Abstract
Exposure of pregnant F0 mouse dams to the obesogen tributyltin (TBT) predisposes unexposed male descendants to obesity and diverts mesenchymal stem cells (MSCs) toward the adipocytic lineage. TBT promotes adipogenic commitment and differentiation of MSCs in vitro. To identify TBT-induced factors predisposing MSCs toward the adipocytic fate, we exposed mouse MSCs to TBT, the peroxisome proliferator activated receptor gamma (PPARγ)-selective agonist rosiglitazone, or the retinoid X receptor (RXR)-selective agonist LG-100268. Then we determined their transcriptomal profiles to determine candidate microRNAs (miR) regulating adipogenic commitment and differentiation. Of the top 10 candidate microRNAs predicted by Ingenuity Pathway Analysis, miR-21, miR-33, and miR-223 were expressed consistent with an ability to differentially regulate target genes during adipogenesis. We found that 24-hour exposure to 50nM TBT caused miR-223 levels in MSCs to increase; expression of its target genes ZEB1, NFIB, and FOXP1 was decreased. Rosiglitazone and TBT increased miR-223 levels. This induction was inhibited by the PPARγ antagonist T0070907 but not by the RXR antagonists HX531 or UVI3003, placing miR-223 downstream of PPARγ. Chromatin immunoprecipitation confirmed TBT-induced binding of PPARγ to regulatory elements in the miR-223 promoter. miR-223 levels were elevated in white adipose tissue of F2 and F3 male descendants of pregnant F0 mouse dams exposed to 50nM TBT throughout gestation. miR-223 levels were potentiated in males fed an increased fat diet. We infer that TBT induced miR-223 expression and increased adipogenesis in MSCs through the PPARγ pathway and that transgenerationally increased expression of miR-223 plays an important role in the development of obesity caused by TBT exposure.
Keywords: microRNA, adipogenesis, obesity, PPARgamma, transgenerational, RXR, MSC, endocrine disruptor, EDC
Obesity is a worldwide public health issue that, although multifactorial in nature, is generally considered to be the result of energy imbalance (1). Compelling evidence suggests that obesity is a critical contributor to the development of adipose tissue inflammation and insulin resistance (2, 3), 2 major causal factors in the pathogenesis of obesity-associated type 2 diabetes (4). Therefore, adipose tissue itself is believed to play an important role in regulating obesity-associated health issues.
A growing number of studies have linked exposure to endocrine-disrupting chemicals and the obesity pandemic (5-8). One subset of endocrine-disrupting chemicals, obesogens, has been found to increase white adipocyte number and/or size and consequently promote adiposity (9). The obesogens tributyltin (TBT) and triphenyltin (TPT) activated the peroxisome proliferator activated receptor gamma (PPARγ), the master regulator of adipogenesis, and its heterodimeric partner, the retinoid X receptor (RXR) (10-13). In vitro studies showed that TBT promoted adipogenesis at low nanomolar levels in both human and mouse multipotent mesenchymal stromal stem cells (mesenchymal stem cells, MSCs) (14, 15). Using a mouse model, we showed that exposure of pregnant F0 mouse dams to TBT predisposed MSCs toward the adipose lineage in F1 offspring (15) and increased lipid accumulation in liver and white adipose depots in F1, F2, and F3 generations (9). Interestingly, male F4 descendants of F0 dams treated with TBT throughout pregnancy (16) or pregnancy and lactation (17) exhibited a large increase in fat storage compared with controls when fed with a diet containing modestly increased fat content (from 13.2% to 21.6% kcal from fat).
MicroRNAs (miRs) are a family of small noncoding RNAs of 21-23 nucleotides in length. miRs typically regulate protein synthesis at the posttranscriptional level by targeting specific mRNAs through perfect or imperfect base-pairing and triggering mRNA degradation or translational repression, respectively (18). Several studies suggested possible roles for miRs in adipogenesis. By targeting adipogenic transcriptional factors, such as PPARγ and CCAAT-enhancer-binding proteins (C/EBPs), miR-155 and miR-130 decreased white adipocyte formation (19, 20). On the other hand, miR-210 targeted TCF7l2 in the Wnt/β-catenin signaling pathway and promoted adipogenesis (21).
miR-223 is a highly conserved microRNA that was first reported to play a role in myeloid differentiation and inflammation (22, 23). miR-223 levels were reduced in hepatocellular carcinomas (24) and leukemias (25, 26), while its overexpression decreased viability of cancer cells (24). miR-223 regulated polarization of adipose-resident macrophages by targeting PBX/Knotted 1 Homeobox 1 (Pknox1). This resulted in suppression of nuclear factor kappa B (NF-κB) and c-Jun N-terminal kinase (JNK) pathways and enhanced adipocyte response to insulin stimulation (27, 28). Increased miR-223 expression was also reported during adipogenesis in mouse bone marrow stromal cells (29). Here, we show that the obesogen TBT induces miR-223 expression through the PPARγ pathway in mouse MSCs, leading to and enhancing adipogenesis. We also provide evidence that treatment of pregnant F0 mouse dams with TBT led to increased miR-223 expression in the white adipose tissue of male descendants through the F3 generation and that miR-223 expression is further elevated when dietary fat is increased. These observations support an important role for miR-223 downstream of PPARγ in the transgenerationally inherited predisposition to obesity.
Materials and Methods
Chemicals and Reagents
TBT, LG-100268 (LG268), HX531, UVI3003, dexamethasone, isobutylmethylxanthine, insulin, Nile Red, and Hoechst 33342 were purchased from Sigma-Aldrich (St. Louis, MO). T0070907 was purchased from Enzo Life Sciences (Farmingdale, NY). Rosiglitazone (ROSI) was purchased from Cayman Chemical (Ann Arbor, MI).
Cell Culture
Bone marrow–derived multipotent MSCs from the long bones of C57BL/6J mice were purchased at passage 6 (OriCell; Cyagen Biosciences, CA) and stored at passage 8 or 9 in liquid N2. Cells were maintained, as previously described (9), in Dulbecco's modified Eagle medium (DMEM) containing 10% calf bovine serum, 10 mM HEPES, 1 mM sodium pyruvate, 100 IU/mL penicillin, and 100 μg/mL streptomycin (13). MSCs were plated at 60 000 cells/cm2 in 12-well cell culture plates for adipogenesis assays. Cells were allowed to attach and acclimate for 24 hours prior to 48 hours of chemical treatment or adipogenesis. Specific ligands, 100nM ROSI, 50nM TBT, and 200nM LG268 were dissolved in DMSO and administered every 3 days throughout the duration of the adipogenesis assay. Antagonist assays were performed in parallel under the same conditions. The antagonist T0070907 (50 nM), HX531 (100 nM), UVI3003 (500 nM), or dimethyl sulfoxide (DMSO) vehicle control was added every 12 hours. The amount of DMSO vehicle was kept at < 0.1% in all assays. miRCURY LNA microRNA inhibitor (anti-miR-223) (Qiagen, Hilden, Germany) is the sequence-specific and chemically modified oligonucleotide that specifically targets and knocks down miR-223 miRNA molecules.
Adipogenesis Assay
Once cells reached 100% confluency in culture plates, cells were induced to differentiate with an adipose induction cocktail (500μM isobutylmethylxanthine, 1μM dexamethasone, and 5 μg/mL insulin) in minimal essential medium α (αMEM) containing 15% fetal bovine serum, 10 mM HEPES, 2 mM l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin as we described (17). Cells were replenished with fresh media, differentiation factors, and chemical ligands every 3 days. Cells were differentiated over the course of 14 days, fixed in buffered 3.7% formaldehyde, followed by one wash of PBS for 1 minutes, and then maintained at 4 °C in PBS overnight to remove residual phenol red. To quantify lipid accumulation, Nile Red (1 μg/mL) was used to stain neutral lipids and Hoechst 33342 (1 μg/mL) to stain DNA. For each biological replicate, Nile Red fluorescence units (excitation 485 nm, emission 590 nm) (RFU) were measured relative to Hoechst RFU (excitation 355 nm, emission 460 nm) using a SpectraMax Gemini XS spectrofluorometer (Molecular Devices, Sunnyvale, CA) using SoftMax Pro (Molecular Devices) (13).
Quantitative Polymerase Chain Reaction
Differentiated adipocytes were lysed with Trizol following the manufacturer's recommended protocol (Thermo Fisher Scientific, MA) and total RNA recovered after isopropanol precipitation (Fisher Chemical, PA). Complementary DNA of microRNAs was synthesized from 5 μg total RNA using microRNA-specific primers (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Gene expression was assessed with real-time quantitative polymerase chain reaction (qPCR) using SYBR Green PCR Master Mix (Thermo Fisher Scientific, MA) on a Roche LightCycler 480 II (Roche, Switzerland). Cycle threshold values were quantified as the second derivative maximum using LightCycler software (Roche, Switzerland). Noncoding small RNA control sno202 served as an endogenous reference gene (30). The 2−ΔΔCt method (31) was used to analyze real-time qPCR data and determine relative quantification. Standard propagation of error was used throughout for each treatment group (32, 33). Error bars represent the standard error of the mean (SEM) from 3 to 4 biological replicates, calculated using standard propagation of error.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using an established method as previously described (13). Briefly, MSCs were plated and treated in 10-cm dishes (Thermo-Nuncalon) to obtain adequate material. At the end of chemical treatment (48 hours), cells were fixed at room temperature for 10 minutes with 1% paraformaldehyde (Fisher Chemical, PA) in DMEM, followed by an ice-cold phosphate-buffered saline wash, and then quenched for 5 minutes with 125 mM glycine at room temperature. Fixed cells were washed and then collected by scraping the plates, centrifuging, and then resuspending in phosphate-buffered saline at 107 cells/mL. Equal numbers of cells were snap-frozen in liquid N2 and stored at −80 °C. To isolate nuclei, cell pellets were lysed at 4 °C for 10 minutes with a gentle detergent recipe consisting of 50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Nonidet P-40, 0.25% Triton X-100, and Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific, MA). Next, nuclei were recovered by centrifugation at 8000g for 15 minutes, washed for 10 minutes at room temperature (10mM Tris-HCl, pH 8.0, 200mM NaCl, 1mM EDTA, 0.5mM EGTA, protease inhibitors [Thermo Fisher Scientific, MA]), and lysed in 300 μL nuclear lysis buffer (10mM Tris-HCl, pH 8.0, 200mM NaCl, 1mM EDTA, 0.5mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, protease inhibitors [Thermo Fisher Scientific, MA]). Chromatin samples were prepared by sonicating in 0.5 mL thin-walled polymerase chain reaction tubes (BrandTech, CT) using a QSonica Q800R2 (QSonica, CT) with the following settings: 30 seconds on/30 seconds off, amplitude 40% repeated for 30 minutes. Triton X-100 (1%) was added to sonicated lysates prior to high-speed, cold centrifugation to remove debris. A total of 5 μg DNA was immunoprecipitated with pre-blocked protein A/G Dynabeads (Thermo Fisher Scientific, MA) complexed to 2.5 μg antibody (anti-PPARγ, ab233218, Abcam, Cambridge, UK). Beads were washed 3 times with LiCl buffer (50 mM HEPES-KOH, pH 7.5, 500 mM LiCl, 1 mM EDTA, 1% Nonidet P-40, 0.7% Na-deoxycholate) and once with Tris-EDTA buffer plus 50 mM NaCl. To release chromatin from beads, pelleted beads were resuspended in elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% sodium dodecyl sulfate) and incubated at 65 °C for 30 minutes. Cross-link reversal was performed overnight at 65 °C. DNA samples were purified using the ChIP DNA Clean & Concentrator kit (Zymo Research, CA) following RNase A (0.2 mg/mL, 2 hours, 37 °C) and proteinase K (0.2 mg/mL, 2 hours, 55 °C) treatment. Input DNA content was determined by spectrophotometry (Nanodrop, Thermo Fisher Scientific, MA).
RNA Sequencing and Bioinformatics
Transcriptomal profiles were generated and published previously (13). In brief, total RNAs integrity was verified using Tapestation high-sensitivity RNA screen tapes (Agilent Technologies, Santa Clara, CA), RNA integrity numbers ranged from 8.0 to 8.6. Strand-specific, barcode-indexed RNA sequencing (RNA-seq) deep-sequencing libraries were synthesized from total RNA with ERCC spike-in controls (Thermo Fisher Scientific, MA) using Ovation RNA-Seq Systems 1-16 for Mouse (NuGen Technologies, CA). Tapestation analysis determined the size distribution of the libraries to be 200 to 800 bp, peaking at 300 bp. KAPA Illumina library quantification kit (KAPA Biosystems, MA) was utilized to quantify RNA-seq libraries, and up to 12 libraries were multiplexed in each run of the Illumina NextSeq500 deep sequencer (75 nt + 75 nt, paired-end) to generate fastq raw read sequence files which were aligned to the mouse genome reference sequence GRCm38/mm10 using the STAR aligner (34), and the resulting bam format aligned reads were subjected to quality control analysis using fastQC (Babraham Institute, Cambridge, UK), followed by extraction of uniquely mapped reads using SAMtools (35). Differential expression was assessed in DESeq2 (36) using the DESeq function with α = 0.01. Differentially expressed genes (DEGs) were defined by Benjamini–Hochberg corrected P values (P-adj < 0.01). Sequencing files have been uploaded and are available on Gene Expression Omnibus (GSE216429) (37). Candidate microRNA enrichments were computed by the microRNA-mRNA correlation feature supported by Ingenuity Pathway Analysis (IPA) (38). P values were corrected for multiple testing using the Benjamini–Hochberg method.
Mouse Treatment
We undertook a new transgenerational experiment based on our previous transgenerational model (17), denoted as T4. Briefly, 148 female and 50 male C57BL/6J mice (5 weeks of age) were purchased from The Jackson Laboratory and randomly assigned to treatment groups and exposed via drinking water to 50 nM TBT or 0.1% DMSO vehicle (all diluted in 0.5% carboxymethyl cellulose in water to maximize solubility) for 7 days prior to mating as we have described (16). Sires were never exposed to the treatment. Treatment was removed during mating, then resumed after males were removed until F1 (first generation) offspring were born. F2 (second generation) offspring were exposed to TBT as germ cells in the exposed F1 embryos. F3 (third generation) offspring were not exposed to TBT. One male and one female were randomly selected per litter for analysis. In diet challenge experiment, designated F2 (14 males and 14 females for each group) and F3 (15 males and 15 females for each group) were switched to a higher-fat diet (PicoLab Rodent Chow, 5058, 21.4% kcal from fat) whereas control groups (F2: 15 males and 15 females; F3: 12 males and 12 females) were maintained on a standard chow diet (PicoLab Rodent Chow, 5053, 13.2% kcal from fat). F2 offspring started diet challenge at 4 weeks age for 8 weeks when a significant fat content increase was confirmed and persisted. F3 offspring started diet challenge at week 17 for 5 weeks when a significant fat content increase was confirmed and persisted. Mice were fasted for 12 hours prior to euthanasia and tissue collection.
Statistical Analysis
GraphPad Prism 7.0 (GraphPad Software, Inc.) was used to perform statistical analysis for all datasets. A one-way analysis of variance (ANOVA) followed by Dunnett's post hoc test was performed to compare the treatment group, ROSI, TBT, or LG268, to DMSO control in adipogenesis assays. In antagonist assays, treatment groups without antagonists were compared to a corresponding group co-treated with T0070907, HX531, or UVI3003 using a Student t test. P ≤ .05 was considered statistically significant.
Results
Expression of miR-21, miR-33, and miR-223 Was Significantly Changed During Adipogenesis of MSCs
We previously analyzed the transcriptomal profiles of MSCs during adipogenesis (13). To predict candidate microRNAs involved in the 14-day process of adipose lineage commitment and differentiation in MSCs, we first utilized Ingenuity Pathway Analysis of differentially expressed genes defined by Benjamini–Hochberg corrected P values (P-adj < 0.01). The top 10 predicted microRNAs were miR-124, miR-30, miR-29, miR-33, miR-8, miR-21, miR-515, miR-223, miR-16, and miR-221 (Supplementary Figs. S1 and S2) (39). We next validated expression of these 10 microRNAs using real-time qPCR and found that 7 of them did not exhibit statistically significant changes in expression during adipogenesis (Supplementary Fig. S3) (39) and were not further considered. miR-21 levels remained unchanged during adipogenesis in vehicle-treated MSCs whereas levels showed a decreasing trend that never reached statistical significance starting at adipogenesis day 4 in MSCs treated with 50 nM TBT (Fig. 1A). miR-33 levels decreased during the 14-day adipogenesis in vehicle-treated MSCs, and TBT treatment partly relieved this suppression (Fig. 1B). miR-223 levels increased by adipogenesis day 7 (2.62- fold compared to day 0) the continued to increase at day 10 (3.5-fold) and day 14 (5.35-fold) compared to day 0) (Fig. 1C). This echoed previous studies of miR-223 expression in obese mice or humans (29, 41). TBT treatment enhanced adipogenesis-induced miR-223 expression from 5.35-fold (DMSO) to 15.84-fold (TBT) by day 14 (Fig. 1C). We next examined the transcriptomal profiles which were used to predict candidate microRNAs and confirmed that mRNA levels of 3 known miR-223 targets—namely, Foxp1, Zeb1, and Nfib—were significantly suppressed by both 50 and 100 nM TBT at days 4, 7, and 14 of adipogenesis (Fig. 1D-1F). We infer that TBT treatment accelerated adipogenesis-induced expression of miR-223, which inhibited expression of these target mRNAs.
Figure 1.
miR-223 expression is induced whereas mi-R223 target genes are suppressed during TBT-enhanced adipogenesis in MSCs. (A–C) qRT-PCR quantitation of 3 microRNAs, miR-21 (A), miR-33 (B), and miR-223 (C) in MSCs at adipogenesis day 0-14. Expression was normalized to sno202 and presented as relative expression to vehicle control at day 0 (mean ± SEM, *P < 0.05, **P < 0.01 and ***P < 0.001). (D-F) RNA-seq determination of mRNA transcripts of miR-223 target genes FOXP1 (D), ZEB1 (E), and NFIB (F) in mMSCs exposed to 0-100 nM TBT at adipogenesis day 0-14. RNA-seq data were described in our previous study (40).
TBT-Induced miR-223 Expression Is Critical for TBT-enhanced Adipogenesis
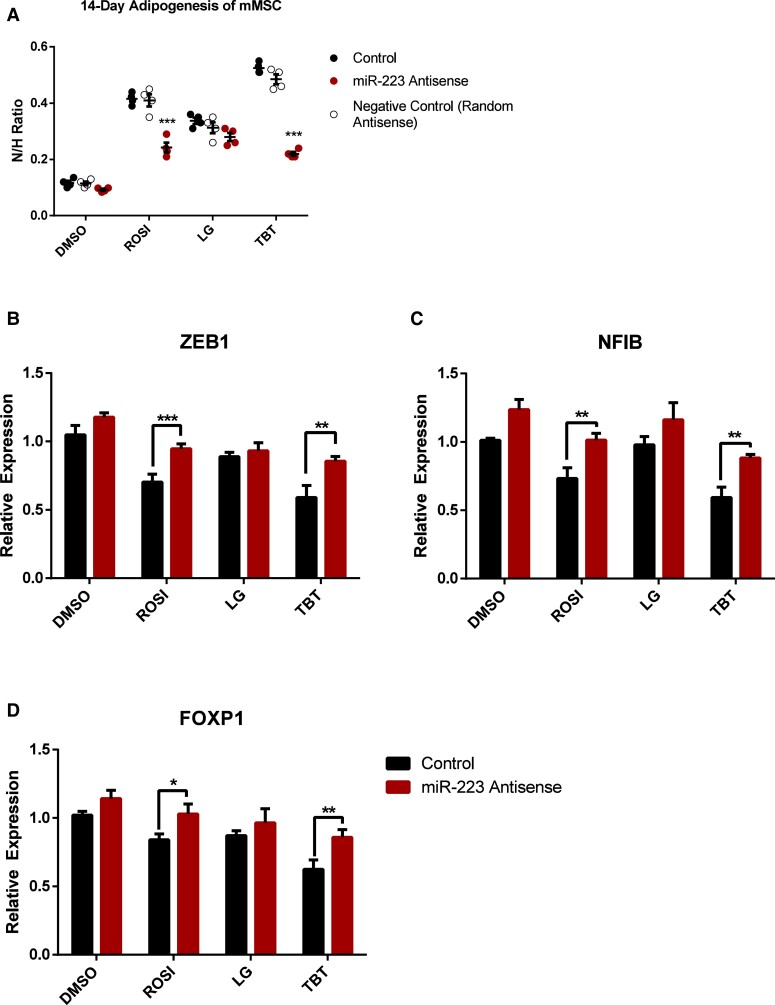
TBT acts through the nuclear receptor PPARγ and its heterodimeric partner RXR to promote adipogenesis in vitro and in vivo (9, 11, 13). To elucidate the role of TBT-induced miR-223 in adipogenesis, we determined effects of 50 nM TBT on adipogenesis of MSCs as described (13). The PPARγ-selective activator ROSI (100 nM) and the selective RXR activator LG268 (100 nM) were used as positive controls for PPARγ or RXR activation, respectively. The microRNA suppression assays employed miR-223 antisense; a random antisense microRNA served as negative control. The treatment of miR-223 antisense RNA significantly attenuated ROSI-induced and TBT-induced lipid accumulation after 14 days of adipogenesis (Fig. 2A). Expression of Zeb1, Nfib, and Foxp1 was decreased by ROSI and TBT after 14 days of adipogenesis while co-treatment with miR-223 antisense reversed these effects (Fig. 2B-2D). We infer that TBT enhances miR-223 expression in MSCs to support in vitro adipogenesis and reduce steady-state levels of 3 target mRNA transcripts of miR-223. These data also support a role for PPARγ in regulation of miR-223 expression.
Figure 2.
TBT-induced miR-223 expression contributes to TBT-enhanced adipogenesis in MSCs. (A) Adipogenesis assay was performed using mMSCs in the presence of adipogenesis cocktail (MDI). Cells were treated with 100nM ROSI, 50nM TBT, or 100nM LG268 in the presence of miR-223 antisense or random antisense as a negative control. All treatment groups were statistically compared to the control (black bars, 0.1% DMSO during 14-day adipogenesis). Lipid accumulation is shown as the ratio between fluorescence units (RFU) of Nile Red (N) and Hoechst (H), which were used to quantify lipid content and the number of cells per well, respectively. Each bar represents average of 4 replicates ± SEM. (B-D) qRT-PCR quantitation of mRNA transcripts of miR-223 target genes ZEB1 (B), NFIB (C), and FOXP1 (D) in mMSC-derived differentiated adipocytes at day 14. Expression was normalized to housekeeping gene Ribosomal Protein Lateral Stalk Subunit P0 (RPLP0) and presented as relative expression to the group of DMSO exposure with negative control, the random antisense oligonucleotides (mean ± SEM, *P < 0.05, **P < 0.01 and ***P < 0.001).
TBT Induced miR-223 Expression Through PPARγ
To assess whether TBT enhanced miR-223 expression via RXR or PPARγ activation, we first asked if TBT alone induces miR-223 expression. We treated naïve MSCs with 50 nM TBT, 100 nM ROSI, or 100 nM LG268 for 24 or 48 hours (Fig. 3). Expression of miR-21 and miR-33 expression was not changed by TBT (Supplementary Fig. S4) (39). Expression of miR-223 was increased in 24 hours by TBT (3.25-fold), ROSI (3.8-fold) and LG268 (2-fold) (Fig. 3A). Expression continued to increase at 48 hours (∼4-fold) in TBT and ROSI groups while expression in the LG268 group plateaued compared to controls (Fig. 3A). Treatment with a selective PPARγ antagonist (T0070907) revealed that TBT-induced expression of miR-223 was strongly attenuated by antagonizing PPARγ (Fig. 3B). In contrast, treatment with the RXR antagonist HX531 decreased miR-223 expression (Fig. 3C) only marginally while treatment with a different RXR antagonist, UVI3003 (Fig. 3D), had no effect (Fig. 3D). Treatment with the RXR activator LG100268 induced expression of miR-223 modestly. Since PPARγ antagonists blunted miR-223 induction by ROSI and TBT, but RXR antagonists had no significant effect, we infer that TBT induced miR-223 expression by activating PPARγ rather than RXR.
Figure 3.
Effects of chemical inhibitors of nuclear receptors on TBT-induced miR-223 expression in MSCs: qRT-PCR determination. (A) miR-223 expression after 0, 24 or 48 hours of exposure to 100nM ROSI, 50nM TBT, or 100nM LG268. (B) Expression after 0, 24, or 48 hours of exposure to 50nM TBT in the presence of 5nM or 50nM T0070907. (C) Expression after 0, 24, or 48 hours of exposure to 50nM TBT in the presence of 10nM or 100nM HX531. (D) Expression after 0, 24 or 48-hours exposure to 50nM TBT in presence of 100-1000nM UVI3003. Expression was normalized to sno202 and presented as relative expression to the day 0 controls (mean ± SEM, **P < 0.01 and ***P < 0.001).
TBT Induces the Binding of PPARγ to Regulatory Elements Upstream of the Pre–miR-223 Coding Region
PPARγ regulatory elements (PPREs) locating upstream of the pre–miR-223 coding region were previously described (28, 42). We used chromatin immunoprecipitation (ChIP) with antibodies against PPARγ to identify which, if any, of these PPREs interacted with PPARγ upon exposure to TBT. Cells were treated for 48 hours with 50nM TBT, 100nM ROSI, or 100nM LG268 in the presence or absence of T0070907 or HX531 in the TBT groups. Enrichment of PPREs was measured by qPCR with primer pairs flanking the PPREs. Compared with the negative control of IgG pull-down groups (Supplementary Fig. S5) (39), our results showed no PPARγ binding enrichment in the PPRE −270 to −275 closest to the transcription start site of miR-223 pre-RNA (Fig. 4A). We found mild, but significant, enrichments in 2 PPREs, −1125 to −1130 and −1305 to −1310 (Fig. 4C and 4D), and strong enrichment in 2 PPREs, −1011 to −1016 and −3919 to −3924 (Fig. 4B and 4E). T0070907 treatment strongly decreased PPARγ enrichments of the 4 positive PPREs (Fig. 4B-4D) whereas HX531 weakly suppressed TBT-induced PPARγ enrichments of 3 PPREs, −1011 to −1016, −1125 to −1130, and −1305 to −1310 (Fig. 4B-4D). Taken together, these ChIP assays demonstrated that TBT induced the binding of PPARγ to 4 out of 5 upstream PPREs of pre–miR-223, suggesting direct transcriptional activation of miR-223 expression by TBT-activated PPARγ.
Figure 4.
TBT-dependent binding of PPARγ to PPREs upstream of the pre–miR-223 transcription initiation site. MSCs were exposed to 100nM ROSI, 50nM TBT, 100nM LG268, or 50nM TBT for 48 hours in presence of 50nM T0070907 or 100nM HX531. PCR-based ChIP assays were performed using an anti-PPARγ antibody for PPREs at the regions indicated in each panel. Each column represents mean ± SEM of 4 replicated assays (**P < 0.01 and ***P < 0.001 compared to control; + P < 0.05, ++ P < 0.01 and +++ P < 0.001 compared to TBT).
miR-223 Expression in Gonadal White Adipose Tissue of F2 and F3 Male Mice
The effects of prenatal TBT treatment on obesity have been reported as transgenerational and detectable in the F1, F2, F3, and F4 descendants of F0 mice exposed during pregnancy (9) or during pregnancy and lactation (17), whereas ROSI was unable to elicit transgenerational obesity (9). The mechanism through which TBT acts to promote these transgenerational effects remains unclear. We next sought to examine the miR-223 levels in the adipose tissues of F2 and F3 animals ancestrally exposed to TBT following the procedure we previously published (17). MicroRNA qPCR revealed a statistically significant increase in miR-223 expression in F2 and F3 adipose tissue of male mice exposed to regular chow diet (Fig. 5A). Male offspring in the TBT group that were exposed to a diet with elevated fat levels (21.6% vs 13.2% in chow) resulted in diet-induced obesity; these TBT group males showed strongly elevated miR-223 expression in both F2 (3.1-fold) and F3 (2.2-fold) generations compared with the vehicle group (Fig. 5B). miR-223 levels were slightly increased in females from the TBT group vs controls in both chow diet and higher-fat diet females, but this increase did not reach statistical significance (Fig. 5C and 5D). These data demonstrate that the transgenerational obesity caused by ancestral exposure to TBT is associated with increased expression of miR-223 in white adipose tissue.
Figure 5.
miR-223 expression in gonadal white adipose tissue (gWAT) of F2 and F3 mice. Amounts of miR-223 mRNA in gWAT of males (A, B) and females (C, D) were determined using qRT-PCR. gWATs were collected before (A, C) or after (B, D) the higher-fat diet challenge. Expression was normalized to sno202 and presented as relative expression to DMSO controls (mean ± SEM, n = 11; *P < 0.05, **P < 0.01 and ***P < 0.001).
Discussion
We previously examined effects of TBT on adipocytic differentiation of mouse MSCs and analyzed the transcriptomal profiles during the course of their in vitro adipogenesis (13). Here, we utilized these transcriptomal data to identify microRNAs that could be involved in adipogenesis in MSCs. We revealed roles of microRNAs in TBT-induced adipogenesis, particularly miR-223. Inhibition of PPARγ action by specific antagonists or microRNA suppression assays using miR-neutralizing antisense RNAs identified a critical role for miR-223 in adipogenesis. Our finding that PPARγ binds to PPREs in the promoter of the gene encoding pre–miR-223 in an agonist-dependent manner supports a direct role of PPARγ in regulating miR-223 expression.
To date, knowledge about miR-223 in obesity has been limited to its role in adipose-resident macrophages (27) as a regulator of myeloid lineage development (23, 43, 44). Here, we identified an axis involving TBT, PPARγ, and miR-223 that enhances differentiation of white adipocytes from MSCs. Overexpression of synthetic miR-223 mimic or lentivirally expressed miR-223 was reported to potentiate adipocyte lipid accumulation by enhancing the expression of PPARγ, C/EBPα, adipsin, and aP2 in both ST2 and C3H10T1/2 cell models (29). Conversely, knockdown of the TBT-induced miR-223 using a specific antisense RNA in our study blunted lipid accumulation in adipocytes. In addition, the 3 miR-223 target genes we measured were reported to play important roles in adipocytes. FOXP1 overexpression impaired adaptive thermogenesis in adipocytes and led to diet-induced obesity (45). ZEB1 was shown to be a critical transcription factor regulating adipose tissue accumulation in C57BL/6 mice (46), A point mutation in ZEB1 also increased body weight in Twirler mice model (47). NFIB is a sequence-specific DNA binding protein that interacts with KDM4D to promote the expression of adipogenic genes (48). These findings support a model in which TBT-activates PPARγ, inducing miR-223 expression and adipocyte differentiation from multipotent MSCs.
Regulation of miR-223 expression has been studied by analysis of cis-acting elements within the proximal promoter. Several potential binding sites for c/EBPs were identified in the miR-223 promoter. In human myeloid precursors, expression of miR-223 was reported to be upregulated by c/EBPs (49), contributing to granulocyte differentiation (50). Studies in human endothelial cells (51) and murine macrophages (52) also suggested that c/EBPs could induce miR-223 expression. Taken together, these studies identified binding of c/EBPs to the proximal promoter of miR-223 as an important transcriptional regulator of miR-223 expression.
Based on our previous findings that TBT bound to both members of the PPARγ-RXR heterodimer to modulate adipogenesis (12, 13), we sought to elucidate molecular mechanisms through which TBT promoted adipogenesis via regulation of miR-223 in MSCs. We examined PPREs known to be important for the expression of pre–miR-223 (28) and identified 4 PPREs to which TBT-activated PPARγ bound in an agonist-dependent manner. These data show that TBT-activated PPARγ upregulated expression of pre–miR-223 via binding to the PPREs in its promoter.
We previously observed that prenatal exposure of pregnant F0 mouse dams to TBT led to heritable effects, including increased white adipose tissue depot sizes, reprogramming of MSC fate toward the adipose lineage, and increased hepatic lipid storage in F1, F2, and F3 generations without further exposure (9). The observed transgenerational predisposition to obesity was strongly male-specific in all 3 generations (9, 16, 17). Here we identified a mechanism through which TBT activated PPARγ, upregulating expression of mir-223 leading to posttranscriptional regulation of important target genes in adipogenesis. miR-223 levels remained higher in F2 and F3 male descendants of F0 dams exposed to TBT throughout pregnancy (17). Elevated miR-223 levels preceded the development of diet-induced obesity in the F2 and F3 males; miR-223 levels were further increased by high-fat diet exposure. Future studies should address the molecular mechanisms underlying the transgenerationally affected expression of miR-223. One possibility suggested by our previous studies is altered higher-order chromatin structure (17, 53). Whether the transgenerationally altered miR-223 expression is part of the vehicle of nongenetic inheritance or a consequence of it, our study demonstrates convincingly that obesogen-induced miR-223 facilitates the increased differentiation of MSCs into white adipocytes. While it has been proposed that obesogen-induced changes in expression of noncoding RNAs can play a role in obesity (54), our current study provides the first evidence that this is indeed the case.
Abbreviations
- C/EBP
CCAAT-enhancer-binding protein
- ChIP
chromatin immunoprecipitation
- DEG
differentially expressed gene
- DMSO
dimethyl sulfoxide
- LG268
LG-100268
- miR
microRNA
- MSC
mesenchymal stem cell
- PPARγ
peroxisome proliferator activated receptor gamma
- PPRE
PPARγ regulatory element
- qPCR
quantitative polymerase chain reaction
- RFU
relative fluorescence units
- RNA-seq
RNA sequencing
- ROSI
rosiglitazone
- RXR
retinoid X receptor
- SEM
standard error of the mean
- TBT
tributyltin
- TPT
triphenyltin
Contributor Information
Richard C Chang, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA.
Erika M Joloya, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA.
Zhuorui Li, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA.
Bassem M Shoucri, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA; Medical Scientist Training Program, University of California, Irvine, CA 92697-2300, USA.
Toshi Shioda, Center for Cancer Research, Massachusetts General Hospital, Charlestown, MA 02129, USA.
Bruce Blumberg, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA; Department of Pharmaceutical Sciences, University of California, Irvine, CA 92697-2300, USA; Department of Biomedical Engineering, University of California, Irvine, CA 92697-2300, USA.
Funding
Supported by grants from the National Institutes of Health (NIH) (ES023316, ES031139) to B.B. and T.S. and the Environmental Protection Agency (STAR FP917800 to B.M.S.).
Disclosures
B.B. is a named inventor on U.S. patents 5,861,274; 6,200,802; 6,815,168; and 7,250,273 related to PPARγ. All other authors declare they have no actual or potential competing financial interests.
Data Availability
Original data generated and analyzed during this study are included in this published article or in the data repositories listed in References. RNA-seq data generated in this study is available from the Gene Expression Omnibus database (accession number GSE216429) (37). Supplementary Figures 1-5 can be found online at Figshare (39).
References
- 1. Hall KD, Heymsfield SB, Kemnitz JW, Klein S, Schoeller DA, Speakman JR. Energy balance and its components: implications for body weight regulation. Am J Clin Nutr. 2012;95(4):989-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest. 1995;95(5):2409-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259(5091):87-91. [DOI] [PubMed] [Google Scholar]
- 4. Burhans MS, Hagman DK, Kuzma JN, Schmidt KA, Kratz M. Contribution of adipose tissue inflammation to the development of type 2 diabetes Mellitus. Compr Physiol. 2018;9(1):1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Janesick AS, Blumberg B. Obesogens: an emerging threat to public health. Am J Obstet Gynecol. 2016;214(5):559-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heindel JJ, Blumberg B, Cave M, et al. Metabolism disrupting chemicals and metabolic disorders. Reprod Toxicol. 2017;68:3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heindel JJ, Blumberg B. Environmental obesogens: mechanisms and controversies. Annu Rev Pharmacol Toxicol. 2019;59(1):89-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Egusquiza RJ, Blumberg B. Environmental obesogens and their impact on susceptibility to obesity: new mechanisms and chemicals. Endocrinology. 2020;161(3):bqaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chamorro-Garcia R, Sahu M, Abbey RJ, Laude J, Pham N, Blumberg B. Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ Health Perspect. 2013;121(3):359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grün F, Watanabe H, Zamanian Z, et al. Endocrine-disrupting organotin compounds are potent inducers of adipogenesis in vertebrates. Mol Endocrinol. 2006;20(9):2141-2155. [DOI] [PubMed] [Google Scholar]
- 11. Kanayama T, Kobayashi N, Mamiya S, Nakanishi T, Nishikawa J. Organotin compounds promote adipocyte differentiation as agonists of the peroxisome proliferator-activated receptor gamma/retinoid X receptor pathway. Mol Pharmacol. 2005;67(3):766-774. [DOI] [PubMed] [Google Scholar]
- 12. Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77(1):289-312. [DOI] [PubMed] [Google Scholar]
- 13. Shoucri BM, Martinez ES, Abreo TJ, et al. Retinoid X receptor activation alters the chromatin landscape to commit mesenchymal stem cells to the adipose lineage. Endocrinology. 2017;158(10):3109-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X, Ycaza J, Blumberg B. The environmental obesogen tributyltin chloride acts via peroxisome proliferator activated receptor gamma to induce adipogenesis in murine 3T3-L1 preadipocytes. J Steroid Biochem Mol Biol. 2011;127(1-2):9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kirchner S, Kieu T, Chow C, Casey S, Blumberg B. Prenatal exposure to the environmental obesogen tributyltin predisposes multipotent stem cells to become adipocytes. Mol Endocrinol. 2010;24(3):526-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chamorro-Garcia R, Poupin N, Tremblay-Franco M, et al. Transgenerational metabolomic fingerprints in mice ancestrally exposed to the obesogen TBT. Environ Int. 2021;157:106822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chamorro-Garcia R, Diaz-Castillo C, Shoucri BM, et al. Ancestral perinatal obesogen exposure results in a transgenerational thrifty phenotype in mice. Nat Commun. 2017;8(1):2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y, Siegel F, Kipschull S, et al. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun. 2013;4(1):1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee EK, Lee MJ, Abdelmohsen K, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;31(4):626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qin L, Chen Y, Niu Y, et al. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics. 2010;11(1):320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vian L, Di Carlo M, Pelosi E, et al. Transcriptional fine-tuning of microRNA-223 levels directs lineage choice of human hematopoietic progenitors. Cell Death Differ. 2014;21(2):290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125-1129. [DOI] [PubMed] [Google Scholar]
- 24. Dong YW, Wang R, Cai QQ, et al. Sulfatide epigenetically regulates miR-223 and promotes the migration of human hepatocellular carcinoma cells. J Hepatol. 2014;60(4):792-801. [DOI] [PubMed] [Google Scholar]
- 25. Agatheeswaran S, Singh S, Biswas S, Biswas G, Chandra Pattnayak N, Chakraborty S. BCR-ABL mediated repression of miR-223 results in the activation of MEF2C and PTBP2 in chronic myeloid leukemia. Leukemia. 2013;27(7):1578-1580. [DOI] [PubMed] [Google Scholar]
- 26. Stamatopoulos B, Meuleman N, Haibe-Kains B, et al. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113(21):5237-5245. [DOI] [PubMed] [Google Scholar]
- 27. Zhuang G, Meng C, Guo X, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125(23):2892-2903. [DOI] [PubMed] [Google Scholar]
- 28. Ying W, Tseng A, Chang RC, et al. MicroRNA-223 is a crucial mediator of PPARgamma-regulated alternative macrophage activation. J Clin Invest. 2015;125(11):4149-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guan X, Gao Y, Zhou J, et al. miR-223 regulates adipogenic and osteogenic differentiation of mesenchymal stem cells through a C/EBPs/miR-223/FGFR2 regulatory feedback loop. Stem Cells. 2015;33(5):1589-1600. [DOI] [PubMed] [Google Scholar]
- 30. Zhou H, Xiao J, Wu N, et al. MicroRNA-223 regulates the differentiation and function of intestinal dendritic cells and macrophages by targeting C/EBPbeta. Cell Rep. 2015;13(6):1149-1160. [DOI] [PubMed] [Google Scholar]
- 31. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402-408. [DOI] [PubMed] [Google Scholar]
- 32. Bevington PR, Keith Robinson D, Morris Blair J, John Mallinckrodt A, McKay S. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill Education; 2003. [Google Scholar]
- 33. Chamorro-Garcia R, Shoucri BM, Willner S, Kach H, Janesick A, Blumberg B. Effects of perinatal exposure to dibutyltin chloride on fat and glucose metabolism in mice, and molecular mechanisms, in vitro. Environ Health Perspect. 2018;126(5):057006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Handsaker B, Wysoker A, et al. Genome project data processing S. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang RC, Shoucri BM, Shioda T, Blumberg B. miR-223 plays a critical role in obesogen-enhanced adipogenesis in mesenchymal stem cells and transgenerational obesity. October 27, 2022 ed. GEO—Gene Expression Omnibus GSE2164292022. https://www.wwpdb.org/pdb?id=pdb_00007nke. Date of deposit 27 October 2022.
- 38. Kramer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang RC, Joloya EM, Li Z, Shoucri BM, Shioda T, Blumberg B. Supplemental materials for miR-223 plays a critical role in obesogen-enhanced adipogenesis in mesenchymal stem cells and in transgenerational obesity. Figshare.com 219218822023. 10.6084/m9.figshare.21921882. Date of deposit 18 January 2023. [DOI] [PMC free article] [PubMed]
- 40. Shoucri BM, Hung VT, Chamorro-Garcia R, Shioda T, Blumberg B. Retinoid X receptor activation during adipogenesis of female mesenchymal stem cells programs a dysfunctional adipocyte. Endocrinology. 2018;159(8):2863-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deiuliis JA, Syed R, Duggineni D, et al. Visceral adipose MicroRNA 223 is upregulated in human and murine obesity and modulates the inflammatory phenotype of macrophages. PLoS One. 2016;11(11):e0165962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ye D, Zhang T, Lou G, Liu Y. Role of miR-223 in the pathophysiology of liver diseases. Exp Mol Med. 2018;50(9):1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun G, Li H, Rossi JJ. Sequence context outside the target region influences the effectiveness of miR-223 target sites in the RhoB 3′UTR. Nucleic Acids Res. 2010;38(1):239-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun W, Shen W, Yang S, Hu F, Li H, Zhu TH. miR-223 and miR-142 attenuate hematopoietic cell proliferation, and miR-223 positively regulates miR-142 through LMO2 isoforms and CEBP-beta. Cell Res. 2010;20(10):1158-1169. [DOI] [PubMed] [Google Scholar]
- 45. Liu P, Huang S, Ling S, et al. Foxp1 controls brown/beige adipocyte differentiation and thermogenesis through regulating beta3-AR desensitization. Nat Commun. 2019;10(1):5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saykally JN, Dogan S, Cleary MP, Sanders MM. The ZEB1 transcription factor is a novel repressor of adiposity in female mice. PLoS One. 2009;4(12):e8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kurima K, Hertzano R, Gavrilova O, et al. A noncoding point mutation of Zeb1 causes multiple developmental malformations and obesity in Twirler mice. PLoS Genet. 2011;7(9):e1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Choi JH, Lee H. Histone demethylase KDM4D cooperates with NFIB and MLL1 complex to regulate adipogenic differentiation of C3H10T1/2 mesenchymal stem cells. Sci Rep. 2020;10(1):3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819-831. [DOI] [PubMed] [Google Scholar]
- 50. Pulikkan JA, Dengler V, Peramangalam PS, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115(9):1768-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shi L, Fisslthaler B, Zippel N, et al. MicroRNA-223 antagonizes angiogenesis by targeting beta1 integrin and preventing growth factor signaling in endothelial cells. Circ Res. 2013;113(12):1320-1330. [DOI] [PubMed] [Google Scholar]
- 52. Fukao T, Fukuda Y, Kiga K, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129(3):617-631. [DOI] [PubMed] [Google Scholar]
- 53. Diaz-Castillo C, Chamorro-Garcia R, Shioda T, Blumberg B. Transgenerational self-reconstruction of disrupted chromatin organization after exposure to an environmental stressor in mice. Sci Rep. 2019;9(1):13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heindel JJ, Howard S, Agay-Shay K, et al. Obesity II: establishing causal links between chemical exposures and obesity. Biochem Pharmacol. 2022;199:115015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data generated and analyzed during this study are included in this published article or in the data repositories listed in References. RNA-seq data generated in this study is available from the Gene Expression Omnibus database (accession number GSE216429) (37). Supplementary Figures 1-5 can be found online at Figshare (39).