Abstract
Recently, hospitals and healthcare providers have made efforts to reduce surgical site infections as they are a major cause of surgical complications, a prominent reason for hospital readmission, and associated with significantly increased healthcare costs. Traditional surveillance methods for SSI rely on manual chart review, which can be laborious and costly. To assist the chart review process, we developed a long short-term memory (LSTM) model using structured electronic health record data to identify SSI. The top LSTM model resulted in an average precision (AP) of 0.570 [95% CI 0.567, 0.573] and area under the receiver operating characteristic curve (AUROC) of 0.905 [95% CI 0.904, 0.906] compared to the top traditional machine learning model, a random forest, which achieved 0.552 [95% CI 0.549, 0.555] AP and 0.899 [95% CI 0.898, 0.900] AUROC. Our LSTM model represents a step toward automated surveillance of SSIs, a critical component of quality improvement mechanisms.
Introduction
Surgical site infections (SSI) are a major cause of complications after surgical procedures, occurring in 3-5% of patients1,2. In addition to the patient morbidity, the development of SSI is one of the most common underlying causes of hospital readmission after surgery and is associated with significantly increased costs and longer hospital stays3. Overall, SSIs contribute an additional $1.6 billion to hospital expenses4. Given the prevalence and contribution to health care costs, SSIs are considered public reportable events by the Centers for Medicare and Medicaid Services and the Centers for Disease Control. The goal of publicly reporting these complications is to drive healthcare systems to develop quality improvement (QI) interventions to target SSI reduction. To accomplish public reporting and QI interventions hospitals rely on quality surveillance programs to identify SSI from the medical records. Such programs include the American College of Surgeons (ACS) National Surgical Quality Improvement Program (NSQIP) and the Centers for Disease Control National Healthcare Safety Network (NHSN) 5,6. Surveillance of SSIs allow hospital and healthcare systems to identify targets for quality improvements, such as perioperative antibiotic prophylaxis, develop interventions to address quality gaps, and measure performance after intervention implementation. Several studies have demonstrated that hospitals who participate in quality surveillance programs can improve the rate of SSI development7,8. Surveillance and quality monitoring are crucial to reducing and preventing SSIs.
The diagnosis of SSI from the medical record is complex, requiring the integration of information located in both structured data fields (laboratory, vital signs, medications, etc.) and unstructured data (clinical notes, microbiology reports, radiology reports, etc.). Given this complexity, quality surveillance programs rely on manual chart review processes to identify SSIs from the medical record. For example, as part of the NSQIP program, surgical clinical reviewers (SCRs) are trained to manually review the electronic health records (EHR) of selected operative events identifying any complications, including SSI, occurring within 30 days of the surgical procedure5. An ACS surgeon then re-reviews the identified complications to ensure NSQIP definitions are met. Disagreements are settled when a consensus is reached. Therefore, the manual chart review process is costly and labor intensive. To address the effort and cost required, most surveillance programs rely on a sampling methodology to decrease the overall chart review burden. This sampling approach limits the ability of healthcare systems to identify targets for quality improvement interventions as it decreases the number of patients and procedures surveyed. New approaches are therefore needed to improve the identification of SSI from the medical record without relying on manual chart review.
Automated or semi-automated methods have the potential to greatly reduce the burden of manual chart review2,9,10. Such methods generally leverage machine leaning models developed to detect SSI from either the EHR, NSQIP data, or a combination of the two. Skube et al. presented a semi-automated method that included a logistic regression model developed using natural language processing (NLP) and EHR data for the identification of possible SSI2. A reviewer then manually determined the final SSI status for these possible cases. They found this semi-automated process decreased manual chart abstraction by more than 90%. Hu et al used multivariate logistic regression and feature engineering to develop a model to classify SSI9. Their data included EHR data as well as some feature engineering to represent the change in laboratory results and vital signs. The model for detecting all SSIs resulted in an area under the receiver operating characteristic curve (AUROC) of 0.896. Colborn et al. developed a generalized linear model to identify SSI, incorporating structured EHR features as well as NSQIP variables10. Their model resulted in 0.89 AUROC. Dos Santos et al. built a model to detect several healthcare-associated infections, including SSI, from vital signs and laboratory results11. When classifying SSI, their model resulted in 0.857 AUROC. Most recently, the top model for SSI detection from Kiser et al. developed using structured EHR data resulted in 0.906 AUROC12. These methods have the potential to greatly reduce the cost of the NSQIP program, lowering a barrier for hospitals to participate and work to improve postoperative complication rates13.
Deep learning is a subcategory of machine learning, capable of learning complex, non-linear relationships in the data14. Several studies have successfully developed deep learning models to identify or phenotype other conditions from EHR data. These efforts include studies that incorporate NLP to extract data from unstructured clinical notes, often in conjunction with structured data when developing the model15-18. Rashidian et al. developed a deep learning model to identify diabetes from the EHR using structured data, resulting in an AUROC of 0.96 and average precision (AP) of 0.9119. The deep learning model from Guo et al. used structured EHR and claims data and incorporated embedding vectors to reduce the dimensionality of the included features20. The model identified high-risk patients for palliative care, outperforming other traditional machine learning models. These previous studies demonstrate the usefulness of deep learning in the identification of clinical conditions from EHR data.
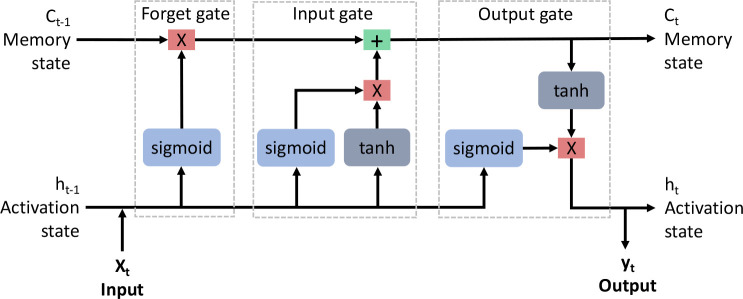
RNN models are a type of deep learning algorithm typically used in sequence prediction, including forecasting in time-series datasets and text prediction in NLP21-23. However, RNNs have difficulty learning information from long sequences as gradients, a derivative of the loss function used to update the model parameters during backpropagation, tend to explode or vanish 24,25. In those cases, the gradients either become exponentially large or vanishingly small. Either case can cause the training to stall out. A long short-term memory (LSTM) network is a variant of an RNN, which includes a forget gate to combat the problem of exploding and vanishing gradients25. An LSTM attempts to remember the important information and forget the uninformative. LSTM networks make use of tanh and sigmoid activation functions to learn sequential information through time. Figure 1 illustrates the basic structure of an LSTM cell.
Figure 1:
Structure of a long short-term memory cell, composed of three gates, the forget gate, input gate, and output gate. In the forget gate, the input (Xt) combines with the previous activation state (ht-1) and passes through a sigmoid function. This result gets passed to a multiplication function with the previous memory state (ct-1). In the input gate, the previous activation state (ht-1) and input (Xt) pass through sigmoid and tanh functions, which combine via a multiplication function. This result gets added to the result from the forget gate and becomes the new memory state (ct). Finally in the output gate, the previous activation state (ht-1) and input (Xt) pass through a sigmoid function. This result is multiplied with the result from the input gate after it passes through a tanh function. These result in the new activation state (ht) as well as the output (yt).
In this study, we developed a deep learning model, an LSTM, to identify SSIs from the EHR. We built our model using structured EHR data collected during routine post-surgical assessments. We chose an LSTM because of its ability to model temporal patterns. Notably, no other studies have developed an LSTM model for SSI identification. We hypothesized that an LSTM model will be able to accurately identify SSI from structured EHR data compared to traditional machine learning models.
Methods
Study Design
We conducted a retrospective study using EHR data obtained from the University of Utah Health system. The institutional review board approved the study, granting a waiver of informed consent.
Participants and Outcomes
We included operative events occurring from January 2016 to June 2021 at the University of Utah Health if they were chosen for manual chart review as part of the local ACS NSQIP program. Shiloach et al. previously described the ACS NSQIP program and reported the overall interrater agreement to be greater than 98%26. All patients who underwent NSQIP SCR review were included in the present study even if the complete documentation was not present for the entire 30-day postoperative period. We used the NSQIP SSI label obtained from manual chart review for each operative event as the gold standard for the study and labeled these as binary values for classification.
Features
The University of Utah has maintained an EHR serviced by Epic since September 1, 2015. Structured EHR data retrieved for each operative event included vital signs, laboratory test results, diagnosis codes, and medications. Clinical notes were not included in this study. We created 3 different datasets, using different levels of temporal aggregation: (1) complete aggregation, where all features from the 30 days were aggregated, (2) daily aggregation, where features were aggregated by day, and (3) hourly aggregation, where features were aggregated by hour. To aggregate, or combine, data into different temporal bins, we found the minimum, maximum, mean, and median for continuous variables, such as vital signs, and the count for categorical variables, such as medications. Aggregating the data shortened the sequences fed into the LSTM, addressing the problem of exploding or vanishing gradients, as the LSTM was able to learn more effectively over shorter sequences. Sequential input of the data allowed the model to learn trends that occurred over time, providing an additional dimension for learning. This is advantageous with healthcare data as we know trends can be important in a patient’s trajectory. While this use case is only looking at classification, in the future a sequential method would be vital to be able to detect SSI early. An analysis of variance (ANOVA) of the completely aggregated dataset revealed the top 50 variables most correlated with SSI. We used those 50 variables as features when developing the models.
Model Development and Testing
For each dataset, we split data into training (70%), validation (10%), and testing (20%) sets by operative events. We imputed missing data with 0 for nominal features and medians – calculated from the training data – for continuous features and normalized the data to have a range from 0 to 127.
With the training and validation sets from the daily and hourly aggregated datasets, we developed LSTM models to classify whether SSI occurred within 30 days after surgery. We employed batch training when developing the LSTM models, padding the time steps to ensure all operative events had the same length. Early stopping, where training is stopped if validation performance has not improved after 10 epochs, prevented overfitting. The final LSTM model architectures resulted from an evaluation of various architectures, which included varying the number of LSTM layers, applying a dropout layer, and training with gradient clipping28,29. We chose the final model architectures that resulted in the highest AP.
To address the class imbalance, we used class weights30. Hyperparameters, tuned for each model using 10-fold cross validation on the training dataset, included batch size and learning rate. Given the severe class imbalance in our data, we chose to optimize the AP, or area under the precision recall curve, as this was a more appropriate metric compared with AUROC31. After finding the optimal hyperparameters, we trained the models using the training and validation datasets. We used the unseen test set to calculate the performance of each model, including the following metrics: AP, sensitivity, specificity, and AUROC.
For comparison, we developed several different traditional machine learning models, including a support vector machine (SVM), random forest, and XGboost, with the completely aggregated dataset32-35. We also developed a feedforward neural network (NN). We used Python (version 3.10.4) and the packages PyTorch (version 1.11.0), scikit-learn (version 1.0.2), and xgboost (version 1.5.0) to develop the models35-37.
Model Architectures
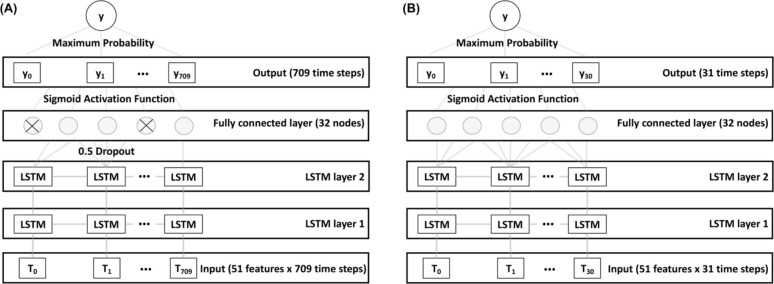
After model architecture selection, we built the hourly LSTM model using 2 stacked LSTM layers and a 0.5 dropout layer before the fully connected linear layer. A sigmoid function processed the output to return a predicted probability between 0 and 1 for each time step. We used the maximum probability as the final predicted probability. We used gradient clipping when updating the gradients to prevent exploding gradients. We built the daily LSTM using 2 stacked LSTM layers and a fully connected linear layer. A sigmoid function processed the output to return a predicted probability between 0 and 1 for each time step with the maximum probability considered the final predicted probability. We did not use gradient clipping to train this model. The hourly and daily LSTM model architectures are represented in Figure 2.
Figure 2.
(A) Architecture of the hourly LSTM model. We trained this model using the dataset aggregated by hour. The top architecture included 2 LSTM layers with a 0.5 dropout layer between the final LSTM layer and linear fully connected layer. (B) Architecture of the daily LSTM model. We trained this model using the dataset aggregated by day. The top architecture included 2 LSTM layers and a linear fully connected layer. LSTM: long short-term memory.
We built the NN with an input layer, 2 hidden layers, and an output layer. We used rectified linear unit (ReLU) activation functions prior to the hidden layers and a sigmoid function to process the final output. The NN architecture is represented in Figure 3.
Figure 3.
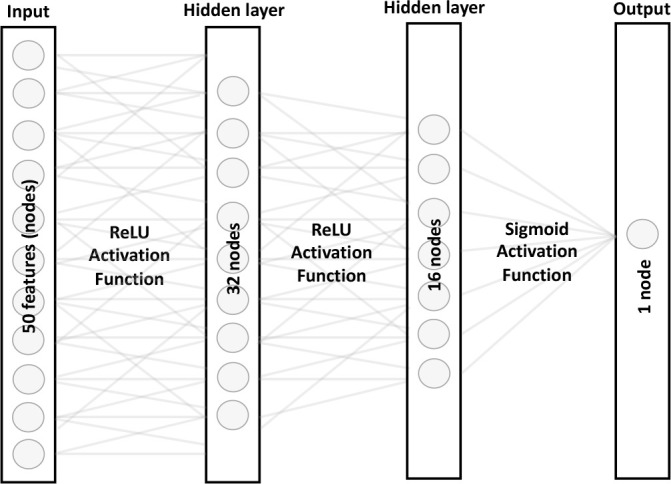
Architecture of the feedforward neural network model. We trained this model using the completely aggregated dataset. The model included an input layer, 2 hidden layers, and an output layer. We used the rectified linear unit (ReLU) function for the hidden layers.
Sensitivity Analysis
As a sensitivity analysis, we investigated the daily predicted probability of SSI between the two cohorts, i.e., SSI and no complication. We looked at the average daily predicted probability and calculated the average time from prediction of SSI to actual diagnosis.
Statistical Analysis
To assess differences in patient demographics, we used the chi-square test of independence for categorical demographics and a 2-sample t-test for continuous demographics. We used bootstrapping to get a distribution of 1000 iterations and find the mean and 95% confidence intervals of the performance metrics. We used the Python package SciPy (version 1.7.3) to perform all statistical analyses38.
Results
Cohort Description
We retrieved data from a total of 9,185 operative events. The resulting prevalence of SSI was 4.7% (N=434). Table 1 describes the demographics of the study patients by cohort. Patients who developed SSI were significantly older (SSI: 56.2 years, No Complication: 52.4 years, p value < 0.001) and inpatient (SSI: 74.9%, No Complication: 39.6%, p value < 0.001) with a significantly greater percentage of gastrointestinal procedures (SSI: 80.9%, No Complication: 73.5%, p value < 0.001) when compared to patients who did not develop complications. Significant comorbidities of patients who developed SSI included disseminated cancer (SSI: 6.5%, No Complication: 2.9%, p value < 0.001), steroid or immunosuppressant use for a chronic condition (SSI: 11.5%, No Complication: 6.3%, p value < 0.001), and > 10% loss of body weight in the 6 months prior to surgery (SSI: 4.6%, No Complication: 1.5%, p value < 0.001).
Table 1.
Demographics of study patients. SSI: surgical site infection.
| Patient Characteristics | SSI (N = 434) | No Complication (N = 8751) | P value |
|---|---|---|---|
| Age at time of surgery (years), mean (SD) | 56.2 (16.0) | 52.4 (16.7) | < 0.001 |
| Gender, male, n (%) | 220 (50.7) | 4247 (48.5) | 0.41 |
| Race, n (%) American Indian or Alaska Native Asian Black or African American Native Hawaiian or Pacific Islander White Unknown or not reported |
7 (1.6) 3 (0.7) 5 (1.2) 8 (1.8) 398 (91.7) 13 (3.0) |
115 (1.3) 133 (1.5) 111 (1.3) 56 (0.6) 8002 (91.4) 334 (3.8) |
0.75 0.23 1 0.008 0.92 0.46 |
| Ethnicity, Hispanic, n (%) | 36 (8.3) | 888 (10.1) | 0.24 |
| Inpatient or outpatient status, inpatient, n (%) | 321 (74.9) | 3467 (39.6) | < 0.001 |
| Current Procedural Terminology code, n (%) 0 – 29999 (skin / soft tissue) 30000 – 39999 (cardiovascular) 40000 – 49999 (gastrointestinal) 50000 – 59999 (genitourinary) 60000 – 69999 (nervous system) |
38 (8.8) 43 (9.9) 351 (80.9) 2 (0.5) 0 (0) |
1403 (16.0) 850 (9.7) 6434 (73.5) 48 (0.5) 16 (0.2) |
< 0.001 0.96 < 0.001 1 0.76 |
| Comorbidities, n (%) Diabetes mellitus Current smoker within 1 year Dyspnea Dependent functional health status Ventilator dependent History of severe chronic obstructive pulmonary disease Ascites within 30 days prior to surgery Congestive heart failure within 30 days prior to surgery Hypertension requiring medication Acute renal failure Currently requiring or on dialysis |
79 (18.2) 69 (15.9) 30 (6.9) 7 (1.6) 3 (0.7) 15 (3.5) 0 (0) 2 (0.5) 162 (37.3) 0 (0) 5 (1.2) |
1130 (12.9) 1089 (12.4) 453 (5.2) 74 (0.8) 13 (0.1) 186 (2.1) 6 (0.07) 20 (0.2) 2745 (31.4) 10 (0.1) 123 (1.4) |
0.002 0.04 0.14 0.16 0.04 0.09 1 0.64 0.01 1 0.82 |
| Disseminated cancer | 28 (6.5) | 252 (2.9) | < 0.001 |
| Open wound with or without infection | 28 (6.5) | 344 (3.9) | 0.01 |
| Steroid / immunosuppressant use for chronic condition | 50 (11.5) | 553 (6.3) | < 0.001 |
| >10% loss of body weight in the 6 months prior to surgery | 20 (4.6) | 128 (1.5) | < 0.001 |
| Bleeding disorder | 5 (1.2) | 152 (1.7) | 0.47 |
Model Training
The ANOVA identified the top 50 features associated with SSI from the structured EHR data. The features are described in Table 2. In the LSTM models, we included an additional feature to represent the time since the surgery. The final hyperparameters for all models are listed in Table 3.
Table 2.
Description of features included in the model. We selected these features using an analysis of variance (ANOVA). CCS: Clinical Classifications Software; CPT: Current Procedural Terminology; LOINC: Logical Observation Identifiers Names and Codes.
| Category | Features |
|---|---|
| Diagnosis codes | CCS 238 – Complications of surgical procedures or medical care; CCS 88 – Glaucoma; CCS 148 – Peritonitis and intestinal abscess; CCS 197 – Skin and subcutaneous tissue infections; CCS 223-Birth trauma |
| Laboratory test results | LOINC 26515-7 – Maximum platelets in blood; LOINC 2862-1 – Minimum, mean, and median albumin in serum or plasma; LOINC 20570-8 – Minimum hematocrit of blood; LOINC 718-7 – Minimum hemoglobin in blood; LOINC 26453-1 – Minimum erythrocytes in blood |
| Medications | Diatrizoate meglumine and sodium; Piperacillin sodium-tazobactam; Iopamidol; Sodium chloride; Magnesium sulfate; Acetaminophen; Dextrose; Parenteral electrolytes; Calcium gluconate; Multiple vitamin; Amino acid infusion; Potassium chloride; Sodium chloride; Trace minerals Cr-Cu-Mn-Se-Zn; Ondansetron HCI; Irrigation solutions physiological; Hydromorphone HCI; Calcium chloride |
| Microbiology results | Negative blood culture; Gram positive cocci wound culture; Negative anaerobic culture; Gram negative rods wound culture; Staphylococcus species wound culture; Negative wound culture |
| Procedure codes | CPT 49406 – Image-guided fluid collection drainage by catheter; CPT 99232 – Subsequent hospital care; CPT 74177 – Abdomen and pelvis CT with contrast; CPT 99152 – Initial moderate sedation services; CPT 99024 – Postoperative follow-up visit; CPT 93010 – Routine electrocardiogram with 12 leads; CPT 99233 – Subsequent hospital care; CPT 97530 – Therapeutic activities; CPT 99153 – Additional moderate sedation services; CPT 99291 – Critical care evaluation and management |
| Visits | Inpatient admission; Inpatient discharge |
| Vital signs | LOINC 8310-5 – Maximum body temperature |
Table 3.
Final hyperparameters for the models, determined from 10-fold cross-validation and used to tune the final models. LSTM: long short-term memory.
| Model | Hyperparameters |
|---|---|
| Hourly LSTM | batch size: 512; learning rate: 0.005; criterion: binary cross entropy; optimizer: adam |
| Daily LSTM | batch size: 256; learning rate: 0.005; criterion: binary cross entropy; optimizer: adam |
| Neural Network | batch size: 256; learning rate: 0.01; criterion: binary cross entropy; optimizer: adam |
| Support vector machine | kernel: linear, C: 1, gamma: scale |
| Random forest | number of estimators: 200; max depth: 10; criterion: entropy |
| XGBoost | number of estimators: 200; max depth: 200; learning rate: 0.1 |
Testing
We tested the models on the unseen test data and built a bootstrap distribution to find the mean and 95% confidence intervals. The daily LSTM resulted in the highest AP (0.570 [95% CI 0.567, 0.573]), sensitivity (0.963 [95% CI 0.961, 0.964]), and AUROC (0.905 [95% CI 0.904, 0.906]). The random forest resulted in the highest specificity (0.995 [95% CI 0.995, 0.996]). Table 4 records the results for the LSTM models, NN, and top traditional machine learning model (RF).
Table 4.
Performance metrics for the final models. The means with 95% confidence intervals in parentheses are represented. RF was the top performing traditional machine learning model. LSTM: long short-term memory; AP: average precision; AUROC: area under the receiver operating characteristic curve.
| Metric | Hourly LSTM | Daily LSTM | Neural Network | Random Forest |
|---|---|---|---|---|
| AP | 0.289 (0.287, 0.292) | 0.570 (0.567, 0.573) | 0.526 (0.523, 0.529) | 0.552 (0.549, 0.555) |
| Sensitivity | 0.962 (0.960, 0.963) | 0.963 (0.961, 0.964) | 0.768 (0.765, 0.771) | 0.334 (0.331, 0.337) |
| Specificity | 0.431 (0.430, 0.431) | 0.238 (0.237, 0.238) | 0.893 (0.892, 0.893) | 0.995 (0.995, 0.996) |
| AUROC | 0.854 (0.852, 0.855) | 0.905 (0.904, 0.906) | 0.901 (0.899, 0.902) | 0.899 (0.898, 0.900) |
Sensitivity Analysis
We reviewed the daily predicted probability of SSI. Figure 4 displays the average daily predicted probability of SSI for operative events correctly classified by the model, i.e. true positives and true negatives. The probability of SSI increased over time for operative events where an SSI occurred yet decreased for operative events with no complications. This result indicated the model performed as expected when predicting probabilities over time.
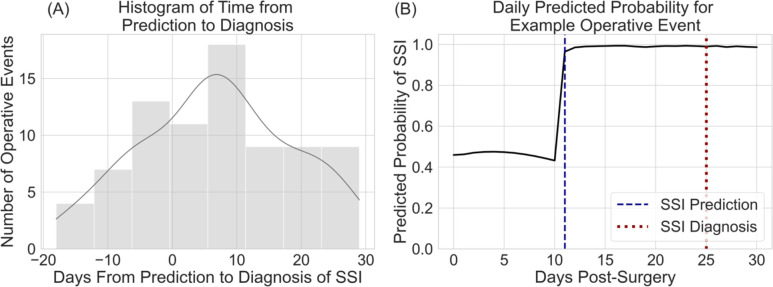
Figure 4.
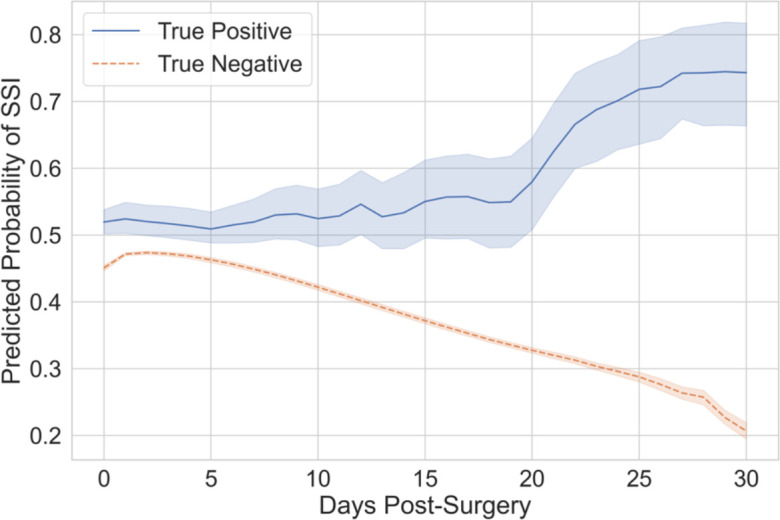
Average predicted probabilities per day for operative events correctly classified by the model. Mean with shaded 95% confidence intervals. The solid blue line represents the true positive cases while the dashed orange line represents the true negative controls.
The average number of days from prediction to diagnosis of SSI was 7 (SD: 12), indicating the model was able to predict the development of SSI an average of 7 days before the actual diagnosis. Figure 5 displays the histogram of days from prediction to diagnosis and illustrates an example of daily predicted probabilities for one operative event.
Figure 5.
(A) displays the histogram of days from prediction to diagnosis of SSI. A positive number of days indicates the model predicted SSI prior to diagnosis. (B) displays the daily predicted probabilities for an operative event in which SSI developed. The dashed blue line represents the day the model predicted SSI. The dotted red line represents the day SSI was diagnosed.
Discussion
We built an LSTM model to classify SSI occurring in operative events and compared this to several machine learning models using various architectures and algorithms. We found an LSTM model developed with data aggregated by day performed better than an LSTM model developed with more granular time steps. When compared to traditional machine learning techniques, including a NN, the LSTM model performed better in AP, sensitivity, and AUROC. Finally, the sensitivity analysis highlighted the ability of the LSTM model to predict the development of SSI before diagnosis. Our daily LSTM model predicted the development of SSI an average of 7 days in advance.
The daily LSTM model developed with data aggregated at a larger time step resulted in better performance compared to the more granular hourly LSTM model, including greater AP and AUROC. It is difficult for LSTM models to learn information over a long sequence29. The hourly data resulted in 709 time steps per operative event compared to only 31 time steps in the daily model. This could be a reason for the worse performance, even after applying gradient clipping and dropout. There are other techniques, like truncated back propagation through time, where the loss is propagated through a chosen number of time steps rather than the whole sequence, that could possibly improve the performance of the hourly LSTM39,40.
While some recent studies have used machine learning methods for the detection of SSI, none have developed an LSTM. Previous models developed included a multivariate logistic regression model, a generalized linear model, a multinomial naïve bayes model, and a support vector machine9-12. We optimized for AP as it has been suggested to do when training a model with a severely imbalanced dataset; however, other studies have not reported AP31. To compare our model performance with other models, we used AUROC. The AUROC of previous studies ranged from 0.857 to 0.906. Our unique LSTM architecture resulted in higher AUROC (0.905 [95% CI 0.904, 0.906]) compared to most of these previous models, achieving comparable AUROC but higher sensitivity (0.963 [95% CI 0.961, 0.964]) when compared to a previous model (AUROC: 0.906 [95% CI 0.904, 0.908]; Sensitivity: 0.804 [95% CI 0.802, 0.807]) developed by this group12.
The sensitivity analysis demonstrated an important advantage of LSTM models. The model was able to provide predicted probabilities at each time step, unlike the random forest where only one probability is given. As shown in Figure 5A, the daily LSTM predicted on average the development of SSI 7 days prior to the diagnosis. In the average trend chart (Figure 4), predicted probabilities for true positive operative events were consistently higher than true negatives as expected. Our future work includes developing an LSTM model to predict SSI prior to diagnosis.
Automated detection of SSI has the potential to improve surgical outcomes. Hospitals require this classification to implement quality improvement programs, assess their effectiveness, and identify shortcomings. While it is currently a manual process, the implementation of machine learning models to assist in identification and extraction of data could cut overhead costs of both time and money. Our daily LSTM model with high AP and AUROC is a step towards this goal of automation.
We acknowledge our study has limitations. Previous studies have successfully incorporated clinical notes when classifying SSI, using NLP techniques1,41. Our study currently only uses structured EHR data; however, we plan to incorporate clinical notes into future models. External validation is important when understanding the performance of a model. We did not have external data to validate our model, instead using an unseen test set to simulate this. Future work should include external validation of the model.
Conclusion
We developed two LSTM models for SSI surveillance with different levels of temporal aggregation. The LSTM model aggregated by day performed the best with an AP of 0.570 [95% CI 0.567, 0.573] and AUROC of 0.905 [95% CI 0.904, 0.906]. When compared with the top traditional machine learning model (random forest), the top LSTM model has improved AP and AUROC. LSTM models have the capability to produce predicted probabilities at each time step, an advantage when looking beyond classification to prediction in the future. Our LSTM model represents a step toward automated surveillance of SSIs, which is a critical component of quality improvement mechanisms.
Acknowledgements
We want to acknowledge the training grant (T15LM007124) from the National Library of Medicine (ACK) and grant (1K08HS025776) from the Agency for Healthcare Research and Quality (BB) that supported this work. The National Institutes of Health and Agency for Healthcare Research and Quality had no role in the design or conduct of the study. The support and resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged. The computational resources used were partially funded by the NIH Shared Instrumentation Grant 1S10OD021644-01A1.
Code Availability
The code for the models and additional results can be found in the public GitHub repository amberkiser/LSTM-for-SSI. The data cannot be shared publicly as it contains protected health information.
Figures & Table
References
- 1.Bucher BT, Shi J, Ferraro JP, et al. Portable automated surveillance of surgical site infections using natural language processing: development and validation. Ann Surg. 2020;272(4):629–36. doi: 10.1097/SLA.0000000000004133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skube SJ, Hu Z, Simon GJ, et al. Accelerating surgical site infection abstraction with a semi-automated machine-learningapproach. Ann Surg. 2022;276(1):180–5. doi: 10.1097/SLA.0000000000004354. [DOI] [PubMed] [Google Scholar]
- 3.Merkow RP, Ju MH, Chung JW, et al. Underlying reasons associated with hospital readmission following surgery in the United States. Jama. 2015;313(5):483–95. doi: 10.1001/jama.2014.18614. [DOI] [PubMed] [Google Scholar]
- 4.de Lissovoy G, Fraeman K, Hutchins V, Murphy D, Song D, Vaughn BB. Surgical site infection: incidence and impact on hospital utilization and treatment costs. Am J Infect Control. 2009;37(5):387–97. doi: 10.1016/j.ajic.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Ko CY, Hall BL, Hart AJ, Cohen ME, Hoyt DB. The American College of Surgeons National Surgical Quality Improvement Program: achieving better and safer surgery. Jt Comm J Qual Patient Saf. 2015;41(5):199–204. doi: 10.1016/s1553-7250(15)41026-8. [DOI] [PubMed] [Google Scholar]
- 6.American College of Surgeons. Frequently asked questions. 2022. [Available from: https://www.facs.org/quality-programs/data-and-registries/acs-nsqip/faq/
- 7.Rosemurgy A, Whitaker J, Luberice K, Rodriguez C, Downs D, Ross S. A cost-benefit analysis of reducing surgical site infections. Am Surg. 2018;84(2):254–61. [PubMed] [Google Scholar]
- 8.Fuglestad MA, Tracey EL, Leinicke JA. Evidence-based prevention of surgical site infection. Surg Clin North Am. 2021;101(6):951–66. doi: 10.1016/j.suc.2021.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, Simon GJ, Arsoniadis EG, Wang Y, Kwaan MR, Melton GB. Automated detection of postoperative surgical site infections using supervised methods with electronic health record data. Stud Health Technol Inform. 2015;216:706–10. [PMC free article] [PubMed] [Google Scholar]
- 10.Colborn KL, Bronsert M, Amioka E, Hammermeister K, Henderson WG, Meguid R. Identification of surgical site infections using electronic health record data. Am J Infect Control. 2018;46(11):1230–5. doi: 10.1016/j.ajic.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 11.dos Santos RP, Silva D, Menezes A, et al. Automated healthcare-associated infection surveillance using an artificial intelligence algorithm. Infection Prevention in Practice. 2021;3(3):100167. doi: 10.1016/j.infpip.2021.100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiser AC, Eilbeck K, Ferraro JP, Skarda DE, Samore MH, Bucher B. Standard vocabularies to improve machine learning model transferability with electronic health record data: retrospective cohort study using health care– associated infection. JMIR medical informatics. 2022;10(8):e39057. doi: 10.2196/39057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman L, Rangel S, Tsao K. Pediatric surgeon perceptions of participation in external patient safety programs: impact on patient safety. Pediatr Qual Saf. 2018;3(6):e124. doi: 10.1097/pq9.0000000000000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteva A, Robicquet A, Ramsundar B, et al. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24–9. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 15.Vincent M, Douillet M, Lerner I, Neuraz A, Burgun A, Garcelon N. Using deep learning to improve phenotyping from clinical reports. Stud Health Technol Inform. 2022;290:282–6. doi: 10.3233/SHTI220079. [DOI] [PubMed] [Google Scholar]
- 16.Moldwin A, Demner-Fushman D, Goodwin TR. Empirical findings on the role of structured data, unstructured data, and their combination for automatic clinical phenotyping. AMIA Jt Summits Transl Sci Proc. 2021;2021:445–54. [PMC free article] [PubMed] [Google Scholar]
- 17.Ni Y, Bachtel A, Nause K, Beal S. Automated detection of substance use information from electronic health records for a pediatric population. J Am Med Inform Assoc. 2021;28(10):2116–27. doi: 10.1093/jamia/ocab116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stemerman R, Arguello J, Brice J, Krishnamurthy A, Houston M, Kitzmiller R. Identification of social determinants of health using multi-label classification of electronic health record clinical notes. JAMIA Open. 2021;4(3) doi: 10.1093/jamiaopen/ooaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rashidian S, Abell-Hart K, Hajagos J, et al. Detecting miscoded diabetes diagnosis codes in electronic health records for quality improvement: temporal deep learning approach. JMIR medical informatics. 2020;8(12):e22649. doi: 10.2196/22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo A, Foraker R, White P, Chivers C, Courtright K, Moore N. Using electronic health records and claims data to identify high-risk patients likely to benefit from palliative care. Am J Manag Care. 2021;27(1):e7–e15. doi: 10.37765/ajmc.2021.88578. [DOI] [PubMed] [Google Scholar]
- 21.Santhanam S. Context based text-generation using lstm networks. arXiv preprint arXiv:200500048. 2020 [Google Scholar]
- 22.Livieris IE, Pintelas E, Pintelas P. A CNN–LSTM model for gold price time-series forecasting. Neural Computing and Applications. 2020;32(23):17351–60. [Google Scholar]
- 23.Shickel B, Tighe PJ, Bihorac A, Rashidi P. Deep EHR: a survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J Biomed Health Inform. 2018;22(5):1589–604. doi: 10.1109/JBHI.2017.2767063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Computation. 1997;9(8):1735–80. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 25.Gers FA, Schmidhuber J, Cummins F. Learning to forget: continual prediction with LSTM. Neural Computation. 2000;12(10):2451–71. doi: 10.1162/089976600300015015. [DOI] [PubMed] [Google Scholar]
- 26.Shiloach M, Frencher SK, Jr., Steeger JE, et al. Toward robust information: data quality and inter-rater reliability in the American College of Surgeons National Surgical Quality Improvement Program. J Am Coll Surg. 2010;210(1):6–16. doi: 10.1016/j.jamcollsurg.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 27.Géron A. Hands-on machine learning with Scikit-Learn, Keras, and TensorFlow: concepts, tools, and techniques to build intelligent systems: “ O’Reilly Media, Inc.”. 2019.
- 28.Greff K, Srivastava RK, Koutník J, Steunebrink BR, Schmidhuber J. LSTM: a search space odyssey. IEEE Transactions on Neural Networks and Learning Systems. 2017;28(10):2222–32. doi: 10.1109/TNNLS.2016.2582924. [DOI] [PubMed] [Google Scholar]
- 29.Pascanu R, Mikolov T, Bengio Y. International conference on machine learning. PMLR; 2013. On the difficulty of training recurrent neural networks. [Google Scholar]
- 30.Naseem U, Rashid J, Ali L, et al. An automatic detection of breast cancer diagnosis and prognosis based on machine learning uusing ensemble of classifiers. IEEE Access. 2022;10:78242–52. [Google Scholar]
- 31.Ozenne B, Subtil F, Maucort-Boulch D. The precision–recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. Journal of clinical epidemiology. 2015;68(8):855–9. doi: 10.1016/j.jclinepi.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM transactions on intelligent systems and technology (TIST) 2011;2(3):1–27. [Google Scholar]
- 33.Platt J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Advances in large margin classifiers. 1999;10(3):61–74. [Google Scholar]
- 34.Breiman L. Random forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 35.Chen T, Guestrin C. Xgboost: A scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. 2016 [Google Scholar]
- 36.Paszke A, Gross S, Massa F, et al. Pytorch: An imperative style, high-performance deep learning library. Advances in neural information processing systems. 2019:32. [Google Scholar]
- 37.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. The Journal of Machine Learning Research. 2011;12:2825–30. [Google Scholar]
- 38.Virtanen P, Gommers R, Oliphant TE, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nature Methods. 2020;17(3):261–72. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fortunato M, Blundell C, Vinyals O. Bayesian recurrent neural networks. arXiv preprint arXiv:170402798. 2017 [Google Scholar]
- 40.Tallec C, Ollivier Y. Unbiasing truncated backpropagation through time. arXiv preprint arXiv:170508209. 2017 [Google Scholar]
- 41.Shi J, Liu S, Pruitt LCC, et al. Using natural language processing to improve EHR structured data-based surgical site infection surveillance. AMIA Annu Symp Proc. 2019;2019:794–803. [PMC free article] [PubMed] [Google Scholar]