Abstract
Correct identification of all introns is necessary to discern the protein-coding potential of a eukaryotic genome. The existence of most of the spliceosomal introns predicted in the genome of Saccharomyces cerevisiae remains unsupported by molecular evidence. We tested the intron predictions for 87 introns predicted to be present in non-ribosomal protein genes, more than a third of all known or suspected introns in the yeast genome. Evidence supporting 61 of these predictions was obtained, 20 predicted intron sequences were not spliced and six predictions identified an intron-containing region but failed to specify the correct splice sites, yielding a successful prediction rate of <80%. Alternative splicing has not been previously described for this organism, and we identified two genes (YKL186C/MTR2 and YML034W) which encode alternatively spliced mRNAs; YKL186C/MTR2 produces at least five different spliced mRNAs. One gene (YGR225W/SPO70) has an intron whose removal is activated during meiosis under control of the MER1 gene. We found eight new introns, suggesting that numerous introns still remain to be discovered. The results show that correct prediction of introns remains a significant barrier to understanding the structure, function and coding capacity of eukaryotic genomes, even in a supposedly simple system like yeast.
INTRODUCTION
Genomic sequencing projects for two eukaryotic organisms have been completed (1,2) and many more are under way (see http://geta.life.uiuc.edu/~nikos/genomes.html ). The massive amount of sequence generated from these projects contains all the information to code for the complete set of proteins needed during the life of the organism. Unfortunately this information is not in a form that can accurately be read directly because introns cloud our view. Some of this information can be recovered by directly comparing sequences in cDNA libraries (ESTs) to the genomic sequence (3,4). This approach is limited by the incomplete representation of mRNA sequences in cDNA libraries, especially with respect to 5′ sequences and rare forms of mRNA. Comparing patterns of conservation between closely related genomes [such as those of mouse and human (5)] may allow exons to be discerned from the more divergent introns, but does not reveal the pattern of exon joining in the mRNA. Given the imminent deluge of genomic sequence from other eukaryotes and the abundance of computational protein predictions for which no validation exists, there is a great need for accurate and rapid methods of intron prediction and verification using raw genomic DNA sequence.
Early attempts to identify introns automatically in yeast DNA sequences exploited the fact that the few introns identified had conserved 5′ splice sites and branchpoint sequences, and were usually near the 5′ end of the gene (6). More than 250 introns have been predicted in yeast, largely on the basis of these features (7–10). Because yeast introns are relatively few and seem easy to recognize, the importance of identifying introns in the yeast genome has been downplayed, to the point that a large fraction of predictions lack experimental support. Recently it has been noted, however, that despite their presence in <4% of genes, introns are found in >25% of pre-mRNA transcripts, due to higher expression levels of the intron-containing gene class (10,11). In addition, more examples of regulation of yeast genes at the level of splicing are accumulating. Two areas of splicing regulation have emerged: one involves autogenous negative regulation of ribosomal protein mRNA levels (12–15), and another involves positive regulation of special introns during meiosis (16–18). In some of these cases, variation from the consensus 5′ splice site or intron position within the gene is important for regulation. This suggests that a search for new introns using criteria less narrowly focused on the features of consensus introns may identify introns with interesting regulation.
In this study, we address two questions. First, how good are the intron predictions made for yeast? Second, how complete is our understanding of the locations of yeast introns? Is it difficult to find new yeast introns given broader search criteria with respect to splice site and branchpoint sequences or intron position? We present a molecular test of 87 intron predictions from the Saccharomyces Genome Database (SGD; 8) and Yeast Protein Database (YPD; 9). The results of this test validate 61 introns (70%) as predicted. Six introns (7%) were found to be present within or near predicted introns, and the specific boundaries of these partially correct predictions have been determined. The results also raise questions concerning the existence of 20 predicted introns (23%) whose splicing could not be detected. An informal search for new introns biased against earlier criteria revealed eight new introns in genes not previously annotated to contain them. During the experimental tests of intron predictions, we uncovered evidence for alternative splicing, additional meiotic regulation of splicing, two novel 5′ splice sites, and a new multiply interrupted gene. Our intron database has been updated to include the results of experiments that validate the existence of true introns and can be found at http://www.cse.ucsc.edu/research/compbio/yeast_introns.html . Our results indicate that any serious attempt to validate genome-wide predictions of splicing in more complex eukaryotic genomes will require significant new technology.
MATERIALS AND METHODS
Yeast strains and plasmids
Saccharomyces cerevisiae HI227 (MATa, leu2-3, 112, ura3-52, trp1, his3Δ, lys2Δ, prb1-1122, pep4-3, prc1-407) represented mating type a cells, IH930 (MATα, trp1, mal1, gal2, prb1-1122, pep4-3, prc1-407) represented mating type α cells, SS330/SS328 (MATa/MATα, ade2-101/ade2-101, his3-d2000/his3-d2000, ura3-52/ura3-52) represented vegetative diploid cells, and NK611 (MATa/MATα, ho::LYS2/ho::LYS2 lys2/lys2 ura3/ura3 leu2::hisG/leu2::hisG), a derivative of SK1 from Nancy Kleckner’s lab (kind gift of Sean Burgess), was used to obtain synchronous populations of diploids undergoing meiosis (19). To test YGR225W/SPO70 splicing for MER1 dependence we cloned a segment of SPO70 spanning the intron into pGAC14, placing it under control of a strong promoter as described previously for other introns (7). This plasmid was introduced into vegetative haploid yeast carrying either a plasmid expressing MER1 under control of the ADH1 promoter, or the same vector lacking MER1 [kind gift of Shirleen Roeder (16)].
Testing predicted introns
To test for splicing of predicted introns, genomic DNA (gDNA) and total RNA was isolated from yeast strains according to standard procedures (20,21). RNA was treated with RNase-free DNase I to remove contaminating genomic DNA, and was reverse transcribed using AMV reverse transcriptase (Life Sciences Inc., St Petersburg, FL) with oligo(dT) as a primer to make cDNA using 12 µg of total RNA per 20 µl reaction as described previously (20). The cDNA product was suspended in 10 µl of dH2O and 1–4 µl was used to seed a standard PCR reaction using Taq DNA polymerase (Promega, Madison, WI) according to the manufacturer’s recommendations. Reactions were subjected to denaturation at 94°C for 5 min followed by 25 temperature cycles consisting of 94°C for 1 min, 50°C for 1 min and 72°C for 1 min, followed by incubation at 72°C for 7 min after the final cycle. In experiments where measurement of unspliced RNA was important, a mock RT–PCR reaction in which reverse transcriptase was left out was performed to ensure that signals were RNA derived. Primer pairs were designed to amplify a fragment of genomic DNA or cDNA containing ∼100–200 bp to either side of the annotated splice sites by PCR using Taq DNA polymerase (21). The sequences of the more than 250 oligonucleotides used in this study are available at our intron database web site (http://www.cse.ucsc.edu/research/compbio/yeast_introns.html ). Restriction sites BamHI, KpnI, SacI or SalI were included in the primers to facilitate cloning of the PCR product into pGEM7zf(+) using Escherichia coli DH5α (21). Amplified products were compared to DNA size markers by agarose gel electrophoresis. Products from cDNA that were smaller than genomic DNA and matched the expected size from correctly spliced RNA (±10 bp) were taken as evidence that the intron prediction was correct. PCR products from cDNA the same size as that from gDNA were taken as evidence for unspliced RNA. PCR products from cDNA amplifications that did not match the size predicted by the intron annotation were gel purified and cloned into pGEM7zf(+). Clones were sequenced across the splice junction to determine the exon junctions and infer splice sites. In cases where oligo(dT) primed cDNA amplification initially produced poor signals (YDR397C, YHR041C, YIL123W, YJL024C, YLR128W, YMR033W, YPL129W), reverse transcription was carried out using the downstream (minus strand) primer specific for the target transcript in place of oligo(dT), in order to increase the specific amount of cDNA derived from the RNAs of interest. In each of these cases, the only abundant RT–PCR products observed matched those expected from a correctly predicted spliced RNA (YDR397C, YHR041C, YJL024C, YLR128W, YMR033W, YPL129W) or from genomic DNA (YIL123W).
RESULTS
Molecular tests of predicted introns
We used our database of yeast introns (7), as well as information at SGD (8) and the YPD at Proteome, Inc. (9) to determine which intron annotations lacked experimental verification. Molecular evidence for 49 introns within genes encoding non-ribosomal proteins and two snRNAs had been documented in the literature. Ribosomal protein genes commonly have introns in yeast, and 35 introns in 32 of these genes have been confirmed. An additional 68 ribosomal protein genes are predicted to have introns, but we did not test these. Considering the prevalence of introns in this class of genes (7,10,11) we felt that these predictions are likely to be correct. In all we attempted to test 88 non-ribosomal intron predictions for which no molecular evidence for splicing was available. One predicted intron in YDR305C was not tested because several primer pair combinations failed to amplify genomic DNA from several strain backgrounds, despite numerous variations in PCR conditions. We do not know the basis for this problem. We compared the length of the PCR product derived from mRNA (cDNA) of haploid cells of different mating types, and diploid cells growing vegetatively or sporulating, to the length of the PCR product from genomic DNA for the remaining 87 predictions. These fell into three main classes. The major class (61 introns) gave a cDNA-derived PCR product of the size (±10 bp) expected from the prediction. This is evidence that these predictions are likely to be correct, and we did not pursue further analysis on this class. The predicted introns we confirmed by this experiment are in or near the open reading frames (ORFs) YAL030W, YBL018C, YBL026W, YBL040C, YBL050W, YBL059W, YBR078W, YBR230C, YDL012C, YDL064W, YDL079C, YDL125C, YDL219W, YDR092W, YDR139C, YDR367W, YDR397C, YER007C-A, YER093C-A, YER133W, YFL034C-A, YFR024C, YGL087C, YGL178W, YGL232W, YGR001C-(2), YHR012W, YHR016C, YHR041C, YHR097C, YHR101C, YHR123W, YIL004C, YJL001W, YJL024C, YJL041W, YJL206C-A, YKL002W, YKL006C-A, YLL050C, YLR078C, YLR128W, YLR275W, YLR426W, YML056C, YML067C, YML094W, YMR033W, YMR116C, YMR201C, YMR292W, YNL044W, YNL050C, YNL147W, YNL246W, YNL265C, YNR053C, YPL129W, YPR028W, YPR063C and YPR187W. The next largest class includes 20 introns for which splicing is not detected because the size of the cDNA-derived PCR product is the same as that from genomic DNA. The ORFs (including Y′ elements in italics) for which we found no evidence for splicing are YBR220C, YCR033W, YEL076C-A, YFL018C, YGR296W, YHL050C, YIL123W, YIL177C, YJL225C, YJR079W, YLR202C, YLR464W, YMR307W, YNL339C, YOR074C, YOR221C, YOR318C, YOR336W, YPL283C and YPR202W. The most interesting class includes six genes that have introns distinct from the annotation: YBR089C-A, YDL189W, YKL157W, YKL186C, YOL047C and YPL175W. Overall this analysis indicates that the annotated intron predictions for non-ribosomal genes are only ∼75% correct. Because we have not tested every possible expression condition in the life of yeast, we cannot exclude the possibility that some predicted introns are spliced under some untested condition.
Intron identification corrects the protein annotation
Predictions based on the most common splice sites can lead to incorrect intron annotation when less common splice sites are used, for example in two ORFs of unknown function YBR090C and YDL189W (Fig. 1). A primer upstream of the annotated YBR090C 5′ splice site fails to give RT–PCR products, suggesting that transcripts do not traverse the YBR090C ORF. A second primer downstream of the annotated 5′ splice site does detect transcripts; sequencing of cloned PCR products shows that these are spliced using a GUAAGU sequence as the 5′ splice site (Fig. 1A). This intron abolishes the YBR090C ORF, yet it does not extend the YBR089C-A ORF. Given this and the apparent position of transcription initiation based on the efficacy of the two 5′ primers for RT–PCR, we conclude that an intron resides in the 5′-UTR of YBR089C-A/NHP6B, which encodes a high-mobility group non-histone chromatin protein, and that YBR090C is a questionable ORF. In another case, RT–PCR of a region spanning an annotated intron in YDL189W produces a product ~50 bp larger than expected. The sequence of cloned RT–PCR products indicates that an AAG 3′ splice site 46 bp upstream of the predicted CAG is the correct 3′ splice site (Fig. 1B). Similarly the annotated intron for YOL047C is incorrect, and the actual intron is much smaller and also has an AAG 3′ splice site. In this case, the ORF is not truncated, but greatly extended at the N-terminus (Fig. 1C). These cases reveal that lengthening the intron in order to find canonical splice sites that extend an ORF can lead to incorrect prediction of introns, and failure to identify protein-coding sequences correctly.
Figure 1.
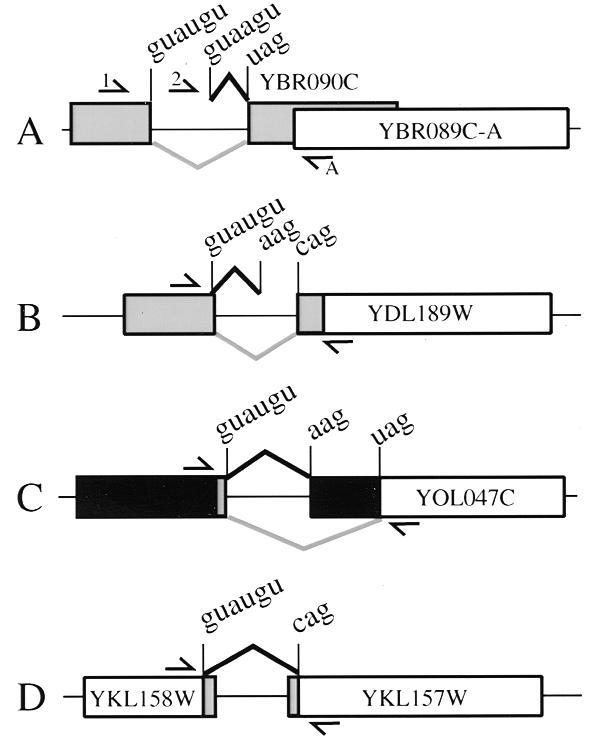
Identification of true splice sites alters predicted proteins. (A) An incorrectly predicted intron required for YBR090C (gray box) uses a 5′ splice site not compatible with YBR090C. In addition, 5′ PCR primer 1 produced no RT–PCR product in combination with the 3′ primer A, suggesting that transcription initiates downstream of primer 1. Thus, YBR090C is questionable and there is an intron in the mRNA leader of YBR089C-A (white box). (B) An incorrectly predicted intron required for the N-terminal segment of YDL189W (gray boxes) uses a 3′ splice site not compatible with the reading frame. Thus, YDL189W is smaller than currently annotated and has an intron in its mRNA leader. (C) An incorrectly predicted intron causes underestimation of the extent of YOL047C. An AAG 3′ splice site is used extending the ORF in the N-terminal direction. (D) An unspecified intron prediction in YKL157W uses splice sites that fuse two ORFs, YKL158W and YKL157W. For all diagrams, protein coding regions and intron predictions that are incorrect are shown in gray, parts of the original ORF annotation that are correct are shown in white and confirmed introns and new protein coding predictions are shown in black.
In two cases, adjacent predicted ORFs are joined by an intron to create a single large ORF. The gene encoding the aminopeptidase YKL157W/APE2 and the upstream gene of unknown function, YKL158W, are both predicted to have introns at YPD, however no specific splice sites are stated. Primers were designed to amplify a region containing the canonical intron signals GUAUGU-UACUAAC-CAG found in the region between the two ORFs. RT–PCR produced a fragment smaller than the genome by an amount consistent with intron removal and the product was cloned and sequenced. Removal of the intron effectively stitches together the two reading frames, extending the N-terminus of YKL157W by 92 amino acids (Fig. 1D). Thus, what was previously considered to be YKL158W is really exon 1 of YKL157W/APE2. The other case involves a newly discovered intron between YML034W and YML033W (described below).
Several predicted introns are required for the existence of ORFs annotated in the yeast genome. Many of these genes encode proteins of unknown function, and the demonstration of splicing for many of these contributes to the analysis of their gene products. Where introns fail to be detected, these ORFs must be called into question. For example YJR079w is a large open reading frame that depends on a predicted intron. We found only unspliced RNA from this region. Another annotated ORF (YJR080C) on the opposite strand of YJR079W does not require splicing. Since oligo(dT) primed RT–PCR could amplify polyadenylated RNA sequences derived from either strand, and no intron appears to be present, we suggest that YJR080C is the correct annotation, and YJR079W is questionable. A predicted intron in YBR219C is not spliced, and since YBR219C partly overlaps YBR220C, it seems likely that YBR219C is a questionable ORF as well. In some cases, predicted introns can be retained without loss of the ORF. Some ORFs (e.g. YMR307W and YOR336W) have predicted introns (annotated at YPD) that are a multiple of 3 nt long and do not carry an in-frame stop codon. Our ability to detect only unspliced RNA supports a longer ORF in these cases.
Our data does not exclude the possibility that predicted introns that fail to splice under the conditions we tested are true introns whose splicing is restricted to a condition we did not test. Of the 20 such cases, 10 represent four different intron predictions in the repeated Y′ elements, which are transcribed and are predicted to encode protein. None of the Y′ element predictions we tested here or previously (7) has shown any evidence for splicing. In all, we have obtained evidence for 61 correctly predicted introns, and identified the correct splice sites for six predictions that were incorrect or unspecified.
Simple searches reveal eight new introns
Given the types of prediction errors observed, we wondered how many additional introns might yet remain hidden in the yeast genome. First, we searched the genome for intron sequences allowing more degenerate splice site and branchpoint sequences. These candidate introns were deleted computationally and the resulting ‘spliced’ sequence was used in a tBLASTn query of the non-redundant protein database to find sequences that generated better BLAST scores than the unspliced genomic sequence. This intron search and BLAST query process was automated and the output was evaluated by visual inspection. One prediction near the end of YGR225W/SPO70 has standard splice sites (Table 1), and its removal would generate a protein with greatly extended homology to the CDC20/fizzy family of proteins than that reported previously (22), to the C-terminal side of the annotated YGR225W ORF (Fig. 2A). The sequence of RT–PCR products indicates that the predicted splice sites are used. YGR226C extensively overlaps the second exon of YGR225W/SPO70 on the other strand and therefore may not be a protein-coding gene (Fig. 2A). This intron was probably missed in previous searches because it is not located near the N-terminus of the ORF (6).
Table 1. Features of new introns.
| ORF | Function | 5′ SS | Branchpoint | 3′ SS | Length | Comments |
|---|---|---|---|---|---|---|
| YBR186W | meiotic checkpoint | GUAUGU | CACUAAC | UAG | 113 | rare bp |
| YDL115C | unknown | GUAUGU | GACUAAC | AAG | 89 | rare bp, 3′ss |
| YGR001C-(1) | unknown | GUAAGU | UACUAAC | UAG | 62 | rare 5′ ss |
| YGR225W | CDC20-like | GUACGU | UACUAAC | CAG | 93 | Mer1-activated |
| YLR093C | v-SNARE | GUAUGU | GACUAAC | UAG | 141 | rare bp |
| YLR211C | unknown | GUAAGU | GACUAAC | UAG | 59 | rare 5′ss, bp |
| YML034W-(1) | unknown | GUGAGU | UACUAAC | UAG | 126 | novel alternative 5′ss |
| YML034W-(2) | unknown | GCAAGU | UACUAAC | UAG | 130 | novel alternative 5′ss |
| YNL012W | phospholipase | GUAAGU | AACUAAC | UAG | 84 | rare 5′ss, bp |
Figure 2.
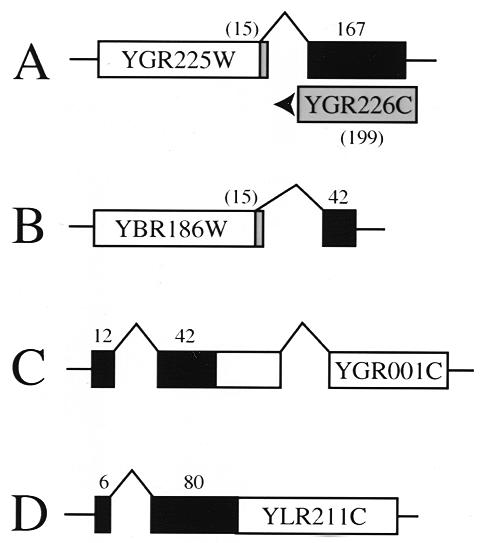
Newly identified introns extend ORFs. (A) An intron near the 3′ end of YGR225W extends the C-terminus and draws an ORF on the opposite strand into question. (B) An intron near the 3′ end of the annotated YBR186W ORF extends the C-terminus. (C) A second intron upstream of the annotated YGR001C ORF extends the N-terminus by 54 amino acids. (D) An intron upstream of the annotated YLR211C ORF extends the N-terminus by 86 amino acids. For all diagrams, protein coding regions that are incorrect are shown in gray, parts of the original ORF annotation that are correct are shown in white and confirmed introns and new protein coding predictions are shown in black. Numbers above the ORF indicate amino acids added or (in parentheses) deleted relative to the original annotation.
In a second approach we searched for introns near genes that are up-regulated in meiosis. Sequences near about 500 meiotically induced genes (22) were scanned using a simple tool that identifies a pattern of degenerate yeast splice sites and branchpoints. Of these 500, 16 intron-like sequences were selected for testing; of those 16, six were bona fide introns (Table 1). An additional intron within YGR001C (Table 1 and Fig. 2C) was identified in a similar search for second introns in genes already known to contain one intron. Several of the new introns alter and extend the predicted protein in either the N- or C-terminal direction (Fig. 2). Nearly all of them have rare splice sites or branchpoint sequences. Thus, this very limited effort to find new yeast introns yielded eight new introns (Table 1). During the course of this study there have appeared in the literature another three introns (17,23). It seems dangerous to conclude that the current intron annotation for the yeast genome is complete. We infer that a significant reason for this is the failure to find introns with variant splice sites and branchpoints.
Alternative splicing creates multiple mRNAs for YKL186C/MTR2 and YML034W
During the test of an intron prediction for YKL186C/MTR2, we uncovered the first example of natural alternative splicing in S.cerevisiae (Fig. 3A). RT–PCR gives rise to a complex pattern of products arising from RNA upstream of the predicted YKL186C ORF. Although one unspliced and three main splicing-derived PCR products are observed, the positions of potential splice sites do not allow determination of which 5′ splice sites are joined to which 3′ splice sites. Sequencing of cloned RT–PCR products provided evidence for use of five of the six possible splice site combinations. Given that splicing is not required to create the ORF, the unspliced RNA may also be one form of the mRNA. Three of the alternatively spliced forms would produce the same protein (Fig. 3A). Two spliced forms observed would code for different proteins, each of which would have a different small peptide sequence at the N-terminus. The unspliced leader contains three short ORFs of 20, 13 and 12 amino acids (uORFs; 24–26), and these are deleted or altered by splicing. Until appropriate mutant alleles are tested, we will not know which of the spliced forms are necessary or sufficient for supplying the function of YKL186C/MTR2. Since Mtr2p is intimately involved in nuclear export of mRNA, the complexity of MTR2 pre-mRNA splicing may have regulatory implications.
Figure 3.
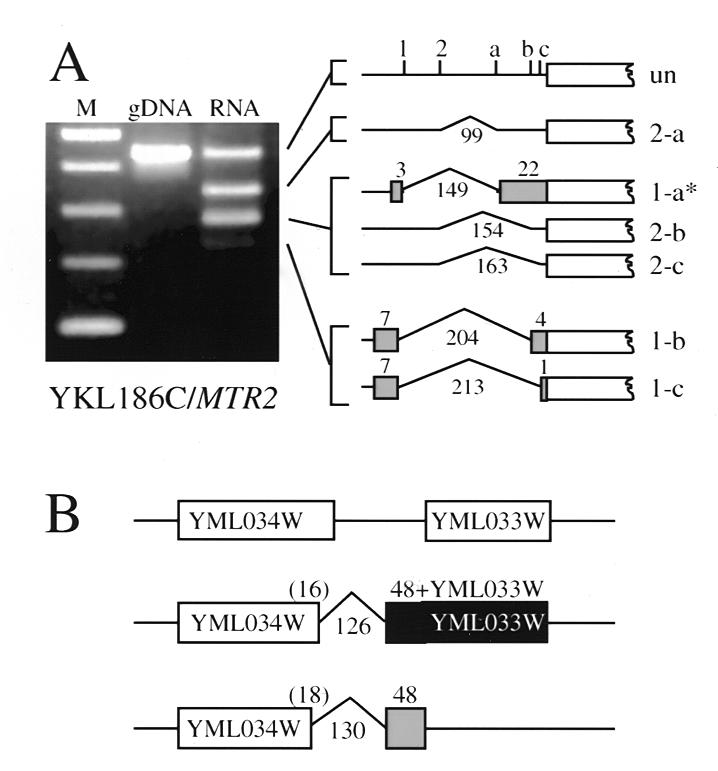
Alternatively spliced mRNAs. (A) At least six forms of mRNA that differ by splicing arise from the YKL186C region. PCR products from genomic DNA (gDNA, lane 1) and RT–PCR products from cDNA (RNA, lane 2) are compared to marker DNA (lane M, 100 bp ladder marker, the fastest migrating band is 100 bp, next fastest is 200 bp, etc.). Positions of the two 5′ splice sites (labeled 1 and 2) and the three 3′ splice sites (labeled a–c) relative to YKL186C are shown on the unspliced RNA (un) to the right of the gel. Different spliced forms of YKL186C mRNA and the migration of the corresponding PCR products are indicated. From top: un, unspliced; 2-a, 5′ splice site 2 joined to 3′ splice site (3′ss) a; 1-a*, 5′ss 1 joined to 3′ss a; 2-b, 5′ss 2 joined to 3′ss b; 2-c, 5′ss 2 joined to 3′ss c; 1-b, 5′ss 1 joined to 3′ss b; 1-c, 5′ss 1 joined to 3′ss c. Shaded boxes indicate additional amino acids encoded at the N-terminus of the YKL186C coding sequence. Each spliced form is identified by sequence of cloned PCR products except for 1-a, as indicated by the asterisk. The numbers above exon segments refer to amino acids encoded, the numbers below the introns refer to nucleotides removed by splicing. (B) Two forms of spliced RNA from the YML034W region. A previously unannotated intron near the 3′ end of YML034W uses two different 5′ splice sites. Top, positions of YML034W and YML033W (white boxes); middle, use of the downstream 5′ss leads to fusion of most of the YML034W (white box) coding sequence to 48 amino acids plus the coding sequence of YML033W (black box); bottom, use of the upstream 5′ss leads to fusion of most of the YML034W coding sequence to a different 48 amino acids (gray box) encoded in a different reading frame.
A second alternatively spliced RNA comes from the YML034W region through alternative use of two novel 5′ splice sites in a newly discovered intron (Table 1 and Fig. 3B). During the sequencing of RT–PCR products obtained in order to identify splice sites, we found that two different 5′ splice sites can be joined to the same 3′ splice site. One form of mRNA is spliced using a novel GUGAGU 5′ splice site that fuses most of the YML034W ORF to a 48 amino acid sequence that reads into the YML033W ORF (Fig. 3B). The other mRNA form is spliced using another novel 5′ splice site GCAAGU 4 nt upstream, which fuses most of YML034W to a different 48 amino acids in an overlapping reading frame. The function of YML034W is not known. The long form of the protein predicted to be produced by splicing at the GUGAGU splice site contains four closely spaced cysteines that could be a zinc binding element. This element is absent from the protein spliced at the GCAAGU splice site. It is possible that the shorter protein could act as a dominant-negative regulator of the full-length protein through control of this alternative splicing event.
Splicing of YGR225W/SPO70 is activated during meiosis by MER1
For the vast majority of introns tested, splicing took place with approximately equivalent efficiencies under the expression conditions we tested. In initial experiments with YGR225W/SPO70, we noted an increase in the amount of PCR product for spliced mRNA from meiotic cells, as compared to vegetative diploid cells. We measured splicing of YGR225W/SPO70 during a meiotic time course using a strain capable of synchronous and efficient meiosis (19,22). During meiosis and sporulation, efficiency of YGR225W/SPO70 splicing is low but increases, as shown by the increase in the ratio of spliced to unspliced RT–PCR product (Fig. 4A). Splicing of two other yeast introns, one in MER2 (16) and another in MER3 (17), is selectively activated by the product of the MER1 gene during meiosis (16,17,27).
Figure 4.
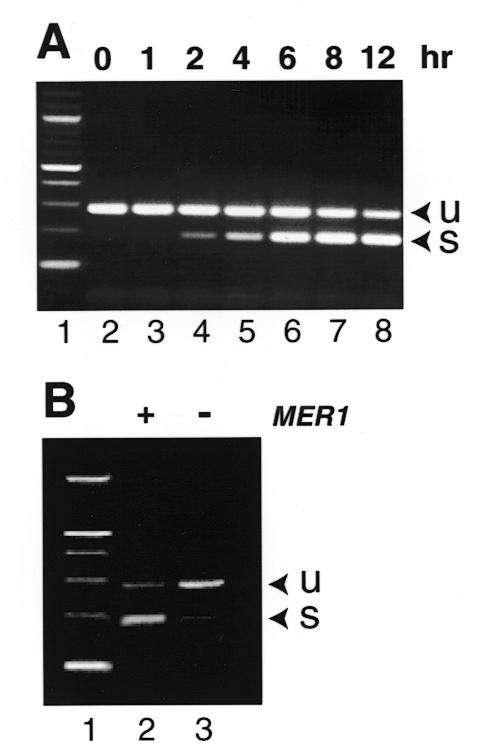
A meiosis-specific intron in YGR225W/SPO70. (A) Splicing efficiency of the intron during meiosis. An SK1 yeast strain was shifted to sporulation medium and RNA was extracted at the indicated times (hours) and subjected to RT–PCR using primers that span the YGR225W/SPO70 intron. Lane 1, 100 bp ladder as for Figure 3; lanes 2–8, 0–12 h after induction of sporulation. Arrow U, PCR signal derived from unspliced RNA; arrow S, PCR signal derived from spliced RNA. (B) Splicing of the YGR225W/SPO70 intron is activated by MER1. Haploid cells were transformed with a plasmid carrying a segment of YGR225W/SPO70 spanning the intron under control of a strong constitutive promoter, and a second plasmid either containing (lane 2, +) or lacking (lane 3, –) the MER1 gene. RNA was extracted and subjected to RT–PCR. Lane 1, 100 bp ladder and arrows are as for (A). The structure of YGR225W is shown in Figure 2A.
To test if YGR225W/SPO70 splicing is also activated by MER1, we cloned a fragment of YGR225W/SPO70 spanning the intron under the control of a constitutive promoter in order to express YGR225W/SPO70 RNA efficiently in vegetative cells. YGR225W/SPO70 splicing efficiency is low in vegetative cells (Fig. 4B, lane 3), but is significantly enhanced by the introduction of a plasmid containing MER1 under control of the ADH1 promoter (16). This and other data (not shown) indicate that the YGR225W/SPO70 intron is MER1-responsive, and that part of the regulation of expression of YGR225W/SPO70 during meiosis involves activation of its splicing (Fig. 4), as well as transcription (22). Splicing of the introns found in four other meiosis-specific genes is not selectively activated in meiosis, although the transcription of these genes is induced (22,28–31). Five other new introns in meiotic genes we have tested (YBR186W/PCH2, YDL115C, YLR093C/NYV1 YLR211C and YNL012W/SPO1; see Table 1) all appear to have similar splicing efficiencies in vegetative and sporulating diploids (data not shown), suggesting that their splicing is not selectively activated during meiosis.
DISCUSSION
We have tested 87 unsubstantiated predictions for non-ribosomal protein genes of Saccharomyces cerevisiae, nearly one-third of all known or suspected introns. Predictions were generated during annotation of the yeast genome (8,9) using computational tools (6). More than 70% of the predictions were correct, several introns were incorrectly placed, and no evidence for splicing could be obtained for numerous annotated introns. In addition to confirming and correcting suspected introns, we also found eight new introns (Table 1). Thus, this work provides the first experimental evidence for 75 yeast introns, 14 of which were not previously known. These and data on other new yeast introns have been incorporated into an updated database at http://www.cse.ucsc.edu/research/compbio/yeast_ introns.html
The information gained from this study allows significant improvement in our understanding of the set of spliceosomal introns in yeast. For example, many of the newly discovered introns are not near the N-terminus of the coding region. Nearly all of the new introns have non-canonical splice sites or branchpoints, extending the known repertoire of splice signals used in this organism. Most errant predictions mistakenly chose a distant canonical splice site over a more local non-canonical splice site. This is most likely due to overemphasis by the program on matches to splice site consensus relative to weak sites in strong context. Better data concerning actual splice site usage and bona fide intron structure, such as those provided here, will aid the development of more accurate and reliable intron prediction methods through a better understanding of splice site context.
Prior to this work, no natural alternatively spliced S.cerevisiae transcripts were known. In some cases, transcripts still retaining an intron have been suggested to produce alternative protein products (32), but thus far the only functional protein isoforms that have been shown to be coded from yeast mRNAs that differ by splicing are found in Schizosaccharomyces pombe (33). There, two isoforms of the SAP155 homolog derived from alternative 3′ splice site usage were shown to be functional. In this study we identified natural mRNAs from two S.cerevisiae genes, YKL186C/MTR2 and YML034W, that differ by alternative splicing events, rather than by simple intron removal or retention. For YKL186C/MTR2, we provide evidence for five different alternatively spliced mRNAs, each generated by a different combination of two alternative 5′ splice sites and three alternative 3′ splice sites, most likely through a common branchpoint region (Fig. 3A). The relative amounts of the different spliced forms, as estimated by the pattern of RT–PCR products, appear similar in all cell types investigated under constant PCR conditions (data not shown).
The introns are near the 5′ end of the YKL186C/MTR2 coding region; however, the predicted protein derived from several of the spliced forms differs, as does the existence of upstream mini-ORFs (uORFs) in the mRNA leader. We do not know whether the different Mtr2 proteins predicted from these mRNAs are functionally distinct, or how the existence of the uORFs might influence the translational efficiency of their mRNAs. Given the influence of uORFs on translation (24–26), it seems likely that the translation efficiency of different MTR2 mRNA isoforms is significantly different. If this is the case, and if alternative splicing of this complex region can be modulated, then expression of Mtr2p isoforms could be subject to multiple levels of regulation. Mtr2p mediates the association of Mex67p with the nuclear pore complex during nuclear export of mRNA (34,35), and thus levels of Mtr2p may generally control the rate of mRNA export. Mtr2p expression could provide the cell with a means of coordinating the linked processes of splicing, mRNA export and translation.
The second case of alternative splicing involves the use of two different 5′ splice sites in an intron in YML034W (Fig. 3B). The expression of this gene is induced during meiosis (22). Due to their proximity, the relative use of these two splice sites is difficult to determine directly using nuclease protection methods, and thus it is not currently possible to determine whether alternative splicing is regulated during meiosis or not. As mentioned above, the shorter version of the protein lacks a putative C4 zinc finger. If this protein requires this C-terminal domain for its function, and the N-terminal domain has a separate function as well, it is possible that the truncated isoform may act in a dominant-negative fashion. If this is the case, the alternative splicing event could be controlled to produce precise levels of activity by producing the active protein and an antagonizing protein in appropriate amounts. As with MTR2, the biological significance of alternative splicing in YML034W is not yet clear.
During the testing of predicted introns, we also explored the possibility that cell type-specific splicing might take place, by using RNA from vegetative haploid cells of different mating types, vegetative diploid cells or diploid cells undergoing meiosis. We identified an intron in YGR225W/SPO70 whose removal is activated by MER1 during meiosis (Fig. 4). Previously only MER2 and MER3 were known to have MER1-responsive introns (16,17). The meiotically transcribed gene MER1 encodes a KH-domain RNA binding protein required for activation of MER2 (16,27), MER3 (17) and SPO70 (Fig. 4B) splicing. Recent results indicate that MER1-responsiveness of the SPO70 intron is mediated through multiple separable elements, some which act negatively to reduce splicing efficiency whether MER1 is present or not, and others which act positively and only in conjunction with MER1 (M.Spingola and M.Ares, unpublished data). Thus, MER2, MER3 and SPO70 represent a splicing regulon in yeast, under the control of the MER1 splicing activator. Although several other meiotic genes have introns, their splicing does not appear to be selectively activated during meiosis (22,28–31). We found the same to be true for five other new introns in meiosis-induced genes (Table 1 and data not shown).
It may seem surprising that the genome sequence of yeast has been completed since 1996 and introns are still being found in 1999. If this scales proportionately to the human genome, it will take at least 750 years, the better part of the next millennium, to identify all human introns and hence the entire protein coding capacity of the human genome. Unfortunately, it is disproportionately harder to identify intron–exon structure in human sequence than in yeast sequence, because human genes have more introns, smaller exons, more degenerate splice sites and branchpoint sequences, as well as more complex and regulated splicing patterns than are known for yeast. With current technologies it is far easier to determine the complete sequence of a eukaryotic genome than it is to determine the complete coding capacity of a eukaryotic genome, due to the intron problem. Even using ESTs (3,4,36), or comparisons between related organisms (5), finding the complete intron set in a genome will require comprehensive prediction and experimental validation, using approaches not yet invented. Such tools will be essential to the understanding of genome function and evolution in eukaryotes.
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank A. H. Igel for expert and timely technical assistance, and R. Perriman, R. Nagel and A. Zahler for comments on the manuscript. This work was supported by NCI grant CA77813.
REFERENCES
- 1.Mewes H.W. et al. (1997) Nature, 387 (suppl.), 7–65.9169865 [Google Scholar]
- 2.Ainscough R. et al. (1998) Science, 282, 2012–2018.9851916 [Google Scholar]
- 3.Deutsch M. and Long,M. (1999) Nucleic Acids Res., 27, 3219–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent W.J. and Zahler,A.M. (2000) Nucleic Acids Res., 28, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansari-Lari M.A., Oeltjen,J.C., Schwartz,S., Zhang,Z., Muzny,D.M., Lu,J., Gorrell,J.H., Chinault,A.C., Belmont,J.W., Miller,W. and Gibbs,R.A. (1998) Genome Res., 8, 29–40. [PubMed] [Google Scholar]
- 6.Kalogeropoulos A. (1995) Yeast, 11, 555–565. [DOI] [PubMed] [Google Scholar]
- 7.Spingola M., Grate,L., Haussler,D. and Ares,M.,Jr (1999) RNA, 5, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chervitz S.A., Hester,E.T., Ball,C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Juvik,G., Malekian,A., Roberts,S., Roe,T., Scafe,C., Schroeder,M., Sherlock,G., Weng,S., Zhu,Y., Cherry,J.M. and Botstein,D. (1999) Nucleic Acids Res., 27, 74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodges P.E., McKee,A.H., Davis,B.P., Payne,W.E. and Garrels,J.I. (1999) Nucleic Acids Res., 27, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez P.J. and Seraphin,B. (1999) RNA, 5, 1135–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ares M. Jr, Grate,L. and Pauling,M.H. (1999) RNA, 5, 1138–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eng F.J. and Warner,J.R. (1991) Cell, 65, 797–804. [DOI] [PubMed] [Google Scholar]
- 13.Li Z., Paulovich,A.G. and Woolford,J.L.,Jr (1995) Mol. Cell. Biol., 15, 6454–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilardell J. and Warner,J.R. (1994) Genes Dev., 8, 211–220. [DOI] [PubMed] [Google Scholar]
- 15.Vilardell J. and Warner,J.R. (1997) Mol. Cell. Biol., 17, 1959–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engebrecht J.A., Voelkel-Meiman,K. and Roeder,G.S. (1991) Cell, 66, 1257–1268. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa T. and Ogawa,H. (1999) EMBO J., 18, 5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nandabalan K., Price,L. and Roeder,G.S. (1993) Cell, 73, 407–415. [DOI] [PubMed] [Google Scholar]
- 19.Padmore R., Cao,L. and Kleckner,N. (1991) Cell, 66, 1239–1256. [DOI] [PubMed] [Google Scholar]
- 20.Ares M. Jr and Igel,A.H. (1990) Genes Dev., 4, 2132–2145. [DOI] [PubMed] [Google Scholar]
- 21.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1987) Current Protocols in Molecular Biology. Greene/John Wiley and Sons, New York, NY.
- 22.Chu S., DeRisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 23.Gerber A.P. and Keller,W. (1999) Science, 286, 1146–1149. [DOI] [PubMed] [Google Scholar]
- 24.Vilela C., Ramirez,C.V., Linz,B., Rodrigues-Pousada,C. and McCarthy,J.E. (1999) EMBO J., 18, 3139–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polymenis M. and Schmidt,E.V. (1997) Genes Dev., 11, 2522–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinnebusch A.G. (1994) Trends Biochem. Sci., 19, 409–414. [DOI] [PubMed] [Google Scholar]
- 27.Nandabalan K. and Roeder,G.S. (1995) Mol. Cell. Biol., 15, 1953–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- 29.Malone R.E., Pittman,D.L. and Nau,J.J. (1997) Mol. Gen. Genet., 255, 410–419. [DOI] [PubMed] [Google Scholar]
- 30.Menees T.M., Ross-MacDonald,P.B. and Roeder,G.S. (1992) Mol. Cell. Biol., 12, 1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leu J.Y. and Roeder,G.S. (1999) Mol. Cell. Biol., 19, 7933–7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ner S.S. and Smith,M. (1989) Mol. Cell. Biol., 9, 4613–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habara Y., Urushiyama,S., Tani,T. and Ohshima,Y. (1998) Nucleic Acids Res., 26, 5662–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos-Rosa H., Moreno,H., Simos,G., Segref,A., Fahrenkrog,B., Pante,N. and Hurt,E. (1998) Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadowaki T., Hitomi,M., Chen,S. and Tartakoff,A.M. (1994) Mol. Biol. Cell, 5, 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gelfand M.S., Dubchak,I., Dralyuk,I. and Zorn,M. (1999) Nucleic Acids Res., 27, 301–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


