Summary
Alfalfa (Medicago sativa L.) is a perennial flowering plant in the legume family that is widely cultivated as a forage crop for its high yield, forage quality and related agricultural and economic benefits. Alfalfa is a photoperiod sensitive long‐day (LD) plant that can accomplish its vegetative and reproductive phases in a short period of time. However, rapid flowering can compromise forage biomass yield and quality. Here, we attempted to delay flowering in alfalfa using multiplex CRISPR/Cas9‐mediated mutagenesis of FLOWERING LOCUS Ta1 (MsFTa1), a key floral integrator and activator gene. Four guide RNAs (gRNAs) were designed and clustered in a polycistronic tRNA–gRNA system and introduced into alfalfa by Agrobacterium‐mediated transformation. Ninety‐six putative mutant lines were identified by gene sequencing and characterized for delayed flowering time and related desirable agronomic traits. Phenotype assessment of flowering time under LD conditions identified 22 independent mutant lines with delayed flowering compared to the control. Six independent Msfta1 lines containing mutations in all four copies of MsFTa1 accumulated significantly higher forage biomass yield, with increases of up to 78% in fresh weight and 76% in dry weight compared to controls. Depending on the harvesting schemes, many of these lines also had reduced lignin, acid detergent fibre (ADF) and neutral detergent fibre (NDF) content and significantly higher crude protein (CP) and mineral contents compared to control plants, especially in the stems. These CRISPR/Cas9‐edited Msfta1 mutants could be introduced in alfalfa breeding programmes to generate elite transgene‐free alfalfa cultivars with improved forage biomass yield and quality.
Keywords: CRISPR/Cas9, genome editing, FLOWERING LOCUS T, alfalfa, quality, biomass
Introduction
Alfalfa (Medicago sativa L.) is cultivated worldwide due to its high forage yield and quality, perennial growth habit, adaptability, nitrogen fixation ability and related agricultural and economic benefits (Bao et al., 2022; Chen et al., 2020; Gao et al., 2018; Lorenzo et al., 2020; Singer et al., 2018; Wolabu et al., 2020a; Zhou et al., 2011). Alfalfa is a fast‐growing forage, both before and after cutting, making it ideal for multiple harvests within one growing season (Du et al., 2021; Kang et al., 2019; Lorenzo et al., 2020; Singer et al., 2018). Flowering time is an important agronomic trait for crops in general as manipulating flowering time helps optimize plant biomass and fitness to different environmental conditions (Aung et al., 2015; Cai et al., 2019; Cheng et al., 2021; Lorenzo et al., 2020; Park et al., 2014; Rajendran et al., 2021; Tadege et al., 2015; Weller and Ortega, 2015; Zhang et al., 2019). Early flowering might be advantageous and preferred for seed production. In forages, the onset of flowering is negatively associated with biomass yield and nutritional quality due to increased lignin accumulation in stems and nutrient recycling from leaves to support seed development (Ball et al., 2001; Fick and Mueller, 1989; Gao et al., 2018; Lacefield, 2004; Lorenzo et al., 2020; Tadege et al., 2015; Zhang et al., 2019). Therefore, in the case of longer favourable growing season, late flowering varieties are advantageous for forage production as the extended growth time enables more vegetative growth and forage production. In forage crop production, high dry matter, digestibility and other nutritional traits are strongly affected by flowering. Consequently, the advantage of late flowering for biomass yield and quality enhancement become the current breeding needs of forage crop production. In general, conflicting breeding programmes for high biomass and efficient seed production should be compromised on the existing situation based on risk assessment. Variable genetic resources with different ranges of flowering time are alternative options. Therefore, delaying flowering may improve biomass production and nutritional quality in forage crops (Aung et al., 2015; Jung and Müller, 2009; Lorenzo et al., 2020; Singer et al., 2018; Tadege et al., 2015; Weller and Ortega, 2015; Zhang et al., 2019). In fact, genetic manipulation of key flowering time genes has increased yield production and nutritional quality in several food and forage crop species (Aung et al., 2015; Cai et al., 2019; Dash and Rai, 2022; Kishchenko et al., 2020; Liu et al., 2021; Lorenzo et al., 2020; Tadege et al., 2015; Weller and Ortega, 2015; Wu et al., 2020; Zhang et al., 2019).
Among these flowering time genes, FLOWERING LOCUS T (FT), which encodes a florigen protein that promotes flowering in annual and perennial plants, has been widely used in cereals and legume species to improve yield potential and related agronomic traits (Cai et al., 2019; Dash and Rai, 2022; Kang et al., 2019; Lorenzo et al., 2020; Shalit et al., 2009; Singer et al., 2018; Wu et al., 2020; Zhang et al., 2019). In Medicago truncatula, the sister species of alfalfa, six FT‐like genes, MtFTa1, MtFTa2, MtFTb1, MtFTb2, MtFTc and Mt6g033040, have been identified (Cheng et al., 2021; Kang et al., 2019; Laurie et al., 2011). MtFTa1 was identified as the key florigen that promotes flowering in M. truncatula and mtfta1 mutants exhibit delayed flowering in all conditions (Cheng et al., 2021; Laurie et al., 2011). Alfalfa MsFTa1 is the closest homologue of MtFTa1 based on the analysis of the exon/intron gene structure in FT genes (Cheng et al., 2021). Other studies also confirmed the biological function of alfalfa MsFTa1 as an FT homologue (Kang et al., 2019; Lorenzo et al., 2020). However, these studies utilized gene over‐expression or down‐regulation methods to investigate the function of MsFTa1 in promoting alfalfa flowering, possibly limiting their agricultural applicability due to the presence of transgenes and/or unstable transgene expression.
The development of the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeat/CRISPR‐associated protein 9) system for genome editing has paved the way for precise engineering of genomes for basic and applied research, including the trait‐driven improvement of important crop species (Cai et al., 2019; Chen et al., 2020; Kumar et al., 2022; Singer et al., 2022; Wolabu et al., 2020a,b). Recently, this system has been used to generate delayed flowering time mutants in different species. For instance, the heading date of rice was delayed by fine‐tuning flowering time genes (Ehd1 and Hd2) using CRISPR/Cas9, which resulted in prolonged vegetative growth period with improved yield potential for flexible cropping systems in diverse environments (Liu et al., 2021; Wu et al., 2020; Zhang et al., 2019). Flowering time in Chinese cabbage (Brassica rapa L.) was also delayed by targeting BrVRN1 using multiple guide RNAs (gRNAs) in the multiplex CRIPSPR/Cas9 construct to generate lines with different flowering times for crop improvement (Hong et al., 2021). Similarly, CRISPR/Cas9 mutagenesis in soybean targeting two flowering genes (GmFT2a and GmFT5a) improved adaptability of soybean to different environments with desirable transgene‐free mutant lines (Cai et al., 2018, 2019).
In this study, we knocked out MsFTa1 in alfalfa line R2336 by employing the previously optimized multiplex CRISPR/Cas9 system (Wolabu et al., 2020a). The line R2336 was developed by Forage Genetic International (FGI) through selection from an elite, high‐yielding, fall‐dormant alfalfa breeding population using a tissue culture screen for callus formation and somatic embryo induction. Several tetra‐allelic homozygous mutants of MsFTa1 were obtained that exhibited delayed flowering up to 21 days. The Msfta1mutant lines accumulated significantly higher biomass compared to control (non‐edited) plants as determined through analyses of fresh and dry weights. Forage quality was significantly improved in the Msfta1 lines, including reduced lignin content, higher crude protein and mineral contents and increased digestibility. These CRISPR/Cas9‐edited Msfta1 mutants could be used in alfalfa breeding programmes to generate transgene‐free elite alfalfa cultivars with improved forage biomass and quality.
Results
CRISPR/Cas9 generates homozygous MsFTa1 knockout mutants with different mutation events at target sites
To generate MsFTa1 CRISPR/Cas9 mutant lines, four guide RNAs (gRNAs) were designed from the conserved coding region of exon 1, exon 2 and exon 3 of MsFTa1 (Figure 1a). The genomic sequence specific to targeted sites were amplified from alfalfa line R2336 and analysed by Sanger sequencing to confirm that there were no genotypic SNPs mismatches across four copies of the gene, which might affect genome editing. The optimized CRISPR/Cas9 vector with high gene‐editing efficiency in alfalfa (Wolabu et al., 2020a) was used to insert the clustered multiplexed MsFTa1 gRNA‐encoding sequences into the alfalfa genome (Figure 1b). Out of 196 transgenic lines generated and genotyped, 96 lines were confirmed by Sanger sequencing to possess at least one mutation signature (double peaks in sequencing chromatograph) at specific target sites (Figure 1c). These putative candidate lines were analysed phenotypically for flowering time difference under long‐day (LD) conditions. Finally, out of 96 lines, 82 lines were subjected to flowering time assessment and half of them showed at least a 10‐day delay in flowering compared to the empty vector control (EV) (Figure 2a). Interestingly, these lines also showed higher vegetative biomass compared to the control (Figure 2b). Twenty‐two lines with flowering time delayed by 21 days or more were further propagated in a replicated experiment to confirm the delayed flowering phenotype. Eventually, six representative lines, namely Msfta1‐002, Msfta1‐015, Msfta1‐043, Msfta1‐074, Msfta1‐094 and Msfta1‐111, were selected for further molecular characterization to identify the nature of the underlying mutations (Figure 3, Figure S1). Sequence analysis revealed diverse mutation events in these lines, including single to multiple nucleotides deletions (1–138 bp deletions), small insertions (1 bp insertion) and substitutions (1–2 bp substitutions) at different target sites. Small deletions (1–11 bp) were the predominant mutation events with 83% frequency (Figure 3, Figure S1). Based on the proportion of mutation occurrence at each target site, we arbitrarily assigned allele numbers (A1–A4) to assess the status of mutation across each gene copy to determine the tetra‐allelic homozygosity/heterozygosity of specific mutations in the line. For instance, Msfta1‐002 showed a 6‐bp deletion in the first allele (A1), 5‐bp deletion in both the second and third alleles (A2 and A3) and a 1‐bp insertion in the fourth allele (A4) at gRNA1, resulting in mutations in all four alleles at the first target site. In addition, Msfta1‐002 also showed mutation events with a 58‐bp deletion in A1 and 2‐bp deletion in A2 and A3 and a 1‐bp substitution in A4 at gRNA2, but no mutation at gRNA3. There was also one mutation event downstream of gRNA4 PAM with 14 bp deletions in A3 and A4 in this line. In general, the overall combined mutations at different targeted sites made line Msfta1‐002 a 100% tetra‐allelic homozygous mutant with delayed flowering phenotype (Figure 3). Remarkably, we also found a large 138‐bp deletion in A1 of the line Msfta1‐074 such that almost all nucleotides between gRNA1 and gRNA2 were deleted (Figure 3). Sequencing analysis of six selected lines (Msfta1‐002, 015, 043, 074, 094 and 111) showed 100% tetra‐allelic homozygous mutations in gRNA1 and gRNA2, indicating the sufficiency of two specific gRNAs to knockout all four allelic copies of the MsFTa1 gene (Figure 3, Figure S1).
Figure 1.
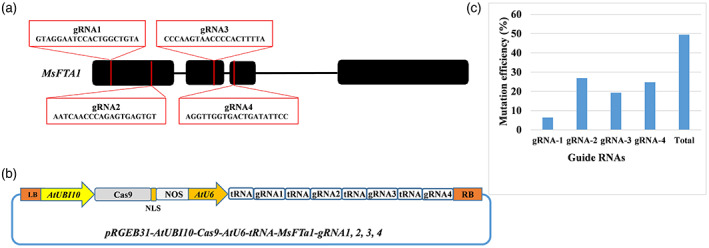
Schematic illustration of alfalfa FLOWERING LOCUS T (MsFTa1) gene structure with designed multiplex gRNAs–CRISPR/Cas9 vector and genome editing efficiency in alfalfa. (a) MsFTa1 gene structure, target sites in the coding region. (b) Illustration of the multiplex MsFTa1–gRNA–CRISPR/Cas9 vector. (c) Mutation efficiency (%) of different gRNAs at different target sites of MsFTa1.
Figure 2.
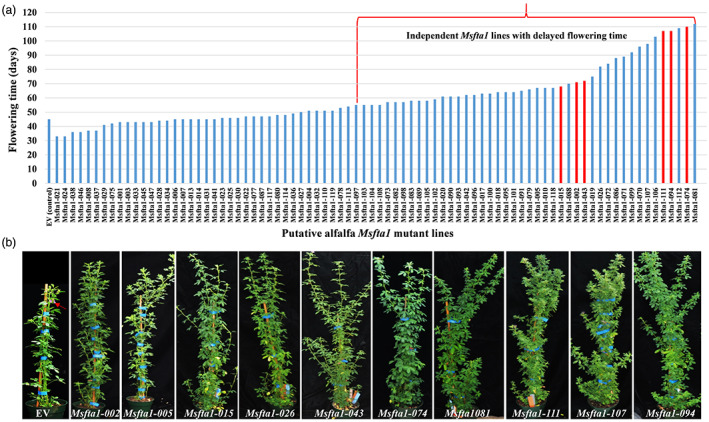
Initial analysis of flowering time in Msfta1 CRISPR/Cas9 mutant lines. (a) Graph representing flowering time (days to flower) in all putative Msfta1 mutant lines (T0) compared to control plants harbouring empty vector (EV). Highlighted in red represents the six lines selected for further analyses. (b) Phenotype of promising putative Msfta1 mutant lines exhibiting delayed flowering and enhanced vegetative growth. Red arrow indicates developed flowers in EV.
Figure 3.
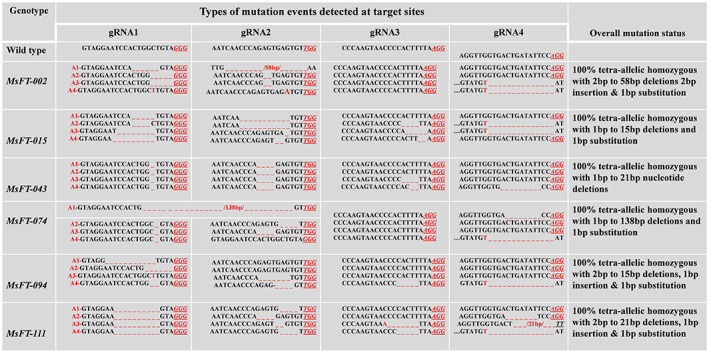
Molecular analysis of MsFTa1 mutation events (deletion/insertion/substation) generated by multiplex gRNA–CRISPR/Cas9 vector. Mutations are shown at the four different target sites: MsFTa1–gRNA1, MsFTa1–gRNA2, MsFTa1–gRNA3 and MsFTa1–gRNA4. Wild‐type (WT) reference sequence is shown for each target site. Deletions are indicated by red dashed lines. Insertions or substitutions are indicated by red letters. PAMs are indicated by red underlined italicized letters. The frequency of mutations in each of the four allelic copies of the MsFTa1 gene is indicated in the right column. The four allelic copies are designated as red allele‐1 (A1), allele‐2 (A2), allele‐3 (A3) and allele‐4 (A4).
Mutation of MsFTa1 delays flowering and increases biomass in alfalfa
The six homozygous lines, Msfta1‐002, 015, 043, 074, 094 and 111, were evaluated in detail for flowering time, forage yield, nutritional quality and other desirable agronomic traits. Flowering time was significantly and substantially delayed in all six homozygous Msfta1 lines compared to the empty vector (EV) control (Figure 4a). Msfta1‐074 showed an extreme delay of 21 days in flowering time, whereas Msfta1‐094 showed intermediate delay of 12 days in flowering time relative to EV. To examine whether delayed flowering affects biomass production, we harvested the above‐ground whole plant organs of each line and measured the fresh and dry weights at flowering time. All six Msfta1 lines showed significantly higher fresh weight than the EV control, with up to 78% increase (Figure 4c). Msfta1‐002, Msfta1‐015, Msfta1‐074 and Msfta1‐094 had the highest fresh weights followed by Msfta1‐043 and Msfta1‐111 (Figure 4c). Similarly, five of the six Msfta1 lines also had significantly higher dry biomass yield with Msfta1‐111 accumulating the highest dry biomass with 76% relative advantage over EV, followed by Msfta1‐015, Msfta1‐074 and Msfta1‐094 (Figure 4d). The plant height of four lines, Msfta1‐0002, Msfta1‐015, Msfta1‐074 and Msfta1‐094, was also significantly greater than that of EV plants (Figure 4b). These results indicate that delayed flowering time in Msfta1 mutant lines led to greater shoot production due to extended vegetative growth (Figure 4a–d).
Figure 4.
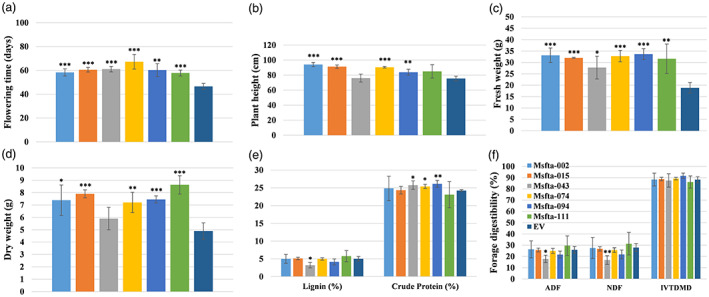
Analysis of agronomic and forage quality traits in selected homozygous Msfta1 mutant lines at the whole plant level. (a) Flowering time analysis of Msfta1 lines versus control plants harbouring empty vector (EV). (b) Analysis of plant height (cm) in Msfta1 lines versus EV plants. (c) Fresh and (d) dry weight analyses of Msfta1 lines versus EV plants. (e) Analysis of crude protein (%) and lignin content (%) in Msfta1 lines versus EV plants. (f) Analysis of forage digestibility parameters: ADF (acid detergent fibre), NDF (neutral detergent fibre) and IVTDMD (in vitro true dry matter digestibility) in Msfta1 lines versus EV plants. Tissue sampling was performed for each line that was harvested when it flowered. Data represent mean values (±SD; n = 6) and were analysed statistically using Student's t test (*P < 0.05; **P < 0.01; ***P < 0.001).
Msfta1 lines with delayed flowering have improved forage quality
Forage quality is determined by intake efficiency, digestibility and nutrient content. Digestibility reflects the feed's net energy, while crude protein (CP) and minerals are the primary nutrients for livestock (Lacefield, 2004). To assess the forage quality of promising Msfta1 lines with delayed flowering, we first examined typical forage quality parameters in the whole shoots. These parameters include lignin and crude protein (CP) contents, acid detergent fibre (ADF), neutral detergent fibre (NDF), in vitro true dry matter digestibility (IVTDMD), total digestible nutrients (TDN) and relative feed value (RFV) (Figure 4e,f, Figure S2a). Forage quality analysis indicated that three lines, Msfta1‐043, Msfta1‐074 and Msfta1‐094, had significantly higher crude protein content than the control (EV) (Figure 4e). Interestingly, Msfta1‐043 also had significantly lower lignin content and ADF and NDF values than the EV control (Figure 4e,f). Among the tested Msfta1 lines, Msfta1‐074 also had significantly higher mineral content (Mg, P, Ca and K) compared to the control, followed by Msfta1‐043 (Mg and Ca) and Msfta1‐094 (Mg) (Figure S2b). The results indicate that line Msfta1‐043 exhibited superior nutritional quality, followed by Msfta1‐074 and Msfta1‐094 in whole plant analyses (Figure 4e,f, Figure S2a,b).
We further analysed the effects of delayed flowering time on forage quality by employing two different harvesting schemes, designated as Batch 1 and Batch 2, to separately collect stem and leaf tissues. In Batch 1, all six promising Msfta1 lines and the EV control were harvested at the time when the control (EV) plants had initiated flowering. In Batch 2, all Msfta1 lines and the control (EV) plants were harvested at the time when Msfta1‐094 (exhibiting intermediate delayed flowering) had started to flower. Interestingly, all six Msfta1 lines in stem Batch 1 and Batch 2 had significantly reduced stem lignin contents compared to control plants (Figure 5a,d). Similarly, stems of three lines, Msfta1‐043, Msfta1‐074 and Msfta1‐111, had significantly higher crude protein content in stem Batch 1 with 7%–20% relative advantages over the control (Figure 5a). In Batch 2, the stem tissues of all six Msfta1 lines had 18%–65% higher crude protein content than the control had (Figure 5d). The stem tissues of all Msfta1 lines, in both Batch 1 and Batch 2, had significantly lower levels of NDF and higher levels of IVTDMD compared to control plants (Figure 5b,e). The ADF levels were also significantly reduced in the stem tissues of all lines from Batch 2 but only Msfta1‐043 showed a significant reduction in Batch 1 (Figure 5b,e). Similarly, all six Msfta1 lines in Batch 2 also had significantly higher levels of TDN and RFV in their stems (Figure S3d). Except for Msfta1‐002 and Msfta1‐015 in Batch 1, the mineral content (Mg, P and K) were significantly higher in stem tissues of the remaining four Msfta1 lines, whereas in Batch 2, the stems of all Msfta1 lines accumulated significantly higher levels of these minerals (Figure 5c,f).
Figure 5.
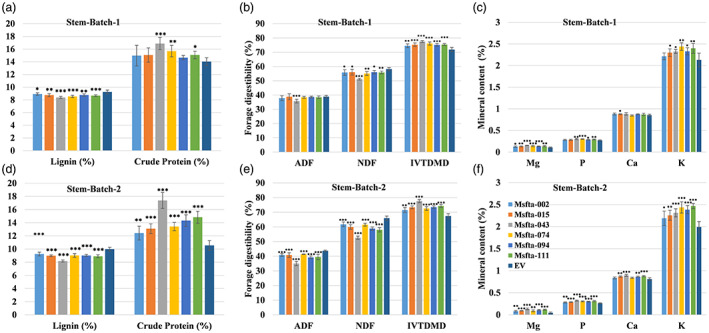
Analysis of forage quality traits in stem tissues of Msfta1 mutant lines harvested under different schemes. Batch 1 tissues were harvested when EV plants started to flower and Batch 2 tissues were harvested when the line Msfta1‐094 (exhibiting intermediate phenotype) started to flower. (a) Analysis of lignin (%) and crude protein content (%) in stem tissues of Msfta1 lines versus EV plants in Batch 1. (b) Analysis of forage digestibility traits including ADF, NDF and IVTDMD in stem tissues of Msfta1 lines versus EV plants in Batch 1. (c) Analysis of minerals content (%) in stem tissues of Msfta1 lines versus EV plants in Batch 1. (d) Analysis of lignin (%) and crude protein content (%) in stem tissues of Msfta1 lines versus EV plants in Batch 2. (e) Analysis of forage digestibility traits including ADF, NDF and IVTDMD in stem tissues of Msfta1 lines versus EV plants in Batch 2. (f) Analysis of minerals content (%) in stem tissues of Msfta1 lines versus EV plants in Batch 2. Data represent mean values (±SD; n = 6) and were analysed statistically using Student's t test (*P < 0.05; **P < 0.01; ***P < 0.001).
Similarly, we also analysed the forage quality parameters in the leaf tissues of all six Msfta1 lines from Batch 1 and Batch 2 harvesting schemes and observed similar trends as stems (Figure 6a–f, Figure S4a–d). All Msfta1 lines in Batch 1 and three lines (Msfta1‐043, Msfta1‐074 and Msfta1‐111) in Batch 2 had significantly higher crude protein content than the control (Figure 6a,d). Again, Msfta1‐043 showed significantly reduced lignin content, ADF and NDF in leaf Batch 1 (Figure 6a,b). Five lines (Msfta1‐002, Msfta1‐043, Msfta1‐074, Msfta1‐094 and Msfta1‐111) showed higher IVTDMD in leaf Batch 1, whereas one line (Msfta1‐094) showed significantly lower IVTDMD levels in leaf Batch 2 (Figure 6b,e). Except for line Msfta1‐043, all lines (Msfta1‐002, Msfta1‐015, Msfta1‐074, Msfta1‐094 and Msfta1‐111) showed significantly elevated levels of NDF but no significant differences for ADF in leaf Batch 2 (Figure 6b,e). The content of certain minerals was found to be higher in some mutants than the control of leaf Batch 1, including Mg in all Msfta1 lines (except Msfta1‐002), K in two lines (Msfta1‐074 and Msfta1‐111) and P in one line (Msfta1‐074), with no significant differences in the Ca content (Figure 6c). All Msfta1 lines accumulated significantly higher P and K, while three lines (Msfta1‐043, Msfta1‐094 and Msfta1‐111) had higher Mg and only one line (Msfta1‐094) had higher Ca content than the control in Batch 2 (Figure 6f).
Figure 6.
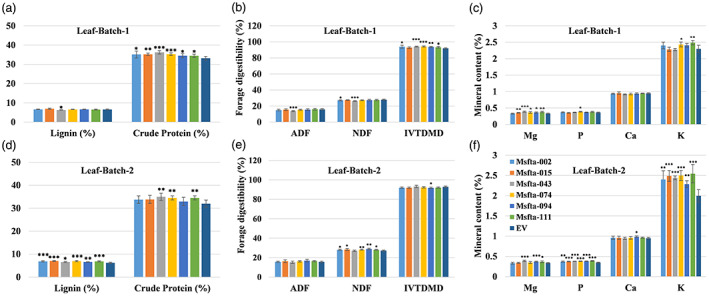
Analysis of forage quality traits in leaf tissues of Msfta1 mutant lines harvested under different schemes: Batch 1 tissues were harvested when EV plants started to flower and Batch 2 tissues were harvested when the line Msfta1‐094 (exhibiting intermediate phenotype) started to flower. (a) Analysis of lignin (%) and crude protein content (%) in leaf tissues of Msfta1 lines versus EV plants in Batch 1. (b) Analysis of forage digestibility traits including ADF, NDF and IVTDMD in leaf tissues of Msfta1 lines versus EV plants in Batch 1. (c) Analysis of minerals content (%) in leaf tissues of Msfta1 lines versus EV plants in Batch 1. (d) Analysis of lignin (%) and crude protein content (%) in leaf tissues of Msfta1 lines versus EV plants in Batch 2. (e) Analysis of forage digestibility traits including ADF, NDF and IVTDMD in leaf tissues of Msfta1 lines versus EV plants in Batch 2. (f) Analysis of minerals content (%) in leaf tissues of Msfta1 lines versus EV plants in Batch 2. Data represent mean values (±SD; n = 6) and were analysed statistically using Student's t test.
Next, we performed correlation analysis among the tested Msfta1 lines in whole plant, stem and leaf samples (Batch 1 and Batch 2) to assess the relationship between forage quality factors including lignin content, crude protein content, digestibility and nutritive values (Table 1, Figures S5–S9). At the whole plant level, the lignin contents were found to be positively correlated with ADF and NDF (R 2 = 0.98; Table 1, Figure S5e, f) and negatively correlated with RFV, TDN and CP with the R 2 value of 0.95, 0.98 and 0.66 respectively (Table 1, Figure S5g–i). Similarly, ADF showed a strong negative correlation with TDN (R 2 = 1) but a strong positive correlation with NDF (R 2 = 0.99) (Table 1, Figure S5j,l). In addition, at the whole plant level, flowering time was positively correlated with plant height and fresh biomass and had a weak positive correlation with dry biomass. Flowering time was weakly negatively correlated with lignin content (Figure S5a–d).
Table 1.
Correlation analyses of forage quality traits of whole plant, stem and leaf tissues of Msfta1 lines of alfalfa representing different harvesting time (leaf and stem Batch 1 and Batch 2)
| Correlation (%) | Whole plant | Stem | Leaf | |||||
|---|---|---|---|---|---|---|---|---|
| (R2 ) | Status | Batch 1 (R 2) | Batch 2 (R 2) | Status | Batch 1 (R 2) | Batch 2 (R 2) | Status | |
| ADF × lignin | 0.98 | (+) | 0.38 | 0.88 | (+) | 0.32 | 0.12 | (+) |
| NDF × lignin | 0.98 | (+) | 0.73 | 0.94 | (+) | 0.37 | 0.31 | (+) |
| RFV × lignin | 0.95 | (−) | 0.62 | 0.9 | (−) | 0.37 | 0.28 | (−) |
| IVTDMD × lignin | 0.11 | (−) | 0.98 | 0.99 | (−) | 0.17 | 0.34 | (−) |
| TDN × ADF | 1.00 | (−) | 1.00 | 1.00 | (−) | 1.00 | 1.00 | (−) |
| IVTDMD × ADF | 0.15 | (−) | 0.44 | 0.88 | (−) | 0.36 | 0.66 | (−) |
| NDF × ADF | 0.99 | (+) | 0.86 | 0.97 | (+) | 0.92 | 0.83 | (+) |
| RFV × TDN | 0.91 | (+) | 0.93 | 0.99 | (+) | 0.96 | 0.89 | (+) |
| CP × lignin | 0.66 | (−) | 0.8 | 0.93 | (−) | 0.05 | 0.42 | (−) |
For plants representing the Batch 1 harvesting scheme, lignin contents were positively correlated with NDF (R 2 = 0.73), but negatively correlated with RFV, IVTDMD and CP content (R 2 = 0.62, 0.98 and 0.80 respectively) in the stem tissues (Table 1, Figure S6b–d). ADF was positively correlated with NDF (R 2 = 0.86) but negatively correlated with TDN (R 2 = 1) and IVTDMD (R 2 = 0.88) (Table 1, Figure S6e–g). A strong positive correlation was observed between RFV and TDN (R 2 = 0.93; Table 1, Figure S6h). Interestingly, similar correlation trends were observed between forage quality traits in the stem tissues of plants representing Batch 2 harvesting scheme (Table 1, Figure S7a–i). For leaf tissues representing Batch 1 and 2 harvesting schemes, weak correlations were observed among quality traits except for a strong positive correlation between ADF and NDF (R 2 = 0.92), RFV and TDN (R 2 = 0.96) and strong negative correlation between ADF and TDN (R 2 = 1) (Table 1, Figures S8 and S9). Taken together, lignin content exhibited a strong positive correlation with ADF and NDF and a strong negative correlation with RFV and TDN at the whole plant level and in the stem tissues of batches 1 and 2 plants, indicating that the nutritional quality of Msfta1 lines was significantly improved compared to the control through flowering time manipulation.
Discussion
MsFTa1 mutation leads to delayed flowering initiation in alfalfa
Forage biomass yield and quality are two important traits to which breeders and ranchers pay special attention. In forage crops, these traits are negatively affected by environmental stresses and internal stimuli, including the onset of flowering. Transition from the vegetative phase to the reproductive phase is associated with various developmental changes including stem lignification, seed set and plant senescence (Ball et al., 2001; Fick and Mueller, 1989; Lacefield, 2004; Singer et al., 2018). These changes result in decreased forage digestibility and nutrient contents including crude protein and minerals in the vegetative tissues. Given the link between flowering time and forage quality, we targeted the alfalfa FLOWERING LOCUS Ta1 (MsFTa1) gene by employing the multiplex‐CRISPR/Ca9 genome editing approach and generated stable Msfta1 mutant lines with delayed flowering and improved forage biomass yield and quality.
Four regions spanning the first, second and third exons of MsFTa1 gene were targeted using a multiplex CRISPR/Cas9 system to produce a single synthetic polycistronic tRNA–gRNA clustering system with four different gRNAs (Xie et al., 2015). We had previously optimized the multiplex CRISPR/Cas9 system in alfalfa to enhance the gene editing efficiency (Wolabu et al., 2020a). Molecular analyses of several putative Msfta1 lines exhibiting delayed flowering revealed that in at least six of the Msfta1 lines (Msfta1‐002, 015, 043, 074, 094 and 111), all four allelic copies of the MsFTa1 gene were mutated at one or more target sites (Figure 3). We identified various mutation events in the target sites of MsFTa1 including deletions (1–138 bp), insertion (1 bp) and substitutions (1–2 bp). In general, small deletions (1–11 bp) were predominant mutations (83% frequency) (Figure 3, Figure S1). CRISPR/Cas9 system has been shown to induce similar mutation events (indels) with large truncations for other genes in alfalfa, soybean and rice (Cai et al., 2018; Chen et al., 2020; Singer et al., 2022; Stritzler et al., 2022; Wolabu et al., 2020a; Wu et al., 2020).
Msfta1 mutants accumulate more biomass
Flowering time is a crucial trait in breeding programmes for forage crop improvement as manipulating floral transition through genetic modification could modulate vegetative growth period and impact biomass yield and quality (Fick and Mueller, 1989; Gou et al., 2019; Jung and Müller, 2009). In this study, mutation of MsFTa1 via CRISPR/Cas9 gene editing resulted in tetra‐allelic homozygous mutant plants with delayed flowering and increased plant height and fresh and dry biomass compared to the control plants (Figures 2, 4). The results of this study are consistent with prior findings on the manipulation of plant genes known to promote flowering in improved agronomic traits, yield and quality in soybean (Cai et al., 2019; Xu et al., 2021), M. truncatula (Tadege et al., 2015), alfalfa (Lorenzo et al., 2020), rice (Cui et al., 2021; Liu et al., 2021; Wu et al., 2020; Zhang et al., 2019), switchgrass (Gou et al., 2019), Chinese cabbage (Hong et al., 2021) and tomato (Rajendran et al., 2021; Shalit et al., 2009). Recently, the role of the FTa1 gene in alfalfa was confirmed by down‐regulating its expression through an artificial microRNA (amiRNA) (Lorenzo et al., 2020). Although down‐regulation of the Msfta1 expression significantly delayed flowering time and increased crude protein content in the vegetative tissues of transgenic plants, other important growth traits such as plant height, height/node ratios and dry biomass accumulation were significantly reduced compared to WT (Lorenzo et al., 2020). However, in our study, the CRISPR/Cas9‐edited Msfta1 mutant lines showed similar morphological features to control plants (EV) but exhibited delayed flowering time up to 21 days with a significant increase in dry biomass (up to 76%) (Figure 4d). It is difficult to determine why MsFTa1 down‐regulation lines exhibited severe phenotype than CRISPR/Cas9‐edited Msfta1 mutant lines. One possibility is that the amiRNA targets other alfalfa FT homologous genes, though the differences in the genetic background of two alfalfa cultivars used in these studies could not be excluded for the observed variations. Nevertheless, the CRISPR/Cas9 gene‐editing approach offers several advantages over other traditional transgenic approaches (down‐regulation or over‐expression), including the ability to obtain stable transgene‐free plants with desired trait by segregating out the Cas9 gene/gRNA construct and overcome issues associated with possible silencing of transgenes over time under the field conditions (Fernandes et al., 2022; Nerkar et al., 2022).
Msfta1 mutants have improved forage quality and nutritive values
Factors like forage palatability, intake and digestibility greatly affect the final performance and/or production of animals (Ball et al., 2001; Capstaff and Miller, 2018; Lacefield, 2004). Forage lignin content, ADF and NDF are key determinants of forage digestibility and intake potential (Ball et al., 2001; Gao et al., 2018; Lacefield, 2004; Lorenzo et al., 2020). The lower the lignin, ADF and NDF values, the better the forage quality. We analysed the lignin content and digestibility parameters in selected Msfta1 lines. The lignin contents were reduced significantly in at least three Msfta1 lines with the most reduction in Msfta1‐043 at the whole plant level (Figure 4e). Similarly, whole plant ADF and NDF levels were lower in three Msfta1 mutant lines compared to the EV control (Figure 4f). Furthermore, all six Msfta1 lines had significantly lower levels of NDF in stem Batch 1 and Batch 2 (Figure 5b,e). All six Msfta1 lines also had lower levels of ADF in stem Batch 2 compared to EV (Figure 5b,e). Since stems contribute most to final biomass accumulation, lower lignin and ADF and NDF values in the stem tissues increase forage digestibility and intake potential (Ball et al., 2001; Bhattarai et al., 2018; Lorenzo et al., 2020).
The higher crude protein (CP) content in forage shoot tissues contributes significantly to successful livestock production (Ball et al., 2001; Capstaff and Miller, 2018). Induction of flowering and subsequent plant maturity lead to reduction in protein content in the vegetative tissues due to increased nutrient run‐off to support fruit and seed development (Bucher and Römermann, 2021; Lacefield, 2004). According to Lacefield (2004), the stage of harvesting has the biggest effect on alfalfa forage quality, with a 45% decrease in protein content when harvesting was postponed from late bud to full bloom. In fact, delaying floral initiation through genetic manipulation has been successful in retaining higher protein content in the vegetative tissues of different forage crops (Aung et al., 2015; Ball et al., 2001; Capstaff and Miller, 2018; Lorenzo et al., 2020). Consistent with these findings, most Msfta1 mutant lines from our study also retained higher crude protein content in the vegetative tissues, regardless of the harvesting scheme used (Batch 1, Batch 2 or whole plant) (Figure 4e, Figure 5a,d, Figure 6a,d).
Mineral contents in forages are also important for livestock production as mineral deficiencies can harm animal health and development. Mineral supplements are expensive, limiting profits in livestock production (Capstaff and Miller, 2018). Similar to the crude protein content, with a few exceptions, the levels of Mg, Ca and K ions were generally higher in the vegetative tissues of most examined Msfta1 lines compared to the control, with some variations depending on the harvesting scheme (Batch 1, Batch 2 or whole plant) (Figure S2b, Figure 5c,f, Figure S4c,f).
Interestingly, the correlation analyses of forage quality parameters showed a positive correlation between lignin content and ADF and NDF and a strong negative correlation between lignin and RFV, IVTDMD and crude protein content (Figure 6a–d, Table 1). These results suggest that the forage nutrition parameters, particularly the crude protein content and digestibility, are directly affected by lignin content. We observed similar negative correlations between lignin content and crude protein content regardless of the harvesting schemes (stem Batch 1 and Batch 2) (Table 1). In general, the correlation analyses suggest that forage nutrition quality is directly affected by lignin and fibre content, which is associated with flowering and maturity, indicating that the forage digestibility and quality could be enhanced by reducing lignin/fibre content through flowering time manipulation. Similar correlation patterns in forage quality parameters were also found in alfalfa with delayed flowering through down‐regulation of MsFTa1 expression (Lorenzo et al., 2020). Tadege et al. (2015) also reported that delaying floral initiation in M. truncatula causes simultaneous improvement in biomass quantity and quality, particularly through reduction in lignin content without any pleiotropic growth defects.
In conclusion, delayed flowering in alfalfa was achieved by knocking out MsFTa1 through a multiplex CRISPR/Cas9 editing approach. The resulting Msfta1 mutant lines accumulated significantly higher biomass and exhibited improved forage quality traits. These lines could serve as important genetic resources for developing transgene‐free alfalfa cultivars that have delayed flowering and other desirable agronomic traits.
Experimental procedures
Plant materials and growth conditions
The alfalfa line, R2336, from Forage Genetic International (FGI), was used as a parent line to generate CRISPR/Cas9‐edited Msfta1 mutants. FGI developed the R2336 line through selection from an elite, high‐yielding, fall‐dormant alfalfa breeding population using a tissue culture screen for callus formation and somatic embryo induction. The CRISPR/Cas9‐generated alfalfa mutant lines and empty vector control (EV) plants were grown under greenhouse conditions of 22/19 °C day/night temperature, 16/8 h day/night photoperiod and 70%–80% relative humidity. All alfalfa plants were vegetatively propagated by cuttings.
Multiplex gRNA design and CRISPR/Cas9 vector construction and expression
To knockout MsFTa1, multiplex gRNAs were designed from the conserved regions of the alfalfa MtFTa1 gene. To avoid any potential genotypic SNPs that might affect genome editing due to mismatch, the genomic DNA fragment of MtFTa1 was amplified from the R2336 by PCR and cloned into the pGEM‐Teasy vector (Promega, Madison, WI). Twenty positive clones were randomly picked and subjected to Sanger sequencing to determine the sequence of the conserved regions of four alfalfa alleles (copies). Four MsFTa1 gRNAs were designed for targeting the first, second and third exons as gRNA1‐GTAGGAATCCACTGGCTGTA, gRNA2‐AATCAACCCA GAGTGAGTGT, gRNA3‐CCCAAGTAACCCCACTTTTA and gRNA4‐AGGTTGGTGACT GATATTCC upstream of PAM (GGG, TGG, AGG and AGG) sites, using the web‐based tool CRISPR‐P (http://cbi.hzau.edu.cn/cgi‐bin/CRISPR) (Lei et al., 2014). All designed gRNAs (spacers) were inserted between tRNA and gRNA scaffolds and clustered in a tandem manner using the Golden Gate assembly method (Engler et al., 2008). The pGTR plasmid, which contains a tRNA–gRNA fragment, was used as a template to synthesize polycistronic tRNA–gRNA (PTG) (Xie et al., 2015) for construction of the multiplex tRNA–MsFTa1 spacer–gRNA. First, the overlapping PCR products were separated and purified using the Spin Column PCR Product Purification Kit (Wizard SV Gel and PCR Clean‐Up System) following the manufacturer's instruction (www.promega.com). The chain of multiplex tRNA–gRNA with four MsFTa1 spacers were then inserted into the optimized vector with AtU6–tRNA–gRNAs–AtUBQ10–Cas9–pRGEB31‐bar backbone by digestion and ligation using Fok I (NEB) and BsaI (Wolabu et al., 2020a). The subsequent multiplex MsFTa1 spacers–gRNAs–CRISPR/Cas9 binary vector was transformed into Agrobacterium tumefaciens strain, AGL1. Agrobacterium‐mediated transformation of alfalfa was carried out following the protocol described by Wolabu et al. (2020a) to generate CRIPR/Cas9‐edited alfalfa Msfta1 mutants.
Genotyping of Msfta1 transgenic lines
CRISPR/Cas9‐edited Msfta1 mutant lines were identified as follows. First, the transgenic lines were screened through the PCR amplification of BAR gene using genomic DNA (gDNAs) with BAR gene‐specific primers (PPT‐F + PPT‐R) (Table S1). The BAR‐gene‐positive transgenic lines were subjected to another round of PCR to amplify the target regions spanning the gRNAs with MsFTa1‐specific primers. The amplified PCR products were sequenced using Sanger sequencing method to determine the mutations (Table S1). The transgenic lines exhibiting delayed flowering phenotype were further characterized for homozygosity by cloning the target regions spanning the gRNAs (gRNA1, gRNA2, gRNA3 and gRNA4) into pGEM‐T Easy vector and 15–20 colonies were randomly picked, and each plasmid DNA was sequenced using Sanger sequencing method to determine the nature of mutations and to identify Msfta1 homozygous lines. Reads were aligned with the reference sequence using the SeqMan Pro 15.0.1 (DNASTAR software for life scientists) (https://www.dnastar.com/quote‐request/).
Flowering time analysis and tissue harvesting
The initial evaluation of flowering time of individual positive transgenic CRISPR/Cas9‐edited lines was carried out by assessing the single plant of each line directly generated from CRISPR/Cas9 transformation (no statistical analysis). However, the initial flowering time evaluation might be influenced by tissue culture effects and/or chimeric mutations. To accurately evaluate the flowering time in selected lines, we conducted statistical assessment using replicated plants (at least six plants for each line) with uniformity within the population. First, the selected lines were subjected to at least six rounds of cutting back in greenhouse to maintain its uniformity as much as possible to overcome or minimize the variations caused tissue culture and chimeric mutation effects. Then, at least six individual plants were vegetatively propagated for each selected line by cuttings from similar shoot source to minimize other source of variation on flowering time that might be caused by environmental effects. In general, when 10% of plants of each line started flowering, whole plant (above‐ground tissues without roots), stem and leaf samples were harvested for yield and nutrition quality analyses. Three schemes were adopted to record the flowering time and to harvest tissues for biomass yield and nutritional quality assessment for each selected line. In the first scheme (whole plant), above‐ground vegetative tissues from each Msfta1 line and the empty vector (EV) plants were harvested when 10% of the plants from each line started to flower. In the second scheme, leaf and stem tissues from all lines (six plants for each line) were harvested separately when 10% of the EV plants had produced their first flowers (referred as Batch 1). In the third scheme, leaf and stem tissues from all plants of each line were harvested separately when 10% of plants of the intermediate late flowering line (Msfta1‐94) had produced their first flowers (referred as Batch 2). After harvesting, fresh and dry weights were recorded prior to forage nutritional analysis.
Forage nutritional quality assessment of Msfta1 mutant lines
Forage nutritional quality assessment was carried out at Ag Services and Resources Core Facility (ASRC), Noble Research Institute, LLC. Sample grinding and processing were performed following procedures described by Gou et al. (2019). Briefly, the plant samples were dried at a fixed temperature of 55 ± 5 °C for 10 days and ground using a Wiley mill. Forage nutritional quality was determined with near infrared reflectance spectroscopy (NIRS) system using a Foss NIRS 6500 monochromator with a scanning range of 1100–2500 nm (Foss NIR System). The following forage nutritional quality parameters were measured: acid detergent fibre (ADF), neutral detergent fibre (NDF) in percentage of dry matter base, crude protein (CP) content, relative feed value (RFV), in vitro dry matter digestibility (IVTDMD), total digestible nutrients (TDN), minerals including magnesium (Mg), potassium (K), calcium (Ca) and phosphorus (P) content. Lignin analysis was carried out using the modified thioacidolysis‐coupled GC–MS method described by Chen et al. (2021).
Statistical analysis
At least six plants for each independent lines were used as biological replicates for statistical analyses. Data shown in graph bars represent the mean ± SD. The analyses were performed using Microsoft Excel software and the asterisks indicate significant differences based on Student's t test (***P < 0.001, **P < 0.01, *P < 0.05). Relative advantage (%) of the tested lines over the control was simply calculated as the performance of tested line minus the control and divided by control and multiplied by 100 (Tested line − EV/EV × 100 = %).
Conflict of interest
The authors declare no competing financial interests.
Authors contribution
TW and JW conceived the research and wrote the article. TW, KM, JY and LC conducted the research. ITJ analysed data. MU and ZW designed the research and edited the article.
Supporting information
Table S1 List of target sequence and primers used in this study.
Figure S1 Molecular analysis of mutation events in the line Msfta1‐015.
Figure S2 Analysis of forage quality traits at whole plant level in Msfta1 mutant lines.
Figure S3 Analysis of dry biomass and forage digestibility parameters in Msfta1 mutant lines representing stem batches 1 and 2.
Figure S4 Analysis of dry biomass and forage digestibility parameters in Msfta1 mutant lines representing leaf batches 1 and 2.
Figure S5 Correlation analysis of forage quantitative and qualitative traits in Msfta1 lines at whole plant level.
Figure S6 Correlation analysis of forage quality traits in stem tissues of Msfta1 lines of alfalfa representing Batch 1.
Figure S7 Correlation analysis of forage quality traits in stem tissues of Msfta1 lines of alfalfa representing Batch 2.
Figure S8 Correlation analysis of forage quality traits in leaf tissues of Msfta1 lines of alfalfa representing Batch 1.
Figure S9 Correlation analysis of forage quality traits in leaf tissues of Msfta1 lines of alfalfa representing Batch 2.
References
- Aung, B. , Gruber, M.Y. , Amyot, L. , Omari, K. , Bertrand, A. and Hannoufa, A. (2015) MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13, 779–790. [DOI] [PubMed] [Google Scholar]
- Ball, D.M. , Collins, M. , Lacefield, G.D. , Martin, N.P. , Mertens, D.A. , Olson, K.E. et al. (2001) Understanding Forage Quality. Park Ridge, IL: American Farm Bureau Federation Publication 1‐01. [Google Scholar]
- Bao, Q. , Wolabu, T.W. , Zhang, Q. , Zhang, T. , Liu, Z. , Sun, J. and Wang, Z.Y. (2022) Application of CRISPR/Cas9 technology in forages. Grassland Res. 1, 244–251. [Google Scholar]
- Bhattarai, K. , Rajasekar, S. , Dixon, R.A. and Monteros, M.J. (2018) Agronomic performance and lignin content of HCT down‐regulated alfalfa (Medicago sativa L.). Bioenergy Res. 11, 505–515. [Google Scholar]
- Bucher, S.F. and Römermann, C. (2021) The timing of leaf senescence relates to flowering phenology and functional traits in 17 herbaceous species along elevational gradients. J. Ecol. 109, 1537–1548. [Google Scholar]
- Cai, Y. , Chen, L. , Liu, X. , Guo, C. , Sun, S. , Wu, C. , Jiang, B. et al. (2018) CRISPR/Cas9‐mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16, 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y. , Wang, L. , Chen, L. , Wu, T. , Liu, L. , Sun, S. , Wu, C. et al. (2019) Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 18, 298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capstaff, N.M. and Miller, A.J. (2018) Improving the yield and nutritional quality of forage crops. Front. Plant Sci. 9, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Zeng, Y. , Yang, Y. , Huang, L. , Tang, B. , Zhang, H. , Hao, F. et al. (2020) Allele‐aware chromosome‐level genome assembly and efficient transgene‐free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 11, 2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F. , Zhuo, C. , Xiao, X. , Pendergast, T.H. and Devos, K.M. (2021) A rapid thioacidolysis method for biomass lignin composition and tricin analysis. Biotechnol. Biofuels, 14, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X. , Li, G. , Krom, N. , Tang, Y. and Wen, J. (2021) Genetic regulation of flowering time and inflorescence architecture by MtFDa and MtFTa1 in Medicago truncatula . Plant Physiol. 185(1), 161–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y. , Xu, Z. and Xu, Q. (2021) Elucidation of the relationship between yield and heading date using CRISPR/Cas9 system‐induced mutation in the flowering pathway across a large latitudinal gradient. Mol. Breed. 41, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash, P.K. and Rai, R. (2022) Green revolution to grain revolution: Florigen in the frontiers. J. Biotechnol. 10, 38–46. [DOI] [PubMed] [Google Scholar]
- Du, J. , Lu, S. , Chai, M. , Zhou, C. , Sun, L. , Tang, Y. and Nakashima, J. (2021) Functional characterization of PETIOLULE‐LIKE PULVINUS (PLP) gene in abscission zone development in Medicago truncatula and its application to genetic improvement of alfalfa. Plant Biotechnol. J. 19, 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler, C. , Kandzia, R. and Marillonnet, S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One, 3, e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, P.M.B. , Favaratto, L. , Fernandes, A.A.R. , Vicien, C. , Capalbo, D.M.F. and Zerbini, F.M. (2022) To become more sustainable organic agriculture needs genome editing technology. Front. Bioeng. Biotechnol. 10, 912793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fick, G.W. and Mueller, S. (1989) Alfalfa: Quality, Maturity, and Mean Stage of Development. Information Bulletin 217. Ithaca, NY: Cornell Cooperative Extension, Cornell University. [Google Scholar]
- Gao, R. , Feyissa, B.A. , Croft, M. and Hannoufa, A. (2018) Gene editing by CRISPR/Cas9 in the obligatory outcrossing Medicago sativa. Planta, 247, 1043–1050. [DOI] [PubMed] [Google Scholar]
- Gou, J. , Tang, C. , Chen, N. , Wang, H. , Debnath, S. , Sun, L. , Flanagan, A. et al. (2019) SPL7 and SPL8 represent a novel flowering regulation Mechanism in switchgrass. New Phytol. 222, 1610–1623. [DOI] [PubMed] [Google Scholar]
- Hong, J.K. , Suh, E.J. , Park, S.R. , Park, J. and Lee, Y.H. (2021) Multiplex CRISPR/Cas9 mutagenesis of BrVRN1 delays flowering time in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Agriculture, 11, 1286. [Google Scholar]
- Jung, C. and Müller, A.E. (2009) Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. [DOI] [PubMed] [Google Scholar]
- Kang, J. , Zhang, T. , Guo, T. , Ding, W. , Long, R. , Yang, Q. and Wang, Z. (2019) Isolation and functional characterization of MsFTa, a FLOWERING LOCUS T homolog from alfalfa (Medicago sativa). Int. J. Mol. Sci. 20, 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishchenko, O. , Zhou1, Y. , Jatayev, J. , Shavrukov, Y. and Borisjuk, N. (2020) Gene editing applications to modulate crop flowering time and seed dormancy. aBIOTECH, 1, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D. , Yadav, A. , Ahmad, R. , Dwivedi, U.N. and Yadav, K. (2022) CRISPR‐based genome editing for nutrient enrichment in crops: a promising approach toward global food security. Front. Genet. 13, 932859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacefield, D.G. (2004) Alfalfa quality: What it is? What can we do about it? And will it pay? In Proceedings, National Alfalfa Symposium, 13–15 December 2004, p. 95616. San Diego, CA: UC Cooperative Extension, University of California, Davis. [Google Scholar]
- Laurie, R.E. , Diwadkar, P. , Jaudal, M. , Zhang, L. , Hecht, V. , Wen, J. , Tadege, M. et al. (2011) The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156, 2207–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y. , Lu, L. , Liu, H.Y. , Li, S. , Xing, F. and Chen, L.L. (2014) CRISPR‐P: a web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Mol. Plant, 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Liu, H. , Zhang, Y. , He, M. , Li, R. , Meng, W. , Wang, Z. et al. (2021) Fine‐tuning flowering time via genome editing of upstream open reading frames of Heading Date 2 in rice. Rice (N Y), 14, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, C.D. , García‐Gagliardi, P. , Antonietti, M.S. , Sánchez‐Lamas, M. , Mancini, E. , Dezar, C.A. , Vazquez, M. et al. (2020) Improvement of alfalfa forage quality and management through the down‐regulation of MsFTa1. Plant Biotechnol. J. 18, 944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerkar, G. , Devarumath, S. , Purankar, M. , Kumar, A. , Valarmathi, R. , Devarumath, R. and Appunu, C. (2022) Advances in crop breeding through precision genome editing. Front. Genet. 13, 880195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.J. , Jiang, K. , Tal, L. , Yichie, Y. , Gar, O. , Zamir, D. , Eshed, Y. et al. (2014) Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46, 1337–1342. [DOI] [PubMed] [Google Scholar]
- Rajendran, S. , Heo, J. , Kim, Y.J. , Kim, D.H. , Ko, K. , Lee, Y.K. , Oh, S.K. et al. (2021) Optimization of tomato productivity using flowering time variants. Agronomy, 11, 285. [Google Scholar]
- Shalit, A. , Rozman, A. , Goldshmidt, A. , Alvarez, J.P. , Bowman, J.L. , Eshed, Y. and Lifschitz, E. (2009) The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA, 106, 8392–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, S.D. , Hannoufa, A. and Acharya, S. (2018) Molecular improvement of alfalfa for enhanced productivity and adaptability in a changing environment. Plant Cell Environ. 41, 1955–1971. [DOI] [PubMed] [Google Scholar]
- Singer, S.D. , Burton, H.K. , Subedi, U. , Dhariwal, G.K. , Kader, K. , Acharya, S. , Chen, G. et al. (2022) The CRISPR/cas9‐ mediated modulation of SQUAMOSA PROMOTER‐BINDING PROTEIN‐LIKE 8 in alfalfa leads to distinct phenotypic outcomes. Front. Plant Sci. 12, 774146. 10.3389/fpls.2021.774146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritzler, M. , Pascuan, C. , Bottero, E. , Gómez, C. , Frare, R. , Puebla, A. , Tajima, H. et al. (2022) Rapid and cloning‐free screening of edited alfalfa via next‐generation sequencing. Plant Cell Tiss. Organ. Cul. 151, 451–456. 10.1007/s11240-022-02358-6 [DOI] [Google Scholar]
- Tadege, M. , Chen, F. , Murray, J. , Wen, J. , Ratet, P. , Udvardi, M.K. , Dixon, R.A. et al. (2015) Control of vegetative to reproductive phase transition improves biomass yield and simultaneously reduces lignin content in Medicago truncatula . Bioenergy Res. 8, 857–867. [Google Scholar]
- Weller, J.L. and Ortega, R. (2015) Genetic control of flowering time in legumes. Front. Plant Sci. 6, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolabu, T.W. , Cong, L. , Park, J.J. , Bao, Q. , Chen, M. , Sun, J. , Xu, B. et al. (2020a) Development of a highly efficient multiplex genome editing system in outcrossing tetraploid alfalfa (Medicago sativa). Front. Plant Sci. 11, 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolabu, T.W. , Park, J.J. , Chen, M. , Cong, L. , Ge, Y. , Jiang, Q. , Debnath, S. et al. (2020b) Improving the genome editing efficiency of CRISPR/Cas9 in Arabidopsis and Medicago truncatula. Planta, 252, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M. , Liu, H. , Lin, Y. , Chen, J. , Fu, Y. , Luo, J. , Zhang, Z. et al. (2020) In‐frame and frame‐shift editing of the Ehd1 gene to develop Japonica rice with prolonged basic vegetative growth periods. Front. Plant Sci. 11, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Minkenberg, B. and Yang, Y. (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proc. Natl. Acad. Sci. USA, 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, K. , Zhang, X.M. , Chen, H. , Zhang, C. , Zhu, J. , Cheng, Z. , Huang, P. et al. (2021) Fine‐tuning florigen increases field yield through improving photosynthesis in soybean. Front. Plant Sci. 12, 710754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhu, S. , Liu, T. , Wang, C. , Cheng, Z. , Zhang, X. , Chen, L. et al. (2019) DELAYED HEADING DATE1 interacts with OsHAP5C/D, delays flowering time and enhances yield in rice. Plant Biotechnol. J. 17, 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C. , Han, L. , Pislariu, C. , Nakashima, J. , Fu, C. , Jiang, Q. , Quan, L. et al. (2011) From model to crop: functional analysis of a STAY‐GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 157, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 List of target sequence and primers used in this study.
Figure S1 Molecular analysis of mutation events in the line Msfta1‐015.
Figure S2 Analysis of forage quality traits at whole plant level in Msfta1 mutant lines.
Figure S3 Analysis of dry biomass and forage digestibility parameters in Msfta1 mutant lines representing stem batches 1 and 2.
Figure S4 Analysis of dry biomass and forage digestibility parameters in Msfta1 mutant lines representing leaf batches 1 and 2.
Figure S5 Correlation analysis of forage quantitative and qualitative traits in Msfta1 lines at whole plant level.
Figure S6 Correlation analysis of forage quality traits in stem tissues of Msfta1 lines of alfalfa representing Batch 1.
Figure S7 Correlation analysis of forage quality traits in stem tissues of Msfta1 lines of alfalfa representing Batch 2.
Figure S8 Correlation analysis of forage quality traits in leaf tissues of Msfta1 lines of alfalfa representing Batch 1.
Figure S9 Correlation analysis of forage quality traits in leaf tissues of Msfta1 lines of alfalfa representing Batch 2.


