Abstract
DNA–substrate conjugates are required for the direct in vitro selection of novel DNA catalysts for reactions between two small reactants. Here we describe the introduction of all necessary features into ssDNA by a novel, multifunctional primer containing a flexible PEG spacer, an o-nitrobenzyl moiety allowing for selective photocleavage, and anthracene as a reactant, a fluorescence label and/or an immobilization tag. These components were checked individually and by a mock selection.
In vitro selection as a very powerful strategy for the identification of novel, non-biological oligonucleotide activities led to the isolation of tight binding DNA molecules (aptamers) as well as to remarkable new DNA catalysts (1). Despite these successes in direct selection, the reactions to be investigated remained confined to oligonucleotide (self-)modification (2,3).
For RNA, these limitations were recently overcome by the use of RNA reactant-conjugates instead of unmodified RNA libraries leading to the isolation of RNAs catalyzing peptide (4) or N-glycosidic bond formation (5) or Diels-Alder reactions (6,7).
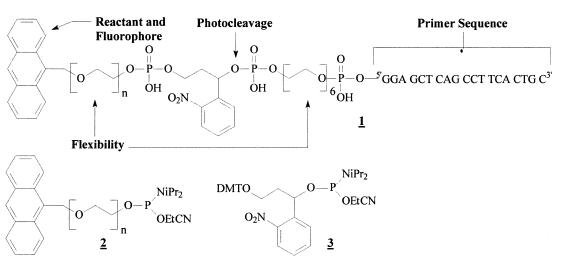
From a synthetic point of view though, DNA has several advantages over RNA such as cheap and convenient synthesis as well as enhanced chemical and biological stability. To extend the concept of in vitro selection with linker coupled reactants (8) to DNA, we designed the multifunctional primer 1 providing the potential reactant anthracene as a diene for Diels-Alder reactions (Fig. 1) (7). It is tethered to the DNA by a polyethylene glycol spacer (PEG) to ensure optimal positioning and further contains an o-nitrobenzyl unit to suppress the selection of side products by regioselective photocleavage (8). The fluorescence of the coupled anthracene can further be used for highly sensitive detection of the resulting DNA conjugates and the Diels-Alder reaction with suitably derivatized maleimides allows site-specific derivatization or immobilization of the PCR products (9). Compound 1 was synthesized on an automated DNA synthesizer in a 15 µmol DMT on DNA mode using commercially available standard phosphoramidites and building blocks 2 and 3 which were synthesized as described (9,10). After deprotection in aqueous NH4OH (33% w/v) at 55°C for 48 h and lyophilization, primer 1 was purified by reversed phase HPLC in a 0.1 M triethyl ammonium acetate (pH 7) gradient with increasing percentage of acetonitrile (0.8–32% v/v in 5 min and 32–48% v/v in 40 min followed by an 80% v/v wash). The product eluting at 16.1 min was quantified by UV spectroscopy, yielded 300 OD255 (~730 nmol, 5%) and showed the characteristic UV maxima of anthracene at λ = 255, 350, 366, 385 nm as well as its typical fluorescence (excitation: 255 nm, emission: 419 nm) (9). Further characterization by MALDI-TOF mass spectroscopy corroborated both the correct polydisperse product (masses calculated [M+H]+ = 7036.2 + n × 44 g/mol; masses found = 7035.1, 7079.6, 7123.4, 7167.2, 7210.7, 7255.3, 7300.0 g/mol) and the expected photoreleased primer fragment (mass calculated [M+H]+ = 6190.0 g/mol; mass found = 6189.9 g/mol).
Figure 1.
Design and synthesis of the multifunctional primer derivative 1 using the polydisperse anthracene–PEG600 (naverage = 13) phosphoramidite 2 and the photocleavable building block 3.
As shown in Figure 2, compound 1 was incorporated into the DNA pool by PCR in a 300 µl reaction containing 50 mM KCl, 20 mM Tris–HCl pH 8.4, 4 mM MgCl2, 0.2 mM of each dNTP (MBI Fermentas), 15 µCi [α32P]dCTP (Amersham), 100 U Taq DNA polymerase (Gibco), ~50 fmol template (120 randomized bases), 5 µM primer 1 and 5 µM of a second primer with a unique 2′, 3′ diol group [5′-d(GTG GAT CCG ACC GTG GTG C) rC-3′]. Since single-stranded DNA conjugates are required for the in vitro selection experiment, this riboprimer was used to facilitate the electrophoretic removal of the complementary strand (11).
Figure 2.
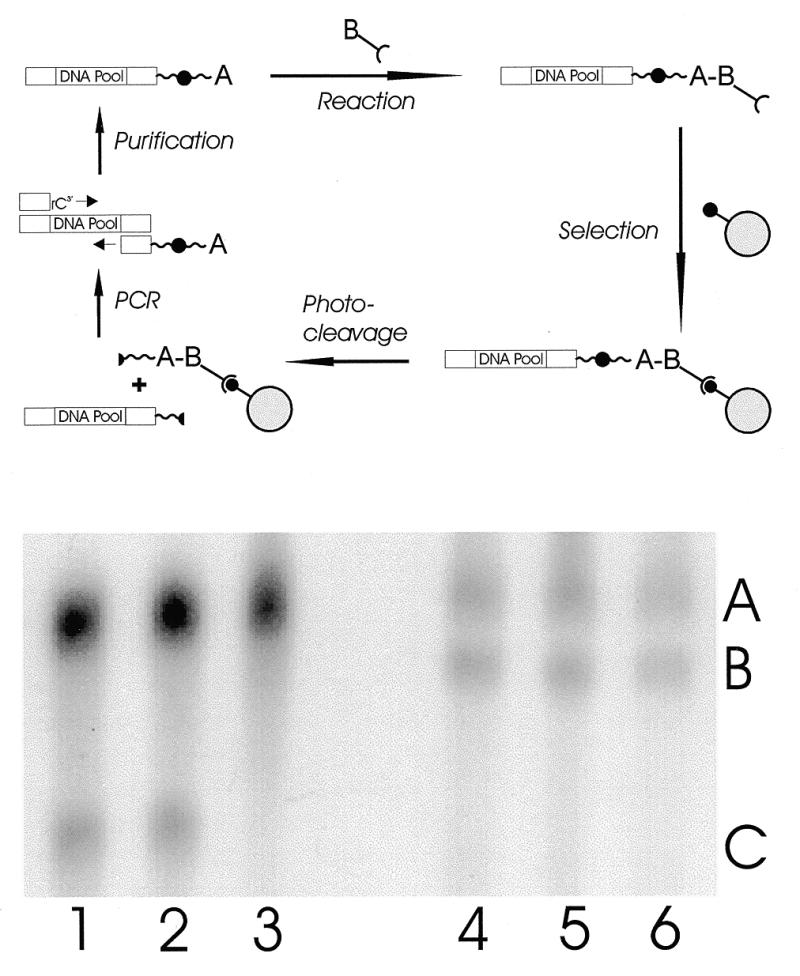
Selection scheme for DNA catalysts with photocleavable primer derivatives. (a) A DNA library is amplified by PCR using a 3′-ribo terminal primer and a primer reactant-conjugate (A, anthracene). ssDNA conjugates were obtained by detaching the riboprimer on the complementary strand with alkaline hydrolysis and PAGE purification. After incubation with a second, free reactant carrying an affinity tag (B, biotin–maleimide), the desired reaction products are purified by affinity immobilization on a solid support (e.g., streptavidin–agarose), released by irradiation and PCR amplified to enter a new cycle of in vitro selection. (b) 5% denaturating PAGE analysis of the PCR amplified DNA conjugates using the anthracene derivatized primer 1 and either a detachable, riboterminal primer (lanes 1–3) or a standard, deoxy primer (lanes 4–6). The ribomoiety containing complementary strand was cleaved by adjusting the PCR mixture to 0.2 M NaOH for 20 min at 95°C (lanes 1 and 4), for 12 h at 37°C (lanes 2 and 5) or kept as a reference (lanes 3 and 6) prior to ethanol precipitation and gel purification. Slower moving bands (A) were assigned to the anthracene modified DNA conjugates by fluorescence spectroscopy and faster moving bands correspond to the unmodified (lanes 4–6, band B) or primer cleaved (lanes 1–3, band C) complementary ssDNA strands.
After 15 PCR cycles for 1 min at 95°C, 1.5 min at 55°C and 2.5 min at 72°C, the complementary strand was cleaved at the riboposition by alkaline hydrolysis in 0.2 M NaOH for 30 min at 80°C, followed by neutralization, ethanol precipitation and separation of the resulting two ssDNA strands by 5% denaturating PAGE (12). As shown in Figure 2b, the alkaline hydrolysis is essentially completed after 20 min and leads to a significant increase in electrophoretic mobility of the complementary strand. The riboprimer was observed to be less efficient in priming than its all-DNA analog, resulting in asymmetric PCR conditions and an overproduction of the desired anthracene-coupled DNA strand (Fig. 2b).
The compatibility of this conjugate with the overall selection scheme was checked by subjecting it to a mock selection. The ssDNA–anthracene conjugates were first reacted with 25 µM biotin–maleimide (Sigma) as the Diels-Alder dienophile for 1 h under typical in vitro selection conditions (6), then precipitated twice and immobilized on 50 µl streptavidin–agarose (twice pre-washed). After washing with 20 ml Urea 7 M, 0.1 M Tris–HCl (pH 7.4) and 2 ml water, 0.3% of the input DNA remained attached to the solid support which corresponded to the independently determined background rate (9). The selected oligonucleotides were recovered by irradiation with a Nd-YAG laser at 355 nm in 400 µl wash buffer with 76% efficiency (8). The released DNA molecules were precipitated and PCR amplified as above to enter the next round of in vitro selection (Fig. 2a).
Since immobilization and photo-release occur under identical chemical conditions, this selection strategy provides extremely high stringency and allows a regiospecific discrimination between possible reaction sites, e.g. between the desired reaction of anthracene and side reactions at internal positions of the DNA which remain attached to the solid support upon irradiation.
To apply this approach to a broader range of reactants, an analog of compound 1 was synthesized with an aminohexyl-hexaethylene glycol (HEG) 5′-terminus replacing the anthracene–PEG part {H2N-C6H12-HEG-X-HEG-d(TCT AAT ACG ACT CAC TAT A), masses calculated [M+H]+ = 6858.7, 6156.1, 703.1 g/mol, masses found 6859.1, 6155.8, 706.0 g/mol}. The 5′ amino-group can selectively be derivatized with various activated N-hydroxysuccinimide esters thereby expanding the strategy to a broad range of organic or biochemical reactants (8).
The presented in vitro selection strategy will aid as a valuable tool in exploring the catalytical potential of DNA and should be especially useful for a detailed comparison with RNA catalysis. The primer derivative 1 can further serve as a fluorescence probe or as a novel immobilization tag if higher concentrations of biotin–maleimide (12.5 mM, overnight) are used. Combined with the photocleavage site this suggests new applications in MALDI-TOF assisted DNA sequencing (13) and/or gene spotting on DNA chips (14) as well as for DNA–protein interaction studies.
Acknowledgments
ACKNOWLEDGEMENTS
We thank C. Schatz and Dr B. Seelig for technical assistance, Dr E. Nordhoff for expertise in MALDI-TOF measurements and Prof. V. A. Erdmann for support. This work was supported by the DFG and the BMBF.
REFERENCES
- 1.Ellington A.D. and Szostak,J.W. (1992) Nature, 355, 850–852. [DOI] [PubMed] [Google Scholar]
- 2.Breaker R.R. (1997) Chem. Rev., 97, 371–390. [DOI] [PubMed] [Google Scholar]
- 3.Sen D. and Geyer,C.R. (1998) Curr. Opin. Chem. Biol., 2, 680–687. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B. and Cech,T.R. (1997) Nature, 390, 96–100. [DOI] [PubMed] [Google Scholar]
- 5.Unrau P.J. and Bartel,D.P. (1998) Nature, 395, 260–263. [DOI] [PubMed] [Google Scholar]
- 6.Seelig B. and Jäschke,A. (1999) Chem. Biol., 6, 167–176. [DOI] [PubMed] [Google Scholar]
- 7.Tarasow T.M., Tarasow,S.L. and Eaton,B.E. (1997) Nature, 389, 54–57. [DOI] [PubMed] [Google Scholar]
- 8.Hausch F. and Jäschke,A. (1997) Bioconjugate Chem., 8, 885–890. [DOI] [PubMed] [Google Scholar]
- 9.Seelig B. and Jäschke,A. (1997) Tetrahedron Lett., 38, 7729–7732. [Google Scholar]
- 10.Ordoukhanian P. and Taylor,J.S. (1995) J. Am. Chem. Soc., 117, 9570–9571. [Google Scholar]
- 11.Silveira M.H. and Orgel,L.E. (1995) Nucleic Acids Res., 23, 1083–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y. and Breaker,R.R. (1999) Proc. Natl Acad. Sci. USA, 96, 2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirpekar F., Nordhoff,E., Larsen,L.K., Kristiansen,K., Roepstorff,P. and Hillenkamp,F. (1998) Nucleic Acids Res., 26, 2554–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreadis J.D. and Chrisey,L.A. (2000) Nucleic Acids Res., 28, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]