoding gene (CP) as previously described (Qiao et al., 2013; Shi et al., 2020). The accumulation of viral RNAs were quantified based on the new sgRNA expression that produced by PsAvh113 and its mutants. Briefly, 6 μg total RNA were loaded into 1.5% agarose gels containing 10% formaldehyde and 1 × MOPS buffer and run at 100 V for 45 min. Following transfer to a nylon membrane, hybridization with a BIOTIN‐labelled probe was performed to detect the 3‐prime regions of PVX genome segments.
Nuclear‐cytoplasmic fractionation assay
The separation of the nuclear and cytoplasmic fractions was carried out as previously described (Qi et al., 2019). About 0.5 g of the infiltrated leaf tissue was ground in liquid nitrogen and homogenized in 2 mL of lysis buffer (20 mm Tris–HCl, pH 7.5, 20 mm KCl, 2 mm EDTA, 2.5 mm MgCl2, 25% glycerol, 250 mm sucrose, 5 mm DTT and one protease inhibitor cocktail tablet (Roche) per 50 mL) for 10 min at 4 °C. The homogenate was filtered through the Miracloth and then centrifuged at 1500 g for 10 min at 4 °C. The centrifuged supernatant was centrifuged at 10 000 g for 10 min at 4 °C and the final supernatant was collected as the cytoplasmic fraction. The centrifuged pellet was washed four times with 5 mL of nuclear resuspension buffer (NRBT buffer, 20 mm Tris–HCl, pH 7.4, 25% glycerol, 2.5 mm MgCl2 and 0.2% Triton X‐100) at 4°C, and then the pellet was resuspended with 500 μL of NRB2 (20 mm Tris–HCl, pH 7.5, 0.25 m sucrose, 10 mm MgCl2, 0.5% Triton X‐100, 5 mm β‐mercaptoethanol and one protease inhibitor cocktail tablet (Roche) per 50 mL). The suspension was laid carefully over 500 μL NRB3 (20 mm Tris–HCl, pH 7.5, 1.7 M sucrose, 10 mm MgCl2, 0.5% Triton X‐100, 5 mm β‐mercaptoethanol and one protease inhibitor cocktail tablet (Roche) per 50 mL), and then centrifuged at 16 000 g for 45 min at 4 °C. Finally, the pellet as the nuclear fraction was resuspended in 400 μL of lysis buffer. As quality controls for the fractionation, Actin/Rubisco (Abmart) and Histone H3 (Abmart) were used as the cytoplasmic and nuclear markers, respectively.
Immunoblot analysis of protein abundance
Protein was extracted from N. benthamiana leaves expressing YFP‐GmDPB‐MYC, YFP‐PsAvh113‐FLAG or other YFP‐FLAG using PEB buffer (25 mm Tris–HCl pH 7.6, 15 mm MgCl2, 150 mm NaCl, 60 mm β‐glycerophosphate, 0.1 mm Na3VO4, 1 mm NaF, 1 mm phenylmethanesulfonyl fluoride, 1% Triton X‐100). Bradford solution was used to quantify protein concentration. GmDPB and the various concentrations of PsAvh113 or YFP protein were co‐incubated at 25 °C for 30 min. The reaction products were detected by SDS‐polyacrylamide gel electrophoresis (PAGE) using anti‐FLAG (MBL, 1 : 5000 dilution) and anti‐MYC (MBL, 1 : 5000 dilution).
Co‐immunoprecipitation and western blotting
Co‐immunoprecipitation was assayed as described previously (Zhang et al., 2019). In brief, the infiltrated leaves of N. benthamiana were harvested at 2 dpi. Then these leaves were powdered equally and mixed in extraction buffer at 4 °C, the suspension was centrifuged and filtered to collect the supernatant. Myc‐beads were added to the supernatant and the mixtures were incubated at 4 °C for 1 h. After washing three times, the protein‐enriched beads were subjected to western blotting. Proteins were separated in 12% SDS‐PAGE gels by electrophoresis, then transferred onto PVDF membrane by semidry electroblotting. The membrane was blocked with TBST (0.01m Tris–HCl, 0.1 m NaCl and 1/1000 Tween 20)‐milk solution and incubated with primary mouse anti‐Myc (MBL, 1 : 5000 dilution), rabbit anti‐HA (MBL, 1 : 1000 dilution) or anti‐Flag (MBL, 1 : 5000 dilution) as the primary antibody and anti‐mouse/rabbit‐HRP (MBL, 1 : 5000 dilution) as the secondary antibody. Signals were developed using Chemiluminescence substrate (Thermo, #34580, USA), then imaged using Amersham Image 600 (GE Healthcare).
Conflict of interest statement
The authors declare no competing interests.
Author contributions
YQ conceived and designed the experiments; XGZ, LG, XYZ, RQZ, SH, RZ, DL and JZ performed the experiments. XGZ analysed the experiment data; YQ and XGZ wrote the manuscript. All authors have read and approved the final manuscript.
Supporting information
Figure S1 PVX‐based recombinant construct and expression pattern of PsAvh113.
Figure S2 Overexpression of PsAvh113 affects plant development and defence response in Arabidopsis.
Figure S3 Analysis of PsAvh113 and its mutated derivatives by subcellular localization, the cytoplasm and the nucleus protein separation.
Figure S4 The nuclear localization of PsAvh113 is required for its virulence activity in plants.
Figure S5 Analysis of interaction between PsAvh113 and GmDPB in vitro and in vivo.
Figure S6 DPB genes in soybean and Arabidopsis positively regulate plant immunity against Phytophthora infection.
Figure S7 PsAvh113 affects the stability of GmDPB protein in cytoplasm and nucleus.
Figure S8 Analysis of subcellular localization and expression of GmCAT1.
Figure S9 Analysis of expression and editing sequence in GmCAT1‐overexpressed and pGmCAT1 or GmCAT1‐edited soybean hairy roots.
Table S1 Primers used in this study.
Acknowledgements
We thank Prof. Yuanchao Wang (Nanjing Agricultural University) for providing the P. Sojae isolate P6497‐RFP. Prof. Hui Zhang for soybean CRISPR/Cas9 vector. This work was supported by grants from the National Natural Science Foundation of China (32001883, 32072502 and 32172359) and the Science and Technology Commission of Shanghai Municipality (18DZ2260500).
References
- Ai, G. , Xia, Q. , Song, T. , Li, T. , Zhu, H. , Peng, H. , Liu, J. et al. (2021) A Phytophthora sojae CRN effector mediates phosphorylation and degradation of plant aquaporin proteins to suppress host immune signaling. PLoS Pathog. 17, e1009388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran, D. , Rickert, J. , Huang, Y. , Steinwand, M.A. , Marr, S.K. and Wildermuth, M.C. (2014) Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe, 15, 506–513. [DOI] [PubMed] [Google Scholar]
- Cheneby, J. , Gheorghe, M. , Artufel, M. , Mathelier, A. and Ballester, B. (2018) ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA‐binding ChIP‐seq experiments. Nucleic Acids Res. 46, D267–D275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Q. , Dong, L. , Gao, T. , Liu, T. , Li, N. , Wang, L. , Chang, X. et al. (2018) The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 69, 2527–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat, J.F. , Pellinen, R. , Beeckman, T. , Van De Cotte, B. , Langebartels, C. , Kangasjarvi, J. , Inze, D. et al. (2003) Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J. 33, 621–632. [DOI] [PubMed] [Google Scholar]
- Datta, A. , Sen, J. , Hagen, J. , Korgaonkar, C.K. , Caffrey, M. , Quelle, D.E. , Hughes, D.E. et al. (2005) ARF directly binds DP1: interaction with DP1 coincides with the G1 arrest function of ARF. Mol. Cell. Biol. 25, 8024–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori, J. and Johnson, D.G. (2006) Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 6, 739–748. [DOI] [PubMed] [Google Scholar]
- Del Pozo, J.C. , Diaz‐Trivino, S. , Cisneros, N. and Gutierrez, C. (2007) The E2FC‐DPB transcription factor controls cell division, Endoreplication and lateral root formation in a SCF‐dependent manner. Plant Signal. Behav. 2, 273–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova, D.K. and Dyson, N.J. (2005) The E2F transcriptional network: old acquaintances with new faces. Oncogene, 24, 2810–2826. [DOI] [PubMed] [Google Scholar]
- Girling, R. , Partridge, J.F. , Bandara, L.R. , Burden, N. , Totty, N.F. , Hsuan, J.J. and La Thangue, N.B. (1993) A new component of the transcription factor DRTF1/E2F. Nature, 365, 468. [DOI] [PubMed] [Google Scholar]
- Gui, X. , Zhang, P. , Wang, D. , Ding, Z. , Wu, X. , Shi, J. , Shen, Q.H. et al. (2022) Phytophthora effector PSR1 hijacks the host pre‐mRNA splicing machinery to modulate small RNA biogenesis and plant immunity. Plant Cell, 34, 3443–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin, K. , Wu, C. , Fattaey, A. , Lees, J. , Dynlacht, B. , Ngwu, C. and Harlow, E. (1993) Heterodimerization of the transcription factors E2F‐1 and DP‐1 leads to cooperative trans‐activation. Genes Dev. 7, 1850–1861. [DOI] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. and Korenaga, T. (1999) Plant cis‐acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y. , Zhai, Y. , Feng, L. , Karimi, H.Z. , Rutter, B.D. , Zeng, L. , Choi, D.S. et al. (2019) A phytophthora effector suppresses trans‐kingdom RNAi to promote disease susceptibility. Cell Host Microbe, 25, 153–165.e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias‐Ara, A. , Osinalde, N. and Zubiaga, A.M. (2018) Detection of E2F‐induced transcriptional activity using a dual luciferase reporter assay. Methods Mol. Biol. 1726, 153–166. [DOI] [PubMed] [Google Scholar]
- Inaba, J. , Kim, B.M. , Shimura, H. and Masuta, C. (2011) Virus‐induced necrosis is a consequence of direct protein‐protein interaction between a viral RNA‐silencing suppressor and a host catalase. Plant Physiol. 156, 2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Z. , Tian, Y. , Cao, Y. , Wang, J. , Zhan, B. , Zhao, Z. , Sun, B. et al. (2021) A novel pathogenicity determinant hijacks maize catalase 1 to enhance viral multiplication and infection. New Phytol. 230, 1126–1141. [DOI] [PubMed] [Google Scholar]
- Jing, M. , Guo, B. , Li, H. , Yang, B. , Wang, H. , Kong, G. , Zhao, Y. et al. (2016) A Phytophthora sojae effector suppresses endoplasmic reticulum stress‐mediated immunity by stabilizing plant binding immunoglobulin proteins. Nat. Commun. 7, 11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jorda, J. and Kajava, A.V. (2010) Protein homorepeats sequences, structures, evolution, and functions. Adv. Protein Chem. Struct. Biol. 79, 59–88. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Furzer, O. , Jones, J.D. , Judelson, H.S. , Ali, G.S. , Dalio, R.J. , Roy, S.G. et al. (2015) The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, L.N. and Leone, G. (2019) The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer, 19, 326–338. [DOI] [PubMed] [Google Scholar]
- Kong, L. , Qiu, X. , Kang, J. , Wang, Y. , Chen, H. , Huang, J. , Qiu, M. et al. (2017) A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Curr. Biol. 27, 981–991. [DOI] [PubMed] [Google Scholar]
- Kosugi, S. and Ohashi, Y. (2002a) E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue‐specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 29, 45–59. [DOI] [PubMed] [Google Scholar]
- Kosugi, S. and Ohashi, Y. (2002b) E2Ls, E2F‐like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J. Biol. Chem. 277, 16553–16558. [DOI] [PubMed] [Google Scholar]
- Kovesdi, I. , Reichel, R. and Nevins, J. (1987) Role of an adenovirus E2 promoter binding factor in E1A‐mediated coordinate gene control. Proc. Natl. Acad. Sci. USA, 84, 2180–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens, T. , Li, J. , Leone, G. and De Veylder, L. (2009) Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol. 19, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdansky, E. , Romano, J. , Shadkchan, Y. , Sharon, H. , Verstrepen, K.J. , Fink, G.R. and Osherov, N. (2007) Coding tandem repeats generate diversity in Aspergillus fumigatus genes. Eukaryot. Cell, 6, 1380–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. , Hu, Q. , Zhou, J. , Yin, W. , Yao, D. , Shao, Y. , Zhao, Y. et al. (2021) Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection. Proc. Natl. Acad. Sci. USA, 118, e2018312118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Ye, W. , Ru, Y. , Yang, X. , Gu, B. , Tao, K. , Lu, S. et al. (2011) Two host cytoplasmic effectors are required for pathogenesis of Phytophthora sojae by suppression of host defenses. Plant Physiol. 155, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Su, J. , Zhu, Y. , Yao, G. , Allan, A.C. , Ampomah‐Dwamena, C. , Shu, Q. et al. (2019) The involvement of PybZIPa in light‐induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G‐boxes in its promoter. Hortic. Res. 6, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q. , Tang, X. , Tian, G. , Wang, F. , Liu, K. , Nguyen, V. , Kohalmi, S.E. et al. (2010) Arabidopsis homolog of the yeast TREX‐2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61, 259–270. [DOI] [PubMed] [Google Scholar]
- Lu, X. , Yang, Z. , Song, W. , Miao, J. , Zhao, H. , Ji, P. , Li, T. et al. (2023) The Phytophthora sojae effector PsFYVE1 modulates immunity‐related gene expression by targeting host RZ‐1A protein. Plant Physiol. 191, 925–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L.S. , Wang, L. , Trippel, C. , Mendoza‐Mendoza, A. , Ullmann, S. , Moretti, M. , Carsten, A. et al. (2018) The Ustilago maydis repetitive effector Rsp3 blocks the antifungal activity of mannose‐binding maize proteins. Nat. Commun. 9, 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, C. , He, J. , Liu, L. , Deng, Q. , Yao, X. , Liu, C. , Qiao, Y. et al. (2020) OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnol. J. 18, 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti, L. , Pellegrini, B. , Cantoni, R. , Stevens, R. , Bergounioux, C. , Cella, R. and Albani, D. (2002) The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277, 9911–9919. [DOI] [PubMed] [Google Scholar]
- Mathioudakis, M.M. , Veiga, R.S. , Canto, T. , Medina, V. , Mossialos, D. , Makris, A.M. and Livieratos, I. (2013) Pepino mosaic virus triple gene block protein 1 (TGBp1) interacts with and increases tomato catalase 1 activity to enhance virus accumulation. Mol. Plant Pathol. 14, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi, A. , Queval, G. , Chaouch, S. , Vanderauwera, S. , Van Breusegem, F. and Noctor, G. (2010) Catalase function in plants: a focus on Arabidopsis mutants as stress‐mimic models. J. Exp. Bot. 61, 4197–4220. [DOI] [PubMed] [Google Scholar]
- Mueller, O. , Kahmann, R. , Aguilar, G. , Trejo‐Aguilar, B. , Wu, A. and de Vries, R.P. (2008) The secretome of the maize pathogen Ustilago maydis. Fungal Genet. Biol. 45(Suppl 1), S63–S70. [DOI] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Atzorn, R. , Miersch, O. and Wasternack, C. (2006) The outcomes of concentration‐specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dushlaine, C.T. , Edwards, R.J. , Park, S.D. and Shields, D.C. (2005) Tandem repeat copy‐number variation in protein‐coding regions of human genes. Genome Biol. 6, R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot‐Rechenmann, C. (2010) Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2, a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, V.D. and Van Breusegem, F. (2012) Hydrogen peroxide—a central hub for information flow in plant cells. AoB PLANTS, 2012:pls014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, T. , Guo, J. , Liu, P. , He, F. , Wan, C. , Islam, M.A. , Tyler, B.M. et al. (2019) Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS‐promoting transcription factor TaLOL2 to defeat ROS‐induced defense in wheat. Mol. Plant, 12, 1624–1638. [DOI] [PubMed] [Google Scholar]
- Qiao, Y. , Liu, L. , Xiong, Q. , Flores, C. , Wong, J. , Shi, J. , Wang, X. et al. (2013) Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45, 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshan, P. , Kulshreshtha, A. , Kumar, S. , Purohit, R. and Hallan, V. (2018) AV2 protein of tomato leaf curl Palampur virus promotes systemic necrosis in Nicotiana benthamiana and interacts with host Catalase2. Sci. Rep. 8, 1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, J.J. , Antoniw, J. , Marshall, R. , Motteram, J. , Fraaije, B. and Hammond‐Kosack, K. (2010) Identification and characterisation of Mycosphaerella graminicola secreted or surface‐associated proteins with variable intragenic coding repeats. Fungal Genet. Biol. 47, 19–32. [DOI] [PubMed] [Google Scholar]
- Shi, J. , Zhu, Y. , Li, M. , Ma, Y. , Liu, H. , Zhang, P. , Fang, D. et al. (2020) Establishment of a novel virus‐induced virulence effector assay for the identification of virulence effectors of plant pathogens using a PVX‐based expression vector. Mol. Plant Pathol. 21, 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, T. , Ma, Z. , Shen, D. , Li, Q. , Li, W. , Su, L. , Ye, T. et al. (2015) An oomycete CRN effector reprograms expression of plant HSP genes by targeting their promoters. PLoS Pathog. 11, e1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, B.M. , Tripathy, S. , Zhang, X. , Dehal, P. , Jiang, R.H. , Aerts, A. , Arredondo, F.D. et al. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science, 313, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Vlieghe, K. , Boudolf, V. , Beemster, G.T. , Maes, S. , Magyar, Z. , Atanassova, A. , de Almeida Engler, J. et al. (2005) The DP‐E2F‐like gene DEL1 controls the endocycle in Arabidopsis thaliana . Curr. Biol. 15, 59–63. [DOI] [PubMed] [Google Scholar]
- Wang, Y. and Wang, Y. (2018) Phytophthora sojae effectors orchestrate warfare with host immunity. Curr. Opin. Microbiol. 46, 7–13. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Gu, Y. , Zebell, S.G. , Anderson, L.K. , Wang, W. , Mohan, R. and Dong, X. (2014) A noncanonical role for the CKI‐RB‐E2F cell‐cycle signaling pathway in plant effector‐triggered immunity. Cell Host Microbe, 16, 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Tyler, B.M. and Wang, Y. (2019) Defense and counterdefense during plant‐pathogenic oomycete infection. Annu. Rev. Microbiol. 73, 667–696. [DOI] [PubMed] [Google Scholar]
- Yang, T. , Qiu, L. , Huang, W. , Xu, Q. , Zou, J. , Peng, Q. , Lin, H. et al. (2020) Chilli veinal mottle virus HCPro interacts with catalase to facilitate virus infection in Nicotiana tabacum . J. Exp. Bot. 71, 5656–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H.M. , Liu, W.C. and Lu, Y.T. (2017) CATALASE2 coordinates SA‐mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe, 21, 143–155. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Li, Q. , Liu, T. , Liu, L. , Shen, D. , Zhu, Y. , Liu, P. et al. (2015) Two cytoplasmic effectors of Phytophthora sojae regulate plant cell death via interactions with plant catalases. Plant Physiol. 167, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Jia, Y. , Shi, J. , Chen, C. , Ye, W. , Wang, Y. , Ma, W. et al. (2019) The WY domain in the Phytophthora effector PSR1 is required for infection and RNA silencing suppression activity. New Phytol. 223, 839–852. [DOI] [PubMed] [Google Scholar]
- Zhu, X. , He, S. , Fang, D. , Guo, L. , Zhou, X. , Guo, Y. , Gao, L. et al. (2020) High‐throughput sequencing‐based identification of Arabidopsis miRNAs induced by Phytophthora capsici infection. Front. Microbiol. 11:1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 PVX‐based recombinant construct and expression pattern of PsAvh113.
Figure S2 Overexpression of PsAvh113 affects plant development and defence response in Arabidopsis.
Figure S3 Analysis of PsAvh113 and its mutated derivatives by subcellular localization, the cytoplasm and the nucleus protein separation.
Figure S4 The nuclear localization of PsAvh113 is required for its virulence activity in plants.
Figure S5 Analysis of interaction between PsAvh113 and GmDPB in vitro and in vivo.
Figure S6 DPB genes in soybean and Arabidopsis positively regulate plant immunity against Phytophthora infection.
Figure S7 PsAvh113 affects the stability of GmDPB protein in cytoplasm and nucleus.
Figure S8 Analysis of subcellular localization and expression of GmCAT1.
Figure S9 Analysis of expression and editing sequence in GmCAT1‐overexpressed and pGmCAT1 or GmCAT1‐edited soybean hairy roots.
Table S1 Primers used in this study.


