Abstract
Place-based exposures, termed “geomarkers”, are powerful determinants of health but are often understudied because of a lack of open data and integration tools. Existing DeGAUSS (Decentralized Geomarker Assessment for Multisite Studies) software has been successfully implemented in multi-site studies, ensuring reproducibility and protection of health information. However, DeGAUSS relies on transporting geomarker data, which is not feasible for high-resolution spatiotemporal data too large to store locally or download over the internet. We expanded the DeGAUSS framework for high-resolution spatiotemporal geomarkers. Our approach stores data subsets based on coarsened location and year in an online repository, and appropriate subsets are downloaded to complete exposure assessment locally using exact date and location. We created and validated two free and open-source DeGAUSS containers for estimation of high-resolution, daily ambient air pollutant exposures, transforming published exposure assessment models into computable exposures for geomarker assessment at scale.
Introduction
Place-based exposures and community characteristics, termed “geomarkers,” are powerful determinants of health but are understudied compared to biomarkers because of a lack of open data and integration tools.[1, 2] Existing high resolution spatiotemporal exposure estimates (e.g. daily, < 1 sq km) are not often available as curated data sources and require specialized computing expertise to link to health data. Additionally, operationalizing exposure data in health studies is difficult because of metadata surrounding the exposure timing, duration, frequency, and latency.[2] A lack of shared standards creates research inefficiencies, including having to continuously develop and recreate exposure assessment models and data workflows, which ultimately prevent data integration at scale.[2] Geomarker data often exist at different spatiotemporal resolutions and extents and integrating these with multi-scale population health data requires new informatic research and development approaches.[3]
One such new development is the concept of a computable exposure that refers to “a representation of exposure data and metadata that can be used by an algorithm or a piece of software to answer a question.”[2] For geomarkers, computable exposures can be considered a tool to estimate and assign exposure data and metadata that can be used by an algorithm, model, or piece of software to answer a question. Assigning exposures necessitates linking an exposure surface to an individual, residence, or population in space and time. Creating an interoperable and portable exposure assessment tool as a computable geomarker also ensures that geomarker data and assessment tools follow FAIR data principles but are designed for privacy and reproducibility.[4, 5] The DeGAUSS (Decentralized Geomarker Assessment of Multi-Site Studies) framework and set of software packages have embraced this approach for geomarker assessment,[6] with successful applications in multi-site studies where the sharing of protected health information (PHI), like addresses and dates, was prohibited.[5, 7] DeGAUSS currently relies on sending and packaging geomarker data in a tool sent to the health data repository, but this approach is not feasible when the geomarker data is too large to store on a local machine or to download over the internet.
Geomarker data size increases exponentially when using high resolution spatiotemporal data common in applications related to climate, land usage, tree canopy, noise, and air pollution. A specific example of such a high resolution exposure common in environmental epidemiology is fine particulate matter (PM2.5) estimated at a daily, < 1 sq. km resolution.[8, 9] Spatiotemporal PM2.5 prediction models use machine learning with high resolution inputs such as land use characterizations, chemical transport modeling simulations, meteorological data, and satellite-based measures calibrated using PM2.5 measurements to accurately predict PM2.5 at locations and times where it was not measured.[10-12] These predictions are used for exposure assessment by linking a geospatial location (e.g., a geocoded residential address) and a time frame of interest (e.g., the fourth and fifth months of pregnancy) with a set of gridded spatiotemporal estimates. Daily estimates from some models of ambient PM2.5 concentration exist for the contiguous United States dating back to 2000 in online repositories and, depending on the file format, across thousands of different files totaling hundreds of GB.[11, 13]
Hosting exposure data online and downloading subject-specific estimates could overcome the data size problems with decentralized approaches like DeGAUSS but require transmission of precise spatiotemporal locations over the internet to third parties. As used in health studies, precise spatiotemporal locations are considered PHI and their sharing is often restricted or prohibited by law, Institutional Review Boards, and institutional data use and sharing policies.[14] The HIPAA Safe Harbor Guidelines specify that spatial boundaries containing fewer than 20,000 residents (e.g., a five-digit ZIP Code) and dates more specific than a calendar year (e.g., date of birth) are considered pseudo-identifiers. For this reason, spatiotemporal locations in datasets that are intended to be deidentified or shared are usually made less specific by (1) coarsening the spatial boundaries until they contain at least 20,000 residents (e.g., the first three digits of a five-digit ZIP Code) and (2) substituting specific dates by their calendar year.
Here, we extended the DeGAUSS platform to deal with high resolution and spatiotemporally gridded geomarkers by implementing an approach where the spatiotemporal precision of PHI is coarsened to query and download spatiotemporal subsets of estimates while the true precision is retained locally for exact spatiotemporal linkage. In addition to detailing our approach, we have created an accompanying DeGAUSS software implementation for two different high-resolution spatiotemporal PM2.5 exposure assessment models and detailed the example implementation of one within the NIH’s Environmental influences on Child Health Outcomes (ECHO) program.[15] Our hope is that this approach will serve as a general framework for using private, FAIR, and reproducible computable exposures for high-resolution and spatiotemporally gridded geomarker assessment in health studies.
Methods
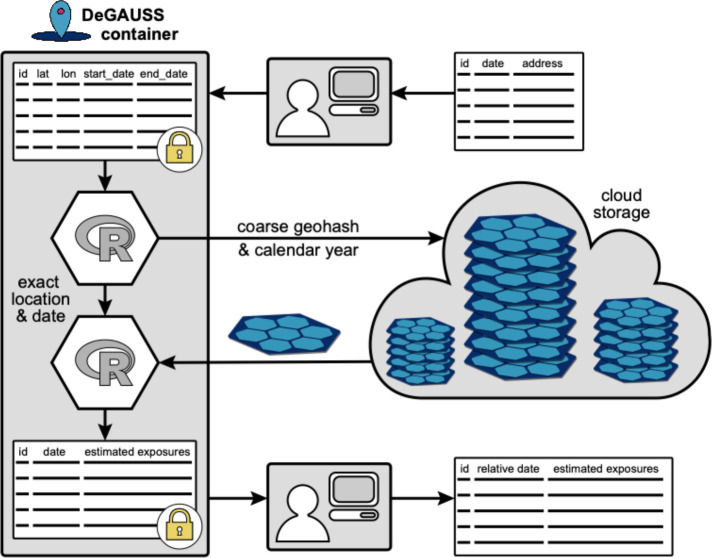
Figure 1 illustrates our extension of the DeGAUSS framework to work with spatiotemporally gridded datasets hosted online by (1) coarsening dates into years and geographic coordinates into Safe Harbor geohashes, (2) using coarsened spatiotemporal location to download appropriate subsets of exposure estimates from an online repository, and (3) completing exposure assessment locally based on exact date and location.
Figure 1.
An overview of the DeGAUSS approach used for large spatiotemporal data. Input start and end dates are expanded into a daily time series for each row in the input data. The calendar year and a coarsened geohash are used to download a spatiotemporal subset of exposure estimates. The files are cached locally, and exposure estimates are merged in based on exact date and location of the input data. Spatiotemporal pseudo-identifiers are removed by the user and exposures can be optionally averaged over specific time periods to reduce the risk of reidentification if sharing data.
Safe Harbor Geohash
Geohashes are hierarchical geospatial indexing systems that can divide space into grid cells. Subdivisions are based on space-filling curves and are identified using strings of letters and digits, termed geohashes. Thus, geographic locations with more similar geohashes are closer together in space. Because they are hierarchical, geohashes can be quickly up-scaled (or down-scaled) to a larger (or smaller) spatial resolution by truncating (or adding) characters. Two of the most widely used geohash systems are Google’s S2 Geometry Library (S2, available at https://s2geometry.io/) and Uber’s H3 Hexagonal Hierarchical Spatial Index (H3, available at https://h3geo.org/). Both systems are available as free and open-source specifications and software products designed for sindexing geospatial data in ways that make it easier to build large distributed spatial databases. S2 is based on square grid cells, while H3 is based on hexagonal cells. Each system has hierarchical levels of resolution, based on individual (square or hexagonal) grid or lattice cells that nest within each other for lower spatial resolution.
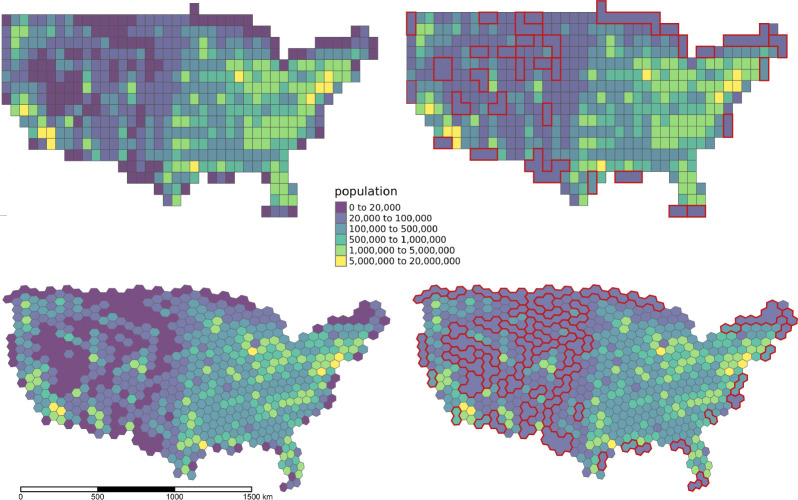
To use for querying spatiotemporal estimates in an online repository, we selected an initial resolution for each system such that most of the geohashes within the contiguous United States had at least 20,000 residents, satisfying HIPAA Safe Harbor Guidelines. The population residing in each geohash was estimated using the area-weighted averages of census tract-level 2018 5-yr ACS population estimates. The left side of Figure 2 shows the S2 and H3 geohashes covering the contiguous United States colored by estimated population. To combine geohashes such that none had a population of less than 20,000, the geohash with the lowest population was repeatedly merged with the neighboring geohash that had the lowest population. This process was repeated until all geohashes had a population greater than 20,000. We term these “Safe Harbor geohashes”, and storing exposure estimate data in flat files organized by these Safe Harbor geohashes creates a system that allows for querying subsets of online data with a lower spatial resolution, while maintaining the exact location for local exposure assessment.
Figure 2.
(Left) S2 and H3 geohashes covering the contiguous United States colored by estimated population. (Right) “Safe Harbor” geohashes aggregated to prevent any single geohash from containing less than 20,000 estimated individuals. Aggregated geohashes are outlined in red.
Harmonizing and Accessing Exposure Data with Safe Harbor Geographies
High resolution spatiotemporal exposure estimates are split into separate files based on the Safe Harbor geohash and calendar year. The matching, hierarchical system used for the geolocations and exposure data ensures efficient extraction of a desired spatiotemporal subset. For these purposes, the exposure estimates from Di et al. [11] were grouped by S2 geohash and calendar year. Brokamp [10] used the H3 geohash directly within the exposure assessment modeling framework and so exposure estimates were further aggregated for storage by Safe Harbor geohash and calendar year. R packages for both sets of exposure estimates were designed to intake spatiotemporal data, perform spatial linkages with the S2 or H3 Safe Harbor geohash, download spatiotemporal subsets of exposure estimates from a cloud service, and locally assign exposures using the exact location and date. R packages and code are free and open source and are available publicly at https://github.com/geomarker-io and https://github.com/degauss-org. In addition to preventing transmission of HIPAA Safe Harbor pseudo-identifiers over the internet to third parties, this approach downloads only spatiotemporal subsets of data that are required locally, decreasing the time and resources to run the software.
Containerization
We then containerized R code for use by non-R users. Also known as “operating-system level virtualization,” containerization is a unique feature of Unix-based operating systems in which the kernel facilitates multiple isolated user-space instances. These containers appear as isolated computers to the programs running them, but also have access to all the host computer’s physical resources and virtual files. As compared to virtual machine software that replicates an entire computer, e.g., “Virtual Box” or “Parallels”, containers require much less overhead because they can rely on the host system’s normal system call interface. Without the need to be run as an emulation or within a virtual machine, containers take advantage of the benefits of these approaches – namely isolation and the use of different operating systems within one host operating system – but also exhibit decreased overhead, increased flexibility, and decreased use of storage.
Specifically, we developed DeGAUSS containers using the Docker containerization platform, in which software is wrapped into a complete file system that contains everything needed to run such as code, system tools and libraries, geographic data, etc. Containers are based on Docker images and run directly on the system infrastructure rather than relying on a guest operating system or virtual machine. Docker has been previously used for reproducible research and solves common challenges in reproducible computational science like managing evolving software dependencies and versions, maintaining code compatibility with changing computing environments, and barriers to adoption and implementation by others.[16]
Software Validation
To validate the ability of the DeGAUSS images for the Di et al. [11] model to estimate exposures, we randomly sampled 5,000 coordinates within the contiguous United States. These locations were matched to the nearest grid coordinate using three methods: (1) the DeGAUSS “Schwartz Grid Lookup” image, version 0.4 (https://degauss.org/schwartz_grid_lookup), (2) ArcGIS (a commercially-available geographic information systems software), and (3) R (a statistical programming language that can use geospatial packages). The ArcGIS and R linkages to grid identifiers were completed by technical geospatial experts (JB, NL) using established methods for linking exposures produced by the Di et al. [11] model. The ArcGIS method consisted of a Spatial Join with the “closest geodesic” match option, while the R method utilized the “geodist” command with the “geodesic” option from the “geodist” R package (v0.0.7).
We validated exposure estimates for three common air pollutants PM2.5, nitrogen dioxide (NO2,) and ozone (O3) for the Cincinnati Childhood Allergy and Air Pollution Study (CCAAPS), one of the birth cohorts contributing data to ECHO. We established a data sharing agreement with investigators in ECHO in order to estimate exposures using existing methods. We estimated exposures using DeGAUSS and compared the results to those obtained using the existing R workflow which extracted estimates from locally stored rds files using the dbplyr and lubridate packages. For consistency, all estimates were rounded to the tenths place before comparison.
Software Usability and Evaluation
In order to track the DeGAUSS user experience, we created a software usability survey in REDCap.[17] We adapted this survey from the System Usability Scale (SUS)[18], a method for quickly determining the general usability of a software system. In our survey, we collected information about users’ backgrounds and previous experiences, details about how DeGAUSS was used, and thoughts on their experience. This included a usability rating and the likelihood that the user would use DeGAUSS again in the future or recommend it to a colleague. We also collected information about how DeGAUSS performs when compared to possible alternative methods and software tools. Each user was requested to respond to the survey while they were using the software.
Results
Safe Harbor Geohash
The final set of S2 and H3 geohashes modified to each have at least 20,000 residents were termed “Safe Harbor geohashes.” Used in combination with the calendar year, this represents the fundamental unit of spatiotemporal resolution by which exposure data is stored and queried in online repositories. The left side of Figure 2 depicts S2 (resolution: 3) and H3 (resolution: 3) geohashes covering the contiguous United States, colored and shaded by population. For the 502 total S2 geohashes, 98 (19.5%) had an estimated population below 20,000 residents. Population among those geohashes ranged from 0 to 19,545 with a median population of 7,515. For the 710 total H3 geohashes, 169 (23.8%) had population less than 20,000. Population among those ranged from 0 to 19,930 and had a median population of 7,116. Geohashes that had less than 20,000 residents were merged with neighboring geohashes, outlined in red and depicted in the right side of Figure 2. Converting between the native geohash and the Safe Harbor geohash resulted in a reduction in the number of total geohashes from 502 to 424 for S2 and from 710 to 578 for H3. A data package with tabular files of S2 and H3 geohashes covering the contiguous United States and their corresponding Safe Harbor geohash is openly available online at https://geomarker-io.s3-us-east- 2.amazonaws.com/sh_geohash/datapackage.json.
DeGAUSS Images
In general, the approach used for high-resolution spatiotemporal exposure estimates in DeGAUSS requires an input CSV file, with each row representing a unique location and time interval by including columns specifying latitude, longitude, a start date, and a stop date. This input data specification was designed to coincide with standard address history collection forms implemented within ECHO and is flexible enough to represent longitudinal residential histories. Optionally, the file may include a column specifying an “index date” that can be used to return exposure estimate dates as relative (e.g., “8 days after the index date”) versus absolute (e.g., “January 2nd, 2015”). This optional addition prevents the necessity of using absolute dates in order to maintain temporal proximity to a significant date, e.g., date of birth, death, or other health outcomes. To assign exposures at a daily level based on input date ranges, the DeGAUSS container expands the start and end dates to a daily time series for each row in the input. Each unique location (i.e., latitude and longitude coordinate) is locally geohashed, and the Safe Harbor geohash (a coarsened version of the geohash) along with the calendar year are used to download a subset of exposure data from an online repository. These subsets of the exposure data are downloaded, and the exact location and date are used locally for precise exposure estimation. Estimates are returned as a CSV file that includes daily predictions for each input location and date range. An overview of this process is depicted in Figure 1.
We used this general approach for high resolution spatiotemporal exposure estimates in DeGAUSS with two different exposure assessment models. The model described in Di et al. [11] predicts daily PM2.5, O3, and NO2 exposures at a roughly 1 km x 1 km spatial resolution across the contiguous United States. We developed a pair of free and open source DeGAUSS images (https://degauss.org/schwartz_grid_lookup and https://degauss.org/schwartz) that use input latitude and longitude coordinates to add an S2 grid cell identifier, and then use start and stop dates to assign spatiotemporal predictions. These steps can also be used independently. For example, the Schwartz Grid Lookup container can be followed up with alternate methods for exposure extraction. Similarly, the model described in Brokamp [10] predicts daily PM2.5 exposures at a roughly 0.75 sq km spatial resolution across the United States using the H3 grid system. We created a single free and open source DeGAUSS image (https://degauss.org/pm) that assigns daily spatiotemporal predictions based on latitude and longitude coordinates as well as start and stop dates.
Validation of Using DeGAUSS Image for Grid Cell Identifier and Exposure Assessment
We considered the ArcGIS and R manual linkage methods to be the “gold standard” for assigning grid identifiers and exposures. Using 5,000 randomly sampled locations, the DeGAUSS “Schwartz Grid Lookup” image assigned the same grid identifier as 99.8% (n = 4,991) of the grid identifiers using the ArcGIS method and 99.8% (n = 4,988) of the grid identifiers using the R method.
We also used DeGAUSS to assign daily PM2.5, O3, and NO2 exposure estimates for 762 children from the CCAAPS cohort. Children had varying lengths of follow-up from 2001 to 2010, resulting in a total of 1,521 unique addresses and 1,691,985 days of assessed exposure. All daily exposure estimates for all three pollutants obtained using DeGAUSS were equal to those obtained using existing methods implemented within ECHO for exposure assessment. Existing exposure estimates are available through the Socioeconomic Data and Applications Center (SEDAC) across compressed GeoTIFF files totaling 212 GB in size. Using DeGAUSS to assign the same exposure estimates to the same CCAAPS cohort required the container to download 4.5 GB of exposure data. This equates to about 2% of the size of the total exposure data hosted on SEDAC used with existing methods.
Implementation of DeGAUSS for Exposure Assessment in ECHO
The DeGAUSS approach for exposure assessment based on the Di et al. [11] model was implemented within the ECHO Program cohorts to estimate daily ambient air pollution concentrations for specific addresses and dates. This decentralized approach allowed us to estimate exposures for 17,587 study participants across 53 cohorts, representing 1,590,931 person-months of follow up time. Critically, this allowed us to conduct exposure assessment using exact addresses and dates in instances where research participants did not or could not consent to sharing their PHI with investigators external to each cohort or with the ECHO Data Analysis Center (DAC). Furthermore, to maintain consistent and reproducible exposure assessment methods, the DeGAUSS approach was also used at the ECHO DAC for study participants’ addresses and dates that were able to be shared with the ECHO DAC.
DeGAUSS Usability Rated by Users
The results from our software usability survey suggest a generally positive experience when using DeGAUSS, though users generally had less ease of use if this was their first experience with command line computing. All respondents (n = 7) agreed that DeGAUSS allowed them to both obtain useful data more quickly than alternative geocoding services and obtain data that would be unavailable elsewhere. All respondents also indicated that they are likely to reuse DeGAUSS for a future project. The average usability rating (0 – 100) was 90.8 and the average likelihood that a user would recommend the software to a colleague was 0.895.
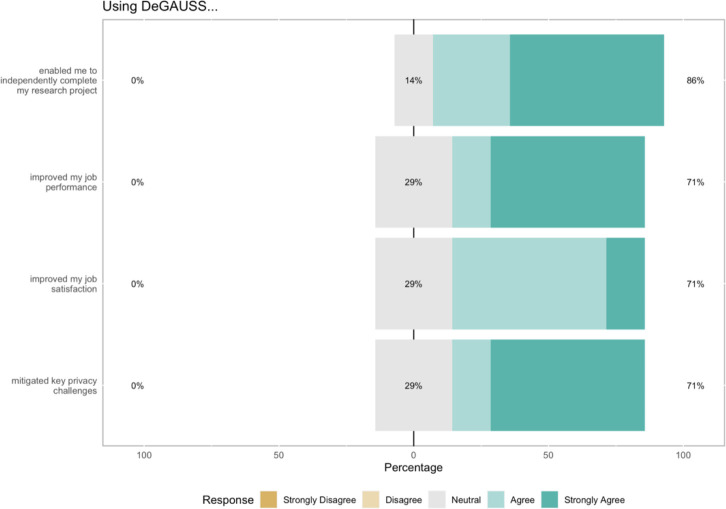
It should, however, be noted that among users without prior command line programming experience (n = 2), the usability was lower than among those with command line programming experience (84.5, 94.0). Further, these respondents indicated that they had some level of difficulty using Docker and/or DeGAUSS. To this point, users generally report back a positive experience when using DeGAUSS and that it may be a preferable alternative to more widely used geocoding and geomarker assessment techniques. Figure 3 shows four additional experience summary responses that indicate an overall positive user experience, with users being able to use DeGAUSS software to help complete their research projects while mitigating key privacy challenges, which was not possible with other approaches.
Figure 3.
System Usability Survey Results. In the four listed questions, respondents summarized their experience using DeGAUSS.
Discussion
Overall, we found that implementing DeGAUSS containers for high-resolution spatiotemporal data allowed for reproducible exposure assessment at scale while overcoming key privacy issues related to geolocation. The validation of our approach with existing methods and a successful implementation in a large, multi-site study suggest that this approach can be generally useful for high-resolution spatiotemporal exposure assessment in health studies, including where sensitive or private spatiotemporal information cannot or should not be shared. Our tool is (1) findable because each piece of software and data uses a persistent identifier and includes standardized, rich metadata; (2) accessible because it is stored in a publicly available repository; (3) interoperable because it uses the widely available CSV format for data and the Open Container Initiative standards for containers; and (4) reusable because it is free and open source and is supported by rich documentation.
The Safe Harbor Geohashes for S2 and H3 are novel tools that can be used to specify subsets of geographic data without disclosing any geographic identifier that might have less than 20,000 residents to satisfy the Safe Harbor provision of HIPAA. We used census population estimates, but other more high-resolution estimates such as the SEDAC (Socioeconomic Data and Applications Center) 1 sq. km grid, could also be used. Compared to the traditional approach of using a three-digit ZIP Code, geohash systems cover the entire globe, are well defined, and could be adapted as necessary based on local population estimates and different regulatory requirements that both change over time. Using a geohash system eliminates the need for specialized spatial software libraries usually required to conduct high-resolution spatiotemporal exposure assessment. In the future, methods and tools should leverage recent advances in privacy and encryption on text to facilitate truly private exposure assessment (e.g., homomorphic encryption).
The general approach of fragmenting data based on spatiotemporal boundaries to ensure no pseudo-identifiers need to be transferred can also be applied to other instances of high-resolution spatiotemporal data sets, including greenness, noise, land use, and climate-related data resources. This approach prevents the downloading and processing of unnecessary spatial and temporal fragments of exposure estimates, which decreases the time and resources needed by a scientist to complete exposure assessment. In contrast to geomasking or jittering methods (e.g., adding random noise to spatial coordinates, restricting to the first three digits of a ZIP Code, shifting all dates) our approach utilizes exact spatiotemporal information for exposure assessment to prevent losses of accuracy or precision common in geomasking methods for individual-level data.[19]
A shareable exposure dataset produced by DeGAUSS does not contain HIPAA Safe Harbor identifiers or pseudo-identifiers, but like any dataset, could possibly be linked to extant data to recover pseudo-identifiers, including a date or geographic extent of an exposure assessment grid/lattice cell. Averaging exposure assessments over at least one day (e.g., weekly exposures averages) would obfuscate the individual daily estimates such that they could not be used for re-identification. If daily estimates are required to be shared, a small amount of random noise can be added to each daily estimate. Although the introduction of random noise will introduce exposure misclassification, if the noise is centered around zero, it will be non-differential and will not cause bias in downstream estimates of health effects.
One limitation of any approach that relies on decentralized methods is that obscuring identifiers to create a shared dataset (e.g., the exact location and date) prevents the use of models that rely on spatial relationships or temporal effects that need to be linked to some period finer than a year (e.g., season, day of week). However, the decentralized approach here could be extended to fit such spatiotemporal statistical models and report shareable model parameter estimates for use in a meta-analysis. Our approach does require exposure estimation data be on some gridded or lattice system that coincides with a file structure, but alternatives for extracting spatiotemporal fragments of data in different file formats (e.g., Cloud Optimized GeoTIFFs, Apache Arrow Multi-File Datasets, Hierarchical Data Format) could be generalized to this approach.
Conclusion
Our approach and DeGAUSS software implementation was found to be highly usable. Because it was designed to be used by both clinical data coordinators and informatics specialists, this tool can be used both in a decentralized and centralized manner within a multi-site study where each site has varying levels of permission to share spatiotemporal PHI. In conclusion, the DeGAUSS approach transforms high-resolution spatiotemporal exposure assessment models into FAIR [4], computable exposures that can be used for private exposure assessment at scale without necessitating the sharing of identifiable information.
Acknowledgements
We thank Bill Wheaton and Jamie Cajka for assisting in the implementation of DeGAUSS software at the ECHO Data Analysis Center. This work was supported by NIH U2COD023375, NIH R01LM013222, UG3 OD023282, and P30-ES000002.
Figures & Table
References
- 1.Vineis P. A self-fulfilling prophecy: Are we underestimating the role of the environment in gene– environment interaction research? International Journal of Epidemiology. 2004;33(5):945–6. doi: 10.1093/ije/dyh277. [DOI] [PubMed] [Google Scholar]
- 2.Thessen AE, Grondin CJ, Kulkarni RD, Brander S, Truong L, Vasilevsky NA, et al. Community approaches for integrating environmental exposures into human models of disease. Environmental Health Perspectives. 2020;128(12):125002. doi: 10.1289/EHP7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin Sanchez F, Gray K, Bellazzi R, Lopez-Campos G. Exposome informatics: Considerations for the design of future biomedical research information systems. Journal of the American Medical Informatics Association. 2013;21(3):386–90. doi: 10.1136/amiajnl-2013-001772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, et al. The fair guiding principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brokamp C, Wolfe C, Lingren T, Harley J, Ryan P. Decentralized and reproducible geocoding and characterization of community and environmental exposures for multisite studies. Journal of the American Medical Informatics Association. 2017;25(3):309–14. doi: 10.1093/jamia/ocx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brokamp C. Degauss: Decentralized geomarker assessment for multi-site studies. Journal of Open Source Software. 2018;3(30):812. [Google Scholar]
- 7.Ryan PH, Brokamp C, Blossom J, Lothrop N, Miller RL, Beamer PI, et al. A distributed geospatial approach to describe community characteristics for multisite studies. Journal of Clinical and Translational Science. 2021;5(1):e86. doi: 10.1017/cts.2021.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Q, Wang Y, Zanobetti A, Wang Y, Koutrakis P, Choirat C, et al. Air pollution and mortality in the medicare population. New England Journal of Medicine. 2017;376(26):2513–22. doi: 10.1056/NEJMoa1702747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brokamp C, Strawn JR, Beck AF, Ryan P. Pediatric psychiatric emergency department utilization and fine particulate matter: A case-crossover study. Environmental Health Perspectives. 2019;127(9):97006-. doi: 10.1289/EHP4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brokamp C. A high resolution spatiotemporal fine particulate matter exposure assessment model for the contiguous united states. Environmental Advances. 2022;7:100155. [Google Scholar]
- 11.Di Q, Amini H, Shi L, Kloog I, Silvern R, Kelly J, et al. An ensemble-based model of PM(2.5) concentration across the contiguous United States with high spatiotemporal resolution. Environment International. 2019;130:104909. doi: 10.1016/j.envint.2019.104909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X, Belle JH, Meng X, Wildani A, Waller LA, Strickland MJ, et al. Estimating PM(2.5) concentrations in the conterminous united states using the random forest approach. Environmental Science & Technology. 2017;51(12):6936–44. doi: 10.1021/acs.est.7b01210. [DOI] [PubMed] [Google Scholar]
- 13.Di Q, Wei Y, Shtein A, Hultquist C, Xing X, Amini H, et al. Daily and annual pm2.5 concentrations for the contiguous United States, 1-km grids, v1 (2000 - 2016) Palisades, NY: NASA Socioeconomic Data and Applications Center (SEDAC); 2021. [Google Scholar]
- 14.VanWey LK, Rindfuss RR, Gutmann MP, Entwisle B, Balk DL. Confidentiality and spatially explicit data: Concerns and challenges. Proceedings of the National Academy of Sciences. 2005;102(43):15337–42. doi: 10.1073/pnas.0507804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillman MW, Blaisdell CJ. Environmental influences on child health outcomes, a research program of the national institutes of health. Current Opinion in Pediatrics. 2018;30(2):260–2. doi: 10.1097/MOP.0000000000000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boettiger C. An introduction to docker for reproducible research. SIGOPS Operating Systems Review. 2015;49(1):71–9. [Google Scholar]
- 17.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (redcap)—a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics. 2009;42(2):377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooke J. Sus: A quick and dirty usability scale. Usability Evaluation in Industry. 1995:189. [Google Scholar]
- 19.Zandbergen PA. Ensuring confidentiality of geocoded health data: Assessing geographic masking strategies for individual-level data. Advances in Medicine. 2014;2014:567049. doi: 10.1155/2014/567049. [DOI] [PMC free article] [PubMed] [Google Scholar]