Abstract
Background:
Carbapenem-resistant Klebsiella pneumoniae, particularly isolates classified as sequence-type 258 (ST258), are multidrug-resistant strains that are strongly associated with poor-prognosis nosocomial infections, as current therapeutic options are limited and ineffective. In recent years, phage therapy has emerged as a promising treatment option for these scenarios.
Methodology and Results:
We report the isolation and characterization of three new phages against Klebsiella pneumoniae ST258 strains recovered from Machángara river wastewater. These new members of the Ackermannviridae family showed stability over a wide temperature and pH range and burst sizes ranging from 6 to 44 plaque-forming units per bacteria. Their genomes were about 157 kilobases, with an average guanine-cytosine content of 46.4% and showed presence of several transfer RNAs, which also allowed us to predict in silico a lytic replicative cycle due to the presence of endolysins and lysozymes.
Conclusion:
Three lytic phages of Ackermannviridae family were recovered against Klebsiella pneumoniae ST258 strains from sewage; however, further characterization is needed for future consideration as therapeutic alternatives.
Keywords: Ackermannviridae, Klebsiella pneumoniae, ST258, wastewater, tRNA, lytic
Introduction
The overuse of antibiotics has led to rapid spread of multidrug-resistant (MDR) Klebsiella pneumoniae virulent lineages, which are associated with high mortality rates especially in infections by isolates belonging to clonal complex 258, where isolates with sequence-type 258 (ST258) are considered as an “high-risk” clonal group causing a large proportion of infections, associated with worse prognosis.1–6 Among the alternative treatments proposed to address this public health crisis, bacteriophage (phage) therapy has demonstrated efficacy in experimental animal models and shows promise in clinical cases involving MDR pathogen infections, including ST258 strains.7–10
Moreover, the most common sources of isolation are relatively easy to access, such as sewage and hospital wastewater.11–14 For example, a recent study published by Hesse et al.8 evaluated the efficacy of early treatment with two phages, termed P1 and P2, in reducing bacterial load in infected mice with refractory bacteremia. The results of the study showed that early treatment with both phages, either individually or in combination, resulted in a significant reduction in bacterial load compared with placebo-treated animals. In addition, an improvement in survival was observed in mice treated with the phages compared with controls.8
Despite some promising results with other high-risk clonal lineages such as ST11, ST15, and ST16, currently available evidence remains insufficient for consideration beyond experimental treatments. Considering the current challenge posed by MDR lineages of K. pneumoniae, this study aims to address the knowledge gap by in vitro characterization of phages recovered from wastewater and evaluation of their lytic potential on ST258 strains for future development of effective therapeutic strategies.
Materials and Methods
Host strains, sample collection and isolation, enrichment, titration, and purification
Two clinical strains of Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae ST258 (14-765 and 14-751) donated by the Instituto Nacional de Investigación en Salud Pública–Leopoldo Izquieta Pérez (INSPI-LIP) from Ecuador were used as host bacteria. The results of antimicrobial susceptibility testing are in Supplementary Table S1. To obtain the exponential phase, 500 μL of a saturated broth (obtained from one or two isolated MacConkey agar colonies mixed in 4.5 mL of Tryptic Soy Broth [TSB] at 37°C and constant shaking at 9 × g overnight) was used beforehand. Next, 500 μL of the aforementioned mixture was added to 4.5 mL of TSB and incubated for 4 h at 180 rpm. A standard inoculum of 0.5 McFarland (1.5 × 108 CFU/mL) was used for all assays.
Samples of wastewater were collected from two different points of Machángara river, Quito-Ecuador. Sample named K0601 was collected from point one, coordinates: 0°12′34.7″ S 78°28′36.6″ W and Sample T0701 from collection point two, coordinates: 0°16′37″ S 78°31′14″ W, all samples were stored at 4°C for 24 h for sedimentation.
Bacteriophage isolation was performed according to the protocol described by Kropinski et al.15 and Clokie and Kropinski16 with certain modifications. In brief, 1 mL of the host strain in exponential phase culture, 49 mL of doubly concentrated TSB supplemented with 10 mM Ca2+ and Mg2+, and 50 mL of supernatant, previously filtered with filter paper (10 μm pore size filter) to separate the suspended solids present in the wastewater samples, were mixed. The mixture was incubated for 24 h at 37°C. The mixture was centrifuged at 1475 × g for 10 min to remove cells, and the supernatant was then filtered through a 0.22 μm pore size filter and stored at 4°C.
Phage presence was verified using the spot test, for which 50 μL of the filtered supernatant was dropped onto the host strain inoculated by extension on Tryptic Soy Agar supplemented with 10 mM Ca2+ (TSAd10) and incubated overnight.
For phage isolation, the double layer agar (DLA) technique was used using nutrient agar supplemented with 10 mM Ca2+. Serial dilutions and the DLA technique were performed with the previously filtered supernatant. In brief, 100 μL of exponential phase culture, 100 μL of dilution, and 5 mL of agar (top layer) were mixed and poured over the base layer and incubated. Procedure was repeated with all serial dilutions. Phage concentration was expressed in plaque-forming units per milliliter (PFU/mL). We selected potential phages with concentrations over 1 × 1010 PFU/mL.
For purification, a lysis plaque (105–108 PFU/mL) was taken, resuspended in 900 μL of buffer (MgSO4∙7H2O 8 mM, Tris-HCl 50 mM, pH 8.4) and shaken vigorously to dislodge the phage from the agar. The mixture was inoculated by extension on TSAd10 and left to stand for 30 min. Later, 500 μL of exponential phase culture was mixed with 5 mL of TSAd10 on top of the aforementioned agar and incubated for 20 h. The best defined and spaced plaques formed after 20 h were picked and stored at 4°C in 1 mL of buffer and 10 μL of chloroform for preservation
In vitro phage activity and stability
Phage characterization was performed by the method described by Manohar et al.17 Thus, 100 μL of exponential phase culture host bacteria and 100 μL of purified phage was mixed and incubated for 25 min. After centrifugation, the pellet was homogenized using 10 mL of TSB, the DLA technique was performed to obtain the number of phages. Burst size was calculated dividing the final number of free phages by the initial number of phages. For pH stability assay we added 100 μL of purified phage in 900 μL of saline solution adjusted to different pH and incubated for 4 h, whereas for thermostability assay a volume of 100 μL of purified phage was maintained at different temperatures for 1 h.
All assays were performed in triplicate and the DLA technique was applied. Adsorption rate was performed according to reference.18 In brief, 100 μL of the exponential phase host strain was mixed with 100 μL of purified phage, allowed to stand for 5 min at room temperature, 100 μL of this mixture was added in 9.9 mL of TSB and incubated for 1 h, every 10 min the DLA method was performed. Adsorption constant was calculated with the following equation:15 , where B is the initial concentration of bacteria, Po the initial titer, P the final titer, and t the time interval between Po and P. The constant was expressed in mL/min.
Genomic DNA extraction
The phage was suspended in buffer, concentrated by centrifugation and filtered through a 0.22 μm pore-size filter. DNA extraction was performed using Wizard Promega protocol and was modified by adding proteinase K and DNase I. 1 mL of concentrated phage was mixed with 10 μL of DNase I, 1 μL of RNase A, 500 μL of lysis solution and 4 μL of proteinase K, shaken and incubated at the following temperatures: 55°C (60 min), 65°C (15 min), then 320 μL isopropanol was added. Six hundred fifty microliters of the mixture was transferred to a spin column and centrifuged (1 min at 6000 g). Wash solution (90% ethanol) was added 400 μL and centrifuged again. Finally, we added 75 μL of rehydration buffer to the column, centrifuged again and obtained phage genomic DNA, which was stored at −20°C.
Complete genome sequencing and analysis
Sequencing was performed by Biosequence (Ecuador) in the Illumina MiSeq system. Raw sequencing data were deposited in the Sequence Reads Archive: BioProject: PRJNA815380, BioSamples: SAMN26549669 (K751), SAMN26549667 (T751), and SAMN26549668 (T765). Reads obtained were assembled de novo using Unicycler19 in PATRIC20. Quality and integrity of the viral metagenome-assembled genomes were evaluated with CheckV21 and subsequently annotated with Prokka22 in Galaxy.23
Genomes were aligned and rearranged with MAUVE.24 Prokaryotic virulence, toxin encoding, and antimicrobial resistance genes were searched for using Bakta.25 Our genomes recovered were deposited in GenBank: ON202820 (K751), ON323462 (T751), and ON399185 (T765). Taxonomic assignment was performed by KRAKEN226 and average nucleotide identity (ANI) was calculated to support it.27 The phylogenomic tree of the whole-genome sequences were generated by VICTOR.28 The replicative cycle was predicted in silico with BACPHLIP29 and PhageAI.30
Results
Phenotypic profile
Phages presence was evidenced by formation of lysis zones and plaques of variable size (ø 1.5–2.0 mm) with a translucent center and a surrounding halo (Supplementary Figs. S1 and S2). Three possible phages were isolated and named according to the recommendations of the International Committee on Taxonomy of Viruses:31 (Table 1) Klebsiella_virus_K751 (K751), Klebsiella_virus_T751 (T751) and Klebsiella_virus_T765 (T765). T751 and T765 were inactivated at temperatures >60°C, whereas K751 was completely inactivated at 80°C (Supplementary Fig. S3).
Table 1.
Characteristics of Phage Isolated Against Carbapenem-Resistant Klebsiella pneumoniae ST258 Strains
| K751 | T765 | T751 | |
|---|---|---|---|
| Host strain | 14–751 | 14–765 | 14–751 |
| Maximum concentration (PFU/mL) | 1.0 × 1011 | 9.0 × 1011 | 2.0 × 1010 |
| Phenotypic profile | |||
| Temperature (°C) | −16 to 70 | −16 to 60 | −16 to 60 |
| pH | 3 to 11 | 3 to 11 | 4 to 11 |
| Adsorption constant, K (mL/min) | 2.1 × 10−10 | 5.1 × 10−10 | 1.1 × 10−10 |
| Adsorption at 10 min (%) | 28.4 | 24.4 | 12.1 |
| Burst size (PFU/bacterial) | 6 | 10 | 44 |
| Latent period (minutes) | 35 | 45 | 35 |
| Taxonomy | |||
| Family | Ackermannviridae | Ackermannviridae | Ackermannviridae |
| Genus | Taipeivirus | Taipeivirus | Taipeivirus |
| Genomic profile | |||
| Size (bp) | 157.100 | 157.384 | 157.384 |
| Depth | 125.1 × | 219.2 × | 274.7 × |
| Content GC (%) | 46.5 | 46.4 | 46.4 |
| Integrity (%) | 94.9 | 94.9 | 94.8 |
| ORF | 197 | 195 | 196 |
| CDS | 204 | 205 | 204 |
| Hypothetical proteins | 188 | 188 | 190 |
| Functional proteins | 23 | 24 | 21 |
| tRNA | 5 | 6 | 6 |
| sRNA | 1 | 1 | 1 |
| In silico replicative cycle (%) | |||
| PhageAI (lytic) | 93.45 | 93.05 | 93.05 |
| BachliB (lytic) | 97.02 | 97.02 | 97.02 |
CDS, coding DNA sequences; GC, guanine-cytosine content; ORFs, open reading frames; PFU, plaque-forming units; sRNA, small RNA; tRNA, transfer RNA.
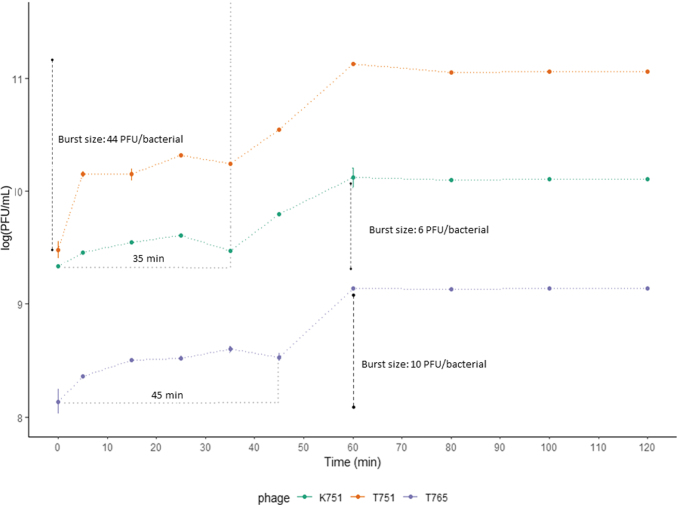
Regarding pH, the candidates survive in an environment of pH 4–11, and are completely inactivated at pH 12. K751 and T765 tolerated pH 3 (Supplementary Fig. S4). K751 and T751 reported a latency period of ∼35 min, whereas T765 was 45 min. Burst sizes (PFU/bacterial) were 6, 44, and 10, respectively (Fig. 1). In addition, at 10 min, it was estimated that 28.4% of K751, 12.1% of T751 and 24.4% of T765 viral particles were adsorbed on the host cell (Table 1).
FIG. 1.
Kinetics of Klebsiella_virus_K751, Klebsiella_virus_T751, and Klebsiella_virus_T765. The relationship between concentration and incubation time with their host cell is shown. Results are based on three replicates. PFU, plaque-forming units.
Genomic profile
Each recovered genome was characterized as double-stranded linear DNA of ∼157 kpb, >94% complete and an average guanine-cytosine (GC) content of 46.4%. The in silico predicted replicative cycle for all three candidates was lytic (Table 1). A multiple alignment of the rearranged genomes showed that the phages shared three syntenic collinear blocks (homologous regions) (Fig. 2), complemented by values >90% of ANI. This allowed us to deduce that our three phages are within the same species. About 190 open reading frames and 200 coding DNA sequences were annotated in each genome. Of the latter, >87% are hypothetical proteins.
FIG. 2.
Progressive multiple alignment performed with MAUVE to compare Klebsiella_virus_K751, Klebsiella_virus_T751, and Klebsiella_virus_T765. Each genome is arranged horizontally with homologous segments (locally collinear blocks) delineated as colored rectangles. Regions inverted with respect to the first genome taken as reference are placed below those that match in forward orientation.
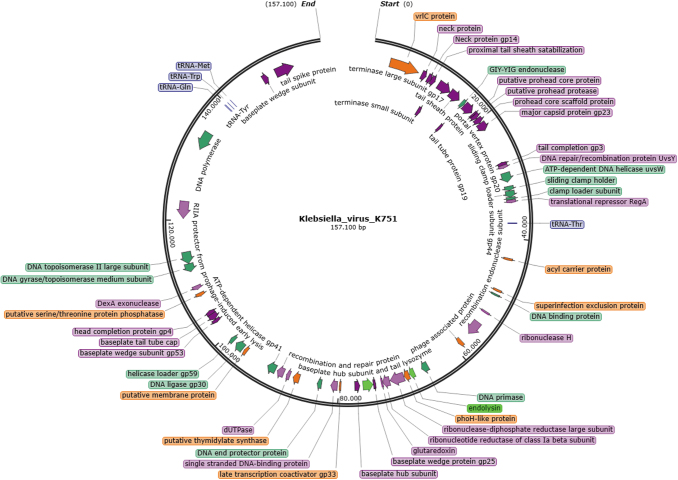
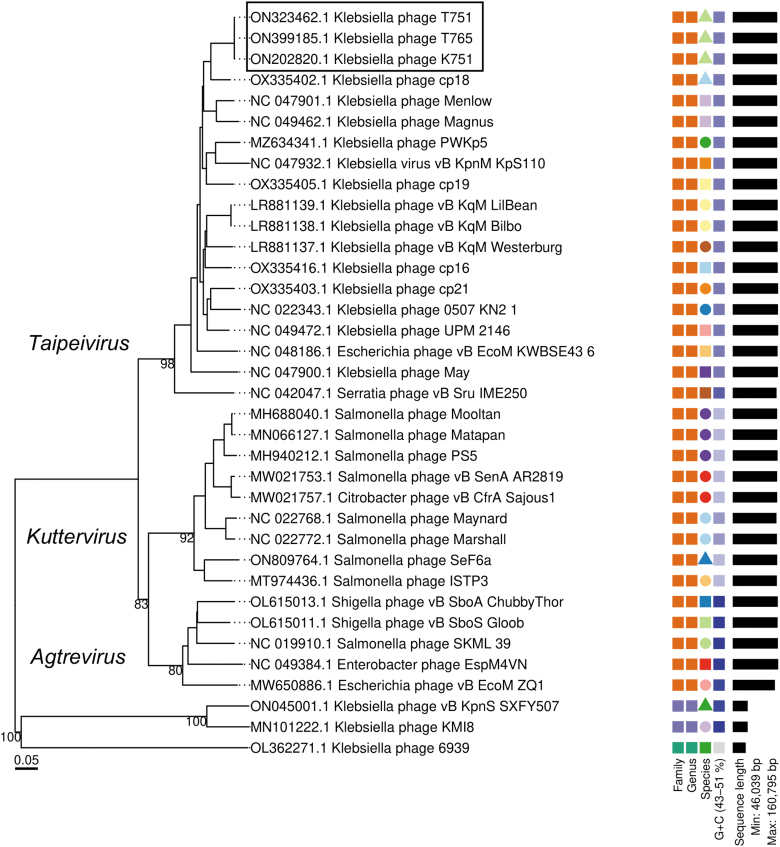
Five common transfer RNAs (tRNAs; tRNAThr, tRNAMet, tRNATrp, tRNATyr, and tRNAGln), and one additional (tRNAAsn) in T751 y T765. One small RNA (STnc100) was found in all three genomes. Prokaryotic virulence, toxin-encoding, and antimicrobial resistance genes were not present, demonstrating the absence of risk of transfer by lysogenization or phage-mediated transduction32 (Fig. 3; Supplementary Fig. S5). K751, T765, and T751 clustered with members of the genus Taipeivirus (Fig. 4), as preliminarily expected, as >90% of the raw reads were taxonomically classified within it.
FIG. 3.
Genomic map of Klebsiella_virus_K751. CDS are colored according to their function such as morphogenesis (purple), replication (green), recombination and repair (pink), lysis (light green), tRNA (blue), and other functions (orange). CDS, coding DNA sequences; tRNA, transfer RNA.
FIG. 4.
Phylogenomic tree generated by VICTOR using the complete genome sequences of Klebsiella_phage_T751, Klebsiella_phage_T765, Klebsiella_phage_K751 and members of the nearest Ackermannviridae according to BLASTn. Members of the families Drexlerviridae and Autographiviridae were used as outgroups.
Discussion
The emergence of carbapenem-resistant Klebsiella pneumoniae ST258 strains has led to strong therapeutic challenges for which some alternatives have been proposed. Among these alternatives is phage therapy, however, for many of these potential candidates, the minimum necessary information is not available or is still in the experimental stage. In the context of our research, we isolated three new lytic phages belonging to the family Ackermannviridae that showed significant genetic similarities despite different times and geographical origins.
Our results indicated that our phages belong to the same species. Reports of phages isolated from wastewater against ST258 strains7,8,12–14,33 have already shown that there is no exclusivity for a single family or taxonomic group.30,34,35 Despite the varied phylogenomic distance between families, mostly aquatic environments shared with their host,36–38 such as wastewater, which due to the presence of human, animal, and hospital waste, are a source rich and varied of these micro-organisms.39–41
After phenotypic analysis and considering the aforementioned, high stability under different temperature and pH conditions, and a latency period ranging from 10 to 40 min were expected.14,42–44 Short latency periods and large burst sizes are usually a combination that increases therapeutic potential; however, our values show a discrete difference, which could indicate that phage-host dynamics are being affected possibly due to the inherent facility of K. pneumoniae to generate spontaneous phage-resistant mutants.32,34,45–48
The presence of these mutants increases bacterial density and affects the adsorption rate49–51 due to poor receptor recognition47,48,52 resulting in slow adhesion and poor initial diffusion,53 in agreement with our report, in which <30% of free phages were adsorbed in the first 10 min, in contrast to Zurabov and Zhilenkov, who recommend that for the selection of therapeutic phages they should adsorb between 70% and 80% in the same period.14,44,54 We describe phages of genus Taipeivirus belong to Ackermannviridae family with lytic activity in KPC-producing K. pneumoniae.
After genomic analysis, the mean GC content (46.4%) of our phages was lower than expected for Klebsiella (57.5%), so it could infect other gram-negative bacteria, as previous studies corroborate that genomic GC content accurately predicts (>95%) potential hosts at the phylum level, but not at lower taxonomic levels.52,55,56 A marked organization of structural and functional genes was evident in each genome, in addition, genes encoding tail proteins (gp5, gp3, and gp17), required for capsid penetration, adsorption,57 and irreversible binding to their receptors,58–60 which have been extensively studied in common models such as enterobacteriophage T4 were also found.61
Furthermore, five similar and one additional tRNA were present in T751 and T765, which is in agreement with Bailly-Bechet et al. and Maganha de Almeida Kumlien et al.62 who state that a lytic phage contains an average of 4 tRNAs and even more, which are useful to compensate the genomic compositional differences with the host, and consequently achieve a more robust integration.61–63 The presence of tRNAs is possibly unique to lytic phages, as they have not been described in lysogenic phages. The tRNAs present correspond to codons abundant in the phage and probably rare in the host, which gives them an advantage over their competitors by translating their proteins more efficiently, reducing latency time and increasing the burst size.64–66 In addition, GC content and tRNAs could indicate the closeness between phage and host, although this relationship has not been explored in detail.66
Despite reporting encouraging results, phenotypic analysis of our phages revealed certain issues that limit their use as therapeutics and need to be further addressed before proposing them for in vivo or clinical trials.
Conclusion
Three lytic phages of the Ackermannviridae family were recovered against Klebsiella pneumoniae ST258 strains from sewage; however, further characterization is needed for future consideration as therapeutic alternatives. Furthermore, our findings support the strategic targeting of phages for infections by MDR pathogens.
Supplementary Material
Acknowledgments
The authors are grateful to the Dirección de Investigación of Universidad Central del Ecuador (UCE), Instituto de Investigación y Zoonosis (CIZ), Centro de Nanociencia y Nanotecnología (CENCINAT) of Universidad de las Fuerzas Armadas ESPE, and Instituto de Microbiología of Universidad San Francisco de Quito (IMUSFQ).
Authors' Contributions
J.A.R. and F.C.M. were responsible for conceptualization. E.T.G. and K.N.R. designed and carried out the methodology, formal analysis, validation, visualization, and research. TEM protocol and observation by K.V. and A.D. The writing—original draft preparation was elaborated by E.T.G. The writing—review and editing article were realized by E.T.G., F.C.M., K.N.R., J.A.R., and L.P. Supervision, project administration, and funding acquisition were conducted by J.A.R., L.P., and I.F. All authors have read and agreed to the published version of the article.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This study is supported by Call for proposals for senior research projects 2019 from Universidad Central del Ecuador, under the Project ID: DI-CONV-2019-052 titled in Spanish: “Alternativa a los antimicrobianos: aislamiento de bacteriófagos a partir de aguas residuales y aplicaciones en el sector salud e industria de alimentos.”
Supplementary Material
References
- 1. Andrey DO, Dantas PP, Martins WBS, et al. An emerging clone, Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae sequence type 16, associated with high mortality rates in a CC258-endemic setting. Clin Infect Dis 2020;71(7):E141–E150; doi: 10.1093/CID/CIZ1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marsh JW, Mustapha MM, Griffith MP, et al. Evolution of outbreak-causing carbapenem-resistant Klebsiella pneumoniae ST258 at a tertiary care hospital over 8 years. mBio 2019;10(5):e01945-19; doi: 10.1128/MBIO.01945-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ebomah KE, Okoh AI. An African perspective on the prevalence, fate and effects of carbapenem resistance genes in hospital effluents and wastewater treatment plant (WWTP) final effluents: A critical review. Heliyon 2020;6(5):e03899; doi: 10.1016/J.HELIYON.2020.E03899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Micozzi A, Gentile G, Santilli S, et al. Reduced mortality from KPC-K. pneumoniae bloodstream infection in high-risk patients with hematological malignancies colonized by KPC-K. pneumoniae. BMC Infect Dis 2021;21(1):1–9; doi: 10.1186/S12879-021-06747-8/TABLES/5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Protonotariou E, Meletis G, Pilalas D, et al. Polyclonal endemicity of carbapenemase-producing Klebsiella pneumoniae in ICUs of a Greek Tertiary Care Hospital. Antibiotics 2022;11(2):149; doi: 10.3390/ANTIBIOTICS11020149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reyes J, Cárdenas P, Tamayo R, et al. Characterization of blaKPC-2-Harboring Klebsiella pneumoniae isolates and mobile genetic elements from outbreaks in a Hospital in Ecuador. Microb Drug Resist 2021;27(6):752–759; doi: 10.1089/MDR.2019.0433 [DOI] [PubMed] [Google Scholar]
- 7. Thiry D, Passet V, Danis-Wlodarczyk K, et al. New bacteriophages against emerging lineages ST23 and ST258 of Klebsiella pneumoniae and efficacy assessment in Galleria mellonella Larvae. Viruses 2019;11(5):411; doi: 10.3390/V11050411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hesse S, Malachowa N, Porter AR, et al. Bacteriophage treatment rescues mice infected with multidrug-resistant Klebsiella pneumoniae ST258. mBio 2021;12(1):1–11; doi: 10.1128/MBIO.00034-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu JY, Lin TL, Chiu CY, et al. Decolonization of carbapenem-resistant Klebsiella pneumoniae from the intestinal microbiota of model mice by phages targeting two surface structures. Front Microbiol 2022;13:3110; doi: 10.3389/FMICB.2022.877074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anand T, Virmani N, Kumar S, et al. Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J Glob Antimicrob Resist 2020;21:34–41; doi: 10.1016/J.JGAR.2019.09.018 [DOI] [PubMed] [Google Scholar]
- 11. Balcão VM, Moreli FC, Silva EC, et al. Isolation and molecular characterization of a novel lytic bacteriophage that inactivates MDR Klebsiella pneumoniae strains. Pharmaceutics 2022;14(7):1412; doi: 10.3390/PHARMACEUTICS14071421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Andrea MM, Marmo P, Henrici De Angelis L, et al. ϕbO1E, a newly discovered lytic bacteriophage targeting carbapenemase-producing Klebsiella pneumoniae of the pandemic Clonal Group 258 clade II lineage. Sci Rep 2017;7(1):2614; doi: 10.1038/s41598-017-02788-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martins WMBS, Cino J, Lenzi MH, et al. Diversity of lytic bacteriophages against XDR Klebsiella pneumoniae sequence type 16 recovered from sewage samples in different parts of the world. Sci Total Environ 2022;839:156074; doi: 10.1016/J.SCITOTENV.2022.156074 [DOI] [PubMed] [Google Scholar]
- 14. Hesse S, Rajaure M, Wall E, et al. Phage resistance in multidrug-resistant Klebsiella pneumoniae st258 evolves via diverse mutations that culminate in impaired adsorption. mBio 2020;11(1):e02530-19; doi: 10.1128/mBio.02530-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kropinski AM, Mazzocco A, Waddell TE, et al. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol 2009;501:69–76; doi: 10.1007/978-1-60327-164-6_7 [DOI] [PubMed] [Google Scholar]
- 16. Clokie MRJ, Kropinski AM. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization and Interactions. 2009:68–74; Humana Press, a part of Springer Science+Business Media. doi: 10.1007/978-1-60327-164-67 [DOI] [Google Scholar]
- 17. Manohar P, Nachimuthu R, Lopes BS. The therapeutic potential of bacteriophages targeting gram-negative bacteria using Galleria mellonella infection model. BMC Microbiol 2018;18(1):97; doi: 10.1186/s12866-018-1234-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rahaman M, Rahman M, Rahman M, et al. Poultry salmonella specific bacteriophage isolation and characterization. Bangladesh J Vet Med 2014;12(2):107–114; doi: 10.3329/BJVM.V12I2.21264 [DOI] [Google Scholar]
- 19. Wick RR, Judd LM, Gorrie CL, et al. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017;13(6):e1005595; doi: 10.1371/JOURNAL.PCBI.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parrello B, Butler R, Chlenski P, et al. A machine learning-based service for estimating quality of genomes using PATRIC. BMC Bioinformatics 2019;20(1):1–9; doi: 10.1186/S12859-019-3068-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nayfach S, Camargo AP, Schulz F, et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol 2020;39(5):578–585; doi: 10.1038/s41587-020-00774-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014;30(14):2068–2069; doi: 10.1093/BIOINFORMATICS/BTU153 [DOI] [PubMed] [Google Scholar]
- 23. The Galaxy Community. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 Update. Nucleic Acids Res 2022;50(W1):W345–W351; doi: 10.1093/nar/gkac247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwengers O, Jelonek L, Dieckmann MA, et al. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom 2021;7(11):000685; doi: 10.1099/MGEN.0.000685/CITE/REFWORKS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol 2019;20(1):1–13; doi: 10.1186/S13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoon SH, Ha SM, Lim J, et al. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017;110(10):1281–1286; doi: 10.1007/S10482-017-0844-4 [DOI] [PubMed] [Google Scholar]
- 27. Meier-Kolthoff JP, Göker M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017;33(21):3396–3404; doi: 10.1093/BIOINFORMATICS/BTX440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hockenberry AJ, Wilke CO. BACPHLIP: Predicting bacteriophage lifestyle from conserved protein domains. PeerJ 2021;9:e11396; doi: 10.7717/PEERJ.11396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krupovic M, Dutilh BE, Adriaenssens EM, et al. Taxonomy of prokaryotic viruses: Update from the ICTV bacterial and archaeal viruses subcommittee. Arch Virol 2016;161(4):1095–1099; doi: 10.1007/S00705-015-2728-0 [DOI] [PubMed] [Google Scholar]
- 30. Townsend EM, Kelly L, Gannon L, et al. Isolation and characterization of Klebsiella phages for phage therapy. Phage (New Rochelle) 2021;2(1):26; doi: 10.1089/PHAGE.2020.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doub JB. Risk of bacteriophage therapeutics to transfer genetic material and contain contaminants beyond endotoxins with clinically relevant mitigation strategies. Infect Drug Resist 2021;14:5629; doi: 10.2147/IDR.S341265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Venturini C, Ben Zakour NL, Bowring B, et al. Fine capsule variation affects bacteriophage susceptibility in Klebsiella pneumoniae ST258. FASEB J 2020;34(8):10801–10817; doi: 10.1096/fj.201902735R [DOI] [PubMed] [Google Scholar]
- 33. Martins W, Cino J, Li M, et al. Lytic bacteriophages against mutidrug-resistant Klebsiella pneumoniae: Development of an effective phage-based approach to combat multidrug resistance. 2021;1–47; doi: 10.21203/RS.3.RS-850585/V1 [DOI] [Google Scholar]
- 34. Nazir A, Qi C, Shi N, et al. Characterization and genomic analysis of a novel drexlervirial bacteriophage IME268 with lytic activity against Klebsiella pneumoniae. Infect Drug Resist 2022;15:1533–1546; doi: 10.2147/IDR.S347110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sime-Ngando T. Environmental bacteriophages: Viruses of microbes in aquatic ecosystems. Front Microbiol 2014;5(JULY):355; doi: 10.3389/FMICB.2014.00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kasman LM, Porter LD. Bacteriophages. In: Brenner's Encyclopedia of Genetics (Second Edition). 2021; pp. 280–283; doi: 10.1016/B978-0-12-374984-0.00131-5 [DOI] [Google Scholar]
- 37. Clokie MRJ, Millard AD, Letarov AV, et al. Phages in nature. Bacteriophage 2011;1(1):31–45; doi: 10.4161/bact.1.1.14942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lawrence D, Baldridge MT, Handley SA. Phages and human health: More than idle hitchhikers. Viruses 2019;11(7):587; doi: 10.3390/V11070587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edham M, Al-Tae Firas, Al-Hammadi ATY. Inhibitory effect of bacteriophages isolated from sewage water in the City of Kirkuk on some types of human pathogenic bacteria. 2017. Available from: https://bsj.uobaghdad.edu.iq/index.php/BSJ/article/view/2416/2347 [Last accessed: October 2, 2022].
- 40. Nepal R, Houtak G, Karki S, et al. Genomic characterization of three bacteriophages targeting multidrug resistant clinical isolates of Escherichia, Klebsiella and Salmonella. Arch Microbiol 2022;204(6):1–9; doi: 10.1007/S00203-022-02948-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ilyas SZ, Tariq H, Basit A, et al. SGP-C: A Broad Host Range Temperate Bacteriophage; Against Salmonella gallinarum. Front Microbiol 2022;12:4042; doi: 10.3389/FMICB.2021.768931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kȩsik-Szeloch A, Drulis-Kawa Z, Weber-Da̧browska B, et al. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol J 2013;10(1):1–12; doi: 10.1186/1743-422X-10-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jamal M, Hussain T, Rajanna Das C, et al. Characterization of siphoviridae phage Z and studying its efficacy against multidrug-resistant Klebsiella pneumoniae planktonic cells and biofilm. J Med Microbiol 2015;64(4):454–462; doi: 10.1099/JMM.0.000040 [DOI] [PubMed] [Google Scholar]
- 44. Zurabov F, Zhilenkov E. Characterization of four virulent Klebsiella pneumoniae bacteriophages, and evaluation of their potential use in complex phage preparation. Virol J 2021;18(1):1–20; doi: 10.1186/S12985-020-01485-W [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kortright KE, Chan BK, Koff JL, et al. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019;25(2):219–232; doi: 10.1016/J.CHOM.2019.01.014 [DOI] [PubMed] [Google Scholar]
- 46. Fang Q, Feng Y, McNally A, et al. Characterization of phage resistance and phages capable of intestinal decolonization of carbapenem-resistant Klebsiella pneumoniae in mice. Commun Biol 2022;5(1):1–14; doi: 10.1038/s42003-022-03001-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Han ML, Nang SC, Lin YW, et al. Comparative metabolomics revealed key pathways associated with the synergistic killing of multidrug-resistant Klebsiella pneumoniae by a bacteriophage-polymyxin combination. Comput Struct Biotechnol J 2022;20:485–495; doi: 10.1016/J.CSBJ.2021.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shao Y, Wang IN. Bacteriophage adsorption rate and optimal lysis time. Genetics 2008;180(1):471; doi: 10.1534/GENETICS.108.090100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gallet R, Shao Y, Wang IN. High adsorption rate is detrimental to bacteriophage fitness in a biofilm-like environment. BMC Evol Biol 2009;9(1):1–12; doi: 10.1186/1471-2148-9-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dennehy JJ, Abedon ST. Adsorption: Phage acquisition of bacteria. In: Bacteriophages. (Harper DR, Abedon ST, Burrowes BH, et al. eds.) Springer: Cham; 2021; pp. 93–117; doi: 10.1007/978-3-319-41986-2_2 [DOI] [Google Scholar]
- 51. Song L, Yang X, Huang J, et al. Phage selective pressure reduces virulence of hypervirulent Klebsiella pneumoniae through mutation of the wzc gene. Front Microbiol 2021;12:2904; doi: 10.3389/FMICB.2021.739319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bonilla BE, Costa AR, van denBerg DF, et al. Genomic characterization of four novel bacteriophages infecting the clinical pathogen Klebsiella pneumoniae. Oxford 2021;28(4):dsab013; doi: 10.1093/dnares/dsab013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bull JJ, Gill JJ. The habits of highly effective phages: Population dynamics as a framework for identifying therapeutic phages. Front Microbiol 2014;5:618; doi: 10.3389/FMICB.2014.00618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pan Y-J, Lin T-L, Chen C-C, et al. Klebsiella Phage ΦK64–1 encodes multiple depolymerases for multiple host capsular types. J Virol 2017;91(6):2457–2473; doi: 10.1128/JVI.02457-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Edwards RA, McNair K, Faust K, et al. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev 2016;40(2):258–272; doi: 10.1093/FEMSRE/FUV048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ploss M, Kuhn A. Membrane insertion and assembly of epitope-tagged gp9 at the tip of the M13 phage. BMC Microbiol 2011;11:211; doi: 10.1186/1471-2180-11-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gorodnichev RB, Volozhantsev NV, Krasilnikova VM, et al. Novel Klebsiella pneumoniae K23-specific bacteriophages from different families: Similarity of depolymerases and their therapeutic potential. Front Microbiol 2021;12:2223; doi: 10.3389/FMICB.2021.669618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li X, Koç C, Kühner P, et al. An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Sci Rep 2016;6:26455; doi: 10.1038/SREP26455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang T, Lin H, Zhang L, et al. Expression and purification of recombinant lyase gp17 from the LSB-1 phage in Escherichia coli. Virol Sin 2015;30(1):69–72; doi: 10.1007/S12250-014-3527-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fokine A, Rossmann MG. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 2014;4(2):e28281; doi: 10.4161/bact.28281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bailly-Bechet M, Vergassola M, Rocha E. Causes for the intriguing presence of tRNAs in phages. Genome Res 2007;17(10):1486–1495; doi: 10.1101/GR.6649807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maganha de Almeida Kumlien AC, Pérez-Vega C, González-Villalobos E, et al. Genome analysis of a new Escherichia phage vB_EcoM_C2–3 with lytic activity against multidrug-resistant Escherichia coli. Virus Res 2022;307:198623; doi: 10.1016/J.VIRUSRES.2021.198623 [DOI] [PubMed] [Google Scholar]
- 63. Yang JY, Fang W, Miranda-Sanchez F, et al. Degradation of host translational machinery drives tRNA acquisition in viruses. Cell Syst 2021;12(8):771.e5–779.e5; doi: 10.1016/J.CELS.2021.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Delesalle VA, Tanke NT, Vill AC, et al. Testing hypotheses for the presence of tRNA genes in mycobacteriophage genomes. Bacteriophage 2016;6(3):e1219441; doi: 10.1080/21597081.2016.1219441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nielsen TK, Carstens AB, Browne P, et al. The first characterized phage against a member of the ecologically important sphingomonads reveals high dissimilarity against all other known phages. Sci Rep 2017;7(1):1–10; doi: 10.1038/s41598-017-13911-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Morgado S, Vicente AC. Global In-silico scenario of tRNA genes and their organization in virus genomes. Viruses 2019;11(2):180; doi: 10.3390/V11020180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.