Key Points
Question
Can an artificial intelligence (AI) model be trained to interpret routine clinical electroencephalograms (EEGs) with accuracy equivalent to that of human experts?
Findings
In this diagnostic study, an AI model (SCORE-AI) was trained on 30 493 EEGs to separate normal from abnormal recordings then classify abnormal recordings as epileptiform-focal, epileptiform-generalized, nonepileptiform-focal, or nonepileptiform-diffuse. The SCORE-AI was validated using 3 independent test data sets consisting of 9945 EEGs not used for training; SCORE-AI achieved diagnostic accuracy similar to human experts.
Meaning
Results of this study suggest that application of SCORE-AI may have utility in improving patient care in underserved areas and efficiency and consistency in specialized centers.
Abstract
Importance
Electroencephalograms (EEGs) are a fundamental evaluation in neurology but require special expertise unavailable in many regions of the world. Artificial intelligence (AI) has a potential for addressing these unmet needs. Previous AI models address only limited aspects of EEG interpretation such as distinguishing abnormal from normal or identifying epileptiform activity. A comprehensive, fully automated interpretation of routine EEG based on AI suitable for clinical practice is needed.
Objective
To develop and validate an AI model (Standardized Computer-based Organized Reporting of EEG–Artificial Intelligence [SCORE-AI]) with the ability to distinguish abnormal from normal EEG recordings and to classify abnormal EEG recordings into categories relevant for clinical decision-making: epileptiform-focal, epileptiform-generalized, nonepileptiform-focal, and nonepileptiform-diffuse.
Design, Setting, and Participants
In this multicenter diagnostic accuracy study, a convolutional neural network model, SCORE-AI, was developed and validated using EEGs recorded between 2014 and 2020. Data were analyzed from January 17, 2022, until November 14, 2022. A total of 30 493 recordings of patients referred for EEG were included into the development data set annotated by 17 experts. Patients aged more than 3 months and not critically ill were eligible. The SCORE-AI was validated using 3 independent test data sets: a multicenter data set of 100 representative EEGs evaluated by 11 experts, a single-center data set of 9785 EEGs evaluated by 14 experts, and for benchmarking with previously published AI models, a data set of 60 EEGs with external reference standard. No patients who met eligibility criteria were excluded.
Main Outcomes and Measures
Diagnostic accuracy, sensitivity, and specificity compared with the experts and the external reference standard of patients’ habitual clinical episodes obtained during video-EEG recording.
Results
The characteristics of the EEG data sets include development data set (N = 30 493; 14 980 men; median age, 25.3 years [95% CI, 1.3-76.2 years]), multicenter test data set (N = 100; 61 men, median age, 25.8 years [95% CI, 4.1-85.5 years]), single-center test data set (N = 9785; 5168 men; median age, 35.4 years [95% CI, 0.6-87.4 years]), and test data set with external reference standard (N = 60; 27 men; median age, 36 years [95% CI, 3-75 years]). The SCORE-AI achieved high accuracy, with an area under the receiver operating characteristic curve between 0.89 and 0.96 for the different categories of EEG abnormalities, and performance similar to human experts. Benchmarking against 3 previously published AI models was limited to comparing detection of epileptiform abnormalities. The accuracy of SCORE-AI (88.3%; 95% CI, 79.2%-94.9%) was significantly higher than the 3 previously published models (P < .001) and similar to human experts.
Conclusions and Relevance
In this study, SCORE-AI achieved human expert level performance in fully automated interpretation of routine EEGs. Application of SCORE-AI may improve diagnosis and patient care in underserved areas and improve efficiency and consistency in specialized epilepsy centers.
This diagnostic study investigates whether an artificial intelligence model can be trained to interpret clinical encephalograms similar to human experts.
Introduction
Electroencephalography (EEG) is the tool most often used in the diagnostic workup of patients with suspected epilepsy.1 In skilled hands, EEG provides essential information to aid diagnosis and classification of epilepsy, important for therapeutic decision-making.1,2,3 EEG helps differentiate epilepsy from other paroxysmal neurological events and nonepileptic causes of impaired consciousness.1,2,3 Although epilepsy is one of the most common serious neurological conditions, with a prevalence of 7.60 per 1000 persons and more than 70 million people affected worldwide,4,5 expertise in reading clinical EEGs is not widely available.6 Even in countries with advanced health care systems, most EEGs are read by physicians without fellowship training in EEG interpretation.7 Misinterpretation of EEG is the most common cause of epilepsy misdiagnosis.8,9,10 Due to the steadily increasing number of EEG referrals, the workload is challenging even in specialized centers.11,12
Artificial intelligence (AI) has the potential for improving the management of epilepsy by addressing unmet clinical needs, providing diagnostic interpretation of EEGs where expertise is scarce, and decreasing excessive workloads placed on human experts interpreting EEGs in specialized centers.13,14,15 To date, AI approaches in clinical EEG have addressed only limited aspects in isolation, such as distinguishing normal from abnormal recordings,16 detecting seizures,17,18,19 or detecting interictal epileptiform discharges.20 Other publications have also claimed that AI achieved human expert performance for spike detectors21,22,23,24 but not for the comprehensive assessment of routine clinical EEGs, equivalent to human expert assessment, which has not yet been reported. Most previously published approaches bear important limitations that are often encountered in AI studies.13 A recently published head-to-head validation study25 of AI models for detection of interictal epileptiform discharges reported that fully automated detection using currently available models had a low specificity precluding their clinical implementation.
Our goal was to develop and validate an AI model for the comprehensive assessment of routine clinical EEGs. Beyond distinguishing abnormal from normal EEG recordings, our aim was to classify abnormal recordings into the major categories that are most relevant for decisions involving patients. These were epileptiform-focal, epileptiform-generalized, nonepileptiform-focal, and nonepileptiform-diffuse abnormalities.1,26,27 We trained a deep learning model on a large data set of highly annotated EEGs, using the Standardized Computer-based Organized Reporting of EEG (SCORE EEG) system.26,27 SCORE EEG is a standardized software tool for annotating EEGs using common data elements. It is endorsed by the International Federation of Clinical Neurophysiology and the International League Against Epilepsy.26,27 Using the SCORE EEG software, human experts label the observed clinically relevant EEG features using standardized data elements. This process generates a clinical report, and at the same time feeds these features into a centralized database. The highly annotated large SCORE EEG database provides a rich source of data for training an AI model.
We conducted a clinical validation study using independent anonymized test data sets consisting of EEGs not used for developing the AI model. We named the AI model Standardized Computer-based Organized Reporting of EEG–Artificial Intelligence (SCORE-AI). eFigure 1 in Supplement 1 summarizes the model’s development and clinical validation algorithm. In this diagnostic accuracy study, the index test was the output of SCORE-AI, using the model and thresholds predetermined in the development phase, with no iterations or adjustments made in the clinical validation phase. All EEGs in the multicenter test data set were independently evaluated by 11 human experts. The majority consensus of the experts was considered the reference standard. The second test data set was a large single-center SCORE EEG data set from a center that did not participate in the development of SCORE-AI. In this data set the reference standard was the clinical evaluation of the human experts from that center (1 physician per recording, in total 14 physicians scoring the EEGs in this data set). In addition, we conducted a benchmarking comparison with 3 previously published AI models using a previously published data set.25 The study was registered (ID ISRCTN14307038). The study protocol is included in eAppendix 2 in Supplement 1.
Methods
We report the study using the Standards for Reporting of Diagnostic Accuracy (STARD) reporting guideline. As the reporting guideline for AI-Centered Diagnostic Accuracy Studies (STARD-AI) is still under development, we included the AI-specific aspects according to the Standard Protocol Items: Recommendations for Interventional Trials-Artificial Intelligence (SPIRIT-AI) extension.
Development of the AI Model
The data set used for development consisted of 30 493 anonymized EEG recordings collected from Haukeland University Hospital, Bergen, Norway, and the Danish Epilepsy Center, Dianalund, Denmark, using NicoletOne equipment (Natus Neuro). A holdout test data set consisting of 2549 EEGs was set aside and not used for development (eFigure 1 and eTable 1 in Supplement 1).
The mean EEG duration was 33 minutes (95% CI, 20-77 minutes). All EEGs had human expert assessments (a total of 17 physicians), using SCORE terminology26,27 implemented in the SCORE EEG Premium software (Holberg EEG). EEG signals from the 19 sensors (10-20 system) and ECG were extracted. The study was approved by the institutional review board and data safety officer at the Danish Epilepsy Centre. As the study used anonymized data sets of previously recorded EEGs, patient consent was not needed.
The SCORE-AI was developed in Python using TensorFlow (eAppendix 1 in Supplement 1) using EEGs recorded between 2014 and 2020. Data were analyzed from January 17, 2022, until November 14, 2022. All the input EEGs and ECG signals were converted into NumPy arrays. The model was configured to access 19 channels of EEG signals, 1 channel of ECG signal as well as the patient age and sex as input. A fixed Fourier resampling was applied to the input data. The final AI model (SCORE-AI) used input frequencies between 0.5 and 128 Hz with a sampling rate of 256 Hz.
The SCORE-AI model was configured to give 5 output scalars, 0<xi<1, where x0 is the normality predictor and x1-4 predicts one of the abnormal categories: epileptiform-focal (x1), epileptiform-generalized (x2), nonepileptiform-diffuse (x3) and nonepileptiform-focal (x4). The values x1 to x4 were mutually independent, but a constraint was placed such that 1 – x0 > max(x1, x2, x3, x4).
The model architecture was determined in the main development phase (eFigure 1 in Supplement 1). No automatic optimization of hyperparameters was performed. The results on the cross-validation data sets are shown in eFigure 2 in Supplement 1. The resulting neural networks are shown in eFigure 7 in Supplement 1.
Once the final model architecture was chosen, the model was retrained on the entire development data set (eFigure 3 in Supplement 1). The development data set was then used to determine the model output threshold (eTable 2 in Supplement 1) yielding the best accuracy estimate and to produce calibration curves (eFigure 4 in Supplement 1), enabling probabilistic interpretation of the model output.
The model was converted into a C++ plugin (dll interface) for the Windows platform. Similar interfaces can be set up for Linux or Mac computers. The output of the model is the assessment of the EEG recording as normal, one of the abnormal categories or a combination of the abnormal categories. The integration of SCORE-AI with the NeuroWorks EEG reader (Natus Neuro), autoSCORE, makes it possible to highlight the abnormal epochs within the EEG recording (eFigure 5 in Supplement 1) so that the expert can adjust the automated assessment, if needed. The SCORE-AI performs a fully automated analysis (ie, no human interaction is needed to obtain the output of the model).
Clinical Validation of the AI Model
The Test Data Sets
For clinical validation, we used independent test data sets consisting of EEGs recorded from patients who were not included in the development phase. We used a fixed and frozen model and threshold values. The index test was the model output. For an expected sensitivity of 75% and specificity of 90%, with a 10% error (±5%) when calculating sample size, we needed at least 85 EEGs.28
Multicenter Test Data Set
To account for the variability in human expert assessment, 11 experts (raters) from 11 different centers, who trained in different institutions (eTable 5 in Supplement 1), independently evaluated a data set of 100 representative routine EEGs, recorded in different centers with different EEG equipment. The raters did not participate in assessing the EEGs in the development data set. The raters independently labeled each EEG using the same categories of EEG abnormalities as described above. The reference standard was majority consensus scoring of the raters.
Inclusion criteria included a targeted distribution of 60 normal recordings vs 40 abnormal recordings. From the holdout data set (eTable 1 in Supplement 1), 75 EEGs (48 adult and 27 pediatric) were randomly selected. The remaining 25 EEGs (17 adult, 8 pediatric) were selected from a data set of 150 anonymized EEGs from the Mayo Clinic. Exclusion criteria included patients aged 3 months or younger (neonatal EEGs) and recordings from intensive care units (ICUs) (EEGs with rhythmic and periodic patterns in critically ill patients were excluded). eTable 6 in Supplement 1 shows the distribution of the 100 patients in the multicenter test data set.
The raters independently evaluated the EEGs. The age and sex of the patient were disclosed for each EEG. The raters were blinded to all other data and to the output of the algorithm. The raters were free to change montages, filters, gain, and time resolution while reviewing EEGs.
Large Single-Center Test Data Set
We compared the output of SCORE-AI with the clinical assessments in a large SCORE EEG data set from Oslo University Hospital (Norway) consisting of 9785 EEGs (5168 male; median age: 38.9 years; 95% CI, 0.6-87.4 years). This center did not participate in the development of the model. Fourteen different physicians assessed the EEGs in this data set, but each EEG was assessed by a single physician in clinical practice. Neonatal and ICU recordings were also excluded from this data set. Mean duration of the recordings was 31 minutes (95% CI, 15-54 minutes). EEGs were recorded with NicoletOne equipment and described using the SCORE EEG Premium software. In this data set 4681 EEG results (47.83%) were abnormal.
Outcome Measures
In the multicenter multirater test data set, we determined the interrater agreement among the 11 human experts, and between SCORE-AI and the human experts (eFigure 6 in Supplement 1). Using majority consensus as reference standard in the multicenter test data set, we determined the diagnostic accuracy measures of SCORE-AI (sensitivity, specificity, accuracy, positive and negative predictive values) using the conventional formulas.
In the large single-center test data set, we calculated the intertest agreement between SCORE-AI and the clinical assessment by the human experts. We hypothesized that the agreement would be within range of the agreement between human experts.
Comparison With Other Approaches
Multicenter Test Data Set
To our knowledge, there are no other commercially available or open-source AI models for comprehensive, fully automated assessment of routine clinical EEGs. We used the spike-detector approach of encevis (Austrian Institute of Technology), software approved by the US Food and Drug Administration and certified in the European Union by Conformité Européen, to compare this specific aspect with the performance of SCORE-AI in the multicenter test data set. For this purpose, the categories epileptiform-focal and epileptiform-generalized were combined, and we compared the accuracy of the 2 approaches for identifying recordings containing epileptiform abnormalities in the multicenter test data set.
Benchmarking With Previously Published AI Models
To compare the performance of SCORE-AI with 3 previously published models (encevis, SpikeNet, Persyst), we used the EEG data set from a previous study.25 The median age in this data set was 36 years (95%CI, 3-77 years). This data set consisted of 20-minute routine clinical EEGs containing sharp transients (epileptiform or not) from 60 patients: 30 with epilepsy (with 340 interictal epileptiform discharges in total) and 30 with nonepileptic paroxysmal events. This data set had an external independent reference standard at the recording level (ie, epilepsy vs no epilepsy), derived from video-EEG recordings of patients obtained during their habitual clinical episodes. As the previously published models were spike detectors, we had to limit the evaluation to the accuracy of detecting epileptiform discharges. We then compared sensitivity, specificity, and overall accuracy.
Statistical Analysis
Gwet AC1 agreement coefficients29,30 were used for measuring interrater agreement, and the strength of agreement beyond chance was interpreted according to Landis and Koch criteria.31 For computation of the 95% CIs, bootstrap resampling (n = 10 000 to 100 000) was used for all metrics except the Gwet AC1 statistic. Bayesian bootstrap resampling with smoothing was used on the multirater multicenter data set of 100 EEGs as well as for the 60 EEGs with external reference standard (raw figures available in eTable 4 of Supplement 1). Otherwise, for the holdout test data set and the encevis comparison, plain bootstrap resampling was used. The smoothing was achieved by stochastically perturbing the confusion matrix by adding random noise from a uniform Dirichlet distribution in each bootstrap sample. For the AC1 statistic we constructed the 95% CIs from the standard deviations as suggested in Gwet29 and Gwet.30 The Python packages SciPy, NumPy, and Pandas were used to compute the results. Inkscape and Pyplot were used to generate figures. Statistical significance was set at 2-sided P < .05. Intrarater agreement comparisons were based on 95% CIs and considered statistically significant if there was no overlap.
Results
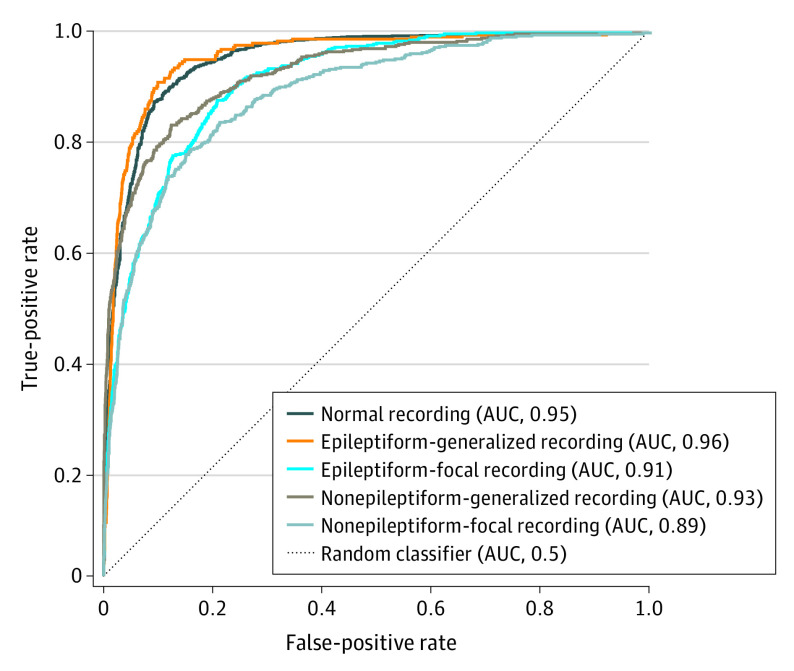
The characteristics of the EEG data sets include development data set (N = 30 493; 14 980 men; median age, 25.3 years [95% CI, 1.3-76.2 years]), multicenter test data set (N = 100; 61 men, median age, 25.8 years [95% CI, 4.1-85.5 years]), single-center test data set (N = 9785; 5168 men; median age, 35.4 years [95% CI, 0.6-87.4 years]), and test data set with external reference standard (N = 60; 27 men; median age, 36 years [95% CI, 3-75 years]). The performance in the holdout EEG data set (n = 2549) is shown in the Figure. The SCORE-AI achieved high accuracy, with an area under the receiver operating characteristic curve between 0.89 and 0.96 for the different categories of EEG abnormalities. For the thresholds predefined in the development data set (eTable 2 in Supplement 1), the accuracy measures in the holdout EEG data set are between 85.4% and 92.3% (eTable 3 in Supplement 1). Recordings shorter than 20 minutes had lower area under the curve; for recordings longer than 20 minutes the area under the curve showed only small relative variations related to the duration of the recording. For duration 0 to less than 20 minutes, the mean area under the curve (AUC) was 0.887. For duration 20 minutes or longer, the mean AUC was 0.903 calculated across all subcategories using both the holdout test set (n = 2549) and the large clinical data set (n = 9785) (eFigure 8 in Supplement 1).
Figure. Receiver Operating Characteristics Curves on the Holdout Test EEG Data Set (n = 2549).
AUC indicates area under the curve.
Table 1 shows the interrater agreement (measured as Gwet agreement coefficient [AC1]) among the 11 human experts and between SCORE-AI and the majority consensus in the multicenter data set (n = 100). There was almost perfect agreement (Gwet AC1 = 0.9) among experts concerning the presence of generalized epileptiform discharges, and substantial agreement (Gwet AC1 of 0.63-0.72) on focal epileptiform discharges, diffuse nonepileptiform abnormalities, and on recordings considered to be normal. The interrater agreement was moderate (Gwet AC1 of 0.50-0.59) for the presence of focal nonepileptiform abnormalities, and for the exact match when several abnormalities were present in the same recording. Beyond-chance agreement between SCORE-AI and the majority consensus of human experts was similar to the agreement among human experts for all aspects except for identifying normal EEG recordings (Gwet AC1 = 0.9) and multiple abnormalities (Gwet AC1 = 0.69) (Table 1), for which SCORE-AI had a significantly higher agreement with the majority consensus compared with the agreement among human experts (almost perfect vs substantial agreement). There was no significant difference in the overall diagnostic accuracy between experts and SCORE-AI in the multicenter data set (Table 2).
Table 1. Gwet AC1 Agreement Coefficients for the 11 Human Experts, SCORE-AI, and the Human Expert Majority Consensus.
| EEG recording category | Agreement coefficient (95% CI) | |
|---|---|---|
| Agreement among the human experts | Agreement between SCORE-AI and majority consensus of human experts | |
| Normal | 0.723 (0.649-0.796)a | 0.903 (0.820-0.987)a |
| Epileptiform-focal | 0.723 (0.643-0.803) | 0.757 (0.634-0.880) |
| Epileptiform-generalized | 0.901 (0.854-0.949) | 0.928 (0.865-0.991) |
| Nonepileptiform-diffuse | 0.630 (0.539-0.721) | 0.738 (0.608-0.868) |
| Nonepileptiform-focal | 0.587 (0.499-0.674) | 0.775 (0.657-0.893) |
| Exact match/multiple abnormalities | 0.497 (0.433-0.561)a | 0.689 (0.611-0.766)a |
Abbreviations: EEG, electroencephalography; SCORE-AI, Standardized Computer-based Organized Reporting of EEG–Artificial Intelligence.
Significant difference. Statistical comparisons were based on the 95% CIs. Significance means there was no overlap between the 95% CIs.
Table 2. Average Accuracy of SCORE-AI and of the Human Experts With Respect to the Human Expert Majority Consensus on 100 EEGs From the Multicenter Test Data Set.
| EEG recording category | Average accuracy (95% CI) | Difference (P value) | |
|---|---|---|---|
| SCORE-AI | Human experts | ||
| Normal | 95.00 (89.61-97.88) | 91.36 (88.04-94.10) | .09 |
| Epileptiform-focal | 84.69 (76.73-90.54) | 88.4 (84.35-91.91) | .12 |
| Epileptiform-generalized | 94.9 (89.41-97.83) | 95.36 (92.51-97.48) | .34 |
| Nonepileptiform-diffuse | 84.69 (76.63-90.83) | 86.09 (81.99-89.66) | .33 |
| Nonepileptiform-focal | 85.71 (77.86-91.41) | 85.25 (81.04-88.78) | .47 |
| Exact match/multiple abnormalities | 65.31 (54.93-73.60) | 66.7 (60.56-72.41) | .33 |
Abbreviations: EEG, electroencephalography; SCORE-AI, Standardized Computer-based Organized Reporting of EEG–Artificial Intelligence.
In the large external single-center test data set (n = 9785), agreement between SCORE-AI and clinical evaluation of the recordings was within the range of the human expert interrater variability for identifying normal recordings (0.74) and recordings with generalized epileptiform abnormalities (0.95), and significantly higher for the remaining categories (0.64-0.87) (Table 3).
Table 3. Gwet AC1 Agreement Coefficients Between SCORE-AI and Clinical Assessment.
| EEG recording category | Agreement between SCORE-AI and the clinical assessment of the EEGs | Difference between SCORE-AI–HE agreement and HE-HE agreementa |
|---|---|---|
| Normal | 0.737 (0.723 to 0.750) | 0.014 (−0.061 to 0.089) |
| Epileptiform-focal | 0.871 (0.862 to 0.879)b | 0.147 (0.067 to 0.228)b |
| Epileptiform-generalized | 0.948 (0.943 to 0.953) | 0.0471 (−0.001 to 0.095) |
| Nonepileptiform-diffuse | 0.737 (0.723 to 0.750)b | 0.106 (0.014 to 0.199)b |
| Nonepileptiform-focal | 0.768 (0.756 to 0.780)b | 0.181 (0.092 to 0.269)b |
| Exact match/multiple abnormalities | 0.637 (0.627 to 0.647)b | 0.140 (0.075 to 0.205)b |
Abbreviations: EEG, electroencephalography; HE, human experts; SCORE-AI, Standardized Computer-based Organized Reporting of EEG–Artificial Intelligence.
HE-HE agreement as detailed in Table 1.
Significant difference. Statistical comparisons were based on the 95% CIs. Significance means there was no overlap between the 95% CIs.
Since none of the currently available AI models provides a comprehensive fully automated assessment of routine clinical EEGs, we limited the benchmarking to the comparison of the ability to identify epileptiform discharges by combining focal and generalized categories (Table 4). In the multicenter test data set compared with encevis software,32 SCORE-AI had significantly higher specificity, positive predictive value, and accuracy, but lower sensitivity.
Table 4. Comparison of Fully Automated Identification of Epileptiform Discharges Between SCORE-AI and encevis on the Same 100 EEGs Used in the Multirater Test Set.
| Algorithm | Fully automated identification of EEG recordings with epileptiform abnormalities, % (95% CI) | ||||
|---|---|---|---|---|---|
| Sensitivity | Specificity | Negative predictive value | Positive predictive value | Accuracy | |
| encevisa | 96.68 (88.89-100.00) | 27.14 (17.19-37.88) | 95.03 (83.33-100.00) | 36.29 (25.93-46.99) | 48.03 (38.00-58.00) |
| SCORE-AIb | 89.94 (77.78-100.00) | 87.13 (78.79-94.29) | 95.28 (89.39-100.00) | 74.98 (60.00-88.57) | 87.97 (81.00-94.00) |
| Difference (P value) | <.001 | <.001 | .49 | <.001 | <.001 |
Abbreviations: EEG, electroencephalography; SCORE-AI, Standardized Computer-based Organized Reporting of EEG–Artificial Intelligence.
For encevis, the detection of one or more spikes was considered as epileptiform classification of the EEG.
For SCORE-AI, either epileptiform-focal and/or epileptiform-generalized was considered as an epileptiform classification of the EEG.
In the previously published data sets25 with external reference standard based on information obtained from epilepsy monitoring units, fully automated detection of epileptiform discharges using the 3 previously published AI models (encevis, SpikeNet, and Persyst) had specificity (3%-63%) too low for clinical implementation (eTable 7 in Supplement 1).25 SCORE-AI demonstrated substantially greater specificity compared with the previously published models (90% vs 3%-63%) and was more specific than the majority consensus of the 3 human experts (73.3%) (eTable 7 in Supplement 1). The sensitivity of SCORE-AI (86.7%) was similar to the sensitivity of the human experts (93.3%), higher than the sensitivity of SpikeNet (66.7%), and lower than encevis (96.7%) and Persyst (100%) (eTable 7 in Supplement 1). The overall accuracy of SCORE-AI (88.3%; 95% CI, 79.2%-94.9%) was similar to the human experts (83.3%; 95% CI, 73%-91.4%) and more accurate (P < .001) than the 3 previously published AI models (eTable 7 in Supplement 1).
Discussion
In this diagnostic accuracy study, we developed and validated the first AI model (SCORE-AI) to date capable of fully automated and comprehensive assessment of routine clinical EEGs. The SCORE-AI achieved human expert performance in automated interpretation of routine clinical EEGs. These results warrant clinical implementation with a potential to improve patient care in remote and underserved areas where EEG expertise is scarce or unavailable. In addition, SCORE-AI may help improve efficiency and reduce excessive workloads for experts in tertiary care centers who regularly interpret high volumes of EEG recordings.
We designed the development and validation of the model with special care to avoid typical errors and sources of bias13 and tailored the process to address specific aspects pertinent to interpretation of clinical EEGs. We used a large data set of 30 493 EEGs, from different centers, to train the AI model. The EEGs were highly annotated by 17 human experts, using a standardized software tool (SCORE-EEG).26,27 For the clinical validation, we used a “fixed and frozen” model and thresholds. To avoid overfitting, no iterations (ie, adjustments) of the AI model were allowed in the clinical validation phase. The test data set was independent from the development data set. To ensure generalizability, we aimed for a test data set with a representative distribution and a large consecutive test data set of nearly 10 000 EEGs. The human experts providing the reference standard in the validation phase of the study were different from those who participated in the development portion of the process. In the multicenter test data set, EEGs were recorded with different EEG equipment. The analysis process was fully automated and blinded to all other data.
The SCORE-AI is the first model to date capable of completing a fully automated and comprehensive clinically relevant assessment of routine EEGs. The output of SCORE-AI provides a more complex classification of EEG abnormalities than previously published AI models. Identifying the presence of epileptiform activity in the EEGs helps in diagnosing epilepsy. Distinguishing focal from generalized epileptiform discharges additionally aids in choosing optimal antiseizure medication.33,34 Distinguishing focal from diffuse nonepileptiform EEG abnormalities directs further diagnostic steps, such as neuroimaging for the former case and a search for systematic etiologies in the latter. Hence, the granularity of classifying abnormal EEGs provides sufficient information for the referring physician to make clinical decisions.
Another remarkable finding in our study is the interrater agreement among human experts. Previous studies found only fair to moderate agreement in EEG reading.35,36 However, those studies assessed short segments of EEG with selected abnormal patterns, and not a complete, continuous recording, as is the case in the clinical setting. In this study, we found better results with comprehensive expert assessment of the entire routine EEG recording: almost perfect agreement for generalized epileptiform abnormalities, substantial agreement for focal epileptiform discharges, diffuse nonepileptiform abnormalities, and for normal EEG recordings, and moderate beyond-chance agreement for the presence of focal nonepileptiform abnormalities. The performance of SCORE-AI was well within the variability present among human experts.
There is no other open-source or commercially available software package for comprehensive assessment of routine clinical EEGs. Several AI-based models have been developed for detection of epileptiform activity on EEG.20 However, this aspect is only a fragment of the complete comprehensive EEG assessment. The other major limitation of the previously published models is the high number of false detections (0.73 per minute) precluding their clinical implementation.37 A recent study reported that the fully automated application of 3 previously published AI models had specificity (3%-63%) that is too low for clinical application.25 Human expert interaction via a semiautomated approach was needed to achieve clinical-level performance when using the previously published AI models.25 The benchmarking in our study confirms this, and shows that as opposed to previously published spike detectors, SCORE-AI reaches high specificity (90%) similar to human expert performance. In that data set, SCORE-AI had similar accuracy to the human raters, and significantly higher accuracy than the 3 previously published AI models. The important improvement in SCORE-AI compared with previous AI models is that our fully automated model achieves high specificity similar to human experts. We believe that the other AI models would probably improve their performance if the current epoch-level output of these algorithms was used to train a recording-level assessment.
The expert-level performance of SCORE-AI warrants its application in remote and underserved areas. Its use has the potential to decrease EEG misinterpretation and circumvent the problem of low interrater agreement in many places where clinical EEG is read by physicians without fellowship training, without access to expert supervision, or with limited experience (often in the general neurology practice setting). Furthermore, SCORE-AI may help reduce the workload in centers where experts are available but overburdened by clinical workloads that include EEG interpretation. Since SCORE-AI appears to identify normal EEGs with nearly 100% precision, experts may decide to spend less time evaluating these recordings and more time on some of the more difficult epilepsy monitoring unit or ICU recordings. The SCORE-AI is currently being integrated with one of the most widely used clinical EEG equipment systems (Natus Neuro). This will promote broad availability of the model in clinical practices because it does not require specialized hardware and it can also be converted into other computer-based interfaces.
Limitations
A limitation of the current version of SCORE-AI is that it was developed and validated on routine EEGs excluding neonates and critically ill patients. Nevertheless, routine EEGs represent the largest volume of EEG recordings performed for clinical purposes and is one of the most important missing clinical tests in underserved areas.1 Another important limitation is that the model was trained to find biomarkers visually identified by human experts. Training the model to predict diagnosis or therapeutic response can potentially circumvent this limitation, and this will be addressed in future studies. Using interpretable or explainable AI, the plan is to identify features used by the model to make the process transparent.
Conclusions
In this diagnostic accuracy study, our convolutional neural network model, SCORE-AI, achieved expert-level performance in reading routine clinical EEGs. Its application may help to provide useful clinical information in remote and underserved areas where expertise in EEG interpretation is minimal or unavailable. Importantly, it may also help reduce the potential for EEG misinterpretation and subsequent mistreatment, improve interrater agreement to optimize routine interpretation by neurologists, and increase efficiency by decompressing excessive workloads for human experts interpreting high volumes of EEGs.
eFigure 1. The Flow Diagram of the AI Model (SCORE-AI) Training and Evaluation
eFigure 2. Receiver Operating Characteristics (ROC) and Area Under the Curve (AUC) on the Cross-Validation Datasets
eFigure 3. Receiver Operating Characteristics (ROC) and ROC Area Under the Curve (AUC) for the Entire Development Set
eFigure 4. Approximate Mapping of the Model Output vs the Estimated Probability of the Condition
eFigure 5. autoSCORE: Integration of SCORE-AI With the Natus NeuroWorks EEG Reader
eFigure 6. Pairwise Comparison Strategy
eFigure 7. SCORE-AI Model Architecture
eFigure 8. Area Under the ROC Curve (AUC) Depending on the Duration of the EEG Recording
eTable 1. A Cross Validation Scheme, Used for Model Development, Partitioning the Development Dataset Into Training and Validation Datasets
eTable 2. Threshold for Optimal Accuracy Based on Training Dataset (Calculated Using Balanced Bootstrap Resampling)
eTable 3. Performance of the Final Model on the Training Dataset.
eTable 4. Raw Figures of Epileptiform Findings in Previously Published Dataset of 60 EEGs
eTable 5. Training and Experience of the Human Experts Who Rated the Multicenter Test Dataset Of EEGs
eTable 6. Demographic Distribution of Patients in the Multicenter Test Dataset of EEGs
eTable 7. Results on Previously Published Dataset of 60 EEGs
eAppendix 1. Installed Packages in the Dev Environment
eAppendix 2. Study Protocol
eReferences
Data Sharing Statement
References
- 1.Tatum WO, Rubboli G, Kaplan PW, et al. Clinical utility of EEG in diagnosing and monitoring epilepsy in adults. Clin Neurophysiol. 2018;129(5):1056-1082. doi: 10.1016/j.clinph.2018.01.019 [DOI] [PubMed] [Google Scholar]
- 2.Pillai J, Sperling MR. Interictal EEG and the diagnosis of epilepsy. Epilepsia. 2006;47(suppl 1):14-22. doi: 10.1111/j.1528-1167.2006.00654.x [DOI] [PubMed] [Google Scholar]
- 3.Engel J Jr. A practical guide for routine EEG studies in epilepsy. J Clin Neurophysiol. 1984;1(2):109-142. doi: 10.1097/00004691-198404000-00001 [DOI] [PubMed] [Google Scholar]
- 4.Fiest KM, Sauro KM, Wiebe S, et al. Prevalence and incidence of epilepsy: a systematic review and meta-analysis of international studies. Neurology. 2017;88(3):296-303. doi: 10.1212/WNL.0000000000003509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thijs RD, Surges R, O’Brien TJ, Sander JW. Epilepsy in adults. Lancet. 2019;393(10172):689-701. doi: 10.1016/S0140-6736(18)32596-0 [DOI] [PubMed] [Google Scholar]
- 6.Kwon CS, Wagner RG, Carpio A, Jetté N, Newton CR, Thurman DJ. The worldwide epilepsy treatment gap: a systematic review and recommendations for revised definitions—a report from the ILAE Epidemiology Commission. Epilepsia. 2022;63(3):551-564. doi: 10.1111/epi.17112 [DOI] [PubMed] [Google Scholar]
- 7.Nascimento FA, Gavvala JR. Education research: neurology resident EEG education: a survey of US neurology residency program directors. Neurology. 2021;96(17):821-824. doi: 10.1212/WNL.0000000000011354 [DOI] [PubMed] [Google Scholar]
- 8.Benbadis SR, Lin K. Errors in EEG interpretation and misdiagnosis of epilepsy: which EEG patterns are overread? Eur Neurol. 2008;59(5):267-271. doi: 10.1159/000115641 [DOI] [PubMed] [Google Scholar]
- 9.Benbadis SR, Tatum WO. Overintepretation of EEGs and misdiagnosis of epilepsy. J Clin Neurophysiol. 2003;20(1):42-44. doi: 10.1097/00004691-200302000-00005 [DOI] [PubMed] [Google Scholar]
- 10.Benbadis SR. Errors in EEGs and the misdiagnosis of epilepsy: importance, causes, consequences, and proposed remedies. Epilepsy Behav. 2007;11(3):257-262. doi: 10.1016/j.yebeh.2007.05.013 [DOI] [PubMed] [Google Scholar]
- 11.Brogger J, Eichele T, Aanestad E, Olberg H, Hjelland I, Aurlien H. Visual EEG reviewing times with SCORE EEG. Clin Neurophysiol Pract. 2018;3:59-64. doi: 10.1016/j.cnp.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng MC, Gillis K. The state of everyday quantitative EEG use in Canada: a national technologist survey. Seizure. 2017;49:5-7. doi: 10.1016/j.seizure.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 13.Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med. 2022;28(1):31-38. doi: 10.1038/s41591-021-01614-0 [DOI] [PubMed] [Google Scholar]
- 14.Beniczky S, Karoly P, Nurse E, Ryvlin P, Cook M. Machine learning and wearable devices of the future. Epilepsia. 2021;62(suppl 2):S116-S124. doi: 10.1111/epi.16555 [DOI] [PubMed] [Google Scholar]
- 15.Abbasi B, Goldenholz DM. Machine learning applications in epilepsy. Epilepsia. 2019;60(10):2037-2047. doi: 10.1111/epi.16333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Leeuwen KG, Sun H, Tabaeizadeh M, Struck AF, van Putten MJAM, Westover MB. Detecting abnormal electroencephalograms using deep convolutional networks. Clin Neurophysiol. 2019;130(1):77-84. doi: 10.1016/j.clinph.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumgartner C, Koren JP. Seizure detection using scalp-EEG. Epilepsia. 2018;59(suppl 1):14-22. doi: 10.1111/epi.14052 [DOI] [PubMed] [Google Scholar]
- 18.Pavel AM, Rennie JM, de Vries LS, et al. A machine-learning algorithm for neonatal seizure recognition: a multicentre, randomised, controlled trial. Lancet Child Adolesc Health. 2020;4(10):740-749. doi: 10.1016/S2352-4642(20)30239-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Japaridze G, Loeckx D, Buckinx T, et al. Automated detection of absence seizures using a wearable electroencephalographic device: a phase 3 validation study and feasibility of automated behavioral testing. Epilepsia. 2022. doi: 10.1111/epi.17200 [DOI] [PubMed] [Google Scholar]
- 20.da Silva Lourenço C, Tjepkema-Cloostermans MC, van Putten MJAM. Machine learning for detection of interictal epileptiform discharges. Clin Neurophysiol. 2021;132(7):1433-1443. doi: 10.1016/j.clinph.2021.02.403 [DOI] [PubMed] [Google Scholar]
- 21.Janmohamed M, Nhu D, Kuhlmann L, et al. Moving the field forward: detection of epileptiform abnormalities on scalp electroencephalography using deep learning-clinical application perspectives. Brain Commun. 2022;4(5):fcac218. doi: 10.1093/braincomms/fcac218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing J, Sun H, Kim JA, et al. Development of expert-level automated detection of epileptiform discharges during electroencephalogram interpretation. JAMA Neurol. 2020;77(1):103-108. doi: 10.1001/jamaneurol.2019.3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheuer ML, Bagic A, Wilson SB. Spike detection: inter-reader agreement and a statistical Turing test on a large data set. Clin Neurophysiol. 2017;128(1):243-250. doi: 10.1016/j.clinph.2016.11.005 [DOI] [PubMed] [Google Scholar]
- 24.Fürbass F, Kural MA, Gritsch G, Hartmann M, Kluge T, Beniczky S. An artificial intelligence-based EEG algorithm for detection of epileptiform EEG discharges: validation against the diagnostic gold standard. Clin Neurophysiol. 2020;131(6):1174-1179. doi: 10.1016/j.clinph.2020.02.032 [DOI] [PubMed] [Google Scholar]
- 25.Kural MA, Jing J, Fürbass F, et al. Accurate identification of EEG recordings with interictal epileptiform discharges using a hybrid approach: artificial intelligence supervised by human experts. Epilepsia. 2022;63(5):1064-1073. doi: 10.1111/epi.17206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beniczky S, Aurlien H, Brøgger JC, et al. Standardized Computer-based Organized Reporting of EEG: SCORE. Epilepsia. 2013;54(6):1112-1124. doi: 10.1111/epi.12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beniczky S, Aurlien H, Brøgger JC, et al. Standardized Computer-based Organized Reporting of EEG: SCORE—second version. Clin Neurophysiol. 2017;128(11):2334-2346. doi: 10.1016/j.clinph.2017.07.418 [DOI] [PubMed] [Google Scholar]
- 28.Jones SR, Carley S, Harrison M. An introduction to power and sample size estimation. Emerg Med J. 2003;20(5):453-458. doi: 10.1136/emj.20.5.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gwet KL. Computing inter-rater reliability and its variance in the presence of high agreement. Br J Math Stat Psychol. 2008;61(Pt 1):29-48. doi: 10.1348/000711006X126600 [DOI] [PubMed] [Google Scholar]
- 30.Gwet KL. Constructing agreement coefficients: AC1 and Aickin’s α. Handbook of Inter-Rater Reliability. 2021. Accessed May 9, 2023. https://www.agreestat.com/books/cac5/chapter5/chap5.pdf
- 31.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159-174. doi: 10.2307/2529310 [DOI] [PubMed] [Google Scholar]
- 32.Kluge T, Skupch A. encevis. Accessed May 9, 2023. https://www.encevis.com/
- 33.Asadi-Pooya AA, Beniczky S, Rubboli G, Sperling MR, Rampp S, Perucca E. A pragmatic algorithm to select appropriate antiseizure medications in patients with epilepsy. Epilepsia. 2020;61(8):1668-1677. doi: 10.1111/epi.16610 [DOI] [PubMed] [Google Scholar]
- 34.Beniczky S, Rampp S, Asadi-Pooya AA, Rubboli G, Perucca E, Sperling MR. Optimal choice of antiseizure medication: agreement among experts and validation of a web-based decision support application. Epilepsia. 2021;62(1):220-227. doi: 10.1111/epi.16763 [DOI] [PubMed] [Google Scholar]
- 35.Halford JJ, Arain A, Kalamangalam GP, et al. Characteristics of EEG interpreters associated with higher interrater agreement. J Clin Neurophysiol. 2017;34(2):168-173. doi: 10.1097/WNP.0000000000000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kural MA, Duez L, Sejer Hansen V, et al. Criteria for defining interictal epileptiform discharges in EEG: a clinical validation study. Neurology. 2020;94(20):e2139-e2147. doi: 10.1212/WNL.0000000000009439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.da Silva Lourenço C, Tjepkema-Cloostermans MC, van Putten MJAM. Efficient use of clinical EEG data for deep learning in epilepsy. Clin Neurophysiol. 2021;132(6):1234-1240. doi: 10.1016/j.clinph.2021.01.035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. The Flow Diagram of the AI Model (SCORE-AI) Training and Evaluation
eFigure 2. Receiver Operating Characteristics (ROC) and Area Under the Curve (AUC) on the Cross-Validation Datasets
eFigure 3. Receiver Operating Characteristics (ROC) and ROC Area Under the Curve (AUC) for the Entire Development Set
eFigure 4. Approximate Mapping of the Model Output vs the Estimated Probability of the Condition
eFigure 5. autoSCORE: Integration of SCORE-AI With the Natus NeuroWorks EEG Reader
eFigure 6. Pairwise Comparison Strategy
eFigure 7. SCORE-AI Model Architecture
eFigure 8. Area Under the ROC Curve (AUC) Depending on the Duration of the EEG Recording
eTable 1. A Cross Validation Scheme, Used for Model Development, Partitioning the Development Dataset Into Training and Validation Datasets
eTable 2. Threshold for Optimal Accuracy Based on Training Dataset (Calculated Using Balanced Bootstrap Resampling)
eTable 3. Performance of the Final Model on the Training Dataset.
eTable 4. Raw Figures of Epileptiform Findings in Previously Published Dataset of 60 EEGs
eTable 5. Training and Experience of the Human Experts Who Rated the Multicenter Test Dataset Of EEGs
eTable 6. Demographic Distribution of Patients in the Multicenter Test Dataset of EEGs
eTable 7. Results on Previously Published Dataset of 60 EEGs
eAppendix 1. Installed Packages in the Dev Environment
eAppendix 2. Study Protocol
eReferences
Data Sharing Statement