Abstract
A novel signal generation principle suitable for real time and end-point detection of specific PCR products in a closed tube is described. Linear DNA probes were labeled at their 5′-ends with a stable, fluorescent terbium chelate. The fluorescence intensity of this chelate is lower when it is coupled to single-stranded DNA than when the chelate is free in solution. The synthesized probes were used in the real time monitoring of PCR using a prototype instrument that consisted of a fluorometer coupled to a thermal cycler. When the probe anneals to a complementary target amplicon, the 5′→3′ exonucleolytic activity of DNA polymerase detaches the label from the probe. This results in an enhanced terbium fluorescence signal. Since terbium has a long excited state lifetime, its fluorescence can be measured in a time-resolved manner, which results in a low background fluorescence and a 1000-fold signal amplification. The detection method is quantitative over an extremely wide linear range (at least 10–107 initial template molecules). The label strategy can easily be combined with existing label technologies, such as TaqMan 5′-exonuclease assays, in order to carry out multiplex assays that do not suffer from overlapping emission peaks of the fluorophores.
INTRODUCTION
The polymerase chain reaction (PCR) (1) is by far the most important nucleic acid diagnostic tool. The first diagnostic tests based on PCR were quite cumbersome and not amenable to large-scale screening methods (2). Several post-PCR steps, such as restriction enzyme analysis, agarose gel electrophoresis or heterogeneous hybridization assays were needed to confirm the identity of the PCR product (3). Recently, the development of new fluorescent techniques has, however, led to novel assay formats that greatly simplify the protocols used for the detection of specific nucleic acid sequences. These methods involve the detection of a specific PCR product in a homogeneous solution without the need to open the amplification tubes after PCR (4). The results can be read in real time as the PCR product accumulates or at the end of the thermal cycling protocol directly from the amplification wells. The choice between real time or end-point measurement modes depends on whether a quantitative or qualitative assay is desired.
At the moment four technologies enabling specific sequence detection in a closed tube are commercially available. All of them (the TaqMan, Molecular Beacons, LightCycler and Amplifluor technologies) are based on fluorescence resonance energy transfer (FRET) (5,6). TaqMan (7,8) uses a linear probe that has a FRET donor in its 5′-end and an acceptor moiety at the 3′-end (9). When the probe is single stranded, the three-dimensional conformation of the probe brings the two labels close enough to each other for the acceptor to quench the donor fluorescence. As the probe hybridizes to its target or when a DNA polymerase that has 5′→3′ exonuclease activity cleaves it, the distance between the two labels increases enough for the donor fluorescence intensity to increase markedly. The difference between TaqMan probes and molecular beacons (10) is that the optimized stem–loop structure of molecular beacons brings the two labels as close together as possible when the probe is not hybridized to a target sequence. This ensures maximal quenching efficiency. Upon hybridization to a target (or when cleaved by a DNA polymerase), the fluorescence intensity of the FRET donor increases as the physical contact between the two labels is disrupted. The Amplifluor technology (11) utilizes hairpin-shaped primers that basically function in the same way as do molecular beacons: when the primer is incorporated into a PCR product, the donor and quencher moieties are separated and donor fluorescence is thus increased. The Hybridization Probe format (12) used in the LightCycler system (13,14) uses two adjacent probes that are labeled such that when both probes are hybridized to a target, the labels are brought close to each other and a FRET occurs between them. The sensitized acceptor emission is measured instead of the donor fluorescence.
All of these methods based on FRET are characterized by relatively high signal-to-noise ratios and a good ability to discriminate between positive and negative reactions. However, they are all limited in the sense that either a dual label probe or primer or two separate probes per target have to be used. This seriously complicates probe design and synthesis. In addition, since they all employ labels with rapidly decaying fluorescence and broad emission peaks, the possibilities for multiplex detection are limited.
We present a novel method for the qualitative and quantitative detection of a specific nucleic acid sequence in a homogeneous solution that relies on the 5′-exonuclease activity of a nucleic acid polymerase and on the use of a stable, fluorescent terbium chelate (Fig. 1). The fluorescence intensity of this chelate is greatly affected by the immediate chemical environment of the label. The chelate is coupled to the 5′-end of an oligonucleotide probe that is used to monitor PCR amplification of a complementary target in a closed tube. An increase in terbium fluorescence is seen as the DNA polymerase detaches the label from the probe hybridized downstream from a primer oligonucleotide. There are two main differences between this assay and the conventional TaqMan 5′-exonuclease assay. First, there is no need for a quencher moiety in the probe. Second, because it is possible to do the measurements in a time-resolved manner, this label technology can easily be combined with conventional, prompt labels in order to carry out a multiplex assay without the need to rely on any spectral resolution software. In this paper we describe the environment-dependent fluorescent properties of the chelate and demonstrate the applicability of this technology to qualitative and quantitative detection of specific target sequences. The feasibility of multi-analyte assays combining this technology with conventional TaqMan detection is also shown here.
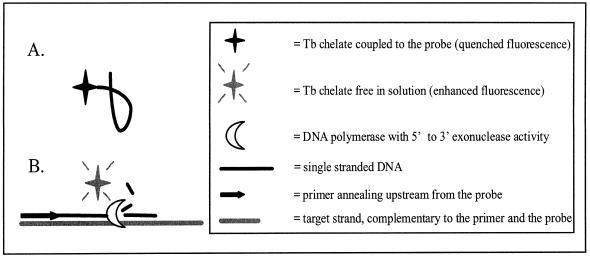
Figure 1.
Assay principle. (A) When the stable, fluorescent terbium chelate used as label is coupled to a single-stranded stretch of DNA, the fluorescence intensity of the chelate is significantly lower than when the chelate is free in solution. (B) Hybridization of a probe labeled at its 5′-end with the chelate to a complementary target amplicon during the annealing/extension step of PCR results in detachment of the label from the probe and a subsequent signal increase. This detachment is catalyzed by the 5′→3′ exonuclease activity of the DNA polymerase used to amplify the target sequence.
MATERIALS AND METHODS
Oligonucleotides and PCR templates
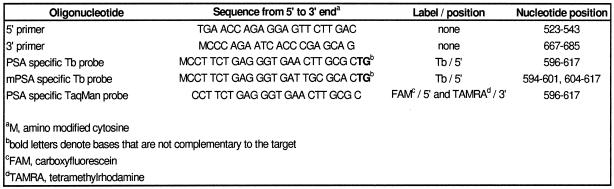
Oligonucleotide primers and probes (Table 1) were synthesized on an ABI392 DNA/RNA Synthesizer (PE Applied Biosystems, USA) using standard phosphoramidite chemistry, except for the conventional prostate-specific antigen (PSA) cDNA-specific TaqMan probe, which was purchased from SGS DNA (Sweden). Primary amino groups were introduced into the 5′-ends of the probes using an amino-modified cytosine phosphoramidite (modC; Perkin Elmer Life Sciences, Finland). Two bases that do not hybridize with the target sequence were added to the 3′-ends of the probes to make sure that the probes cannot act as primers during PCR.
Table 1. Oligonucleotides used in this study.
PSA- and mutant PSA (mPSA)-specific probes were labeled with a 100-fold molar excess of an isothiocyanate-modified stable, fluorescent terbium chelate [2,2′,2′′,2′′′-{{6,6′-{4′′-[2-(4-isothiocyanatophenyl)ethyl]-1H-pyrazole-1′′,3′′-diyl}bis(pyri-dine)-2,2′-diyl}bis(methylenenitrilo)}tetrakis(acetato) terbium(III); Perkin Elmer Life Sciences] by dissolving the chelate and the DNA in 50 mM sodium carbonate buffer (pH 9.8). The DNA concentration was adjusted to ~2 µg µl–1 and the reactions were incubated at 37°C overnight. Excess chelate was removed using a Nick gel filtration column (Pharmacia Biotech, Sweden). The probes were HPLC purified using a Sephasil Peptide C8 4.6/100 column (Pharmacia Biotech) and a 16.5 min linear gradient of 2–70% acetonitrile in 50 mM triethylammonium acetate buffer (pH 6.8). The probes were vacuum dried (Hetovac, Heto Holten, Denmark), dissolved in 50 mM Tris–HCl (pH 7.5), quantified spectrophotometrically and stored in small aliquots at –20°C.
PSA and mPSA double-stranded cDNAs excised from plasmids pGEM3-PSA and pGEM3-IS (15,16) were used as templates during the development of the technology. Mutant PSA was used as an internal standard in a quantitative PSA reverse transcription PCR assay (16). It differs from PSA by a 2 bp deletion situated in the middle of the amplicon. Therefore, PSA and mPSA are good model analytes for a detection system that is to be applied to quantitative and qualitative sequence analysis. The template concentrations of stock solutions were determined spectrophotometrically at 260 nm. In the dual end-point assay the templates were mixed with a 108-fold excess of human genomic DNA from a healthy donor.
Characterization of the fluorescent properties of the probes
In order to investigate the effect of probe cleavage on terbium signal intensity, 1012 molecules of terbium-labeled PSA or mPSA probes were cleaved with 25 U Benzon nuclease (Merck, Germany). The probes were incubated with the enzyme in 50 µl of 50 mM Tris–HCl (pH 7.5) containing 1 mM MgCl2 at 37°C for 30 min, after which the terbium signals were measured using a Victor™ 1420 Multilabel Counter (Perkin Elmer Life Sciences) and compared to signals obtained from intact probes. The following settings were used in all terbium measurements in this study: excitation filter 340 nm; emission filter 545 nm; delay 500 µs; window time 1400 µs; cycle 2000 µs.
To confirm that the probes are in fact cleaved during PCR, PSA and mPSA double-stranded cDNAs were amplified in 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 3.5 mM MgCl2, 0.1 mM dNTPs (Pharmacia Biotech), 0.1 µM 3′-primer, 0.2 µM 5′-primer, 33 nM PSA or mPSA terbium probe and 1.25 U AmpliTaq Gold DNA polymerase (PE Applied Biosystems). Each 50 µl reaction contained 10 000 copies of either PSA or mPSA template or no template at all. The thermal cycling consisted of a 10 min initial denaturation and polymerase activation step at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 61.5°C. The reactions were done in a PTC-200 DNA Engine (MJ Research, USA) in black polypropylene wells (catalog no. 6885; Corning Costar, UK) and sealed with Cycleseal® PCR Plate Sealer (Robbins Scientific Corp., USA). The terbium signals were measured directly from the amplification wells after thermal cycling at room temperature. To confirm that the detected terbium signal increase in positive reactions was due to probe cleavage, POPO™-3 iodide DNA binding dye (Molecular Probes, USA) was added to each well to a concentration of 100 nM. Time-resolved terbium and POPO™-3 iodide emissions at 545 and 572 nm were measured. In the measurement at 572 nm, the following settings were used: excitation filter 340 nm; emission filter 572 nm; delay 60 µs; window time 100 µs; cycle 1000 µs. POPO™-3 iodide is excited at the terbium maximum emission wavelength (545 nm) and emits at 570 nm. The binding of POPO™-3 iodide to a terbium-labeled probe will therefore result in a FRET between terbium and the dye (12). This can be seen as a decrease in terbium emission at 545 nm and, on the other hand, as an increase in the sensitized emission of POPO™-3, which can be recorded at 572 nm. If the probe has been cleaved, however, there will be no detectable energy transfer between the two fluorophores.
Real time quantitative PCR
To demonstrate the quantitative nature of terbium signal generation during thermal cycling, PSA and mPSA were amplified so that the amount of starting template was varied from 0 to 107 molecules. The thermal cycling parameters were as described above. Real time quantitative PCR was performed on a prototype instrument that consisted of a Victor™ 1420 Multilabel Counter measurement unit coupled to a PTC-100 thermal cycler (MJ Research) through an in-house hot lid that was kept at 105°C. The terbium signals were measured at the end of each annealing/extension step to determine the threshold cycles (Ct) of individual reactions. The threshold cycle is defined as the PCR cycle where the fluorescence signal-to-noise ratio (S/N) of a given amplification reaction exceeds the threshold S/N value. This threshold value is set by the user. In this study, threshold values of 1.3 for the PSA and 1.35 for the mPSA assay were used. PSA and mPSA standard curves were made by plotting the threshold cycle as a function of initial PSA or mPSA starting copy number.
Dual assay using time-resolved and prompt fluorescence
Aliquots of 1000 molecules of PSA and/or mPSA double-stranded cDNA were amplified in the presence of 250 ng of human genomic DNA. Human genomic DNA is not amplified in this assay since the primer sequences represent exon–intron junctions (16). The reaction conditions were identical to those used in the real time quantitative PCR, only this time two probes, the PSA-specific TaqMan probe and the mPSA-specific terbium probe were used, both at a concentration of 33 nM. The FAM and terbium signals were measured directly from the amplification wells after thermal cycling with a Victor™ 1420 Multilabel Counter at room temperature. In the FAM measurement, 485 nm excitation and 535 nm emission filters were used. Excitation energy was set at 24 064 and the measurement time was 1.0 s. The results were analyzed by calculating the average signals (M) and standard deviation (SD) from reactions containing only genomic DNA and no specific template. A threshold value was set at M + 2SD. All reactions with a terbium or FAM signal greater than the threshold were considered positive for the corresponding template.
RESULTS
Fluorescent properties of the terbium chelate
It has been noted in our laboratory that the fluorescence intensity of the chelate used in this study is very sensitive to the immediate chemical environment of the label moiety. Therefore, it was envisioned that detachment of the chelate from a DNA probe would result in a detectable change in terbium fluorescence. By placing the label at the 5′-end of a probe, the 5′→3′ exo-nuclease activity of a thermostable DNA polymerase could be used to detach the chelate from a probe hybridized downstream from a primer during the annealing/extension step of PCR. The resulting signal change could be used to monitor nucleic acid amplification. The idea was first tested by digesting the probe with a nuclease. Enzymatic cleavage of terbium-labeled probes resulted in an ~3- to 4-fold terbium signal increase (2.9 for the PSA- and 3.8 for the mPSA-specific probe) when compared to an intact probe.
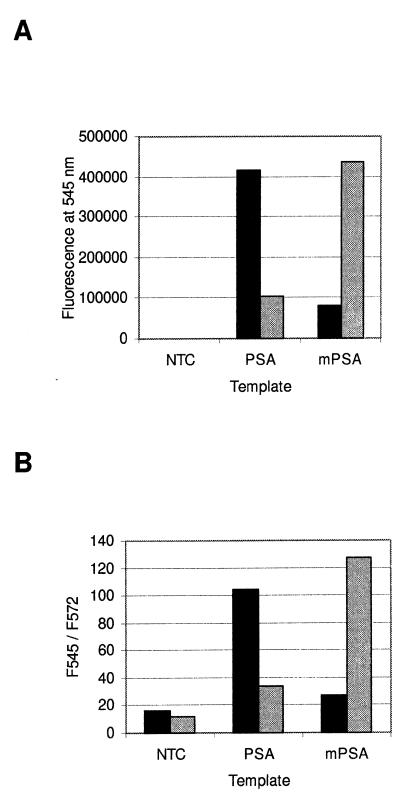
Figure 2A illustrates how amplification of a target sequence in the presence of a complementary probe results in a clear terbium signal increase. After 40 cycles, terbium fluorescence is clearly greater in reactions containing a PSA-specific probe and 10 000 initial copies of PSA cDNA than in reactions containing no template or a non-specific mPSA template. Accordingly, in reactions containing a mPSA-specific probe, only mPSA amplification results in a significant signal increase.
Figure 2.
Homogeneous end-point detection of PSA and mPSA amplification. Background subtracted terbium fluorescence of reactions containing no template (NTC), 10 000 molecules of PSA or 10 000 molecules of mPSA cDNA template before addition of POPO™-3. Black and gray columns denote reactions containing a PSA- or mPSA-specific probe, respectively. (B) Fluorescence intensity ratios after addition of POPO™-3. When PSA is used as template, the PSA-specific probe (black column) is cleaved and thus the addition of POPO™-3 does not affect the terbium signal at 545 nm. Accordingly, the sensitized POPO™-3 emission at 572 nm is less significant than in negative reactions. The result is the same when mPSA is used as template in an amplification reaction containing a mPSA-specific probe (gray column).
Figure 2B shows that this signal increase is indeed due to probe cleavage. The addition of a DNA-binding dye that is excited at the maximum emission wavelength of terbium results in a clear terbium fluorescence decrease in reactions that did not contain a target complementary to the probe. Also, in the negative reactions, the sensitized long-lived emission of the DNA-binding dye is detectable at 572 nm. The shift in emission intensities can be illustrated by dividing the emission signal at 545 nm by the signal at 572 nm. The ratio is ~10-fold greater in reactions that did contain a target complementary to the probe, indicating detachment of the label from the probe in these reactions.
Real time quantitative PCR
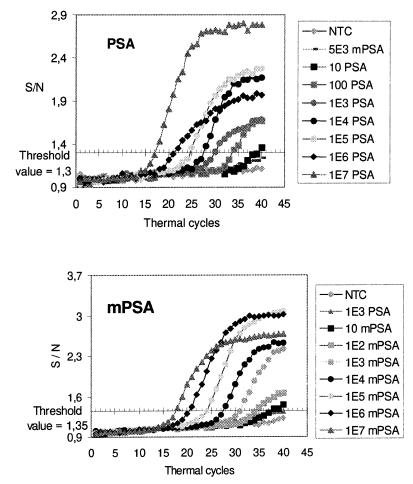
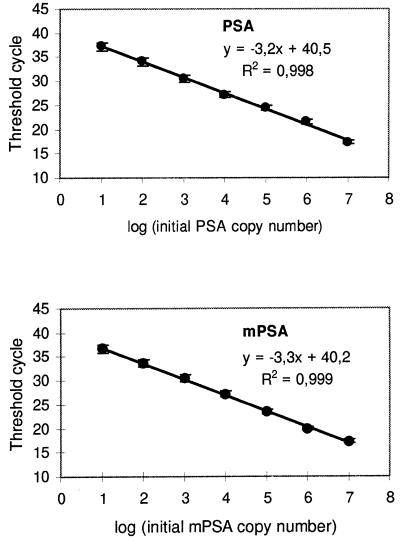
The amplification plots for different amounts of PSA and mPSA cDNA are shown in Figure 3. The signals have been converted to S/N ratios. The noise value was determined individually for each reaction by calculating the average terbium signal of cycles 6–10. A threshold value to indicate the difference between positive and negative reactions was set at S/N 1.3 for the PSA and 1.35 for the mPSA assay. A higher threshold value had to be used in the mPSA assay because of a slight cross-reactivity between the mPSA probe and PSA target at the annealing temperature used. The Ct values were plotted as a function of the initial target copy number to produce standard curves for each assay (Fig. 4). There is a very precise inverse log-linear relationship between the starting copy number and the threshold cycle in both assays. The average coefficient of variation in replicate measurements was 2.3 ± 2.5%.
Figure 3.
Real time monitoring of PSA and mPSA amplifications. The reactions contained either a PSA- or a mPSA-specific terbium-labeled probe and 0–107 molecules of the respective target. Terbium fluorescence rises above background when the amount of synthesized target exceeds the detection limit of the assay. The position of the x-axis shows the threshold value of each assay (1.3 in the PSA and 1.35 in the mPSA assay).
Figure 4.
PSA and mPSA standard curves. Threshold cycles were determined at initial template amounts ranging from 10 to 107 molecules.
Dual end-point assay
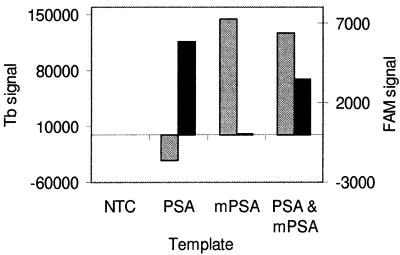
Time-resolved fluorometry and labels with prompt fluorescence were combined in a model assay that detects PSA and mPSA separately in one tube. Figure 5 shows the background subtracted FAM and terbium fluorescence signals (specific for PSA and mPSA, respectively) of reactions that contained 1000 initial copies of PSA, mPSA or both together. The signals were measured after 40 PCR cycles. Threshold values (mean of negative controls + 2SD) for FAM and terbium were 260 and 46 500 counts, respectively. The assay gives correct results (a positive FAM signal in the presence of PSA and a positive terbium signal in the presence of mPSA) even when the target molecules were mixed with a large excess of non-specific DNA.
Figure 5.
Dual end-point assay. Aliquots of 1000 copies of PSA, mPSA or both double-stranded cDNAs were amplified in the presence of a PSA-specific TaqMan probe and a mPSA-specific terbium probe. Black and gray columns represent the background subtracted FAM and terbium signals, respectively, after 40 PCR cycles. There is no cross-talk between the two labels: a positive FAM signal (>260 counts) is detected only in the presence of a PSA target and a positive terbium signal (>46 500 counts) in the presence of a mPSA target. All reactions contained 250 ng of human genomic DNA.
DISCUSSION
Multiple techniques exist that allow simultaneous nucleic acid amplification and detection in a closed tube (10,11,12,17). All these techniques need either a dual label probe or primer or two separate probes labeled with an energy transfer couple. Despite the possibility of coupling several different kinds of label moieties into a probe during automated oligonucleotide synthesis, the synthesis of such dual label probes is still somewhat difficult. Since any probe impurities easily lead to a high fluorescence background and reduced assay performance, there is a need for a more straightforward approach. Although one can avoid the problems associated with dual label probes by designing two probes, as in the Hybridization Probe format, this is not an ideal solution. The more probes one has to use for each target, the more complex the amplification reaction becomes and undesired artifacts, such as probe–primer dimers etc., may accumulate (18). Ideally, one would only have to design one linear probe with one label and with no extra secondary structures that might interfere with probe hybridization efficiency. The detection limit of such a method should not, however, be any higher than in the existing methods, where one can easily reach a linear range of at least 10–107 molecules of initial template, when real time detection is used in conjunction with target amplification.
We have described a label system that does not require a separate quencher moiety, since the mere proximity of single-stranded DNA quenches the time-resolved fluorescence of the reporter label. This quenching is based on the environment-sensitive properties of the light absorbing moiety. It has been previously reported that the coupling of some lanthanide chelates to biomolecules, especially to proteins, has a profound effect on the decay time and thus on the quantum yield of the label (19). The probe has the simplest possible design since no extra secondary structures are needed. Signal generation is based on enzymatic, template-dependent probe cleavage. Cleavage of the probe results in a signal increase that can be used for qualitative and quantitative detection of a target nucleic acid in a homogeneous solution. A maximum S/N ratio of 4 was obtained by digesting a terbium-labeled probe with a nuclease. Although this signal increase is quite modest when compared to results obtained with, for example, molecular beacon probes (20), it is however sufficient: a large dynamic range (at least 10–107 molecules of initial template) was obtained. This is probably due to the fact that in a time-resolved fluorescence measurement the non-specific background fluorescence is almost insignificant (21) and therefore very subtle changes in the specific signal can be detected. The actual detection limit of a real time PCR assay refers to the number of target amplicons synthesized when a detectable fluorescence signal occurs. An estimation of this detection limit can be obtained by extrapolating a standard curve to the point where Ct = 0. With our detection technology this detection limit is ~1012 synthesized target amplicons, which is very similar to results obtained with other techniques (22,23). It is possible that this detection limit can be further improved by increasing the labeling degree of the probe and/or by optimizing the position of the label moiety in the probe. Further studies are needed to determine the effect of probe sequence on quenching efficiency, albeit according to preliminary results probe length and GC-content do not seem to have a significant effect on quenching efficiency (data not shown).
We had to label our probes following synthesis because a phosphoramidite that would enable direct coupling of the chelate to the probe during synthesis does not exist. Different chelates have, however, been derivatized into phosphoramidites (24). Therefore, we assume that a phosphoramidite reagent that would contain the chelate used in this study is feasible. Once such a reagent is available, probe synthesis will be simplified even further.
The benefits of real time quantitative PCR (25) have been widely described (26,27). These include improved reproducibility, reduced contamination risks and higher assay throughput when compared to traditional quantitative PCR assays that include several post-PCR steps. Quantitative PCR assays based on end-point determination of the amount of PCR product are not only laborious but the reliability of the results is often questionable because the amount of PCR product at the end of thermal cycling is only poorly correlated with the amount of initial template (Fig. 3). However, determination of the threshold cycle (the point in time when enough PCR product has been synthesized for a detectable fluorescence signal to occur) enables a very precise and reproducible quantification, because detection is done before the amplification reaction reaches the plateau phase.
Currently, all available homogeneous nucleic acid detection methods employ labels with rapidly decaying or prompt fluorescence. Because of the wide, overlapping emission peaks of the labels used, multiparametric measurements always require the use of powerful spectral resolution software, i.e. the results cannot be read directly from the fluorescence counts at different wavelengths. However, even with the best software it is sometimes impossible to distinguish the emissions of two labels from one another, which may result in false positive results (23). A better approach would be the use of fluorescent labels with long excited state lifetimes (such as stable, fluorescent lanthanide chelates) and time-resolved fluorimetry (21) together with prompt labels. In a time-resolved fluorescence measurement the signal is not recorded immediately after the excitation pulse but after a certain delay. During this delay non-specific background fluorescence and fluorescence from any prompt labels, such as fluorescein, disappear. The measurement is repeated several times in a short period of time, which results in signal amplification. Since lanthanides are excited by light in the UV range, they do not cause any background signal for prompt labels that are excited by light in the visible range. Therefore, by combining these label technologies in one assay one can totally discriminate between the signals from different labels and maximize the specificity of a homogeneous multi-analyte assay. In this study we detected two very similar analytes separately in one tube. This was done to demonstrate that by combining labels with different fluorescence lifetimes one can achieve multi-analyte systems that do not suffer from any cross-talk between labels. The emission spectra of the labels that were used (terbium and FAM) overlap considerably. However, their signals were completely resolved from each other. We expect it to be fairly straightforward to increase the number of analytes in one tube further without compromising assay specificity by choosing labels with different fluorescence lifetimes and different excitation wavelengths and by using a variable wavelength excitation light source in the measurements.
In addition to allele discrimination and other qualitative assays, the possibility of carrying out multiplex detection is of importance to quantitative assays as well. Despite the fact that an internal standard is not needed to compensate for changes in amplification efficiency when quantification is based on real time detection (28), an internal standard is still needed to compensate for the errors due to sample processing and possible reverse transcription steps (27). The best way to quantify the internal standard, in terms of assay throughput, simplicity and reliability, is in the same tube with the actual target. We have shown this approach to be feasible by detecting PSA and mPSA, an internal standard that is amplified by the same primers as PSA, in one sealed tube.
We have demonstrated the applicability of this new signal generation principle to both quantitative and qualitative nucleic acid assays. Although a qualitative assay is not needed for our model analytes, PSA cDNA and an in vitro produced mutant form of PSA, their detection in the presence of a large excess of unrelated DNA shows that our label technology can be applied to various kinds of mutation or pathogen detection systems. We conclude that by using a probe labeled at its 5′-end with a stable, fluorescent terbium chelate one can easily construct a nucleic acid assay that is as user friendly and reliable as other corresponding methods with the additional attributes of simpler probe design and a far better suitability to multiplex assays.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Petri Aronkytö, Jari Hovinen and Hannu Kojola (Perkin Elmer Life Sciences, Finland) for providing the necessary equipment and oligonucleotide synthesis reagents.
REFERENCES
- 1.Saiki R.K., Scharf,S., Faloona,F., Mullis,K.B., Horn,G.T., Erlich,H.A. and Arnheim,N. (1985) Science, 230, 1350–1354. [DOI] [PubMed] [Google Scholar]
- 2.Isaksson A. and Landegren,U. (1999) Curr. Opin. Biotechnol., 10, 11–15. [DOI] [PubMed] [Google Scholar]
- 3.Kricka L.J. (1999) Clin. Chem., 45, 453–458. [PubMed] [Google Scholar]
- 4.Williams P.M., Giles,T., Tucker,A., Winer,A. and Heid,C. (1998) In Ferré,F. (ed.), Gene Quantification. Birkhäuser, Boston, MA, pp. 313–325.
- 5.Clegg R.M. (1992) Methods Enzymol., 211, 353–388. [DOI] [PubMed] [Google Scholar]
- 6.Selvin P.R. (1995) Methods Enzymol., 246, 300–334. [DOI] [PubMed] [Google Scholar]
- 7.Holland P.M., Abramson,R.D., Watson,R. and Gelfand,D.H. (1991) Proc. Natl Acad. Sci. USA, 88, 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee G.L., Connell,C.R. and Bloch,W. (1993) Nucleic Acids Res., 21, 3761–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak K., Flood,S., Marmaro,J., Giusti,W. and Deetz,K. (1995) PCR Methods Appl., 4, 357–362. [DOI] [PubMed] [Google Scholar]
- 10.Tyagi S. and Kramer,F.R. (1996) Nature Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 11.Nazarenko I.A., Bhatnagar,S.K. and Hohman,R.J. (1997) Nucleic Acids Res., 25, 2516–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison L.E. (1995) In Kricka,L.J. (ed.), Nonisotopic Probing, Blotting, and Sequencing. Academic Press, San Diego, CA, pp. 430–471.
- 13.Wittwer C.T., Herrman,M.G., Moss,A.A. and Rasmussen,R.P. (1997) Biotechniques, 22, 130–131; 134–138. [DOI] [PubMed] [Google Scholar]
- 14.Wittwer C.T., Ririe,K., Andrew,R., David,D., Gundry,R. and Balis,U. (1997) Biotechniques, 22, 176–181. [DOI] [PubMed] [Google Scholar]
- 15.Lundwall,Å. and Lilja,H. (1987) FEBS Lett., 214, 317–322. [DOI] [PubMed] [Google Scholar]
- 16.Ylikoski A., Sjöroos,M., Lundwall,Å., Karp,M., Lövgren,T., Lilja,H. and Iitiä,A. (1999) Clin. Chem., 45, 1397–1407. [PubMed] [Google Scholar]
- 17.Whitcombe D., Theaker,J., Guy,S.P., Brown,T. and Little,S. (1999) Nature Biotechnol., 17, 804–807. [DOI] [PubMed] [Google Scholar]
- 18.Kreuzer K.A., Lass,U., Bohn,A., Landt,O. and Schmidt,C.A. (1999) Cancer Res., 59, 3171–3174. [PubMed] [Google Scholar]
- 19.Takalo H., Mukkala,V.-M., Meriö,L., Rodríguez-Ubis,J.C., Sedano,R., Juanes,O. and Brunet,E. (1997) Helv. Chim. Acta, 80, 372–387. [Google Scholar]
- 20.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Nature Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 21.Soini E. and Lövgren,T. (1987) CRC Crit. Rev. Anal. Chem., 18, 105–154. [Google Scholar]
- 22.Nitsche A., Steuer,N., Schmidt,C.A., Landt,O. and Siegert,W. (1999) Clin. Chem., 45, 1932–1937. [PubMed] [Google Scholar]
- 23.Vet J.A.M., Majithia,A.R., Marras,S.A.E., Tyagi,S., Dube,S., Poiesz,B.J. and Kramer,F.R. (1999) Proc. Natl Acad. Sci. USA, 96, 6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwiatkowski M., Samiotaki,M., Lamminmäki,U., Mukkala,V.M. and Landegren,U. (1994) Nucleic Acids Res., 22, 2604–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higuchi R., Dollinger,G., Walsh,P.S. and Griffith,R. (1992) Biotechnology, 10, 413–417. [DOI] [PubMed] [Google Scholar]
- 26.Orlando C., Pinzani,P. and Pazzagli,M. (1998) Clin. Chem. Lab. Med., 36, 255–269. [DOI] [PubMed] [Google Scholar]
- 27.Freeman W.M., Walker,S.J. and Vrana,K.E. (1999) Biotechniques, 26, 112–122; 124–125. [DOI] [PubMed] [Google Scholar]
- 28.McBride L., Livak,K., Lucero,M., Goodsaid,F., Carlson,D., Stevens,J., Allen,T., Wyatt,P., Thiel,D., Honebein,P. et al. (1998) In Ferré,F. (ed.), Gene Quantification. Birkhäuser, Boston, MA, pp. 97–110.