Abstract
The integration of electronic health records (EHRs) with social determinants of health (SDoH) is crucial for population health outcome research, but it requires the collection of identifiable information and poses security risks. This study presents a framework for facilitating de-identified clinical data with privacy-preserved geocoded linked SDoH data in a Data Lake. A reidentification risk detection algorithm was also developed to evaluate the transmission risk of the data. The utility of this framework was demonstrated through one population health outcomes research analyzing the correlation between socioeconomic status and the risk of having chronic conditions. The results of this study inform the development of evidence-based interventions and support the use of this framework in understanding the complex relationships between SDoH and health outcomes. This framework reduces computational and administrative workload and security risks for researchers and preserves data privacy and enables rapid and reliable research on SDoH-connected clinical data for research institutes.
Introduction
Social Determinants of Health (SDoH), encompassing economic and social factors, is one of the five interconnected categories influencing health outcomes1. Assessing the impact of population health requires considering various factors, including socio-demographic conditions, medical care, genetics, environmental and physical factors, and behavioral information. However, gathering and analyzing this comprehensive data can be a daunting task.2. Studies have indicated the correlation between Socioeconomic Status (SES) and health outcomes and life expectancy1,3. Lower SES intertwined with poor health outcomes leading to diminished income and which eventually backpropagates to the reason for having lower SES4. To improve health care from this vicious cycle of health and poverty and to provide health equity, primary health care should go hand in hand with SDoH5. Health research, promotion, and prevention initiatives can decrease health disparities6. However, conducting a population health outcomes-related study needs both SDoH and clinical data. Risk factors of health conditions (Cancer7, Post-induction cesarean delivery8, Readmission risk9, etc10, 11, 12) can be determined using a specific type of SDoH or by combining multiple SDoH with clinical data.
The paucity of renowned studies and standardized data presents a formidable challenge for researchers seeking information on social determinants of health. One notable exception is the American Community Survey (ACS) undertaken by the US Census Bureau, which enforces mandatory participation through legal means. The Bureau aids the nation by providing copious amounts of population data through this survey program, which is disseminated in a variety of formats annually, and amenable to multiple forms of analysis13. ACS dataset comprises manifold variables beneficial to business, healthcare, research, and government entities. Indices of social and neighborhood deprivation are used in the US and other countries to gauge the impact of social determinants on public health9,14–17, specifically geographic indices to enhance health quality18,19. A frequently used proxy measure of socioeconomic status is the Area Deprivation Index (ADI)14, provided by the University of Wisconsin School of Medicine and Public Health Department of Medicine.
Publicly available SDoH data are mostly connected to US Census Bureau-defined geocodes. Thus, for population and area-based studies, the researcher must collect the SDoH data, geocode the patient addresses20,21, and connect SDoH data to clinical data from electronic health records (EHR). However, access to clinical data from EHR linked with publicly available SDoH data has some restrictions because the privacy of personal health information (PHI) is as important as protecting and promoting public health7. The federal common rule governing human subjects research, the Health Insurance Portability and Accountability Act (HIPAA), sets rules for researchers regarding the nature of the research or limits the availability of identifiable health data based on need22. To conduct research with SDoH- connected EHR data in an institution, the researcher needs to obtain an Institutional Review Board’s (IRB) approval to support the research activity and use identified or limited datasets. As identified and limited dataset contains PHI, the researcher needs to provide the data management and security plan to get approval from the IRB. All these processes are repeated for each SDoH-connected EHR data study in a research institution (Figure 1).
Figure 1.
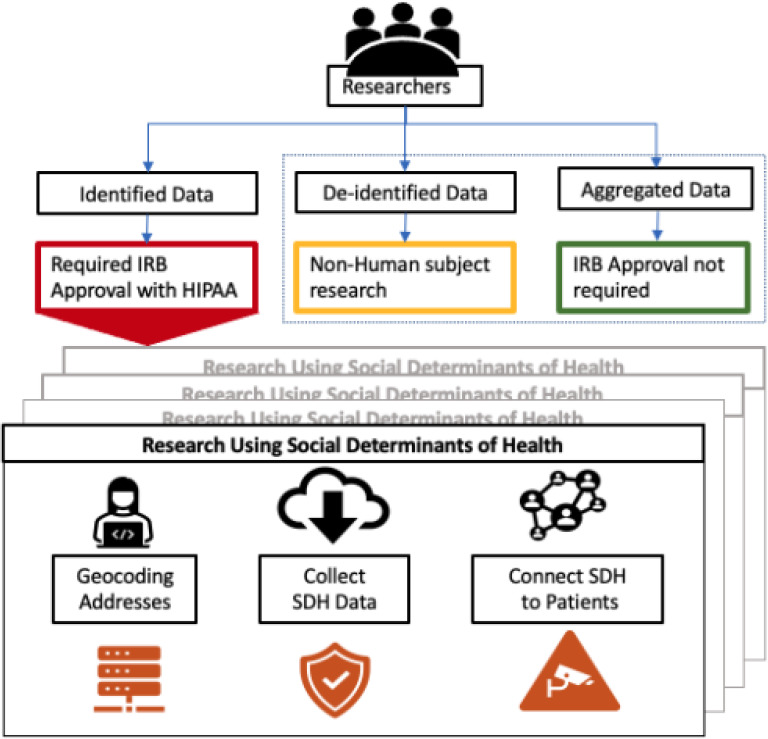
The existing procedure for research using SDoH-connected EHR data
The process of phenotyping patient conditions is paramount for research as it facilitates the identification of patient groups based on specific clinical characteristics. This enables researchers to study specific patient populations and gain insight into their unique health needs and outcomes. The Center for Medicare and Medicaid Services (CMS)23 is a valuable resource for phenotyping algorithms as it is responsible for administering the Medicare and Medicaid programs, which cover a significant proportion of the US population. As such, CMS has access to a vast amount of patient health data, making it an ideal source for developing and implementing phenotyping algorithms. Furthermore, CMS’s emphasis on providing patient-centered care aligns with the goal of phenotyping, which aims to improve our understanding of specific patient populations and their health outcomes.
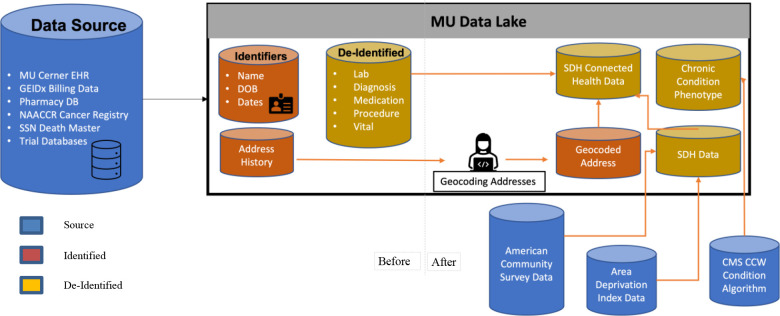
This study presents a novel framework (Figure 2) for supporting population health outcomes research by leveraging socio-demographic and health-related data integration. The developed framework encompasses geocoding addresses, phenotyping of diagnostic conditions or risk factors, and staging of socio-demographic data. It aims to streamline the process of conducting such studies by reducing repetitive steps typically undertaken by researchers. By leveraging the expertise of a clinical research institution that uses the framework, researchers can access de-identified data without the need to connect EHR data with socio-demographic data themselves. Additionally, the framework standardizes the process of providing phenotypic data generated using standardized algorithms to identify patient cohorts based on clinically relevant characteristics.
Figure 2.
Before and after the enrichment of Data Lake to support SDoH-connected EHR data
The framework was implemented initially on the University of Missouri’s (MU) Data Lake, which features an integrated clinical data repository. MU’s Data Lake converts MU Cerner Millennium, MU billing, and MU pharmacy data into a common data model (CDM). A non-profit institute, Patient-Centered Outcomes Research Institute (PCORI)24, was developed through the Patient Protection and Affordable Care Act (ACA) to provide patient-centered care. PCORI gave rise to the National Patient-Centered Clinical Research Network (PCORnet), which offers a well-established ecosystem for research platforms. PCORnet implemented a CDM to enhance patient-centered research among the Clinical Data Research Networks (CDRN) and other contributors. This CDM was necessary to achieve a universally compatible and system-integrated data model. MU’s Data Lake uses this CDM definition to create the clinical research data repository as part of Greater Plain Collaborative (GPC), a PCORnet CDRN.
This study endeavors to enrich the clinical research Data Lake by facilitating a more efficient and effective approach for population health outcomes research. Furthermore, a population health outcomes study (correlation analysis between chronic conditions and socioeconomic status) has been conducted to demonstrate the utility of this framework.
Methods
Data Acquisition
The MU Data Lake comprises a total of 1,897,412 unique patient records, of which 983,015 have been diagnosed, 901,659 have undergone treatment procedures, and 237,335 have been reported as deceased. Within this data repository, 27,206 providers administered medication to 405,306 patients. Of the total patient population, only 1,044,417 reported having encounters, with a total of 18,878,445 encounters captured, indicating that certain patients underwent multiple visits. Additionally, the Data Lake contains a total of 52,385,174 diagnoses and 39,353,994 treatment procedures. The mean and median age of patients in the CDM is 49. Age was subsequently grouped into five categories, with 0.4% of patients having missing information regarding their age. The most significant percentage of patients falls within the age range of 22-64 years, while the smallest percentage is observed among patients aged 0-5 years. The gender distribution of patients in the CDM is 51.1% female and 48.2% male, with 0.6% missing values. A significant proportion of patients’ ethnicity is either missing or refused, and nearly 40% of data regarding patients’ race is missing or refused.
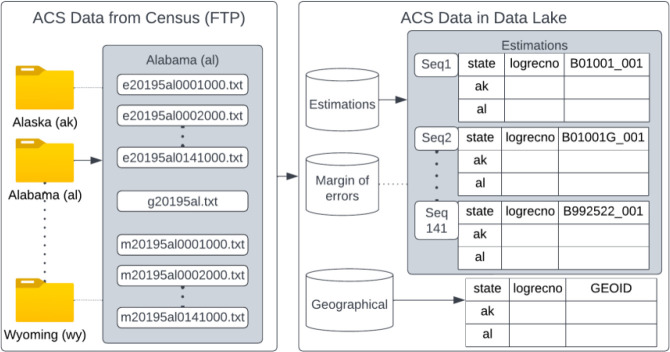
US Census Bureau’s official website13 provides Geographic Identifiers (GEOIDs) and their survey data for each year and five years on average. We utilized the 2015-2019 American Community Survey (ACS) 5-year estimates data in our Data Lake. This data is provided in three different datasets, including 5-year estimates of survey values, the margin of error, and geographical information. The data is organized into 52 state folders, each containing 141 sequences. Not all the records are at the block group level; some are at the state, county, or tract level. In total, there are 294,334 block group IDs and 27,045 distinct variables. The state-level folders contain estimates, the margin of error, and geographical information in comma-separated text files. ACS also provides separate excel files for variable names and definitions for estimates and margin of error, as well as for geographical data.
The University of Wisconsin School of Medicine and Public Health’s Department of Medicine provide the area deprivation index data. 2019’s ADI data is based on a 5-year average of a selective set of ACS variables obtained from 2015-201925. 2019’s ADI has both nationally and state-ranked ADI. The national-ranked ADI ranged from 1 to 100, whereas the state-ranked ADI ranged from 1 to 10. This means 1 is the less deprived area, and 100 or 10 is the most deprived area.
The CMS created a Chronic Condition Data Warehouse (CCW) with 27 common chronic conditions and their respective identifying algorithm26. Most of these are based on ICD codes from Medicare and Medicaid claims, reference periods, and the count and type of encounters. Most of the CCW data was extracted or collected from CMS beneficiaries, and to have a record linkage across its sources, CMS connected the records using unique identifiers. Claims records have the diagnosis and procedural codes, and CMS uses this information to build its algorithms. These algorithms are predefined to identify the beneficiary association with chronic diseases. The total number of conditions in the CCW is 67, among which 27 are the most common, and the rest indicate potentially disabling or other chronic diseases. These most common 27 chronic conditions are Acquired Hypothyroidism, Acute Myocardial Infraction, Alzheimer’s Disease, Alzheimer’s Disease and Related Disorders or Senile Dementia, Anemia, Asthma, Atrial Fibrillation, Benign Prostatic Hyperplasia, Cataract, Chronic Kidney Disease, Chronic Obstructive Pulmonary Disease and Bronchiectasis, Depression, Diabetes, Glaucoma, Heart Failure, Hip/Pelvic Fracture, Hyperlipidemia, Hypertension, Ischemic Heart Disease, Osteoporosis, Rheumatoid Arthritis/ Osteoarthritis (RA/OA), Stroke/Transient Ischemic Attack, Female/Male Breast Cancer, Colorectal Cancer, Prostate Cancer, Lung Cancer, Endometrial Cancer.
Geocoding Addresses
The first application for retrieving geographic identifiers and assigning them to geographic space was developed for health care27. In literature, there are many methods28–30 of geocoding but mostly adding census geographic codes to health records is the goal of this study. A decentralized method for geocoding called DeGAUSS can derive geographic identifiers and maintain the privacy of protected health information. A critical benefit of using DeGAUSS is that it can be executed in a local machine, requiring less computational resources. The most vital part of using DeGAUSS for health data is not exposing Protected Health Information (PHI) to a third party or the Internet.
To obtain geocoded data from DeGAUSS and connect each patient to a Census block-level group ID, we followed the steps outlined in Figure 3. In step 1, as addresses were not consistently stored with the same granularity and some patients were from the same household, duplicate address details were removed based on five attributes (street, city, county, state, and zip code). In step 2, to conform to the format expected by the DeGAUSS geocoder program, certain records were modified to convert attributes to a single-column format. This process involved correcting records with default values (e.g., "insert the county", "city name", empty strings, zeroes) to an empty value and then utilizing attributes with proper values to generate the final address string. This was done to ensure that DeGAUSS could geocode an address, even if only the five-digit zip code was provided, in order to prevent data loss. In step 3, all addresses were geocoded using the DeGAUSS geocoder docker image version 3.0.1.12. No geocoding score threshold was set, thus resulting in geocoding for all records. In the final step, the geocoded file with all the information about street, city, state, zip, latitude, longitude, score, and precision was provided as input to the DeGAUSS’s census_block_group:0.3 image to generate the block group id for all addresses. US Census Bureau’s block group shapefiles for 2020 were used to populate the block group ids.
Figure 3.
Geocoding steps with DeGAUSS geocoder
Staging SDoH Data
In this framework, 2015-2019 ACS 5 years data was selected to stage from the "Data from FTP" option. Fifty states’ information was merged horizontally per sequence and thus created 141 tables from 141 sequences. Vertically merging 141 sequences was not feasible as there were 27,045 data points. Five years estimations (file starting with e), the margin of errors (file starting with m), and geographical (file starting with g) data are stored into separate schemas to avoid complications. As a final output, the database should contain two schemas with 141 tables (27,045 columns in total) and a geographical table to link these data to geographical units (Figure 4). ADI data was already mapped to the block group level. Both nationally-ranked and state-ranked ADI values per block group were staged.
Figure 4.
Staging ACS data into Data Lake by merging horizontally
Calculating Reidentification Risks of PHI
In an effort to preserve patient privacy, the reidentification risks of public socioeconomic data (SDoH) were evaluated by connecting it to de-identified clinical data on a geographical unit basis. Specifically, the study focused on the block group level and utilized sixteen variables from the American Community Survey (ACS) - associated with a total of 63 variables - to generate the Area Deprivation Index (ADI)31,32. These variables included demographic, socioeconomic, housing-related information, and data on the cost of housing. The uniqueness of these sixteen variables at the block group level was analyzed to identify potential reidentification risks within the combined clinical and SDoH data. Figure 5 shows an example of how a unique public SDoH variable combination can reveal patients’ GEOID.
Figure 5.
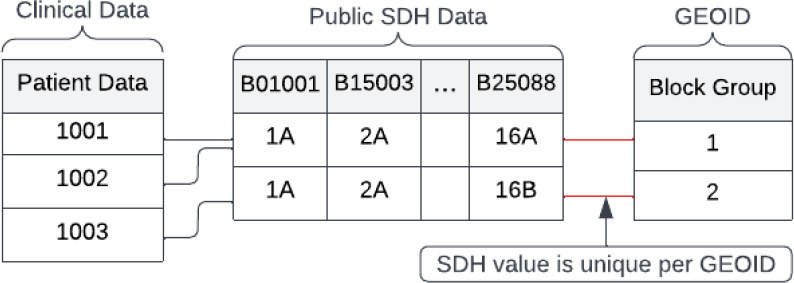
Reidentification of PHI when SDoH value is unique per GEOID
Phenotyping Chronic Condition
Twenty-seven chronic conditions phenotyping algorithms from CCW are primarily descriptive in nature and not programmable without preprocessing. The reference period is not needed as the primary intention of the reference period is for claim purposes23. In order to adapt these algorithms for research purposes, we refined them with structural values and modified them to fit all 27 algorithms into a single algorithm. This involved creating 16 variables to granularize the data and simplify the inclusion and exclusion criteria, as detailed in Table 1. To phenotype patient conditions, the single algorithm was applied to the CDM of the MU Data Lake, which contains patient records, including information on diagnoses, treatment procedures, and encounters.
Table 1.
Definition of Structured Variables from CMS Descriptive Algorithms
| Variable | Details | Remarks |
|---|---|---|
| cc | Chronic Condition | |
| icd9 (C1) | ICD9 Codes for all types of Inclusion Criteria | Effective dates for ICD-9 codes vary but are valid through 09/2015 |
| icd9_exc_a (C2) | ICD9 Codes for any of the diagnosis’s positions for exclusion criteria | Contains logic and code |
| icd9_exc_p | ICD9 Codes for primary diagnoses position for exclusion criteria | Contains only logic |
| icd10 | ICD10 Codes for all types of Inclusion Criteria | ICD-10 codes are effective 10/2015; |
| icd10_exc_a | ICD10 Codes for any of the diagnosis’s positions for exclusion criteria | Contains only code |
| icd10_exc_p | ICD10 Codes for primary diagnoses position for exclusion criteria | Contains only code |
| inc_a | Any DX on the claim | 1=Yes, 0=No |
| inc_12 | INCLUSION: Only the first or second DX on the claim | 1=Yes, 0=No |
| inc_p | INCLUSION: Only principal DX on the claim | 1=Yes, 0=No |
| exc_a | EXCLUSION: If any of the qualifying claims have these DX Codes in any DX position | 1=Yes, 0=No |
| exc_p | EXCLUSION: If any of the qualifying claims have these DX Codes in principle, the DX position | 1=Yes, 0=No |
| ip | InPatient | Minimum visit frequency |
| snf | SNF refers to the skilled nursing facility | Minimum visit frequency |
| hha | HHA refers to the home health agency | Minimum visit frequency |
| hop | HOP refers to hospital outpatient | Minimum visit frequency |
Correlation Analysis Between Chronic Conditions And Socioeconomic Status
To evaluate the effectiveness of our proposed framework, we conducted a population health outcomes study to investigate the relationship between chronic conditions and socioeconomic status. Using the Data Lake, we gathered clinical data of adult patients (aged 18 or older) who had at least one encounter between 2015 and 2019. Additionally, we obtained the national and state-ranked Area Deprivation Index (ADI) values of the patients’ block groups as a socioeconomic indicator and pre-calculated phenotypic data for chronic conditions. We calculated the percentage of patients with a specific chronic condition among the 27 chronic conditions and lived in block groups with similar ADI values. We then used univariate logistic regression to determine the odds of having a specific chronic condition among the 27 conditions in relation to the ADI values of the patients’ block groups.
Results
1,673,145 patients had 900,268 unique addresses based on five attributes (street, city, county, state, and zip code). Many address records did not have one or more attributes, whereas many attributes had meaningless default values. DeGAUSS decoder could not geocode 936 records as either the ZIP code was missing or the address was not a physical address. DeGAUSS could not assign a block group ID to 101 geocoded addresses, but it generated block group IDs for 899,231 addresses.
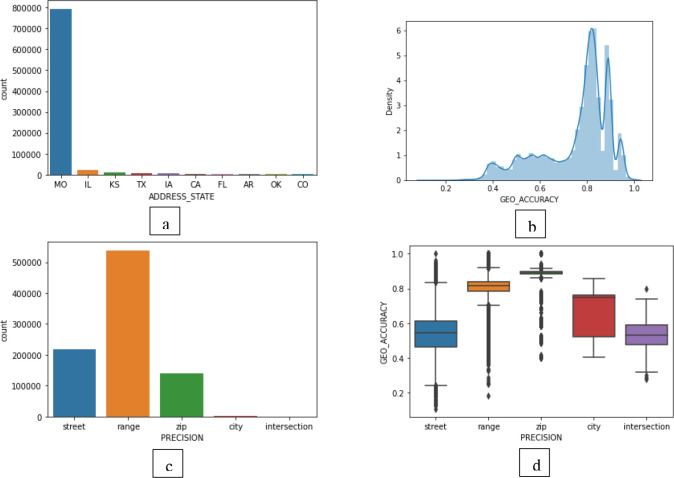
As stated in Figure 6(a), most patients in Data Lake are from Missouri. A fair number of patients are from Illinois, Kansas, and Texas. Some patients are from US Armed Forces – Americas/ Europe/ Pacific. Most of the addresses geocoded by DeGAUSS show high accuracy. If we plot the density of geo accuracy, most addresses were geocoded with around 80% accuracy. But the accuracy depends upon the precision type of the geocode. There are five precision types. They are range, street, intersection, zip, and city. These names are mentioned from most granular to least granular kind of precision. That means range-level precision is the most granular, and city-level precision is the least granular geocoding. Even though our overall geocoding accuracy is high, we needed to investigate accuracy distribution amongst the precision types. Most of the addresses were geocoded with the precision of "range" (Figure 6(c)). The mean and standard deviation of geocoding accuracy for the precision "range" is comparatively high as well (Figure 6(d)). Any researcher can make decisions on their inclusion criteria by giving a threshold using the combination of precision type and accuracy score based on these statistics.
Figure 6.
(a) Distribution of patients per state, (b) Density of addresses with respect to their geocoding accuracy, (c) Distribution of addresses based on their geocoding precision type, (d) Box plot of geocoding accuracy of all addresses per precision type
Our analysis of ACS data revealed a potential for the reidentification of patients due to variations in data at the block group level. Through a uniqueness check per block-level ID and modifications to the original data, we were able to mitigate this issue effectively. Our findings, as presented in Table 2, indicate that our proposed data modification strategy was successful in reducing the risk of reidentification from 100% to 97.4% when converting 16 SES variables into percentiles and further to 77% and 13% when converting into deciles and quartiles respectively.
Table 2.
The uniqueness of the combination of ACS data per block group level
| Category | Number of Re-Identifiable Block Group Level (Percentage) |
|---|---|
| Unmodified 63 variables to generate 16 SES variables | 294334 (100 %) |
| Converted the value of 16 SES variables from 63 variables | 294334 (100 %) |
| Percentile form of 16 SES Variables | 286901 (97.4 %) |
| Decile form of 16 SES Variables | 226715 (77 %) |
| Quartile form of 16 SES Variables | 5299 (13 %) |
After applying the CCW-derived single algorithm, patients with at least one of the 27 chronic conditions were generated. A Boolean per patient per chronic condition is used to represent this phenotypic data. The total number of patients in MU Data Lake per chronic condition is given in Table 3. Most patients we have is for Hypertension and Depression. The highest mean age is 82.17 years for Alzheimer’s Disease. The lowest mean age is for Asthma (42.5 years). Based on the criteria described in the method, 52,964 patients have at least one of the 27 chronic conditions. Of all the population, people suffering from hypertension showed a higher number with a count of 15377. Depression followed hypertension with a count of 10048 patients. Endometrial cancer showed fewer counts than other conditions, with a count of 167.
Table 3.
Number of adult patients, mean and standard deviation of their age and Odds Ratio (OR), P-value (P), and 95% Confidence Interval (95% CI) of univariate analysis between Area Deprivation Index (ADI) and percentage of patients living in an area with similar ADI and having the same chronic condition (red= P<0.05 and OR>1; green=P<0.05 and OR<1)
| Predictor (ADI vs.) | # Of Patient | Age mean±SD | National Ranked ADI | State Ranked ADI | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | P | 95% CI | OR | P | 95% CI | |||||
| Acquired Hypothyroidism | 4950 | 63.0±19.2 | 0.9993 | 0.357 | 0.9979 | 1.0007 | 0.9939 | 0.2604 | 0.9834 | 1.0045 |
| Acute Myocardial Infraction | 569 | 66.5±14.2 | 1.0125 | 0 | 1.0081 | 1.017 | 1.091 | 0 | 1.0574 | 1.1257 |
| Alzheimers Disease | 358 | 82.2 ±10.2 | 0.9991 | 0.746 | 0.994 | 1.0043 | 0.9921 | 0.69 | 0.954 | 1.0317 |
| Alzheimers Disease and Related Disorders or Senile Dementia | 1400 | 77.4±15.6 | 1.0025 | 0.0653 | 0.9998 | 1.0052 | 1.0174 | 0.0882 | 0.9974 | 1.0377 |
| Anemia | 7370 | 52.7±23.3 | 1.0076 | 0 | 1.0064 | 1.0088 | 1.0573 | 0 | 1.0481 | 1.0666 |
| Asthma | 3505 | 42.6±23.2 | 1.0061 | 0 | 1.0044 | 1.0079 | 1.0458 | 0 | 1.0327 | 1.059 |
| Atrial Fibrillation | 1427 | 72.6±13.9 | 1.0039 | 0.0035 | 1.0013 | 1.0066 | 1.0361 | 0.0004 | 1.016 | 1.0566 |
| Benign Prostatic Hyperplasia | 1320 | 73.3±11.5 | 0.9988 | 0.3632 | 0.9961 | 1.0014 | 0.9904 | 0.3561 | 0.9704 | 1.0109 |
| Cataract | 1678 | 66.9±15.5 | 0.9963 | 0.0021 | 0.9939 | 0.9987 | 0.9695 | 0.0009 | 0.952 | 0.9873 |
| Chronic Kidney Disease | 5054 | 60.8±21.4 | 1.0101 | 0 | 1.0086 | 1.0115 | 1.0751 | 0 | 1.0638 | 1.0865 |
| Chronic Obstructive Pulmonary Disease and Bronchiectasis | 2780 | 64.4±15.1 | 1.0135 | 0 | 1.0115 | 1.0155 | 1.0941 | 0 | 1.0786 | 1.1098 |
| Depression | 10048 | 43.8±19.5 | 1.0042 | 0 | 1.0032 | 1.0052 | 1.0318 | 0 | 1.024 | 1.0396 |
| Diabetes | 9580 | 61.1±17.6 | 1.0106 | 0 | 1.0095 | 1.0117 | 1.077 | 0 | 1.0687 | 1.0853 |
| Glaucoma | 1676 | 62.8±18.7 | 1 | 0.9858 | 0.9976 | 1.0024 | 0.9994 | 0.9521 | 0.9815 | 1.0177 |
| Heart Failure | 2952 | 67.2±16.5 | 1.0122 | 0 | 1.0102 | 1.0141 | 1.0892 | 0 | 1.0742 | 1.1043 |
| Hip/Pelvic Fracture | 468 | 58.1±25.3 | 1.0078 | 0.0012 | 1.0031 | 1.0125 | 1.0589 | 0.0011 | 1.0232 | 1.0958 |
| Hyperlipidemia | 1084 | 62.4±15.5 | 0.9933 | 0 | 0.9904 | 0.9962 | 0.9555 | 0.0001 | 0.934 | 0.9774 |
| Hypertension | 15377 | 63.6±16.8 | 1.0052 | 0 | 1.0044 | 1.006 | 1.0378 | 0 | 1.0315 | 1.0442 |
| Ischemic Heart Disease | 5967 | 66.7±14.6 | 1.0103 | 0 | 1.009 | 1.0117 | 1.0752 | 0 | 1.0647 | 1.0857 |
| Osteoporosis | 942 | 75.3±14.1 | 0.9992 | 0.6007 | 0.996 | 1.0023 | 0.9954 | 0.7054 | 0.9716 | 1.0197 |
| Rheumatoid Arthritis/ Osteoarthritis (RA/OA) | 0 | - | - | - | - | - | - | - | - | - |
| Stroke/Transient Ischemic Attack | 1051 | 64.8±18.0 | 1.0096 | 0 | 1.0064 | 1.0128 | 1.068 | 0 | 1.0438 | 1.0928 |
| Female/Male Breast Cancer | 857 | 62.6±14.3 | 1.0031 | 0.0756 | 0.9997 | 1.0065 | 1.0227 | 0.0817 | 0.9972 | 1.0489 |
| Colorectal Cancer | 251 | 66.4±13.9 | 1.0103 | 0.0019 | 1.0038 | 1.0169 | 1.0746 | 0.0026 | 1.0253 | 1.1261 |
| Prostate Cancer | 309 | 69.8±11.0 | 0.9958 | 0.133 | 0.9904 | 1.0013 | 0.9693 | 0.1494 | 0.9292 | 1.0113 |
| Lung Cancer | 313 | 65.4±11.9 | 1.0118 | 0.0001 | 1.0059 | 1.0177 | 1.0864 | 0.0001 | 1.0416 | 1.1331 |
| Endometrial Cancer | 167 | 65.1±12.5 | 1.01 | 0.0136 | 1.0021 | 1.0181 | 1.0593 | 0.0489 | 1.0003 | 1.1218 |
As shown in Table 3, for national-ranked and state-ranked ADI, we have significant p-value for Acute Myocardial Infraction, Anemia, Asthma, Atrial Fibrillation, Cataract, Chronic Kidney Disease, Chronic Obstructive Pulmonary Disease and Bronchiectasis, Depression, Diabetes, Heart failure, Hyperlipidemia, Hypertension, Heart Disease, Stroke/Transient Ischemic Attack, Colorectal Cancer, and Lung Cancer. Out of these chronic conditions, Cataract and Hyperlipidemia showed decreasing odds with an increase in ADI. But for the rest of them, it was an increasing trend with an increase in ADI, which means the chances of having chronic conditions increase with the increase in deprivation index.
Discussion
The use of the DeGAUSS geocoder to geocode the addresses of patients within the Data Lake yielded promising results, with nearly 99.8% of unique addresses successfully assigned a block-group ID. However, it was observed that a significant proportion of addresses, precisely 11% of total patient addresses, were geocoded with a geocoding accuracy of less than 50%. Additionally, 10.6% of total patient addresses were geocoded with a geocoding precision type of either ZIP or City. These findings indicate that further examination of patient addresses and potential strategies to enhance geocoding accuracy and precision type beyond ZIP and City is necessary.
In addition, socioeconomic data from the ACS and ADI were collected from public repositories and linked with patient data using the block group ID for each patient. The linkage of ACS data raised concerns regarding the potential for reidentification. As such, appropriate steps must be taken to ensure obfuscation of the data before classifying it as fully de-identified. The staging of social determinants should be considered a continuous process, with future plans to incorporate additional publicly available social determinants into the dataset and to conduct extensive research to address the issue of reidentification in order to facilitate data science and population health research using privacy-preserved de-identified data.
Furthermore, phenotypic data were also staged in conjunction with electronic health record (EHR) data, allowing researchers to take advantage of preprocessed data. However, it is acknowledged that the use of the CMS algorithm for phenotyping is limited to the use of diagnosis codes only. As such, future investigations should consider the implementation of other validated algorithms that utilize various forms of clinical information, such as laboratory results, procedures, medications, and vital signs, such as the SUPREME-DM algorithm for diabetes. To further enhance the dataset, consideration may also be given to utilizing a two-step crowd-sourcing approach for phenotyping, which includes a sharing source code mechanism that works in conjunction with the dataset and a common data model, allowing researchers to run the codes to obtain phenotypic data, as well as periodically using pre-computed phenotypes for direct access by researchers within the dataset.
The findings from the univariate analysis suggest that individuals living in areas with higher levels of deprivation may be at an increased risk for most of the chronic conditions (16). Our developed framework provided a comprehensive approach to this population health outcomes study by utilizing a large sample size and integrating various data sources, including de-identified clinical data, socioeconomic data, and pre-computed phenotypic data for chronic conditions.
Any individual researcher conducting a population health outcomes study can leverage the pre-computed de-identified data with minimal administrative processing.
Conclusion
Integrating publicly available data into a clinical research data repository and linking it to patient information while preserving privacy can significantly enhance the accessibility and efficiency of health research. By eliminating the administrative burden and challenges associated with storing identified data in individual research workspaces, researchers will have improved access to a wide range of data for their studies. Additionally, by utilizing algorithms to generate modified or phenotypic data from patient data, the research process can be streamlined, reducing overall computational effort, and increasing research productivity. The goal of this framework is to foster a collaborative and dynamic environment where researchers can not only utilize preprocessed data but also contribute their own algorithms and feedback to enrich the data repository further.
Figures & Table
References
- 1.Marmot M, Friel S, Bell R, Houweling TA, Taylor S. Closing the gap in a generation: health equity through action on the social determinants of health. Lancet. 2008;372(9650):1661–1669. doi: 10.1016/S0140-6736(08)61690-6. doi:10.1016/S0140-6736(08)61690-6. [DOI] [PubMed] [Google Scholar]
- 2.Cole BL, Fielding JE. Health impact assessment: A tool to help policy makers understand health beyond health care. Annu Rev Public Health. 2007;28:393–412. doi: 10.1146/annurev.publhealth.28.083006.131942. doi:10.1146/annurev.publhealth.28.083006.131942. [DOI] [PubMed] [Google Scholar]
- 3.Beard JR, Lincoln D, Donoghue D, et al. Socioeconomic and maternal determinants of small-for-gestational age births: Patterns of increasing disparity. Acta Obstet Gynecol Scand. 2009;88(5):575–583. doi: 10.1080/00016340902818170. doi:10.1080/00016340902818170. [DOI] [PubMed] [Google Scholar]
- 4.Wagstaff A. Poverty and health sector inequalities. Bull World Health Organ. 2002;80(2):97–105. /pmc/articles/PMC2567730/?report=abstract. Accessed March 8, 2022. [PMC free article] [PubMed] [Google Scholar]
- 5.Marmot M. Achieving health equity: from root causes to fair outcomes. Lancet. 2007;370(9593):1153–1163. doi: 10.1016/S0140-6736(07)61385-3. doi:10.1016/S0140-6736(07)61385-3. [DOI] [PubMed] [Google Scholar]
- 6.Koh HK, Piotrowski JJ, Kumanyika S, Fielding JE. Healthy people: A 2020 vision for the social determinants approach. Heal Educ Behav. 2011;38(6):551–557. doi: 10.1177/1090198111428646. doi:10.1177/1090198111428646. [DOI] [PubMed] [Google Scholar]
- 7.Rushton G, Armstrong MP, Gittler J, et al. Geocoding in cancer research: A review. Am J Prev Med. 2006;30(2 SUPPL.) doi: 10.1016/j.amepre.2005.09.011. doi:10.1016/j.amepre.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Meeker JR, Burris HH, Bai R, Levine LD, Boland MR. Neighborhood deprivation increases the risk of Post-induction cesarean delivery. J Am Med Inform Assoc. 2022;29(2):329–334. doi: 10.1093/jamia/ocab258. doi:10.1093/jamia/ocab258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, Kind AJH, Nerenz D. Area Deprivation Index Predicts Readmission Risk at an Urban Teaching Hospital. Am J Med Qual. 2018;33(5):493–501. doi: 10.1177/1062860617753063. doi:10.1177/1062860617753063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wadhwani SI, Brokamp C, Rasnick E, Bucuvalas JC, Lai JC, Beck AF. Neighborhood socioeconomic deprivation, racial segregation, and organ donation across 5 states. Am J Transplant. 2021;21(3):1206–1214. doi: 10.1111/ajt.16186. doi:10.1111/ajt.16186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadhwani SI, Bucuvalas JC, Brokamp C, et al. Association between neighborhood-level socioeconomic deprivation and the medication level variability index for children following liver transplantation. Transplantation. 2020;104(11):2346–2353. doi: 10.1097/TP.0000000000003157. doi:10.1097/TP.0000000000003157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metcalfe A, Lail P, Ghali WA, Sauve RS. The association between neighbourhoods and adverse birth outcomes: A systematic review and meta-analysis of multi-level studies. Paediatr Perinat Epidemiol. 2011;25(3):236–245. doi: 10.1111/j.1365-3016.2011.01192.x. doi:10.1111/j.1365-3016.2011.01192.x. [DOI] [PubMed] [Google Scholar]
- 13.US Census Bureau. Understanding Geographic Identifiers (GEOIDs) US Census. https://www.census.gov/programs-surveys/geography/guidance/geo-identifiers.html. Published 2018. Accessed July 16, 2021.
- 14.Kind AJH, Buckingham WR. Making Neighborhood-Disadvantage Metrics Accessible — The Neighborhood Atlas. N Engl J Med. 2018;378(26):2456–2458. doi: 10.1056/NEJMp1802313. doi:10.1056/nejmp1802313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig J, Sanbonmatsu L, Gennetian L, et al. Neighborhoods, Obesity, and Diabetes — A Randomized Social Experiment. N Engl J Med. 2011;365(16):1509–1519. doi: 10.1056/NEJMsa1103216. doi:10.1056/nejmsa1103216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kind AJH, Jencks S, Brock J, et al. Neighborhood socioeconomic disadvantage and 30-day rehospitalization: A retrospective cohort study. Ann Intern Med. 2014;161(11):765–774. doi: 10.7326/M13-2946. doi:10.7326/M13-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lantos PM, Hoffman K, Permar SR, et al. Neighborhood Disadvantage is Associated with High Cytomegalovirus Seroprevalence in Pregnancy. J Racial Ethn Heal Disparities. 2018;5(4):782–786. doi: 10.1007/s40615-017-0423-4. doi:10.1007/s40615-017-0423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler DC, Petterson S, Phillips RL, Bazemore AW. Measures of Social Deprivation That Predict Health Care Access and Need within a Rational Area of Primary Care Service Delivery. Health Serv Res. 2013;48(2 Pt 1):539. doi: 10.1111/j.1475-6773.2012.01449.x. doi:10.1111/J.1475-6773.2012.01449.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofer TP, Wolfe RA, Tedeschi PJ, McMahon LF, Griffith JR. Use of community versus individual socioeconomic data in predicting variation in hospital use. Health Serv Res. 1998;33(2 Pt 1):243. [PMC free article] [PubMed] [Google Scholar]
- 20.Pitts C, McKissack H, Alexander B, et al. Do Geographic Region, Pathologic Chronicity, and Hospital Affiliation Affect Access to Care among Medicaid-And Privately Insured Foot and Ankle Surgery Patients? South Med J. 2021;114(1):35–40. doi: 10.14423/SMJ.0000000000001198. doi:10.14423/SMJ.0000000000001198. [DOI] [PubMed] [Google Scholar]
- 21.Rushton G, Armstrong MP, Gittler J, et al. Geocoding in cancer research: A review. Am J Prev Med. 2006;30(2 SUPPL.) doi: 10.1016/j.amepre.2005.09.011. doi:10.1016/j.amepre.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Hodge JGJ, Gostin LO. Public health practice vs research: A report for Public Health Practitioners Including Cases and Guidance for Making Distinctions. Rep funded by Counc State Territ Epidemiol Atlanta, Georg. 2008;2(3):185–191. http://www.ncbi.nlm.nih.gov/pubmed/20698919. Accessed March 8, 2022. [Google Scholar]
- 23.Gorina Y, Kramarow EA. Identifying chronic conditions in medicare claims data: Evaluating the chronic condition data warehouse algorithm. Health Serv Res. 2011;46(5):1610–1627. doi: 10.1111/j.1475-6773.2011.01277.x. doi:10.1111/j.1475-6773.2011.01277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.PCORI |. https://www.pcori.org/. Accessed July 15, 2021.
- 25.University of Wisconsin School of Medicine and Public Health. 2019. Area Deprivation Index V3.1. Downloaded from. https://www.neighborhoodatlas.medicine.wisc.edu/. Accessed July 15, 2021.
- 26.Services C for M and M. Chronic conditions among Medicare beneficiaries, chart book. Baltimore, MD: http://www.cms.gov/Research-Statistics-Data-and-Systems/Statistics-Trends-and-Reports/Chronic-Conditions/Downloads/2012Chartbook.pdf. Published 2012. [Google Scholar]
- 27.Cromer SJ, Lakhani CM, Wexler DJ, Burnett-Bowie S-AM, Udler M, Patel CJ. Geospatial Analysis of Individual and Community-Level Socioeconomic Factors Impacting SARS-CoV-2 Prevalence and Outcomes. medRxiv Prepr Serv Heal Sci. 2020;1636870 doi:10.1101/2020.09.30.20201830. [Google Scholar]
- 28.Faure E, Danjou AMN, Clavel-Chapelon F, Boutron-Ruault MC, Dossus L, Fervers B. Accuracy of two geocoding methods for geographic information system-based exposure assessment in epidemiological studies. Environ Heal A Glob Access Sci Source. 2017;16(1):1–12. doi: 10.1186/s12940-017-0217-5. doi:10.1186/s12940-017-0217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh H, Fortington L V., Thompson H, Finch CF. An overview of geospatial methods used in unintentional injury epidemiology. Inj Epidemiol. 2016;3(1) doi: 10.1186/s40621-016-0097-0. doi:10.1186/s40621-016-0097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira VM, Howard GJ, Gallagher LG, Fletcher T. Geocoding rural addresses in a community contaminated by PFOA: A comparison of methods. Environ Heal A Glob Access Sci Source. 2010;9(1):1–7. doi: 10.1186/1476-069X-9-18. doi:10.1186/1476-069X-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knighton AJ, Savitz L, Belnap T, Stephenson B, VanDerslice J. Introduction of an Area Deprivation Index Measuring Patient Socioeconomic Status in an Integrated Health System: Implications for Population Health. eGEMs. 2016;4(3):9. doi: 10.13063/2327-9214.1238. doi:10.13063/2327-9214.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh GK. Area Deprivation and Widening Inequalities in US Mortality, 1969–1998. Am J Public Health. 2003;93(7):1137. doi: 10.2105/ajph.93.7.1137. doi:10.2105/AJPH.93.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]