Abstract
We describe a simple procedure for photolithographic patterning of streptavidin on silicon substrates. Long wavelength UV (365 nm) light was used to direct the covalent attachment of photoactivatable biotin onto silylated silicon wafers. Fluorescently labeled streptavidin was found to bind only in areas exposed to the light. We used this procedure to selectively pattern streptavidin inside microwells etched in silicon, and we investigated the binding characteristics of biotinylated oligonucleotides of lengths, n = 16, 54 and 99 bases. The binding curves were found to fit the functional form of the Langmuir isotherm, with binding saturation proportional to n–3/4.
INTRODUCTION
Immobilization of biomolecules on solid surfaces has considerable applications in biosensor design. Numerous techniques have been developed to anchor proteins and nucleic acids onto silicon and glass, but few procedures exist to selectively pattern the biomolecules at specific sites on the substrate. Fodor et al. have used a light-directed chemical approach to directly synthesize peptide and oligonucleotide sequences on glass substrates (1,2). The sequence length is limited to short peptides and oligonucleotides because the failed sequences cannot be removed from the substrate, and they accumulate after each round of coupling. Chrisey et al. describe two procedures for the self-assembly of DNA oligonucleotides from solution onto patterned organosilanes (3). This approach is attractive in that oligonucleotides can be purified before surface attachment.
In this paper we present a simple approach to biomolecular patterning which requires fewer steps than the approach of Chrisey et al., and relatively unsophisticated technology. Our method involves the use of photoactivatable biotin, a biotin analog that covalently attaches to nearby organic moieties upon irradiation with 365 nm light. A photolithographic mask was used to pattern photoactivatable biotin onto silylated silicon surfaces. Streptavidin was discretely immobilized onto the surface through interactions with the immobilized biotin; half of the biotin-binding sites of streptavidin interact with the immobilized biotin, leaving two additional free sites to bind biotinylated ligands such as oligonucleotides. We used this method to pattern streptavidin inside 2 mm × 2 mm × 200 µm wells etched in silicon. Biotinylated oligonucleotides could successfully bind to the patterned streptavidin, and the oligonucleotide binding capacity was found to be length dependent. The data presented here should be very useful to those interested in immobilizing single-stranded DNAs such as oligonucleotides or PCR amplicons less than 100 bases long.
MATERIALS AND METHODS
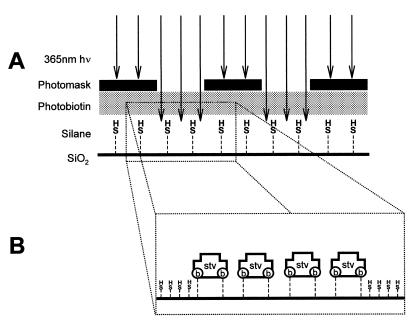
All references to H2O refer to 0.22 µm filtered, deionized (>18 MΩ-cm) water. A thermal oxide layer was grown on <100> silicon wafers by heating to 1000°C for 24 h. The wafers were cleaned prior to silanization by immersion in H2O: H2O2:NH4OH (5:1:1) heated to 70–75°C for 1 h, followed by an H2O rinse, boiled in H2O for ∼5 min, then allowed to air dry. A 2% solution of (3-mercaptopropyl)-trimethoxysilane (MPTS; Sigma, St Louis, MO) in 95% aqueous methanol acidified to pH 5.0 with acetic acid was deposited onto the wafers and allowed to evaporate. The wafers were rinsed in anhydrous methanol then baked at 110°C for 4 h. 1-[4-azidosalicylicylamido]-6-[biotinamido]-hexane (Pierce, Rockford, IL), herein referred to as photoactivatable biotin, was covalently linked to the MPTS layer upon exposure to long wavelength UV light. Photoactivatable biotin was taken up in anhydrous methanol to a final concentration of 2 µg/ml; 1 ml of this solution was deposited onto the MPTS-modified wafer and left to evaporate (this step was carried out in the dark). A standard photolithographic mask used in the microelectronics industry was placed over the wafer and exposed for 5 min to 365 nm light from a transilluminator (Fig. 1A). The wafer was washed for 15 min in dimethyl sulfoxide, followed by a brief H2O rinse, then incubated at 37°C overnight in blocking solution [PBS (137 mM NaCl, 2.68 mM KCl, 10.1 mM Na2HPO4, 1.76 mM KH2PO4, pH 7.4), 0.1% sodium dodecyl sulfate (SDS), 0.1 mg/ml bovine serum albumin]. Following a wash in H2O, the wafer was incubated in 10 µg/ml fluorescently labeled (Cy2) streptavidin (Amersham Pharmacia Biotech, Arlington Heights, IL) in PBS for 30 min in the dark, washed extensively with PBS/0.1% SDS, followed by a rinse in H2O, and then allowed to air dry. The tetrameric structure of streptavidin allowed for protein immobilization via half of its biotin binding sites, leaving the remaining half accessible to biotinylated molecules. A schematic representation of micropatterned streptavidin is shown in Figure 1B. Figure 2A shows the results of the Cy2-streptavidin patterning process imaged with a Zeiss Axiovert microscope equipped with an air cooled back fin illuminated CCD camera (Princeton Instruments, Trenton, NJ).
Figure 1.
(A) Side view of scheme used for photochemical attachment of photoactivatable biotin on MPTS-functionalized SiO2. A photomask is used to direct UV light to specific areas of the substrate. (B) Proposed orientation of immobilized streptavidin.
Figure 2.
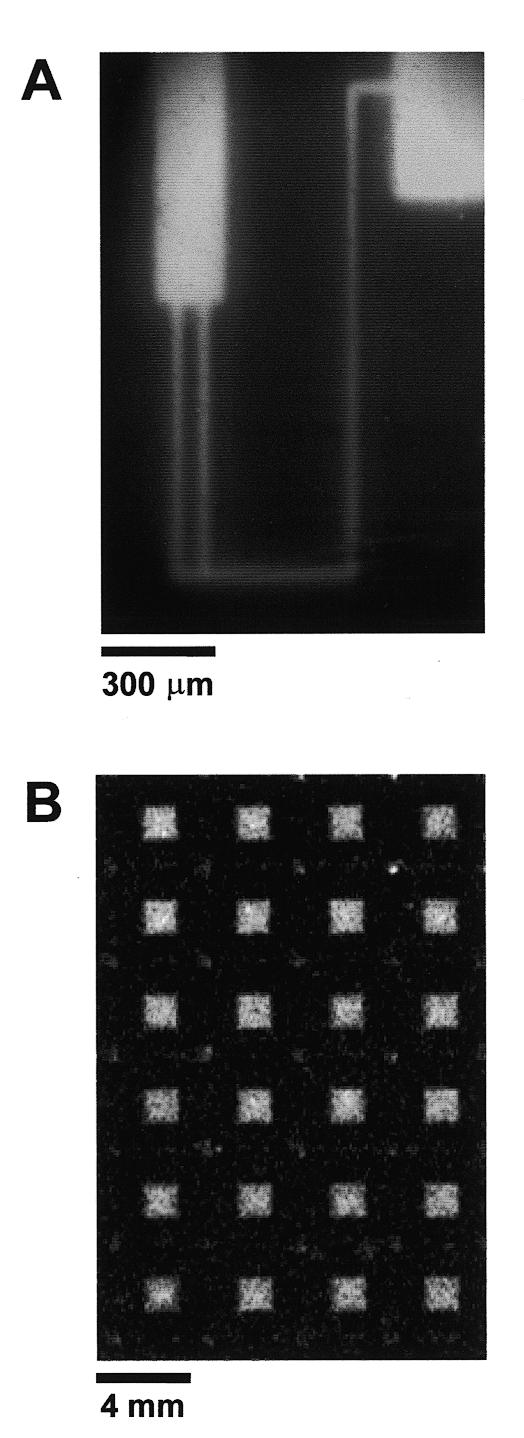
Micropatterned streptavidin surfaces. (A) Fluorescence micrograph of Cy2-labeled streptavidin patterned on a flat MPTS-functionalized SiO2 surface. Fluorescent streptavidin is shown in white. (B) Fluorescent scan of Cy2-streptavidin-coated microwells. Each microwell is 2 × 2 mm2 × 200 µm deep, and spaced 2 mm apart.
The binding properties of the immobilized streptavidin were investigated by measuring its ability to bind radiolabeled biotinylated oligonucleotides. Because the feature size used to create the pattern in Figure 2A would be too small to resolve using isotopic imaging methods, we characterized the oligonucleotide binding capacity of the streptavidin surfaces by creating streptavidin-coated microwells. The microwells measured 2 mm × 2 mm × 200 µm deep and were etched in crystalline <100> silicon by standard wet chemical methods (4). After etching, a thermal oxide layer was grown on the wafers by heating to 1000°C for 24 h. The etched wafers were patterned with streptavidin as described above, utilizing the same photolithographic mask used to fabricate the microwells. The Cy2-streptavidin-coated microwells were scanned on a Molecular Dynamics STORM 860 imager (Fig. 2B).
The binding capacity of the streptavidin microwells was determined using three different length oligonucleotides containing a biotin moiety at the second nucleotide position from the 5′ terminus (Operon Technologies, Alameda, CA). The sequences are: 16mer 5′-A[biotin-A]GTACGGACGCTTCG-3′; 52mer 5′-A[biotin-A]GAACACTCAAATCAATGAACTGGAAACTGAAGAGGAAAACACTGATGACA-3′; 99mer 5′-A[biotin-A]GAACACTCAAATCAATGAACTGGACGTACAATAGACGCACATCGCCTACTAACAGAATCACA-TGACACACCAACTGAAGAGGAAAACACTGATGACA-3′. T4 polynucleotide kinase (New England Biolabs, Beverly, MA) was used to label the 5′ termini with [γ-32P]ATP (3000 Ci/mmol; Amersham Pharmacia Biotech). A 50 µl kinase reaction consisted of 10 pmol of oligonucleotide, 50 µCi [γ-32P]ATP and 20 U of kinase taken up in buffer supplied by the manufacturer. The reaction was incubated at 37°C for 1 h, followed by heat inactivation of the kinase at 95°C for 15 min. Oligonucleotides were purified from unincorporated [γ-32P]ATP using a spin column (ChromaSpin –10 + TE). Various concentrations of 32P-labeled oligonucleotides were suspended in TE (10 mM Tris–HCl, pH 8/1 mM EDTA) to a final specific activity of 5000 c.p.m./pmol. Additionally, oligonucleotides were suspended in TE supplemented with 0.1 and 1.0 M NaCl to test the influence of salt on oligonucleotide immobilization.
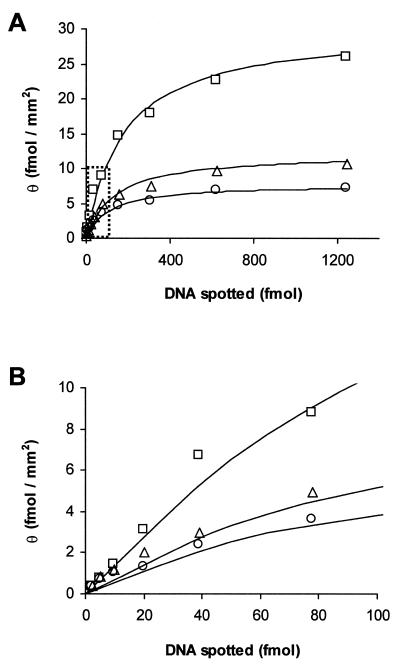
The three different lengths of 32P-labeled oligonucleotides were immobilized inside the microwells under different printing concentrations ranging from 5 to 2500 fmol/µl. Printing was performed by depositing 500 nl of oligonucleotide solutions in each of the microwells using a standard 2 µl pipetman. After evaporation, the unoccupied biotin binding sites were saturated by placing the microwells upside down for 15 min in 50 µM biotin suspended in SPE (0.1 M Na-phosphate, pH 6.6/1 mM EDTA/1 M NaCl). Non-specifically bound oligonucleotides were removed by three 15 min washes in SPE buffer at 37°C. A standard curve was determined by printing similar solutions in additional microwells, but without washing. The wafers containing the samples were exposed to a phosphorimaging plate and imaged using a Molecular Dynamics STORM 860 imager. The oligonucleotide binding capacities of the washed microwells were determined by comparison with the standard curve using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). Figure 3 shows that the binding capacity of oligonucleotides in the streptavidin-coated microwells increases as oligonucleotide length decreases. The salt concentration did not affect the binding characteristics of any of the oligonucleotides studied (data not shown).
Figure 3.
(A) Binding capacity of oligonucleotides versus amounts added for 99 base (circles), 54 base (triangles) and 16 base (squares) lengths. (B) Expanded view of dotted section of (A). The solid curves are Langmuir isotherm functions fitted to the experimental data (Discussion).
DISCUSSION
Micropatterned streptavidin surfaces should have utility in a wide variety of applications including DNA and antibody arrays, and nanofabrication. There are three main advantages of printing biotinylated molecules onto streptavidin supports as opposed to a direct synthesis approach. Because the coupling efficiency of even the best peptide or oligonucleotide synthesis is typically between 98 and 99%, it is desirable to remove failed sequences prior to arraying, particularly for long chain lengths. Printed oligonucleotides may be immobilized by either their 5′- or 3′-biotinylated ends. Our methods are also compatible with printing complex structures such as DNA duplexes and triplexes, nucleic acid hybrids, antibodies, etc., whereas only single chain molecules can be synthesized directly on a support.
In this investigation we determined the binding characteristics of oligonucleotides inside streptavidin-coated microwells. The binding curves given in Figure 3 were fitted to the functional form of the Langmuir isotherm
θ = θmax kN/(1+kN)
where θ is the number of bound oligonucleotides (fmol/mm2), N is the total number of oligonucleotides added to each well (fmol) and k is a measure of the effectiveness of the attachment reaction under the conditions used. The maximum oligonucleotide binding capacity, θmax, is dependent on n, the oligonucleotide length in bases by,
θmax = 242 n–3/4 (fmol/mm2)
and k is dependent on n by
k = 5 × 10–5 n + 0.0047 (fmol–1).
The relation between θmax and n is probably due to steric crowding as the chain length increases. The increase in k as a function of n is not expected. It may reflect weak non-specific binding that facilitates the ultimate biotin-specific attachment.
In a previous study we investigated the binding of biotin and biotinylated oligonucleotides onto homogeneous (i.e., not patterned) streptavidin surfaces in solution (unpublished data), and note two major differences between oligonucleotide binding from solution versus the procedures used here. For binding experiments in solution, there appeared to be no chain length dependence on θmax; in fact, there were no discernable differences between the binding capacity of streptavidin supports for free biotin and a 99 base biotinylated oligonucleotide. In addition, we noted a strong salt dependence on oligonucleotide binding in solution; 1 M NaCl was required to achieve maximum oligonucleotide binding, and minimal binding was observed when salt was omitted from the solution (<2% of θmax), most likely due to charge repulsion from the first bound oligonucleotides preventing additional oligonucleotides from approaching the surface. In sharp contrast, we did not observe any salt effects when oligonucleotides were printed onto the surface; however it must be kept in mind that the salt concentrations tend towards saturation immediately prior to dessication. Thus salt is not required as a supplement for DNA array printing, but should be used when hybridizing target DNAs to the arrayed probes.
CONCLUSIONS
The methods outlined in this paper were given to illustrate a general process to make and use micropattern streptavidin supports. Because photobiotin is not chemically very specific, other silanes (e.g., aminosilane or alkylsilane) could potentially be used; poly-L-lysine functionalized supports, plastics and painted surfaces should also be compatible with the procedures given here. Also, we used a photolithographic mask to make micron and millimeter sized streptavidin features, but in principle, submicron resolution could be obtained by using a proximal probe to deliver the light such as that used in near field optical microscopy. The resulting photo-patterned biotin can be used to immobilize phages and cells expressing surface-bound streptavidin, or alternatively, streptavidin can be used as a linker molecule to immobilize a variety of biotinylated molecules such as nucleic acids, proteins, antibodies, lipids and polysaccharides. In principle, by simply repeating the patterning process one could create complex surfaces with localized areas each comprising of different physical or chemical properties, such as hydrophobic or hydrophilic regions, and affinity, hybridization and enzymatic pads.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Drs Jan Smits and Weichun Yuan from Boston University’s Department of Electrical Engineering for fabricating the silicon microwells, and Dr Joel Graber for insightful scientific discussions. This work was supported by grants from the DOA (DAMD17-94-J-414) and NIH (IP50 HL55001-01) to C.L.S. C.R.S. was funded by a training grant from the NHGRI (T32 HG00041-03) and Sequenom, Inc.
REFERENCES
- 1.Fodor S.P.A, Read,J.L., Pirrung,M.C., Stryer,L., Tsai Lu,A. and Solas,D. (1991) Science, 251, 767–773. [DOI] [PubMed] [Google Scholar]
- 2.Pease A.C., Solas,D., Sullivan,E.J., Cronin,M.T., Holmes,C.P. and Fodor,S.P.A. (1994) Proc. Natl Acad. Sci. USA, 91, 5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chrisey L.A., O’Ferrall,C.E., Sparfo,B.J., Dulcey,C.S. and Calvert,J.M. (1996) Nucleic Acids Res., 24, 3040–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi W.C. and Smits,J.G. (1993) J. Microelectromechan. Syst., 2, 82–86. [Google Scholar]