Abstract
Cytosine-5 DNA methylation occurs in the context of CpG dinucleotides in vertebrates. Aberrant methylation of CpG islands in human tumors has been shown to cause transcriptional silencing of tumor-suppressor genes. Most methods used to analyze cytosine-5 methylation patterns require cumbersome manual techniques that employ gel electrophoresis, restriction enzyme digestion, radiolabeled dNTPs or hybridization probes. The development of high-throughput technology for the analysis of DNA methylation would significantly expand our ability to derive molecular information from clinical specimens. This study describes a high-throughput quantitative methylation assay that utilizes fluorescence-based real-time PCR (TaqMan®) technology that requires no further manipulations after the PCR step. MethyLight is a highly sensitive assay, capable of detecting methylated alleles in the presence of a 10 000-fold excess of unmethylated alleles. The assay is also highly quantitative and can very accurately determine the relative prevalence of a particular pattern of DNA methylation. We show that MethyLight can distinguish between mono-allelic and bi-allelic methylation of the MLH1 mismatch repair gene in human colorectal tumor specimens. The development of this technique should considerably enhance our ability to rapidly and accurately generate epigenetic profiles of tumor samples.
INTRODUCTION
The gene expression profile of a cancer specimen is considered a valuable source of biological information with potential clinical utility (1). However, the recovery of sufficient quality and quantity of RNA from clinical samples can present problems. The analysis of cytosine-5 DNA methylation patterns may provide an alternative strategy, since methylation patterns in a tumor cell are thought to reflect, at least in part, the gene expression profile of that cell (2). Cytosine-5 DNA methylation occurs in the context of CpG dinucleotides in vertebrates, and is often associated with transcriptional repression. Interest in DNA methylation has increased with the growing understanding of its involvement in cancer. Transcriptional inactivation of CpG island-containing promoters of tumor suppressor genes by DNA hypermethylation has been well documented in many human cancers (2).
Considerable advances have been made in high-throughput technology for mutation screening and expression profiling. However, the analysis of cytosine-5 methylation patterns still requires cumbersome manual techniques. The development of high-throughput technology for the analysis of this fifth base of the genome would significantly expand our ability to extract molecular information from clinical specimens. The analysis of DNA methylation patterns has been significantly hampered by the fact that methylation information is not retained during amplification steps that form the basis of most standard molecular biology techniques. This includes biochemical amplification, such as PCR, biological amplification by cloning in Escherichia coli, and signal amplification by probe hybridization. Therefore, DNA methylation analysis methods generally rely on a methylation-dependent modification of the original genomic DNA before any amplification step.
The first generation of methylation detection assays employed the digestion of genomic DNA with a methylation-sensitive restriction enzyme followed by either Southern blot analysis or PCR (3). Although these techniques are relatively straightforward, problems such as the limited availability of informative restriction sites, the occurrence of false positive results due to incomplete digestion and the requirement of large amounts of high molecular weight DNA have restricted their use.
A second generation of techniques resulted from the demonstration that treatment of genomic DNA with sodium bisulfite followed by alkaline treatment converts unmethylated cytosines to uracil, while leaving methylated cytosine residues intact (4). Sequence variants at a particular locus can sub-sequently be analyzed by PCR amplification with primers designed to anneal with bisulfite-converted DNA. The sequence differences resulting from various DNA methylation patterns can then be revealed in two principally different ways. Either the discrimination is made at the PCR amplification step by the use of primers that anneal specifically with either the converted methylated or converted unmethylated sequence, or the discrimination is left until after the PCR reaction by the use of PCR primers that do not themselves cover any CpG dinucleotides sites in the original genomic DNA. The first strategy is referred to as methylation-specific PCR (MSP) (5). The latter approach is used by all other bisulfite-based methods. This strategy results in the simultaneous amplification of all sequence variants that may have arisen due to various patterns of DNA methylation in the region located in between the two primers. The prevalence of each of these sequence variants in the pool of PCR products can then be assessed using a variety of standard methods (4,6,7).
The benefit of sodium bisulfite-based assays is that they require very small amounts of DNA and consequently, are compatible with DNA obtained from microdissected paraffin-embedded tissue samples (4–7). However, the existing DNA methylation detection assays require gel electrophoresis and many of them also employ restriction enzyme digestion, radiolabeled dNTPs or hybridization probes. These labor-intensive steps limit the use of these methods for high-throughput analyses. We describe a new methylation assay referred to as MethyLight that is not only highly specific, sensitive and reproducible, but is also compatible with very small amounts of template DNA and allows for rapid analysis of many samples at multiple gene loci (8).
MATERIALS AND METHODS
Sample collection
Twenty-five paired tumor and normal mucosal tissue samples were obtained from 25 patients with primary colorectal adenocarcinoma. The patients comprised 16 males and 9 females, ranging in age from 39 to 88 years, with a mean age of 68.8. Approximately 2 g of the surgically removed tissue was immediately frozen in liquid nitrogen and stored at –80°C until RNA and DNA isolation.
Nucleic acid isolation
Genomic DNA was isolated by the standard method of proteinase K digestion and phenol–chloroform extraction (9). Total RNA was isolated by the single-step guanidinium isothiocyanate method (10).
Sodium bisulfite conversion and COBRA analysis
Sodium bisulfite conversion of genomic DNA and COBRA analysis was performed as previously described (6). The primers and a probe were designed to generate a 174-bp PCR product within the 5′ UTR of the human ESR1 locus. The following are the forward primer, probe and reverse primer sequences: TCCTAAAAACTACACTTACTCCC, GGGTTA-TTTGGAAAAAGAGTATAGTT, GTAGGGTAGAAGGTTTAGAA.
MethyLight reactions
After sodium bisulfite conversion, genomic DNA is amplified by fluorescence-based, real-time quantitative PCR (11,12). In brief, bisulfite-converted genomic DNA is amplified using locus-specific PCR primers flanking an oligonucleotide probe with a 5′ fluorescent reporter dye (6FAM) and a 3′ quencher dye (TAMRA) (13). The 5′ to 3′ nuclease activity of Taq DNA polymerase cleaves the probe and releases the reporter, whose fluorescence can be detected by the laser detector of the ABI Prism 7700 Sequence Detection System (Perkin-Elmer, Foster City, CA). After crossing a fluorescence detection threshold, the PCR amplification results in a fluorescent signal proportional to the amount of PCR product generated. Initial template quantity can be derived from the cycle number at which the fluorescent signal crosses a threshold in the exponential phase of the PCR reaction (8). Serial dilutions of a control sample are included on each plate to generate a standard curve. Several reference samples are included on each assay plate to verify plate-to-plate consistency. Plates are normalized to each other using these reference samples. The PCR amplification is performed using a 96-well optical tray and caps with a final reaction mixture of 25 µl consisting of 600 nM each primer, 200 nM probe, 200 µM each dATP, dCTP and dGTP, 400 µM dUTP, 3.5 mM MgCl2, 1× TaqMan® Buffer A containing a reference dye, and bisulfite-converted DNA or unconverted DNA at the following conditions: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min.
MethyLight primer and probe sequences
Five sets of PCR primers and probes, designed specifically for bisulfite converted DNA sequences, were used: a set representing fully methylated and fully unmethylated DNA for the ESR1 gene, a fully methylated set for the MLH1 gene, and an internal reference set for the MYOD1 and ACTB genes to control for input DNA. The methylated and unmethylated primers and the probe were designed to overlap one to five potential CpG dinucleotides sites. Parallel TaqMan® PCR reactions were performed with primers specific for the bisulfite-converted methylated and/or unmethylated sequences of ESR1 or MLH1 and with MYOD1 or ACTB reference primers. The primer and probe sequences are listed below. In all cases, the first primer listed is the forward PCR primer, the second is the TaqMan® probe and the third is the reverse PCR primer. ESR1 Methylated (GGCGTTCGTTTTGGGATTG, 6FAM5′-CGATAAAACC-GAACGACCCGACGA-3′TAMRA, GCCGACACGCGAA-CTCTAA); ESR1 Unmethylated (ACACATATCCCACCAA-CACACAA, 6FAM5′-CAACCCTACCCCAAAAACCTAC-AAATCCAA-3′TAMRA, AGGAGTTGGTGGAGGGTGTTT); MLH1 Methylated (CTATCGCCGCCTCATCGT, 6FAM5′-CG-CGACGTCAAACGCCACTACG-3′TAMRA, CGTTATA-TATCGTTCGTAGTATTCGTGTTT); MYOD1 (CCAACTCCAAATCCCCTCTCTAT, 6FAM5′ TCCCTTCCTATTCCT-AAATCCAACCTAAATACCTCC-3′TAMRA, TGATTAA-TTTAGATTGGGTTTAGAGAAGGA); ACTB (TGGTGATGGAGGAGGTTTAGTAAGT, 6FAM5′ACCACCACCCAA-CACACAATAACAAACACA-3′TAMRA, AACCAATAA-AACCTACTCCTCCCTTAA).
Quantitative RT–PCR and microsatellite instability analysis
The quantitation of mRNA levels was carried out using real-time fluorescence detection. The TaqMan® reactions were performed as described above for the MethyLight assay, but with the addition of 1 U AmpErase uracil N-glycosylase). After RNA isolation, cDNA was prepared from each sample as previously described (14). Contamination of the RNA samples by genomic DNA was excluded by analysis of all RNA samples without prior cDNA conversion. Relative gene expression was determined based on the threshold cycles of the MLH1 gene and of the internal reference gene ACTB. The forward primer, probe and reverse primer sequences, respectively, of the ACTB and MLH1 genes are listed below. ACTB (TGAGCGCGGCTACAGCTT, 6FAM5′-ACCACCA-CGGCCGAGCGG-3′TAMRA, CCTTAATGTCACACACG-ATT); MLH1 (GTTCTCCGGGAGATGTTGCATA, 6FAM5′-CCTCAGTGGGCCTTGGCACAGC-3′TAMRA, TGGTGG-TGTTGAGAAGGTATAACTTG). Microsatellite instability (MSI) was determined by PCR of the BAT25 and BAT26 loci as previously described (15).
Bisulfite genomic sequencing
The MLH1 promoter region spanning the entire MLH1 MethyLight amplicon was analyzed by bisulfite genomic sequencing as described (4,16). The following primers were used for the PCR amplification: MLH1 forward primer: TTAGGAGTGAAGGAGG, MLH1 reverse primer: GAATTAAACCCTATACCTAA.
RESULTS
The basis of MethyLight technology
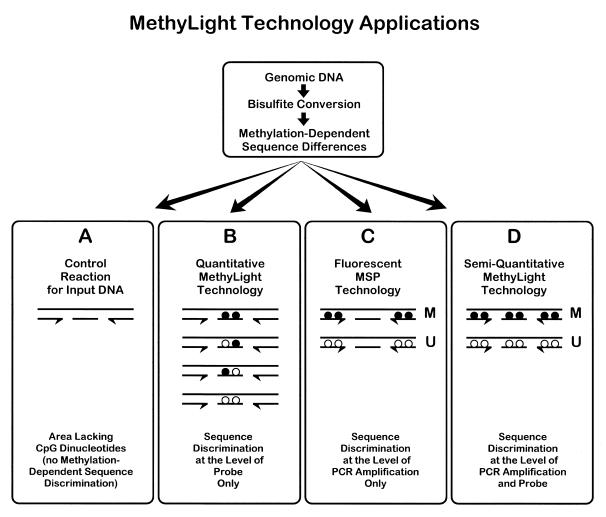
Sodium bisulfite treatment of genomic DNA converts unmethylated cytosines to uracil, while methylated cytosine residues remain unaffected. This modification creates methylation-dependent sequence differences in the genomic DNA. These sequence differences could potentially be detected by fluorescence-based quantitative PCR (12). Various instruments are capable of real-time fluorescence measurement during PCR reactions, including the Roche/Boehringer Mannheim LightCycler (Roche Molecular Biochemicals, Indianapolis, IN) and the PE Biosystems ABI PRISM 7700 and 5700 GeneAmp Sequence Detection Systems (PE Biosystems, Foster City, CA). We used the ABI PRISM 7700 system in combination with TaqMan® technology, which is based on the cleavage of a dual-labeled fluorogenic hybridization probe by the 5′ nuclease activity of Taq polymerase during PCR amplification (8,13,17,18). The use of three different oligonucleotides in the TaqMan® technology (forward and reverse PCR primers and the fluorogenic hybridization probe) offers the opportunity for several sequence detection strategies. The sequence discrimination can occur at the level of the PCR amplification process and/or at the level of the fluorogenic probe hybridization (Fig. 1). In both steps, the discrimination is based on the differential annealing of the perfectly matched, versus mismatched oligonucleotides.
Figure 1.
Schematic of the theoretical basis of MethyLight technology. Genomic DNA is first chemically modified by sodium bisulfite. This generates methylation-dependent sequence differences at CpG dinucleotides by converting unmethylated cytosine residues (locations indicated by white circles) to uracil, while methylated cytosine residues (locations indicated by black circles) are retained as cytosine. Fluorescence-based PCR is then performed with primers that either overlap CpG methylation sites or that do not overlap any CpG dinucleotides. Sequence discrimination can occur either at the level of the PCR amplification process or at the level of the probe hybridization process, or both. Sequence discrimination at the PCR amplification level requires the primers and probe (application D), or just the primers (application C), to overlap potential methylation sites (CpG dinucleotides). Only two [fully methylated (M) and fully unmethylated (U)] of the many theoretical methylation permutations are shown. The MethyLight assay can also be designed such that sequence discrimination does not occur at the PCR amplification level. If neither the primers nor the probe overlap sites of CpG dinucleotides (application A), then no methylation-dependent sequence discrimination occurs at the PCR amplification or probe hybridization level. This reaction represents amplification of the converted genomic DNA without bias to methylation status, which can serve as a control for the amount of input DNA. When just the probe overlaps methylation sites (application B), then sequence discrimination can occur through probe hybridization. The design of separate probes for each sequence variant resulting from different methylation patterns (22 = 4 probes in the case of two CpGs, as illustrated) can potentially serve as a quantitative version of the MethyLight technology.
In the MethyLight technology, sequence discrimination at the PCR amplification level occurs by designing the primers and probe (Fig. 1, application D), or just primers (Fig. 1, application C), to overlap potential sites of DNA methylation (CpG dinucleotides). Application C in Figure 1 represents a fluorescence-based version of the MSP technique (5). Each oligonucleotide (primers and probe) can cover anywhere from zero to multiple CpG dinucleotides. Each CpG dinucleotide can result in two different sequence variations following bisulfite conversion, depending on whether that particular site was methylated (mCpG) or unmethylated (UpG). For example, in application D, if an oligo overlaps two CpG dinucleotides, then the number of possible sequence variants in the genomic DNA within the region covered by that oligo is 22 = 4. If both of the primers and the probe each overlap two CpGs, then the total number of variants contained within the sequence covered by the oligonucleotides is 4 × 4 × 4 = 64. In theory, one could design separate PCR reactions to analyze the relative amounts of each of these potential 64 sequence variants. However, significant methylation information can be derived from the analysis of a much smaller number of variants by designing reactions for the fully methylated and fully unmethylated molecules, which represent the two most extreme sequence variants in our hypothetical example (Fig. 1, application D). The ratio between these two reactions or, more reliably, the ratio between the methylated reaction and a control reaction would provide a measure for the prevalence of methylated molecules at this locus.
The MethyLight technology can also be modified to avoid sequence discrimination at the PCR amplification level (Fig. 1, applications A and B). If the neither the primers nor the probe overlie any CpG dinucleotides, then the reaction represents unbiased amplification and can serve as a control for the amount of input DNA (Fig. 1, application A). The ideal control reaction is one in which the entire amplicon is devoid of any CpG dinucleotides in the unconverted genomic sequence.
When just the probe is designed to cover CpG dinucleotides (Fig. 1, application B), then sequence discrimination occurs solely at the level of probe hybridization. In this version, all sequence variants resulting from the sodium bisulfite conversion step are amplified with equal efficiency, as long as there is no amplification bias (19). In this case, the design of separate probes for each of the different sequence variants associated with a particular methylation pattern (22 = 4 probes in the case of two CpGs) would allow a quantitative determination of the relative prevalence of each sequence permutation in the mixed pool of PCR products.
Validation of MethyLight technology
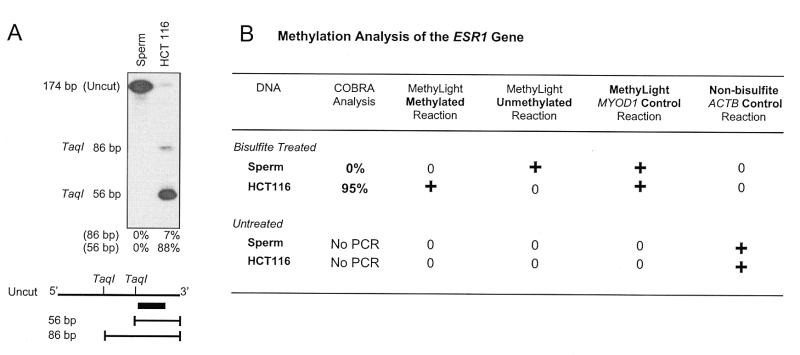
We first used an independent method to determine the methylation status of the CpG island associated with the estrogen receptor (ESR1) in the human colorectal cell line HCT116 and in human sperm DNA. This CpG island has been reported to be highly methylated in HCT116 and unmethylated in human sperm DNA (6,20). We used an established method (COBRA; 6) to confirm this. COBRA (COmbined Bisulfite Restriction Analysis) relies on restriction digestion to reveal sequence differences resulting from the bisulfite conversion of methylated, versus unmethylated CpG dinucleotides (6). COBRA analysis of two TaqI sites within the ESR1 CpG island showed a lack of methylation in the sperm DNA and nearly complete methylation in HCT116 DNA (Fig. 2A).
Figure 2.
Comparison of the MethyLight assay to a conventional COBRA assay. (A) COBRA gel used to quantitatively determine the level of DNA methylation at the ESR1 locus in DNAs of known methylation status [sperm (unmethylated) and HCT116 (methylated)] The relative amounts of the cleaved products are indicated below the gel. The 56-bp fragment represents DNA molecules in which the TaqI site proximal to the hybridization probe (black box) is methylated in the original genomic DNA. The 86-bp fragment represents DNA molecules in which the proximal TaqI site is unmethylated and the distal site is methylated. (B) A summary of the COBRA results comparing them to the absolute results obtained with the methylated and unmethylated version of the MethyLight assay (application D in Fig. 1). +, a positive absolute value determined by the MethyLight assay; 0, a lack of detectable MethyLight amplification. For the bisulfite-treated samples, the MYOD1 MethyLight assay was performed in parallel to control for the amount of input DNA (application A in Fig. 1). For the untreated samples, the ACTB primers described for the RT–PCR reactions were used as a control to verify the input of unconverted DNA samples. (The ACTB primers do not span an intron.) No PCR indicates that, as expected, no PCR product was obtained on unconverted genomic DNA with COBRA primers designed to amplify bisulfite-converted DNA sequences.
We selected two of the four possible MethyLight applications (applications A and D in Fig. 1) to determine the methylation status of the same 5′ region within the ESR1 gene in the previously analyzed sperm and HCT116 DNA. We designed the forward and reverse primers and the fluorogenic probe to discriminate between either fully methylated or fully unmethylated molecules of bisulfite converted DNA (application D in Fig. 1). We also designed oligos for a stretch of the MYOD1 gene completely devoid of CpG dinucleotides as a control reaction for the amount of input DNA (application A in Fig. 1). We performed three separate MethyLight reactions using either the methylated, unmethylated or control oligos on both sperm and HCT116 DNA. Sperm DNA yielded a positive MethyLight reaction with the unmethylated primers and probe, while there was no detectable value for the methylated reaction, consistent with its unmethylated status. In contrast, HCT116 DNA with predominantly methylated ESR1 alleles gave a positive reaction in the methylated reaction, but not in the unmethylated reaction (Fig. 2B). Both the sperm and HCT116 DNA were positive for their MYOD1 reactions, indicating that there was sufficient input DNA in each sample. Neither bisulfite-treated DNA sample gave any detectable signal with oligos designed to recognize a non-bisulfite converted ACTB control sequence. This confirms that the bisulfite conversion process had been successful. As expected, we were unable to amplify non-bisulfite converted DNA with either the methylated, the unmethylated, or with the control oligonucleotides (Fig. 2B). These results are consistent with the COBRA findings above, suggesting that the MethyLight assay can clearly discriminate between the methylated and unmethylated alleles of the ESR1 gene. In addition, the MethyLight reactions are specific to bisulfite-converted DNA, which precludes the generation of false positive results caused by incomplete bisulfite conversion.
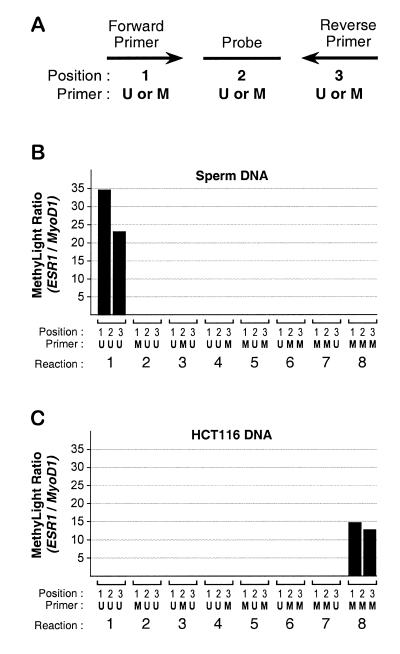
We tested all possible combinations of primers and probes to further examine the specificity of the methylated and unmethylated oligonucleotides. The ESR1 methylated and unmethylated forward and reverse primers and probe were tested in different combinations in MethyLight assays on sperm and HCT116 DNA in duplicate. All reaction values were normalized to the MYOD1 control reaction run in parallel. The resulting ratios are shown in the bar chart in Figure 3. We found that only the fully unmethylated (reaction 1) or fully methylated combinations (reaction 8) resulted in positive MethyLight ratios for the sperm and HCT116, respectively. The other combinations were negative, suggesting that the PCR conditions do not allow for weak annealing of the mismatched oligonucleotides. This selectivity indicates that the MethyLight technology can discriminate between fully methylated or unmethylated alleles with a high degree of specificity.
Figure 3.
Determination of the specificity of the MethyLight oligonucleotides. Eight different combinations of forward primer, probe and reverse primer were tested on DNA samples with known methylation or lack of methylation at the ESR1 locus. (A) The nomenclature used for the combinations of the ESR1 oligos. U refers to the oligo sequence that anneals with bisulfite-converted unmethylated DNA, while M refers to the methylated version. Position 1 indicates the forward PCR primer, position 2 the probe and position 3 the reverse primer. The combinations used for the eight reactions are shown below each pair of bars, representing duplicate experiments. The MethyLight results are expressed as ratios between the ESR1 values and the MYOD1 control values. (B) An analysis of human sperm DNA. (C) An analysis of DNA obtained from the human colorectal cancer cell line HCT116.
Sensitivity and quantitative accuracy of MethyLight technology
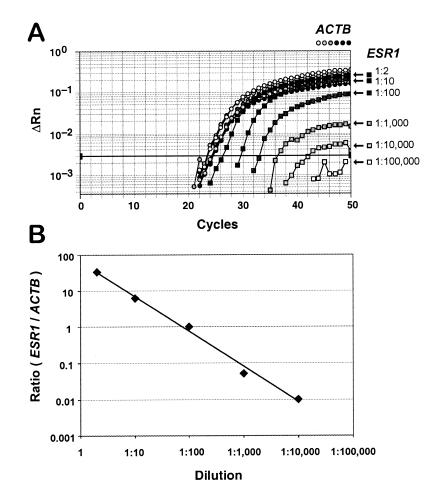
We performed a dilution experiment to determine the detection limits and quantitative accuracy of the MethyLight assay. Human sperm DNA that had been fully methylated by treatment with SssI methyltransferase in vitro was serially diluted with untreated, unmethylated human sperm DNA. An ACTB (β-actin) MethyLight control reaction was included to determine the total amounts of input DNA in each dilution. The ESR1 methylated reaction was used to track the decreasing amount of methylated DNA in the dilution series. The amplification plot resulting from this experiment is shown in Figure 4. All of the samples contained approximately equal amounts of DNA, as is evident from the overlapping ACTB curves indicated by the circles. On the other hand, the ESR1 reaction shows a decreasing detection of methylated alleles, as indicated by the increasing cycle number at which each reaction crosses the threshold. ESR1 methylation can be detected reliably in the presence of a 10 000-fold excess of unmethylated alleles. This is at least 10-fold more sensitive than reported for the highly sensitive MSP technology (5). This greater sensitivity results in an improved ability to detect aberrant methylation patterns in human samples with substantial contamination of normal DNA, such as non-microdissected, heterogeneous tissue samples. We calculated the ESR1/ACTB ratios obtained for each dilution to determine the quantitative accuracy of the MethyLight technology. These ratios are shown in Figure 4B, plotted against the dilution factor. It is clear from this figure that the technique is linearly quantitative over four orders of magnitude. Once again, this is a far greater range of quantitative accuracy than reported for any of the other DNA methylation analysis methods.
Figure 4.
Test of the sensitivity and quantitative accuracy of the MethyLight technique. Human sperm DNA that had been fully methylated by treatment with SssI methyltransferase in vitro was serially diluted in 10-fold increments up to 1:100 000 with untreated, unmethylated human sperm DNA. (A) Subsequent ESR1 MethyLight analysis of these samples, in which the increasing dilutions are indicated by decreasing shades of gray squares, as shown on the right. An ACTB MethyLight control reaction (circles) was included to determine the total amounts of input DNA in each dilution. The shade of gray of the circles corresponds to the dilutions shown for the squares on the right. The relative fluorescence (ΔRn) is plotted as a function of cycle number. The threshold used for the calculation of initial template amounts is indicated by the dark horizontal line (Materials and Methods). (B) ESR1/ACTB ratios obtained for each dilution, plotted against the dilution factor.
Reproducibility of the MethyLight assay on heterogeneous samples
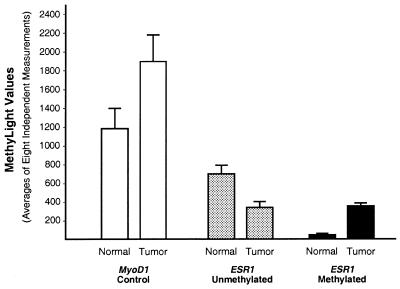
We analyzed the methylation status of the ESR1 locus in DNA samples derived from a colorectal adenocarcinoma and matched normal mucosa derived from the same patient in order to study a heterogeneous population of methylated and unmethylated alleles. In addition, we tested the reproducibility of the MethyLight assay by performing eight independent reactions for each assay. The results for the ESR1 reactions and for the MYOD1 control reaction, shown in Figure 5, represent absolute values obtained for these reactions, rather than ratios, so that the standard errors of the individual reactions can be evaluated. The MYOD1 control reaction was designed to be impervious to the methylation status of the various different genomic DNA samples (Fig. 1, application A), and thus reflect the amount and integrity of input genomic DNA. We found that the mean value for the methylated reaction was higher in the tumor compared to the normal tissue whereas the unmethylated reaction showed the opposite result. The standard errors observed for the eight independent measurements are relatively modest and are comparable to those reported for other studies utilizing TaqMan® technology (18). Some of the variability of the MethyLight assay may be a result of stochastic PCR amplification, which can occur at low template concentrations (19). In summary, these results suggest that the MethyLight assay can yield reproducible results for complex, heterogeneous DNA samples.
Figure 5.
Test of the reproducibility of the Methylight reactions. MethyLight assays were performed in eight independent reactions to determine the reproducibility on samples of complex origin. A primary human colorectal adenocarcinoma and matched normal mucosa was used for this purpose (samples 10N and 10T shown in Fig. 6). The results shown in this figure represent the raw values obtained in the MethyLight reaction. The values have been plate-normalized (Materials and Methods), but not corrected for input DNA. The bars indicate the mean values obtained for the eight separate reactions. The error bars represent the standard error of the mean.
Application of the MethyLight assay
The main benefit of the MethyLight technology would be the ability to rapidly screen human tumors for the methylation state of a particular locus. We tested this by interrogating the methylation status of the MLH1 promoter in human colorectal adenocarcinomas. The mismatch repair gene MLH1 plays a pivotal role in the development of sporadic cases of mismatch repair-deficient colorectal tumors (21). It has been reported that MLH1 can become transcriptionally silenced by DNA hypermethylation of its promoter region, leading to MSI (22–26).
We analyzed 50 samples consisting of 25 matched pairs of human colorectal adenocarcinomas and normal mucosa for the methylation status of the MLH1 CpG island. We also performed real-time quantitative RT–PCR (TaqMan®) analyses of the expression levels of MLH1 normalized to ACTB. Furthermore, we analyzed the microsatellite instability (MSI) status of each sample by PCR of the BAT25 and BAT26 loci (15). Figure 6A shows the correlation between MLH1 gene expression, MSI status and promoter methylation of MLH1, as determined by the MethyLight assay. In most tissue samples, we observed a varying amount of MLH1 expression, which appears to be unrelated to the associated levels of MLH1 methylation, and is presumably due to sample variables, such as cell-type composition, mitotic index, grade, stage, anatomic subsite, age of the subject, etc. Four colorectal tumors have significantly elevated methylation levels compared to the corresponding normal tissue. One of these (tumor 17) exhibits a particularly high degree of MLH1 methylation, as scored by the MethyLight assay. Tumor 17 is the only sample that is both MSI positive (black circles) and that shows transcriptional silencing of MLH1. The remaining methylated tumors express MLH1 at modest levels and are MSI negative (white circles). These results suggest that MLH1 may be biallelically methylated (or mono-allelically methylated with LOH) in tumor 17, resulting in epigenetic silencing and consequent microsatellite instability, whereas the other tumors showing lesser degrees of MLH1 promoter hypermethylation could have just one methylated allele, allowing expression from the unaltered allele. We investigated whether this was indeed the case by performing bisulfite genomic sequencing on samples 12T, 17T and 19T. The results are shown in Figure 6B. It is clear that all alleles appear to be hypermethylated in tumor 17T, and all alleles unmethylated in tumor 19T, as predicted from the MethyLight assay, the expression analysis and the MSI assay. Approximately half of the sequenced alleles were heavily methylated (five out of eight), and half were unmethylated (three out of eight) in sample 12T. This result is consistent with either a mono-allelic methylation pattern of the MLH1 promoter or with a substantial amount of stromal contamination of this tumor sample. We consider the latter explanation to be less likely, since a tumor with bi-allelic methylation of MLH1 that has substantial stromal infiltration would still be expected to show detectable microsatellite instability. We conclude that the MethyLight assay is capable of rapidly and reliably generating significant biological information, such as promoter CpG island hypermethylation in human tumors, which can be associated with the transcriptional silencing of genes relevant to the cancer process.
Figure 6.
Comparison of MLH1 expression, microsatellite instability and MLH1 promoter methylation of 25 matched-paired human colorectal samples. (A) (Upper) MLH1 expression levels measured by quantitative, real time RT–PCR (TaqMan®) in matched normal (hatched bars) and tumor (solid black bars) colorectal samples. The expression levels are displayed as a ratio between MLH1 and ACTB measurements. MSI status is indicated by the circles located between the two charts. A black circle denotes MSI positivity, while an open circle indicates that the sample is MSI negative, as determined by analysis of the BAT25 and BAT26 loci. (Lower) Methylation status of the MLH1 locus as determined by MethyLight assay. The methylation levels are represented as the ratio between the MLH1 methylated reaction and the MYOD1 reaction. (B) Summary of the results of bisulfite genomic sequencing of the 12 CpG sites covered by the three MLH1 MethyLight oligonucleotides for the three tumor samples (12T, 17T and 19T) indicated in (A). Two CpG dinucleotides within the MethyLight amplicon are not covered by any of the three oligos. Eight clones are shown for each sample. Black circles denote methylated cytosines while white circles denote unmethylated cytosines at the indicated CpG dinucleotides. The third CpG in clone 8 of sample 12T could not be conclusively read in that sequence and is indicated by a gray circle.
DISCUSSION
This study reports a novel high-throughput methylation assay that utilizes highly sensitive and accurate fluorescence-based real-time PCR (TaqMan®). We have shown that MethyLight is not only highly specific, sensitive and reproducible, but that it also can rapidly detect biologically relevant information in patient samples. MethyLight is a PCR-based method that requires only minute amounts of DNA of modest quality, making it compatible with small biopsies and paraffin-embedded tissues.
In this initial study, we have not explored all variants of the MethyLight assay as outlined in Figure 1. In particular, application B promises to be a powerful method, since it has the potential to provide quantitative information on the relative prevalence of different sequence variants, representing different methylation patterns, in a pool of PCR products. However, a truly quantitative application of version B in Figure 1 will require a systematic exploration of hybridization efficiencies of probes with varying numbers of CpG mismatches. We expect some cross-reactivity to occur in cases of single mismatches in relatively long oligonucleotide probes. The relatively simple version of MethyLight technology explored here (applications A and D), though less comprehensive, is more cost-effective and capable of rapidly generating biologically relevant information with a minimal amount of manual labor, as shown in Figure 6.
It should be emphasized that the MethyLight technique was not designed to yield high-resolution methylation information, such as the pattern information obtainable with bisulfite genomic sequencing (Fig. 6) or the accurate methylation percentage determination at single CpGs obtainable with COBRA (Fig. 2). Rather, its strongest features are its high-throughput capabilities and its high degree of sensitivity. The assay as we have explored it here is also highly quantitative, but in a different way than traditional methylation analysis methods are. Rather than capturing all methylation occurences of a CpG dinucleotide in a heterogeneous genomic DNA sample, the MethyLight technique can very accurately determine the relative prevalence of a particular pattern of DNA methylation. However, in doing so, the technique is oblivious to all other methylation permutations. MethyLight is similar in this respect to MSP (5), but differs in that it determines the relative amounts of a particular methylation pattern with quantitative accuracy. MSP is an endpoint analysis technique, whereas the quantitative nature of MethyLight is based on the cycle number at which the fluorescent signal crosses a threshold in the exponential phase of the PCR reaction. This allows quantitative conclusions to be drawn concerning methylation levels relative to a control reaction as shown for MLH1 in Figure 6. This would not have been possible with a standard MSP reaction.
The most striking advantage of MethyLight, as compared to existing techniques, is its potential to allow the rapid screening of hundreds to thousands of samples. Unlike other techniques, the MethyLight assay is completed at the PCR step, without the need for further gel electrophoretic separation or hybridization. This reduces the chance of sample contamination and error, and dramatically decreases the amount of labor involved in DNA methylation analysis. The technique is also extremely rapid. We recently reported a relatively small-scale MethyLight analysis of four CpG islands in 50 human tissue samples, for a total of 200 methylation analyses (8). The methylation analysis for this study was accomplished in <3 days, including the bisulfite conversion step. Once the sodium bisulfite conversion step has been performed, a complete determination of the methylation status can be obtained in <2 h of 96 separate gene loci for a single sample, or of a single locus in 96 separate DNA samples. The development of this technique should considerably enhance our ability to generate epigenome maps of tumor samples. As such, it should extend and complement ongoing efforts to determine molecular profiles of tumor samples using high-throughput genomic and RNA-based technologies.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dennis Salonga and Ji Min Park for help in generating cDNAs and in designing TaqMan® oligonucleotides. This work was supported by NIH/NCI grants R01 CA 71716 (P.V.D) and R01 CA 75090 (P.W.L.). P.W.L. is a founding shareholder of ORCA Biosciences, Inc., Seattle, WA, which has a commercial interest in DNA methylation research. The research described in this paper was not supported by ORCA Biosciences.
REFERENCES
- 1.Kononen J., Bubendorf,L., Kallioniemi,A., Barlund,M., Schraml,P., Leighton,S., Torhorst,J., Mihatsch,M.J., Sauter,G. and Kallioniemi,O.P. (1998) Nat. Med., 4, 844–847. [DOI] [PubMed] [Google Scholar]
- 2.Jones P.A. and Laird,P.W. (1999) Nat. Genet., 21, 163–167. [DOI] [PubMed] [Google Scholar]
- 3.Singer-Sam J., Le Bon,J.M., Tanguay,R.L. and Riggs,A.D. (1990) Nucleic Acids Res., 18, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frommer M., McDonald,L.E., Millar,D.S., Collis,C.M., Watt,F., Grigg,G.W., Molloy,P.L. and Paul,C.L. (1992) Proc. Natl Acad. Sci. USA, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herman J.G., Graff,J.R., Myohanen,S., Nelkin,B.D. and Baylin,S.B. (1996) Proc. Natl Acad. Sci. USA, 93, 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong Z. and Laird,P.W. (1997) Nucleic Acids Res., 25, 2532–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalgo M.L. and Jones,P.A. (1997) Nucleic Acids Res., 25, 2529–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eads C.A., Danenberg,K.D., Kawakami,K., Saltz,L.B., Danenberg,P.V. and Laird,P.W. (1999) Cancer Res., 59, 2302–2306. [PubMed] [Google Scholar]
- 9.Wolff R.K., Frazer,K.A., Jackler,R.K., Lanser,M.J., Pitts,L.H. and Cox,D.R. (1992) Am. J. Hum. Genet., 51, 478–485. [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski P. and Sacchi,N. (1987) Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 11.Gibson U.E., Heid,C.A. and Williams,P.M. (1996) Genome Res., 6, 995–1001. [DOI] [PubMed] [Google Scholar]
- 12.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 13.Livak K.J., Flood,S.J., Marmaro,J., Giusti,W. and Deetz,K. (1995) PCR Methods Appl., 4, 357–362. [DOI] [PubMed] [Google Scholar]
- 14.Bender C.M., Pao,M.M. and Jones,P.A. (1998) Cancer Res., 58, 95–101. [PubMed] [Google Scholar]
- 15.Parsons R., Myeroff,L.L., Liu,B., Willson,J.K., Markowitz,S.D., Kinzler,K.W. and Vogelstein,B. (1995) Cancer Res., 55, 5548–5550. [PubMed] [Google Scholar]
- 16.Gonzalgo M.L., Bender,C.M., You,E.H., Glendening,J.M., Flores,J.F., Walker,G.J., Hayward,N.K., Jones,P.A. and Fountain,J.W. (1997) Cancer Res., 57, 5336–5347. [PubMed] [Google Scholar]
- 17.Lee L.G., Connell,C.R. and Bloch,W. (1993) Nucleic Acids Res., 21, 3761–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fink L., Seeger,W., Ermert,L., Hanze,J., Stahl,U., Grimminger,F., Kummer,W. and Bohle,R.M. (1998) Nat. Med., 4, 1329–1333. [DOI] [PubMed] [Google Scholar]
- 19.Warnecke P.M., Stirzaker,C., Melki,J.R., Millar,D.S., Paul,C.L. and Clark,S.J. (1997) Nucleic Acids Res., 25, 4422–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Issa J.P., Ottaviano,Y.L., Celano,P., Hamilton,S.R., Davidson,N.E. and Baylin,S.B. (1994) Nat. Genet., 7, 536–540. [DOI] [PubMed] [Google Scholar]
- 21.Thibodeau S.N., Bren,G. and Schaid,D. (1993) Science, 260, 816–819. [DOI] [PubMed] [Google Scholar]
- 22.Kane M.F., Loda,M., Gaida,G.M., Lipman,J., Mishra,R., Goldman,H., Jessup,J.M. and Kolodner,R. (1997) Cancer Res., 57, 808–811. [PubMed] [Google Scholar]
- 23.Ahuja N., Mohan,A.L., Li,Q., Stolker,J.M., Herman,J.G., Hamilton,S.R., Baylin,S.B. and Issa,J.P. (1997) Cancer Res., 57, 3370–3374. [PubMed] [Google Scholar]
- 24.Cunningham J.M., Christensen,E.R., Tester,D.J., Kim,C.Y., Roche,P.C., Burgart,L.J. and Thibodeau,S.N. (1998) Cancer Res., 58, 3455–3460. [PubMed] [Google Scholar]
- 25.Herman J.G., Umar,A., Polyak,K., Graff,J.R., Ahuja,N., Issa,J.P., Markowitz,S., Willson,J.K., Hamilton,S.R., Kinzler,K.W., Kane,M.F., Kolodner,R.D., Vogelstein,B., Kunkel,T.A. and Baylin,S.B. (1998) Proc. Natl Acad. Sci. USA, 95, 6870–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veigl M.L., Kasturi,L., Olechnowicz,J., Ma,A.H., Lutterbaugh,J.D., Periyasamy,S., Li,G.M., Drummond,J., Modrich,P.L., Sedwick,W.D. and Markowitz,S.D. (1998) Proc. Natl Acad. Sci. USA, 95, 8698–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]