Abstract
Rapid construction of high-resolution physical maps requires accurate information about overlap between DNA clones and the size of gaps between clones or clone contigs. We recently developed a procedure termed ‘quantitative DNA fiber mapping’ (QDFM) to help construct physical maps by measuring the overlap between clones or the physical distance between non-overlapping contigs. QDFM is based on hybridization of non-isotopically labeled probes onto DNA molecules that were bound to a solid support and stretched homogeneously to ~2.3 kb/µm. In this paper, we describe the design of probes that bind specifically to the cloning vector of DNA recombinants to facilitate physical mapping. Probes described here delineate the most frequently used cloning vectors such as BACs, P1s, PACs and YACs. As demonstrated in representative hybridizations, vector-specific probes provide valuable information about molecule integrity, insert size and orientation as well as localization of hybridization domains relative to specifically-marked vector sequences.
INTRODUCTION
Quantitative DNA fiber mapping (QDFM) is a versatile, reliable and inexpensive way to construct high-resolution physical maps at near kilobase resolution (1). The method relies on binding of probes to DNA molecules (called ‘fibers’) that have been immobilized on a modified glass surface and stretched homogeneously to ~2.3 kb/µm. Non-isotopically labeled DNA probes are hybridized to the stretched DNA fibers. Taking advantage of the homogeneous stretching, the extent and location of the bound probes allow the direct visualization of target DNA sequences along the fiber when viewed in the fluorescence microscope.
There are numerous applications for QDFM. The precise location of cloned DNA fragments in a larger genomic interval and the extent of overlap between different clones as determined by QDFM accelerate high-resolution physical map assembly (1). QDFM also allows rapid measurement of the contig extent and map position, as well as overlap between contigs with an accuracy of ~1 kb (1,2). This information reduces redundant sequencing of overlapping clones and can identify chimeric clones. Previously, QDFM resolved ambiguities in maps constructed by STS content mapping and DNA clone fingerprinting (2). Two other study generated high-resolution physical maps of the human apolipoprotein B gene region of human chromosome 2 and immunoglobubin λ variant gene cluster (3,4).
In general, QDFM has mostly been used to analyze fragments of genomic DNA that had been propagated in cloning systems such as BACs, P1s, PACs and YACs. One method to increase the information content of a QDFM hybridization experiment is to include probes specific for the vector in the hybridization mixture. These vector-specific probes serve as ‘landmarks’ for the entire fiber and allow the researcher to deduce the localization and orientation of the hybridization domains relative to the vector sequences in a large insert clone. Landmark probes can also mark the ends of linear DNA molecules to determine if a fiber is a full-length linear molecule. We have developed highly specific probes to visualize the vector sequences of widely used cloning systems for genomic DNA. Here, we present QDFM experiments that illustrate how these probes can accelerate the construction of genomic maps.
MATERIALS AND METHODS
Preparation of DNA fibers
High molecular weight DNA from cosmid, BAC and P1 clones is routinely isolated in our laboratory using an alkaline lysis protocol (1,5). This included the BAC clone #97 [library coordinate 133E8, mapping to chromosome 20q13.2, clone RMC20B4097 (6)], P1 clone #41 (maps the same location as BAC #97 on 20q13.2) and PAC clone 11C11 (RPCI-1-11C11) used in this experiment. YAC DNA from clone 141G6 (1) was isolated from whole yeast cultures following standard procedures (7). The size of its human DNA insert is ~475 kb (8). Following extraction and precipitation, BAC DNA was separated by pulsed field gel electrophoresis to separate bands corresponding to circular and randomly broken linear DNA molecules and bands were excised. Subsequent purification of DNA, glass slide cleaning, silane modification and application of the DNA molecules to the slides were performed as described previously (1,9).
Probe preparation and labeling
High molecular weight DNA from P1 clones #1107 and #1143 of a human genomic P1 library (10) was isolated in the course of assembly of a physical map based on STS content (1). All BAC, P1 or PAC probes were generated from DNA prepared from overnight cultures by an alkaline lysis protocol (1). The YAC DNA for probe preparation was isolated from whole yeast cultures following standard procedures (7). The DNA from plasmid clone P1 #41-2d8, the recombinant cloning vector pAd10SacBII (~17 kb; Genome Systems, Inc., St Louis, MO) and the plasmid pYAC3 (11) (used to mark both arms of YAC) were prepared by alkaline lysis followed by phenol:chloroform extraction (12). For the vector part-specific DNA probes, five BAC vector-specific and one P1 vector-specific primer pairs (Genset Corp., La Jolla, CA) were used in in vitro DNA amplification reactions to highly enrich fragments between 1 and 2 kb in length. The sequences of these primers, their estimated melting temperature (Tm), position along the respective vector and the PCR product sizes are summarized in Table 1.
Table 1. PCR primers for vector-specific probes.
| Primer name | Primer sequence | Tm | Position in vector | Vector | Product size (bp) |
|---|---|---|---|---|---|
| F1-pBELO | TTAACTATGCGGCATCAGAGCAG | 57 | 52–74 | BAC | 1476 |
| B3-pBELO | TGATCGGCACGTAAGAGGTTCC | 60 | 1507–1528 | ||
| F7-pBELO | TCTTACGTGCCGATCAACGTC | 58 | 1513–1533 | BAC | 1324 |
| B14-pBELO | GAGGTCGTTTGACTGGACGATTC | 60 | 2814–2836 | ||
| F12-pBELO | CGGTTTACGCAGTTTCGGCTTAG | 60 | 3197–3219 | BAC | 1229 |
| B17-pBELO | CGGTGGAAATACGTCTTCAGCAC | 60 | 4406–4428 | ||
| F18-pBELO | ATTCGAGGACGGGTTGAGCAAC | 62 | 4321–4342 | BAC | 1406 |
| B21-pBELO | CCAATCTGGATAATGCAGCCATC | 57 | 5705–5727 | ||
| F25-pBELO | GGCTGCATTATCCAGATTGGG | 57 | 5142–5162 | BAC | 2058 |
| B27-pBELO | CATATCACAACGTGCGTGGAGG | 60 | 7178–7199 | ||
| F1-P1 | TACCCCATTTAGGACCACCCAC | 56 | 10 054–10 075 | P1/ | 1444 |
| B1-P1 | CAGCCGAAGCCATTAAGGTTC | 55 | 11 477–11 497 | PAC |
BAC clone #978A1 [mapping to chromosome 2 band q2 (3), SK #1241R1, California Institute of Technology BAC Library] was used as a template in PCR reactions to generate most of the BAC vector-specific DNA fragments. The primer pair F1/B3 was used in conjunction with a circular BAC vector pFOS1 (California Institute of Technology). The P1 clone pAd10SacBII (Genome Systems) containing vector DNA without insert was linearized with BamHI before serving as PCR template for generating P1 vector-specific probe. 200 µl of PCR buffer (1,9,13,14) was used for each reaction, and the PCR parameters were 30 cycles of 95°C (1 min), 53°C (1 min) and 72°C (2 min) followed by 10 min at 72°C for elongation. After validation by agarose gels, the PCR products were separated from the overlaying mineral oil by chloroform extraction. The DNA was then precipitated with 2 vol of ethanol, washed with 70% ethanol and concentrations were determined with Hoechst fluorometry (TKO100, Amersham Pharmacia Biotech, Piscataway, NJ). 420 ng of DNA was used for 50 µl labeling reactions. Labeling was performed by random priming reactions using a commercial kit (BIOPRIME, Life Technologies, Gaithersburg, MD) incorporating biotin (bio)-14-dCTP. To label with digoxigenin (dig)-dUTP (Boehringer Mannheim, Indianapolis, IN) or FITC-dUTP (Boehringer Mannheim), we used a previously published protocol (13). The labeling and hybridization schemes for the probes are summarized in Table 2.
Table 2. Probe labeling scheme.
| Clone (fiber) | Whole fiber probe | Insert probe | Vector-specific probe |
|---|---|---|---|
| BAC #97 (133E8) | BAC #97-biotin | BAC #131-dig | F1/B3-FITC, F7/B14-FITC |
| F12/B17-FITC, F18/B21-dig | |||
| F25/B27-FITC | |||
| P1 #41 | P1 #41-biotin | Plasmid P1 #41-2d8-dig | P1 vector pAd10SacBII-FITC |
| F1/B1-dig | |||
| PAC 11C11 | PAC 11C11-biotin | BAC 97A23-dig | P1 vector pAd10SacBII-FITC |
| F1/B1-dig | |||
| YAC 141G6 | YAC 141G6-biotin | P1 #1107-dig | YAC vector pYAC3-dig |
| P1 #1143-dig |
Fluorescence in situ hybridization and image analysis
Two slightly different hybridization schemes were used in this study. For YAC mapping, a two-color scheme (biotin-green, digoxigenin-red) was used. Biotinylated probes were detected in green using avidin-FITC (Vector, Burlingame, CA) while digoxigenin-labeled probes were detected with rhodamine-conjugated sheep antibody against digoxigenin (Boehringer Mannheim) (14). Hybridization signals were amplified with biotinylated goat-anti-avidin (Vector) followed by a second layer of avidin-FITC or a Texas Red-labeled rabbit antibody against sheep IgG (Vector) (15).
For BAC, P1 and PAC mapping, a three-color scheme (biotin-blue, digoxigenin-red, FITC-green) was used. Biotinylated probes were detected in blue using avidin-AMCA (Vector) and signals amplified with biotinylated goat-anti-avidin (Vector) followed by a second layer of avidin-AMCA. Digoxigenin-labeled probes were detected and amplified as in the two-color scheme. Detection of FITC-labeled probes was performed with a mouse anti-FITC antibody (DAKO, Carpintera, CA) followed by incubation with an FITC-conjugated horse-anti-mouse antibody (Vector). Slides were washed twice in 2× SSC and mounted in 1% PPD (p-phenylenediamine, Sigma Chemicals, St Louis, MO) antifade solution in glycerol for microscopic inspection.
Images were acquired on a quantitative image processing system based on a Zeiss fluorescence microscope equipped with 63×, 1.25 NA and 40×, 1.3 NA oil objectives, a Photometrics cooled CCD camera, multiband pass filters for simultaneous observation of FITC, Texas Red or AMCA/DAPI (Chroma Technology, Brattleboro, VT), and a SUN SPARC workstation (16). Images were recorded on the SUN system, converted to 24-bit tiff-format images, imported to a PC-based system and processed with PhotoShop 4.0 (Adobe Systems Inc., Seattle, WA) and PowerPoint 97 (Microsoft Corporation, Redmond, WA).
RESULTS
Vector-specific probes help determine the orientation and extent of overlap between linked BAC clones
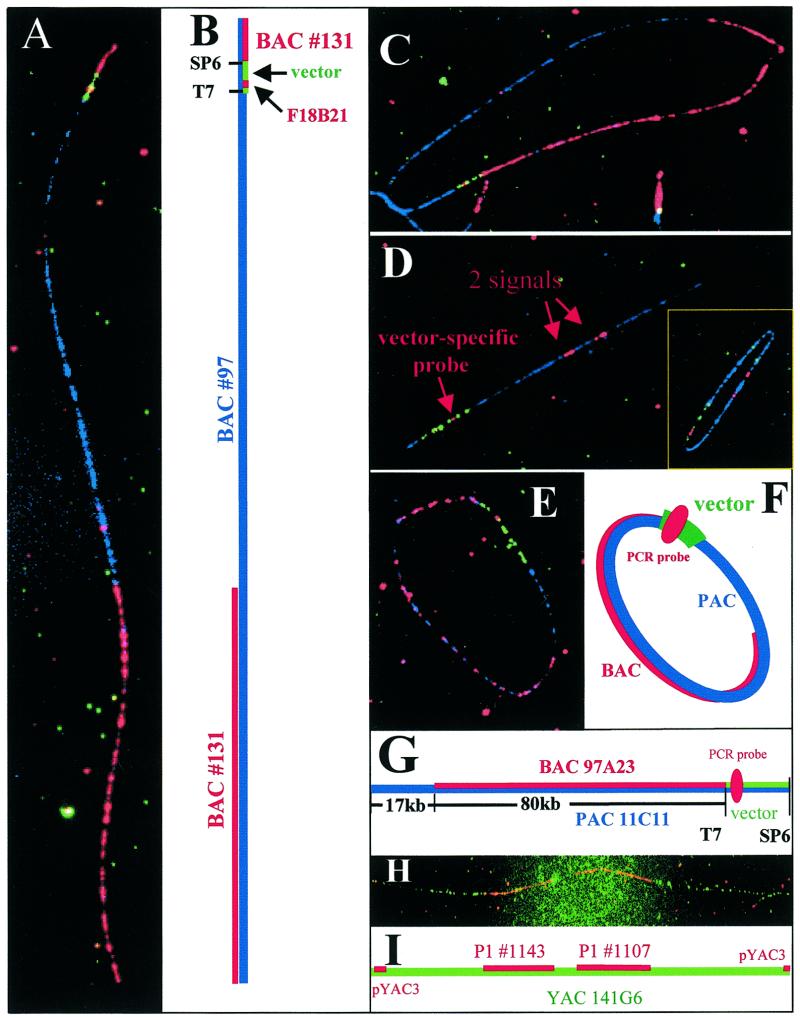
Individual DNA molecules from BAC clone #97 were immobilized on APS slides and hybridized with a second overlapping BAC probe (#131, overlaps with BAC #97 in chromosome 20q13) (6). Hybridization of the BAC clone #131 probes (red) to clone #97 was observed on all full-length BAC clone #97 molecules (Fig. 1A). In addition to labeled probe #131, five vector-specific probes (red and green) as well as BAC #97 probe (blue) were hybridized to DNA fiber from clone #97. The vector sequences were divided into five segments (Table 1): segment 1 amplified by the F1/B3 primer pair, segment 2 by F7/B14, segment 3 by F12/B17, segment 4 by F18/B21 and segment 5 by F25/B27. The adjacent PCR amplified products had short overlaps except segment 2 and 3 (see Table 1). Segment 4 probe was labeled in red and segments 1, 2, 3 and 5 in green. This yielded asymmetric labeling pattern on the 7.3 kb vector such that a red signal (F18/B21) was flanked by green signals and appeared off-centered toward the T7 side in the vector (Fig. 1B).
Figure 1.
(A–C) BAC clone #131 mapped to BAC clone #97. (A) A linear DNA fiber of BAC #97 fiber (blue), BAC #131 hybridization domain (red) and the PCR-synthesized vector-specific probes hybridization signals (green/red). (B) A schematic illustration of the different hybridization domains for the linear BAC molecule. (C) A circular molecule from the same hybridization mixture with all the hybridization domains as explained in (B). (D) A plasmid clone hybridized to a P1 clone (#41, blue). The hybridization shows two domains (red) indicating chimerism of the clone. The vector-specific probes (green/red) appeared on the lower left corner of the picture. (Insert) A comparison of hybridization signals recorded from either randomly broken linear or circular DNA molecules. (E–G) Mapping of a BAC clone 97A23 onto a PAC clone 11C11. (E) A circular PAC clone 11C11 DNA fiber (blue) was hybridized with BAC clone 97A23 (red) and PAC vector-specific probes (green/red). (F) A schematic drawing of each hybridization domain and the extent of overlap. (G) A schematic of hybridization domains. The PCR synthesized vector-specific probe (red) is located near the T7 end of the vector pAd10SacBII (green). (H–I) Hybridization of two P1 clones #1107 and #1143 onto YAC clone 141G6. (H) Probes of two P1 clones #1107 and #1143 (red) and the pYAC3 vector (red) were hybridized onto a YAC clone 141G6 DNA fiber (green). The hybridization of the vector probe pYAC3 (red dots at both ends) indicates a complete YAC DNA fiber. (I) A schematic drawing showing the extent of the hybridization domains and the pYAC3 vector probe signals (red) on both ends.
From the position of the vector-specific probes on the BAC #97 DNA fiber, it could be determined that the BAC #131 mapped close to the Sp6 end of the vector in BAC clone #97 (Fig. 1A and B). The overlap between BAC #131 and BAC #97 starts from the SP6 end and extends 73 kb as observed on the circular fiber (Fig. 1C). The hybridization domain of BAC #131 (red) is bordered by the BAC vector (green) on one side, leaving no gap (blue) between BAC #131 and vector-specific signals. Blue signals from the insert of BAC #97 are seen in the region of the non-overlapping domain (Fig. 1C). By comparing the length of the DNA fiber and extent of overlap of the linear DNA molecule (Fig. 1A) to those of the circular one (Fig. 1C), it was concluded that the linear molecule represents the entire length of the BAC #97 DNA fiber with a nick on the insert near the SP6 end of the vector.
QDFM can minimize sequencing efforts by mapping plasmids onto P1 molecules
When applied to sequencing templates comprised of large insert DNA clones, QDFM can yield information regarding the map position of contigs at a resolution of ~1 kb, i.e. a resolution surpassing most other mapping techniques (1). The information includes overlapping clones identification, assessment of the extent of overlap, and identification and characterization of the extent of gaps between adjacent but non-overlapping contigs. With the incorporation of designed vector-specific probes labeled in different colors, these tasks can be achieved more easily. In the following example, a P1 clone (#41) was used for DNA fiber mapping of sequencing templates comprised of plasmid clones. Individual plasmid clones were analyzed and one clone (P1 #41-2d8) showed two separate hybridization signal domains on P1 #41 (Fig. 1D). By incorporating a 1.4 kb PCR-amplified vector-specific probe (F1/B1 fragment, red) and a 17 kb P1 vector probe (pAd10SacBII, green), the relative position and location of the two hybridization domains could be easily determined. The combined size of both hybridization domains is ~5 kb. Figure 1D shows a side-by-side comparison of hybridization of the plasmid and vector probes onto a circular P1 #41 molecule (insert) and a randomly broken P1 #41 molecule. While all circular DNA molecules are full length, the stretching of the linearized molecules is more reproducible allowing a more precise measurement of map position relative to the vector probes. In this example, the two hybridization domains, i.e. the chimerism of the plasmid clone, are readily identified. Information obtained from these hybridization experiments (e.g. chimeric clones or overlapping contigs) can be used to reduce sequencing efforts by identifying mini-mally overlapping clones or contigs and rejecting chimeric clones.
Vector probes improve the accuracy of overlap calculation between BAC and PAC clones
We determined the extent and location of overlap between a BAC clone 97A23 (Clone ID CIT978SKB-97A23, ~130 kb and maps on chromosome 1q) (Fig. 1E, red) and the insert of a PAC molecule (PAC11C11, Fig. 1E, blue). We included a 17 kb vector-specific probe (pAd10SacBII, green) and a PCR-generated probe of ~1.4 kb (F1/B1, red) that hybridized close to the T7 end of the vector (Fig. 1E). Figure 1F shows a schematic representation of the probes and their relative map positions along the ~114 kb PAC molecule. An overlap region of ~80 kb was found near the T7 promoter in the PAC vector. About 17 kb of the PAC insert did not overlap with the BAC and appears blue in Figure 1E. The size of the vector pAd10SacBII (~17 kb, green) probe was used as a crosscheck to confirm how evenly the DNA fibers were stretched. These results are depicted in Figure 1G. The location of the overlap of the PAC with the BAC insert was determined in a separate experiment whereby the PAC insert was mapped onto BAC DNA fibers (data not shown).
Vector-specific probes identify complete versus fragmented DNA molecules and determine the relative position of P1 clones along YAC DNA fibers
To extend the utility of the QDFM to map longer pieces of DNA, two P1 clones (#1107 and #1143) were labeled with digoxigenin and hybridized onto YAC DNA fibers labeled with biotinylated probes. A representative image of a hybridized full-length YAC (clone 141G6, ~490 kb) in Figure 1H shows the locations of the two P1 probes, the extent of the gap between them and the terminal pYAC3 signals. Separate hybridizations of the two P1 probes revealed that clone #1143 mapped close to one end of YAC141G6, while clone #1107 mapped close to the center of the YAC molecule (see illustration in Fig. 1I). These results agree well with the map based on STS content mapping (1).
DISCUSSION
The physical distance between markers on a larger DNA fragment or two DNA clones from the same chromosome and the extent of overlap between two clones are important values for physical map assembly. QDFM has proven to be uniquely able to address these questions in a precise, reliable and inexpensive fashion (1,2,9).
The digoxigenin-labeled, red-visualized PCR-derived BAC vector probe, which we selected to map closer to the T7 than the Sp6 promoter in the BAC vector, allowed us to determine the orientation of the insert of BAC #97 and thus the location of hybridization between two overlapping BAC clones (Fig. 1A–C).
DNA fiber mapping slides may contain a mixture of linear and circular molecules. The stretching of DNA molecules in QDFM varies, and the most reproducible results are obtained with linear molecules. These linear molecules are typically stretched to ~2.3 kb/µm, eliminating the need for internal controls to scale each experiment (1). In a typical fiber mapping, circular DNA molecules appear in a wide range of diameters representing different degrees of stretching. The largest circles stretched to ~2.3 kb/µm, a degree similar to randomly broken linear molecules (9). However, smaller, more condensed molecules can also be analyzed using vector probe as a molecular ruler. As demonstrated in the third example, the 17 kb vector probe pAd10SacBII (green) hybridization domain served as standard for normalization in the calculation of clone overlap for a circular PAC molecule (Fig. 1E–G).
When applied in the hybridization of high molecular weight DNA such as a YAC in QDFM, the vector probes provide additional visual markers across different fields of view. The large size of YAC usually does not permit them in one field of view. Thus, the addition of YAC plasmid vector pYAC3 probes (which binds to both arms of YAC vector) in the hybridization becomes more important for the identification of the entire fiber and hybridization domains. In the last example, pYAC3 vector probe bound to both ends of YAC DNA molecule and enabled the differentiation between complete and fragmented molecules. The distance between two clones became readily apparent by hybridizing probes from both clones onto the DNA fiber. This way, gaps between contigs can be quantitated accurately and rapidly (Fig. 1H and I). Similar to the procedures described above, primers for PCR amplification of vector-specific probes can be designed that anchor on either side of the YAC cloning sites. Such probes for the YAC vectors can then be labeled in different colors and applied in the hybridization mixture to allow rapid DNA fiber orientation and clone identification.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a grant from the Director, Office of Energy Research, Office of Health and Environmental Research, US Department of Energy, under contract DE-AC-03-76SF00098, a training grant from the U.C. Systemwide Biotechnology Research and Education Program (S96-25) and a postdoctoral fellowship from the Cancer Research Foundation of America to H.B.H.
REFERENCES
- 1.Weier H.-U.G., Wang,M., Mullikin,J.C., Zhu,Y., Cheng,J.-F., Greulich,K.M., Bensimon,A. and Gray,J.W. (1995) Hum. Mol. Genet., 4, 1903–1910. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J.-F. and Weier,H.-U.G. (1997) In Fox,C.F. and Connor,T.H. (eds), Biotechnology International. Universal Medical Press, San Francisco, pp. 149–157.
- 3.Duell T., Nielsen,L.B., Jones,A., Wang,M., Young,S.G. and Weier,H.-U.G. (1998) Cytogenet. Cell Genet., 79, 64–70. [DOI] [PubMed] [Google Scholar]
- 4.Duell T., Wang,M., Wu,J., Kim,U.-J. and Weier,H.-U.G. (1997) Genomics, 45, 479–486. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim H.C. and Doly,J. (1979) Nucleic Acids Res., 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins C., Rommens,J.M., Kowbel,D., Godfrey,T., Tanner,M., Hwang,S.-I., Polikoff,D., Nonet,G., Cochran,G., Myambo,K., Jay,K.E., Froula,J., Cloutier,T., Kuo,W.L., Yaswen,P., Dairkee,S., Giovanola,J., Hutchinson,G.B., Isola,J., Kallioniemi,O.-P., Palazzolo,M., Martin,C., Ericsson,C., Pinkel,D., Albertson,D., Li,W.-B. and Gray,J.W. (1998) Proc. Natl Acad. Sci. USA, 95, 8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherman F., Fink,G.R. and Hicks,J.B. (1986) Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 8.Patil N., Peterson,A., Rothman,A., DeJong,P.J., Myers,R.M. and Cox,D.R. (1994) Hum. Mol. Genet., 3, 1811–1817. [DOI] [PubMed] [Google Scholar]
- 9.Wang M., Duell,T., Gray,J.W. and Weier,H.-U.G. (1996) Bioimaging, 4, 1–11. [Google Scholar]
- 10.Shepherd N.S., Pfrogner,B.D., Coulby,J.N., Ackerman,S.L., Vaidyanathan,G., Sauer,R.H., Balkenhol,T.C. and Sternberg,N. (1994) Proc. Natl Acad. Sci. USA, 91, 2629–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke D.T., Carle,G.F. and Olson,M.V. (1987) Science, 236, 806–812. [DOI] [PubMed] [Google Scholar]
- 12.Maniatis T., Fritsch,E.F. and Sambrook,J. (1986) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Weier H.-U., Pinkel,D. and Gray,J. (1995) In Meyers,R.A. (ed.), Molecular Biology and Biotechnology. VCH Verlagsgesellschaft, Weinheim, Germany, pp. 965–968.
- 14.Weier H.-U., Rosette,C., Matsuta,M., Zitzelsberger,H., Matsuta,M. and Gray,J. (1994) Methods Mol. Cell. Biol., 4, 231–248. [Google Scholar]
- 15.Weier H.-U., Polikoff,D., Fawcett,J.J., Greulich,K.M., Lee,K.-H., Cram,S., Chapman,V.M. and Gray,J.W. (1994) Genomics, 24, 641–644. [DOI] [PubMed] [Google Scholar]
- 16.Mascio L.N., Verbeek,P.W., Sudar,D., Kuo,W.L. and Gray,J.W. (1995) Cytometry, 19, 51–69. [DOI] [PubMed] [Google Scholar]