Abstract
Osteosarcoma (OS) is the most common primary bone cancer in children and adolescents. In clinical treatments, the insensitivity of OS to conventional radiotherapy regimens significantly contributes to poor patient prognosis and survival. EXO1 is responsible for DNA repair pathways and telomere maintenance. Meanwhile, ATM and ATR are considered switches because they can regulate the expression of EXO1. However, their expression and interaction in OS cells under irradiation (IR) remain unclear. This study aims to investigate the roles of FBXO32, ATM, ATR and EXO1 in OS radiotherapy insensitivity and poor patient prognosis and explore potential pathogenic mechanisms. Bioinformatics is employed to analyse differential gene expression and correlations with prognosis in OS. Cell counting kit 8 assay, clone formation assay, and flow cytometry are used to evaluate cell survival and apopotosis under IR. Co-IP assay is used to detect protein‒protein interactions. Bioinformatics analysis reveals that EXO1 is closely related to survival, apoptosis and poor prognosis in OS. Silencing of EXO1 suppresses cell proliferation and increases the sensitivity of OS cells. Molecular biological experiments show that ATM and ATR act as switches to regulate EXO1 expression under IR. Higher expression of EXO1, which is closely correlated with IR insensitivity and poorer prognosis, might be used as a prognostic indicator for OS. Phosphorylated ATM enhances the expression of EXO1, and phosphorylated ATR induces the degradation of EXO1. More importantly, FBXO32 degrades ATR via ubiquitination in a time-dependent manner. Our data may provide a reference for future research in the mechanisms, clinical diagnosis, and treatment of OS.
Keywords: osteosarcoma, EXO1, ATM, ATR, FBXO32, irradiation
Introduction
Osteosarcoma (OS), a highly hereditary and unstable cancer, is the most common primary malignant bone tumor in children and adolescents, accounting for approximately 20% of all bone tumors [1]. The 5-year survival rate of patients with nonmetastatic OS was found to be only 60%–70% after combination treatment with surgery and chemotherapy [2]. Moreover, OS is prone to local invasion and early metastasis, mainly affecting the lung and skeleton. Additionally, local recurrence, metastasis, and chemotherapy resistance are associated with poor prognosis [ 3, 4] . Despite significant improvements in treatment strategies in recent years, treatment outcomes for patients with metastatic or recurrent OS remain pessimistic, with a rather grim prognosis and greatly reduced 5-year overall survival rate of only 20%–30% [ 5, 6] . Current surgical treatment has a serious impact on limb function, and chemotherapy can induce adverse side effects [7]. Furthermore, OS insensitivity to traditional radiotherapy increases the difficulty of its treatment [ 8– 11] . Therefore, new effective treatments and early diagnosis strategies are urgently needed.
Exonuclease 1 (EXO1), a member of the Rad2/XPG nuclease family, plays an important role in the regulation of cell cycle checkpoints, replication fork maintenance, and DNA repair after replication [12]. Previous studies revealed that EXO1 is associated with various cancers, including Lynch syndrome and ovarian, gastric, lung, breast, and pancreatic cancers [13]. Overexpression of the EXO1 gene disturbs the relative stability of the genome. Dai et al. [14] reported that EXO1 knockdown reduced the proliferation of hepatocellular carcinoma cells in vitro, inhibited the survival of cancer clone cells and was associated with poor prognosis in patients with hepatocellular carcinoma. To date, studies on EXO1 and OS are still lacking, and the role of EXO1 in OS is unclear. The protein kinases ataxia-telangiectasia mutated (ATM) and ataxia-telangiectasia-mutated-and-Rad3-related kinase (ATR) act as DNA damage sensors, which activate checkpoint signaling upon double-strand breaks, apoptosis and genotoxic stresses, such as ultraviolet light and irradiation. They are critical nexuses of the DNA damage response, which induces cell cycle arrest and facilitates DNA repair via downstream targets. Meanwhile, a wide variety of cancers are highly dependent on DNA damage response pathways for survival [15]. Studies have reported that there is obvious coexpression of ATM and EXO1 in bladder cancer, and nuclear translocation of phosphorylated-ATM (p-ATM) can enhance the expression of EXO1 [ 14, 16] . On the other hand, it was demonstrated that ATR promoted EXO1 degradation through phosphorylation-dependent ubiquitination [17]. FBXO32 (MAFbx/Atrogin-1) is an E3 ubiquitin ligase that was reported to be able to degrade c-Myc protein in HCT116 cells [18]. Yang et al. [19] reported that FBXO32 is also involved in DNA damage repair in pancreatic cancer. However, its role in OS remains unclear.
In this study, we aimed to investigate the roles of EXO1, ATM, ATR and FBXO32 in OS irradiation insensitivity and poor prognosis to explore the potential pathogenic mechanisms.
Materials and Methods
Microarray data information
The microarray dataset GSE94805 was obtained from the Gene Expression Omnibus (GEO) database ( https://www.ncbi.nlm.nih.gov/geo/). GSE94805 was based on the GPL13252 platform (Agilent-027114 Genotypic Technology designed Custom Human Whole Genome 8×60 k Microarray). The dataset was used to analyse gene expression in OS, as it included the data of normal vs G0-sorted and normal vs doxorubicin (DOX)-treated OS samples, which helped in screening for the growth-dependent genes that influence OS. The GEO2R online tool was used to identify differentially expressed genes (DEGs) in the dataset based on log fold change>2 and an adjusted P value<0.05.
Protein-protein interaction (PPI) network and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
The PPI network of the coexpressed DEGs was constructed using the retrieval tool STRING ( https://string-db.org/). The MCODE plug-in of Cytoscape (version 3.7.1) was used to select the densely connected modules in the PPI network and determine the key genes. Using the KOBAS 3.0 database, KEGG pathway analysis of the differentially expressed genes was carried out to observe their roles in possible signaling pathways.
Expression and survival analysis of EXO1
To validate the expression of EXO1 in OS, Gene Expression Profiling Interactive Analysis ( http://gepia.cancer-pku.cn) was performed using RNA sequencing expression data based on thousands of samples from the Genotype-Tissue Expression and The Cancer Genome Atlas (TCGA) projects. The prognostic indicators, including overall survival, progression-free interval, disease-free interval, and disease-specific survival, were evaluated using Sangerbox software ( http://www.sangerbox.com). Survival analysis was evaluated using the Kaplan–Meier method and log-rank test ( P<0.05). Every median core gene expression was chosen as the cut-off value for the human cancer dichotomy, thus dividing respective patients into high- and low-risk groups.
Cell culture and transfection
The OS U2OS and MG63 cell lines were purchased from the National Collection of Authenticated Cell Cultures (Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Waltham, USA) containing 10% fetal bovine serum (FBS; Gibco) and a penicillin/streptomycin mixture (Invitrogen, Carlsbad, USA) at 37°C in a humidified chamber at 5% CO 2.
Cells at the logarithmic growth phase were seeded in a 6-well plate and grown to 30%–50% confluence. Subsequently, the cells were transfected with overexpression plasmid DNA, short hairpin RNA (shRNA), or empty vectors (pLVX) (the aforementioned plasmids had all been purchased from GenePharma, Shanghai, China) using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions [20]. The gene silencing efficiency of the shRNA was confirmed using quantitative real-time polymerase chain reaction (qRT-PCR). The shRNA sequences are shown in Supplementary Table S1.
qRT-PCR analysis
Total RNA was extracted using RNA extraction kits (Invitrogen) according to the manufacturer’s instructions. Reverse transcription for cDNA synthesis and qRT-PCR was performed using a SYBR PrimeScript RT Kit (Takara, Shiga, Japan) on an ABI 7500 RT-PCR detection system (Thermo Fisher Scientific, Waltham, USA) following the manufacturer’s instructions. The conditions for qPCR amplification were as follows: 95°C for 30 s; 40 cycles of 95°C for 30 s, 60°C for 60 s; 95°C for 15 s; 60°C for 60 s; and 95°C for 15 s. The qPCR amplification average efficiencies were 97%. The corresponding qPCR efficiencies were calculated according to the equation: E (%) = (10 (–1/slope)–1) × 100%. Actin was used as the internal reference, and the comparative cycle threshold method (2 –ΔΔCt) was used to analyse differences in the levels of each transcript [21]. The primer sequences are shown in Supplementary Table S2.
Western blot analysis
Protein samples were collected from OS cells using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) according to the manufacturer’s instructions. A total of 30 μg protein was loaded in each well, and 10% SDS-PAGE was performed. Then, proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Invitrogen), which were subsequently blocked with 5% skim milk for 1 h, followed by incubation with primary antibodies including EXO1 (ab155553; Abcam, Cambridge, USA), ATM (ab199726; Abcam), p-ATM (ab81292; Abcam), ATR (#13934S; CST, Danvers, USA), p-ATR (#30632S; CST), p-CHK1 (#12302; CST), FBXO32 (ab168372; Abcam), γ-H2A.X (ab26350; Abcam), HA (AF5057; Beyotime), Flag (AF5051; Beyotime), β-Actin (#2D4H5; Proteintech, Chicago, USA), P53 (#60283-2-Ig; Proteintech), p-P53 (#67826-1-Ig; Proteintech), and CHK1 (#60277-1-Ig; Proteintech) overnight at 4°C. After being washed with Tris-buffered saline with Tween-20 (TBS-T) three times, PVDF membranes were incubated at 37°C for 1 h with the goat anti-mouse IgG(H+L) HRP (70-GAM0072; MULTISCIENCES, Hangzhou, China) or goat anti-rabbit IgG(H+L) HRP (70-GAR0072; MULTISCIENCES) as corresponding secondary antibody. Finally, chemiluminescence (ECL) luminous fluid (Beyotime) was added onto the PVDF membrane, and protein bands were detected using a chemiluminescence imaging instrument (Bio-Rad, Hercules, USA).
Coimmunoprecipitation
MG63 cells were transfected with plasmids (EXO1-HA, ATR-Flag, shFBXO32, ATR-HA, FBXO32-Flag, and Ub-His) for 48 h, followed by treatment with 20 μM MG132 for 8 h. Next, MG63 cells were collected and lysed by lysate from plates, and 50 μL of cell lysate was saved for use as input. The remaining lysates were incubated with 20 μL HA magnetic beads (#P2181M and #P2185S; Beyotime) overnight at 4°C on a rotating shaker. The mixture was placed on the magnetic rack for 1 min, and then the supernatant was discarded, followed by two washes with 500 μL lysis buffer. Subsequently, 100 μL SDS-PAGE Sample Loading Buffer was added to HA magnetic beads and incubated at 95°C for 5 min, and then the mixture was placed on a magnetic rack for 1 min. Finally, the supernatant was collected for western blot analysis. The above experimental operation was based on previously described procedures [21].
Ionizing radiation
A single dose of irradiation was delivered using a Siemens 6 MV X-ray linear accelerator (small-animal radiation research platform; XStrahl Gulmay, Suwanee, USA) at 24°C at a dose rate of 1.177 Gy/min. Cells were treated with different dosages of ionizing radiation (0, 1, 2, 4, and 8 Gy). The cell survival rate and clonal growth were evaluated. Cells were irradiated with 4 Gy irradiation and cultured for 12 h, 24 h, and 48 h. Then, total protein was extracted, and western blot analysis was performed as previously described.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was measured using a CCK-8 kit (Dojindo, Shanghai, China) according to the manufacturer’s instructions. Briefly, cells were seeded in 96-well plates at a concentration of 1 × 10 4 cells per well. After 4 Gy irradiation, cells were cultured under normal conditions for 24 h. Then, CCK-8 solution (10 μL) was added to each well, and the cells were incubated at 37 °C for 4 h. Optical density (OD) values were measured at 450 nm using a microplate reader (3925; Costar Company, Bethesda, USA).
Clonogenic assay
After 4 Gy irradiation, cell growth was assessed by observing clone formation in soft agar during 14 days of incubation as previously described [22]. Briefly, 1×10 3 cells were suspended in 1 mL of DMEM supplemented with 10% FBS containing 0.2% agarose and plated in 35-mm tissue culture dishes with 1 mL of a 0.4% agarose underlay containing DMEM. After seven days, 0.2 mL of DMEM with 10% FBS was added to the culture. Clones with a diameter > 125 μm were scored as positive.
Flow cytometry assay
After 4 Gy irradiation, cells were cultured under normal conditions for 24 h. Then, cell apoptosis was measured using an annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) kit (Beyotime) according to the manufacturer’s instructions. Cells were cultured in 6-well plates and incubated overnight. After the indicated treatment, cells were collected and stained with annexin V-FITC for 5 min and 5 μL PI for 5 min at 4 °C. Samples were analysed by flow cytometry on a BD FACS Aria flow cytometry system (BD Biosciences, Franklin Lakes, USA).
Inhibitor experiment
The ATM inhibitor KU55933 was purchased from Calbiochem (Darmstadt, Germany). For ATM inhibitor experiments, OS cells were pretreated with 10 μM KU55933. The ATR inhibitor VE822 was purchased from Selleckchem (Selleckchem, Houston, USA) and used at a final concentration of 10 μM. Both inhibitors were added 2 h before irradiation.
Statistical analysis
Statistical analysis was performed using Graph Pad Prism 7 (GraphPad Software, La Jolla, USA) and IBM SPSS Statistics for Windows version 23.0 (IBM Corp., Armonk, USA). Data are expressed as the mean ± standard deviation. The F test was used to analyse the homogeneity of variance among quantitative data groups, and Student’s t test and nonparametric test were used to compare the data between groups. A P value < 0.05 was selected as the threshold of statistical significance.
Results
EXO1 influenced OS proliferation and prognosis
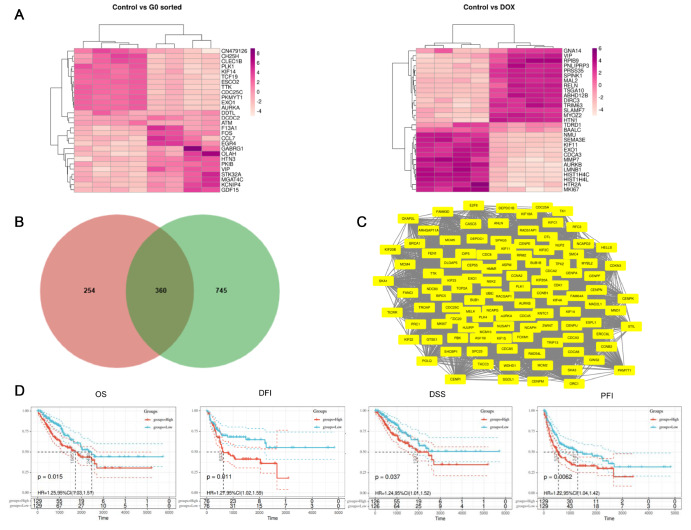
According to the GSE94805 dataset, 614 and 1,105 upregulated DEGs were identified in control (cells undergoing normal mitosis) vs G0-sorted OS (cells in the G0 phase) and control vs doxorubicin (DOX)-treated OS (cells entering senescence after DOX treatment). We considered genes highly expressed in the normal group to be expected to have positive associations with tumor proliferation and survival. Therefore, a total of 360 upregulated DEGs shared by the two DEG sets were selected for further analysis ( Figure 1A,B). The PPI network corresponding to the 360 intersecting DEGs was analysed using STRING and Cytoscape and contained 112 gene nodes ( Figure 1C). By KEGG analysis of gene nodes, we chose the genes RFC5, MCM4 and EXO1 that were enriched in mismatch repair and DNA replication ( Table 1, Pathway ID: hsa03030). We know that DNA repair is an important way for tumor cells to evade medical therapy and radiotherapy. To further evaluate the prognostic value of the genes ( RFC5, MCM4 and EXO1) in OS, survival outcomes were assessed. As shown in Figure 1D, high EXO1 expression was associated with poor prognosis.
Figure 1 .
EXO1 is highly expressed in patients with osteosarcoma (OS) and is associated with poor prognosis
(A) Heat map analysis of differentially expressed genes (DEGs) in the control vs G0-sorted OS cells (left) and in the control vs doxorubicin (DOX)-treated OS cells (right). (B) Venn diagram of DEGs. Gene list A indicates upregulated DEGs in normal vs quiescent OS; Gene list B indicates upregulated DEGs in normal vs senescent OS. (C) Node degree distribution of the 360 intersecting DEGs’ PPI network, including 112 nodes. (D) TCGA database analysis shows that high EXO1 expression is significantly associated with poor prognosis in OS patients. Overall survival (OS), P=0.015, groups for EXO1 high expression n=211; groups for EXO1 low expression n=239. Disease-free interval (DFI), P=0.011, groups for EXO1 high expression n=108; groups for EXO1 low expression n=132. Disease-specific survival (DSS), P=0.0062, groups for EXO1 high expression n=208; groups for EXO1 low expression n=229. Progression-free interval (PFI), P=0.037, groups for EXO1 high expression n=172; groups for EXO1 low expression n=200.
Table 1 KEGG analysis of gene nodes
|
Pathway ID |
Name |
Count |
% |
FDR |
Genes |
|
hsa04110 |
Cell cycle |
5 |
4.1 |
8.09e-06 |
RBL1, SKP2, CCNA2, MCM4, PLK1 |
|
hsa04914 |
Progesterone-mediated oocyte maturation |
3 |
3.2 |
0.0023 |
CCNA2, PLK1, AURKA |
|
hsa03430 |
Mismatch repair |
2 |
8.7 |
0.0031 |
RFC5, EXO1 |
|
hsa04218 |
Cellular senescence |
2 |
1.9 |
0.0049 |
RBL1, CCNA2, |
|
hsa03030 |
DNA replication |
3 |
5.6 |
0.0049 |
RFC5, MCM4, EXO1 |
|
hsa05203 |
Viral carcinogenesis |
3 |
1.6 |
0.0052 |
SKP2, CCNA2, RBL1 |
|
hsa04114 |
Oocyte meiosis |
3 |
1.7 |
0.0287 |
AURKA, MYBL2A, PLK1 |
|
hsa04068 |
FoxO signaling pathway |
2 |
1.5 |
0.0312 |
SKP2, PLK1 |
EXO1 contributed to cellular resistance to irradiation
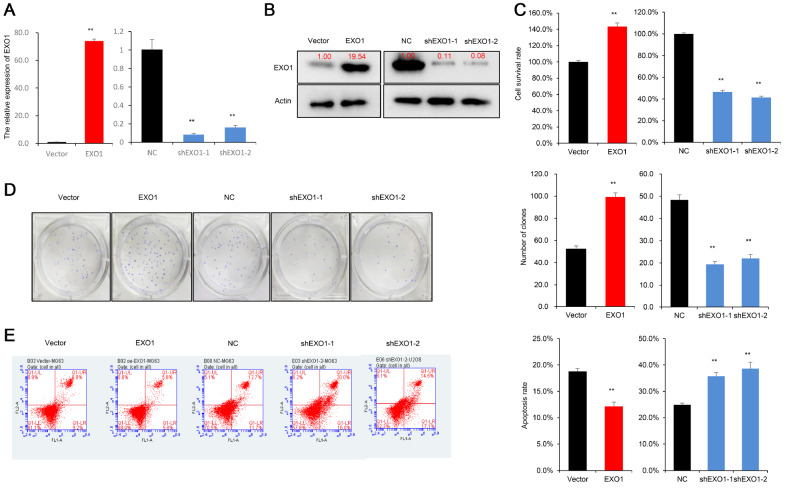
Radiation therapy is widely used to kill tumor cells, and the radiation tolerance capacity of OS cells directly influences cell survival. Therefore, validating the role of EXO1 in the radiation tolerance of OS cells requires further investigation. We tested the mRNA and protein levels of four OS cell lines (MG63, 143B, U2OS, HOS) and chose two higher expressing EXO1 lines (MG63 and U2OS) ( Supplementary Figure S1A,B). The overexpression and knockdown of EXO1 using shRNA were confirmed by qRT-PCR and western blot analysis ( Figure 2A,B and Supplementary Figure S1C,D). First, the CCK-8 assay and clonogenic assay were used to detect the cytotoxic effect of irradiation treatment on OS cells. The results showed that OS cell survival decreased with increasing irradiation doses, as cell survival was significantly lower following irradiation with 4 Gy and 8 Gy than following irradiation with 1 Gy and 2 Gy ( P<0.01; Supplementary Figure S1E,F). Next, we assessed the effect of EXO1 on the OS cellular response to irradiation, and a dose of 4 Gy was selected as the experimental condition. The CCK-8 assay showed that EXO1 overexpression significantly increased OS cell survival ( P<0.01), whereas EXO1 gene silencing reduced OS cell survival after 4 Gy radiotherapy ( Figure 2C and Supplementary Figure S2A). The clonogenic assay demonstrated that after 4 Gy radiotherapy, EXO1 overexpression significantly increased clone numbers, while EXO1 gene silencing reduced clone numbers formed by U2OS and MG63 cells after 14 days of incubation, indicating the inhibition of OS cell growth ( Figure 2D and Supplementary Figure S2B). Moreover, the results of cell apoptosis assays revealed that EXO1 overexpression significantly inhibited the cell apoptosis rate in response to 4 Gy irradiation. In contrast, EXO1 gene silencing accelerated the cell apoptosis rate in U2OS and MG63 cells in response to 4 Gy irradiation ( Figure 2E and Supplementary Figure S2C).
Figure 2 .
EXO1 promotes tolerance of OS cells to radiotherapy
(A) Overexpression and knockdown of EXO1 in MG63 cells were detected by qRT-PCR. (B) Overexpression and knockdown of EXO1 in MG63 cells were confirmed by western blot analysis. (C) CCK-8 assay demonstrates that EXO1 promotes OS cell survival after 4 Gy radiotherapy. (D) Clonogenic assay reveals that EXO1 promotes clone formation ability after 4 Gy radiotherapy. (E) Apoptosis assays show that EXO1 inhibits cells apoptosis after 4 Gy radiotherapy. NC (negative control) vs shEXO1 ( EXO1 knockdown); empty vector vs EXO1 (overexpressed EXO1); ** P<0.01. CCK-8, clonogenic and apoptosis assays were repeated three times.
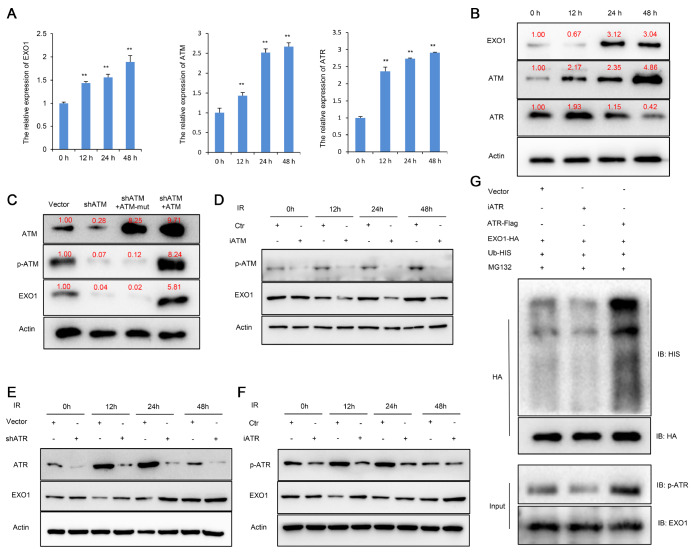
Phosphorylation-ATM/ATR functions as a regulatory switch of the EXO1 protein
A previous study revealed that ATM and ATR are involved in the DNA damage response and are also mutually regulated by EXO1 [17]. However, their mechanism in OS needs to be further demonstrated. First, we validated the effect of irradiation on EXO1, ATM and ATR expressions in MG63 cells. We performed the experiments under a 4 Gy irradiation dose at different time points. The qPCR results showed that EXO1, ATM and ATR mRNA levels increased over time ( Figure 3A). Western blot analysis results indicated that the level of ATM protein in OS increased with time. However, after irradiation in MG63 cells, the expression of EXO1 protein decreased at 12 h and increased at 24 h. More interestingly, the ATR protein changes were the opposite of those in EXO1 ( Figure 3B). These results demonstrate that irradiation can upregulate the mRNA levels of ATM, ATR and EXO1, but the regulation of these proteins is apparently more complex. To further investigate the regulatory mechanism of this process, we used shRNA to knockdown ATM, and a phosphorylation-deficient ATM (ATM-mut) cell line was generated by performing site-directed mutagenesis of Ser1981 in MG63 cells. As shown in Figure 3C, ATM silencing inhibited EXO1 protein expression. The expression levels of p-ATM and EXO1 were significantly diminished in the ATM mutation group ( Figure 3C and Supplementary Figure S2D). Furthermore, Ku-55933 (an ATM-specific inhibitor) was used to inhibit the activity of ATM and prevent phosphorylation of downstream targets. The results showed that decreased phosphorylation of ATM significantly inhibited EXO1 expression ( Figure 3D). Simultaneously, we infected MG63 cells with ATR-specific shRNA or control shRNA. The results showed that ATR knockdown markedly increased the level of the EXO1 protein ( Figure 3E). These results suggested that EXO1 was regulated in the opposite direction by ATM and ATR. Next, VE822 ( ATR-specific inhibitor) was used to inhibit the activity of ATR and prevent phosphorylation of downstream targets. As shown in Figure 3F, the decreased phosphorylation of ATR elevated EXO1 expression at 12 h. Furthermore, we cotransfected MG63 cells with plasmids expressing HIS‐tagged ubiquitin (Ub‐HIS), ATR-FLAG, iATR and ATR-HA. The Co-IP assay demonstrated that iATR reduced the ubiquitination of EXO1, while high p-ATR expression increased the ubiquitination of EXO1 ( Figure 3G). These results indicated that ATR phosphorylation could promote EXO1 degradation.
Figure 3 .
Phosphorylation-ATM/ATR functions as a regulatory switch of EXO1 protein
(A) qPCR assays were used to detect the mRNA levels of EXO1, ATM and ATR at different time points. The results showed that EXO1, ATM and ATR mRNA increased over time. (B) Western blot analysis result indicated that the level of ATM protein in OS increased with time, and the expression of EXO1 protein decreased at 12 h, and increased at 24 h. Simultaneously, the ATR protein increased at 12 h, and decreased at 24 h. (C) Western blot analysis showed that phosphorylated (p)-ATM and EXO1 were significantly reduced in the ATM mutation group. ATM overexpression markedly increased EXO1 expression in MG63 cells. NC (normal control) vs shATM ( ATM knockdown); empty vector vs ATM (overexpressed ATM). (D) Ku-55933 (iATM) was used to inhibit the activity of ATM. Western blot analysis showed that iATM reduced p-ATM and EXO1 protein expressions. (E) Western blot analysis showed that ATR knockdown significantly increased EXO1 expression. (F) VE822 (iATR) was used to inhibit the activity of ATR. Western blot analysis showed that iATR reduced p-ATR protein expression and rescued EXO1 protein expression. (G) MG63 cells were transfected with plasmids (EXO1-HA, ATR-Flag and Ub-His) for 48 h followed by treatment with 20 μM MG132 for 8 h. Western blot analysis showed that p-ATR promoted EXO1 ubiquitination and degradation. ** P<0.01.
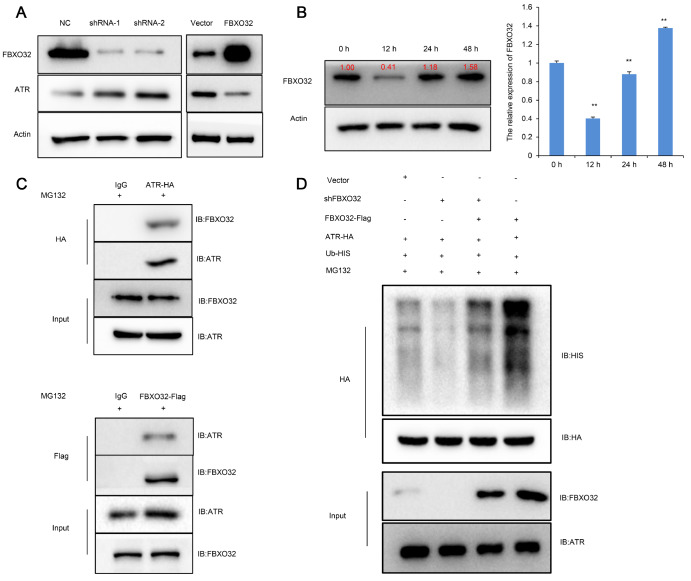
E3 ubiquitin-protein ligase FBXO32 promotes ATR ubiquitination and degradation
Tomimatsu et al. [17] reported that ATR induced phosphorylation-dependent EXO1 degradation in HeLa cells. Our experimental results also showed that ATR promoted EXO1 degradation in OS cells. Meanwhile, we found that the change trend of ATR and EXO1 protein with irradiation time was obvious and regular. Therefore, it is important to determine the regulator of ATR to explain the variation in ATR and EXO1 protein. We were interested in the E3 ubiquitin-protein ligase FBXO32, which downregulated ATR expression in pancreatic cancer [19]. Using a lentiviral vector, we overexpressed FBXO32 in OS cells. On the other hand, shRNA was used to downregulate the expression of FBXO32 in OS cells. Then, we examined the mRNA and protein expression of ATR in OS cells by using qPCR and western blot analysis. The results showed that ATR protein was increased in OS cells expressing reduced levels of FBXO32. A similar result showed that elevated expression of FBXO32 decreased the protein expression of ATR in OS cells ( Figure 4A). However, the mRNA detection results showed that the change in FBXO32 did not exert its effects on the expression of ATR ( Supplementary Figure S3A,B).
Figure 4 .
FBXO32 promoted ATR ubiquitination and degradation
(A) Overexpression and knockdown of the FBXO32 gene in MG63 cells were confirmed by western blot analysis. Results showed that FBXO32 knockdown significantly increased ATR expression and high expression of FBXO32 suppressed the expression of ATR. (B) Western blot analysis and qPCR results indicated that the level of FBXO32 in OS cells decreased at 12 h and increased at 24 h after irradiation. (C) Co-IP assay demonstrated the binding of FBXO32 with ATR (MG63 cells were transfected with ATR-HA and FBXO32-Flag plasmids; IgG served as a negative control). (D) MG63 cells were transfected with plasmids (shFBXO32, ATR-HA, FBXO32-Flag and Ub-His) for 48 h followed by treatment with 20 μM MG132 for 8 h. Western blot analysis showed that FBXO32 promoted ATR ubiquitination and degradation. ** P<0.01.
These results demonstrated that the ATR protein level was regulated posttranslationally by FBXO32 in OS cells. Next, we detected FBXO32 mRNA and protein expression changes over time. We found that FBXO32 mRNA and protein expression were decreased at 12 h after irradiation (4 Gy), whereas FBXO32 expression substantially increased between 24 and 48 h after irradiation ( Figure 4B). This result showed that the expression of FBXO32 correlated well with the change in irradiation time. Interestingly, FBXO32, which encodes the E3 ubiquitin ligase Atrogin-1, can promote the ubiquitination degradation of substrates [23]. Thus, Co-IP assays were performed to detect the association between FBXO32 and ATR, and the results showed that there was an apparent interaction between FBXO32 and ATR ( Figure 4C). To further investigate whether ATR is degraded via ubiquitination, we cotransfected MG63 cells with plasmids expressing HIS‐tagged ubiquitin (Ub‐HIS), FBXO32-FLAG, shFBXO32 and ATR-HA. The Co-IP assay demonstrated that overexpression of FBXO32 increased the ubiquitination of ATR, while FBXO32 knockdown decreased the ubiquitination of ATR ( Figure 4D). Collectively, these results suggested that ATR was a degradation substrate of the E3 ubiquitin ligase FBXO32 in OS cells.
ATM promoted cellular resistance to irradiation
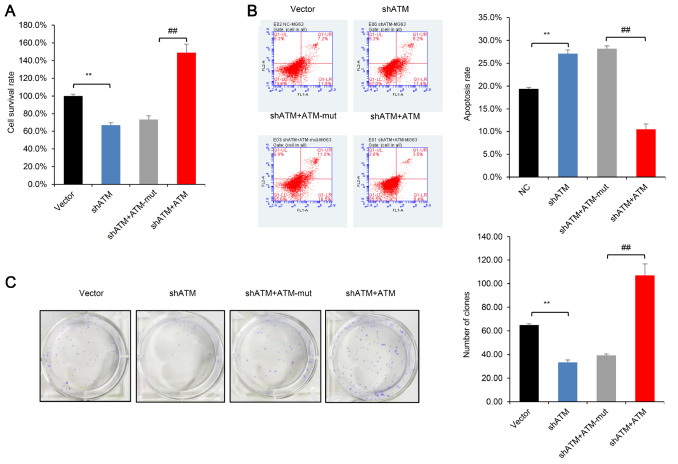
The role of ATM was further validated in U2OS and MG63 cells. The CCK-8 assay demonstrated that in contrast to the ATM-WT (wild type) overexpression group, the cell survival rate after 4 Gy radiotherapy could not be rescued in the ATM-mut group ( Figure 5A and Supplementary Figure S3C). Furthermore, the results of cell apoptosis assays showed that contrary to the ATM-WT overexpression group, the cell apoptosis rate of U2OS and MG63 cells was not rescued in the ATM-mut group ( Figure 5B and Supplementary Figure S3D). Moreover, clone formation assays showed that ATM phosphorylation played an important role in clone formation by U2OS and MG63 cells after 4 Gy irradiation, indicating that ATM phosphorylation reduced OS cell sensitivity to irradiation ( Figure 5C and Supplementary Figure S3E). These results revealed that ATM phosphorylation promoted OS cellular resistance to radiotherapy.
Figure 5 .
ATM phosphorylation promotes tolerance of OS cells to irradiation
(A) CCK-8 assays showed that ATM, but not ATM-mut, promoted cell survival under conditions of 4 Gy irradiation. (B) Apoptosis assays revealed that if ATM couldn’t be phosphorylated and activated, it wouldn’t prevent OS cells apoptosis under irradiation. (C) Clonogenic assay demonstrated that compared with ATM overexpression, ATM-mut couldn’t rescue the clone formation ability of OS cells. Empty vector vs shATM ( ATM knockdown), ** P<0.01; ## P<0.01. CCK-8, clonogenic and apoptosis assays were repeated three times.
Discussion
Osteosarcoma is the most common primary bone tumor in children and adolescents, and patients with advanced osteosarcoma with signs of metastasis have a poor prognosis and apparent radiotherapy tolerance [24]. In the present study, we analysed the GSE94805 dataset through bioinformatics analysis. Normal vs G0-sorted is here to identify the genes that influence the mitosis of OS cells. Cells in the normal vs DOX-treated group were screened to identify the genes that are necessary for cell survival. We compared the two groups of genes to identify key genes that deeply influence the proliferation and survival of OS ( Figure 1B). We analysed the interrelationships among the screened genes by STRING and Cytoscape, and the results of KEGG analysis helped us further understand the functions and pathways of the genes. Such efforts will enable the identification of key genes that could influence the proliferation and survival of OS. Ultimately, we found that EXO1 had a significant impact on radiotherapy resistance in OS cells and that disease-progression prognostic indicators were consistently associated with poor OS prognosis. Therefore, EXO1 may be a potential core gene that serves as a biomarker for radiotherapy resistance and poor prognosis of OS. Radiotherapy has evolved into an important therapeutic scheme in cancer therapy owing to its ability to kill tumor cells, resulting in tumor growth inhibition and prolonged survival of OS patients [25]. However, as tumor cells develop tolerance to radiotherapy, the therapeutic effects of radiotherapy on OS are often less than ideal. Hence, we believe that finding a route of radiotherapy resistance for OS cells is a promising therapeutic target. Relevant treatment strategies may potentially improve treatment efficacy while reducing common side effects.
The exonuclease function allows EXO1 to resect damaged DNA, which is advantageous for DNA repair and recombination [26]. Thus, the hyperactivity of EXO1 may be detrimental to the efficacy of chemoradiotherapy. Multiple EXO1 gene site mutations and single nucleotide polymorphisms are associated with colorectal, small intestinal, breast, pancreatic, stomach, lung, ovarian, and cervical cancer, melanoma, and glioma [27]. He et al. [28] found that EXO1 deficiency increases the sensitivity of ovarian cancer cells to drugs and radiotherapy. A study indicated that EXO1 plays important roles in astrocytoma tumorigenesis, progression and therapy resistance through the DNA double-strand break repair pathway [29]. To further study the function of EXO1, we performed CCK-8 assay and apoptosis assay in nonirradiated OS cells. The results showed that neither EXO1 overexpression nor EXO1 knockdown had an effect on OS cell apoptosis in the absence of radiation. Interestingly, changes in EXO1 expression caused differences in cell proliferation ( Supplementary Figure 4SA,B). This finding suggested that EXO1 was involved in the regulation of OS cell growth cycles, although the differences were not very significant. To explore the effects of EXO1 expression on cellular DNA repair, we referred to the study of Bolderson et al. [30]. Our results showed that compared with the control group, the protein expression of γH2A. X was continually upregulated in the EXO1-knockdown group ( Supplementary Figure 4SC). This illustrates that the efficiency of DNA repair in the EXO1-knockdown group was lower than that in the control group, which is consistent with the literature.
According to the latest study, ATM is activated by DSBs resulted from IR, and the activation of ATR occurs through the RPA-coated single-stranded DNA generated after end resection at DSBs [ 31, 32] . In our study, we demonstrated that ATM and ATR proteins acted as molecular switches that regulated the expression of EXO1. Its potential mechanism may be that irradiation triggers DSBs to activate ATM and thus initiate DNA repair programs that upregulate EXO1 expression. The mechanism may be that activated ATM blocks the degradation of EXO1 and enhances EXO1 recruitment. However, when EXO1 generates single-stranded DNA via a process called DNA end resection, ATR expression is activated to promote EXO1 degradation and prevent excessive EXO1 activity ( Figure 6). In addition, our study revealed that the expression of EXO1 in OS cells increased slightly but not significantly with increasing radiation dose. Tomimatsu et al. [17] found that the level of EXO1 was decreased in HEK-293 cells after ionizing radiation (10 Gy, 8 h). Bologna et al. [33] also suggested that the level of EXO1 was decreased after CPT treatment. Tomimatsu et al. [17] reported that at the start of DNA damage, cells downregulate EXO1 expression by ubiquitination degradation to prevent hyperresection and maintain DNA stability. We think this may be the cellular short-duration stimulus response. However, after that, cells undergoing the DNA repair process require an increase in the expression of EXO1. Thus, EXO1 expression was rapidly elevated. Interestingly, our results showed that the protein change trend of ATR was opposite to that of EXO1. Tomimatsu et al. [17] found that phosphorylation of EXO1 by ATR triggers its degradation in HEK293 cells. To further verify this conclusion, shRNA and VE822 (ATR-specific inhibitor) were used to inhibit the expression and activity of ATR, respectively. We demonstrated that inhibition of ATR expression or its activity led to elevated levels of EXO1 at early time points after irradiation. Moreover, the Co-IP assay also demonstrated that ATR promotes the ubiquitination and degradation of EXO1, which agrees with reports in the literature [17]. In contrast, both switching off ATM expression and inhibiting its activity decreased EXO1 expression in MG63 cells. In this study, we found that ATM can regulate the expression of EXO1 as a downstream target gene. Importantly, ATM phosphorylation deficiency significantly restrained the expression of EXO1. From the above results, we observed that elevated expression of ATR is a key reason for the downregulation of EXO1 expression at early time points after irradiation. However, ATR mRNA was increased immediately after irradiation, which indicated the degradation of ATR protein after 24 h of irradiation. These results further illustrate that ATM, as a monitor of the cell cycle, makes a response to DSBs to elevate its own expression and activate the downstream checkpoint protein, which is beneficial for DNA repair and cell survival. However, excessive DNA end resection and generation of an extended ssDNA moiety are detrimental to DNA repair. Thus, the expression of ATR increases rapidly to avoid the overreaction of exonuclease in the early stage of DSBs, and ATR degrades in steps with the processes of DNA damage repair changes in OS cells. Yang et al. [19] reported that FBXO32 acts as a ubiquitin E3 ligase, which targets ATR protein and leads to protein degradation in pancreatic cancer. In the data (GSE50532) of Kansara et al. [34], we found a decrease in FBXO32 mRNA expression within 4 to 16 hours after irradiation, but the expression rose rapidly after 24 h of irradiation. This is consistent with our experimental results. In this study, we confirmed that FBXO32 can bind with ATR to induce its ubiquitination degradation in OS cells. Moreover, the examination of ATR downstream checkpoint factors (P53, p-P53, CHK1, p-CHK, NBS1, BRCA1) indicated that FBXO32 inhibited the phosphorylation and activation of P53 and CHK1 ( Supplementary Figure S5A,B), and IR induced the initial increases in P53, p-P53, CHK1, p-CHK, NBS1 but decreased gradually with prolonged time ( Supplementary Figure S5C,D). These results were generally as expected and consistent with the literature [ 35, 36] . Furthermore, FBXO32 has also been reported to be involved in tumorigenesis and tumor progression [ 19, 37, 38] . Taken together, we mainly illustrated a mechanism by which the FBXO32/ ATR/ ATM/ EXO1 axis mediates radiotherapy resistance ( Figure 6).
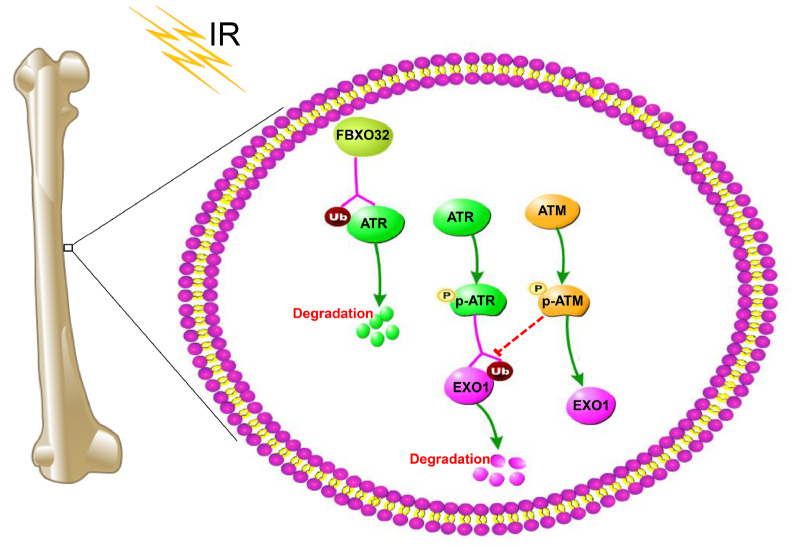
Figure 6 .
FBXO32/ATR/ATM/EXO1 axis induces OS cells to anti-radiotherapy
Activated ATM blocks the degradation of EXO1 and enhances EXO1 recruitment. On the other hand, when EXO1 generates single-stranded DNA via a process called DNA end resection, ATR expression is activated to promote EXO1 degradation and prevent excessive EXO1 activity.
Nevertheless, there are several limitations in this study. First, EXO1 is a common downstream regulator of ATM and ATR, which are modulated in opposite directions in the two different phosphorylation states. Different combinations of the phosphorylation site mutations in EXO1 should be performed to validate the respective mechanisms of action of ATR and ATM. Second, animal experiments are needed to determine whether and to what extent ATM/ EXO1 silencing enhances the efficacy of radiotherapy in OS. High doses of radiotherapy can adversely affect other organs, and thus, if ATM/ EXO1 silencing can reduce the dose required for radiotherapy, it will extend its clinical application. To validate these issues, OS cells should be transplanted orthotopically into the bones of mice as described [39]. Then, the adenovirus suspension is injected by pipetting into the tumor to knock down ATM/ EXO1. Right after, radiotherapy for orthotopic mouse tumors is performed at different doses. Third, OS is treated mainly by surgical resection combined with radiotherapy and chemotherapy. Therefore, in future trials, we plan to study it in combination with traditional chemotherapeutic agents and evaluate the effect of ATM/ EXO1 silencing on cells subjected to the combined treatment.
Supporting information
Supplementary Data
Supplementary data is available at Acta Biochimica et Biphysica Sinica online.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the Jiangxi Provincial Department of Education Fund (No. GJJ211526) and the National College Students Innovation Entrepreneurship Training Plan Program of China (No. 202210413009).
References
- 1.Namlos HM, Kresse SH, Muller CR, Henriksen J, Holdhus R, Saeter G, et al. Global gene expression profiling of human osteosarcomas reveals metastasis-associated chemokine pattern. Sarcoma. 2012, 2012: 639038. . [DOI] [PMC free article] [PubMed]
- 2.Bacci G, Ferrari S, Bertoni F, Ruggieri P, Picci P, Longhi A, Casadei R, et al. Long-term outcome for patients with nonmetastatic osteosarcoma of the extremity treated at the istituto ortopedico rizzoli according to the istituto ortopedico rizzoli/osteosarcoma-2 protocol: an updated report. J Clin Oncol. . 2000;18:4016–4027. doi: 10.1200/JCO.2000.18.24.4016. [DOI] [PubMed] [Google Scholar]
- 3.Simpson S, Dunning MD, de Brot S, Grau-Roma L, Mongan NP, Rutland CS. Comparative review of human and canine osteosarcoma: morphology, epidemiology, prognosis, treatment and genetics. Acta Vet Scand. . 2017;59:71. doi: 10.1186/s13028-017-0341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu Y, Song T, Xue X, Cao G, Huang P. Kinesin superfamily proteins: roles in osteosarcoma. Front Biosci. 2021, 26: 370–378 . [DOI] [PubMed]
- 5.Mialou V, Philip T, Kalifa C, Perol D, Gentet JC, Marec-Berard P, Pacquement H, et al. Metastatic osteosarcoma at diagnosis. Cancer. . 2005;104:1100–1109. doi: 10.1002/cncr.21263. [DOI] [PubMed] [Google Scholar]
- 6.Kelleher FC, O′Sullivan H. Monocytes, macrophages, and osteoclasts in osteosarcoma. J Adolesc Young Adult Oncol. . 2017;6:396–405. doi: 10.1089/jayao.2016.0078. [DOI] [PubMed] [Google Scholar]
- 7.Ruggieri P, Mavrogenis AF, Mercuri M. Quality of life following limb-salvage surgery for bone sarcomas. Expert Rev Pharmacoeconomics Outcomes Res. . 2011;11:59–73. doi: 10.1586/erp.10.91. [DOI] [PubMed] [Google Scholar]
- 8.Nytko KJ, Thumser-Henner P, Russo G, Weyland MS, Rohrer Bley C. Role of HSP70 in response to (thermo)radiotherapy: analysis of gene expression in canine osteosarcoma cells by RNA-seq. Sci Rep. . 2020;10:12779. doi: 10.1038/s41598-020-69619-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed AA, Zia H, Wagner L. Therapy resistance mechanisms in Ewing’s sarcoma family tumors. Cancer Chemother Pharmacol. . 2014;73:657–663. doi: 10.1007/s00280-014-2392-1. [DOI] [PubMed] [Google Scholar]
- 10.Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. . 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 11.de Jong Y, Ingola M, Briaire-de Bruijn IH, Kruisselbrink AB, Venneker S, Palubeckaite I, et al. Radiotherapy resistance in chondrosarcoma cells; a possible correlation with alterations in cell cycle related genes. Clin Sarcoma Res. . 2019;9:9. doi: 10.1186/s13569-019-0119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. . 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 13.Keijzers G, Bakula D, Petr M, Madsen N, Teklu A, Mkrtchyan G, Osborne B, et al. Human exonuclease 1 (EXO1) regulatory functions in DNA replication with putative roles in cancer. Int J Mol Sci. . 2019;20:74. doi: 10.3390/ijms20010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai Y, Tang Z, Yang Z, Zhang L, Deng Q, Zhang X, Yu Y, et al. EXO1 overexpression is associated with poor prognosis of hepatocellular carcinoma patients. Cell Cycle. . 2018;17:2386–2397. doi: 10.1080/15384101.2018.1534511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther. . 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Fan J, Zhao Y, Yuan H, Yang J, Li T, He Z, et al. Phospholipase C-epsilon regulates bladder cancer cells via ATM/EXO1. Am J Cancer Res. 2020, 10: 2319–2336 . [PMC free article] [PubMed]
- 17.Tomimatsu N, Mukherjee B, Harris JL, Boffo FL, Hardebeck MC, Potts PR, Khanna KK, et al. DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J Biol Chem. . 2017;292:10779–10790. doi: 10.1074/jbc.M116.772475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei Z, Zhang D, Hu B, Wang J, Shen X, Xiao W. FBXO32 targets c-myc for proteasomal degradation and inhibits c-myc activity. J Biol Chem. . 2015;290:16202–16214. doi: 10.1074/jbc.M115.645978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C, Fan P, Zhu S, Yang H, Jin X, Wu H. 3F‑Box protein 32 degrades ataxia telangiectasia and Rad3‑related and regulates DNA damage response induced by gemcitabine in pancreatic cancer. Oncol Lett. . 2018;15:8878. doi: 10.3892/ol.2018.8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y, Cao G, Lan H, Liao H, Hu Y, Feng H, Liu X, et al. Chondrocyte-derived exosomal miR-195 inhibits osteosarcoma cell proliferation and anti-apoptotic by targeting KIF4A in vitro and in vivo. Transl Oncol. . 2022;16:101289. doi: 10.1016/j.tranon.2021.101289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang P, Liao R, Chen X, Wu X, Li X, Wang Y, Cao Q, et al. Nuclear translocation of PLSCR1 activates STAT1 signaling in basal-like breast cancer. Theranostics. . 2020;10:4644–4658. doi: 10.7150/thno.43150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatzka M, Prisco M, Baserga R. Stabilization of the Ras oncoprotein by the insulin-like growth factor 1 receptor during anchorage-independent growth. Cancer Res. 2000, 60: 4222–4230 . [PubMed]
- 23.Habel N, El-Hachem N, Soysouvanh F, Hadhiri-Bzioueche H, Giuliano S, Nguyen S, Horák P, et al. FBXO32 links ubiquitination to epigenetic reprograming of melanoma cells. Cell Death Differ. . 2021;28:1837–1848. doi: 10.1038/s41418-020-00710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. . 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacob JA. Researchers turn to canine clinical trials to advance cancer therapies. Jama. . 2016;315:1550. doi: 10.1001/jama.2016.0082. [DOI] [PubMed] [Google Scholar]
- 26.Desai A, Qing Y, Gerson SL. Exonuclease 1 is a critical mediator of survival during DNA double strand break repair in nonquiescent hematopoietic stem and progenitor cells. Stem Cells. . 2014;32:582–593. doi: 10.1002/stem.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dherin C, Gueneau E, Francin M, Nunez M, Miron S, Liberti SE, Rasmussen LJ, et al. Characterization of a highly conserved binding site of mlh1 required for exonuclease I-dependent mismatch repair. Mol Cell Biol. . 2009;29:907–918. doi: 10.1128/MCB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He D, Li T, Sheng M, Yang B. Exonuclease 1 (Exo1) participates in mammalian non-homologous end joining and contributes to drug resistance in ovarian cancer. Med Sci Monit. . 2020;26:e918751. doi: 10.12659/MSM.918751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sousa JF, Serafim RB, Freitas LM, Fontana CR, Valente V. DNA repair genes in astrocytoma tumorigenesis, progression and therapy resistance. Genet Mol Biol. . 2020;43(1 Suppl 1): e20190066 doi: 10.1590/1678-4685-GMB-2019-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK, Burma S, et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. . 2010;38:1821–1831. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soni A, Duan X, Stuschke M, Iliakis G. ATR contributes more than ATM in intra-S-phase checkpoint activation after IR, and DNA-PKcs facilitates recovery: evidence for modular integration of ATM/ATR/DNA-PKcs functions. Int J Mol Sci. . 2022;23:7506. doi: 10.3390/ijms23147506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mladenov E, Fan X, Dueva R, Soni A, Iliakis G. Radiation-dose-dependent functional synergisms between ATM, ATR and DNA-PKcs in checkpoint control and resection in G2-phase. Sci Rep. . 2019;9:8255. doi: 10.1038/s41598-019-44771-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bologna S, Altmannova V, Valtorta E, Koenig C, Liberali P, Gentili C, Anrather D, et al. Sumoylation regulates EXO1 stability and processing of DNA damage. Cell Cycle. . 2015;14:2439–2450. doi: 10.1080/15384101.2015.1060381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kansara M, Leong HS, Lin DM, Popkiss S, Pang P, Garsed DW, Walkley CR, et al. Immune response to RB1-regulated senescence limits radiation-induced osteosarcoma formation. J Clin Invest. . 2013;123:5351–5360. doi: 10.1172/JCI70559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X, Wang Y, Ji K, Liu Y, Kong Y, Nie S, Li N, et al. NRF2 preserves genomic integrity by facilitating ATR activation and G2 cell cycle arrest. Nucleic Acids Res. . 2020;48:9109–9123. doi: 10.1093/nar/gkaa631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karnitz LM, Zou L. Molecular pathways: targeting ATR in cancer therapy. Clin Cancer Res. . 2015;21:4780–4785. doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chou JL, Su HY, Chen LY, Liao YP, Hartman-Frey C, Lai YH, Yang HW, et al. Promoter hypermethylation of FBXO32, a novel TGF-β/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab Invest. . 2010;90:414–425. doi: 10.1038/labinvest.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo W, Zhang M, Shen S, Guo Y, Kuang G, Yang Z, Dong Z. Aberrant methylation and decreased expression of the TGF-β/Smad target gene FBXO32 in esophageal squamous cell carcinoma. Cancer. . 2014;120:2412–2423. doi: 10.1002/cncr.28764. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Qin G, Liang X, Wang W, Wang Z, Liao D, Zhong L, et al. Targeting the CK1α/CBX4 axis for metastasis in osteosarcoma. Nat Commun. . 2020;11:1141. doi: 10.1038/s41467-020-14870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.