Abstract
Fusarium species are important cereal pathogens that cause severe production losses to major cereal crops such as maize, rice, and wheat. However, the causal agents of Fusarium diseases on cereals have not been well documented because of the difficulty in species identification and the debates surrounding generic and species concepts. In this study, we used a citizen science initiative to investigate diseased cereal crops (maize, rice, wheat) from 250 locations, covering the major cereal-growing regions in China. A total of 2 020 Fusarium strains were isolated from 315 diseased samples. Employing multi-locus phylogeny and morphological features, the above strains were identified to 43 species, including eight novel species that are described in this paper. A world checklist of cereal-associated Fusarium species is provided, with 39 and 52 new records updated for the world and China, respectively. Notably, 56 % of samples collected in this study were observed to have co-infections of more than one Fusarium species, and the detailed associations are discussed. Following Koch’s postulates, 18 species were first confirmed as pathogens of maize stalk rot in this study. Furthermore, a high-confidence species tree was constructed in this study based on 1 001 homologous loci of 228 assembled genomes (40 genomes were sequenced and provided in this study), which supported the “narrow” generic concept of Fusarium (= Gibberella). This study represents one of the most comprehensive surveys of cereal Fusarium diseases to date. It significantly improves our understanding of the global diversity and distribution of cereal-associated Fusarium species, as well as largely clarifies the phylogenetic relationships within the genus.
Taxonomic novelties: New species: Fusarium erosum S.L. Han, M.M. Wang & L. Cai, Fusarium fecundum S.L. Han, M.M. Wang & L. Cai, Fusarium jinanense S.L. Han, M.M. Wang & L. Cai, Fusarium mianyangense S.L. Han, M.M. Wang & L. Cai, Fusarium nothincarnatum S.L. Han, M.M. Wang & L. Cai, Fusarium planum S.L. Han, M.M. Wang & L. Cai, Fusarium sanyaense S.L. Han, M.M. Wang & L. Cai, Fusarium weifangense S.L. Han, M.M. Wang & L. Cai.
Citation: Han SL, Wang MM, Ma ZY, Raza M, Zhao P, Liang JM, Gao M, Li YJ, Wang JW, Hu DM, Cai L (2023). Fusarium diversity associated with diseased cereals in China, with an updated phylogenomic assessment of the genus. Studies in Mycology 104: 87–148. doi: 10.3114/sim.2022.104.02
Keywords: Cereal pathogens, citizen science, co-infection, new taxa, pathobiome, phylogeny, species complexes, systematics
INTRODUCTION
Cereal crops, including maize (Zea mays), rice (Oryza sativa) and wheat (Triticum aestivum), are primary staple foods worldwide (http://www.fao.org/faostat/en). The total grain output of China in 2020 was 658 million tons, of which 595 million tons were maize, rice and wheat (Ma 2020). The significant factors diminishing the value and yield of these crops were phytopathogens and pests, especially fungi (e.g., Puccinia striiformis and Fusarium graminearum) (Trail 2009, Matny 2015, Savary et al. 2019). Fusarium pathogens are notorious, and not only cause yield losses, but also produce mycotoxins threatening human and animal health (Desjardins 2006, Leslie & Summerell 2006, Renev et al. 2021). For instance, the annual loss due to wheat scab in China was up to 3.41 million tons from 2000 to 2018 (Su et al. 2021), and from 1993 to 2001, Fusarium diseases on crops caused economic losses up to $7.7 billion in the United States (Nganje et al. 2004). Therefore, the investigation of species diversity, distribution patterns, host range and pathogenicity of Fusarium is of great significance for cereal disease diagnosis and management.
Accurate species identification is the first step in Fusarium disease diagnosis and management (O’Donnell et al. 2015, 2018, 2022). However, the taxonomic framework of Fusarium has undergone numerous significant changes based on different criteria (Crous et al. 2021, Wang et al. 2022a), including eras dominated by morphology (Booth 1971, Nelson et al. 1983), or phylogenetic inference (Gräfenhan et al. 2011, O’Donnell et al. 2018, Lombard et al. 2019a). These different taxonomic frameworks are not necessarily consistent in inferring Fusarium species diversity. Historically, species within the F. graminearum clade (Fg clade) have for long been considered as a single species using the morphological species concept, until the application of the genealogical concordance phylogenetic species recognition in the last two decades (O’Donnell et al. 2000a). Furthermore, for many decades users had favoured systems with fewer species, such as the system proposed by Snyder & Hansen (1940, 1941, 1945, 1954), which reduced the number of Fusarium species to nine with many formae speciales. In addition, there are several dilemmas in the delimitation of Fusarium species: i) the morphological characteristics of Fusarium species are greatly influenced by environmental factors, and often overlap with each other; ii) the fungal universal barcode, the internal transcribed spacer region cistron (ITS) has a low resolution of Fusarium species; iii) most of the current Fusarium sequences in the public database are still under the species complex name or even under wrong species or genus names (Aoki et al. 2014, Lombard et al. 2019b, Wang et al. 2019a, Xia et al. 2019, Crous et al. 2021, Yilmaz et al. 2021). These dilemmas, together with the changes in the taxonomic system have resulted in difficulties in the diagnosis and management of Fusarium diseases of cereal crops.
To solve the species delimitation and identification dilemma, a polyphasic approach has gradually been applied and several online databases (Fusarium-ID, Fusarium MLST and FUSARIOID-ID) have been established (O’Donnell et al. 2012, Crous et al. 2021, Torres-Cruz et al. 2022). Evolutionary relationships of several important Fusarium species complexes, e.g., F. fujikuroi species complex (FFSC), F. incarnatum-equiseti species complex (FIESC), and F. oxysporum species complex (FOSC) were also published (Aoki et al. 2014, Lombard et al. 2019b, Wang et al. 2019a, Xia et al. 2019, Crous et al. 2021, Yilmaz et al. 2021). Despite these significant contributions, debates surrounding the generic delimitation of Fusarium, especially whether the genus Neocosmospora (also known as F. solani species complex, FSSC) belongs to Fusarium was disputed (Crous et al. 2021, Geiser et al. 2021, Wang et al. 2022a).
According to the USDA fungal database (Farr & Rossman 2022), more than 20 % of cereal-associated Fusarium species belong to the F. sambucinum species complex (FSAMSC). Nevertheless, the species relationships within this complex remain to be further clarified, as several sister species cannot be distinguished based on only RNA polymerase second largest subunit (rpb2) and translation elongation factor 1-alpha (tef1) phylogenetic analyses (Laraba et al. 2021). Recently, the phylogenomic approach has been increasingly shown to be able to provide more information towards understanding species evolution and boundaries (Haridas et al. 2020, Liu et al. 2022a). In the case of Fusarium, a phylogenomic approach has also been used to assess generic boundaries (Geiser et al. 2021), but still suffered from imbalanced generic sampling, and lack of data derived from ex-type cultures.
Previously, Fusarium associated with cereals in China were identified mainly based on morphological features, preliminary nucleotide BLAST, or phylogenetic analyses using single locus datasets (either ITS or tef1) (Huang et al. 2011, Zhang et al. 2012, 2015, Xu et al. 2016, Hao et al. 2017). These studies, particularly those applying subspecies, varieties and formae speciales names, proved to be insufficient to identify Fusarium species (Lombard et al. 2019b, Xia et al. 2019, Yilmaz et al. 2021, Wang et al. 2022a). For instance, the pathogen of rice bakanae diseases in China has been identified as three Fusarium species and varieties, i.e., F. moniliforme, F. moniliforme var. subglutinans and F. moniliforme var. intermedium, based on morphological features (Booth 1971, Ye et al. 1990). However, these epithets have been revealed to be synonyms of F. fujikuroi (O’Donnell et al. 1998a, Crous et al. 2021). Thus, species diversity and distribution of this group require amendment in accordance with the currently used taxonomic system and names.
The purpose of this study was therefore to examine the Fusarium species associated with diseased cereals in China. Specifically to: i) understand the diversity, distribution patterns, host preference and pathogenicity of Fusarium species associated with diseased cereals; ii) reassess the boundaries of Fusarium s. str. and allied genera, and assess species boundaries within the FSAMSC; iii) sequence the genomes of new and several known species and provide an updated phylogenomic overview of Fusarium.
MATERIALS AND METHODS
Sample collection
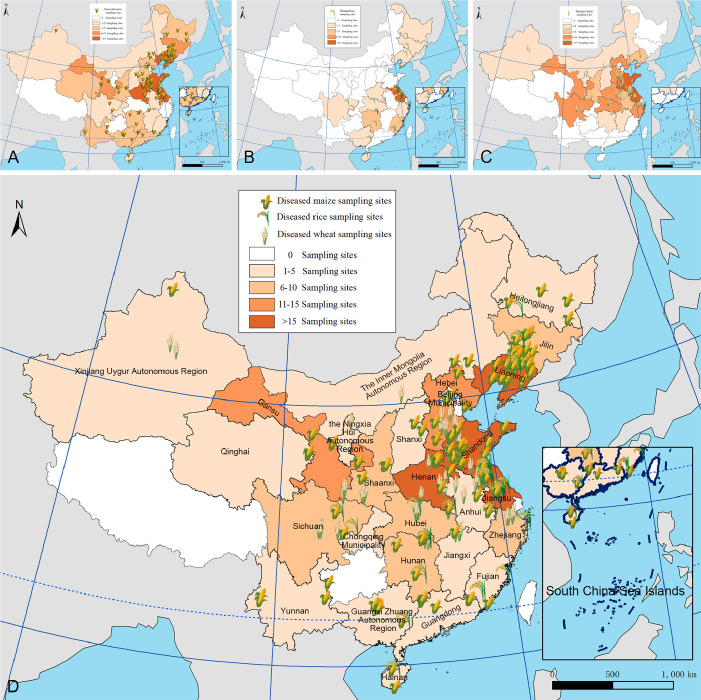
Diseased cereals associated with Fusarium spp. were investigated in China from July 2020 to October 2021, including maize ear rot (Fig. 1), maize stalk rot (Fig. 2), wheat scab (Fig. 3), wheat crown rot (Fig. 4), rice spikelet rot and rice bakanae disease (Fig. 5). Geographically, 315 diseased samples (172 diseased maize samples, Fig. 6A; 105 diseased wheat samples, Fig. 6B; and 38 diseased rice samples, Fig. 6C) were collected from 250 sampling sites (several diseased samples were collected from various hosts at the same location) which covered 21 provinces, four autonomous regions and two municipalities (Fig. 6D). Parts of these samples were collected by local farmers (Supplementary Fig. S1, Supplementary Table S1) who followed a directive protocol in our Citizen Science Initiative of “Collecting Diseased Cereals” posted on social media. Diseased samples in good condition were dried and stored in the Fungarium of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS).
Fig. 1.

Field symptoms of maize ear rot (MER). A. Kernels covered with salmon-coloured powdery molds and accompanied by insect injury. B. Kernels covered with white moulds. C. Kernels covered with orchid-coloured mycelia. D, F. Ears covered with whitish or a mixture of pinkish and blackish powdery moulds coexist with symptoms caused by other fungi. E. Kernels covered with white moulds or violet mycelia. G. Kernels covered with salmon-coloured powdery moulds. Note: Maize ear rot caused by Fusarium was referred to as Fusarium ear rot of maize (Zhang et al. 2014a), Fusarium maize ear rot (Zhang et al. 2014b), Gibberella/red ear rot (Lana et al. 2022) and Fusarium/pink ear rot (Zhang et al. 2016) in various previous studies. To avoid confusion, in this study we refer to this disease as maize ear rot (MER).
Fig. 2.

Field symptoms of maize stalk rot (MSR). A, B. Maize ear drooping without shedding. C. Stalks turn grey green; internodes turn straw-coloured or dark brown. D. Stalks covered with whitish mycelia. E, F. Stalks were covered with salmon-coloured powdery moulds and snapped at the nodes. G. Rotted and brownish stalks. Note: Maize stalk rot caused by Fusarium, known as Fusarium stalk rot (Jiang et al. 2021), Gibberella stalk rot in maize (Ye et al. 2013) in previous literature, is herein universally referred to as maize stalk rot (MSR) in this paper.
Fig. 3.

Field symptoms of Fusarium head blight of wheat (FHB). A–E. Head blight of wheat accompanied by pinkish mould and/or whitish mycelia. Note: The disease name Fusarium head blight (FHB) is adopted in this paper, which was also called wheat head blight and wheat scab in literature (O’Donnell et al. 2000a).
Fig. 4.

Field symptoms of Fusarium crown rot of wheat (FCR). A, B. “White-head” of wheat tillers. C, D. Reddish-brown discoloration on the lower stems. E. Dark brown to black discolouration on the crowns and roots. Note: The disease name Fusarium crown rot of wheat (FCR) is adopted in this paper, which was also referred to as wheat crown rot (Zhang et al. 2015), crown rot of wheat in previous literature (Li et al. 2016).
Fig. 5.

Field symptoms of Fusarium diseases on rice. A. Rice bakanae disease (RBD), caused by F. fujikuroi, showing elongated barren seedlings. B–E. Rice spikelet rot (RSR), caused by multiple Fusarium species, showing reddish or brown discoloration on the glumes, sometimes with salmon-coloured and/or whitish powdery mould. Note: The disease name rice spikelet rot (RSR) is adopted in this paper, which was also known as Fusarium head blight in rice in literature (Liu et al. 2022d).
Fig. 6.
Maps showing sampling sites in China, generated by ArcGIS v. 10.5 software (Esri, Redlands, CA, USA). A. Map showing the distribution of 172 diseased maize samples collected in this study. B. Map showing the distribution of 38 diseased rice samples collected in this study. C. Map showing the distribution of 105 diseased wheat samples collected in this study. D. Map showing the overall distribution of a total of 315 diseased samples collected in this study.
Strain isolation, preservation and selection
A total of 2 600 fungal strains were isolated from symptomatic tissues (i.e., kernels, stems, roots; Fig. 1–5) of diseased cereals using single spore isolation or direct hyphal isolation (Zhang et al. 2013, Raza et al. 2019). Of these, 2 020 strains were assigned to Fusarium based on colony morphology and tef1 sequences. Strains were further selected following three steps: firstly, for strains with 100 % tef1 similarity from the same symptomatic tissue of the same host species in each location, only one strain was retained for subsequent analyses. Secondly, the genus level phylogenetic analysis was conducted using tef1-rpb2 sequences, and the above selected strains were assigned to six Fusarium species complexes. For each complex, a tef1-rpb2 phylogeny was re-analysed, and only one isolate in each subclade (without sequence variation) was selected if multiple isolates from the same symptomatic tissue of each host species in each location were available. Thirdly, different loci were amplified for the above selected strains, and phylogenetic analyses of Fusarium species complexes were conducted using different multi-locus datasets, i.e., calmodulin (CaM), RNA polymerase largest subunit (rpb1), rpb2, tef1 and beta-tubulin (tub2) for FFSC; CaM, rpb2 and tef1 for the FIESC and FOSC; histone (H3), rpb1, rpb2 and tef1 for the FSAMSC; rpb1, rpb2 and tef1 for the FNSC and FTSC. Only one isolate of a particular species from each symptomatic tissue of the same host species in each location was selected. After the above selection steps, 608 representative strains were included (Supplementary Table S1). The type specimens of new species described in this study were deposited in the Fungarium of the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), and the living ex-type cultures were deposited in the China General Microbiological Culture Collection Centre (CGMCC). All isolates examined in this study were deposited in Lei Cai’s personal culture collection (LC), the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China. Taxonomic novelties, descriptions and nomenclature were deposited in MycoBank (Crous et al. 2004).
DNA extraction and amplification
Total genomic DNAs were extracted from fresh fungal mycelia grown on potato dextrose agar (PDA; DifcoTM, Becton, Dickinson and Company, Sparks, MD, USA) using the cetyltrimethylammonium bromide (CTAB) method (Porebski et al. 1997) and stored at -20 °C until polymerase chain reaction (PCR). PCR amplifications were performed in a reaction mixture consisting of 12.5 μL 2 × Taq PCR Master Mix (Vazyme Biotech Co., Ltd, Nanjing, China), 1 μL each of 10 μM primers, 2 μL of the undiluted genomic DNA, adjusted to a final volume of 25 μL with distilled deionised water. Seven loci, including ITS (White et al. 1990), tef1 (O’Donnell et al. 1998b), CaM (O’Donnell et al. 2000b), rpb1 (O’Donnell et al. 2010), rpb2 (Liu et al. 1999; Reeb et al. 2004), tub2 (O’Donnell & Cigelnik 1997), and H3 (Roux et al. 2001) were amplified and sequenced. The PCR primer pairs and amplification conditions are listed in Table 1. The PCR products were visualised using 1 % agarose electrophoresis gels. Sequencing was done by the Tianyi Huiyuan Company (Beijing, China) and the SinoGenoMax Company (Beijing, China).
Table 1.
Primers used in this study, with originating loci, sequences program and references.
| Gene/DNA regions | Primers | PCR amplification procedures | References | |||
|---|---|---|---|---|---|---|
|
| ||||||
| Name | Abbreviation | Name | Direction | Sequence (5’→3’) | ||
| Beta tubulin | tub | T1 | Forward | AACATGCGTGAGATTGTAAGT | 95 °C 3 min; 35 cycles of 94 °C 30 s, 54 °C 45 s, 72 °C 15 s; 72 °C 10 min; 10 °C soak | O’Donnell & Cigelnik (1997) |
| T2 | Reverse | TAGTGACCCTTGGCCCAGTTG | ||||
| Calmodulin | CaM | CL1 | Forward | GARTWCAAGGAGGCCTTCTC | 95 °C 1 min; 35 cycles of 94 °C 30 s, 55 °C 30 s, 72 °C 15 s; 72 °C 10 min; 10 °C soak | O’Donnell et al. (2000b) |
| CL2A | Reverse | TTTTTGCATCATGAGTTGGAC | ||||
| Histone | H3 | H3-la | Forward | ACTAAGCAGACCGCCCGCAGG | 96 °C 2 min; 30 cycles of 92 °C 1 min, 60 °C 1 min, 72 °C 10 s; 72 °C 5 min; 10 °C soak | Roux et al. (2001) |
| H3-lb | Reverse | GCGGGCGAGCTGGATGTCCTT | ||||
| Internal transcribed spacer region of the rDNA | ITS | ITS1 | Forward | CCGTAGGTGAACCTGCGG | 95 °C 5 min; 35 cycles of 95 °C 30 s, 52 °C 30 s, 72 °C 10 s; 72 °C 5 min; 10 °C soak | White et al. (1990) |
| ITS4 | Reverse | TCCTCCGCTTATTGATATGC | ||||
| RNA polymerase largest subunit | rpb1 | F7 | Forward | CRACACAGAAGAGTTTGAAGG | 95 °C 5 min; 5 cycles of 95 °C 2 min, 58 °C 45 s, 72 °C 20 s; 5 cycles of 95 °C 2 min, 57 °C 45 s, 72 °C 20 s; 35 cycles of 95 °C 2 min, 56 °C 45 s, 72 °C 20 s; 72 °C 10 min; 10 °C soak | O’Donnell et al. (2010) |
| G2R | Reverse | GTCATYTGDGTDGCDGGYTCDCC | ||||
| RNA polymerase second largest subunit | rpb2 | 5f2 | Forward | GGGGWGAYCAGAAGAAGGC | 94 °C 90 s; 35 cycles of 94 °C 45 s, 57 °C 45 s, 72 °C 20 s; 72 °C 5 min; 10 °C soak | Reeb et al. (2004) |
| 7cf | Forward | ATGGGYAARCAAGCYATGGG | Liu et al. (1999) | |||
| 7cr | Reverse | CCCATRGCTTGYTTRCCCAT | ||||
| 11ar | Reverse | GCRTGGATCTTRTCRTCSACC | ||||
| translation elongation factor 1-alpha | tef1 | EF-1 | Forward | ATGGGTAAGGARGACAAGAC | 94 °C 90 s; 35 cycles of 94 °C 45 s, 55 °C 45 s, 72 °C 15 s; 72 °C 10 min; 10 °C soak | O’Donnell et al. (1998b) |
| EF-2 | Reverse | GGARGTACCAGTSATCATG | ||||
Phylogenetic analyses
Consensus sequences were obtained using MEGA v. 7 software (Kumar et al. 2016), and sequences for each locus were aligned using MAFFT v. 7 (Katoh & Standley 2013). Misalignments were corrected manually where necessary. Phylogenetic analyses were performed based on individual and combined datasets, using Maximum-Likelihood (ML) and Bayesian Inference (BI) methods through the CIPRES Science Gateway portal (https://www.phylo.org/; Miller et al. 2012).
The ML analyses were carried out using RAxML-HPC v. 8.2.12 (Stamatakis 2014), with 1 000 replicates under the GTR+GAMMA model. The Bayesian analyses were carried out using MrBayes v. 3.2.7a (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003), incorporating the best evolutionary models for each marker as determined by MrModelTest v. 2.4 (Nylander 2004). Bayesian analyses were computed with four simultaneous Markov Chain Monte Carlo chains, 10 M generations, and a sampling frequency of 1 000 generations for the first datasets and 100 generations for the other two datasets, ending the run automatically when the standard deviation of split frequencies fell below 0.01. The burn-in fraction was set to 0.25, after which the 50 % majority rule consensus trees and posterior probability (PP) values were calculated.
The clade is supported when its RAxML Bootstrap support value is ≥ 70 %, and the Bayesian PP value is ≥ 0.9. The resulting trees were plotted using FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree). All sequences and their alignments generated from 2 020 Fusarium strains in this study were deposited in GenBank (Supplementary Table S1) and TreeBASE (submission ID 30020), respectively.
Phylogenetic analyses of different Fusarium species complexes were performed using different multi-locus datasets in accordance with previous studies (Sarver et al. 2011, O’Donnell et al. 2013, Gräfenhan et al. 2016, Laurence et al. 2016, Sandoval-Denis et al. 2018b, Torbati et al. 2018, Lombard et al. 2019c, Maryani et al. 2019b, Wang et al. 2019a, Xia et al. 2019, Yilmaz et al. 2021). The composition of the multi-locus datasets, outgroup taxa, number of characters and the best models are listed in Supplementary Table S2. Specifically, the combined CaM, rpb1, rpb2, tef1 and tub2 datasets were constructed for phylogenetic analyses for the FFSC, rooted with F. nirenbergiae CBS 744.97. Phylogenetic analyses of the FIESC were performed by using the combined CaM, rpb2 and tef1 datasets, rooted with F. concolor NRRL 13459. Phylogenetic analyses of the F. nisikadoi species complex (FNSC) were performed by using the combined rpb1, rpb2 and tef1 datasets, rooted with F. concolor NRRL 13994. Phylogenetic analyses of the FOSC were performed by using the combined CaM, rpb2 and tef1 datasets, rooted with F. udum CBS 177.31. Phylogenetic analyses of the FSAMSC were performed by using the combined H3, rpb1, rpb2 and tef1 datasets, rooted with F. nelsonii NRRL 13338. Phylogenetic analyses of the F. tricinctum species complex (FTSC) were performed by using the combined ITS, rpb1, rpb2 and tef1 datasets, and rooted with F. concolor NRRL 13459.
Morphological observations
Fungal isolates were studied morphologically based on their macroscopic and microscopic features (Crous et al. 2021). Petri plates were incubated for 7 d at 25 °C. Agar pieces of approximately 5 × 5 mm were taken from edge of colonies on synthetic nutrient-poor agar (SNA; Nirenberg 1976) and transferred onto media for morphological characterisation. For morphological observations, PDA and oatmeal agar (OA) media were used. The culture characteristics of the colony, including pigmentation and odour, were observed after 7 d of incubation in the dark on PDA and OA (Crous et al. 2021). Colours were rated according to the colour charts of Kornerup & Wanscher (1978).
For morphological comparisons, carnation leaf agar (CLA; Fisher et al. 1982) was used. Micromorphological characteristics, including sporodochia, conidiophores, phialides, conidia (sporodochial and aerial conidia) and chlamydospores, were observed after 7–14 d of incubation under a 12/12 h near-ultraviolet light/dark cycle at 25 °C (Leslie & Summerell 2006, Crous et al. 2021). Morphological characteristics were examined and photo-documented with water as mounting medium under a Nikon 80i compound microscope with differential interference contrast (DIC) optics, and a Nikon SMZ1500 dissecting microscope. For each species, 30 phialides and chlamydospores, and 50 conidia were randomly measured to calculate the mean value, standard deviation, and minimum–maximum values.
Whole-genome sequencing, assembly and gene annotation
Whole-genome sequences were generated for eight ex-type strains of new species (i.e., F. erosum, F. fecundum, F. jinanense, F. mianyangense, F. nothincarnatum, F. planum, F. sanyaense, and F. weifangense) and 32 strains of known species (Supplementary Table S3). All strains were purified using a single-spore isolation method (Zhang et al. 2013). Hyphae of 7-d-old colonies growing on PDA were collected and then stored at -80 °C. Genome extraction and sequencing were done by the Annoroad Gene Technology Company (Beijing, China). The DNA libraries were sequenced as 150 bp pair-end reads using Illumina NovaSeq 6000 platform. Reference genomes were retrieved from the public database (Supplementary Table S3).
The genome assembly and gene annotation were conducted following the protocol of Ma et al. (2021). Specifically, read quality was assessed by using FastQC v. 0.11.8 (Andrews & Babraham 2010). Clean reads were assembled with SPAdes v. 3.12.0 (Bankevich et al. 2012), using the “careful” mode and various kmers (21, 33, 55, 77, 99). Genome assembly quality was assessed using QUAST v. 5.0.2 (Alexey et al. 2013). Genome completeness was assessed using genome mode in BUSCO v. 5.3.2 (Seppey et al. 2019), with the Sordaromyceta_odb9 gene set. Gene predictions were made using the Funannotate pipeline v. 1.7.0 (Palmer 2016); repetitive elements were initially soft-masked using default parameters; funannotate predict was implemented using Sordariomycetes BUSCO database and Fusarium graminearum as seed species. Genome assemblies were deposited in the National Microbiology Data Centre (NMDC) under BioProject NMDC10018280.
Phylogenomic tree construction
Predicted proteins were clustered into orthologous groups by using Orthofinder v. 2.3.3 (Emms & Kelly 2015, 2019). Amino acid sequences of single-copy orthologs were aligned using MAFFT v. 7 (Katoh & Standley 2013). Conserved sites in the alignment were extracted using Gblocks v. 0.91b (Castresana 2000). The substitution model and maximum-likelihood tree were inferred by IQ-TREE v. 2.2.0.8 (Kalyaanamoorthy et al. 2017, Hoang et al. 2018, Minh et al. 2020a), and the clade is supportive when its ultrafast bootstrap (UFBoot) support value is ≥ 95 %. In addition, gene concordance factors (gCF) and site concordance factors (sCF) were calculated using the “–gcf” and “–scf” options in IQ-TREE v. 2.2.0.8 (Minh et al. 2020b), to quantify genealogical concordance. For every branch of a species tree, gCF is defined as the percentage of “decisive” gene trees containing that branch, and sCF is defined as the percentage of decisive alignment sites supporting that branch (Minh et al. 2020b).
Diversity and distribution analyses
The richness of Fusarium species was defined as recorded species number associated with different hosts and climate regions (hosts: maize, rice, wheat; climate regions: regions affected by plateau mountain, subtropical monsoon, temperate continental, temperate monsoon, and tropical monsoon climate). Co-infection was defined as a single diseased cereal sample (e.g., a single wheat head, maize ear) from which more than one Fusarium species was isolated. The relative percentage was calculated by counting the number of each Fusarium species presented in different groups, and was visualised by the ggplot2 package in R v. 4.1.0 (Wickham 2016). The diversity of Fusarium species and distribution patterns across different hosts and geographic regions were performed using GraphPad Prism v. 8.0.2.
Pathogenicity assays
Pathogenicity tests were performed to determine whether the species isolated from the symptomatic tissues of diseased cereals are pathogens. The Fusarium species isolated from maize stalk rot were used as examples, because maize is the largest grain crop in China (http://www.stats.gov.cn/), and maize stalk rot has been one of the primary diseases that threaten maize production in recent years (Wang et al. 2010). In brief, we completed pathogenicity tests fulfilling Koch’s postulates of all Fusarium species isolated from maize, which have not yet been reported as causal agents of maize stalk rot in previous studies.
Specifically, representative isolates were inoculated on seedlings of the maize cultivar Zhengdan 958, as Ye et al. (2013) and Han et al. (2022) described but with slight modifications. Hyphae of 7-d-old colonies growing on SNA were transferred to 400 mL of carboxymethylcellulose sodium fluid medium, and cultivated for 3–7 d at 25 °C at 180 rpm. Then the conidial concentration (CC) of Fusarium suspension were adjusted to 106~107 conidia/mL by haemocytometer, and the suspension were stored at 4 °C until maize seedlings became three-leaf-old. The maize kernels were soaked in water for 24 h, moisturised in a wet chamber for 48 h, and then incubated in sterilised soil under 16 h of light and 8 h of darkness at 25 °C. After 7 d, the roots (three-leaf-old seedlings, Fig. 7A) were immersed into the prepared suspension and incubated for 6 h at 25 °C, then transferred to soil (25 °C, 16 h light/25 °C, 8 h dark). As a negative control, seedlings were inoculated with sterile water. For each tested fungal strain, eight plant replicates were used for the pathogenicity test. Pathogenicity was evaluated based on leaf symptoms of the tested maize plants 3 d post-inoculation. Koch’s postulates were fulfilled by re-isolating the fungus from symptomatic plants.
Fig. 7.

Pathogenicity test of maize stalk rot (MSR) fulfilling Koch’s postulates. A. Diagram of a three-leaf-old seedlings of maize. B. Typical symptoms of disease severity (0–4). C. Blank control treated with sterile water. D–U. Maize seedlings treated with different Fusarium suspensions. The conidial concentration (CC) of Fusarium strains and disease severity index (DSI) of infected plants were listed. DSI (%) = [sum (class frequency × score of rating class)/(total number of plants × maximal disease severity score)] × 100.
Disease severity was estimated visually on a 0 to 4 scale as described in previous studies (Jin et al. 1994). Disease severity was displayed in Fig. 7B and categorised as follows: 0, no visible symptoms; 1, the dry area of the first leaf under 50 %, and the dry area of the second leaf under 25 %; 2, the dry area of the first leaf over 50 % but under 100 %, and the dry area of the second leaf under 50 %; 3, the first leaf was completely withered, the dry area of the second leaf over 50 % but under 100 %, and the dry area of the third leaf was under 25 %; 4, the three leaves of inoculation plants were completely withered. The disease severity index (DSI) was calculated based on Chiang et al. (2017). Specifically, DSI (%) = [sum (class frequency × score of rating class)/(total number of plants × maximal disease severity score)] × 100.
RESULTS
Phylogenetic analyses
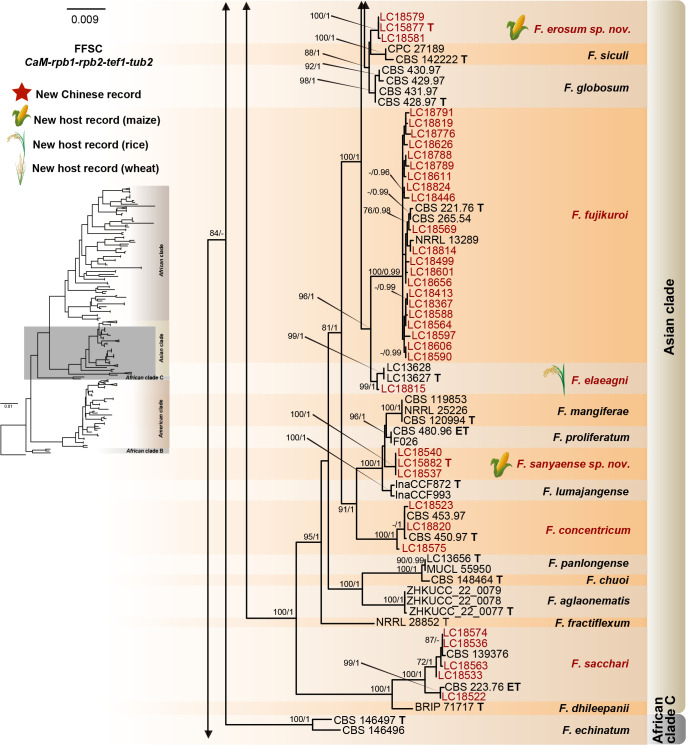
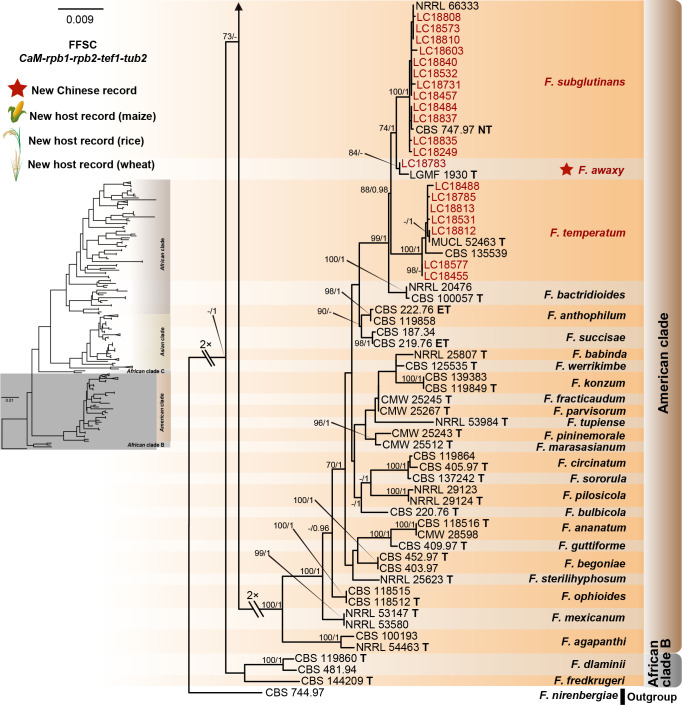
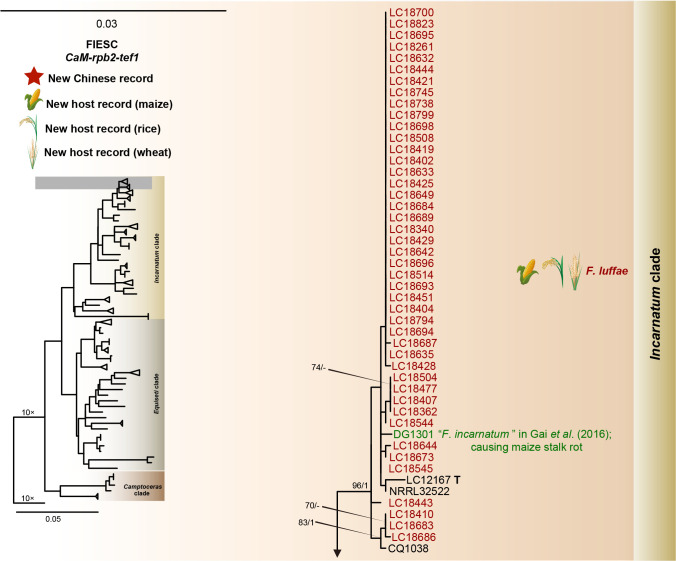
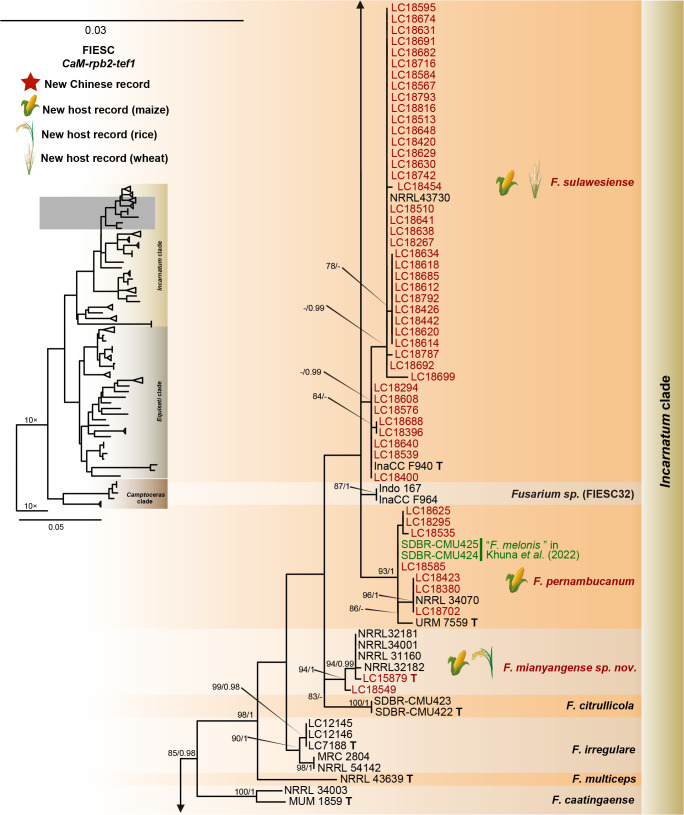
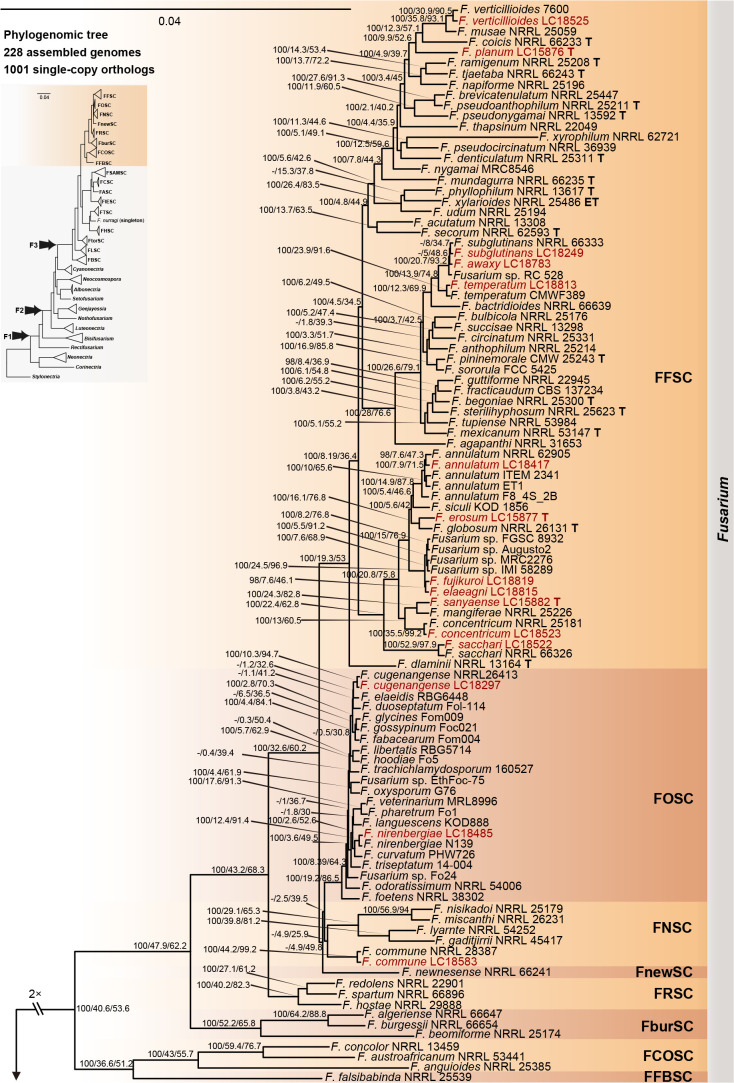
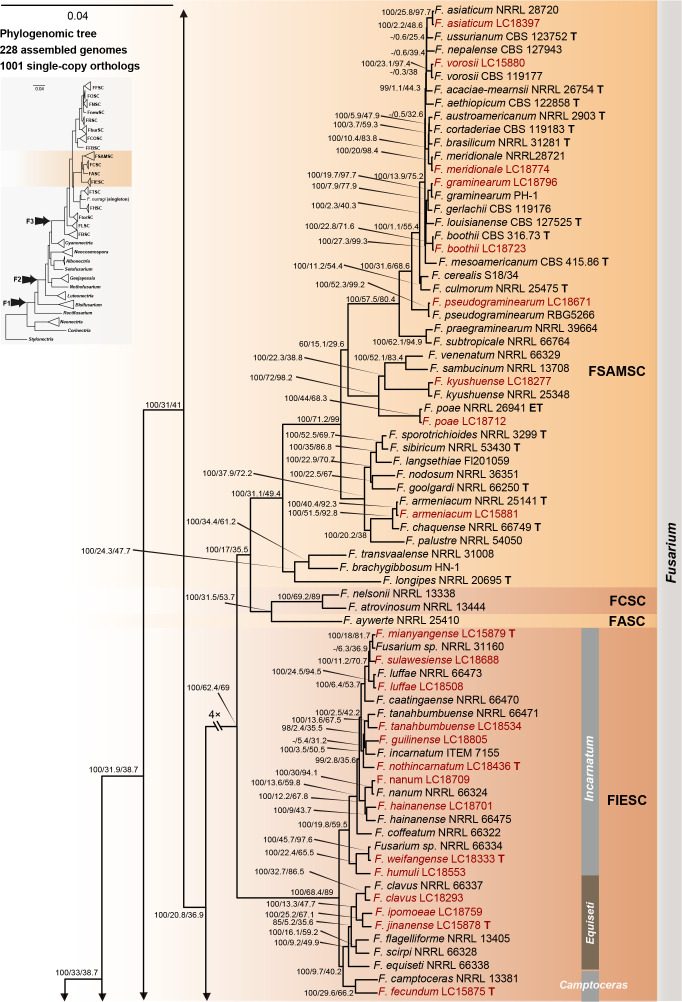
The preliminary phylogenetic analyses based on combined rpb2 and tef1 loci revealed that the 608 representative isolates were distributed over six Fusarium species complexes (Supplementary Fig. S2). Phylogenetic analyses were performed respectively for each Fusarium species complex using different datasets. Combined with morphological characters, these isolates were identified to 12 species in the FFSC (Fig. 8), 17 species in the FIESC (Fig. 9), one species in the FNSC (Fig. 10), two species in the FOSC (Fig. 11), nine species in the FSAMSC (Fig. 12), and two species in the FTSC (Fig. 13). In summary, the 608 selected strains were identified to 43 species, including 35 known and eight novel species.
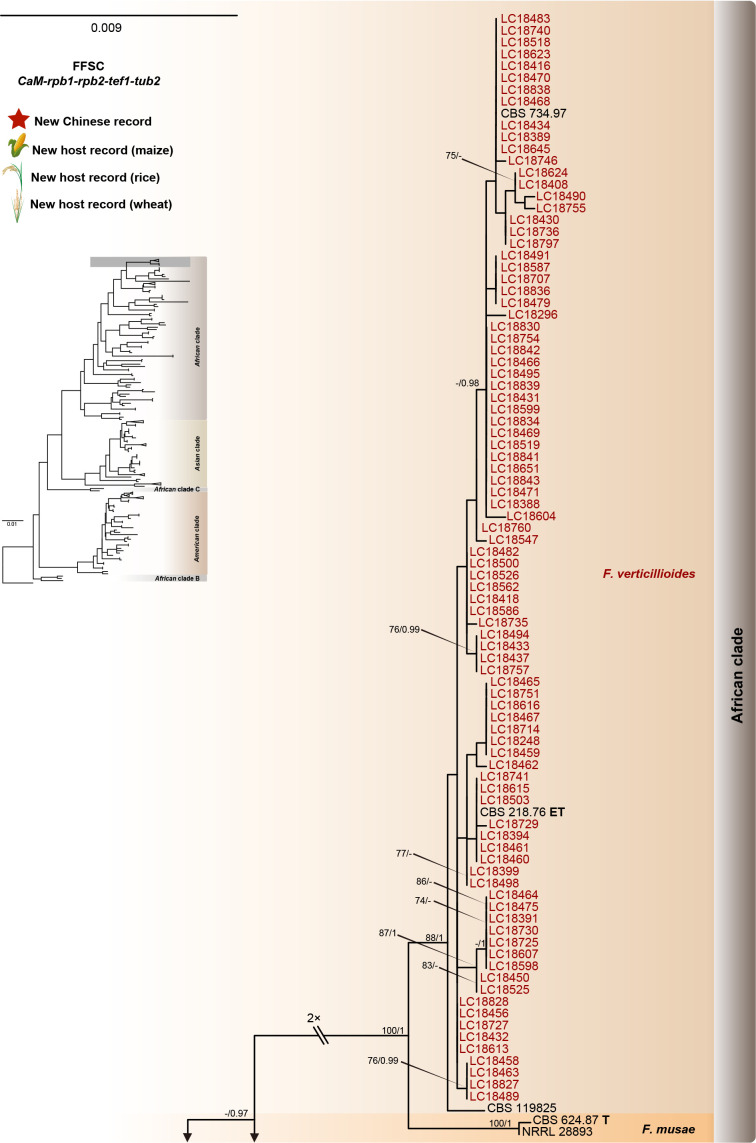
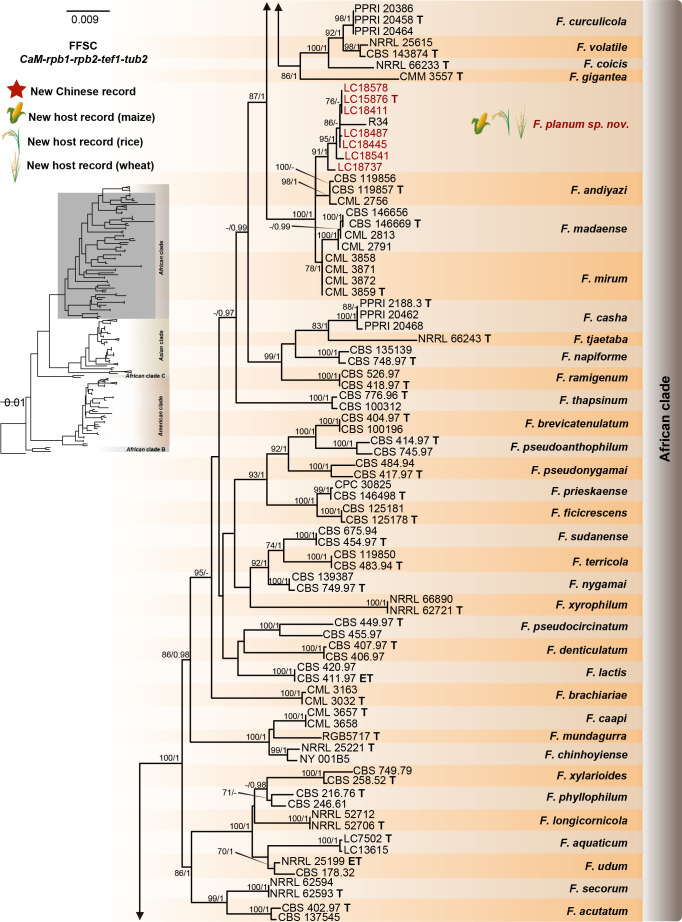
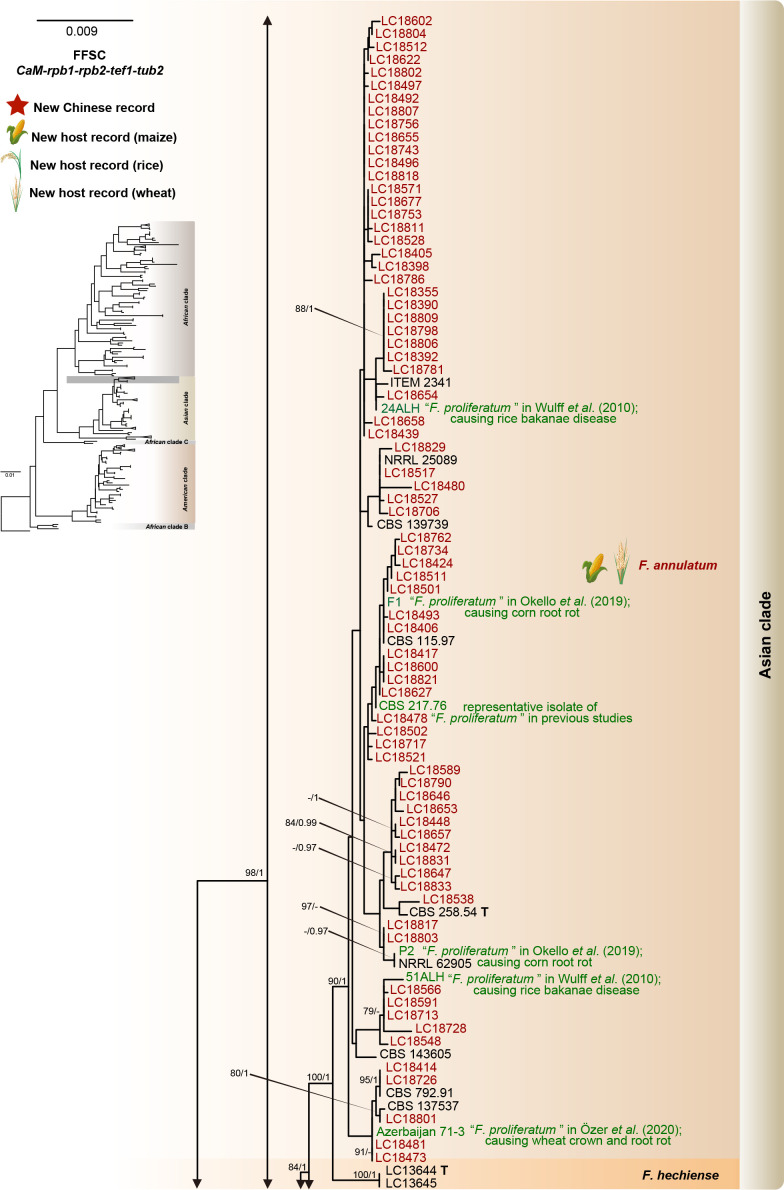
Fig. 8.
Phylogeny inferred based on the combined CaM-rpb1-rpb2-tef1-tub2 gene regions of the Fusarium fujikuroi species complex (FFSC). Fusarium nirenbergiae (CBS 744.97) was used as an outgroup. Strains isolated in this study were indicated in red. Pathogenetic strains from previous studies were indicated in green. The RAxML Bootstrap support values (ML-BS > 70 %) and Bayesian posterior probabilities (BI-PP > 0.9) were displayed at the nodes (ML-BS / BI-PP). Ex-type, ex-epitype and ex-neotype strains were indicated in bold with T, ET, and NT, respectively.
Fig. 9.
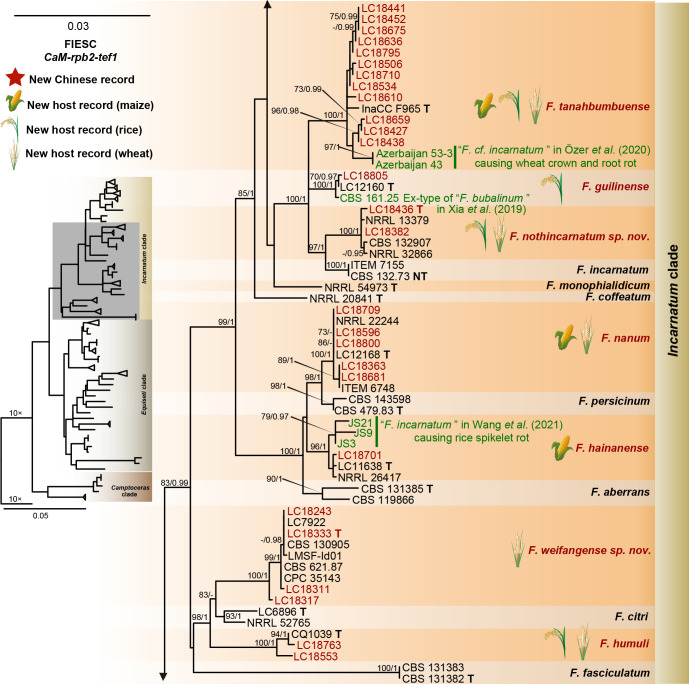
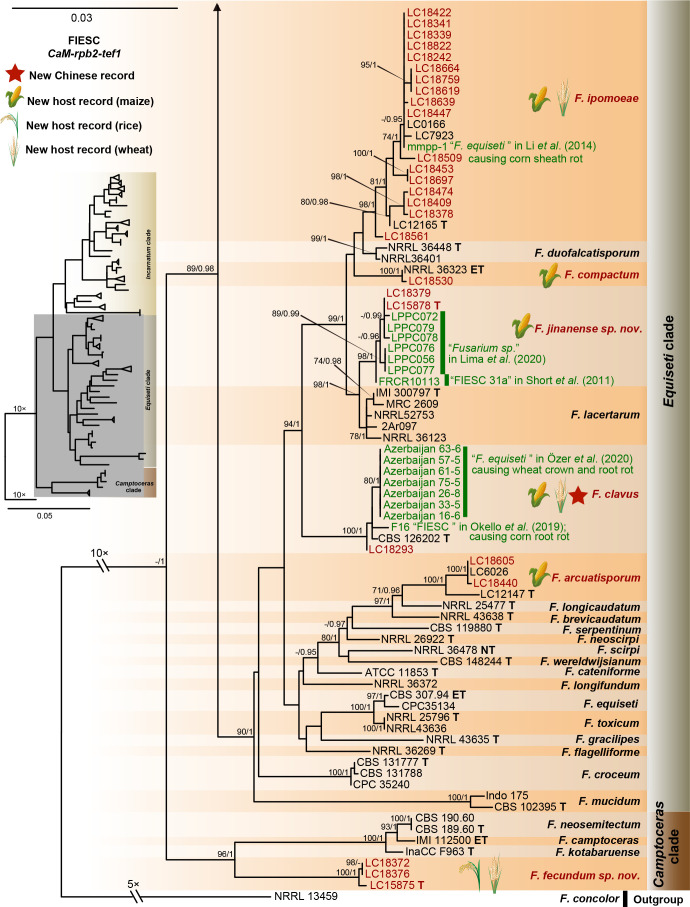
Phylogeny inferred based on the combined CaM-rpb2-tef1 gene regions of the Fusarium incarnatum-equiseti species complex (FIESC). Fusarium concolor (NRRL 13459) was used as an outgroup. Strains isolated in this study were indicated in red. Pathogenetic strains from previous studies were indicated in green. The RAxML Bootstrap support values (ML-BS > 70 %) and Bayesian posterior probabilities (BI-PP > 0.9) were displayed at the nodes (ML-BS / BI-PP). Ex-type, ex-epitype and ex-neotype strains were indicated in bold with T, ET, and NT, respectively.
Fig. 10.
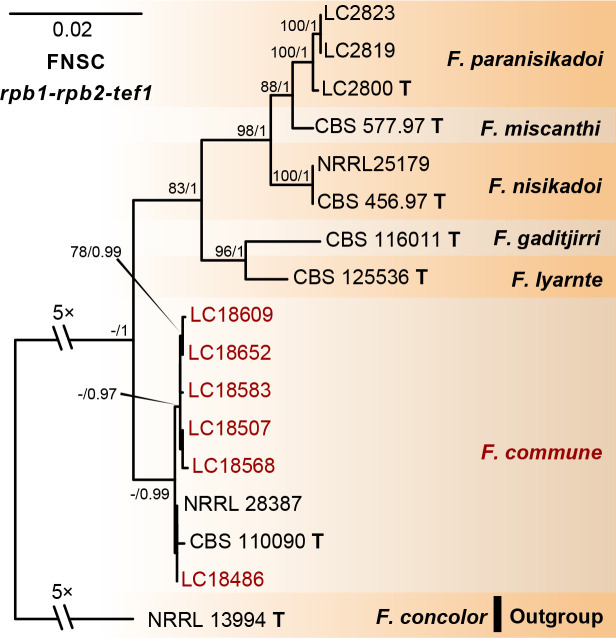
Phylogeny inferred based on the combined rpb1-rpb2-tef1 gene regions of the Fusarium nisikadoi species complex (FNSC). Fusarium concolor (NRRL 13994) was used as an outgroup. Strains isolated in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) and Bayesian posterior probabilities (BI-PP > 0.9) were displayed at the nodes (ML-BS / BI-PP). Ex-type strains were indicated in bold with T.
Fig. 11.
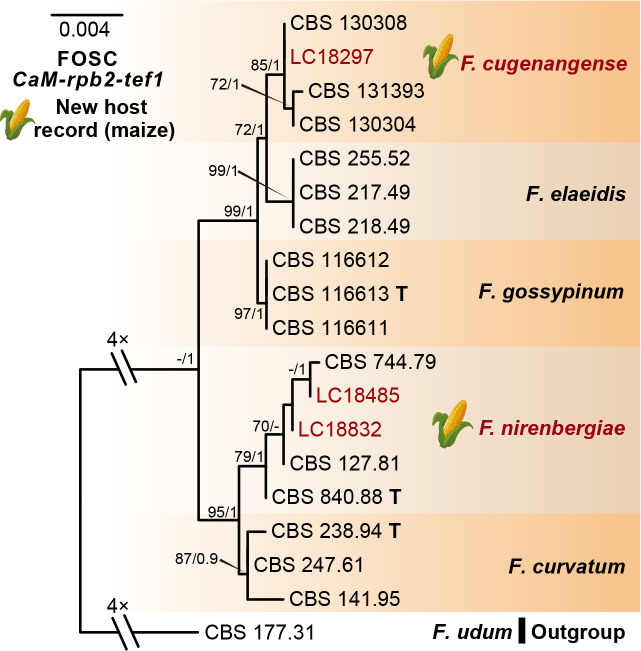
Phylogeny inferred based on the combined CaM-rpb2-tef1 gene regions of the Fusarium oxysporum species complex (FOSC). Fusarium udum (CBS 177.31) was used as an outgroup. Strains isolated in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) and Bayesian posterior probabilities (BI-PP > 0.9) were displayed at the nodes (ML-BS / BI-PP). Ex-type strains were indicated in bold with T.
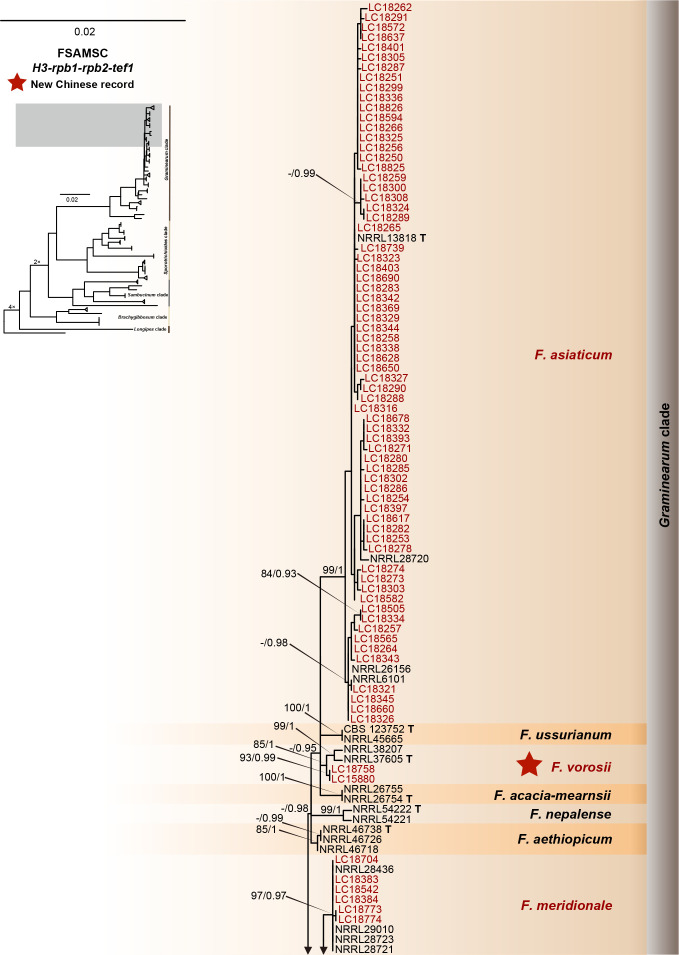
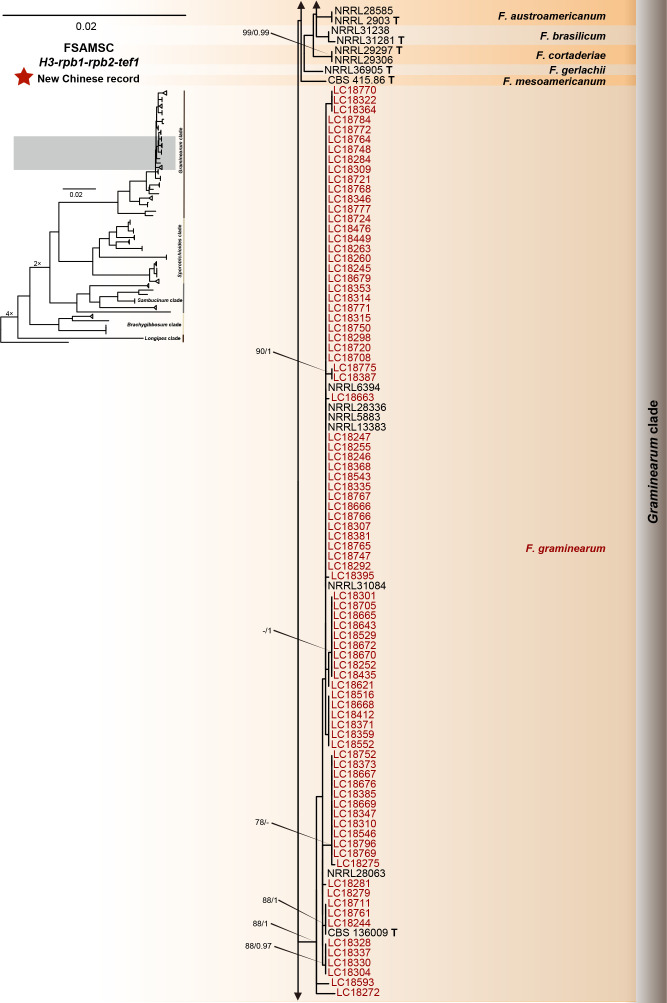
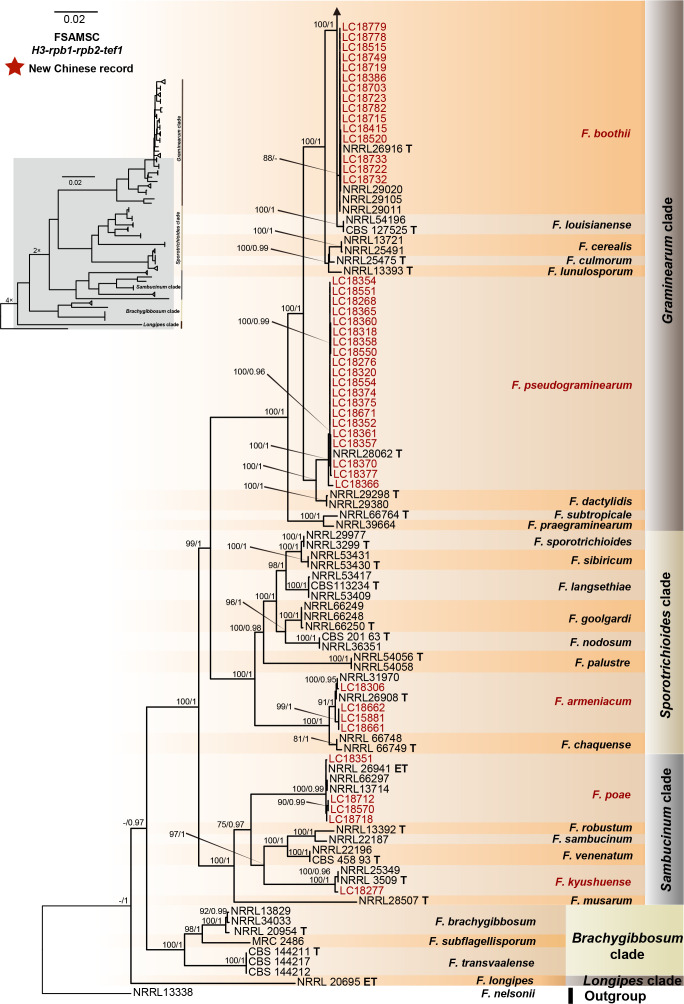
Fig. 12.
Phylogeny inferred based on the combined H3-rpb1-rpb2-tef1 gene regions of the Fusarium sambucinum species complex (FSAMSC). Fusarium nelsonii (NRRL 13338) was used as an outgroup. Strains isolated in this study were indicated in red. The RaxML Bootstrap support values (ML-BS > 70 %) and Bayesian posterior probabilities (BI-PP > 0.9) were displayed at the nodes (ML-BS / BI-PP). Ex-type and ex-epitype strains were indicated in bold with T and ET, respectively.
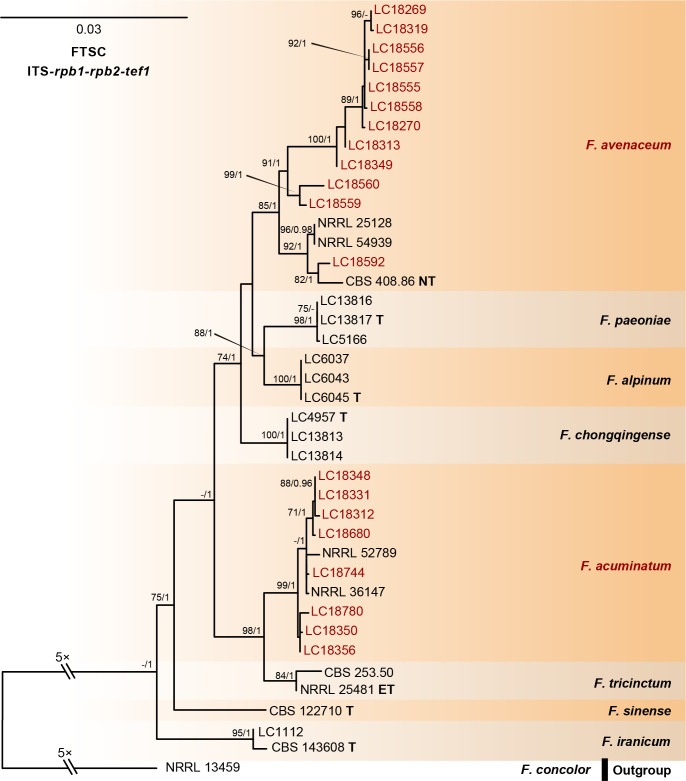
Fig. 13.
Phylogeny inferred based on the combined ITS-rpb1-rpb2-tef1 gene regions of the Fusarium tricinctum species complex (FTSC). Fusarium concolor (NRRL 13459) was used as an outgroup. Strains isolated in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) and Bayesian posterior probabilities (BI-PP > 0.9) were displayed at the nodes (ML-BS / BI-PP). Ex-type, ex-epitype, and ex-neotype strains were indicated in bold with T, ET, and NT, respectively.
Furthermore, single gene trees were evaluated respectively for FFSC (Supplementary Fig. S3), FIESC (Supplementary Fig. S4), FNSC (Supplementary Fig. S5), FOSC (Supplementary Fig. S6), FSAMSC (Supplementary Fig. S7) and FTSC (Supplementary Fig. S8). The phylogenetic analyses based on single genes showed that the rpb2 locus provided a better resolution in species recognition for the six species complexes. Using the three most species-rich complexes (FFSC, FIESC, FSAMSC) as examples, the rpb2 locus was able to recognise 59, 33, and 35 species among the FFSC (with 84 species), FIESC (with 44 species), and FSAMSC (with 41 species), respectively.
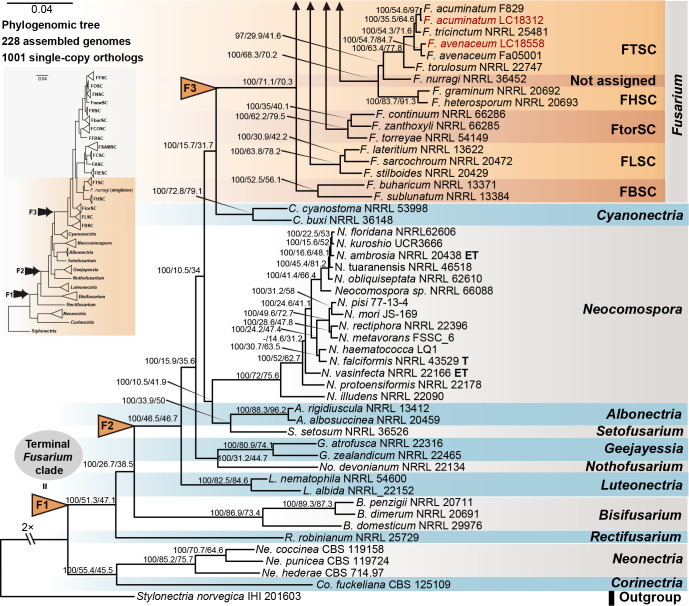
Phylogenomic assessment of Fusarium and allied genera
Employing 228 assembled genomes covering 17 Fusarium species complexes, 11 Fusarium allied genera and one outgroup (Stylonectria norvegica IHI 201603), a high-confidence and genome-scale phylogenetic tree was generated (Fig. 14). The Gblocks filtered alignment of 1 001 single-copy orthologs (Supplementary Table S4) consisted of 223 451 characters, including alignment gaps. The genome sizes of the four cereal-associated species-rich complexes (i.e., FFSC, FIESC, FOSC and FSAMSC) differed significantly (P < 2e-16, Supplementary Fig. S9), with FOSC having the largest genome size (av. ± SD: 52.6 ± 4.2 Mb) and FSAMSC having the smallest genome size (av. ± SD: 37.5 ± 3.7 Mb). In our phylogenomic tree, 193 nodes (86 %) received 100 % bootstrap support, including the nodes F1, F2, and F3 (Fig. 14). Meanwhile, the gCF and sCF values resolved higher in node F3 (gCF 71.1 %, sCF 70.3 %) than in F2 (gCF 46.5 %, sCF 46.7 %) and F1 (gCF 51.3 %, sCF 47.1 %).
Fig. 14.
Maximum likelihood phylogenomic tree of Fusarium and allied genera. A total of 1 001 single-copy orthologs were employed in the analysis. Stylonectria norvegica IHI 201603 was used as an outgroup. Strains sequenced in this study were indicated in red. The IQ-TREE ultrafast bootstrap support values (UFBoot ≥ 95 %), gCF and sCF values were displayed at the nodes (UFBoot / gCF / sCF). Arrows “F1” (= “Terminal Fusarium clade”), “F2” and “F3” indicate the three alternative Fusarium generic hypotheses sensu Geiser et al. (2013). Ex-type, ex-epitype and ex-neotype strains were indicated in bold with T, ET, and NT, respectively. Subdivision of the Fusarium clade represents the recognised species complexes, including F. aywerte SC (FASC), F. buharicum SC (FBSC), F. burgessii SC (FburSC), F. chlamydosporum SC (FCSC), F. concolor SC (FCOSC), F. falsibabinda SC (FFBSC), F. fujikuroi SC (FFSC), F. heterosporum SC (FHSC), F. incarnatum-equiseti SC (FIESC), F. lateritium SC (FLSC), F. newnesense SC (FnewSC), F. nisikadoi SC (FNSC), F. oxysporum SC (FOSC), F. redolens SC (FRSC), F. sambucinum SC (FSAMSC), F. torreyae SC (FtorSC) and F. tricinctum SC (FTSC).
There are only two subtle topology differences between our phylogenomic tree (Fig. 14) and the multi-locus trees in previous studies (Xia et al. 2019, Crous et al. 2021, Wang et al. 2022a). First, in our phylogenomic tree, F. camptoceras (previously included in F. camptoceras species complex, FCAMSC) clustered within the Equiseti clade of the FIESC (Fig. 14), while the FCAMSC formed a separate sister clade to the FIESC in previous phylogenetic studies (Xia et al. 2019, Crous et al. 2021, Wang et al. 2022a). This is not surprising, as when considering the FIESC and the FCAMSC as two separate species complexes, the clade representing the FIESC was not statistically supported in previous phylogenetic trees (Xia et al. 2019, Crous et al. 2021, Wang et al. 2022a), indicating that previous multi-locus analyses could not provide sufficient phylogenetic resolution to reveal the evolutionary relationship between the two species complexes. However, when the FCAMSC and the FIESC were considered together as a whole, this larger group is clearly a distinct evolutionary lineage that is highly supported in the phylogenomic tree (support value = 100 %; Fig. 14), as well as in previous multi-locus trees (Crous et al. 2021). Therefore, we herein include the FCAMSC in FIESC as the Camptoceras clade (Fig. 14). Second, Bisifusarium (also known as F. dimerum species complex) and Rectifusarium (also known as F. ventricosum species complex), appearing basal in the tree, were paraphyletic in our phylogenomic tree (Fig. 14), in agreement with Lombard et al. (2015) and Crous et al. (2021), but differed from O’Donnell et al. (2013) and Geiser et al. (2021) in which Bisifusarium and Rectifusarium clustered together forming a separate clade. However, in both publications, the clade including Bisifusarium and Rectifusarium was not statistically supported (O’Donnell et al. 2013, Geiser et al. 2021). Overall, our phylogenomic tree is essentially similar with that of Crous et al. (2021), and our data resolved several remaining conflicts among phylogenies inferred from different studies.
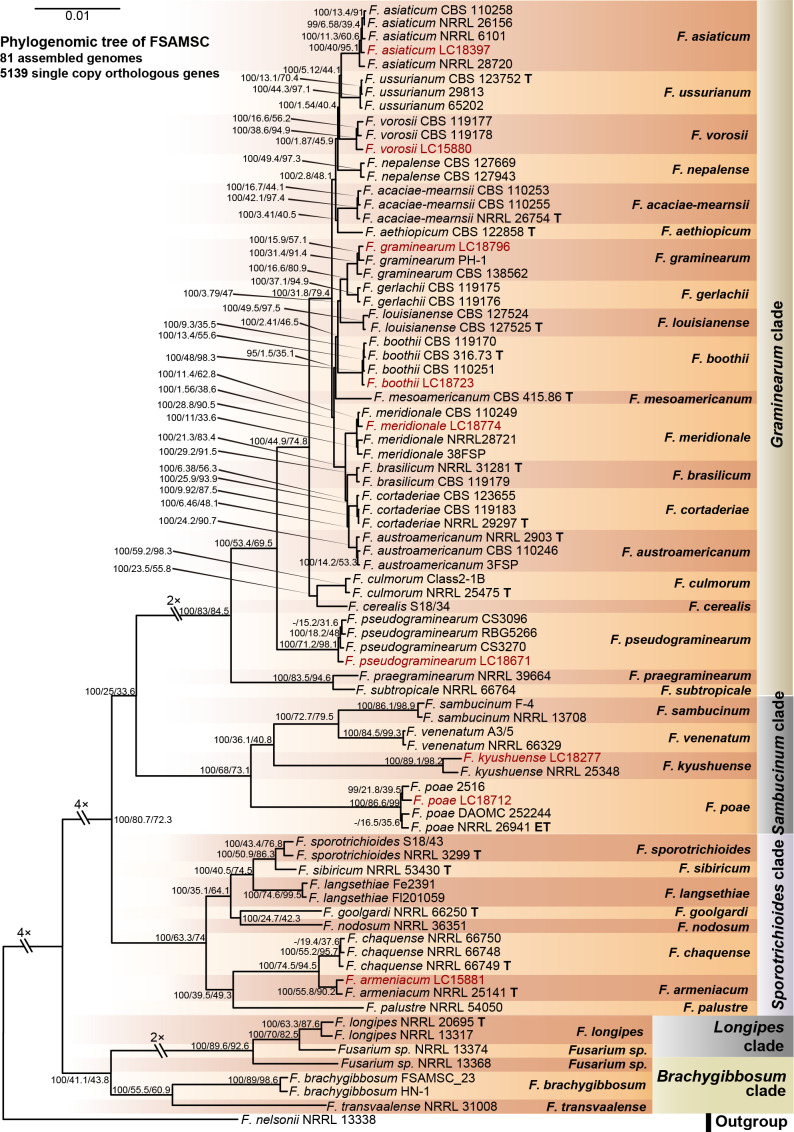
Phylogenomic analyses within the FSAMSC
FSAMSC is one of the most species-rich complexes containing 41 accepted species, 35 of which have available genome data. Species within the FSAMSC particularly the Fg clade, however, exhibit almost indistinguishable morphological differences (Sarver et al. 2011) and few nucleotide differences (Laraba et al. 2021), although they have been recognised as different species in previous studies (O’Donnell et al. 2000a, 2004, Starkey et al. 2007). Therefore, to better understand the evolutionary relationship of species within the FSAMSC, especially the Fg clade, a phylogenomic tree of the FSAMSC (Fig. 15) was inferred employing 5 139 single-copy orthologs (Supplementary Table S5) obtained from 81 assembled genomes, with Fusarium nelsonii NRRL 13338 as an outgroup (Fig. 15). Our phylogenomic tree is strongly supported by UFBoot values, but weakly supported by gCF and sCF values (Supplementary Fig. S10B), suggesting strongly conflicting signals among genes (Minh et al. 2020b). Meanwhile, the conflicting topologies were further confirmed by the comparison of phylogenomic and multi-locus phylogenetic trees (see Supplementary Fig. S11 for more details). This discordance is potentially due to incomplete lineage sorting (ILS), horizontal gene transfer (HGT), gene duplication and loss, hybridisation, or recombination (Degnan & Rosenberg 2009). In general, both phylogenomic and multi-locus phylogenetic trees revealed five major clades (Graminearum, Sporotrichioides, Sambucinum, Brachygibbosum and Longipes clades), and both provide effective resolution for species delimitation within the FSAMSC.
Fig. 15.
Maximum likelihood phylogenomic tree of the Fusarium sambucinum species complex (FSAMSC). A total of 5 139 single copy orthologs were employed in the analysis. F. nelsonii (NRRL 13338) was used as an outgroup. Strains sequenced in this study were indicated in red. The IQ-TREE ultrafast bootstrap support values (UFBoot ≥ 95 %), gCF and sCF values were displayed at the nodes (UFBoot / gCF / sCF). Ex-type and ex-epitype strains were indicated in bold with T and ET, respectively.
Distribution and pathogenicity of Fusarium spp. on diseased cereals
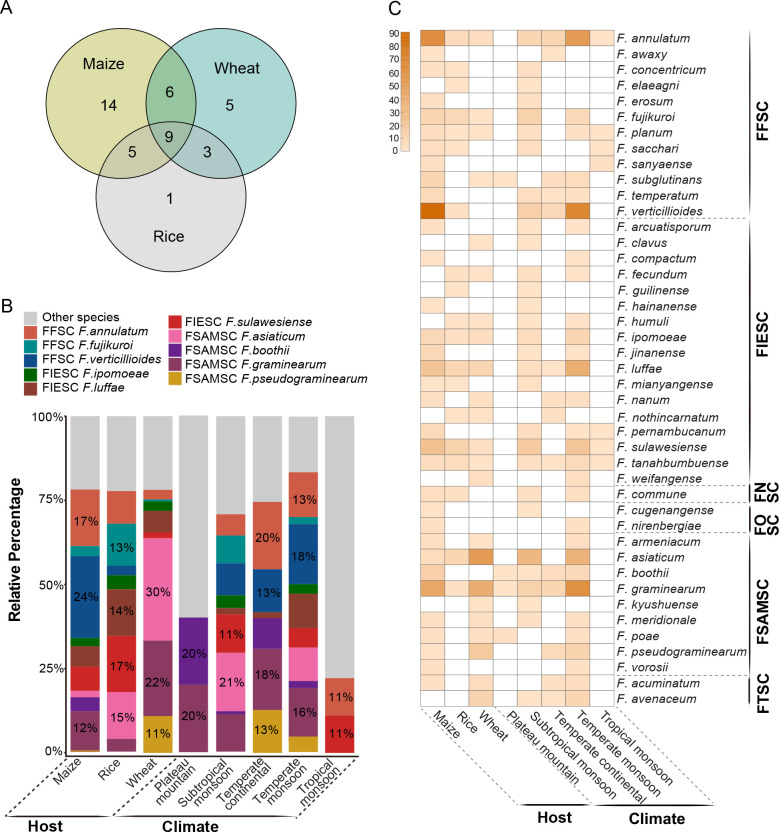
Based on multi-locus phylogenetic analyses, the 608 representative Fusarium strains were identified as 43 species. Further analyses showed that species richness is higher on maize (34 species), followed by wheat (23 species) and rice (18 species), and only nine species were shared by all three kinds of cereals (Fig. 16A, C). Species with high proportion of isolation in maize samples were F. verticillioides (24 %) and F. annulatum (17 %), compared to F. asiaticum (30 %) and F. graminearum (22 %) in wheat samples and F. sulawesiense (17 %) in rice samples (Fig. 16B). The majority of Fusarium species collected in this study (98 % species from 90 % samples) were from regions affected by a monsoon climate (Fig. 16C, Supplementary Table S1).
Fig. 16.
A. The number of unique and shared Fusarium species among different hosts. B. The relative percentage of Fusarium species in different groups (host: maize, rice, wheat; climate regions: regions affected by plateau mountain, subtropical monsoon, temperate continental, temperate monsoon, and tropical monsoon climate). The smaller proportion strains of Fusarium were defined as “Other species”, including F. acuminatum, F. arcuatisporum, F. armeniacum, F. awaxy, F. clavus, F. commune, F. compactum, F. concentricum, F. cugenangense, F. elaeagni, F. erosum, F. fecundum, F. guilinense, F. hainanense, F. humuli, F. ipomoeae, F. jinanense, F. kyushuense, F. meridionale, F. mianyangense, F. nanum, F. nirenbergiae, F. nothincarnatum, F. pernambucanum, F. planum, F. poae, F. sacchari, F. sanyaense, F. subglutinans, F. tanahbumbuense, F. temperatum, F. vorosii, and F. weifangense. C. Detailed information on Fusarium composition among different hosts and climate regions. The number of isolates (0–90) was represented by the shade of colour (light to dark).
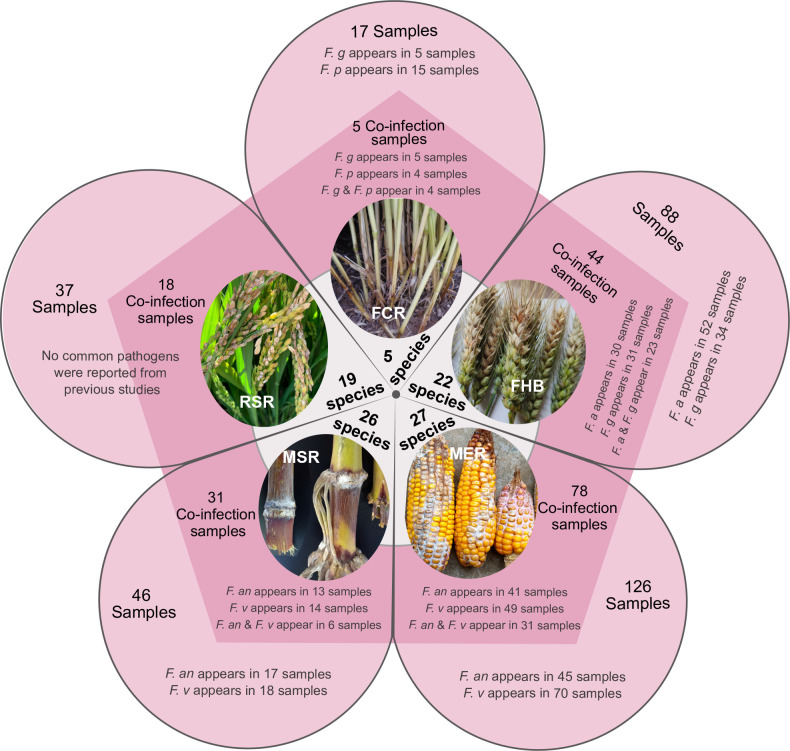
Interestingly, a total of 315 diseased samples were used for fungal isolation. Of these, 176 samples (56 %) were observed to have a co-infection (Fig. 17), half of which contain previously reported dominant pathogens, including F. annulatum (previously usually known as “F. proliferatum”), F. asiaticum, F. graminearum, F. pseudograminearum, and F. verticillioides (Fig. 17, Supplementary Table S6). In addition, co-infection by two species can be observed from 59 % of the samples (Supplementary Fig. S12).
Fig. 17.
Summary of co-infections discovered in this study. The white circle represents the number of species isolated from different disease samples (only one species was isolated from the rice bakanae disease sample, thus information was not shown in this figure). The pentagon represents the number of co-infection samples, also marked the occurrence of common pathogens, including F. a, F. an, F. g, F. p and F. v. The outside sectors of pentagon represent the number of whole samples, also marked the occurrence of common pathogens as previously mentioned.
Abbreviations: Fusarium crown rot of wheat (FCR), Fusarium head blight of wheat (FHB), maize ear rot (MER), maize stalk rot (MSR), rice spikelet rot (RSR), F. asiaticum (F. a), F. annulatum (F. an), F. graminearum (F. g), F. pseudograminearum (F. p), F. verticillioides (F. v).
Furthermore, we summarised the world records of Fusarium species associated with cereals (Table 2). The data were retrieved from the USDA fungal database (more than 1 600 records) (Farr & Rossman 2022), previous studies (168 peer-reviewed papers, Table 2) and this study (2 020 strains, Supplementary Table S1). According to the current taxonomy updated by Crous et al. (2021) and this study, these records correspond to an estimated 97 Fusarium species, belonging to 13 species complexes and one singleton. Among them, 39 and 52 host records for Fusarium species for the world and China were updated, respectively (Table 2). A total of 62 species have been recorded as cereal pathogens (Table 2), of which 18 species (F. acuminatum, F. annulatum, F. arcuatisporum, F. armeniacum, F. awaxy, F. concentricum, F. erosum, F. ipomoeae, F. jinanense, F. luffae, F. mianyangense, F. nirenbergiae, F. pernambucanum, F. planum, F. sacchari, F. sanyaense, F. sulawesiense and F. tanahbumbuense) were proven as pathogens of maize stalk rot for the first time in this study (Fig. 7). Notably, the DSI varies among seedlings infected by different Fusarium species in this study (Fig. 7). For instance, the DSI of seedlings infected by F. annulatum was 68.75 % even under low conidium concentrations (1 × 106 conidia/mL) (Fig. 7E), while those infected by F. awaxy was 6.25 % under low conidium concentrations (1 × 106 conidia/mL), but appeared 59.38 % under higher concentrations (1 × 107 conidia/mL) (Fig. 7H).
Table 2.
Summary of Fusarium species reported from cereals in literatures and the present study.
| Current name1 | Reported name1 | Complex1 | Pathogens1 | References of species publication and subsequent adjustment |
Host records in China2
|
Host records worldwide (except China)2
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maize | Rice | Wheat | References3 | Maize | Rice | Wheat | References3 | |||||
| F. algeriense | F. algeriense | FburSC | FCR | Laraba et al. (2017) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Özer et al. (2020) |
| F. beomiforme | F. beomiforme | FburSC | FCR | Nelson et al. (1987) | n/a | n/a | n/a | n/a | √ | n/a | n/a | Laraba et al. (2017) |
| F. anguioides | F. anguioides | FCOSC | n/a | Sherbakoff (1915) | n/a | n/a | n/a | n/a | √ | √ | √ | Mulenko et al. (2008) |
| F. concolor | F. concolor | FCOSC | n/a | Reinking (1934) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Ebbels & Allen (1979) |
| F. atrovinosum | F. atrovinosum | FCSC | n/a | Lombard et al. (2019c) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Lombard et al. (2019c) |
| F. chlamydosporum | F. chlamydosporum | FCSC | FCR | Wollenweber & Reinking (1925) | n/a | n/a | n/a | n/a | n/a | √ | √ | Chehri et al. (2010) |
| F. nelsonii | F. nelsonii | FCSC | MSR | Marasas et al. (1998) | √ | n/a | n/a | Zhang et al. (2021b) | √ | n/a | √ | Marasas et al. (1998), Chehri et al. (2010) |
| F. andiyazi | F. andiyazi | FFSC | MER; RBD | Marasas et al. (2001) | √ | √ | n/a | Zhang et al. (2014a), Qiu et al. (2020) | √ | √ | n/a | Wulff et al. (2010), Leyva-Madrigal et al. (2015), Venturini et al. (2017) |
| F. annulatum | F. annulatum | FFSC | FCR; FHB; MSR; MER; RBD | Bugnicourt (1952), Yilmaz et al. (2021) | * | * | * | Huang et al. (2011), Qiu et al. (2020), present study | * | √ | * | O’Donnell et al. (1998), Okello et al. (2019), Özer et al. (2020) |
| F. anthophilum | F. anthophilum | FFSC | FCR; FHB | Wollenweber (1916) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Chehri et al. (2010) |
| F. awaxy | F. awaxy | FFSC | MSR | Crous et al. (2019) | * | n/a | n/a | present study | √ | n/a | n/a | Crous et al. (2019) |
| F. brevicatenulatum | F. brevicatenulatum, F. pseudoanthophilum | FFSC | MER; RBD | Nirenberg et al. (1998) | n/a | n/a | n/a | n/a | √ | √ | n/a | Amatulli et al. (2010), Tsehaye et al. (2016) |
| F. concentricum | F. concentricum | FFSC | MER; MSR; RBD | Nirenberg & O’Donnell (1998) | √√ | * | n/a | Du et al. (2020), present study | √ | √ | √ | Aoki et al. (2001), Choi et al. (2018) |
| F. dlaminii | F. dlaminii | FFSC | n/a | Marasas et al. (1985) | n/a | n/a | n/a | n/a | √ | n/a | n/a | Scauflaire et al. (2011) |
| F. elaeagni | F. elaeagni | FFSC | n/a | Wang et al. (2022a) | n/a | * | n/a | present study | n/a | n/a | n/a | n/a |
| F. erosum | F. erosum | FFSC | MSR | present study | * | n/a | n/a | present study | n/a | n/a | n/a | n/a |
| F. fujikuroi | F. fujikuroi, F. moniliforme, F. sacchari var. subglutinans, F. moniliforme var. majus, Gibberella fujikuroi, Lisea fujikuroi | FFSC | FCR; FHB; MER; MSR; RBD; RSR | Nirenberg (1976) | √√ | √√ | √√ | Duan et al. (2020), Qiu et al. (2020), present study | √ | √ | √ | McGuire & Crandall (1967), Byrnes & Carroll (1986), Adikaram & Yakandawala (2020) |
| F. globosum | F. globosum | FFSC | n/a | Rheeder et al. (1996) | n/a | n/a | n/a | n/a | √ | n/a | √ | Nirenberg & O’Donnell (1998); Moses et al. (2010) |
| F. madaense | F. madaense | FFSC | MSR | Ezekiel et al. (2020) | n/a | n/a | n/a | n/a | √ | √ | n/a | Costa et al. (2022) |
| F. napiforme | F. napiforme | FFSC | RBD | Marasas et al. (1987) | n/a | n/a | n/a | n/a | n/a | √ | n/a | Amatulli et al. (2010) |
| F. nygamai | F. nygamai | FFSC | FHB; MSR | Burgess & Trimboli (1986) | n/a | n/a | n/a | n/a | √ | √ | √ | Balmas et al. (2000), Leyva-Madrigal et al. (2015) |
| F. proliferatum | F. proliferatum, F. proliferatum var. minus | FFSC | ? | Nirenberg (1976) | ? | ? | ? | Huang et al. (2010), Qiu et al. (2020) | ? | ? | ? | Wulff et al. (2010), Yasuhara-Bell et al. (2018), Özer et al. (2020) |
| F. pseudoanthophilum | F. pseudoanthophilum | FFSC | n/a | Nirenberg et al. (1998) | n/a | n/a | n/a | n/a | √ | n/a | n/a | Tsehaye et al. (2016) |
| F. pseudonygamai | F. pseudonygamai | FFSC | RBD | Nirenberg & O’Donnell (1998) | n/a | n/a | n/a | n/a | n/a | √ | n/a | Bashyal et al. (2016) |
| F. planum | F. planum | FFSC | MSR | present study | * | * | * | present study | n/a | n/a | n/a | n/a |
| F. sacchari | F. sacchari, Cephalosporium sacchari | FFSC | FHB; MSR; MER; RBD | Gams (1971) | √√ | * | √ | Wang et al. (2015), Duan et al. (2019), present study | √ | √ | √ | Mulenko et al. (2008), Bashyal et al. (2016) |
| F. sanyaense | F. sanyaense | FFSC | MSR | present study | * | n/a | n/a | present study | n/a | n/a | n/a | n/a |
| F. subglutinans | F. subglutinans, F. moniliforme var. subglutinans, Gibberella fujikuroi var. subglutinans | FFSC | MER; MSR; RBD | Nelson et al. (1983) | √√ | n/a | * | Gai et al. (2017), present study | √ | √ | √ | Byrnes & Carroll (1986), Pak et al. (2016) |
| F. temperatum | F. temperatum | FFSC | MER; MSR | Scauflaire et al. (2011) | √√ | n/a | n/a | Zhang et al. (2014c), Gai et al. (2017), present study | √ | n/a | n/a | Varela et al. (2013) |
| F. thapsinum | F. thapsinum | FFSC | MER; MSR; RBD | Klittich et al. (1997) | √ | √ | n/a | Dong et al. (2020b), Zhang et al. (2021a) | √ | √ | n/a | Rahjoo et al. (2008), Bashyal et al. (2016) |
| F. verticillioides | F. verticillioides, Fusarium moniliforme var. majus, Gibberella moniliform, Gibberella moniliformis | FFSC | FCR; FHB; MSR; MER; RBD | Nirenberg (1976), Yilmaz et al. (2021) | √√ | √√ | n/a | Gai et al. (2017), Dong et al. (2020b), Qiu et al. (2020), present study | √ | √ | √ | Chehri et al. (2010), Wulff et al. (2010), Gräfenhan et al. (2011) |
| F. graminum | F. avenaceum var. graminum | FHSC | n/a | Corda (1837) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Mulenko et al. (2008) |
| F. heterosporum | F. heterosporum | FHSC | FCR | Nees von Esenbeck (1818) | n/a | n/a | n/a | n/a | √ | √ | √ | Tunali et al. (2008), Mulenko et al. (2008) |
| F. aberrans | F. aberrans | FIESC | n/a | Xia et al. (2019) | n/a | n/a | n/a | n/a | n/a | √ | n/a | Xia et al. (2019) |
| F. arcuatisporum | F. arcuatisporum | FIESC | MSR | Wang et al. (2019a) | * | n/a | n/a | present study | n/a | n/a | n/a | n/a |
| F. camptoceras | F. camptoceras | FIESC | n/a | Wollenweber & Reinking (1925) | n/a | n/a | n/a | n/a | n/a | √ | √ | Chehri et al. (2010) |
| F. clavus (as ‘clavum’) | F. clavus (as ‘clavum’) | FIESC | n/a | Xia et al. (2019) | n/a | n/a | * | present study | * | n/a | * | Okello et al. (2019), Özer et al. (2020) |
| F. compactum | F. compactum | FIESC | FCR | Raillo (1950) | * | n/a | n/a | present study | n/a | n/a | √ | Tunali et al. (2008) |
| F. nothincarnatum | F. nothincarnatum | FIESC | n/a | present study | n/a | * | * | present study | n/a | * | n/a | O’Donnell et al. (2009) |
| F. equiseti | F. equiseti, Gibberella intricans | FIESC | ? | Saccardo (1886) | ? | n/a | ? | Li et al. (2014), Xu et al. (2016), Gai et al. (2017) | ? | ? | ? | Aguín et al. (2014), Barkat et al. (2016) |
| F. fasciculatum | F. fasciculatum | FIESC | n/a | Xia et al. (2019) | n/a | n/a | n/a | n/a | n/a | √ | n/a | Xia et al. (2019) |
| F. guilinense | F. guilinense | FIESC | n/a | Wang et al. (2019a) | n/a | * | n/a | present study | n/a | n/a | n/a | n/a |
| F. hainanense | F. hainanense | FIESC | n/a | Wang et al. (2019a) | * | * | n/a | Wang et al. (2021), present study | n/a | √ | n/a | Xia et al. (2019) |
| F. humuli | F. humuli | FIESC | n/a | Wang et al. (2019a) | n/a | * | * | present study | n/a | n/a | n/a | n/a |
| F. incarnatum | F. incarnatum, F. semitectum, F. semitectum var. majus, F. diversisporum, F. pallidoroseum, F. semitectum | FIESC | ? | Saccardo (1886) | ? | n/a | ? | Gai et al. (2016, 2017) | ? | ? | ? | Tsehaye et al. (2016), Choi et al. (2018), Özer et al. (2020) |
| F. ipomoeae | F. ipomoeae | FIESC | MSR | Wang et al. (2019a) | * | √ | * | Li et al. (2014), Wang et al. (2019a), present study | n/a | n/a | n/a | n/a |
| F. jinanense | F. jinanense | FIESC | MSR | present study | * | n/a | n/a | present study | n/a | n/a | n/a | n/a |
| F. luffae | F. luffae | FIESC | MSR | Wang et al. (2019a) | * | * | * | Gai et al. (2016), present study | n/a | n/a | n/a | n/a |
| F. mianyangense | F. mianyangense | FIESC | MSR | present study | * | * | n/a | present study | n/a | n/a | n/a | n/a |
| F. nanum | F. nanum | FIESC | n/a | Wang et al. (2019a) | * | n/a | * | present study | n/a | n/a | n/a | n/a |
| F. pernambucanum | F. pernambucanum | FIESC | MSR | Santos et al. (2019) | * | n/a | n/a | present study | n/a | n/a | n/a | n/a |
| F. sulawesiense (as ‘sulawense’) | F. sulawesiense (as ‘sulawense’) | FIESC | MSR | Maryani et al. (2019b) | * | √√ | * | Wang et al. (2019a), present study | n/a | n/a | n/a | n/a |
| F. tanahbumbuense | F. tanahbumbuense | FIESC | MSR | Maryani et al. (2019b) | * | * | * | present study | n/a | n/a | * | Özer et al. (2020) |
| F. weifangense | F. weifangense | FIESC | n/a | present study | n/a | n/a | * | present study | n/a | n/a | n/a | n/a |
| F. fecundum | F. fecundum | FIESC | n/a | present study | n/a | * | * | present study | n/a | n/a | n/a | n/a |
| F. lateritium | F. lateritium | FLSC | FCR | Nees von Esenbeck (1817) | n/a | n/a | n/a | n/a | √ | √ | √ | Richardson (1979), Mulenko et al. (2008), Tunali et al. (2008) |
| F. sarcochroum | F. sarcochroum | FLSC | n/a | Saccardo (1879) | n/a | n/a | n/a | n/a | √ | n/a | n/a | Mulenko et al. (2008) |
| F. miscanthi | F. miscanthi | FnewSC | MER | Gams et al. (1999) | √ | n/a | n/a | Shang et al. (2020) | n/a | n/a | n/a | n/a |
| F. nisikadoi | F. nisikadoi | FnewSC | n/a | Nirenberg (1997) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Nirenberg (1997) |
| F. commune | F. commune | FNSC | MSR; RBD | Skovgaard et al. (2003) | √√ | * | n/a | Xi et al. (2019), present study | √ | √ | n/a | Choi et al. (2018), Husna et al. (2020), Mezzalama et al. (2021) |
| F. carminascens | F. carminascens | FOSC | n/a | Lombard et al. (2019b) | n/a | n/a | n/a | n/a | √ | n/a | n/a | Lombard et al. (2019b) |
| F. cugenangense | F. cugenangense | FOSC | n/a | Maryani et al. (2019a) | * | n/a | n/a | present study | n/a | n/a | n/a | n/a |
| F. fabacearum | F. fabacearum | FOSC | n/a | Lombard et al. (2019b) | n/a | n/a | n/a | n/a | √ | n/a | n/a | Lombard et al. (2019b) |
| F. inflexum | F. inflexum | FOSC | FCR | Schneider & Dalchow (1975) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Tunali et al. (2008) |
| F. languescens | F. languescens | FOSC | n/a | Lombard et al. (2019b) | n/a | n/a | n/a | n/a | √ | n/a | n/a | Lombard et al. (2019b) |
| F. nirenbergiae | F. nirenbergiae | FOSC | MSR | Lombard et al. (2019b) | * | n/a | n/a | present study | n/a | n/a | n/a | n/a |
| F. oxysporum | F. oxysporum, F. aurantiacum, F. oxysporum, F. vasinfectum, F. orthoceras | FOSC | FCR; FHB; MER; MSR; RBD | von Schlechtendal (1824) | n/a | n/a | n/a | n/a | √ | √ | √ | Amatulli et al. (2010), Barkat et al. (2016), Abdul Rahm et al. (2020) |
| F. hostae | F. hostae | FRSC | FCR | Geiser et al. (2001) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Özer et al. (2019) |
| F. redolens | F. redolens, F. oxysporum var. redolens | FRSC | FCR | Wollenweber (1913) | n/a | √ | √ | Tai (1979), Wang et al. (2019b) | √ | n/a | √ | Mulenko et al. (2008), Esmaeili Taheri et al. (2011) |
| F. acaciae-mearnsii | F. acaciae-mearnsii | FSAMSC | n/a | O’Donnell et al. (2004) | n/a | n/a | √ | Liu et al. (2018) | n/a | n/a | √ | Beukes et al. (2017) |
| F. armeniacum | F. armeniacum, F. acuminatum subsp. armeniacum | FSAMSC | FHB; MSR | Burgess et al. (1993) | * | n/a | * | present study | √ | n/a | √ | Burgess et al. (1993), Krone et al. (2020) |
| F. asiaticum | F. asiaticum | FSAMSC | FCR; FHB; MER; MSR; RSR | O’Donnell et al. (2004) | √√ | √√ | √√ | Zhang et al. (2007, 2015, 2016), Hao et al. (2017), Dong et al. (2020b, c), present study | √ | √ | √ | Gomes et al. (2015), Beukes et al. (2017), Khaledi et al. (2017) |
| F. boothii | F. boothii | FSAMSC | FHB; MER; MSR | O’Donnell et al. (2004) | √√ | n/a | n/a | Gai et al. (2017), present study | √ | n/a | √ | Beukes et al. (2017) |
| F. brachygibbosum | F. brachygibbosum | FSAMSC | FCR; MSR | Padwick (1945) | √ | n/a | n/a | Shan et al. (2017) | n/a | n/a | √ | Özer et al. (2020) |
| F. cerealis | F. cerealis, F. crookwellense | FSAMSC | FHB; MSR | Saccardo (1886) | √ | n/a | n/a | Shan et al. (2018) | √ | n/a | √ | Castañares et al. (2019), Xue et al. (2019) |
| F. cortaderiae | F. cortaderiae | FSAMSC | MER; MSR | O’Donnell et al. (2004) | n/a | n/a | n/a | n/a | √ | √ | √ | Gomes et al. (2015), Kuhnem et al. (2016), Beukes et al. (2017) |
| F. culmorum | F. culmorum | FSAMSC | FCR; FHB; MSR; MER | Saccardo (1892) | √ | n/a | √ | Tai (1979), Zhuang (2005), Li et al. (2016), Xu et al. (2016), Xia et al. (2021) | √ | n/a | √ | Aguín et al. (2014), Touati-Hattab et al. (2016) |
| F. gerlachii | F. gerlachii | FSAMSC | FHB | Starkey et al. (2007) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Starkey et al. (2007) |
| F. graminearum | F. graminearum, Gibberella roseum f. cerealis, Gibberella zeae | FSAMSC | FCR; FHB; MSR; MER; RBD; RSR | Schwabe (1839) | √√ | √√ | √√ | Tai (1979), Zhuang (2005), Zhang et al. (2007, 2015), Dong et al. (2020b), present study | √ | √ | √ | Devay et al. (1957), Pennycook (1989), Cho & Shin (2004) |
| F. kyushuense | F. kyushuense | FSAMSC | FHB; MER; MSR | Aoki & O’Donnell (1998) | √ | √ | * | Zhao & Lu (2007), Wang et al. (2014), Cao et al. (2021), present study | n/a | n/a | √ | Sandoval-Denis et al. (2018b) |
| F. langsethiae | F. langsethiae | FSAMSC | n/a | Torp & Nirenberg (2004) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Lukanowski et al. (2008) |
| F. louisianense | F. louisianense | FSAMSC | FHB | Sarver et al. (2011) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Yasuhara-Bell et al. (2018) |
| F. meridionale | F. meridionale | FSAMSC | FHB; MER; MSR; RSR | O’Donnell et al. (2004) | √√ | √ | * | Zhang et al. (2014b), Gai et al. (2017), Dong et al. (2020a), present study | √ | √ | √ | Gomes et al. (2015), Kuhnem et al. (2016), Khaledi et al. (2017) |
| F. nepalense | F. nepalense | FSAMSC | RSR | Sarver et al. (2011) | n/a | n/a | n/a | n/a | n/a | √ | n/a | Sarver et al. (2011) |
| F. nodosum | F. nodosum | FSAMSC | n/a | Lombard et al. (2019c) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Lombard et al. (2019c) |
| F. poae | F. poae | FSAMSC | FHB; MER | Wollenweber (1914) | √√ | n/a | √√ | Tai (1979), Xu et al. (2020), present study | √ | n/a | √ | Logrieco et al. (2002a), Yli-Mattila et al. (2004) |
| F. pseudograminearum | F. pseudograminearum | FSAMSC | FCR; FHB | Aoki & O’Donnell (1999) | * | n/a | √√ | Li et al. (2012), Xu et al. (2015), Zhang et al. (2015), Ji et al. (2016), present study | √ | n/a | √ | Obanor et al. (2010), Abdallah-Nekache et al. (2019) |
| F. sambucinum | F. sambucinu, Gibberella pulicaris, F. roseum | FSAMSC | n/a | Link (1809), Gams (1997) | n/a | n/a | n/a | n/a | √ | √ | √ | Mulenko et al. (2008), Chehri et al. (2010) |
| F. sporotrichioides | F. sporotrichioides, F. sporotrichioides var. sporotrichioides | FSAMSC | FHB; MER; RSR | Sherbakoff (1915) | √ | n/a | n/a | Wang et al. (2020) | √ | √ | √ | Tekauz et al. (2004), Amatulli et al. (2010), Wang et al. (2020) |
| F. venenatum | F. venenatum | FSAMSC | FCR | Nirenberg (1995) | n/a | n/a | n/a | n/a | √ | n/a | √ | Sandoval-Denis et al. (2018a, b) |
| F. vorosii | F. vorosii | FSAMSC | FHB; MSR; MER | Starkey et al. (2007) | * | n/a | n/a | present study | √ | √ | √ | Starkey et al. (2007), Lee et al. (2016) |
| F. acuminatum | F. acuminatum, F. scirpi, F. scirpi var. acuminatum | FTSC | FCR; FHB; MSR | Sherbakoff (1915) | * | n/a | √√ | Pan et al. (2015), Zhang et al. (2015), present study | √ | √ | √ | Desjardins et al. (2000), Mao et al. (1998), Shrestha et al. (2021) |
| F. avenaceum | F. avenaceum, Gibberella avenacea | FTSC | FCR; FHB; MER | Saccardo (1886) | √ | n/a | √√ | Zhuang (2005), Zhang et al. (2013, 2015), Ma et al. (2019), present study | √ | √ | √ | Logrieco et al. (2002b); Amatulli et al. (2010) |
| F. flocciferum | F. flocciferum | FTSC | n/a | Sturm (1829) | n/a | n/a | n/a | n/a | √ | n/a | √ | Gerlach & Ershad (1970), Sandoval-Denis et al. (2018a) |
| F. torulosum | F. torulosum | FTSC | n/a | Nirenberg (1995) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Kristensen et al. (2005) |
| F. tricinctum | F. tricinctum | FTSC | n/a | Saccardo (1886) | n/a | √ | n/a | Li et al. (2019b) | √ | n/a | √ | Mulenko et al. (2008), Castañareset al. (2011) |
| F. trichothecioides | F. trichothecioides | n/a | FCR | Jamieson & Wollenweber (1912) | n/a | n/a | n/a | n/a | n/a | n/a | √ | Chehri et al. (2010) |
1 Abbreviations: F. = Fusarium. FburSC = F. burgessii species complex. FCSC = F. chlamydosporum species complex. FCOSC = F. concolor species complex. FFSC = F. fujikuroi species complex. FHSC = F. heterosporum species complex. FIESC = F. incarnatum-equiseti species complex. FLSC = F. lateritium species complex. FnewSC = F. newnesense species complex. FNSC = F. nisikadoi species complex. FOSC = F. oxysporum species complex. FRSC = F. redolens species complex. FSAMSC = F. sambucinum species complex. FTSC = F. tricinctum species complex. FCR = Fusarium crown rot of wheat. FHB = Fusarium head blight of wheat. MSR = maize stalk rot. MER = maize ear rot. RBD = Rice bakanae disease. RSR = Rice spikelet rot. Maize = Zea mays. Rice = Oryza sp. Wheat =Triticum sp.
2 n/a: No record found. √: Species previously reported from cereals in literatures. √√: Species also reported on cereals in the present study. *: New records on cereals in China or updated records on cereals by re-identification. ?: The records on cereals need to be further confirmed.
3 If there are more than three references for a particular record, only several representative references were listed here.
Taxonomy
Fusarium fujikuroi species complex (FFSC)
As the largest and best-studied species complex in the genus Fusarium, the FFSC comprises 84 described species separated into five major clades (African clade, African clade B, African clade C, American clade, and the Asian clade) (Yilmaz et al. 2021, Wang et al. 2022a). This species complex includes many important plant pathogens that cause various diseases and produce numerous mycotoxins (Choi et al. 2018, Duan et al. 2020, Qiu et al. 2020, Yilmaz et al. 2021). The FFSC can be distinguished from other complexes by its typical Fusarium macroconidia, abundant microconidia and rare chlamydospore formation. In the present study, 227 strains clustered into three major clades (African clade, American clade, and Asian clade) and 12 subclades, representing nine known and three novel species (Fig. 8).
Fusarium annulatum Bugnic., Rev. Gén. Bot. 59: 13. 1952.
Materials examined: (See Supplementary Table S1).
Notes: Fusarium annulatum was proposed by Bugnicourt (1952), with CBS 258.54 as ex-type strain. Later, a phylogenetic analysis based on LSU-SSU-tub2 suggested that CBS 258.54 clustered with the representative strains (CBS 217.76, NRRL 25089, etc.) of F. proliferatum (O’Donnell et al. 1998a). Given that F. annulatum was rarely reported and has only one collection (CBS 258.54), whereas F. proliferatum represented by CBS 217.76 has been widely used, O’Donnell et al. (1998a) recommended treating F. annulatum as synonym of F. proliferatum, but did not formalise the proposal. Recently, a multi-locus phylogenetic analyses including the epitype of F. proliferatum (CBS 480.96) and additional sequences of CBS 258.54 (CaM, rpb1, rpb2, tef1) demonstrated that CBS 217.76 clustered together with CBS 258.54, but clustered distant from CBS 480.96. Therefore, CBS 217.76 and CBS 480.96 represent two distinct species. In the current study, based on the taxonomic treatment of Yilmaz et al. (2021), we further confirmed that many cereals pathogenic strains (including F1, P2, 24ALH, 51ALH, Azerbaijan 71-3) previously identified as F. proliferatum (Wulff et al. 2010, Okello et al. 2019, Özer et al. 2020), are actually F. annulatum (Fig. 8). Meanwhile, our results also showed that F. annulatum has a broad host range and geographical distribution (Fig. 16C). In addition, our inoculation test showed its high pathogenicity to maize seedlings (DSI = 68.75 % when CC = 1 × 106 conidia/mL) (Fig. 7E).
Fusarium awaxy Petters-Vandresen et al., Persoonia 43: 363. 2019.
Material examined: (See Supplementary Table S1).
Notes: Fusarium awaxy was originally described from rotten stalks of Zea mays in Brazil (Crous et al. 2019), and we herein reported it for the first time from China (Fig. 8). This species is closely related to the cosmopolitan species F. subglutinans (Figs 8, 14), and both species have been confirmed for causing maize stalk rot (Leslie & Summerell 2006: Fig. 7H). Morphologically, they differ from each other in their sporodochia colour (Crous et al. 2019). In addition, F. subglutinans has been reported for causing human keratitis (Al-Hatmi et al. 2014), but F. awaxy does not grow above 32 °C, suggesting the inability to cause human infection (Crous et al. 2019).
Fusarium concentricum Nirenberg & O’Donnell, Mycologia 90: 442. 1998.
Material examined: (See Supplementary Table S1).
Notes: Fusarium concentricum was originally described from Musa sapientum in Costa Rica, and from Nilaparvata lugens in Korea (Nirenberg & O’Donnell 1998). It has since been reported from other hosts, including maize, rice and wheat from central America and Asia (Aoki et al. 2001, Choi et al. 2018, Du et al. 2020). The pathogenicity of this species in causing maize ear rot, rice bakanae disease and maize stalk rot has been confirmed by Du et al. (2020), Choi et al. (2018) and this study (Fig. 7I).
Fusarium elaeagni M.M. Wang & L. Cai, Persoonia 48: 35 2022.
Material examined: (See Supplementary Table S1).
Notes: Fusarium elaeagni was originally described from Elaeagnus pungens (Wang et al. 2022a), and subsequently from rice in this study. This species is closely related to F. fujikuroi (Figs 8, 14) but could be distinguished by morphological characters, i.e., sporodochial colour, macroconidial septa, microconidial shape, and the type of aerial phialides (Wang et al. 2022a).
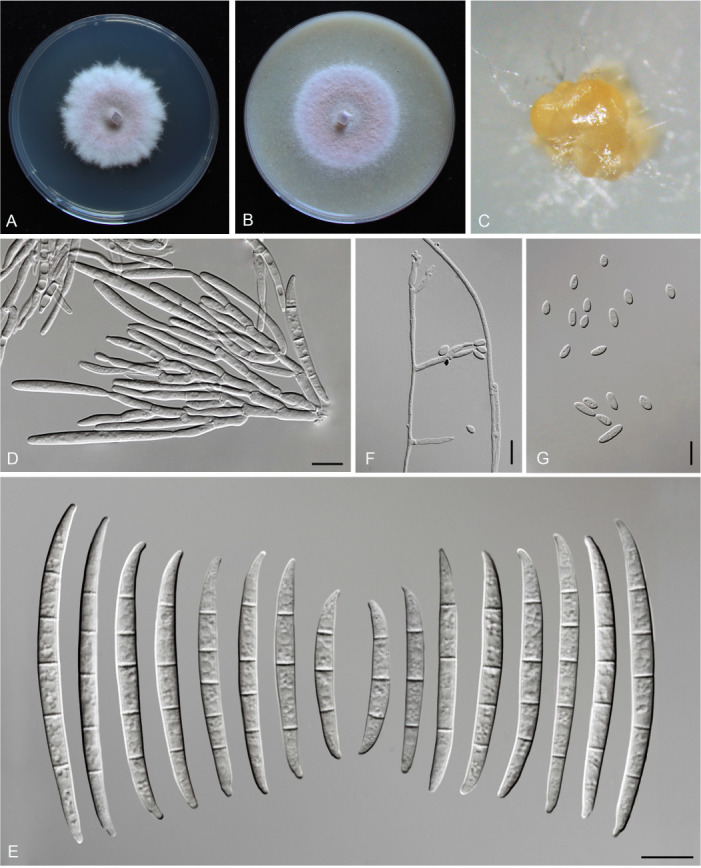
Fusarium erosum S.L. Han, M.M. Wang & L. Cai, sp. nov. MycoBank MB 847021. Fig. 18.
Fig. 18.
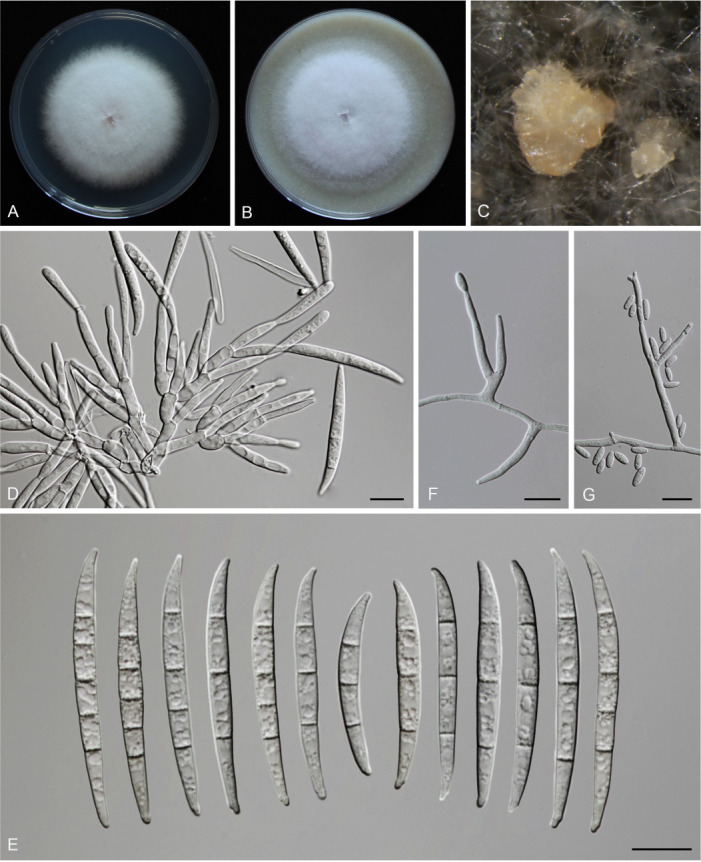
Morphology of Fusarium erosum (CGMCC3.23518, ex-type culture). A. Colony on PDA. B. Colony on OA. C. Sporodochia. D. Sporodochial conidiophores and phialides. E. Sporodochial conidia. F. Aerial conidiophores and phialides. G. Phialides and aerial conidia. Scale bars = 10 μm.
Etymology: Refers to its colony margin on PDA, erose.
Sporodochia cream (4A3), formed frequently on carnation leaves, and often covered by aerial sparse mycelium. Sporodochial conidiophores densely, irregularly and verticillate branched, bearing apical whorls of 2–4 phialides. Sporodochial phialides subulate to subcylindrical, smooth, thin-walled, 12.4–19.8 × 2.0–3.8 μm. Sporodochial conidia falcate, slender, straight to slightly curved dorsiventrally, with a curved to pointed apical cell and a well-developed foot-shaped basal cell, hyaline, thin- and smooth-walled, 3(–5)-septate: 3-septate conidia: 29.3–46.6 × 3.5–4.7 μm (av. ± SD: 39.8 ± 5.8 × 4.0 ± 0.4 μm); 4-septate conidia: 42.7–50.6 × 3.5–4.7 μm (av. ± SD: 46.4 ± 2.3 × 4.0 ± 0.3 μm); 5-septate conidia: 39.4–50.5 × 3.5–5.1 μm (av. ± SD: 46.8 ± 2.8 × 4.2 ± 0.4 μm). Aerial conidiophores borne on aerial mycelium, straight or flexuous, erect or prostrate, smooth- and thin-walled, bearing terminal or lateral phialides. Aerial phialide mono- and polyphialides, subulate to subcylindrical, smooth- and thin-walled, periclinal thickening inconspicuous or absent, 9.7–32.1 × 1.9–3.5 μm. Aerial conidia single on the tips of phialides, ovoid, hyaline, smooth- and thin-walled, aseptate, 3.7–11.3 × 1.8–3.5 μm (av. ± SD: 7.2 ± 1.7 × 2.7 ± 0.4 μm). Chlamydospores not observed.
Culture characteristics: Colonies on PDA incubated at 25 °C in the dark reaching 72–82 mm diam in 7 d; convex, with abundant aerial mycelium, colony margin lightly erose; surface purplish white (14A2), reverse pinkish white (10A2) in the centre, white (–A1) at the margin; odour absent. On OA in the dark reaching 66–67 mm in 7 d; convex, with abundant aerial mycelium, margin entire; surface pinkish white (10A2), reverse greyish red (10C4); odour absent.
Typus: China, Guangdong Province, Meizhou City (E116.3, N24.52), from the symptomatic tissues of maize stalk rot, 7 Jun. 2021, Y.M. Wu (holotype HMAS 351950, ex-type living culture CGMCC3.23518 = LC15877 = HSL2912).
Additional materials examined: (See Supplementary Table S1).
Notes: The three isolates representing F. erosum were resolved as a strongly supported genealogically exclusive lineage in the phylogeny inferred from combined CaM, rpb1, rpb2, tef1, and tub2 loci (Fig. 8). Fusarium erosum is closely related to F. siculi and F. globosum, but differs by 20 bp and 12 bp from the latter two species respectively in the combined dataset. Morphologically, F. erosum differs in the types of aerial conidia production and the number of septa. For example, F. erosum produces aseptate, ovoid aerial conidia; F. siculi produces 0–1-septate, subcylindrical to clavate aerial conidia; while three types of aerial conidia were found in F. globosum: clavate with a truncate base (0–3-septate), napiform/pyriform, and globose (0–1-septate) which often have a distinct papilla (Rheeder et al. 1996, Leslie & Summerell 2006, Sandoval-Denis et al. 2018a). The pathogenicity test fulfilling Koch’s postulates confirmed its pathogenicity, causing maize stalk rot (Fig. 7J).
Fusarium fujikuroi Nirenberg, Mitt. Biol. Bundesanst. Land-Forstw. Berlin-Dahlem 169: 32. 1976.
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: As a pathogen causing rice bakanae disease, F. fujikuroi has been widely studied, but some are under different synonyms (e.g., F. moniliforme var. subglutinans, F. moniliforme var. intermedium, F. moniliforme and Gibberella fujikuroi) based on previous morphological identification systems (Booth 1971). With the application of multi-locus analyses, F. fujikuroi was gradually revealed to be a pathogen of various crops worldwide (Farr & Rossman 2022), causing maize ear rot, maize stalk rot, wheat scab, wheat crown rot and rice spikelet rot (Wulff et al. 2010, Duan et al. 2020, Qiu et al. 2020, this study).
Fusarium planum S.L. Han, M.M. Wang & L. Cai, sp. nov. MycoBank MB 847022. Fig. 19.
Fig. 19.
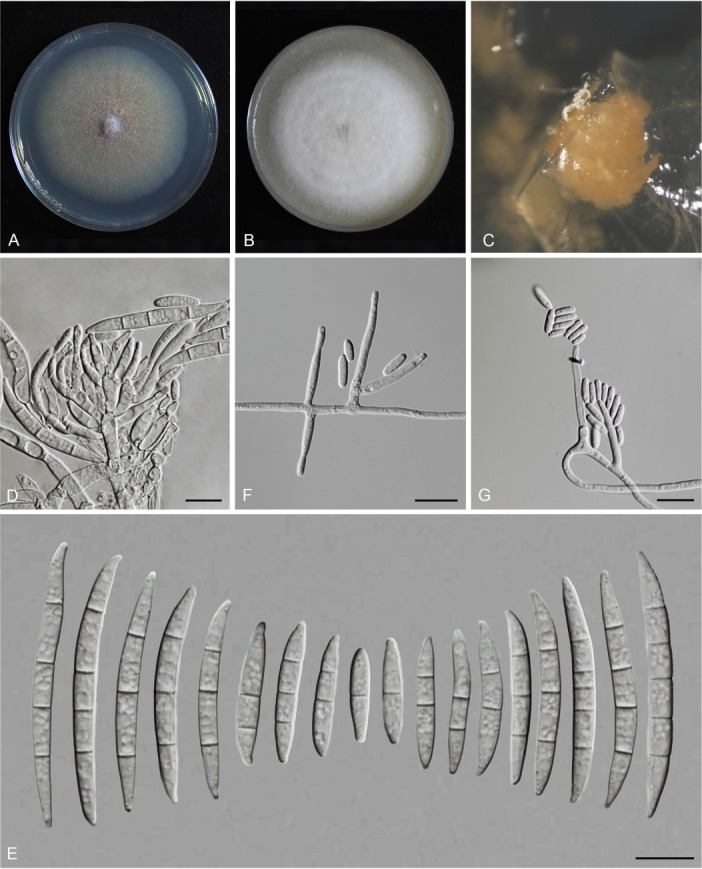
Morphology of Fusarium planum (CGMCC3.23517, ex-type culture). A. Colony on PDA. B. Colony on OA. C. Sporodochia. D. Sporodochial conidiophores and phialides. E. Sporodochial conidia. F. Aerial conidiophores and phialides. G. Aerial conidia. Scale bars = 10 μm.
Etymology: Referring to its colony elevation in PDA, flat.
Sporodochia cadmium orange (5A8), formed infrequently inside agar. Sporodochial conidiophores densely and irregularly branched, bearing apical whorls of 3–5 phialides. Sporodochial phialides subulate to subcylindrical, smooth, thin-walled, 9.0–17.5 × 2.3–3.9 μm. Sporodochial conidia falcate, slender, straight to slightly curved dorsiventrally, with a conical to slightly papillate apical cell and obtuse to barely notched basal cell, hyaline, thin- and smooth-walled, (1–)3–4(–5)-septate: 1-septate conidia: 14.4–22.1 × 3.1–3.9 μm (av. ± SD: 17.6 ± 2.3 × 3.5 ± 0.3 μm); 2-septate conidia: 21.5–28.5 × 3.2–4.3 μm (av. ± SD: 25 ± 2.4 × 3.7 ± 0.4 μm); 3-septate conidia: 25–43 × 3.0–4.5 μm (av. ± SD: 35.8 ± 5.7 × 3.6 ± 0.4 μm); 4-septate conidia: 41.8–49.4 × 3.3–5.0 μm (av. ± SD: 44.5 ± 2.8 × 4.2 ± 0.5 μm); 5-septate conidia: 40.5–48.3 × 3.4–4.8 μm (av. ± SD: 43.7 ± 4.1 × 4.2 ± 0.5 μm). Aerial conidiophores borne on aerial mycelium, straight or flexuous, erect or prostrate, smooth- and thin-walled, bearing terminal or lateral phialides. Aerial phialides monophialide, subulate to subcylindrical, smooth- and thin-walled, periclinal thickening inconspicuous or absent, 12.6–25 × 2–2.8 μm. Aerial conidia often forming false heads, subcylindrical to clavate, hyaline, smooth- and thin-walled, aseptate, 5–10.6 × 1.7–2.3 μm (av. ± SD: 7.7 ± 1.3 × 2.1 ± 0.2 μm). Chlamydospores not observed.
Culture characteristics: Colonies on PDA incubated at 25 °C in the dark reaching 68–70 mm diam in 7 d; flat, aerial mycelia scant, colony margin entire; surface greyish orange (6B5) in the centre, pale (2A2) at the margin; reverse peach (7A4) in the centre, pale (2A2) at the margin; odour absent. On OA in the dark reaching 69–71 mm in 7 d; convex, with abundant aerial mycelium, margin entire; surface white (–A1), reverse pastel pink (11A4) in the centre, white (–A1) at the margin; odour absent.
Typus: China, Guangdong Province, Qingyuan City (E113.42, N24.19), from the symptomatic tissues of maize ear rot, 21 Nov. 2020, S.Q. Wang (holotype HMAS 351949, ex-type living culture CGMCC3.23517 = LC15876 = HSL2645).
Additional materials examined: (See Supplementary Table S1).
Notes: The isolates representing F. planum were resolved as a strongly supported genealogically exclusive lineage in the phylogeny inferred from combined CaM, rpb1, rpb2, tef1, and tub2 loci (Fig. 8). Fusarium planum is closely related to F. andiyazi, F. madaense and F. mirum, but differs by 23 bp and 31 bp from F. andiyazi and F. madaense in the 5-locus (CaM-rpb1-rpb2-tef1-tub2) dataset and 11 bp from F. mirum in the 3-locus (rpb2-tef1-tub2) dataset (CaM, rpb1 and the latter half of rpb2 sequence were not available for F. mirum). Morphologically, this species is distinguished based on the number of septa in sporodochial conidia (1–5-septate in F. planum vs 3–6-septate in F. andiyazi, 0–6-septate in F. madaense, 1–6-septate in F. mirum); and the number of septa in aerial conidia (aseptate in F. planum vs 0–2-septate in F. andiyazi and F. mirum, 0–3-septate in F. madaense) (Marasas et al. 2001, Ezekiel et al. 2020, Costa et al. 2022). The pathogenicity test fulfilling the Koch’s postulates confirmed its pathogenicity and ability to cause maize stalk rot (Fig. 7Q).
Fusarium sacchari (E.J. Butler) W. Gams, Cephalosporium-artige Schimmelpilze: 218. 1971.
Basionym: Cephalosporium sacchari E.J. Butler, Mem. Dept. Agric. India, Bot. Ser. 6: 185. 1913.
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: Fusarium sacchari is a common pathogen of diverse crops worldwide (Farr & Rossman 2022). In China, its correlation with maize ear rot, wheat scab, rice spikelet rot and maize stalk rot was confirmed by Duan et al. (2019), Wang et al. (2015) and this study (Fig. 7R).
Fusarium sanyaense S.L. Han, M.M. Wang & L. Cai, sp. nov. MycoBank MB 847023. Fig. 20.
Fig. 20.
Morphology of Fusarium sanyaense (CGMCC3.23523, ex-type culture). A. Colony on PDA. B. Colony on OA. C. Sporodochia. D. Sporodochial conidiophores and phialides. E. Sporodochial conidia. F. Aerial conidiophores and phialides. G. Aerial conidia. Scale bars = 10 μm.
Etymology: Named after the city, Sanya, where the holotype was collected.
Sporodochia champagne (4B4), formed frequently on carnation leaves, and infrequently on agar. Sporodochial conidiophores densely and irregularly branched, bearing apical whorls of 2–3 phialides. Sporodochial phialides subulate to subcylindrical, smooth, thin-walled, 9.3–19 × 2.5–4.3 μm. Sporodochial conidia falcate, slightly slender, straight to slightly curved dorsiventrally, with a conical to slightly papillate apical cell and a well-developed and foot-shaped basal cell, hyaline, thin- and smooth-walled, 3–5-septate: 3-septate conidia: 25.3–50.6 × 2.7–4.3 μm (av. ± SD: 38.3 ± 6.2 × 3.6 ± 0.5 μm), 4-septate conidia: 42.4–61.1 × 2.6–5.1 μm (av. ± SD: 50.6 ± 4.8 × 3.9 ± 0.6 μm), 5-septate conidia: 45.7–67.2 × 3.4–4.5 μm (av. ± SD: 55.3 ± 5.2 × 3.9 ± 0.4 μm). Aerial conidiophores borne on aerial mycelium, straight or flexuous, erect or prostrate, smooth- and thin-walled, bearing terminal or lateral phialides. Aerial phialides mono- and polyphialides, subulate to subcylindrical, smooth- and thin-walled, periclinal thickening inconspicuous or absent, 11–27.6 × 2.2–3.4 μm. Aerial conidia single on the tips of phialides, ovoid, hyaline, smooth- and thin-walled, aseptate: 4.4–11 × 1.9–4.2 μm (av. ± SD: 6.5 ± 1.6 × 2.7 ± 0.5 μm). Chlamydospores not observed.
Culture characteristics: Colonies on PDA incubated at 25 °C in the dark reaching 46–55 mm diam in 7 d; raised, aerial mycelia lightly scant, colony margin undulate; surface flamingo (12A4) in the centre, white (–A1) at the margin, reverse greyish ruby (12D5) in the centre, white (–A1) at the margin; odour absent. On OA in the dark reaching 47–58 mm in 7 d; flat, aerial mycelia scant, margin undulate; surface amaranth (14E7) in the centre, reverse orange white (5A2); odour absent.
Typus: China, Hainan Province, Sanya City (E109.75, N18.39), from the symptomatic tissues of maize stalk rot, 6 Feb. 2021, Y.J. Li (holotype HMAS 351955, ex-type living culture CGMCC3.23523 = LC15882 = HSL2737).
Additional materials examined: (See Supplementary Table S1).
Notes: The three isolates representing F. sanyaense were resolved as a strongly supported genealogically exclusive lineage in the phylogeny inferred from combined CaM, rpb1, rpb2, tef1, and tub2 loci (Fig. 8). Fusarium sanyaense is closely related to F. lumajangense, F. mangiferae and F. proliferatum, but differed by 18 bp from F. mangiferae in the 5-locus (CaM-rpb1-rpb2-tef1-tub2) dataset, 13 bp from F. lumajangense in the 3-locus (rpb2-tef1-tub2) dataset (CaM and rpb1 sequences are not available for F. lumajangense) and 10 bp from F. proliferatum in the 4-locus (CaM-rpb2-tef1-tub2) dataset (rpb1 sequence is not available for F. proliferatum). Morphologically, they could be distinguished based on the size of sporodochial conidia (25.3–67.2 × 2.7–4.5 μm in F. sanyaense vs 30.0–56.0 × 3.0–4.5 μm in F. lumajangense, 43.1–61.4 × 1.9–3.4 μm in F. mangiferae, and 16.5–60.5 × 1.5–4 μm in F. proliferatum), and the number of septa in sporodochial conidia (3–5-septate in F. sanyaense, F. lumajangense and F. madaense vs 1–4-septate in F. proliferatum) (Britz et al. 2002, Maryani et al. 2019b, Yilmaz et al. 2021). Pathogenicity tests fulfilling the Koch’s postulates confirmed its ability to cause maize stalk rot (Supplementary Fig. 7S).
Fusarium subglutinans (Wollenw. & Reinking) P.E. Nelson et al., Fusarium species. An illustrated manual for identification: 135. 1983.
Basionym: Fusarium moniliforme var. subglutinans Wollenw. & Reinking, Phytopathology 15: 163. 1925.
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: Fusarium subglutinans is known for causing cereal, animal and human diseases (e.g., maize ear rot, animal rapid death and human keratitis) and producing various mycotoxins (e.g., moniliformin, fusaproliferin) (Kriek et al. 1977, Bacon & Hinton 1996, Lew et al. 1996, Al-Hatmi et al. 2014, Farr & Rossman 2022). Despite its widely recognised threats to agricultural and public health, its taxonomy was unstable as it lacked a living ex-type culture and holotype specimen, which was resolved by Yilmaz et al. (2021), who designated CBS 747.97 as its neotype. Twelve strains obtained from this study clustered together with the neotype, of which one strain was isolated from wheat, as a new host record in China (Table 2).
Fusarium temperatum Scaufl. & Munaut, Mycologia 103: 593. 2011.
Material examined: (See Supplementary Table S1).
Notes: Fusarium temperatum was first reported as a pathogen on maize in Belgium (Scauflaire et al. 2011), and subsequently on humans in Mexico (Al-Hatmi et al. 2014). In China, its ability to cause maize ear rot and maize stalk rot have been confirmed by Zhang et al. (2014c) and Liu et al. (2022b). Notably, the existence of sexual reproduction of F. temperatum in the field (Scauflaire et al. 2011), together with the semipermeable species boundary with its relative species F. subglutinans (Fumero et al. 2021), may facilitate the genetic recombination and further contribute to the high genetic diversity of F. temperatum. This species appears to be highly adaptable and would potentially be widely distributed.
Fusarium verticillioides (Sacc.) Nirenberg, Mitt. Biol. Bundesanst. Land-Forstw. 169: 26. 1976.
Basionym: Oospora verticillioides Sacc., Fung. Ital., Fasc. 17–28: pl. 879. 1881.
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: Fusarium verticillioides was first isolated from maize in Italy as Oospora verticillioides (Saccardo 1881). The controversy on the taxonomy of this species was recently summarised and resolved (Yilmaz et al. 2021). Fusarium verticillioides is cosmopolitan and notorious, being predominately a pathogen of maize, causing a massive quality and yield reduction (Murillo-Williams & Munkvold 2008, Blacutt et al. 2018, Schoeman et al. 2018). In our study, Fusarium verticillioides is also one of the dominant species on maize, with a total of 88 representative strains from diseased maize and from 88 locations (Supplementary Table S1).
Fusarium incarnatum-equiseti species complex (FIESC)
Species within the FIESC are cosmopolitan in various ecological types. The species relationships within this group have only recently been clarified. Previously, members of this group were in most cases identified as F. equiseti or F. incarnatum, based on morphological similarities or ITS/tef1 sequences (Khoa et al. 2004, Leslie & Summerell 2006, Marín et al. 2012). With the application of genealogical concordance phylogenetic species recognition, FIESC has been revealed to include 32 phylogenetic species separated in the Incarnatum and Equiseti clades (O’Donnell et al. 2009, 2012, Villani et al. 2016). By employing morphological characters and multi-locus phylogenetic relationships, the species within the FIESC were clarified and delimitated, and the majority of the cryptic phylo-species have been provided with Latin binomials (Wang et al. 2019a, Xia et al. 2019). Furthermore, our phylogenomic tree (Fig. 14) support the merger of FCAMSC into the FIESC (as the Camptoceras clade). Thus, the FIESC now includes 44 species, which is characterised by a dorsiventral curvature of its macroconidia, abundant chlamydospores, and mostly lacking microconidia (Wang et al. 2019a). To date, about half of the species in this complex have been reported from cereals (Table 2). However, the pathogenicity of several species remains suspicious due to uncertainty of the identities of isolates used, e.g., F. incarnatum (Fig. 9). In this study, 147 strains clustered into 19 distinct clades (13 in Incarnatum clade, five in Equiseti clade, and one in Camptoceras clade), representing 14 known and five novel species (Fig. 9).
Fusarium arcuatisporum M.M. Wang et al., Persoonia 43: 78. 2019.
Material examined: (See Supplementary Table S1).
Notes: Fusarium arcuatisporum was first described based on isolation from Nelumbo nucifera, Brassica campestris, and Oryza sp. from China (Wang et al. 2019a). Previously, this species has been isolated from a human toenail, as an unnamed phylogenetic species (FIESC 7) (Leslie & Summerell 2006). Here we update a new host record (maize), and confirm its ability to cause maize stalk rot (Fig. 7F).
Fusarium clavus J.W. Xia et al., [as ‘clavum’], Persoonia 43: 199. 2019.
Material examined: (See Supplementary Table S1).
Notes: Fusarium clavus was first recognised by O’Donnell et al. (2009) as an unnamed phylogenetic species (FIESC 5), and later formally named and described by Xia et al. (2019). To date, F. clavus has been reported from humans, insects, plants and various environments (Xia et al. 2019, Matic et al. 2020). Notably, several isolates of this species have been reported from maize (Okello et al. 2019) and wheat (Özer et al. 2020), but inaccurately identified as FIESC or F. equiseti, based on tef1 phylogenetic analysis (Okello et al. 2019, Özer et al. 2020, Fig. 9). These inaccurate identification results are common, because species within the FIESC could not be distinguished based on only tef1 sequences, especially when using the previously wrongly named reference sequences from databases. In this study, we reported it as a new Chinese record.
Fusarium compactum (Wollenw.) Raillo, Fungi of the Genus Fusarium: 180. 1950.
Basionym: Fusarium scirpi var. compactum Wollenw., Fusaria Autogr. Delin. 3: no. 924. 1930.
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: The phylogenetic relationship of Fusarium compactum was revealed by O’Donnell et al. (2009), which represents a well-supported phylogenetic species (FIESC 3). Later, Xia et al. (2019) designated an epitype for F. compactum to stabilise the use of this name. To date, this species has been reported from at least 18 substrates/hosts, e.g., Austrostipa aristiglumis, Gossypium barbadense, Triticum aestivum (Bentley et al. 2007, Tunali et al. 2008, Schroers et al. 2011), and maize (this study).
Fusarium fecundum S.L. Han, M.M. Wang & L. Cai, sp. nov. MycoBank MB 847024. Fig. 21.
Fig. 21.
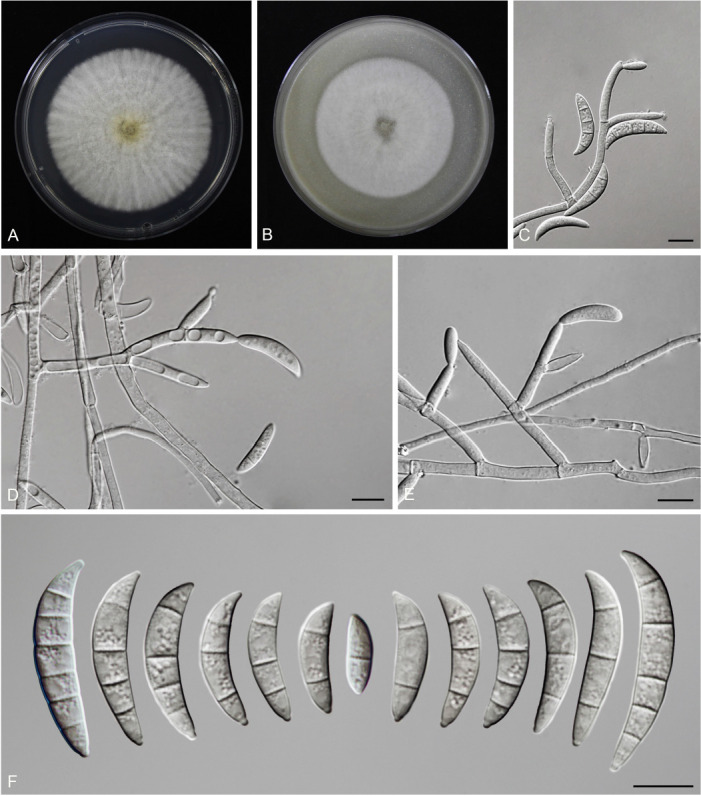
Morphology of Fusarium fecundum (CGMCC3.23516, ex-type culture). A. Colony on PDA. B. Colony on OA. C–E. Aerial conidiophores and phialides. F. Aerial conidia. Scale bars = 10 μm.
Etymology: Refers to its high fecundity of aerial conidia in CLA.
Sporodochia not observed. Aerial conidiophores borne on aerial mycelium, straight or flexuous, erect or prostrate, smooth- and thin-walled, bearing terminal or lateral phialides. Aerial phialides mono- and polyphialides, subulate to subcylindrical, smooth- and thin-walled, periclinal thickening inconspicuous or absent, 8–44 × 2.5–3.7 μm. Aerial conidia falcate, the dorsal side curved than the ventral; with a blunt, conical to pointed apical cell and papillate basal cell, hyaline, smooth- and thin-walled, (1–)2–4(–6)-septate: 1-septate conidia: 13–14 × 3.5–4 μm (av. ± SD: 13.6 ± 0.4 × 3.8 ± 0.2 μm); 2-septate conidia: 12.6–26 × 3.6–5.4 μm (av. ± SD: 18 ± 4.6 × 4.3 ± 0.6 μm); 3-septate conidia: 24.1–33.1 × 4.5–6.5 μm (av. ± SD: 29.6 ± 2.5 × 5.5 ± 0.7 μm); 4-septate conidia: 30.9–38.5 × 5.4–5.9 μm (av. ± SD: 33.4 ± 3.1× 5.7 ± 0.2 μm); 5-septate conidia: 30.5–40.1 × 5.4–6.4 μm (av. ± SD: 36.7 ± 5.8 × 6.0 ± 0.4 μm); 6-septate conidia: 33.6–35.8 × 6.1–6.8 μm (av. ± SD: 34.4 ± 1 × 6.4 ± 0.3 μm). Chlamydospores not observed.
Culture characteristics: Colonies on PDA incubated at 25 °C in the dark reaching 84–90 mm diam in 7 d; raised, felty to velvety, radiate, with abundant aerial mycelia, colony margin entire; surface greyish yellow (2B5) in the centre, white (–A1) at the margin; reverse milk white (1A2) in the centre, white (–A1) at the margin; odour absent. On OA in the dark reaching 76–84 mm in 7 d; raised, felty to dusty, with abundant aerial mycelium, margin entire; surface white, reverse mimosa (2B8); odour absent.
Typus: China, Shaanxi Province, Hanzhong city (E107.632; N33.211), obtained from the symptomatic tissues of wheat scab, 13 May 2021, Y.J. Chen (holotype HMAS 351948, ex-type living culture CGMCC3.23516 = LC15875 = HSL1587).
Additional materials examined: (See Supplementary Table S1).
Notes: The three isolates representing F. fecundum were resolved as a strongly supported genealogically exclusive lineage in the phylogenies inferred from combined CaM, rpb2, and tef1 loci (Fig. 9), and genomic datasets (Fig. 14). Phylogenetically, F. fecundum is closely related to the species within the previous FCAMSC (F. kotabaruense, F. camptoceras and F. neosemitectum), but differs by 83 bp, 84 bp and 84 bp in the combined dataset, respectively. Morphologically, this species is distinguishable from closely related species based on its conidial size (13–40.1 × 3.5–6.8 μm in F. fecundum vs 21–45 ×5–7.5 μm in F. kotabaruense, 15–51 × 4–7 μm in F. camptoceras, 17–41 × 3–6 μm in F. neosemitectum) and the number of conidial septa (1–6-septate in F. fecundum vs 2–7-septate in F. kotabaruense, 0–7-septate in F. camptoceras, 1–5-septate in F. neosemitectum) (Marasas et al. 1998, Maryani et al. 2019b, Xia et al. 2019).
Fusarium guilinense M.M. Wang et al., Persoonia 43: 80. 2019.
New synonym: Fusarium bubalinum J.W. Xia et al., Persoonia 43: 199. 2019.
Material examined: (See Supplementary Table S1).
Notes: Fusarium guilinense was first reported from China from leaves of Musa nana (Wang et al. 2019a), and in this study from rice glumes (Supplementary Table S1). In addition, Xia et al. (2019) in their analyses, did not include the ex-type isolate of F. guilinense (also F. caatingaense, F. citri, F. hainanense, F. humuli, F. ipomoeae, F. irregulare, F. luffae, F. nanum and F. pernambucanum), thus could not reflect on species relationships in this group. Here our multi-locus phylogenetic analyses clearly showed that “F. bubalinum” (Xia et al. 2019) clustered within the F. guilinense clade (Fig. 9). Therefore, we consider F. bubalinum as a synonym of F. guilinense.
Fusarium hainanense M.M. Wang et al., Persoonia 43: 82. 2019.
Material examined: (See Supplementary Table S1).
Notes: Fusarium hainanense was first reported from Musa nana and Oryza sp. (Wang et al. 2019a), and subsequently from Acacia sp., Musa acuminata, Oryza australiensis (Xia et al. 2019), and maize (this study). Three isolates (JS21, JS3, JS9) of rice spikelet rot clustered with the ex-type strain of F. hainanense in our multi-locus analyses (Fig. 9).
Fusarium humuli M.M. Wang et al., Persoonia 43: 83. 2019.
Material examined: (See Table Supplementary S1).
Notes: Fusarium humuli was first reported from China on 12 different hosts, including Humulus scandens (Wang et al. 2019a), and is herein reported from rice and wheat as new host records. Phylogenetically, this species clusters in the Incarnatum clade of FIESC, closely related to F. citri, F. fasciculatum and F. weifangense (Wang et al. 2019a, Xia et al. 2019, this study). Morphologically, these four species could be distinguished based on the morphology of their sporodochial conidia (Wang et al. 2019a, Xia et al. 2019, this study).
Fusarium ipomoeae M.M. Wang et al., Persoonia 43: 83. 2019.
Material examined: (See Table Supplementary S1).
Notes: Fusarium ipomoeae has been isolated from multiple hosts (Wang et al. 2019a, Xia et al. 2019, Zhou et al. 2020, Xu et al. 2021b). Notably, one strain of this species has been reported as a pathogen of maize but previously misidentified as F. equiseti through tef1 BLASTn in GenBank (Li et al. 2014, Fig. 9). Here, we isolated F. ipomoeae from maize stalks rot (Fig. 2E), maize ear rot (Fig. 1A), wheat scab (Fig. 3D) and rice spikelet rot (Fig. 5E), updating two new host (maize and wheat) records (Table 2), and confirmed its pathogenicity to maize (Fig. 7K).
Fusarium jinanense S.L. Han, M.M. Wang & L. Cai, sp. nov. MycoBank MB 847025. Fig. 22.
Fig. 22.
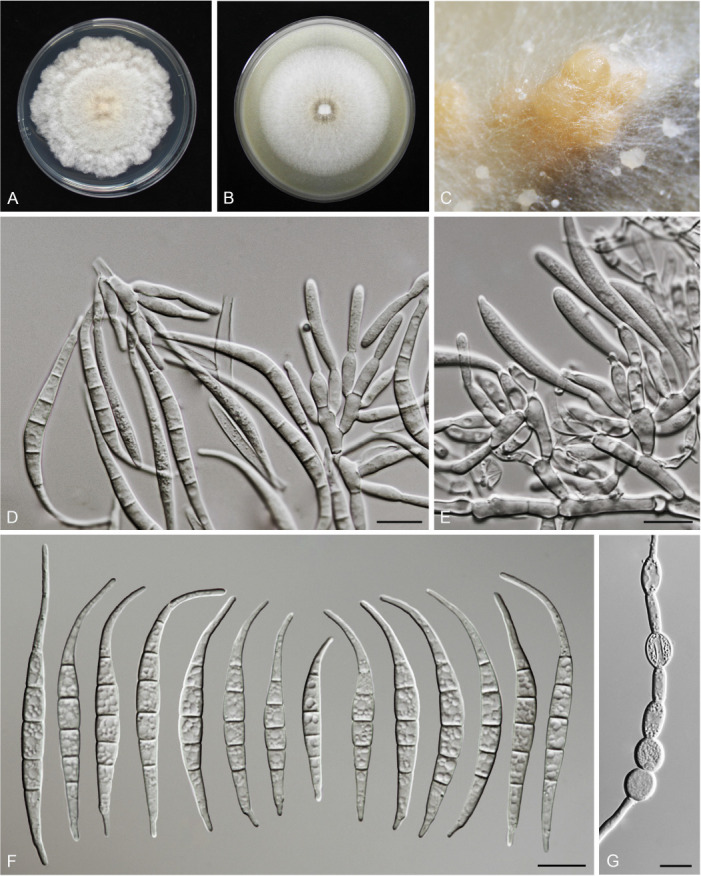
Morphology of Fusarium jinanense (CGMCC3.23519, ex-type culture). A. Colony on PDA. B. Colony on OA. C. Sporodochia. D, E. Sporodochial conidiophores and phialides. F. Sporodochial conidia. G. Chlamydospores. Scale bars = 10 μm.
Etymology: Named after the city, Jinan, where the holotype was collected.
Sporodochia melon (5A6), formed infrequently on carnation leaves or agar. Sporodochial conidiophores densely and irregularly branched, bearing apical whorls of 3–6 phialides. Sporodochial phialides subulate to subcylindrical, smooth, thin-walled, 8–14 × 2.5–4 μm. Sporodochial conidia falcate, curved dorsoventrally, tapering towards both ends, with an elongated or whip-like curved apical cell and a well-developed to elongate foot-shaped basal cell, hyaline, thin- and smooth-walled, (4–)5(–6)-septate: 4-septate conidia: 34.5–43.5 × 3.7–4.7 μm (av. ± SD: 34.6 ± 0.1 × 3.9 ± 0.2 μm); 5-septate conidia: 50–58 × 3.9–5.2 μm (av. ± SD: 54.5 ± 3.5 × 4.6 ± 0.6 μm); 6-septate conidia: 58.7–63.8 × 4.1–4.5 μm (av. ± SD: 61.3 ± 2.8 × 4.2 ± 0.2 μm). Aerial conidia not observed. Chlamydospores abundant, globose, subglobose to ovoid, subhyaline, smooth or rough-walled, terminal or intercalary, solitary, in pairs or forming long chains, 6.2–10.3 μm diam.
Culture characteristics: Colonies on PDA incubated at 25 °C in the dark reaching 67–73 mm diam in 7 d; flat, felty to velvety, radiate, with abundant aerial mycelium, colony margin undulate; surface cream (4A3) in the centre, white (–A1) at the margin; reverse flame yellow (4A8) in the centre, white (–A1) at the margin; odour absent. On OA in the dark reaching 98–101 mm in 7 d; flat, felty to dusty, with abundant aerial mycelium, margin entire; surface white, reverse curry yellow (4C8) in the centre, cream (4A3) at the margin; odour absent.
Typus: China, Shandong Province, Jinan City (E117.1; N36.4), obtained from the symptomatic tissues of maize ear rot, 29 Sep. 2020, X.Y. Liu (holotype HMAS 351951, ex-type living culture CGMCC3.23519 = LC15878 = HSL751).
Additional materials examined: (See Supplementary Table S1).
Notes: The isolates representing F. jinanense were resolved as a strongly supported genealogically exclusive lineage in the phylogeny inferred from combined CaM, rpb2, and tef1 loci (Fig. 9). Fusarium jinanense is closely related to F. lacertarum, but differs by 14 bp in the three loci dataset. Morphologically, F. jinanense is distinguished from F. lacertarum based on the morphologies of conidia (4–6-septate, dorsiventral curvature conidia in F. jinanense vs 2–4-septate, typical Fusarium conidia in F. lacertarum); the presence of sporodochia (which are absent in F. lacertarum), and the absence of aerial conidia (present in F. lacertarum) (Subrahmanyam 1983, Leslie & Summerell 2006). Pathogenicity tests confirmed its ability to cause maize stalk rot (Fig. 7L).
Fusarium mianyangense S.L. Han, M.M. Wang & L. Cai, sp. nov. MycoBank MB 847026. Fig. 23.
Fig. 23.
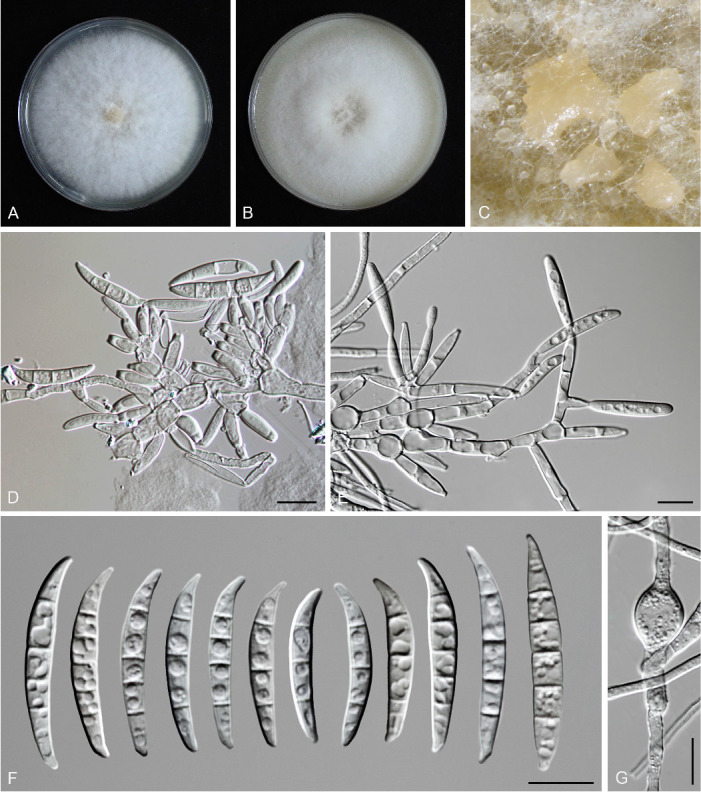
Morphology of Fusarium mianyangense (CGMCC3.23520, ex-type culture). A. Colony on PDA. B. Colony on OA. C. Sporodochia. D, E. Sporodochial conidiophores and conidiogenous cells. F. Sporodochial conidia. G. Chlamydospores. Scale bars = 10 μm.
Etymology: Named after the city, Mianyang, where the holotype was collected.
Sporodochia cream (4A3), formed frequently on carnation leaves, and infrequently on agar. Sporodochial conidiophores densely and irregularly branched, bearing apical whorls of 3–5 phialides. Sporodochial phialides subulate to subcylindrical, smooth, thin-walled, 7.5–13.6 × 1.7–3.8 μm. Sporodochial conidia falcate, the dorsal side curved than the ventral, with a blunt apical cell and well-developed foot-shaped basal cell, hyaline, smooth- and thin-walled, 3(–5)-septate: 3-septate conidia: 24.5–29.9 × 2.8–4.7 μm (av. ± SD: 27.3 ± 1.6 × 3.6 ± 0.4 μm); 4-septate conidia: 27.6–34.1 × 2.5–4.5 μm (av. ± SD: 30.9 ± 1.8 × 3.6 ± 1.4 μm); 5-septate conidia: 29.5–36.6 × 3.0–4.9 μm (av. ± SD: 33.0 ± 2.3 × 3.9 ± 0.5 μm). Aerial conidia not observed. Chlamydospores abundant, globose to subglobose, subhyaline, smooth or rough-walled, intercalary, solitary or forming long chains, 6.0–14.0 μm diam.
Culture characteristics: Colonies on PDA incubated at 25 °C in the dark reaching 74–80 mm diam in 7 d; flat, felty to velvety, with abundant aerial mycelium, colony margin entire; surface white; reverse orange white (5A2) in the centre, white (A1) at the margin; odour absent. On OA in the dark reaching 73–80 mm in 7 d; convex, with abundant aerial mycelium, margin entire; surface white, reverse apricot (5B6) in the centre, cream (4A3) at the margin; odour absent.
Typus: China, Sichuan Province, Mianyang City (E105.15, N31.78), obtained from the symptomatic tissues of Oryza sativa spikelet rot, 4 Oct. 2020, M.L. Feng (holotype HMAS 351952, ex-type living culture CGMCC3.23520 = LC15879 = HSL859).
Additional materials examined: (See Supplementary Table S1).
Notes: Isolates representing F. mianyangense were resolved as a strongly supported genealogically exclusive lineage in the phylogenies inferred from combined CaM, rpb2, and tef1 loci (Fig. 9). Fusarium mianyangense is closely related to F. citrullicola, but differs by 15 bp in the three loci dataset. Morphologically, they are distinguished from each other in the number of septa in sporodochial conidia (3–5-septate in F. mianyangense vs 1–5-septate in F. citrullicola) and the size of sporodochial conidia (24.5–36.6 × 2.5–4.9 μm in F. mianyangense vs 8–39 × 2–4.9 μm in F. citrullicola) (Khuna et al. 2022). Pathogenicity tests fulfilling the Koch’s postulates confirmed its ability to cause maize stalk rot (Fig. 7N).
Fusarium luffae M.M. Wang et al., Persoonia 43: 85. 2019.
Material examined: (See Supplementary Table S1).
Notes: Fusarium luffae has been reported as endophyte of Luffa aegyptiaca, Humulus scandens and Setaria verticillata in previous studies (Wang et al. 2019a, Xia et al. 2019). In addition, one pathogenic strain isolated from maize stalk rot was previously misidentified as F. incarnatum through tef1 BLASTn (Gai et al. 2016), but it clustered with F. luffae in our multi-locus (CaM-rpb2-tef1) phylogenetic analyses (Fig. 9). We herein confirmed the pathogenicity of F. luffae (Fig. 7M) and confirmed three new host records (maize, rice and wheat).
Fusarium nanum M.M. Wang et al., Persoonia 43: 85. 2019.
Material examined: (See Supplementary Table S1).
Notes: Fusarium nanum appears to be a broad host range species, which has been reported from Musa nana, Oryza sp., Solanum lycopersicum, Glycine max, Musa nana, Triticum sp., maize and wheat (Wang et al. 2019a, Xia et al. 2019, this study). Phylogenetically, it clustered in the Incarnatum clade, which includes 19 species, with 16 of them reported from cereals (Wang et al. 2019a, Xia et al. 2019, this study).
Fusarium nothincarnatum S.L. Han, M.M. Wang & L. Cai, sp. nov. MycoBank MB 847027. Fig. 24.
Fig. 24.
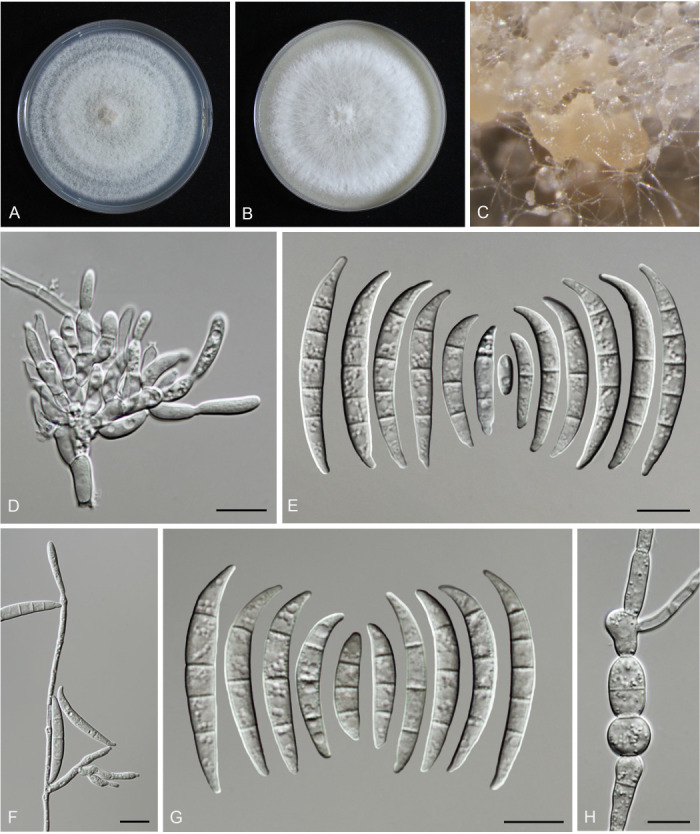
Morphology of Fusarium nothincarnatum (CGMCC3.24286, ex-type culture). A. Colony on PDA. B. Colony on OA. C. Sporodochia. D. Sporodochial conidiophores and conidiogenous cells. E. Sporodochial conidia. F. Aerial conidiophores and phialides. G. Aerial conidia. H. Chlamydospores. Scale bars = 10 μm.
Etymology: Noth = nothus in Greek, fake, close but different; incarnatum = incarnatum-like morphology.
Sporodochia cream (4A3), formed abundantly on carnation leaves and the surface of the medium. Sporodochial conidiophores densely and irregularly branched, bearing apical whorls of 3–5 phialides. Sporodochial phialides subulate to subcylindrical, with a short-flared apical collarette, smooth, thin-walled, 6.1–17.8 × 2.3–4.0 μm. Sporodochial conidia falcate, the dorsal side curved than the ventral, with a curved apical cell and poorly-developed foot-shaped basal cell, hyaline, thin- and smooth-walled, (0–)3–5-septate; 0-septate conidia: 8.7–23.3 × 1.9–2.9 μm (av. ± SD: 16.5 ± 6.3 × 2.5 ± 0.4 μm), 1-septate conidia: 14.6–19.0 × 2.3–2.8 μm (av. ± SD: 16.9 ± 1.7 × 2.6 ± 0.2 μm), 2-septate conidia: 13.6–26.4 × 2.6–4.0 μm (av. ± SD: 20.1 ± 3.9 × 3.0 ± 0.4 μm), 3-septate conidia: 25.8–37.7 × 3.3–4.8 μm (av. ± SD: 32.9 ± 3.3 × 3.9 ± 0.4 μm), 4-septate conidia: 34.7–41.8 × 3.2–4.6 μm (av. ± SD: 38.2 ± 2.1 × 4.1 ± 0.3 μm), 5-septate conidia: 32.5–41.5 × 2.9–4.9 μm (av. ± SD: 37.3 ± 2.2 × 3.7 ± 0.4 μm). Aerial conidiophores borne on aerial mycelium, straight or flexuous, erect or prostrate, smooth- and thin-walled, unbranched, sympodial or irregularly branched, bearing terminal or lateral phialides; Aerial phialides monophialidic, subulate to subcylindrical, smooth- and thin-walled, periclinal thickening inconspicuous or absent, sometimes proliferating percurrently, 8.2–21.6 × 1.4–3.5 μm. Aerial conidia falcate, curved dorsiventrally, with curved apical cell and blunt to barely notched basal cell, hyaline, smooth- and thin-walled, 3(–5)-septate: 3-septate conidia: 17.7–34.8 × 3.0–4.5 μm (av. ± SD: 26.4 ± 4.2 × 3.7 ± 0.4 μm); 4-septate conidia: 30.6–39.9 × 3.2–7.4 μm (av. ± SD: 35.9 ± 2.4 × 4.0 ± 0.7 μm); 5-septate conidia: 32.7–42.4 × 3.2–4.5 μm (av. ± SD: 36.2 ± 3.0 × 3.7 ± 0.4 μm). Chlamydospores abundant, globose, subglobose to ovoid, subhyaline, smooth-walled, intercalary, solitary, in pairs or forming chains, 7.3–12.4 μm diam.
Culture characteristics: Colonies on PDA incubated at 25 °C in the dark reaching 77–79 mm diam in 7 d; flat, felty to velvety, with abundant aerial mycelium, colony margin entire; surface white (A1); reverse orange white (5A2) in the centre, white (A1) at the margin; odour absent. On OA in the dark reaching 73–76 mm in 7 d; raised, with relatively sparse aerial mycelium, margin entire; surface white (A1), reverse milk white (1A2); odour absent.
Typus: China, Heilongjiang Province, Daqing City (E124.81, N46.04), obtained from the symptomatic tissues of Oryza sativa spikelet rot, 7 Oct. 2020, Y.J. Li (holotype HMAS 352343, ex-type living culture CGMCC3.24286 = LC18436 = HSL221).
Additional materials examined: (See Supplementary Table S1).
Notes: Isolates representing F. nothincarnatum were resolved as a strongly supported genealogically exclusive lineage in the phylogenies inferred from combined CaM, rpb2, and tef1 (Fig. 9), and genomic datasets (Fig. 14). Fusarium nothincarnatum is closely related to F. incarnatum, but differs by 23 bp in the three loci dataset. Morphologically, F. nothincarnatum is distinguished from F. incarnatum based on the number of septa in sporodochial conidia (0–5-septate in F. nothincarnatum vs 1–6-septate in F. incarnatum); the morphology of sporodochial conidia (the dorsal side more curved than the ventral, with more curved apical cell in F. nothincarnatum); the type of aerial phialides (monophialides in F. nothincarnatum vs mono- and polyphialides in F. incarnatum); and the morphologies of aerial conidia (F. nothincarnatum produces only falcate aerial conidia with 3–5 septa; while two types of aerial conidia were found in F. incarnatum: ellipsoidal to fusoid-shaped with 0–3 septa, and falcate shaped with 1–7 septa) (Xia et al. 2019).
Fusarium pernambucanum A.C.S. Santos et al., Mycologia 111: 253. 2019.
New synonym: Fusarium melonis S. Khuna et al., J. Fungi 8: 1135. 2022, nom. inval., Art. 40.8 (Shenzen).
Material examined: (See Supplementary Table S1).
Notes: Fusarium pernambucanum was originally reported from Aleurocanthus woglumi (Santos et al. 2019), and subsequently from Cucumis melo (Medeiros Araújo et al. 2020). This study expands its host range (maize) with confirmed pathogenicity (Fig. 7P). In addition, the inclusion of a larger sampling of F. pernambucanum isolates for the multi-locus phylogenetic analysis clearly showed that the ex-type isolate (SDBR-CMU424) of F. melonis (Khuna et al. 2022) clustered within the F. pernambucanum clade (Fig. 9). Therefore, we consider F. melonis as a later synonym of F. pernambucanum.
Fusarium sulawesiense Maryani et al. (as ‘sulawense’), Persoonia 43: 65. 2019.
Material examined: (See Supplementary Table S1).
Notes: Fusarium sulawesiense was originally reported from Musa acuminata (Maryani et al. 2019b), and subsequently from rice (Wang et al. 2019a). This study expands its host range (maize and wheat) with confirmed pathogenicity (Fig. 7T).
Fusarium tanahbumbuense Maryani et al., Persoonia 43: 63. 2019.
Material examined: (See Table S1).
Notes: Fusarium tanahbumbuense was first obtained from infected pseudostem of Musa sp. var. Pisang Hawa (ABB), but when conducting pathogenicity trials, it only caused a slight discoloration in the corm without further disease development (Maryani et al. 2019b). Later, two isolates of this species were recorded as a pathogen of wheat, but misidentified as F. incarnatum based on tef1 phylogenetic analysis (Özer et al. 2020, Fig. 9). Here we add two new host records (maize and rice), and confirm its ability to cause maize stalk rot (Fig. 7U).
Fusarium weifangense S.L. Han, M.M. Wang & L. Cai, sp. nov. MycoBank MB 847028. Fig. 25.
Fig. 25.
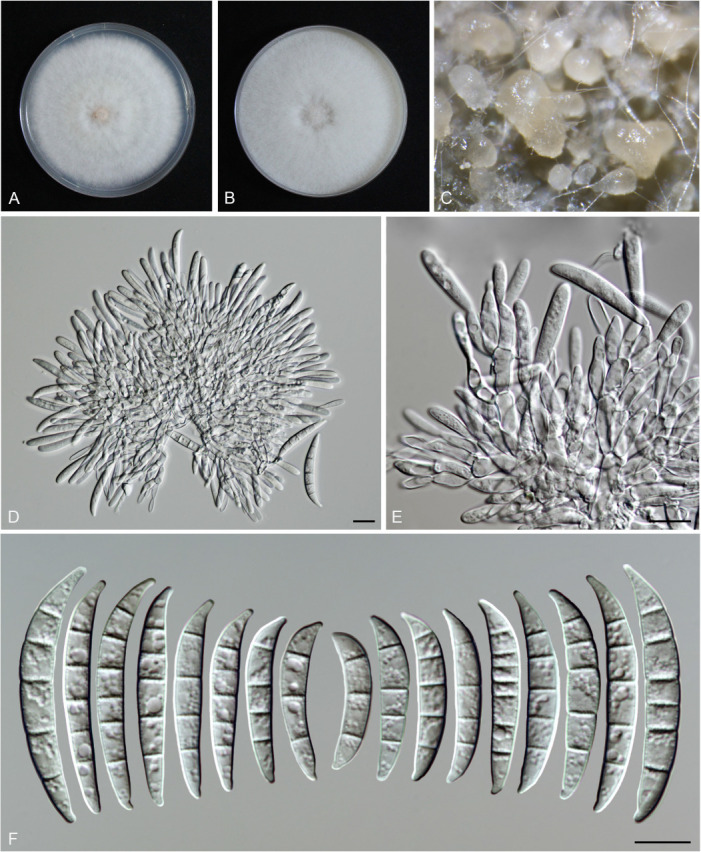
Morphology of Fusarium weifangense (CGMCC3.24285, ex-type culture). A. Colony on PDA. B. Colony on OA. C. Sporodochia. D, E. Sporodochial conidiophores and conidiogenous cells. F. Sporodochial conidia. Scale bars = 10 μm.
Etymology: Named after the city, Weifang, where the holotype was collected.
Sporodochia milk white (1A2), formed frequently on carnation leaves. Sporodochial conidiophores densely and irregularly branched, bearing apical whorls of 3–4 phialides. Sporodochial phialides subulate to subcylindrical, smooth, thin-walled, 8.8–29.9 × 3.1–5.1 μm. Sporodochial conidia falcate, the dorsal side curved than the ventral, with a blunt apical cell and poorly-developed foot-shaped basal cell, hyaline, smooth- and thin-walled, (3–)5(–7)-septate: 3-septate conidia: 26.5–30.9 × 4.5–5.8 μm (av. ± SD: 28.8 ± 1.7 × 4.9 ± 0.4 μm); 4-septate conidia: 28.0–35.4 × 4.1–5.5 μm (av. ± SD: 31.0 ± 2.1 × 4.7 ± 0.3 μm); 5-septate conidia: 30.0–49.4 × 4.9–6.7 μm (av. ± SD: 37.2 ± 4.3 × 5.6 ± 0.4 μm); 6-septate conidia: 37.7–49.2 × 5.1–7.1 μm (av. ± SD: 44.5 ± 3.7 × 5.9 ± 0.7 μm); 7-septate conidia: 35.5–36 × 4.8–5.6 μm (av. ± SD: 35.7 ± 0.3 × 5.2 ± 0.6 μm). Aerial conidia and chlamydospores not observed.
Culture characteristics: Colonies on PDA incubated at 25 °C in the dark reaching 69–79 mm diam in 7 d; flat, felty to velvety, with abundant aerial mycelium, colony margin entire; surface white (A1); reverse milk white (1A2); odour absent. On OA in the dark reaching 76–83 mm in 7 d; crateriform, with abundant aerial mycelium, margin entire; surface white (A1), reverse cream (4A3); odour absent.
Typus: China, Shandong Province, Weifang City (E119.79, N36.23), from the symptomatic tissues of wheat scab, 18 May 2021, P. Shen (holotype HMAS 352342, ex-type living culture CGMCC3.24285 = LC18333 = HSL1800).
Additional materials examined: (See Supplementary Table S1).
Notes: The isolates representing F. weifangense were resolved as a strongly supported genealogically exclusive lineage in the phylogeny inferred from combined CaM, rpb2, and tef1 loci (Fig. 9). Fusarium weifangense is closely related to F. citri and F. humuli, but differs by 37 bp and 51 bp in the three loci dataset, respectively. Morphologically, F. weifangense could be distinguished from the latter two species in the number of septa in sporodochial conidia (3–7-septate in F. weifangense vs 3–5-septate in F. citri and F. humuli), the size of sporodochial conidia (26.5–49.4 × 4.1–7.1 μm in F. weifangense vs 25.5–40.5 × 3–5.5 μm in F. citri vs 21–35 × 2–3 μm in F. humuli) (Wang et al. 2019a).
Fusarium nisikadoi species complex (FNSC)
FNSC now includes six species (F. commune, F. gaditjirrii, F. lyarnte, F. miscanthi, F. nisikadoi and F. paranisikadoi), all of which are associated with plants (Nirenberg 1997, Gams et al. 1999, Skovgaard et al. 2003, Phan et al. 2004, Crous et al. 2021, Wang et al. 2022a). In this study, all six obtained strains belonging to this complex were identified as F. commune based on multi-locus analyses (Fig. 10).
Fusarium commune K. Skovg. et al., Mycologia 95: 632. 2003.
Material examined: (See Supplementary Table S1).
Notes: Fusarium commune has been reported as pathogens of rice wilt, rice root rot, and maize stalk rot by Husna et al. (2020) and Xi et al. (2019). This species is pathologically similar to species in FOSC, causing root rot and wilt diseases on a variety of plants (Ma et al. 2010, Maryani et al. 2019a, McTaggart et al. 2021).
Fusarium oxysporum species complex (FOSC)
Members of FOSC are mostly soil-borne fungi causing root and vascular wilt diseases (Ma et al. 2010, Maryani et al. 2019a, McTaggart et al. 2021). Previously, the FOSC has been classified as one species which included more than 100 formae speciales, based on subspecific classification systems (Snyder & Hansen 1940, Gordon 1965, Lombard et al. 2019b). The F. oxysporum strains belonging to the same forma specialis, however, have been shown to represent multiple species based on the multi-locus and phylogenomic analyses (Zhang & Ma 2017, Lombard et al. 2019b, McTaggart et al. 2021). For instance, Fusarium oxysporum f. sp. lycopersici, has been shown to be an assemblage of two distinct species, namely F. nirenbergiae and F. languescens (Lombard et al. 2019b). With the continuous clarification of the phylogenetic relationships in this species complex, up to 31 cryptic species have been recognised and formally named (Lombard et al. 2019b, Maryani et al. 2019a, Wang et al. 2022a). Recently, population genomics analyses based on 410 genomes within the FOSC suggested that long-term sexual or parasexual reproduction have contributed to the species diversity of this complex, stepping away from the previous hypothesis suggesting strictly clonal propagation (McTaggart et al. 2021).
Fusarium cugenangense Maryani et al., Stud. Mycol. 92: 181. 2018 [2019].
Material examined: (See Supplementary Table S1).
Notes: The banana Fusarium wilt pathogen, previously called F. oxysporum f. sp. cubense has been confirmed to include nine independent phylogenetic lineages (Maryani et al. 2019a). Among them, the lineage referred to as Foc Lineage L7 by Fourie et al. (2009), was recently formally described as a new species Fusarium cugenangense (Maryani et al. 2019a). According to multi-locus analyses of available data, F. cugenangense appears to have a very wide host range, including Acer palmatum, Crocus sp., Gossypium barbadense, Hordeum vulgare, Solanum tuberosum, Smilax sp., Tulipa gesneriana, Musa nana, Musa sp., Vicia faba and Zea mays (Maryani et al. 2019a, Wang et al. 2022a, this study).
Fusarium nirenbergiae L. Lombard & Crous, Persoonia 43: 29. 2018 [2019].
Material examined: (See Supplementary Table S1).
Notes: Fusarium nirenbergiae has been isolated from at least 20 host genera (Lombard et al. 2019b, Wang et al. 2022a), and has been recorded to contain isolates which were previously classified in 14 different forma specialis, e.g., F. oxysporum f. sp. bouvardiae, F. oxysporum f. sp. cubense, F. oxysporum f. sp. lycopersici, etc. (Lombard et al. 2019b, McTaggart et al. 2021). In this study, we reported a new host record of F. nirenbergiae and confirmed its pathogenicity (Fig. 7O).
Fusarium sambucinum species complex (FSAMSC)
According to the multi-locus phylogenetic (Fig. 12) and phylogenomic support (Fig. 15), the FSAMSC now includes 41 described species (Crous et al. 2021, Laraba et al. 2021, Lombard et al. 2022), and almost half of the species in this complex have been reported from cereals (Table 2). The 205 isolates obtained in this study were identified as nine known species (Fig. 12).
Fusarium armeniacum (G.A. Forbes et al.) L.W. Burgess & Summerell, Stud. Mycol. 98: 86. 2021.
Basionym: Fusarium acuminatum subsp. armeniacum G.A. Forbes et al., Mycologia 85: 120. 1993.
Material examined: (See Supplementary Table S1).
Notes: Fusarium armeniacum has been recorded as causal agent of wheat scab, root rot and seed rot of various plants (Wing et al. 1993, Leslie & Summerell 2006, Summerell et al. 2010, Krone et al. 2020). Here we update two new host records (maize and wheat) in China, and confirm its ability to cause maize stalk rot (Fig. 7G).
Fusarium asiaticum O’Donnell et al., Fungal Genet. Biol. 41: 619. 2004.
Material examined: (See Supplementary Table S1).
Notes: Fusarium asiaticum is one of the main pathogens of wheat scab in the world, especially in Asia (Suga et al. 2008, Zhang et al. 2012, Castañares et al. 2016), and it is also a notorious pathogen of wheat crown rot, maize ear rot, maize stalk rot and rice spikelet rot (Desjardins & Proctor 2011, Gomes et al. 2015, Zhang et al. 2016, Dong et al. 2020b, Xi et al. 2021). In our study, most strains of this species were isolated from blighted wheat spikes (wheat scab) from southern China, supporting earlier findings that it is a dominant agent of wheat scab in China (Zhang et al. 2012, Yang et al. 2018, Xu et al. 2021a).
Fusarium boothii O’Donnell et al., Fungal Genetics Biol. 41: 618. 2004.
Material examined: (See Supplementary Table S1).
Notes: Fusarium boothii has been reported as a pathogen of wheat scab, maize ear rot and stalk rot (Malihipour et al. 2012, Zhang et al. 2016, Beukes et al. 2017, Gai et al. 2017). The hybrid of F. graminearum × F. boothii has been reported from maize in France and South Africa (Boutigny et al. 2011, 2013). Future comparative genomic analyses of F. graminearum, F. boothii, F. graminearum × F. boothii would be useful for understanding the speciation of these closely related species.
Fusarium graminearum Schwabe, Fl. Anhalt. 2: 285. 1839.
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: Fusarium graminearum, with the smallest genome in the whole genus (Supplementary Table S3), has been considered as one of the top 10 most economically harmful fungal pathogens (Dean et al. 2012). This species is widely distributed on different hosts with different lifestyles (pathogens, endophytes, saprophytes) (Cuomo et al. 2007, Starkey et al. 2007, Talas & McDonald 2015, Lofgren et al. 2018, Sarowar et al. 2019). In our study, F. graminearum was also one of the most frequently isolated species (85 strains) and, in many cases, co-occurred with other pathogens in the same diseased plant, leading to co-infection (Supplementary Table S6).
Fusarium kyushuense O’Donnell & T. Aoki, Mycoscience 39: 2. 1998.
Material examined: (See Supplementary Table S1).
Notes: Fusarium kyushuense clustered within the Sambucinum clade, closely related to F. venenatum and F. poae (Fig. 12), all of which have been reported as cereal pathogens (Table 2). In China, F. kyushuense has been reported as pathogen causing maize ear and stalk rot (Wang et al. 2014, Cao et al. 2021). Here we report F. kyushuense for the first time from diseased wheat (Fig. 3C).
Fusarium meridionale T. Aoki et al., Fungal Genet. Biol. 41: 618. 2004.
Material examined: (See Supplementary Table S1).
Notes: Fusarium meridionale was originally known from the Southern Hemisphere, but is now known to have a global distribution (O’Donnell et al. 2004, Farr & Rossman 2022). To date, this species has been reported as pathogens of maize ear rot, maize stalk rot, wheat scab and rice spikelet rot (Zhang et al. 2014b, 2016, Dong et al. 2020a). Herein we newly record this species from wheat in China.
Fusarium poae (Peck) Wollenw., in Lewis, Bull. Maine. Agric. Exp. Sta. 219: 254. 1913 [1914].
Basionym: Sporotrichum poae Peck, Bull. New York State Mus. 67: 29. 1904 [1903].
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: Fusarium poae produces a series of mycotoxins, such as diacetoxyscirpenol, beauvericin and nivalenol (Laraba et al. 2021). This species has been widely recorded from cereals, but appears to be pathologically less aggressive compared with F. graminearum (Bottalico & Perrone 2002, Xu et al. 2020, Laraba et al. 2021). In our investigation, F. poae appeared to be more frequently recorded from alpine and dry areas (Fig. 16C; Supplementary Table S1).
Fusarium pseudograminearum O’Donnell & T. Aoki, Mycologia 91: 604. 1999.
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: Fusarium pseudograminearum is a well-known mycotoxin (3-acetyldeoxynivalenol, zearalenone, nivalenol and culmorin) producing fungus (Farr & Rossman 2022, Laraba et al. 2021). It is also a major causal agent of wheat crown rot in Northern China (Li et al. 2012, Xu et al. 2015), which was further confirmed in our study (Supplementary Table S1).
Fusarium vorosii B. Tóth et al., Fungal Genet. Biol. 44: 1202. 2007.
Material examined: (See Supplementary Table S1).
Notes: Fusarium vorosii was first described as a pathogen of wheat scab by Starkey et al. (2007). Later, it has been isolated from other cereals, including Hordeum vulgare, Oryza sativa and Zea mays (Lee et al. 2016). To date, this species has been reported from five countries (Farr & Rossman 2022), including Hungary, Serbia, Japan, Korea and China (this study).
Fusarium tricinctum species complex (FTSC)
Species in FTSC are widely distributed in various grains and produce a broad range of mycotoxins (Leslie & Summerell 2006). Currently, 14 cryptic species have been identified and described in this complex (Wang et al. 2022a), among which F. acuminatum and F. avenaceum have the widest host range (more than 250 different hosts). Coincidentally, all 20 strains of this complex isolated in this study were identified as F. acuminatum or F. avenaceum (Fig. 13).
Fusarium acuminatum Ellis & Everh., Proc. Acad. Nat. Sci. Philadelphia 47: 441. 1895.
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: Fusarium acuminatum was usually reported from temperate regions as a soil saprobe or pathogen of root and crown diseases of various plants (Leslie & Summerell 2006, Zhang et al. 2015, this study: Fig. 7D). In this study, eight strains isolated from diseased cereals clustered with the currently widely used reference strain of Fusarium acuminatum (NRRL 52789) (O’Donnell et al. 2012, Li et al. 2019a, Wang et al. 2022a).
Fusarium avenaceum (Fr.) Sacc., Syll. Fung. 4: 713. 1886.
Basionym: Fusisporium avenaceum Fr., Syst. Mycol. 2: 238. 1822, nom. sanct. [Fr., l.c.].
Synonyms: see Crous et al. (2021).
Material examined: (See Supplementary Table S1).
Notes: Fusarium avenaceum is a cosmopolitan species and has been recorded from more than 250 hosts (Leslie & Summerell 2006, Ma et al. 2019, Crous et al. 2021, Farr & Rossman 2022). On cereals it has been associated with maize ear rot, wheat scab and wheat crown rot (Logrieco et al. 2002b, Vogelgsang et al. 2008a, b, Zhang et al. 2015), but due to the lack of sequence data we were unable to confirm the species identify in previous studies. In this investigation, 12 representative strains from wheat scab clustered together with the ex-neotype of F. avenaceum (Crous et al. 2021).
DISCUSSION
“Broad” and “narrow” generic concept of Fusarium
Fusarium species have attracted much attention from plant pathologists, food chemists and taxonomists because of their widespread distribution, pathogenicity and mycotoxin production (Marasas et al. 1984, Aoki et al. 2012, Yang et al. 2018, Torbati et al. 2019, Crous et al. 2021, Tralamazza et al. 2021). However, especially in the past decade, there have been several controversial proposals on whether Fusarium should remain broadly defined to keep most of the well-known pathogens under the generic name Fusarium, or circumscribe Fusarium in a narrower sense, but monophyletic, morphologically meaningful and as a definable unit (Crous et al. 2021, Geiser et al. 2021). The “broad” concept emphasised that the “terminal Fusarium clade” (F1) is monophyletic (Fig. 14), with the same synapomorphy (fusarium-like macroconidia) (Geiser et al. 2021). However, fusarium-like asexual macroconidia occur in many other genera (e.g., Atractium, Fusicolla, Macroconia) outside the “terminal Fusarium clade” (Crous et al. 2021). Under the “narrow” concept, a set of synapomorphies on both sexual and asexual characters could be used to distinguish the genus Fusarium from allied genera (Crous et al. 2021). Moreover, recent genomic analyses have revealed distinct divergence patterns between Fusarium s. str. and other fusarioid taxa in natural selection, translational selection and codon usage bias, which further support the “narrow” Fusarium concept (Hill et al. 2022).
In our phylogenomic tree, 193 nodes (86 %) received 100 % bootstrap support, including the nodes F1, F2, and F3 (Fig. 14). Thus, we calculated concordance factors (gCF and sCF) as complementary information which may help to improve the accuracy of our interpretations of phylogenetic reconstructions (Fig. 14, Supplementary Fig. S10A). As we had previously expected, the gCF and sCF values resolved higher in node F3 (gCF 71.1 %, sCF 70.3 %) than in F2 (gCF 46.5 %, sCF 46.7 %) and F1 (gCF 51.3 %, sCF 47.1 %). Notably, low concordance values do not mean that the phylogenetic tree is unresolved, but rather gives us further insights on how related or congruent the genes are in resolving the species phylogeny (Minh et al. 2020b). In our case, among the 1 001 single-copy orthologous gene trees, 712 of them support node F3 (Fusarium s.str.), while only 465 and 514 of them support node F2 and F1, respectively. Our phylogenomic analyses (Fig. 14) thus further support the rationality of the “narrow” Fusarium concept (assigning Fusarium to node F3) as that of Gräfenhan et al. (2011), Lombard et al. (2015) and Crous et al. (2021). Although the selection of a “narrow” concept has inevitably brought in more name changes, in this scheme the well-supported genealogically exclusive Fusarium species were clearly associated with synapomorphies, including sexual morphs (dark purple to black perithecia and subhyaline, smooth, 1–3-septate ascospores), asexual morphs (macroconidia with variously shaped apical and basal cells), and trichothecene mycotoxin production (Lombard et al. 2015, Crous et al. 2021). Despite the sampling still being unbalanced (e.g., few from Bisifusarium, and Rectifusarium), our 1 001 homologous loci of 228 assembled genomes provided much more phylogenetic information and a higher resolution than previous multi-locus and genomic data (Crous et al. 2021, Geiser et al. 2021). It is hoped that in the future more type material-derived genomes will be sequenced and made publicly available (Crous et al. 2022), in turn establishing more robust and natural classifications, with species and generic names being more meaningful with regards to their evolutionary relationships and phenotypic traits.
Co-infections of Fusarium species on cereals
In the present study, there is a notable phenomenon that various Fusarium species were isolated from the same cereal disease, and these co-infection samples account for 56 % of the whole 315 samples (Fig. 17, Supplementary Table S6). Almost all species isolated from co-infection samples have been previously reported as pathogens (Table 2, Supplementary Table S6), and notably, it is impossible to distinguish the co-infection species only through the disease symptoms on the host. In an extreme example of this study, four different species (i.e., F. luffae, F. sulawesiense, F. tanahbumbuense, and F. verticillioides) were isolated from the same diseased maize ear sample (Fig. 1F) collected from Lianyungang city (E119.09, N34.5), Jiangsu Province. This type of co-infection (or mixed infection) phenomenon has been noted and discussed in previous plant or human diseases (Waner 1994, Tarashi et al. 2017, Strauss et al. 2021, Wang et al. 2022b, Zhang et al. 2022). Using wheat scab as an example, a recent study showed that multiple Fusarium species have been isolated from the same diseased wheat head samples (Wang et al. 2022b).
Previous studies suggested that pathogens occur more frequently in an antagonistic or synergistic manner (van Kan et al. 2014, Bashyal et al. 2016, Gao et al. 2021, Mesny et al. 2021, de Faria Ferreira 2022). For instance, both F. aglaonematis and F. elaeidis could cause stem rot in Aglaonema modestum independently, but co-infection enhanced plant disease severity (Zhang et al. 2022). In contrast, F. graminearum causes more serious wheat scab than co-infection of F. graminearum and F. poae (Tan et al. 2020). The concepts of the pathobiome appear to provide a more holistic perspective to understand the occurrence of diseases, rather than simply employing the “one pathogen–one disease” theory (Vayssier-Taussat et al. 2014, 2015, Sweet & Bulling 2017, Bass et al. 2019). First, the pathobiome reflects a more natural state of interaction between the host and its microbes, as the boundaries between pathogens, endophytes and saprophytes are somewhat ambiguous and relative (Álvarez-Loayza et al. 2011, van Kan et al. 2014, Lofgren et al. 2018, Hill et al. 2022). Many so-called “pathogens” only cause plant symptoms at higher conidial concentrations (Fig. 7), and in many cases they present different lifestyles on different plants (Varga et al. 2015, Lofgren et al. 2018). Secondly, the co-occurrence of different species may promote the emergence of new pathogenic varieties through interspecific hybridization (Boutigny et al. 2011, 2013) or gene transfer (Ma 2014, Vlaardingerbroek et al. 2016, Simbaqueba et al. 2018), thus serve as a potential driver of pathogen evolution. For example, F. nirenbergiae (Fo47) assimilated the pathogenicity genes from F. languescens (Fol4287) via horizontal gene transfer (Vlaardingerbroek et al. 2016, McTaggart et al. 2021). Thirdly, the pathobiome vision provides a new perspective to explain why the higher richness of fungal pathogens is usually associated with a more harmful threat to plant health (Liu et al. 2022c), because co-infection may enhance the reproduction and transmission capacity of pathogens (Susi et al. 2015). The co-infection widely detected in this study suggested that Fusarium pathogens may have close interaction with each other in the process of the disease development but the specific mechanisms in different diseases/hosts remain to be revealed.
There is no doubt that co-infection will pose diagnostic challenges for plant pathologists and quarantine officials. The traditional isolation and culture-based protocols may often underestimate the diversity of pathogens, ignoring one or more other pathogens. Given the vast species number and the difficulty in pathogen recognition, there is an urgent need to develop rapid and efficient diagnosis protocols for the identification of Fusarium species. The gene chip is likely a useful technical option for the recognition of known species (Shi et al. 2012, Yin et al. 2020, Yu et al. 2022), and Ward et al. (2008) has made a successful attempt in the genus Fusarium. Given the considerable number of Fusarium species causing a same disease as noted in this study (Supplementary Table S6), more specific primers need to be designed in future studies to diagnose these cereal pathogens more accurately. The high-throughput sequencing is also potentially useful but currently, widely used 2nd generation sequencing could only reveal the pathogens at the generic level in most cases. The 3rd generation sequencing represented by PacBio technology enabling longer reads of sequences would be potentially useful in revealing the diversity of the pathobiome. Meanwhile, considering the complex interactions among pathogenic Fusarium species, it is necessary to analyse the functional complexities of the pathobiome based on genome plasticity, metabolomics and microbial interactions. A better understanding of the Fusarium diversity and related pathobiome may revolutionize our understanding of Fusarium pathogenicity and allow us to more effectively prevent and control these diseases.
Need for universal barcode
Based on currently available data (Table 2), the Fusarium diseases associated with cereals are mainly caused by members of FFSC, FIESC, and FSAMSC. Our results were in agreement with previous studies that the combination of CaM, rpb1, rpb2, tef1, and tub2 datasets could recognise all species within the FFSC (Yilmaz et al. 2021); and the combination of CaM, rpb2, and tef1 datasets could recognise all species within the FIESC (Wang et al. 2019a, Xia et al. 2019). As for the FSAMSC, we recommend to use concatenated H3-rpb1-rpb2-tef1 loci for species identification. In addition to these three species-rich complexes, other cereal-related Fusarium species could be discerned by rpb2 and tef1 datasets (Crous et al. 2021, Wang et al. 2022a, this study). As noted above, it is quite frustrating that different complexes require different combinations of molecular fragments to achieve correct identification, making the identification process cumbersome and difficult for trainees and plant pathologists. Taxonomists should continue to search for more universal and useful barcodes to facilitate rapid and accurate identification of Fusarium species. For the currently widely sequenced loci, even the most effective species recognition barcode (rpb2) could only recognize 59, 33 and 35 species from the FFSC (with 84 species), FIESC (with 43 species), and FSMSC (with 41 species), respectively.
Host preference of complexes and species
Similar to previous studies, several Fusarium species complexes present certain host preferences (Gale et al. 2011, Zhang et al. 2012, 2015, Leyva-Madrigal et al. 2015, Yang et al. 2018, Duan et al. 2020). For example, in this study, 90 % of diseased wheat samples were associated with the FSAMSC, while 83 % of diseased maize samples were associated with the FFSC (Supplementary Table S1). These host preferences are usually explained as the results of the co-evolution of Fusarium species with angiosperms in warm and humid regions (Leslie & Summerell 2006, Ploetz 2006, Przemieniecki et al. 2014, Aamot et al. 2015, Lofgren et al. 2018, Vorob’eva & Toropova 2020). Taking Fusarium oxysporum f. sp. cubense as an example, it has been identified and delimited as nine independent species (O’Donnell et al. 1998b, Fourie et al. 2009, Maryani et al. 2019a), and these species are believed to have long co-evolved with their banana hosts in Asia (Ploetz & Pegg 1997, Mostert et al. 2017, Maryani et al. 2019a).
Checklist of cereal-associated Fusarium species
In this study, we updated 39 and 52 host records for Fusarium species for the world and China, respectively, and for the first time (Table 2), confirmed the pathogenicity of 18 species causing maize stalk rot (Fig. 7). According to the updated records presented in this study, about 79 % of the new host records in China were from new species published since 2019 (Table 2), indicating that the description rate of new species in Fusarium has significantly increased, along with the establishment of the new classification scheme for the genus. The previous classification was confusing, resulting in many sequences in public databases being assigned to incorrect names or widely used complex names. For example: i) most strains historically identified as F. proliferatum (a well-known pathogen of maize) in previous studies (Leslie & Summerell 2006, Qiu et al. 2020) have been shown to be F. annulatum based on the updated taxonomic system (Yilmaz et al. 2021); ii) many pathogenic isolates previously recognised as F. incarnatum were re-identified as different species (F. clavus, F. hainanense, F. luffae, and F. tanahbumbuense) in the current study (Fig. 9). Considering the large number of cryptic Fusarium species, there will be more cereal-associated Fusarium species reported in the future. For instance, there are 74 genealogically exclusive phylogenetically distinct species in the FSAMSC, whereas only 41 of them were formally named to date (Laraba et al. 2021).
Acknowledgments
We are very grateful to all who helped us to contact farmers, collect diseased samples in their farmland, or provide photographs of diseased cereals (Supplementary Fig. S1). The name of all participants are listed in Supplementary Table S7. Funding for this study has been provided by National Science and Technology Fundamental Resources Investigation Program of China (2021FY100900) for fungal genome sequencing, NSFC (31725001, 32070023) and Biological Resources Programme, CAS (KFJ-BRP-009) for fungal diversity survey, the Major Scientific Research Focus Project of Henan Academy of Science (210105002) for academic visiting of JWW to IMCAS, and the Two Thousand Plan Foundation of Jiangxi Province (jxsq2019102026) for LC to visit Jiangxi Agricultural University. We are very grateful to Prof. P.W. Crous for inspiring our idea of sample collection through a citizen science approach, as well as spending time in editing our manuscript.
DECLARATION ON CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Supplementary Material: https://studiesinmycology.org/
Sample collection by us and other people from various regions in China. A. S.L. Han collecting rice spikelet rot samples at Jiangsu Province, Yangzhou city. B. Y.J. Li collecting diseased maize samples at Shanxi Province, Yangquan city. C. M.Y. Wang collecting wheat crown rot samples at Shandong Province, Taian city. D. S.Q. Wang collecting diseased maize samples at Guangdong Province, Qingyuan city. E. X.Y. Liu collecting diseased maize samples at Shandong Province, Jinan city. F. Y.M. Wu collecting maize ear rot samples at Guangdong Province, Meizhou city. G. M.L. Feng collecting rice spikelet rot samples at Sichuan Province, Mianyang city. H. G.J. Han collecting maize ear rot samples at Shandong Province, Rizhao city. I–N. Local collectors showing photos of diseased cereals to us to confirm the occurrence of cereal diseases. O–S. We received the diseased samples and information from local collectors.
Phylogeny inferred based on the combined rpb2-tef1 gene regions of 608 representative Fusarium strains. Neocosmospora nelsonii (CBS 309.75) was used as an outgroup. Ex-type, ex-epitype and ex-neotype strains were indicated with T, ET, and NT, respectively. Subdivision of the Fusarium clade represent the recognised species complexes, including F. fujikuroi SC (FFSC), F. incarnatum-equiseti SC (FIESC), F. nisikadoi SC (FNSC), F. oxysporum SC (FOSC), F. sambucinum SC (FSAMSC), and F. tricinctum SC (FTSC), which were shown in different colours.
Phylogeny of the Fusarium fujikuroi species complex (FFSC) inferred based on the CaM (A), rpb1 (B), rpb2 (C), tef1 (D), and tub2 (E) loci, respectively. Fusarium nirenbergiae (CBS 744.97) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type, ex-epitype and ex-neotype strains were indicated with T, ET, and NT, respectively.
Phylogeny of the Fusarium incarnatum-equiseti species complex (FIESC) inferred based on the CaM (A), rpb2 (B), and tef1 (C) loci, respectively. Fusarium concolor (NRRL 13459) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type, ex-epitype and ex-neotype strains were indicated with T, ET, and NT, respectively.
Phylogeny of the Fusarium nisikadoi species complex (FNSC) inferred based on the rpb1 (A), rpb2 (B), and tef1 (C) loci, respectively. Fusarium concolor (NRRL 13994) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type strains were indicated with T.
Phylogeny of the Fusarium oxysporum species complex (FOSC) inferred based on the CaM (A), rpb2 (B), and tef1 (C) loci, respectively. Fusarium udum (CBS 177.31) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type strains were indicated with T.
Phylogeny of the Fusarium sambucinum species complex (FSAMSC) inferred based on the H3 (A), rpb1 (B), rpb2 (C), tef1 (D), and H3-rpb1-rpb2-tef1 loci (E), respectively. Fusarium nelsonii (NRRL 13338) was used as an outgroup. GCP clade in Sarver et al. (2011) were indicated in green. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type, ex-epitype and ex-neotype strains were indicated with T, ET, and NT, respectively.
Phylogeny of the Fusarium tricinctum species complex (FTSC) inferred based on the ITS (A), rpb1 (B), rpb2 (C), and tef1 (D) loci, respectively. Fusarium udum (CBS 177.31) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type, ex-epitype, and ex-neotype strains were indicated with T, ET, and NT, respectively.
Genome sizes of the four cereal-associated species-rich complexes (i.e., FFSC, FIESC, FOSC and FSAMSC) differs significantly (P < 2e-16), average and standard deviation (av. ± SD) were annotated on the corresponding boxplots.
The scatter plot of gCF values against sCF values for all branches of the phylogenetic tree of Fig. 14 (A) and Fig. 15 (B) is shown. The shade of colour (bright to dark) indicated the support values of UFBoot (0–100).
The topology conflicts within the Fg clade, between the phylogenomic tree and the muli-locus phylogenetic tree.
The total number of species discovered in samples collected from various climate zones. The number on the diagram indicates the species number detected from different climate zone, and the percentage indicates the proportion of samples collected. The number of species co-occurrence on one sample was represented by different colour.
Collection details and GenBank accession numbers of 2 020 Fusarium strains isolated in this study. The 608 representative Fusarium strains are in bold.
Number of characters/model for BI of each locus in the phylogenetic analyses of different Fusarium species complexes.
Fusarium species for which whole-genome sequences are retrieved from public databases, or generated in the current study (indicated in bold).
The number of genes in each orthogroup for each species in Fig. 14.
The number of genes in each orthogroup for each species in Fig. 15.
Detail information of co-infection.
Acknowledgement of participants.
REFERENCES
- Aamot HU, Ward TJ, Brodal G, et al. (2015). Genetic and phenotypic diversity within the Fusarium graminearum species complex in Norway. European Journal of Plant Pathology 142: 501–519. [Google Scholar]
- Abdallah-Nekache N, Laraba I, Ducos C, et al. (2019). Occurrence of Fusarium head blight and Fusarium crown rot in Algerian wheat: identification of associated species and assessment of aggressiveness. European Journal of Plant Pathology 154: 499–512. [Google Scholar]
- Abdul Rahm M, Mohamed Gh K, Essia Reha NA. (2020). Prevalence and transmission of seed-borne fungi of maize and their control by phenolic antioxidants. Plant Pathology Journal 19: 176–184. [Google Scholar]
- Adikaram NKB, Yakandawala DMD. (2020). A checklist of plant pathogenic fungi and Oomycota in Sri Lanka. Ceylon Journal of Science 49: 93–123. [Google Scholar]
- Aguín O, Cao A, Pintos C, et al. (2014). Occurrence of Fusarium species in maize kernels grown in north western Spain. Plant Pathology 63: 946–951. [Google Scholar]
- Al-Hatmi AM, Bonifaz A, de Hoog GS, et al. (2014). Keratitis by Fusarium temperatum, a novel opportunist. BMC Infectious Diseases 14: 588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexey G, Vladislav S, Nikolay V, et al. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Loayza P, White JF, Jr, Torres MS, et al. (2011). Light converts endosymbiotic fungus to pathogen, influencing seedling survival and niche-space filling of a common tropical tree, Iriartea deltoidea. PLoS ONE 6: e16386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatulli MT, Spadaro D, Gullino ML, et al. (2010). Molecular identification of Fusarium spp. associated with bakanae disease of rice in Italy and assessment of their pathogenicity. Plant Pathology 59: 839–844. [Google Scholar]
- Andrews S, Babraham B. (2010). FastQC: a quality control tool for high throughput sequence data. <http://www.bioinformatics.babraham.ac.uk/projects/fastqc>. [Google Scholar]
- Aoki T, O’Donnell K. (1998). Fusarium kyushuense sp. nov. from Japan. Mycoscience 39: 1–6. [Google Scholar]
- Aoki T, O’Donnell K. (1999). Morphological and molecular characterization of Fusarium pseudograminearum sp. nov., formerly recognized as the Group 1 population of F. graminearum. Mycologia 91: 597–609. [Google Scholar]
- Aoki T, O’Donnell K, Geiser DM. (2014). Systematics of key phytopathogenic Fusarium species: current status and future challenges. Journal of General Plant Pathology 80: 189–201. [Google Scholar]
- Aoki T, O’Donnell K, Ichikawa K. (2001). Fusarium fractiflexum sp. nov. and two other species within the Gibberella fujikuroi species complex recently discovered in Japan that form aerial conidia in false heads. Mycoscience 42: 461–478. [Google Scholar]
- Aoki T, Ward TJ, Kistler HC, et al. (2012). Systematics, phylogeny and trichothecene mycotoxin potential of Fusarium head blight cereal pathogens. Mycotoxins 62: 91–102. [Google Scholar]
- Bacon CW, Hinton DM. (1996). Symptomless endophytic colonization of maize by Fusarium moniliforme. Canadian Journal of Botany 74: 1195–1202. [Google Scholar]
- Balmas V, Corda P, Marcello A, et al. (2000). Fusarium nygamai associated with Fusarium foot rot of rice in Sardinia. Plant Disease 84: 807–807. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkat EH, Hardy GESJ, Ren Y, et al. (2016). Fungal contaminants of stored wheat vary between Australian states. Australasian Plant Pathology 45: 621–628. [Google Scholar]
- Bashyal BM, Aggarwal R, Sharma S, et al. (2016). Single and combined effects of three Fusarium species associated with rice seeds on the severity of bakanae disease of rice. Journal of Plant Pathology 98: 405–412. [Google Scholar]
- Bass D, Stentiford GD, Wang HC, et al. (2019). The pathobiome in animal and plant diseases. Trends in Ecology and Evolution 34: 996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley AR, Petrovic T, Griffiths SP, et al. (2007). Crop pathogens and other Fusarium species associated with Austrostipa aristiglumis. Australasian Plant Pathology 36: 434–438. [Google Scholar]
- Beukes I, Rose LJ, van Coller GJ, et al. (2017). Disease development and mycotoxin production by the Fusarium graminearum species complex associated with South African maize and wheat. European Journal of Plant Pathology 150: 893–910. [Google Scholar]
- Blacutt AA, Gold SE, Voss KA, et al. (2018). Fusarium verticillioides: advancements in understanding the toxicity, virulence, and niche adaptations of a model mycotoxigenic pathogen of maize. Phytopathology 108: 312–326. [DOI] [PubMed] [Google Scholar]
- Booth C. (1971). The genus Fusarium. Commonwealth Mycological Institte, Kew, Surrey, England. [Google Scholar]
- Bottalico A, Perrone G. (2002). Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. European Journal of Plant Pathology 108: 611–624. [Google Scholar]
- Boutigny AL, Ward TJ, Ballois N, et al. (2013). Diversity of the Fusarium graminearum species complex on French cereals. European Journal of Plant Pathology 138: 133–148. [Google Scholar]
- Boutigny AL, Ward TJ, Van Coller GJ, et al. (2011). Analysis of the Fusarium graminearum species complex from wheat, barley and maize in South Africa provides evidence of species-specific differences in host preference. Fungal Genetics and Biology 48: 914–920. [DOI] [PubMed] [Google Scholar]
- Britz H, Steenkamp ET, Coutinho TA, et al. (2002). Two new species of Fusarium section Liseola associated with mango malformation. Mycologia 94: 722–730. [DOI] [PubMed] [Google Scholar]
- Bugnicourt F. (1952). Une espèce fusarienne nouvelle, parasite du riz. Revue Geìneìrale de Botanique 59: 13–18. [Google Scholar]
- Burgess LW, Forbes GA, Windels C, et al. (1993). Characterization and distribution of Fusarium acuminatum subsp. armeniacum subsp. nov. Mycologia 85: 119–124. [Google Scholar]
- Burgess LW, Trimboli D. (1986). Characterization and distribution of Fusarium nygamai sp. nov. Mycologia 78: 223–229. [Google Scholar]
- Byrnes KJ, Carroll RB. (1986). Fungi causing stalk rot of conventional-tillage and no-tillage corn in Delaware. Plant Disease 70: 238–239. [Google Scholar]
- Cao Y, Zhang J, Han S, et al. (2021). First report of maize stalk rot caused by Fusarium kyushuense in China. Plant Disease 105: 3759–3759. [DOI] [PubMed] [Google Scholar]
- Castañares E, Dinolfo MI, Del Ponte EM, et al. (2016). Species composition and genetic structure of Fusarium graminearum species complex populations affecting the main barley growing regions of South America. Plant Pathology 65: 930–939. [Google Scholar]
- Castañares E, Martínez M, Cristos D, et al. (2019). Fusarium species and mycotoxin contamination in maize in Buenos Aires Province, Argentina. European Journal of Plant Pathology 155: 1265–1275. [Google Scholar]
- Castañares E, Stenglein SA, Dinolfo MI, et al. (2011). Fusarium tricinctum associated with head blight on wheat in Argentina. Plant Disease 95: 496–496. [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chehri K, Salleh B, Yli-mattila T, et al. (2010). Occurrence, pathogenicity and distribution of Fusarium spp. in stored wheat seeds Kermanshah Province, Iran. Pakistan Journal of Biological Sciences 13: 1178–1186. [DOI] [PubMed] [Google Scholar]
- Chiang KS, Liu HI, Bock CH. (2017). A discussion on disease severity index values. Part I: warning on inherent errors and suggestions to maximise accuracy. Annals of Applied Biology 171: 139–154. [Google Scholar]
- Cho WD, Shin HD. (2004). List of plant diseases in Korea. Korean Society of Plant Pathology, Korea. [Google Scholar]
- Choi HW, Hong SK, Lee YK, et al. (2018). Taxonomy of Fusarium fujikuroi species complex associated with bakanae on rice in Korea. Australasian Plant Pathology 47: 23–34. [Google Scholar]
- Corda ACJ. (1837). Icones fungorum hucusque cognitorum. J.G. Calve, Austria. [Google Scholar]
- Costa MM, Saleh AA, Melo MP, et al. (2022). Fusarium mirum sp. nov., intertwining Fusarium madaense and Fusarium andiyazi, pathogens of tropical grasses. Fungal Biology 126: 250–266. [DOI] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Lombard L, Sandoval-Denis M, et al. (2021). Fusarium: more than a node or a foot-shaped basal cell. Studies in Mycology 98: 1–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Sandoval-Denis M, Costa MM, et al. (2022). Fusarium and allied fusarioid taxa (FUSA). 1. Fungal Systematics and Evolution 9: 161–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Lombard L, et al. (2019). Fungal planet description sheets: 951–1041. Persoonia 43: 223–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, Gueldener U, Xu JR, et al. (2007). The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317: 1400–1402. [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JA, Pretorius ZA, et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology & Evolution 24: 332–340. [DOI] [PubMed] [Google Scholar]
- Desjardins AE. (2006). Fusarium mycotoxins: chemistry, genetics and biology. American Phytopathological Society, USA. [Google Scholar]
- Desjardins AE, Manandhar HK, Plattner RD, et al. (2000). Fusarium species from nepalese rice and production of mycotoxins and gibberellic acid by selected species. Applied and Environmental Microbiology 66: 1020–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins AE, Proctor RH. (2011). Genetic diversity and trichothecene chemotypes of the Fusarium graminearum clade isolated from maize in Nepal and identification of a putative new lineage. Fungal Biology 115: 38–48. [DOI] [PubMed] [Google Scholar]
- Devay JE, Covey RP, Nair PN. (1957). Corn diseases and their importance in Minnesota in 1956. Plant Disease Reporter 41: 505–507. [Google Scholar]
- de Faria Ferreira M, Brito-Santos F, Henrique Nascimento Theodoro P, et al. (2022). Mixed infection by Cryptococcus neoformans and Cryptococcus gattii and coinfection with paracoccidioidomycosis in PLHIV. Medical Mycology Case Reports 35: 48–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Xu JH, Shi JR, et al. (2020a). First report of Fusarium head blight caused by Fusarium meridionale in rice in China. Plant Disease 104: 2726–2726. [Google Scholar]
- Dong F, Zhang X, Xu JH, et al. (2020b). Analysis of Fusarium graminearum species complex from freshly harvested rice in Jiangsu Province (China). Plant Disease 104: 2138–2143. [DOI] [PubMed] [Google Scholar]
- Dong HY, Qin PW, Gao ZG, et al. (2020c). First report of seedling blight of maize caused by Fusarium asiaticum in Northeast China. Plant Disease 105: 1206–1026. [DOI] [PubMed] [Google Scholar]
- Du Q, Duan CX, Li SC, et al. (2020). First report of maize ear rot caused by Fusarium concentricum in China. Plant Disease 104: 1539–1540. [Google Scholar]
- Duan CX, Du Q, Tang ZL, et al. (2019). First report of maize ear rot caused by Fusarium sacchari in China. Plant Disease 103: 2674–2674. [Google Scholar]
- Duan CX, Wang BB, Sun FF, et al. (2020). Occurrence of maize ear rot caused by Fusarium fujikuroi in China. Plant Disease 104: 587–587. [Google Scholar]
- Ebbels DL, Allen DJ. (1979). A supplementary and annotated list of plant diseases, pathogens and associated fungi in Tanzania. Commonwealth Mycological Institute, UK. [Google Scholar]
- Emms DM, Kelly S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology 16:157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20: 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili Taheri A, Hamel C, Gan Y, et al. (2011). First report of Fusarium redolens from Saskatchewan and its comparative pathogenicity. Canadian Journal of Plant Pathology 33: 559–564. [Google Scholar]
- Ezekiel CN, Kraak B, Sandoval-Denis M, et al. (2020). Diversity and toxigenicity of fungi and description of Fusarium madaense sp. nov. from cereals, legumes and soils in north-central Nigeria. MycoKeys 67: 95–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr DF, Rossman AY. (2022). Fungal Databases, U.S. National Fungus Collections, ARS, USDA. <https://nt.ars-grin.gov/fungaldatabases/>. Accessed on 13 August 2022. [Google Scholar]
- Fisher NL, Burgess LW, Toussoun TA, et al. (1982). Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72: 151–153. [Google Scholar]
- Fourie G, Steenkamp ET, Gordon TR, et al. (2009). Evolutionary relationships among the Fusarium oxysporum f. sp. cubense vegetative compatibility groups. Applied and Environmental Microbiology 75: 4770–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumero MV, Yue W, Chiotta ML, et al. (2021). Divergence and gene flow between Fusarium subglutinans and F. temperatum isolated from maize in Argentina. Phytopathology 111: 170–183. [DOI] [PubMed] [Google Scholar]
- Gai XT, Xuan YH, Gao ZG. (2017). Diversity and pathogenicity of Fusarium graminearum species complex from maize stalk and ear rot strains in northeast China. Plant Pathology 66: 1267–1275. [Google Scholar]
- Gai XT, Yang RX, Pan XJ, et al. (2016). First report of Fusarium incarnatum causing stalk rot on maize in China. Plant Disease 100: 1010–1010. [Google Scholar]
- Gale LR, Harrison SA, Ward TJ, et al. (2011). Nivalenol-type populations of Fusarium graminearum and F. asiaticum are prevalent on wheat in southern Louisiana. Phytopathology 101: 124–134. [DOI] [PubMed] [Google Scholar]
- Gams W. (1971). Cephalosporium-artige schimmelpilze (Hyphomycetes). Gustav Fischer Verlag, Stuttgart, Germany. [Google Scholar]
- Gams W, Klamer M, O’Donnell K. (1999). Fusarium miscanthi sp. nov. from Miscanthus litter. Mycologia 91: 263–268. [Google Scholar]
- Gams W, Nirenberg HI, Seifert KA, et al. (1997). Proposal to conserve the name Fusarium sambucinum (Hyphomycetes). Taxon 46: 111–113. [Google Scholar]
- Gao M, Xiong C, Gao C, et al. (2021). Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome 9: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser DM, Al-Hatmi AMS, Aoki T, et al. (2021). Phylogenomic analysis of a 55.1 kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani species complex. Phytopathology 111: 1064–1079. [DOI] [PubMed] [Google Scholar]
- Geiser DM, Juba JH, Wang B, et al. (2001). Fusarium hostae sp. nov., a relative of F. redolens with a Gibberella teleomorph. Mycologia 93: 670–678. [Google Scholar]
- Gerlach W, Ershad D. (1970). Beitrag zur kenntnis der Fusarium-und Cylindrocarpon-arten in Iran. Nova Hedwigia 20: 725–784. [Google Scholar]
- Gomes LB, Ward TJ, Badiale-Furlong E, et al. (2015). Species composition, toxigenic potential and pathogenicity of Fusarium graminearum species complex isolates from southern Brazilian rice. Plant Pathology 64: 980–987. [Google Scholar]
- Gordon WL. (1965). Pathogenic strains of Fusarium oxysporum. Canadian Journal of Botany 43: 1309–1318. [Google Scholar]
- Gräfenhan T, Johnston PR, Vaughan MM, et al. (2016). Fusarium praegraminearum sp. nov., a novel nivalenol mycotoxin-producing pathogen from New Zealand can induce head blight on wheat. Mycologia 108: 1229–1239. [DOI] [PubMed] [Google Scholar]
- Gräfenhan T, Schroers HJ, Nirenberg HI, et al. (2011). An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Studies in Mycology 68: 79–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SB, Cao YY, Zhang J, et al. (2022). First report of Fusarium cf. longipes associated with maize stalk rot in China. Plant Disease 106: 1064–1064. [DOI] [PubMed] [Google Scholar]
- Hao JJ, Xie SN, Sun J, et al. (2017). Analysis of Fusarium graminearum species complex from wheat-maize rotation regions in Henan (China). Plant Disease 101: 720–725. [DOI] [PubMed] [Google Scholar]
- Haridas S, Albert R, Binder M, et al. (2020). 101 Dothideomycetes genomes: A test case for predicting lifestyles and emergence of pathogens. Studies in Mycology 96: 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R, Buggs RJA, Vu DT, et al. (2022). Lifestyle transitions in fusarioid fungi are frequent and lack clear genomic signatures. Molecular Biology and Evolution 39: msac085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, et al. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SW, Wang L, Liu LM, et al. (2011). Rice spikelet rot disease in China–1. Characterization of fungi associated with the disease. Crop Protection 30: 1–9. [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001). MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Husna A, Zakaria L, Mohamed Nor NMI. (2020). Fusarium commune associated with wilt and root rot disease in rice. Plant Pathology 70: 123–132. [Google Scholar]
- Jamieson CO, Wollenweber HW. (1912). An external dry rot of potato tubers caused by Fusarium trichothecioides. Journal of the Washington Academy of Sciences 2: 146–152. [Google Scholar]
- Ji LJ, Kong LX, Li QS, et al. (2016). First report of Fusarium pseudograminearum causing Fusarium head blight of wheat in Hebei Province, China. Plant Disease 100: 220–220. [Google Scholar]
- Jiang W, Han W, Wang R, et al. (2021). Development of an inoculation technique for rapidly evaluating maize inbred lines for resistance to stalk rot caused by Fusarium spp. in the field. Plant Disease 105: 2306–2313. [DOI] [PubMed] [Google Scholar]
- Jin QM, Lu ZZ, Pan SF, et al. (1994). Study on pathogenicities of pathogenic fungi of corn stalk rot in corn seedling stage. Maize Science 1: 73–75. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaledi N, Taheri P, Falahati Rastegar M. (2017). Identification, virulence factors characterization, pathogenicity and aggressiveness analysis of Fusarium spp., causing wheat head blight in Iran. European Journal of Plant Pathology 147: 897–918. [Google Scholar]
- Khoa LV, Hatai K, Aoki T. (2004). Fusarium incarnatum isolated from black tiger shrimp, Penaeus monodon Fabricius, with black gill disease cultured in Vietnam. Journal of Fish Diseases 27: 507–515. [DOI] [PubMed] [Google Scholar]
- Khuna S, Kumla J, Thitla T, et al. (2022). Morphology, molecular identification, and pathogenicity of two novel Fusarium species associated with postharvest fruit rot of cucurbits in northern Thailand. Journal of Fungi 8: 1135–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klittich CJR, Leslie JF, Nelson PE, et al. (1997). Fusarium thapsinum (Gibberella thapsina): a new species in section Liseola from sorghum. Mycologia 89: 643–652. [Google Scholar]
- Kornerup A, Wanscher JH. (1978). Methuen handbook of colour. 3rd edn. Eyre Methuen, London. [Google Scholar]
- Kriek NPJ, Marasas WFO, Steyn PS, et al. (1977). Toxicity of a moniliformin-producing strain of Fusarium moniliforme var. subglutinans isolated from maize. Food and Cosmetics Toxicology 15: 579–587. [DOI] [PubMed] [Google Scholar]
- Kristensen R, Torp M, Kosiak B, et al. (2005). Phylogeny and toxigenic potential is correlated in Fusarium species as revealed by partial translation elongation factor 1 alpha gene sequences. Mycological Research 109: 173–186. [DOI] [PubMed] [Google Scholar]
- Krone MJ, Salazar M, Mideros S. (2020). First report of Fusarium armeniacum causing Fusarium head blight on soft red winter wheat in Illinois. Plant Disease 105: 1199–1199. [DOI] [PubMed] [Google Scholar]
- Kuhnem PR, Ward TJ, Silva CN, et al. (2016). Composition and toxigenic potential of the Fusarium graminearum species complex from maize ears, stalks and stubble in Brazil. Plant Pathology 65: 1185–1191. [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lana FD, Madden LV, Carvalho CP, et al. (2022). Impact of Gibberella ear rot on grain quality and yield components in maize as influenced by hybrid reaction. Plant Disease 106: 3061–3075. [DOI] [PubMed] [Google Scholar]
- Laraba I, Keddad A, Boureghda H, et al. (2017). Fusarium algeriense, sp. nov., a novel toxigenic crown rot pathogen of durum wheat from Algeria is nested in the Fusarium burgessii species complex. Mycologia 109: 935–950. [DOI] [PubMed] [Google Scholar]
- Laraba I, McCormick SP, Vaughan MM, et al. (2021). Phylogenetic diversity, trichothecene potential, and pathogenicity within Fusarium sambucinum species complex. PLoS ONE 16: e0245037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence MH, Walsh JL, Shuttleworth LA, et al. (2016). Six novel species of Fusarium from natural ecosystems in Australia. Fungal Diversity 77: 349–366. [Google Scholar]
- Lee T, Paek JS, Lee KA, et al. (2016). Occurrence of toxigenic Fusarium vorosii among small grain cereals in Korea. The Plant Pathology Journal 32: 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie JF, Summerell BA. (2006). The Fusarium Laboratory Manual. Blackwell Publishing Professional, USA. [Google Scholar]
- Lew H, Chelkowski J, Pronczuk P, et al. (1996). Occurrence of the mycotoxin moniliformin in maize (Zea mays L.) ears infected by Fusarium subglutinans (Wollenw. & Reinking) Nelson et al. Food Additives and Contaminants 13: 321–324. [DOI] [PubMed] [Google Scholar]
- Leyva-Madrigal KY, Larralde-Corona CP, Apodaca-Sánchez MA, et al. (2015). Fusarium species from the Fusarium fujikuroi species complex involved in mixed infections of maize in Northern Sinaloa, Mexico. Journal of Phytopathology 163: 486–497. [Google Scholar]
- Li HL, He XL, Ding SL, et al. (2016). First report of Fusarium culmorum causing crown rot of wheat in China. Plant Disease 100: 2532–2532. [Google Scholar]
- Li HL, Yuan HX, Fu B, et al. (2012). First report of Fusarium pseudograminearum causing crown rot of wheat in Henan, China. Plant Disease 96: 1065–1065. [DOI] [PubMed] [Google Scholar]
- Li PP, Cao ZY, Wang K, et al. (2014). First report of Fusarium equiseti causing a sheath rot of corn in China. Plant Disease 98: 998–998. [DOI] [PubMed] [Google Scholar]
- Li YG, Jiang WY, Jiang D, et al. (2019a). First report of fruit rot on postharvest pumpkin caused by Fusarium acuminatum in China. Plant Disease 103: 1035–1035. [Google Scholar]
- Li YG, Zhang X, Zhang R, et al. (2019b). Occurrence of seedling blight caused by Fusarium tricinctum on rice in China. Plant Disease 103: 1789-1790. [Google Scholar]
- Lima EN, Oster AH, Bordallo PN, et al. (2020). A novel lineage in the Fusarium incarnatum-equiseti species complex is one of the causal agents of Fusarium rot on melon fruits in northeast Brazil. Plant Pathology 70: 133–143. [Google Scholar]
- Link HF. (1809). Observationes in ordines plantarum naturals, Dissetatio I. Gesellschaft Naturforschender Freunde zu Berlin, Magazin 3: 3–42. [Google Scholar]
- Liu F, Ma ZY, Hou LW, et al. (2022a). Updating species diversity of Colletotrichum, with a phylogenomic overview. Studies in Mycology 101: 1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Han Y, Li W, et al. (2022b). Identification of pathogens and evaluation of resistance and genetic diversity of maize inbred lines to stalk rot in Heilongjiang Province, China. Plant Disease: 10.1094/PDIS-03-22-0525-RE. [DOI] [PubMed] [Google Scholar]
- Liu S, García-Palacios P, Tedersoo L, et al. (2022c). Phylotype diversity within soil fungal functional groups drives ecosystem stability. Nature Ecology and Evolution 6: 900–909. [DOI] [PubMed] [Google Scholar]
- Liu X, Fang X, Yu F, et al. (2022d). Improved whole-genome sequence of Fusarium meridionale, the fungal pathogen causing Fusarium head blight in rice. Molecular Plant-Microbe Interactions 35: 85–89. [DOI] [PubMed] [Google Scholar]
- Liu XX, Wang Y, Qiu JF, et al. (2018). Analysis of population, mycotoxin chemotypes and mycotoxin pollution of Fusarium from spring wheat in northern China. Jiangsu Agricultural Sciences 46: 199–202. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lofgren LA, LeBlanc NR, Certano AK, et al. (2018). Fusarium graminearum: pathogen or endophyte of North American grasses?. New Phytologist 217: 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logrieco A, Mule G, Moretti A, et al. (2002a). Toxigenic Fusarium species and mycotoxins associated with maize ear rot in Europe. European Journal of Plant Pathology 108: 597–609. [Google Scholar]
- Logrieco A, Rizzo A, Ferracane R, et al. (2002b). Occurrence of beauvericin and enniatins in wheat affected by Fusarium avenaceum head blight. Applied and Environmental Microbiology 68: 82–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Sandoval-Denis M, Cai L, et al. (2019a). Changing the game: resolving systematic issues in key Fusarium species complexes. Persoonia 43: i–ii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, Sandoval-Denis M, Lamprecht SC, et al. (2019b). Epitypification of Fusarium oxysporum – clearing the taxonomic chaos. Persoonia 43: 1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, van der Merwe NA, Groenewald JZ, et al. (2015). Generic concepts in Nectriaceae. Studies in Mycology 80: 189–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, van Doorn R, Crous PW. (2019c). Neotypification of Fusarium chlamydosporum – a reappraisal of a clinically important species complex. Fungal Systematics and Evolution 4: 183–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L, van Doorn R, Groenewald JZ, et al. (2022). Fusarium diversity associated with the Sorghum-Striga interaction in Ethiopia. Fungal Systematics and Evolution 10: 177–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukanowski A, Lenc L, Sadowski C. (2008). First report on the occurrence of Fusarium langsethiae isolated from wheat kernels in Poland. Plant Disease 92: 488–488. [DOI] [PubMed] [Google Scholar]
- Ma LJ. (2014). Horizontal chromosome transfer and rational strategies to manage Fusarium vascular wilt diseases. Molecular Plant Pathology 15: 763–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LJ, van der Does HC, Borkovich KA, et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Haseeb HA, Xing F, et al. (2019). Fusarium avenaceum: a toxigenic pathogen causing ear rot on maize in Yunnan Province, China. Plant Disease 103: 1424–1425. [Google Scholar]
- Ma WF. (2020). Analysis of China’s food security situation. Food Processing 46: 1–4. [Google Scholar]
- Ma ZY, Wu Q, Zhou X, et al. (2021). Assembly and annotation of fungal genome based on Illumina sequencing. Bio-protocol: e2003704. [Google Scholar]
- Malihipour A, Gilbert J, Piercey-Normore M, et al. (2012). Molecular phylogenetic analysis, trichothecene chemotype patterns, and variation in aggressiveness of Fusarium isolates causing head blight in wheat. Plant Disease 96: 1016–1025. [DOI] [PubMed] [Google Scholar]
- Mao W, Carroll RB, Whittington DP. (1998) Association of Phoma terrestris, Pythium irregulare, and Fusarium acuminatum in causing red root rot of corn. Plant Disease 82: 337–342. [DOI] [PubMed] [Google Scholar]
- Marasas WFO, Nelson PE, Toussoun TA. (1984). Toxigenic Fusarium species: identity and mycotoxicology. Pennsylvania State University, USA. [Google Scholar]
- Marasas WFO, Nelson PE, Toussoun TA. (1985). Fusarium dlamini, a new species from southern Africa. Mycologia 77: 971–975. [Google Scholar]
- Marasas WFO, Rabie CJ, Lübben A, et al. (1987). Fusarium napiforme, a new species from millet and sorghum in southern Africa. Mycologia 79: 910–914. [Google Scholar]
- Marasas WFO, Rheeder JP, Lamprecht SC, et al. (2001). Fusarium andiyazi sp. nov., a new species from sorghum. Mycologia 93: 1203–1210. [Google Scholar]
- Marasas WFO, Rheeder JP, Logrieco A, et al. (1998). Fusarium nelsonii and F. musarum: two new species in Section Arthrosporiella related to F. camptoceras. Mycologia 90: 505–513. [Google Scholar]
- Marín P, Moretti A, Ritieni A, et al. (2012). Phylogenetic analyses and toxigenic profiles of Fusarium equiseti and Fusarium acuminatum isolated from cereals from southern Europe. Food Microbiology 31: 229–237. [DOI] [PubMed] [Google Scholar]
- Maryani N, Lombard L, Poerba YS, et al. (2019a). Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Studies in Mycology 92: 155–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryani N, Sandoval-Denis M, Lombard L, et al. (2019b). New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Persoonia 43: 48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic S, Tabone G, Gullino ML, et al. (2020). Emerging leafy vegetable crop diseases caused by the Fusarium incarnatum-equiseti species complex. Phytopathologia Mediterranea 59: 303–317. [Google Scholar]
- Matny ON. (2015). Fusarium head blight and crown rot on wheat & barley: losses and health risks. Advances in Plants & Agriculture Research 2: 38–43. [Google Scholar]
- McGuire JU, Crandall BS. (1967). Survey of insect pests and plant diseases of selected food crops of Mexico, central America and Panama. USDA Int. agric. Development Service, USA. [Google Scholar]
- McTaggart AR, James TY, Shivas RG, et al. (2021). Population genomics reveals historical and ongoing recombination in the Fusarium oxysporum species complex. Studies in Mycology 99: 100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros Araújo MB, Moreira GM, Nascimento LV, et al. (2020). Fusarium rot of melon is caused by several Fusarium species. Plant Pathology 70: 712–721. [Google Scholar]
- Mesny F, Miyauchi S, Thiergart T, et al. (2021). Genetic determinants of endophytism in the Arabidopsis root mycobiome. Nature Communications 12: 7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzalama M, Guarnaccia V, Martino I, et al. (2021). First report of Fusarium commune causing root and crown rot on maize in Italy. Plant Disease 105: 4156. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st conference of the extreme science and engineering discovery environment: Bridging from the extreme to the campus and beyond. Association for Computing Machinery, USA: 1–8. [Google Scholar]
- Minh BQ, Hahn MW, Lanfear R. (2020b). New methods to calculate concordance factors for phylogenomic datasets. Molecular Biology and Evolution 37: 2727–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, et al. (2020a). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses LM, Marasas WF, Vismer HF, et al. (2010). Molecular characterization of Fusarium globosum strains from South African maize and Japanese wheat. Mycopathologia 170: 237–249. [DOI] [PubMed] [Google Scholar]
- Mostert D, Molina AB, Daniells J, et al. (2017). The distribution and host range of the banana Fusarium wilt fungus, Fusarium oxysporum f. sp. cubense, in Asia. PLoS ONE 12: e0181630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulenko W, Majewski T, Ruszkiewicz-Michalska M. (2008). A preliminary checklist of micromycetes in Poland. W. Szafer Institute of Botany, Polish Academy of Sciences, Poland. [Google Scholar]
- Murillo-Williams A, Munkvold GP. (2008). Systemic infection by Fusarium verticillioides in maize plants grown under three temperature regimes. Plant Disease 92: 1695–1700. [DOI] [PubMed] [Google Scholar]
- Nees von Esenbeck CGD. (1817). Das System der Pilze und Schwämme. Stahel, Germany. [Google Scholar]
- Nees von Esenbeck CGD, Nees von Esenbeck TFL. (1818). De plantis nonnullis e mycetoidearum regno tum nuper detectis, tum minus cognitis commentatio prior doctoris Nees ab Esenbeck et Friderici Nees fratrum. Nova Acta Physico-Medica Academiae Caesareae Leopoldino-Carolinae Naturae Curiosorum 9: 226–262. [Google Scholar]
- Nelson PE, Toussoun TA, Burgess LW. (1987). Characterization of Fusarium beomiforme sp. nov. Mycologia 79: 884–889. [Google Scholar]
- Nelson PE, Toussoun TA, Marasas WFO. (1983). Fusarium species: an Illustrated manual for identification. Pennsylvania State University Press, USA. [Google Scholar]
- Nganje WE, Kaitibie S, Wilson WW, et al. (2004). Economic impacts of Fusarium head blight in wheat and barley: 1993-2001. Agribusiness & Applied Economics Report 23627, North Dakota State University, Department of Agribusiness and Applied Economics. [Google Scholar]
- Nirenberg H. (1976). Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. [Google Scholar]
- Nirenberg HI. (1995). Morphological differentiation of Fusarium sambucinum Fuckel sensu stricto, F. torulosum (Berk. & Curt.) Nirenberg comb. nov. and F. venenatum Nirenberg sp. nov. Mycopathologia 129: 131–141. [DOI] [PubMed] [Google Scholar]
- Nirenberg HI. (1997). Fusarium nisikadoi, a new species from Japan. Mycoscience 38: 329–333. [Google Scholar]
- Nirenberg HI, O’Donnell K. (1998). New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 90: 434–458. [Google Scholar]
- Nirenberg HI, O’Donnell K, Kroschel J, et al. (1998). Two new species of Fusarium: Fusarium brevicatenulatum from the noxious weed Striga asiatica in Madagascar and Fusarium pseudoanthophilum from Zea mays in Zimbabwe. Mycologia 90: 459–464. [Google Scholar]
- Nylander JAA. (2004). MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Sweden. [Google Scholar]
- O’Donnell K, Cigelnik E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Cigelnik E, Nirenberg HI. (1998a). Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90: 465–493. [Google Scholar]
- O’Donnell K, Humber RA, Geiser DM, et al. (2012). Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia 104: 427–445. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistlerr HC, Cigelnik E, et al. (1998b). Multiple evolutionary origins of the fungus causing panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95: 2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Tacke BK, et al. (2000a). Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proceedings of the National Academy of Sciences of the United States of America 97: 7905–7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, McCormick SP, Busman M, et al. (2018). Marasas et al. 1984 “Toxigenic Fusarium Species: Identity and Mycotoxicology” revisited. Mycologia 110: 1058–1080. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg H, Aoki T, et al. (2000b). A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. [Google Scholar]
- O’Donnell K, Rooney AP, Proctor RH, et al. (2013). Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genetics and Biology 52: 20–31. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, et al. (2009). Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. Journal of Clinical Microbiology 47: 3851–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, et al. (2010). Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. Journal of Clinical Microbiology 48: 3708–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Ward TJ, Geiser DM, et al. (2004). Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genetics and Biology 41: 600–623. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Ward TJ, Robert VARG, et al. (2015). DNA sequence-based identification of Fusarium: current status and future directions. Phytoparasitica 43: 583–595. [Google Scholar]
- O’Donnell K, Whitaker B, Laraba I, et al. (2022). DNA sequence-based identification of Fusarium: a work in progress. Plant Disease 106: 1579–1609 [DOI] [PubMed] [Google Scholar]
- Obanor F, Erginbas-Orakci G, Tunali B, et al. (2010). Fusarium culmorum is a single phylogenetic species based on multilocus sequence analysis. Fungal Biology 114: 753–765. [DOI] [PubMed] [Google Scholar]
- Okello PN, Petrović K, Kontz B, et al. (2019). Eight species of Fusarium cause root rot of corn (Zea mays) in South Dakota. Plant Health Progress 20: 38–43. [Google Scholar]
- Özer G, Ýmren M, Bayraktar H, et al. (2019). First report of Fusarium hostae causing crown rot on wheat in Azerbaijan. Plant Disease 103: 3278–3278. [Google Scholar]
- Özer G, Paulitz TC, Imren M, et al. (2020). Identity and pathogenicity of Fungi associated with crown and root rot of dryland winter wheat in Azerbaijan. Plant Disease 104: 2149–2157. [DOI] [PubMed] [Google Scholar]
- Padwick GW. (1945). Notes on Indian fungi. III. Mycological Papers 12: 1–12. [Google Scholar]
- Pak D, You MP, Lanoiselet V, et al. (2016). Reservoir of cultivated rice pathogens in wild rice in Australia. European Journal of Plant Pathology 147: 295–311. [Google Scholar]
- Palmer JM. (2016). Funannotate: pipeline for genome annotation. <https://funannotate.readthedocs.io/en/latest/commands.html>. [Google Scholar]
- Pan XJ, Chen N, Yao Y, et al. (2015). Population and pathogenicity of Fusarium spp. causing wheat head blight in Northeast of China. Acta Agriculturae Boreali-Sinica 30: 205–210. [Google Scholar]
- Pennycook SR. (1989). Plant diseases recorded in New Zealand (Vol. 3). Plant Diseases Division, DSIR, New Zealand. [Google Scholar]
- Phan HT, Burgess LW, Summerell BA, et al. (2004). Gibberella gaditjirrii (Fusarium gaditjirrii) sp. nov., a new species from tropical grasses in Australia. Studies in Mycology 50: 261–272. [Google Scholar]
- Ploetz RC. (2006). Fusarium-induced diseases of tropical, perennial crops. Phytopathology 96: 648–652. [DOI] [PubMed] [Google Scholar]
- Ploetz R, Pegg K. (1997). Fusarium wilt of banana and Wallace’s line: was the disease originally restricted to his Indo-Malayan region?. Australasian Plant Pathology 26: 239–249. [Google Scholar]
- Porebski S, Bailey LG, Baum BR. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter 15: 8–15. [Google Scholar]
- Przemieniecki SW, Kurowski TP, Korzekwa K. (2014). Chemotypes and geographic distribution of the Fusarium graminearum species complex. Environmental Biotechnology 10: 45–54. [Google Scholar]
- Qiu JB, Lu YA, He D, et al. (2020). Fusarium fujikuroi species complex associated with rice, maize, and soybean from Jiangsu Province, China: phylogenetic, pathogenic, and toxigenic analysis. Plant Disease 104: 2193–2201. [DOI] [PubMed] [Google Scholar]
- Rahjoo V, Zad J, Javan-Nikkhah M, et al. (2008). Morphological and molecular identification of Fusarium isolated from maize ears in Iran. Journal of Plant Pathology 90: 463–468. [Google Scholar]
- Raillo A. (1950). Fungi of the genus Fusarium. Publication State Agricultural Literature, Russia. [Google Scholar]
- Raza M, Zhang ZF, Hyde KD, et al. (2019). Culturable plant pathogenic fungi associated with sugarcane in southern China. Fungal Diversity 99: 1–104. [Google Scholar]
- Reeb V, Lutzoni F, Roux C. (2004). Contribution of RPB2 to multilocus phylogenetic studies of the Euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Molecular Phylogenetics and Evolution 32: 1036–1060. [DOI] [PubMed] [Google Scholar]
- Reinking OA. (1934). Interesting new Fusaria. Zentralblatt für Bakteriologie und Parasitenkunde, Abteilung 2 89: 509–514. [Google Scholar]
- Renev E, Zargaryan NY, Kekalo AY, et al. (2021). Infection of grain crops with fungi of the genus Fusarium. BIO Web of Conferences 36: 04008–04015. [Google Scholar]
- Rheeder JP, Marasas WFO, Nelson PE. (1996). Fusarium globosum, a new species from corn in southern Africa. Mycologia 88: 509–513. [Google Scholar]
- Richardson MJ. (1979). An annotated list of seed-borne diseases. 4th edn. International Seed Testing Association, Switzerland. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Roux J, Steenkamp ET, Marasas WFO, et al. (2001). Characterization of Fusarium graminearum from Acacia and Eucalyptus using β-tubulin and histone gene sequences. Mycologia 93: 704–711. [Google Scholar]
- Saccardo PA. (1881). Fungi Italici Autographice Delineati. Patavii, Italy. [Google Scholar]
- Saccardo PA. (1879). Fungi Gallici lecti a cl. viris P. Brunaud, C.C. Gillet et Abb. Letendre. Michelia 1: 500–538. [Google Scholar]
- Saccardo PA. (1886). Sylloge Hyphomycetum. Sylloge Fungorum 4: 1–807. [Google Scholar]
- Saccardo PA. (1892). Supplementum Universale, Pars II. Discomyceteae-Hyphomyceteae. Sylloge Fungorum 10: 1–964. [Google Scholar]
- Sandoval-Denis M, Guarnaccia V, Polizzi G, et al. (2018a). Symptomatic citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 40: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Swart WJ, Crous PW. (2018b). New Fusarium species from the Kruger National Park, South Africa. MycoKeys 34: 63–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A, Trindade JVC, Lima CS, et al. (2019). Morphology, phylogeny, and sexual stage of Fusarium caatingaense and Fusarium pernambucanum, new species of the Fusarium incarnatum-equiseti species complex associated with insects in Brazil. Mycologia 111: 244–259. [DOI] [PubMed] [Google Scholar]
- Sarowar S, Alam ST, Makandar R, et al. (2019). Targeting the pattern-triggered immunity pathway to enhance resistance to Fusarium graminearum. Molecular Plant Pathology 20: 626–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver BA, Ward TJ, Gale LR, et al. (2011). Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genetics and Biology 48: 1096–1107. [DOI] [PubMed] [Google Scholar]
- Savary S, Willocquet L, Pethybridge SJ, et al. (2019). The global burden of pathogens and pests on major food crops. Nature Ecology and Evolution 3: 430–439. [DOI] [PubMed] [Google Scholar]
- Scauflaire J, Gourgue M, Munaut F. (2011). Fusarium temperatum sp. nov. from maize, an emergent species closely related to Fusarium subglutinans. Mycologia 103: 586–597. [DOI] [PubMed] [Google Scholar]
- Schneider R, Dalchow J. (1975). Fusarium inflexum spec. nov., als erreger einer welkekrankheit an Vicia faba L. in Deutschland. Journal of Phytopathology 82: 70–82. [Google Scholar]
- Schoeman A, Flett BC, Janse van Rensburg B, et al. (2018). Pathogenicity and toxigenicity of Fusarium verticillioides isolates collected from maize roots, stems and ears in South Africa. European Journal of Plant Pathology 152: 677–689. [Google Scholar]
- Schroers HJ, Gräfenhan T, Nirenberg HI, et al. (2011). A revision of Cyanonectria and Geejayessia gen. nov., and related species with Fusarium-like anamorphs. Studies in Mycology 68: 115–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe SH. (1839). Flora Anhaltina. Apud Ge. Reimerum, Germany. [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. (2019). BUSCO: assessing genome assembly and annotation completeness. Methods in Molecular Biology 1962: 227–245. [DOI] [PubMed] [Google Scholar]
- Shan L, Zhang J, Ma N, et al. (2018). First report of maize stalk rot disease caused by Fusarium cerealis in Yunnan, China. Plant Disease 102: 444–444. [Google Scholar]
- Shan LY, Cui WY, Zhang DD, et al. (2017). First report of Fusarium brachygibbosum causing maize stalk rot in China. Plant Disease 101: 837–837. [Google Scholar]
- Shang G, Yu H, Yang J, et al. (2020). First report of Fusarium miscanthi causing ear rot on maize in China. Plant Disease 105: 1565–1565. [DOI] [PubMed] [Google Scholar]
- Sherbakoff CD. (1915). Fusaria of potatoes. New York (Ithaca) Agriculture 6: 87–270. [Google Scholar]
- Shi XC, Liu XQ, Xie XL, et al. (2012). Gene chip array for differentiation of mycobacterial species and detection of drug resistance. Chinese Medical Journal 125: 3292–3297. [PubMed] [Google Scholar]
- Short DP, O’Donnell K, Zhang N, et al. (2011). Widespread occurrence of diverse human pathogenic types of the fungus Fusarium detected in plumbing drains. Journal of Clinical Microbiology 49: 4264–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, Poudel RS, Zhong S. (2021). Identification of fungal species associated with crown and root rots of wheat and evaluation of plant reactions to the pathogens in North Dakota. Plant Disease 105: 3564–3572. [DOI] [PubMed] [Google Scholar]
- Simbaqueba J, Catanzariti AM, Gonzalez C, et al. (2018). Evidence for horizontal gene transfer and separation of effector recognition from effector function revealed by analysis of effector genes shared between cape gooseberry- and tomato-infecting formae speciales of Fusarium oxysporum. Molecular Plant Pathology 19: 2302–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard K, Rosendahl S, O’Donnell K, et al. (2003). Fusarium commune is a new species identified by morphological and molecular phylogenetic data. Mycologia 95: 630–636. [PubMed] [Google Scholar]
- Snyder WC, Hansen HN. (1940). The species concept in Fusarium. American Journal of Botany 27: 64–67. [Google Scholar]
- Snyder WC, Hansen HN. (1941). The species concept in Fusarium with reference to section Martiella. American Journal of Botany 28: 738–742. [Google Scholar]
- Snyder WC, Hansen HN. (1945). The species concept in Fusarium with reference to Discolour and other sections. American Journal of Botany 32: 657–666. [Google Scholar]
- Snyder WC, Hansen HN. (1954). Variation and speciation in the genus Fusarium. Annals of the New York Academy of Sciences 60: 16–23. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey DE, Ward TJ, Aoki T, et al. (2007). Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genetics and Biology 44: 1191–1204. [DOI] [PubMed] [Google Scholar]
- Strauss AT, Bowerman L, Porath-Krause A, et al. (2021). Mixed infection, risk projection, and misdirection: interactions among pathogens alter links between host resources and disease. Ecology and Evolution 11: 9599–9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm J. (1829). Deutschlands Flora, Abt. III. Die Pilze Deutschlands. Nabu Press, Germany. [Google Scholar]
- Su PS, Ge WY, Wang HW, et al. (2021). Advances in understanding the mechanisms of wheat-Fusarium graminearum interactions. Scientia Sinca (Vitae) 51: 1493–1507. [Google Scholar]
- Subrahmanyam A. (1983). Fusarium laceratum. Mykosen 26: 478–480. [PubMed] [Google Scholar]
- Suga H, Karugia GW, Ward T, et al. (2008). Molecular characterization of the Fusarium graminearum species complex in Japan. Phytopathology 98: 159–166. [DOI] [PubMed] [Google Scholar]
- Summerell BA, Leslie JF, Liew ECY, et al. (2010). Fusarium species associated with plants in Australia. Fungal Diversity 46: 1–27. [Google Scholar]
- Susi H, Barrès B, Vale PF, et al. (2015). Co-infection alters population dynamics of infectious disease. Nature Communications 6: 5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet MJ, Bulling MT. (2017). On the importance of the microbiome and pathobiome in coral health and disease. Frontiers in Marine Science 4: 1–11. [Google Scholar]
- Tai FL. (1979). Sylloge Fungorum Sinicorum Science Press, Academia Sinica, China. [Google Scholar]
- Talas F, McDonald BA. (2015). Genome-wide analysis of Fusarium graminearum field populations reveals hotspots of recombination. BMC Genomics 16: 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Ameye M, Landschoot S, et al. (2020). At the scene of the crime: New insights into the role of weakly pathogenic members of the Fusarium head blight disease complex. Molecular Plant Pathology 21: 1559-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarashi S, Fateh A, Mirsaeidi M, et al. (2017). Mixed infections in tuberculosis: the missing part in a puzzle. Tuberculosis (Edinb) 107: 168–174. [DOI] [PubMed] [Google Scholar]
- Tekauz A, McCallum B, Ames N, et al. (2004). Fusarium head blight of oat – current status in western Canada. Canadian Journal of Plant Pathology 26: 473–479. [Google Scholar]
- Torbati M, Arzanlou M, Sandoval-Denis M, et al. (2019). Multigene phylogeny reveals new fungicolous species in the Fusarium tricinctum species complex and novel hosts in the genus Fusarium from Iran. Mycological Progress 18: 119–133. [Google Scholar]
- Torp M, Nirenberg HI. (2004). Fusarium langsethiae sp. nov. on cereals in Europe. International Journal of Food Microbiology 95: 247–256. [DOI] [PubMed] [Google Scholar]
- Torres-Cruz TJ, Whitaker BK, Proctor RH, et al. (2022). FUSARIUM-ID v.3.0: an updated, downloadable resource for Fusarium species identification. Plant Disease 106: 1610–1616. [DOI] [PubMed] [Google Scholar]
- Touati-Hattab S, Barreau C, Verdal-Bonnin MN, et al. (2016). Pathogenicity and trichothecenes production of Fusarium culmorum strains causing head blight on wheat and evaluation of resistance of the varieties cultivated in Algeria. European Journal of Plant Pathology 145: 797–814. [Google Scholar]
- Trail F. (2009). For blighted waves of grain: Fusarium graminearum in the postgenomics era. Plant Physiology 149: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tralamazza SM, Piacentini KC, Savi GD, et al. (2021). Wild rice (O. latifolia) from natural ecosystems in the Pantanal region of Brazil: Host to Fusarium incarnatum-equiseti species complex and highly contaminated by zearalenone. International Journal of Food Microbiology 345: 109127. [DOI] [PubMed] [Google Scholar]
- Tsehaye H, Brurberg MB, Sundheim L, et al. (2016). Natural occurrence of Fusarium species and fumonisin on maize grains in Ethiopia. European Journal of Plant Pathology 147: 141–155. [Google Scholar]
- Tunali B, Nicol JM, Hodson D, et al. (2008). Root and crown rot Fungi associated with spring, facultative, and winter wheat in Turkey. Plant Disease 92: 1299–1306. [DOI] [PubMed] [Google Scholar]
- van Kan JA, Shaw MW, Grant-Downton RT. (2014). Botrytis species: relentless necrotrophic thugs or endophytes gone rogue?. Molecular Plant Pathology 15: 957–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela CP, Casal OA, Padin MC, et al. (2013). First report of Fusarium temperatum causing seedling blight and stalk rot on maize in Spain. Plant Disease 97: 1252–1253. [DOI] [PubMed] [Google Scholar]
- Varga E, Wiesenberger G, Hametner C, et al. (2015). New tricks of an old enemy: isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environmental Microbiology 17: 2588–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayssier-Taussat M, Albina E, Citti C, et al. (2014). Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Frontiers in Cellular and Infection Microbiology 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayssier-Taussat M, Kazimirova M, Hubalek Z, et al. (2015). Emerging horizons for tick-borne pathogens: from the ‘one pathogen-one disease’ vision to the pathobiome paradigm. Future Microbiology 10: 2033–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini G, Toffolatti SL, Quaglino F, et al. (2017). First report of Fusarium andiyazi causing ear rot on maize in Italy. Plant Disease 101: 839–839. [Google Scholar]
- Villani A, Moretti A, De Saeger S, et al. (2016). A polyphasic approach for characterization of a collection of cereal isolates of the Fusarium incarnatum-equiseti species complex. International Journal of Food Microbiology 234: 24–35. [DOI] [PubMed] [Google Scholar]
- Vlaardingerbroek I, Beerens B, Rose L, et al. (2016). Exchange of core chromosomes and horizontal transfer of lineage-specific chromosomes in Fusarium oxysporum. Environmental Microbiology 18: 3702–3713. [DOI] [PubMed] [Google Scholar]
- Vogelgsang S, Sulyok M, Banziger I, et al. (2008a). Effect of fungal strain and cereal substrate on in vitro mycotoxin production by Fusarium poae and Fusarium avenaceum. Food Additives and Contaminants: Part A 25: 745–757. [DOI] [PubMed] [Google Scholar]
- Vogelgsang S, Sulyok M, Hecker A, et al. (2008b). Toxigenicity and pathogenicity of Fusarium poae and Fusarium avenaceum on wheat. European Journal of Plant Pathology 122: 265–276. [Google Scholar]
- von Schlechtendal DFL. (1824). Flora Berolinensis, Pars secunda: Cryptogamia. Ferdinandi Dümmler, Germany. [Google Scholar]
- Vorob’eva I, Toropova E. (2020). Fungi ecological niches of the genus Fusarium Link. BIO Web of Conferences 24: 00095. [Google Scholar]
- Waner JL. (1994). Mixed viral infections: detection and management. Clinical Microbiology Reviews 7: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Guo C, Sun SL, et al. (2020). First report of maize ear rot caused by Fusarium sporotrichioides in China. Plant Disease 104: 567–568. [Google Scholar]
- Wang JH, Li HP, Zhang JB, et al. (2014). First report of Fusarium maize ear rot caused by Fusarium kyushuense in China. Plant Disease 98: 279–280. [DOI] [PubMed] [Google Scholar]
- Wang JH, Peng XD, Lin SH, et al. (2015). First report of Fusarium head blight of wheat caused by Fusarium sacchari in China. Plant Disease 99: 160–160. [DOI] [PubMed] [Google Scholar]
- Wang L, Ge SL, Zhao KH, et al. (2021). First report of Fusarium incarnatum causing spikelet rot on rice in China. Plant Disease 105: 3306–3306. [DOI] [PubMed] [Google Scholar]
- Wang MM, Chen Q, Diao YZ, et al. (2019a). Fusarium incarnatum-equiseti complex from China. Persoonia 43: 70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MM, Crous PW, Sandoval-Denis M, et al. (2022a). Fusarium and allied genera from China: species diversity and distribution. Persoonia 48: 1–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Song R, Fan S, et al. (2022b). Diversity of Fusarium community assembly shapes mycotoxin accumulation of diseased wheat heads. Molecular Ecology: 10.1111/mec.16618. [DOI] [PubMed] [Google Scholar]
- Wang S, Sun L, Li WQ, et al. (2019b). First report of seedling blight caused by Fusarium redolens on rice in northeast China. Plant Disease 103: 1418–1418. [Google Scholar]
- Wang XM, Shi J, Jin QM, et al. (2010). Field handbook of corn disease and insects. Chinese Agricultural Science Technology Press, China. [Google Scholar]
- Ward TJ, Clear RM, Rooney AP, et al. (2008). An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genetics and Biology 45: 473–484. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds). Academic Press, San Diego (California): 315–322. [Google Scholar]
- Wickham H. (2016). ggplot2: elegant graphics for data analysis. R package version 3.4.0. <https://ggplot2.tidyverse.org/>. [Google Scholar]
- Wing N, Lauren DR, Bryden WL, et al. (1993). Toxicity and trichothecene production by Fusarium acuminatum subsp. acuminatum and Fusarium acuminatum subsp. armeniacum. Natural Toxins 1: 229–234. [DOI] [PubMed] [Google Scholar]
- Wollenweber HW. (1913). Studies on the Fusarium problem. Phytopathology 3: 24–50. [Google Scholar]
- Wollenweber HW. (1914). Identification of species of Fusarium occurring on the sweet potato. Journal of Agricultural Research 2: 251–286. [Google Scholar]
- Wollenweber HW. (1916). Fusaria autographice delineata. Selbstverlag, Berlin, Germany. [Google Scholar]
- Wollenweber HW, Reinking OA. (1925). Aliquot Fusaria tropicalia nova vel revisa. Phytopathology 15: 155–169. [Google Scholar]
- Wulff EG, Sorensen JL, Lubeck M, et al. (2010). Fusarium spp. associated with rice bakanae: ecology, genetic diversity, pathogenicity and toxigenicity. Environmental Microbiology 12: 649–657. [DOI] [PubMed] [Google Scholar]
- Xi K, Haseeb HA, Shan L, et al. (2019). First report of Fusarium commune causing stalk rot on maize in Liaoning Province, China. Plant Disease 103: 773–773. [Google Scholar]
- Xi K, Shan L, Yang Y, et al. (2021). Species diversity and chemotypes of Fusarium species associated with maize stalk rot in Yunnan Province of Southwest China. Frontiers in Microbiology 12: 652062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia JW, Sandoval-Denis M, Crous PW, et al. (2019). Numbers to names – restyling the Fusarium incarnatum-equiseti species complex. Persoonia 43: 186–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia LK, Cao YY, Wang J, et al. (2021). First report of Fusarium culmorum causing maize stalk rot in China. Plant Disease 106: 1521–1521. [DOI] [PubMed] [Google Scholar]
- Xu F, Liu W, Song Y, et al. (2021a). The distribution of Fusarium graminearum and Fusarium asiaticum causing Fusarium head blight of wheat in relation to climate and cropping system. Plant Disease 105: 2830–2835. [DOI] [PubMed] [Google Scholar]
- Xu F, Song YL, Yang GQ, et al. (2015). First Report of Fusarium pseudograminearum from wheat heads with Fusarium head blight in North China Plain. Plant Disease 99: 156–156. [DOI] [PubMed] [Google Scholar]
- Xu F, Wang JM, Yang GQ, et al. (2020). Occurrence of Fusarium poae from wheat heads with Fusarium head blight in Henan Province of China. Plant Protection 46: 60–64. [Google Scholar]
- Xu M, Zhang X, Yu J, et al. (2021b). First report of Fusarium ipomoeae causing peanut leaf spot in China. Plant Disease 105: 3754–3754. [DOI] [PubMed] [Google Scholar]
- Xu X, Yang GQ, Wang JM, et al. (2016). Composition and variation in aggressiveness of Fusarium populations causing wheat head blight in Henan Province. Acta Phytopathologica Sinica 46: 294–303. [Google Scholar]
- Xue AG, Chen Y, Seifert K, et al. (2019). Prevalence of Fusarium species causing head blight of spring wheat, barley and oat in Ontario during 2001–2017. Canadian Journal of Plant Pathology 41: 392–402. [Google Scholar]
- Yang MX, Zhang H, Kong XJ, et al. (2018). Host and cropping system shape the Fusarium population: 3ADON-producers are ubiquitous in wheat whereas NIV-producers are more prevalent in rice. Toxins 10: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara-Bell J, Pedley KF, Farman M, et al. (2018). Specific detection of the wheat blast pathogen (Magnaporthe oryzae Triticum) by loop-mediated isothermal amplification. Plant Disease 102: 2550–2559. [DOI] [PubMed] [Google Scholar]
- Ye J, Guo Y, Zhang D, et al. (2013). Cytological and molecular characterization of quantitative trait locus qRfg1, which confers resistance to Gibberella stalk rot in maize. Molecular Plant-Microbe Interactions 26: 1417–1428. [DOI] [PubMed] [Google Scholar]
- Ye QM, Wang GC, Chen HK. (1990). Identification of Fusarium moniliforme and its similar species. Jiangxi Plant Protection 3: 13–15. [Google Scholar]
- Yilmaz N, Sandoval-Denis M, Lombard L, et al. (2021). Redefining species limits in the Fusarium fujikuroi species complex. Persoonia 46: 129–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G, Bie S, Gu H, et al. (2020). Application of gene chip technology in the diagnostic and drug resistance detection of Helicobacter pylori in children. Journal of Gastroenterology and Hepatology 35: 1331–1339. [DOI] [PubMed] [Google Scholar]
- Yli-Mattila T, Paavanen-Huhtala S, Parikka P, et al. (2004). Molecular and morphological diversity of Fusarium species in Finland and North-Western Russia. European Journal of Plant Pathology 110: 573–585. [Google Scholar]
- Yu JJ, Zhang X, Tian JH, et al. (2022). Research and development of butterfly microfluidic gene chip for 19 common pathogenic microorganisms of nosocomial infection. Journal of Clinical and Nursing Research 6: 80–85. [Google Scholar]
- Zhang H, Brankovics B, Luo W, et al. (2016). Crops are a main driver for species diversity and the toxigenic potential of Fusarium isolates in maize ears in China. World Mycotoxin Journal 9: 701–715. [Google Scholar]
- Zhang H, Luo W, Pan Y, et al. (2014a). First report of Fusarium ear rot of maize caused by Fusarium andiyazi in China. Plant Disease 98: 1428–1428. [DOI] [PubMed] [Google Scholar]
- Zhang H, Luo W, Pan Y, et al. (2014b). First report of Fusarium maize ear rot caused by Fusarium meridionale in China. Plant Disease 98: 1156–1156. [DOI] [PubMed] [Google Scholar]
- Zhang H, Luo W, Pan Y, et al. (2014c). First report of Fusarium temperatum causing Fusarium ear rot on maize in Northern China. Plant Disease 98: 1273–1273. [DOI] [PubMed] [Google Scholar]
- Zhang H, Van der Lee T, Waalwijk C, et al. (2012). Population analysis of the Fusarium graminearum species complex from wheat in China show a shift to more aggressive isolates. PLoS ONE 7: e31722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JB, Li HP, Dang FJ, et al. (2007). Determination of the trichothecene mycotoxin chemotypes and associated geographical distribution and phylogenetic species of the Fusarium graminearum clade from China. Mycological Research 111: 967–975. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cao YY, Han SB, et al. (2021a). First report of Fusarium thapsinum causing maize stalk rot in China. Plant Disease 105: 2722–2722. [DOI] [PubMed] [Google Scholar]
- Zhang K, Su YY, Cai L. (2013). An optimized protocol of single spore isolation for fungi. Cryptogamie, Mycologie 34: 349–356. [Google Scholar]
- Zhang XJ, Guo C, Wang CM, et al. (2021). First report of maize stalk rot caused by Fusarium nelsonii in China. Plant Disease 105: 4168–4168. [DOI] [PubMed] [Google Scholar]
- Zhang XX, Sun HY, Shen CM, et al. (2015). Survey of Fusarium spp. causing wheat crown rot in major winter wheat growing regions of China. Plant Disease 99: 1610–1615. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen C, Mai Z, et al. (2022). Co-infection of Fusarium aglaonematis sp. nov. and Fusarium elaeidis causing stem rot in Aglaonema modestum in China. Frontiers in Microbiology 13: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma LJ. (2017). Deciphering pathogenicity of Fusarium oxysporum from a phylogenomics perspective. Advances in Genetics 100: 179–209. [DOI] [PubMed] [Google Scholar]
- Zhao ZH, Lu GZ. (2007). Fusarium kyushuense, a newly recorded species from China. Mycotaxon 102: 119–126. [Google Scholar]
- Zhou LY, Yang SF, Wang SM, et al. (2020). Identification of Fusarium ipomoeae as the causative agent of leaf spot disease in Bletilla striata in China. Plant Disease 105: 1204–1204. [DOI] [PubMed] [Google Scholar]
- Zhuang WY. (2005). Fungi of northwestern China. Mycotaxon Ltd. Ithaca, China. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample collection by us and other people from various regions in China. A. S.L. Han collecting rice spikelet rot samples at Jiangsu Province, Yangzhou city. B. Y.J. Li collecting diseased maize samples at Shanxi Province, Yangquan city. C. M.Y. Wang collecting wheat crown rot samples at Shandong Province, Taian city. D. S.Q. Wang collecting diseased maize samples at Guangdong Province, Qingyuan city. E. X.Y. Liu collecting diseased maize samples at Shandong Province, Jinan city. F. Y.M. Wu collecting maize ear rot samples at Guangdong Province, Meizhou city. G. M.L. Feng collecting rice spikelet rot samples at Sichuan Province, Mianyang city. H. G.J. Han collecting maize ear rot samples at Shandong Province, Rizhao city. I–N. Local collectors showing photos of diseased cereals to us to confirm the occurrence of cereal diseases. O–S. We received the diseased samples and information from local collectors.
Phylogeny inferred based on the combined rpb2-tef1 gene regions of 608 representative Fusarium strains. Neocosmospora nelsonii (CBS 309.75) was used as an outgroup. Ex-type, ex-epitype and ex-neotype strains were indicated with T, ET, and NT, respectively. Subdivision of the Fusarium clade represent the recognised species complexes, including F. fujikuroi SC (FFSC), F. incarnatum-equiseti SC (FIESC), F. nisikadoi SC (FNSC), F. oxysporum SC (FOSC), F. sambucinum SC (FSAMSC), and F. tricinctum SC (FTSC), which were shown in different colours.
Phylogeny of the Fusarium fujikuroi species complex (FFSC) inferred based on the CaM (A), rpb1 (B), rpb2 (C), tef1 (D), and tub2 (E) loci, respectively. Fusarium nirenbergiae (CBS 744.97) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type, ex-epitype and ex-neotype strains were indicated with T, ET, and NT, respectively.
Phylogeny of the Fusarium incarnatum-equiseti species complex (FIESC) inferred based on the CaM (A), rpb2 (B), and tef1 (C) loci, respectively. Fusarium concolor (NRRL 13459) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type, ex-epitype and ex-neotype strains were indicated with T, ET, and NT, respectively.
Phylogeny of the Fusarium nisikadoi species complex (FNSC) inferred based on the rpb1 (A), rpb2 (B), and tef1 (C) loci, respectively. Fusarium concolor (NRRL 13994) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type strains were indicated with T.
Phylogeny of the Fusarium oxysporum species complex (FOSC) inferred based on the CaM (A), rpb2 (B), and tef1 (C) loci, respectively. Fusarium udum (CBS 177.31) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type strains were indicated with T.
Phylogeny of the Fusarium sambucinum species complex (FSAMSC) inferred based on the H3 (A), rpb1 (B), rpb2 (C), tef1 (D), and H3-rpb1-rpb2-tef1 loci (E), respectively. Fusarium nelsonii (NRRL 13338) was used as an outgroup. GCP clade in Sarver et al. (2011) were indicated in green. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type, ex-epitype and ex-neotype strains were indicated with T, ET, and NT, respectively.
Phylogeny of the Fusarium tricinctum species complex (FTSC) inferred based on the ITS (A), rpb1 (B), rpb2 (C), and tef1 (D) loci, respectively. Fusarium udum (CBS 177.31) was used as an outgroup. Strains sequenced in this study were indicated in red. The RAxML Bootstrap support values (ML-BS > 70 %) were displayed at the nodes. Ex-type, ex-epitype, and ex-neotype strains were indicated with T, ET, and NT, respectively.
Genome sizes of the four cereal-associated species-rich complexes (i.e., FFSC, FIESC, FOSC and FSAMSC) differs significantly (P < 2e-16), average and standard deviation (av. ± SD) were annotated on the corresponding boxplots.
The scatter plot of gCF values against sCF values for all branches of the phylogenetic tree of Fig. 14 (A) and Fig. 15 (B) is shown. The shade of colour (bright to dark) indicated the support values of UFBoot (0–100).
The topology conflicts within the Fg clade, between the phylogenomic tree and the muli-locus phylogenetic tree.
The total number of species discovered in samples collected from various climate zones. The number on the diagram indicates the species number detected from different climate zone, and the percentage indicates the proportion of samples collected. The number of species co-occurrence on one sample was represented by different colour.
Collection details and GenBank accession numbers of 2 020 Fusarium strains isolated in this study. The 608 representative Fusarium strains are in bold.
Number of characters/model for BI of each locus in the phylogenetic analyses of different Fusarium species complexes.
Fusarium species for which whole-genome sequences are retrieved from public databases, or generated in the current study (indicated in bold).
The number of genes in each orthogroup for each species in Fig. 14.
The number of genes in each orthogroup for each species in Fig. 15.
Detail information of co-infection.
Acknowledgement of participants.