Abstract
Over 150,000 Americans are diagnosed with colorectal cancer (CRC) every year, and annually >50,000 individuals are estimated to die of CRC, necessitating improvements in screening, prognostication, disease management, and therapeutic options. CRC tumors are removed en bloc with surrounding vasculature and lymphatics. Examination of regional lymph nodes at the time of surgical resection is essential for prognostication. Developing alternative approaches to indirectly assess recurrence risk would have utility in cases where lymph node yield is incomplete or inadequate. Spatially dependent, immune cell–specific (eg, tumor-infiltrating lymphocytes), proteomic, and transcriptomic expression patterns inside and around the tumor—the tumor immune microenvironment—can predict nodal/distant metastasis and probe the coordinated immune response from the primary tumor site. The comprehensive characterization of tumor-infiltrating lymphocytes and other immune infiltrates is possible using highly multiplexed spatial omics technologies, such as the GeoMX Digital Spatial Profiler. In this study, machine learning and differential co-expression analyses helped identify biomarkers from Digital Spatial Profiler–assayed protein expression patterns inside, at the invasive margin, and away from the tumor, associated with extracellular matrix remodeling (eg, granzyme B and fibronectin), immune suppression (eg, forkhead box P3), exhaustion and cytotoxicity (eg, CD8), Programmed death ligand 1–expressing dendritic cells, and neutrophil proliferation, among other concomitant alterations. Further investigation of these biomarkers may reveal independent risk factors of CRC metastasis that can be formulated into low-cost, widely available assays.
Colorectal cancer (CRC) has the fourth highest cancer incidence rate in the United States and carries a lifetime risk of roughly 4%. The incidence of colorectal cancer is correlated with increasing age, with most colorectal cancer cases occurring in patients aged >50 years. However, in the United States, the incidence of colorectal cancer in patients aged <50 years is increasing. The reason for this sudden increase is not known, as the disease etiology is thought to be a complex interaction of dietary patterns, environmental exposures, and genetic influences. In addition, CRC incidence varies widely around the world, with the highest rates reported in Australia, New Zealand, Europe, and North America, further supporting the complexity of the disease. According to the National Cancer Institute's Surveillance Epidemiology and End Results program, disparities exist by age, sex, and ethnicity in disease incidence nationwide. Despite varying incidence rates, CRC is the third largest contributor to cancer deaths worldwide, indicating a significant unmet need to improve curative-intent therapy for adequately identifying and treating CRC to prevent death.1, 2, 3
Early-stage colorectal cancer that has not spread to lymph nodes or other distant sites has a 5-year survival rate of 91%; however, if it has spread to the regional lymph nodes, the 5-year survival rate decreases to 72%, an important factor in patient outcomes. Pathologic tumor, node, and metastasis (pTNM) stage at presentation is considered the most important factor for ascertaining CRC prognosis. CRC staging elements include the following: the level of tumor invasion (T0-4b), regional lymph node metastasis (N0-2b), and distant metastasis (M0-1). Distant metastasis at diagnosis is associated with a 5-year survival rate of only 14%. Approximately 37% of patients are diagnosed with localized disease, 36% are diagnosed with regional lymph node spread, and 22% are diagnosed with distant metastasis. Identification of regional nodal disease often indicates the usage of adjuvant therapies (eg, chemotherapy), as regional nodal positivity is a risk factor for tumor recurrence.4,5
Processes governing cellular migration and metastasis involve cellular biochemical alterations related with primary tumor formation. Through rapid mitotic activity and the accumulation of genomic instability, cells gain the capacity to invade mesenchymal tissue and vasculature. In circulation, tumor cells migrate through the intravascular and extravascular systems, evade DNA damage response pathways, prime sites for metastasis, and establish a favorable micro-environment for metastasis, including activation of T-regulatory cells and angiogenesis. Outside of the standard pTNM staging, some hypothesize that tumor border morphology is a strong, independent prognostic factor (eg, tumor budding). Other specific pathologic findings of poor prognosis include a poorly defined border, invasion through mucosal layers without expected stromal reaction, and focal dedifferentiation. Metastatic potential has also been shown to vary by primary tumor site (eg, left sided).6 These findings may be specific to common progression pathways [eg, adenomatous polyposis coli, microsatellite instability (MSI), and CpG island methylator phenotype].
Several studies concur on the importance of the tumor immune microenvironment in CRC prognosis. Specifically, high densities of tumor-infiltrating lymphocytes (TILs) improve prognosis and the 5-year survival rate of patients with CRC metastases. In addition to spatial distribution and density, previous studies have shown that the type, activation state, and location of infiltrating lymphocytes determine the tumor microenvironment's immune response and its antitumoral effectiveness. In general, the presence of TILs confers a favorable prognosis. TIL cells appear to have more potent and directed anti-tumor effects than peripheral blood circulating lymphocytes.7, 8, 9 Various tumor-specific characteristics, including mismatch repair alterations, determine TILs’ effect on the tumor microenvironment (TME) and prognosis.10, 11, 12 Although ample evidence suggests that while the overall presence of immune infiltration carries a favorable prognosis, not all immune lineages support these favorable findings.
Current methods of prognosticating recurrence and survival are crude and need improving. Specimen inadequacy and inadequate lymph node yield are important limitations to current prognostication methods.13, 14, 15, 16, 17 This incomplete or inadequate assessment can affect the accuracy of tumor staging and subsequent disease management options, such as whether a patient should receive adjuvant chemotherapy. Patients who receive extended lymphadenectomies have better outcomes; however, this is not the standard of care and can cause increased morbidity. Multiplexed genomic, proteomic, and transcriptomic assays of tumors have revealed an incredible variation at the level of the host, the tumor, and the tumor's micro-environment, along with complex regulatory networks and interactions. However, most of these findings are still in the discovery phase and have not been validated clinically, with the weak link of such multiplexed methods being the origin of the examined cells. New evidence strongly suggests that cells' origin and anatomic location play a significant role in prognostication. Multiplexed spatial molecular cancer tools have recently been developed, generating a new frontier in cancer diagnosis, treatment, and prognosis. In CRC specifically, a lot remains to be learned, such as information on cell type–specific molecular alterations (eg, transcriptome expression) within unique spatial arrangements related to colon cancer metastasis.18 The development of spatial omics technologies, such as Spatial Transcriptomics (10x Genomics, Pleasanton, CA) or GeoMX Digital Spatial Profiling (DSP; Nanostring, Seattle, WA), has enabled multiplexing findings (eg, whole transcriptome) at incredible spatial resolution. The GeoMX platform first deposits an RNA/protein barcode for expression profiling across an entire tissue slide. Fluorescent antibodies that highlight various tissue types stain the slide to highlight relevant structures for the selection of regions of interest (ROIs). Within selected regions of interest (which can vary from single cell sized to nearly a centimeter squared), ultraviolet light is used to cleave barcodes from substrate selectively, and these barcodes are retrieved for quantitation using nCounter and next-generation sequencing. Analysis of such data may reveal spatially variable gene expression, characteristic spatial patterns of expression (which may inform spatially variable cell type proportions), or information about how cells communicate to elicit an immune response.
As an application of these techniques, GeoMX platform has been used to study the differences between TILs and nontumor/stromal lymphocytes in tumors driven by microsatellite instability.19 This study uncovered expression patterns in intratumoral T cells and extratumoral T cells related to cytolytic activity and cell-cell interactions. Another study found a correlation between spatial statistics from 55 fluorescently tagged antibodies and the 5-year risk of CRC progression.20,21 Other studies include the role of neutrophils in poor prognosis22 and how the spatial distance between key TIL lineages may be influenced by programmed death ligand 1 (PD-L1) expression.20,21 Finally, the spatial relationship between lymphocytes and tumor budding is a prognostic predictor.23,24 Few studies have attempted to use information on tissue context-specific immune cell expression to predict the metastatic potential of CRC tumors.
Several factors complicate assessments for the presence and prevention/inhibition of tumor metastasis, chiefly considering the capacity to surgically resect the positive lymph nodes as well as establish etiological models of metastasis that are targetable through commensurate emerging therapeutics (eg, immunotherapies). For instance, an incomplete lymph node dissection can potentially result in a false-negative finding, inaccurately determining recurrence risk. These deficiencies necessitate the development of tools to reduce the potential for inadequate assessments or to fill in the missing information. This study aims to characterize spatial immunologic correlates of nodal and distant metastasis from the primary tumor site through the application of digital spatial profiling.
Materials and Methods
Methods Overview
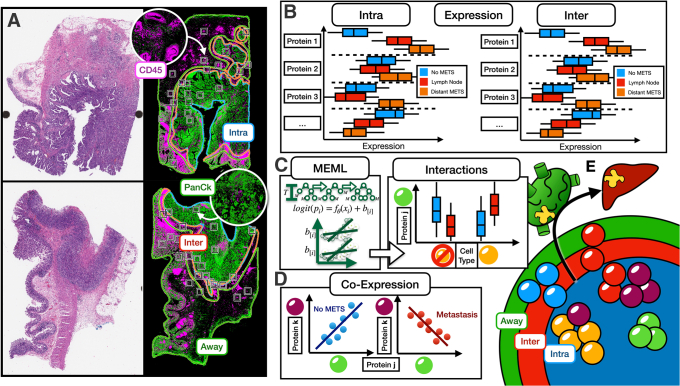
Figure 1 shows a graphical overview of the methods used herein. In brief, we: i) collected 36 resected colorectal tumor specimens; ii) using a spatial proteomics assay, profiled immune cells within three spatial architectures (Figure 1E); and iii) identified biomarkers/lineages associated with nodal and distant metastasis within these architectures, controlling for local invasiveness. Biomarkers were inferred through differential expression analyses, and the potential for two biomarkers at a time to carry additional predictive value (as compared with individual markers) for metastasis was assessed through the following assessments: i) relative expression between the biomarkers (ie, CD8/FOXP3); ii) interactions (ie, conditional on one cell type, what is the association of another and metastasis); and iii) differential co-expression.
Figure 1.
Study overview: A: Hematoxylin and eosin– and immunofluorescence-stained slide used to help place region of interest for profiling within three distinct architectures: intratumoral (Intra; blue), interface (Inter; red; invasive margin or peritumoral), and away (green); colors for outlined macroarchitectures correspond with colors used in Figure 1E to denote separate tissue architectures. B: Within distinct architectures, expression of specific lineages/protein markers predictive of metastasis (METS) status; box plots used to communicate center and spread of hypothetical expression of canonical protein markers correspondent to immune cell sublineages; box plots are compared for patients without metastasis (blue), patients with nodal metastasis (red), or patients with distant spread (orange); expression differences reported for each architecture (here intratumoral and interface). C: Mixed-effects machine learning (MEML) models uncover statistical interactions between specific immune cell types (ie, different risk of metastasis for specific cell type conditional on another cell type); illustrates how interaction (two crossing lines) are uncovered, invariant to batch/patient; association of protein j (corresponding to the green cell type, as denoted by the green sphere) with metastasis is reported separately for yellow cell types and for cell types that are not yellow. D: Differential co-expression patterns identify correlations between markers (proteins j and k corresponding to the green and purple cell types, respectively) that are metastasis specific. E: Identifying predictive protein biomarkers (ie, cell lineages, with different lineages denoted by different color spheres) for nodal and distant metastasis within these regions: intratumoral (blue), interface (red; invasive margin), and away (green); color assigned to each macroarchitecture similar to colors in Figure 1A. Intra, Inter, and Away are described in Data Acquisition and Preprocessing. PanCk, pancytokeratin.
Data Acquisition and Preprocessing
Thirty-six colon adenocarcinoma resections performed at Dartmouth Hitchcock Medical Center from 2016 to 2019 were selected for digital spatial profiling with institutional review board approval. Approximately half of them showed local invasion but no nodal or distant metastasis, and the other half showed nodal and/or distant metastasis. Of the cases with concurrent metastasis, all cases exhibited local lymph node involvement—about half of these cases metastasized to distant sites. Sample size was determined on the basis of the feasibility of this pilot study and through an empirical power analysis, which simulated data from the statistical models introduced in subsequent sections. The cohort was restricted to stage pT3 assignments under the pTNM staging system, which balances the impacts of local invasion and nodal and distant metastasis for prognostication. T stage refers to the degree of invasion at the local site. By restricting the T stage, the authors sought to identify markers that provide prognostic value (ie, predictive of metastasis) beyond that offered through the current prognostic staging system (based on T stage; future studies will assess recurrence risk biomarkers with predictiveness above and beyond pTNM staging). Cases were matched between the nonmetastatic and metastatic groups based on tissue size (measured through connected component analysis of whole slide images), tumor grade, mismatch repair (MMR) dysregulation status [MMR deficient (dMMR) and MMR proficient (pMMR); as assessed through immunohistochemistry], site of the tumor (eg, descending or ascending colon), age, and sex. Matching and randomization were achieved by conducting Fisher exact tests and two-sample t-tests after iterative resampling (Table 1). dMMR reflects the loss of staining in at least one of four mismatch repair genes [MutL homolog 1 (MLH1), postmeiotic segregation increased 2 (PMS2), MutS homolog 2 (MSH2), and MSH6]. As MSH2 and MSH6 alterations were relatively rare for cases within the queried time periods, dMMR status was reported from alterations to either MLH1 or PMS2 (MSH2 and MSH6 alterations were not present in this cohort).25,26
Table 1.
Patient Cohort Characteristics
| Characteristic | No metastasis | Metastasis | P value | Lymph node only | Distant + lymph node | P value |
|---|---|---|---|---|---|---|
| N | 16 | 19 | 8 | 11 | ||
| pMMR, n % | 10 (62.5) | 12 (63.2) | 1 | 6 (75.0) | 6 (54.5) | 0.667 |
| Grade, n (%) | 0.789 | 0.408 | ||||
| 1 | 8 (50.0) | 11 (57.9) | 6 (75.0) | 5 (45.5) | ||
| 2 | 4 (25.0) | 3 (15.8) | 1 (12.5) | 2 (18.2) | ||
| 3 | 4 (25.0) | 5 (26.3) | 1 (12.5) | 4 (36.4) | ||
| Site, n (%) | 0.936 | 0.142 | ||||
| Cecum | 6 (37.5) | 5 (26.3) | 1 (12.5) | 4 (36.4) | ||
| Hepatic flexure | 1 (6.2) | 1 (5.3) | 0 (0.0) | 1 (9.1) | ||
| Descending colon | 2 (12.5) | 2 (10.5) | 1 (12.5) | 1 (9.1) | ||
| Rectum | 1 (6.2) | 3 (15.8) | 3 (37.5) | 0 (0.0) | ||
| Ascending colon | 2 (12.5) | 3 (15.8) | 2 (25.0) | 1 (9.1) | ||
| Sigmoid | 1 (6.2) | 3 (15.8) | 0 (0.0) | 3 (27.3) | ||
| Splenic flexure | 1 (6.2) | 1 (5.3) | 1 (12.5) | 0 (0.0) | ||
| Transverse colon | 2 (12.5) | 1 (5.3) | 0 (0.0) | 1 (9.1) | ||
| Male sex, n (%) | 9 (56.2) | 11 (57.9) | 1 | 6 (75.0) | 5 (45.5) | 0.414 |
| Age, mean (SD), years | 71.06 (12.87) | 66.21 (16.29) | 0.342 | 67.75 (17.53) | 65.09 (16.10) | 0.736 |
| N stage, n (%) | N/A | 0.074 | ||||
| 0 | 16 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| 1a | 0 (0.0) | 9 (47.4) | 2 (25.0) | 7 (63.6) | ||
| 1b | 0 (0.0) | 4 (21.1) | 2 (25.0) | 2 (18.2) | ||
| 1c | 0 (0.0) | 2 (10.5) | 0 (0.0) | 2 (18.2) | ||
| 2a | 0 (0.0) | 2 (10.5) | 2 (25.0) | 0 (0.0) | ||
| 2b | 0 (0.0) | 2 (10.5) | 2 (25.0) | 0 (0.0) | ||
Stratified by metastasis status (yes/no). For patients with metastasis, stratified by whether there was only nodal involvement or both nodal and distant involvement.
N/A, not applicable; pMMR, mismatch repair proficient.
Tissue blocks were sectioned (5 μm thick) and stained with fluorescent-labeled antibodies [highlighting tumor (pancytokeratin [PanCk]), immune cells (CD45), and nuclei (SYTO13)]. Fluorescent antibodies were covalently linked to photocleavable oligonucleotide tags associated with a targeted panel of immune cell profiling and tumor immune environment protein markers (40 total markers). Sections were visualized using the GeoMX DSP instrument, which displayed immunofluorescence (IF) images. Subsequent sections were stained with hematoxylin and eosin and scanned using the Aperio-AT2 scanner (Leica, Wetzlar, Germany) at ×20 magnification. Hematoxylin and eosin–stained images were stored in SVS format (eight-bit color channels). IF whole slide images were stored in TIFF format (16-bit unsigned color channels; one channel per stain).
A gastrointestinal tract pathologist (L.J.V.) used the ASAP annotation software version 2.1 (https://computationalpathologygroup.github.io/ASAP) to view the hematoxylin and eosin and IF whole slide images side by side to annotate sections and spatially resolved immune populations based on three distinct macroarchitectural regions: intratumoral (intra), tumor-immune interface (inter), and away from the tumor (away) (Figure 1A).27 These regions were outlined using polygonal/spline annotations. Eight ROIs (square grids of maximal spatial dimensions allowable by the GeoMx DSP instrument) were placed within each annotated region per slide (24 ROIs per slide) using a semi-autonomous placement system. The ROIs were uploaded and registered to the DSP IF images. An automated process initially selected a random distribution of eight potential ROIs per annotated region. When ROIs were suboptimal (eg, insufficient CD45 staining, incorrect region; determined through a visual assessment), a pathologist (L.J.V.) manually adjusted to the nearest appropriate region. Immune cells were isolated within each ROI via image segmentation of the CD45 stain (to establish pixel-wise locations with CD45+SYTO13+PanCk staining), followed by a connected component analysis. Segmented ROIs were profiled through targeted ultraviolet cleavage of attached oligo tags. The Nanostring nCounter was used to quantify immune cell protein expression across 40 immuno-oncology markers. These markers were selected from three GeoMx DSP immuno-oncology protein panels offered by NanoString Technologies (Seattle, WA). Specifically, the following three panels were selected of eight potential options because they were specific to immune lineage profiling and previous studies7, 56, 57 demonstrating association of some of these markers with metastasis (see Introduction; other panels were less relevant):
-
1.
GeoMx Immune Cell Profiling Panel: Includes 18 targets for human immune cell profiling, with positive and negative controls. This panel includes markers for immune cells (CD45), proliferation (Ki-67), antigen presentation (B2M), and vasculature (CD31) and the controls needed to run any GeoMx DSP experiment.
-
2.
GeoMx Immune Activation Status Panel: Includes eight targets for human immune activation status. This panel is run in tandem with the GeoMx Immune Cell Profiling Panel and includes additional checkpoint molecules and other markers corresponding to activated or memory T cells.
-
3.
GeoMx Immune Cell Typing Panel: Includes seven targets for human immune cell typing, AbMix, and Probe R4. This panel is run in tandem with the GeoMx Immune Cell Profiling Panel and includes key immuno-oncology targets and markers of immune cell types, including T cells, B cells, macrophages, natural killer (NK) cells, and stroma.
The specificity of the antibodies used in the DSP protein assay was validated by NanoString through immunohistochemistry staining patterns and their performance in cell pellet arrays and tissue microarrays, along with positive and negative controls. This validation process is in line with recent guidelines for antibody validation from the Society for Immunotherapy of Cancer. Additional information on the validation of these markers can be found in several previous white papers and conference abstracts.30,31
Four slides were profiled per DSP batch. Tissue lifting after cover-slipping procedures led to additional case inclusion/exclusion. Because of issues with de-coverslipping one of the tissue slides (eg, tear), one sample was removed from the set (n = 35). Returned data included protein expression measurements for each ROI, tagged with positional x and y coordinates, an ROI-specific nuclei count, and coregistered hematoxylin and eosin and IF slides from the same section. ROIs were filtered on the basis of expression relative to the negative control (825 ROIs remaining after removal of one case and 15 additional ROIs), normalized to IgG isotype controls [rabbit (Rb)-IgG, mouse (Ms)-IgG1; Ms IgG2a demonstrated significant batch effects] across batches after comparison to other normalization methods (External RNA Controls Consortium, nuclei count/area, and housekeepers), and log2 transformed. ROIs were further labeled with mismatch repair deficiency status (using MLH1/PMS2 deficiency as assessed through immunohistochemistry, as a proxy), age and sex, site of origin/metastasis, tumor grade, nodal and distant metastasis status, and macro-architectural region (intra, inter, or away).
Differential Expression to Establish Clinical Markers of Metastasis
The following bayesian hierarchical linear regression models were fit to predict log2-transformed protein expression to establish associations with metastasis (mets used to separately indicate nodal metastasis, distant metastasis, and nodal or distant metastasis): An interaction term between mets and TME allowed for the evaluation of metastasis conditioned on the macroarchitecture ( {intra, inter, away}), adjusting for potential confounding (dMMR, age, or sex). Batch- and case-level variations were captured with random intercepts, . Residual technical variation from the isotype controls was modeled using . Effect estimates were communicated using the median posterior sample of the effect estimate, 95% high-density posterior credible interval (similar to the CI), posterior probability of direction (pd), transformed into a value correlated to the P value () to communicate the significance of the effect, with post hoc comparisons via estimated marginal means through the emmeans software package (R statistical programming language version 4.1; https://cran.r-project.org/web/packages/emmeans/index.html) to report metastasis-related markers by macroarchitecture.32,33 A weakly informative prior centered around 0 was chosen as type I error control in favor of a multiple comparisons adjustment given the nature of this exploratory analysis, although the exploratory nature of this assessment was emphasized.34,35
These associations were also reported separately for patients with/without MSI via the following statistical model:
The three-way interaction between metastasis, MSI status, and TME architecture was interrogated using estimated marginal means (emmeans) to report metastasis-related markers, conditional on macroarchitecture and MSI status.
The effect estimates for each analysis for each protein and their corresponding P values were displayed using volcano plots using the EnhancedVolcano R package version 3.16 (https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html). Expression for markers with large effect sizes and significant P values (given an α significance level of 0.05) were visualized using box plots (Figure 1B). Statistical analyses were performed using the brms package version 2.16 (https://paul-buerkner.github.io/brms) from the R version 4.1 statistical programming language, which leverages Hamiltonian Monte Carlo techniques in Stan to compute bayesian effect estimates.34, 35, 36 Hamiltonian Monte Carlo models were run using the Discovery computing cluster at Dartmouth College (https://rc.dartmouth.edu/index.php/discovery-overview).
Relative Expression Between Proteins as Additional Markers of Metastasis
In addition to evaluating specific cellular subsets independently, the relative abundance of different immune cell subsets could also point to metastasis-related factors. The relative expression between two protein markers was modeled using the following statistical models:
Where the offset term, is used to model the relative abundances between immune cell lineages j and k. Similar post hoc comparisons and displays (ie, volcano plots and box plots) were constructed on the basis of the relative proportion between marker expression: .
Machine Learning Classifiers to Report Salient Effect Modifiers
A set of classifiers to estimate the probability of tumor metastasis (lymph node, distant, or any) was developed based on all markers () within distinct architectures (intra, inter, or away) by fitting nine tree-boosting models, , in a mixed effects machine learning modeling framework, which leveraged gaussian process boosting decision trees39,40: . Salient cross-level interactions were identified from the mixed effects machine learning modeling framework method using the interactiontransformer package in Python version 3.7, which scores interactions using Shapley Additive Explanations (SHAP) and selects the top interactions using a knee locator method. Interactions were applied to a bayesian hierarchical logistic regression model to report pertinent effect modifiers (eg, effect of CD20, conditional on age; interactions encapsulated in )27,41: . As many interactions were initially selected, features were selected using the Horseshoe Least Absolute Shrinkage and Selection Operator (LASSO) method, and the remaining features were fit with weakly informative priors.42,43 Effect estimates for salient effect modifiers were reported similar to the previous sections. Box plots were generated for select interactions, reporting differences in expression of one protein marker by metastasis, conditional on the presence of another protein marker (dichotomized into high or low expression by median expression) (Figure 1C).
Statistically significant interactions were extracted from all multivariable regression models. To determine whether these interactions conferred additional predictive value beyond individual protein biomarkers alone (ie, two biomarkers studied in conjunction are additionally predictive of metastasis over either marker alone), for each identified interaction, models were refit with protein interactions and without interactions (only keeping main effects from two proteins), adjusting for age, sex, and MMR status. These two models were compared using leave-one-out cross-validation posterior likelihood testing. The proportion of times the extracted interactions outperformed the individual markers (via leave-one-out cross-validation metrics), p, was recorded and compared with the null hypothesis that the extracted interactions only outperformed the individual biomarkers half of the time (H0: P = 0.5; H1: P > 0.5) via a one-sample proportions test with continuity correction.
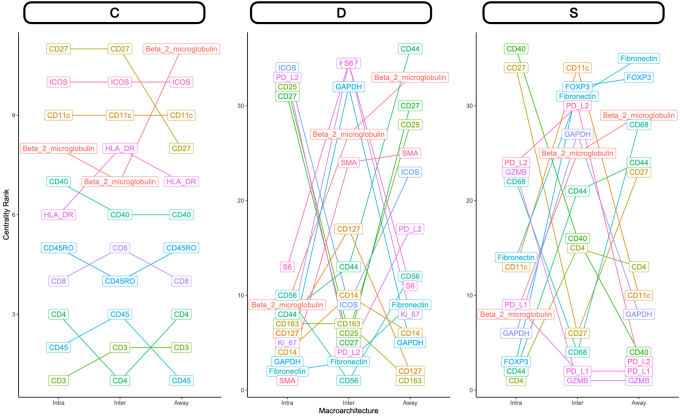
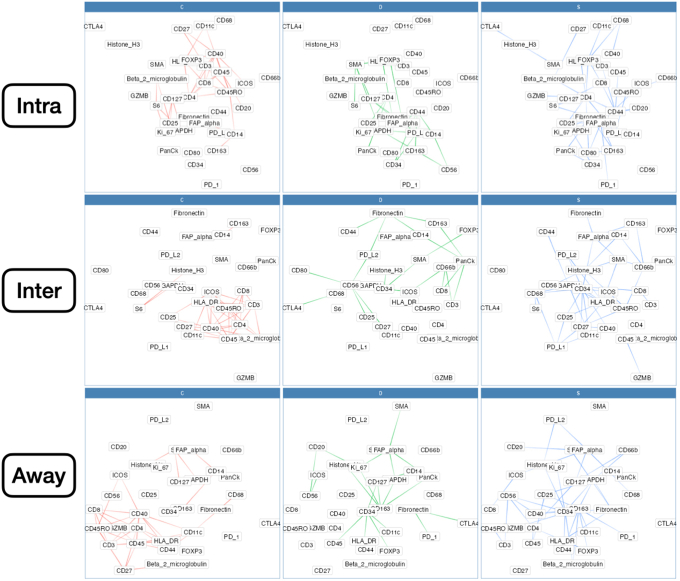
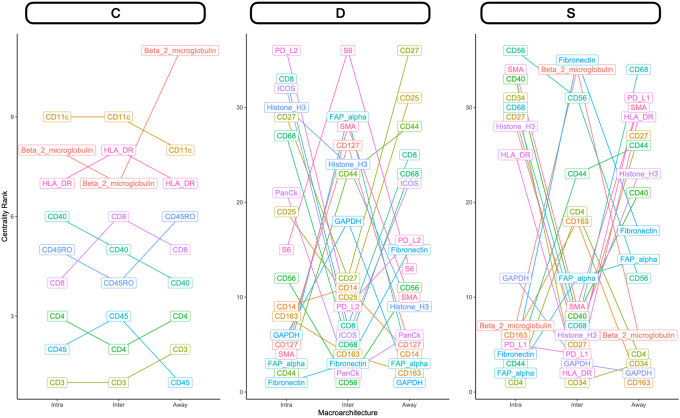
Differential Co-Expression Networks
Differential co expression networks were constructed to identify sets of genes whose co-expression differed by metastasis status. Co-expression between all pairs of genes was assessed using repeated measure correlation (via the rmcorr R package). The CDS framework was adapted for this analysis, which evaluates whether log2-transformed protein co-expression was conserved (C), specific (S), or differential (D) between patient cohorts with and without metastasis.44,45 Each pair of proteins was scored for each of these criteria, which was used to generate networks that pointed to co-expressed proteins that were metastasis related (Figure 1D). A total of nine networks were constructed for combinations of outcomes (lymph node metastasis, distant, or any) and distinct architectures (intra, inter, or away). Important markers for each network were identified through the calculation of the eigenvector centrality on a weighted adjacency matrix.
Web Application for Result Viewing
Because only a small subset of comparisons made for this study are discussed, an RShiny web application (https://levylab.shinyapps.io/ViewColonDSPResults, last accessed October 15, 2021) was developed for viewing the study findings.46 This application features the ability to view volcano plots, box plots, differential co-expression, and classifier results from all study comparisons and all numerical findings, including those featured outside of the present work.
Code and Data Availability
Example analysis code used to generate article results can be found in the following GitHub repository (https://github.com/jlevy44/Colon_Protein_DSP_Study, last accessed November 15, 2022). Because of patient privacy restrictions and ongoing efforts to expand the study cohort for public access, the spatial proteomics data featured in this study can be made available on reasonable request. Raw expression data can also be explored using the RShiny web application (see Web Application for Result Viewing).
Results
This section reports metastasis associations identified using the Digital Spatial Profiler for individual tissue architectures (intra, inter, and away). The first set of experiments sought to establish individual protein markers that correlated with tumor metastasis. The next set of experiments sought to report disease associations by assessing the following markers in tandem: i) relative abundance/expression, ii) protein interactions, and iii) differential co-expression.
Report of Clinical Characteristics for Cohort
Patient demographic characteristics, stratified by metastasis status and restricted to patients with metastases, whether the involvement was local or distant, are included in Table 1. Results indicate that the cohort is well matched based on MMR status, grade, primary site, sex, age, and N stage.
Differential Expression Results
Intratumoral
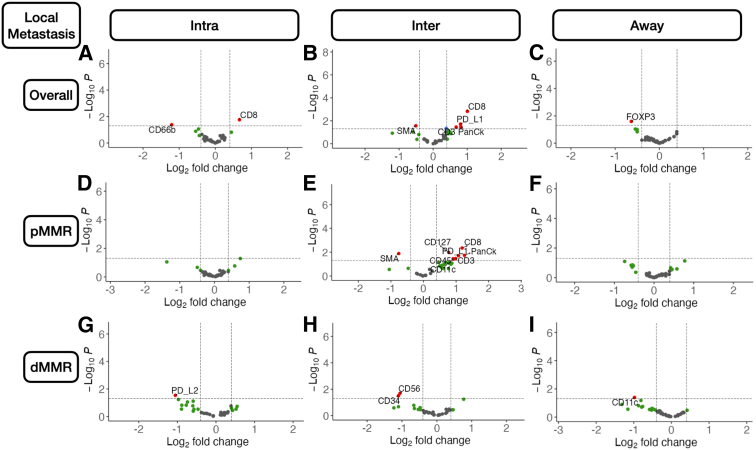
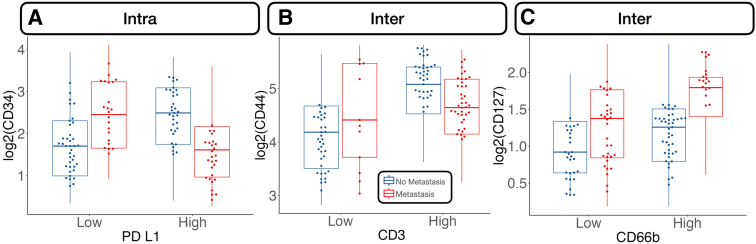
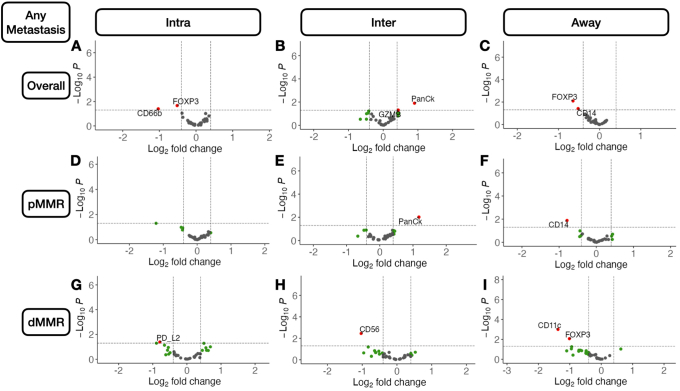
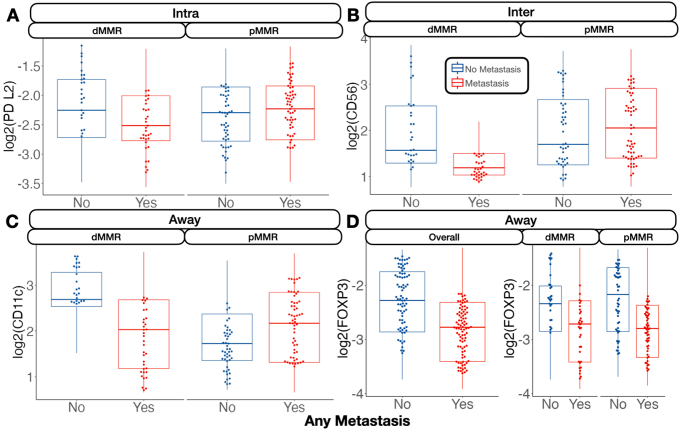
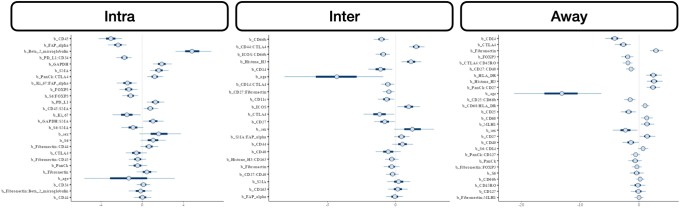
Metastasis-related markers within distinct macroarchitectures were investigated. In general, expression of forkhead box P3 (FOXP3) and CD66b was reduced inside the tumor for patients with metastasis (Supplemental Figures S1 and S2). When restricted to patients with MMR deficiency, PD-L2 expression was negatively associated with metastasis in the intratumoral region. CD8 was positively associated with lymph node metastasis in the intratumoral regions (Figure 2), regardless of MMR status, whereas CD44 expression was positively associated with distant metastasis. Granzyme B (GZMB), PD-L1, and β-2-microglobulin in patients with MMR deficiency were associated with distant metastasis (Supplemental Figure S3).
Figure 2.
Differentially expressed protein markers of local metastasis. Results stratified by tissue architecture: intratumoral (Intra; A, D, and G); interface/peritumoral (Inter; B, E, and H); and away/stroma (Away; C, F, and I); results also stratified by mismatch repair (MMR) status: MMR proficient (pMMR; D–F) and MMR deficient (dMMR; G–I); statistical significance cutoff at ; x-axis indicates effect size and directionality (positive x-value indicates metastasis-related marker; negative indicates decreased metastasis risk); y-axis indicates effect significance (positive y-value indicates lower P value). Intra, Inter, and Away are described in Data Acquisition and Preprocessing. FOXP3, forkhead box P3; PD-L, programmed death ligand; SMA, smooth muscle actin.
Interface
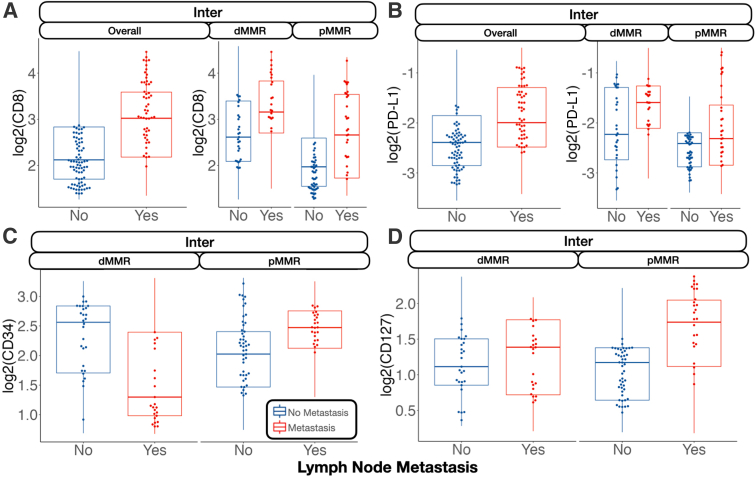
In the tumor-immune invasive interface, increased expression of GZMB was associated with any metastasis. For patients with MMR deficiency, CD56 expression was negatively correlated with metastasis. Meanwhile, CD8, PD-L1, and CD3 were positively associated with lymph node metastasis (Figure 3). In addition to these markers, CD127, CD3, and CD11c were also positively associated with lymph node metastasis for microsatellite-stable tumors. For MMR-deficient patients, CD34 and CD56 expression were reduced for patients with lymph node metastasis. The presence of GZMB in patients with MMR deficiency was positively associated with distant metastasis.
Figure 3.
Select protein marker expression for biomarkers predictive of nodal metastasis, stratified by mismatch repair (MMR) status: CD8 at the interface (Inter; A); programmed death ligand 1 (PD-L1) at the interface (B); CD34 at the interface (C); and CD127 at the interface (D). Marker expression plotted in beeswarm plots was filtered on the basis of the detection of outliers using a modified Tukey outlier test—after this initial filtering, only points between the 10% and 90% quantiles for each stratum were included. Inter is described in Data Acquisition and Preprocessing. dMMR, MMR deficient; pMMR, MMR proficient.
Away
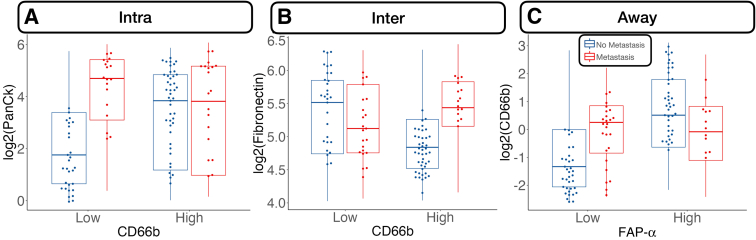
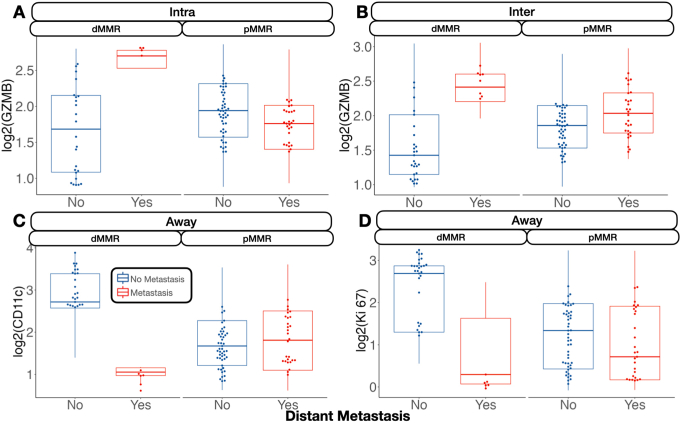
Away from the tumor, several factors were associated with metastasis. In general, FOXP3 and CD14 expression were negatively associated with metastasis. Although CD14 expression was especially relevant for patients without microsatellite instability, FOXP3 was salient for MMR-deficient patients. In these patients, CD11c was associated with lack of tumor metastasis. FOXP3 and CD11c were associated with lack of lymph node metastasis regardless of MMR status and in MMR-deficient patients, respectively. CD11c, Ki-67, and FOXP3 were associated with no distant metastasis, and GZMB expression was associated with distant metastasis for patients with MMR deficiency (Supplemental Figure S4).
A complete listing of differentially expressed markers and the relevant statistical findings can be found in the Shiny application.
Relative Expression Results
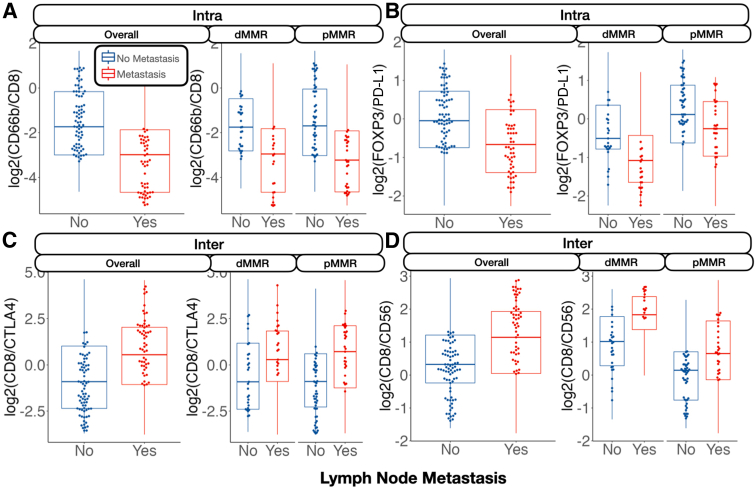
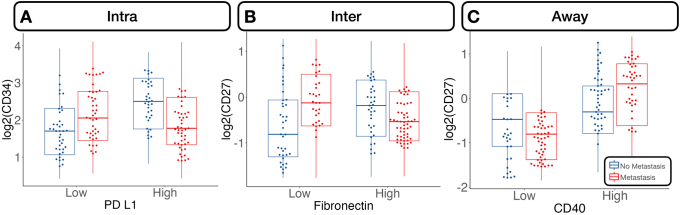
Nodal Metastasis
The relative abundance between specific immune cell lineages was highly predictive of metastasis. For instance, the ratio between CD66b/CD8 expression in the intratumoral region was negatively associated with lymph node metastasis (Figure 4). This trend was similar for the ratio between FOXP3/PD-L1, but only for microsatellite-stable patients. The relative expressions between CD8/cytotoxic T-lymphocyte–associated antigen 4 (CTLA4) and CD8/CD56 were associated with lymph node metastasis at the tumor interface.
Figure 4.
Relative protein expression between markers predictive of nodal metastasis, stratified by mismatch repair (MMR) status: CD66b/CD8 inside the tumor (Intra; A); forkhead box P3 (FOXP3)/programmed death ligand 1 (PD-L1) inside the tumor (B); CD8/cytotoxic T-lymphocyte–associated antigen 4 (CTLA4) at the interface (Inter; C); and CD8/CD56 at the interface (D). Marker expression plotted in beeswarm plots was filtered on the basis of the detection of outliers using a modified Tukey outlier test—after this initial filtering, only points between the 10% and 90% quantiles for each stratum were included. Intra and Inter are described in Data Acquisition and Preprocessing. dMMR, MMR deficient; pMMR, MMR proficient.
Distant Metastasis
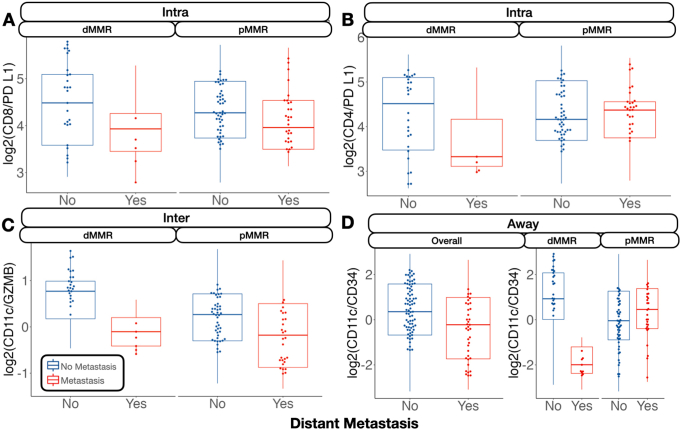
For distant metastasis, the relative expression between CD8 and CD4 compared with PD-L1 inside the tumor was negatively associated with tumor metastasis for microsatellite-stable patients (Supplemental Figures S5 and S6). At the interface, the relative expression between CD11c and GZMB was associated with no metastasis, whereas away from the tumor, CD11c compared with CD34 was heavily negatively associated with distant metastasis.
A complete listing of relative abundance differences can be found in the Shiny application.
Salient Effect Modifiers
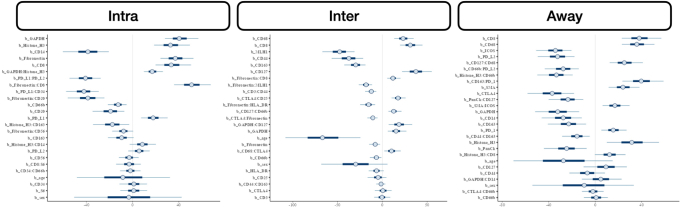
Overall Metastasis
Several protein interactions were identified using the classifiers. Of interest, immune cells in the intratumoral region expressing the PD-L1 surface antigen were at reduced risk of metastasis with higher CD34 expression (marker for immune cell stemness and vascular endothelial cells). Cells lacking this antigen demonstrated an increased risk of metastasis with higher CD34 expression. Similar relationships were noted between fibronectin and CD27 at the tumor interface. While CD27+ cells demonstrated increased metastasis risk compared with CD27– cells for low levels of fibronectin, the opposite relationship was noted for fibronectin-positive cells. Away from the tumor, tumor necrosis factor receptor family member cells (CD40+), which also expressed CD27 (another tumor necrosis factor family cell), demonstrated an increased risk of metastasis compared with CD27– cells, whereas the opposite relationship held for CD40– cells.
Nodal Metastasis
Several noteworthy interactions for nodal metastasis were noted and included the following: i) CD34–PD-L1 interaction in the intratumoral region (CD34 positively associated with metastasis for PD-L1– cells and negatively associated for PD-L1+ cells), ii) an interaction between CD3 and CD44 at the tumor interface [high CD44 expression related to nodal metastasis for tumors with cytotoxicity (eg, CD3+/CD8+) at the interface], and iii) strengthened positive association between CD127 and nodal metastasis for CD66b+ cells at the interface (Figure 5).
Figure 5.
Select protein marker expression, conditional on cell type (stratified by median expression), predictive of nodal metastasis: CD34 expression stratified by programmed death ligand 1 (PD-L1)–expressing cells inside the tumor (Intra; A), CD44 stratified by CD3 at the interface (Inter; B), and CD127 stratified by CD66b at the interface (C). Marker expression plotted in beeswarm plots was filtered on the basis of the detection of outliers using a modified Tukey outlier test—after this initial filtering, only points between the 10% and 90% quantiles for each stratum were included. Intra and Inter are described in Data Acquisition and Preprocessing.
Distant Metastasis
Increased PanCk expression for CD66b– cells was positively associated with distant metastasis. At the interface, increased expression of fibronectin was associated with metastasis, but only for CD66b+ cells. Away from the tumor, CD66b expression was correlated with metastasis for immune cells deficient in fibroblast activation protein (FAP)-α expression (Figure 6).
Figure 6.
Select protein marker expression predictive of distant metastasis, conditional on cell type (stratified by median expression): pancytokeratin (PanCk) expression stratified by CD66b-expressing cells inside the tumor (Intra; A), fibronectin stratified by CD66b at the interface (Inter; B), and CD66b stratified by fibroblast activation protein (FAP)-α away from the tumor (Away; C). Marker expression plotted in beeswarm plots was filtered on the basis of the detection of outliers using a modified Tukey outlier test—after this initial filtering, only points between the 10% and 90% quantiles for each stratum were included. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
The identified metastasis-related interactions were additionally predictive beyond considering the individual protein biomarkers alone [proportion of times interactions outperformed individual biomarkers: P = 0.85 (95% CI, 0.68–0.94); P < 0.0001]. A complete listing of relevant metastasis-related interactions can be found in the Shiny application and supplementary table (Supplemental Table S1) and figures (Supplemental Figures S7–S10).
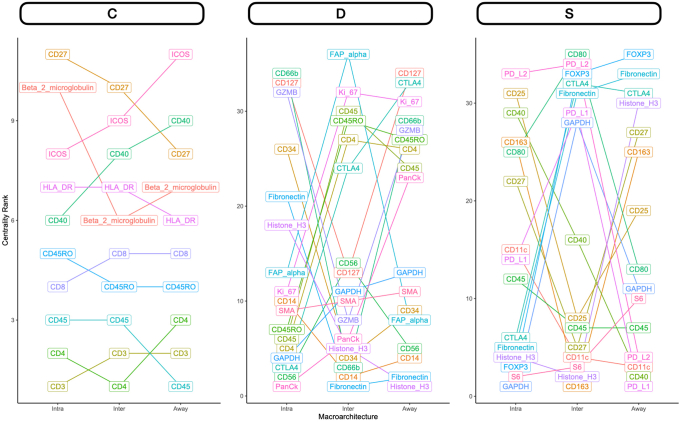
Differential Co-Expression
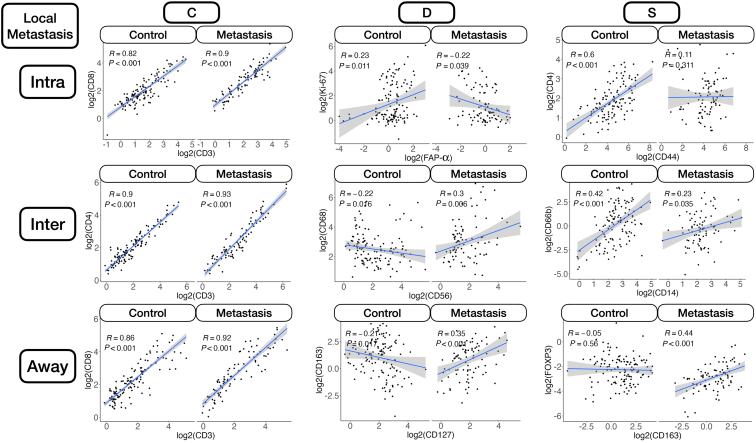
Nodal Metastasis
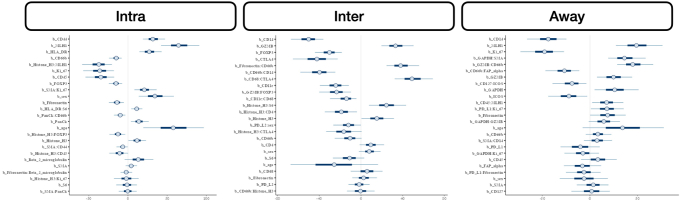
Intratumoral: An analysis of differential co-expression within architectures revealed conserved co-expression/colocalization among helper, cytotoxic T cells and their co-activators (eg, CD3, CD4, CD8, and CD40) within the intratumoral region between patients with or without nodal metastasis. Important co-expressed genes did not change across tissue architectures. Fibronectin, CD44, FAP-α, and CD127 exhibited significant differential intratumoral co-expression between patients based on their lymph node status. Interface/away: Interestingly, co-expression with fibronectin, FAP-α, and CD44 became increasingly less relevant (as defined by either differential or specific co-expression) for nodal metastasis at the interface and away from the tumor, whereas CD163 demonstrated the opposite relationship (Figure 6 and Supplemental Figures S11–S15).
Distant Metastasis
Patterns of conserved co-expression were similar between patients with nodal and distant metastasis compared with the controls. Intratumoral: Co-expression with CD40 and inducible T-cell costimulator (ICOS) was increasingly less relevant as a function of distance to the tumor for patients with distant metastasis, whereas CD27 co-expression was increasingly relevant with distance. Differential co-expression with fibronectin in the tumor's periphery (inter and away) was associated more with distant metastasis than co-expression inside the tumor. Interface/away: Co-expression with fibronectin and FOXP3 at the periphery and beyond was not specific to metastasis/controls compared with intratumoral regions (Figure 7 and Supplemental Figures S16–S18).
Figure 7.
Differential co-expression between select protein markers, stratified by lymph node metastasis status within three tissue architectures (Intra, Inter, and Away). C indicates whether co-expression was conserved between patients with and without metastasis; D indicates whether co-expression differed between patients with and without metastasis; and S indicates whether significant co-expression was specific to either patients with or without metastasis. Intra, Inter, and Away are described in Data Acquisition and Preprocessing. FAP, fibroblast activation protein.
Quantitative findings can be found in the Shiny application. Significant markers were additionally predictive when taken together using hierarchical clustering (Supplemental Figure S19).
Discussion
Examination of regional lymph nodes at the time of surgical resection is essential for CRC prognostication through accurate TNM staging. Although it is important to maximize the number of lymph nodes assessed, recent population-based studies have shown that examination and processing of lymph node involvement is usually incomplete or inadequate. This incomplete or inadequate assessment can impact the accuracy of tumor staging and downstream disease management options, such as whether the patient should receive adjuvant chemotherapy. Instead, assessing lymph nodes (eg, submit higher percentage of resected fat) can increase lymph node yield. To complement these increasingly thorough assessments, developing alternative methods that assess lymph node involvement through indirect molecular mechanisms could be useful in cases where lymph node examination/processing is inadequate.
Multiplexed spatial transcriptomics and proteomics methods help identify novel predictive -omics signatures of metastasis through indirect observation from the primary site. In a set of stage pT3 tumors with or without nodal and/or distant metastases, the study sought to identify spatial proteomic markers of metastasis with digital spatial profiling of immune cells. Furthermore, compared with previous studies7, 56 with limited multiplexing capacity, machine learning technologies were leveraged to evaluate multiple markers in conjunction with increased capacity to determine metastasis status. Three distinct architectures (intra, inter, and away) were assessed within the primary site for i) relative abundance (ratio tests), ii) interactions (classifier), and iii) co-expression (differential co-expression analysis). Potential nodal and distant metastasis biomarkers were identified separately and together for patients with or without tumor metastasis, which will be pursued in the future using additional data collection and orthogonal validation using assays less onerous and subject to variation compared with the DSP.
Although the limited sample of this study size precludes any firm conclusions, it demonstrated important trends of concordant expression levels across the distinct tumor architectures. Several proteins (eg, GZMB, CD8, PD-L1, CD3, FOXP3, CD56, fibronectin, and CD66b) appeared to be important in metastasis. The study identified several emergent trends that will motivate future work such as: i) architectural differences, ii) the role of GZMB and the dual role of extracellular remodeling, iii) the role of PD-L1–expressing dendritic cells, iv) T-cell exhaustion in tumors with mismatch repair deficiencies, v) paradoxical role of immune suppression in CRC, vi) predictive value of NK cells in a nonimmunosuppressive environment, and vii) neutrophil infiltrates and fibroblast activation.
Architectural Differences
The findings differed between the three distinct architectures. For instance, a CD34–PD-L1 interaction was observed in the intratumoral region. An interaction between CD3 and CD44 was found at the tumor interface. A positive association between CD127 and nodal metastasis was strengthened for CD66b+ cells at the interface. Fibronectin, CD44, FAP-α, and CD127 exhibited significant differential intratumoral co-expression between patients based on their lymph node status, whereas several helper, cytotoxic T cells and their co-activators, such as CD3, CD4, CD8, and CD40, exhibited conserved co-expression within the intratumoral region between patients with and without nodal metastasis. These are examples of many architectural differences, reaffirming the importance of evaluating the tumor invasive margin/interface.
Role of GZMB
GZMB is a potent molecule used by CD8 T cells to induce cytotoxicity when detecting certain antigens. As an extracellular matrix agent that induces apoptosis, GZMB is involved in cleaving certain target proteins, leading to DNA fragmentation and loss of membrane integrity.47 Its role in colorectal cancer was recently elucidated by Daemen et al.48 The current study showed that GZMB is associated with a better prognosis of CRC in both MSI-positive and MSI-negative clones. Its function was not only linked to cytotoxic T cells but was also found to be elicited by tumor cells independent of CD8, with a role in cleaving vitronectin, fibronectin, and laminin.48 This hypothesis is supported by previous experiments of pretreating the laminin matrix with GZMB with significant inhibition of cell spreading in the LIM1215 colon cancer cell line.49 In dMMR patients, GZMB-expressing immune cells were associated with distant metastasis, which contradicts these prior findings. Additionally, fibronectin expression was positively associated with distant metastasis in immune cells, which did not express GZMB. Although GZMB may assist in extracellular matrix remodeling, allowing for the transmigration of T cells, similar remodeling of the extracellular matrix could potentially facilitate the migration of tumor cells, suggesting a dual role for this protein.50
Role of PD-L1–Expressing Dendritic Cells
PD-L1 is a protein found in the cell surface of tumor cells that couples with the PD-1 protein of T cells, causing inhibition of the T cells' immune functions against the tumor. Therefore, the current results, showing increased PD-L1 expression near immune cells at the invasive margin for local lymph node involvement and inside the tumor in dMMR patients for distant metastasis, agree with previous studies that link this molecule to immune evasion.51 The current study also agrees with a prior study of 221 patients with stage pT3 colon cancer that investigated cancer tissues immunostained to examine the prognostic impact of CD11c+ dendritic cell co-expressing PD-L1 and their spatial relationship with CD8+ T cells. Significant survival benefits for patients with intratumoral CD8+ cell density, stromal CD11c+ cell density, intratumoral CD11c+ PD-L1+ cell density, and stromal CD11c+ PD-L1+ cell density were found. CD8+ cell density was positively correlated with both CD11c+ cell density and CD11c+ PD-L1+ cell density in tumor epithelium and stromal compartments. CD11c is a member of the integrin family (adhesion molecule), and is particularly expressed in dendritic cells.52 Dendritic cells are antigen-presenting cells that play an important role in adaptive immunity by attracting naïve T cells to activate, differentiate, and finally infiltrate tumors. Mature tumor-infiltrating dendritic cells are associated with better prognosis.53 CD11c dendritic cells play an important role in cancer control in various types of cancer. Patients with gastric cancer with high CD11c expression levels have, on average, a better survival and significantly reduced risk of relapse. Lee et al54 studied tissue micro-arrays from 681 pretreated patients with triple-negative breast cancer. Microscopically, CD11c cells are concentrated in areas with high numbers of TILs. Tumors with high expression of CD11c also had higher histologic grades. More importantly, those with lymph node metastasis and high CD11c expression showed a trend of increased recurrence-free survival and significantly better overall survival (P = 0.047) compared with those with low CD11c expression.54 These results are also corroborated by findings herein, where PD-L1–expressing CD11c+ cells (dendritic cells) were negatively associated with distant metastasis.
T-Cell Exhaustion in Tumors with Mismatch Repair Deficiencies
Stimulating the recruitment of cytotoxic T cells has been clinically explored in immunotherapy. One example of a molecule that has been studied as a potential therapeutic target is CD3, a protein found on the surface of T cells, with downstream signaling resulting in the activation of T cells. CD3 is an important target as it can redirect T cells to attack tumors by secreting inflammatory cytokines and cytolytic molecules, but its overproduction may cause a cytokine storm, associated with worse outcomes. CD3 is an important pan-T-cell antigen; direct targeting of CD3 cells can redirect other T cells to attack the tumors through secreting inflammatory cytokines and cytolytic molecules.52, 53, 54 Herein, expression of CD3 and CD8 inside the tumor and at the invasive margin was positively associated with local metastasis, which contradicts the general notion that the presence of these biomarkers suggests a favorable diagnosis. This is not the first time contradictions of these predictive effects have been discussed, as the predictive value of these markers can vary widely based on mismatch repair status.57 This is partly because a mismatch repair–deficient tumor can exhibit a T-cell exhaustion phenotype, even before metastasis, which can indicate a diminished capacity to impede metastasis (eg, exhaustion or functional suppression from other TME immune cells). In particular, the association with CD8 was statistically significant for MMR-proficient patients and marginally significant for dMMR patients. As tumors were not profiled before metastasis, reverse causality is also possible (ie, infiltration at the primary site as instigated by metastasis). As the Immunoscore, a digital pathology assay that assesses immune cells at the tumor's core and invasive margin, is derived from these CD3 and CD8 measurements, further exploration of the interaction between the Immunoscore and mismatch repair for colon cancer prognostication is warranted.7,28 Patients with dMMR also exhibit heterogeneous immune activation (eg, COLD and HOT tumors), which are related to different prognostic outcomes and can be elucidated through whole transcriptomic characterization.29
Paradoxical Role of Immune Suppression in CRC
The FOXP3 gene, located on the X chromosome, is a transcription factor that plays an important role in the production of CD4+ CD25+ regulatory T cells.58 However, its exact role in cancer development and metastasis remains elusive. Although in the past FOXP3 expression had been associated only with regulatory T cells and studied almost exclusively in lymphoid tissue, FOXP3 expression in nearby neighboring tumor cells can lead to immune suppression by curbing the immune response, further complicating the conclusion of previous research results.59 Although for most human carcinomas, FOXP3 is typically associated with poor prognosis, paradoxically, the opposite holds true for CRC.60,61 FOXP3 can also function as a tumor suppressor, with its expression linked to better outcomes.62 However, the opposite has also been reported in other cancers [namely, its action as an oncogene through activation of the adenomatous polyposis coli and epithelial-to-mesenchymal pathways through fibroblast differentiation (release of GZMB)].63 Further complicating the analysis of FOXP3 is the fact that it has four isoforms, each being functional but interacting with different molecules that play a role in cancer progression. For instance, further evidence suggests that FOXP3-expressing T-regulatory cells could be further fractionated into lineages, which are nonsuppressive but secrete proinflammatory cytokines.64 Several studies also suggest that the protective effects of FOXP3 may be mediated through, interact with, or be impacted by the abundance of certain host microbiota by suppressing the inflammatory response to gut microbiota. The current study noted the protective effects of FOXP3 away from the tumor, particularly for dMMR patients. Regardless, more studies are needed, controlling for FOXP3's isoforms and their various interactions in the tumor and its microenvironment.65
Prognostic Value of NK Cells in a Nonimmunosuppressive Environment
NK cells infiltrate solid tumors to lyse cancerous cells. The current study identified CD56+ NK cells as a significant protective factor against metastasis in the peritumoral region, but only for dMMR patients. This is consistent with the observation of substantial inhibition of NK in an immunosuppressive TME, where presence, despite the inhibition, would prove favorable. Sconocchia et al66 studied NK cells in 1410 CRC specimens and other solid tumors and mostly confirmed that NK cells are minimally detectable. However, an interesting subgroup of patients were identified who exhibited a high degree of NK cell infiltration along with their CD8+ T cells. CD56 was used as the antigenic biomarker of NK cells. Using CD56 expression to track the degree of NK cell infiltration in colorectal tumors and further characterizing CD8+ lymphocytes, they found that CD56+ CD8+ patients presented significantly higher survival (80%) versus 55% for the CD56– CD8+ group.66 Similar findings have demonstrated that CD56+ patients with rectal cancer experience significantly better overall survival, with CD56+ as a signature for clinical decision-making and treatment duration. Others have explored how NK cells may be activated after treatment with cetuximab plus IL-2 or IL-15.67 The current study identified a potential interaction between CD56 and MMR status.
Neutrophil Infiltrates and Fibroblast Activation
CD66b is an antigenic marker of tumor-infiltrating neutrophils, representing proinflammatory myeloid infiltrates.68,69 Infiltrating neutrophils seem to increase cancer invasion, lymph node metastasis, and tumor stage.70,71 The neutrophil/lymphocyte ratio in the intratumoral regions commonly serves as a prognostic indicator. Prior studies have suggested that a high neutrophil/lymphocyte ratio could suggest a poor prognosis. In CRC, the prognostic role of tumor-associated neutrophils has not been established because various authors report conflicting results, ranging from poor prognosis, to no association, to improved prognosis. For instance, similar to the current study findings, a prior study suggested that absence of CD66b+ immune cell infiltrates could be predictive of local metastasis.72 It has been implied that favorable antitumoral effects could be mediated through costimulation of other cell lineages (eg, CD3+/CD8+). Cross talk between cytotoxic T cells and neutrophils can be further substantiated by studying factors pertaining to neutrophil recruitment. Interestingly, recent publications report neutrophils as a protective factor, whereas older articles demonstrate a negative role for neutrophils.73,74 Interaction between CD66b+ cells, fibronectin, and other fibroblast activation proteins (eg, FAP-α) have been noted. Fibronectin has been previously implicated as means to promote invasion and metastasis and is a crucial structural component of angiogenic tumors. At the invasive margin, fibronectin's impact on local metastasis was specific to CD66+ cells compared with CD66– cells. These effects were not identified away from the tumor. However, the opposite effect was noted for FAP-α: non–fibroblast-activating (FAP-α–) CD66+ cells away from the tumor exhibited a higher risk of metastasis compared with FAP-α– CD66– cells.75 For CD66b+ cells in the away region (eg, stroma), FAP-α appeared to be negatively associated with local involvement. Colocalization between these two cell types was noticed away from the tumor.76 These findings coincide with prior research suggesting that cancer-associated fibroblasts can regulate the recruitment of myeloid cells.77,78
Limitations and Future Directions
There are several study limitations worth mentioning. The sample size was limited because of the high cost of spatial proteomics assays on a per-slide basis, making it challenging to control for tumor site. The sample size also limits both the certainty of the current study findings as well as the power to reveal additional biomarkers of metastasis. Tumors with distant but not local metastasis were not assessed, which may derive separate predictive cell lineages as the notion of lymph node metastasis serving as a staging ground for distant metastasis has been disputed as the exclusive progression.79, 80, 81, 82 Potential batch effects were controlled for by balancing the batches with an even number of patients with and without metastasis and further adjustments via mixed effects machine learning and statistical models. Even so, there may be technical factors introducing heterogeneous expression, which were uncontrolled. Prior treatment (eg, chemotherapy) and comorbidities may have interfered with the assessment of the primary, in spite of it being exclusively searched and filtered for as inclusion criteria. The nCounter proteomics assay does not account for different protein isoforms and post-translational modifications (eg, phosphorylation), which could prove additionally (ie, more biomarkers) predictive of metastasis. The number of protein markers profiled on the basis of immune-relevant panels available for the GeoMX DSP platform were limited. Additional protein panels (eg, immune-oncology drug target, pan-tumor, mitogen-activated protein kinase signaling, phosphatidylinositol 3-kinase/AKT, and cell death panel) could have been selected for analysis. Additional custom targets could have been selected though the three panels selected were germane to immune-related pathogenesis/metastasis while targets in these other panels were less related to the research objective and would have been underutilized. In addition, nonimmune cell lineages could have been selected through adoption of additional morphology markers that highlight areas to be profiled using the DSP. Furthermore, although many of the ROIs placed away from the tumor were placed in the stroma, additional tissue architectural differences (eg, peritumoral fat, immune nests, and benign epithelium) were not explicitly accounted for. Although a semi-autonomous workflow was developed for placement of ROIs, biased placement of DSP ROIs may impact study findings, warranting the exploration of assays that do not experience these effects (eg, Visium Spatial Transcriptomics).83 The Visium assay could also provide the capacity for untargeted characterization of the whole transcriptome at 50-μm resolution. Pairing these assays with a proteomics assessment will be explored in future work. Promising biomarkers established from these studies will be orthogonally validated through immunostaining.
Despite these limitations, leveraging the Digital Spatial Profiler for a highly multiplexed assessment of colorectal tumors has allowed for the simultaneous exploration of several emergent pathways and immunomodulatory effects that characterize the potential for tumor metastasis. Consulting the known literature on tumor immune microenvironment to corroborate our study conclusions has demonstrated that CRC tumor immunology is still riddled with contradictory findings that continue to perplex researchers on this important subject. Spatial profiling provides an opportunity to further disaggregate the tumor immune microenvironment. Although the current study sought to demonstrate associations with tumor metastasis, future works will explore how these assessments can be put into practice to complement incomplete lymph node assessment and demonstrate additional predictive value for ascertaining the risk of recurrence, above and beyond pTNM staging. This will be accomplished by either restricting by pathologic stage or controlling for pTNM stage as a covariate in a follow-up study where enough follow-up time has passed to collect meaningful recurrence/survival information. The current study identifies factors pertaining to metastasis beyond local invasiveness by restricting to pT3 staged tumors. With the advent of multimodal spatial assays, which allow for greater multiplexing at higher resolution, future iterations of these studies may further elucidate the precise mechanisms of metastasis as means to develop low-cost diagnostic/prognostic tests that complement traditional assessments (eg, Immunoscore and lymph node assessment) and inform novel therapeutics.
Conclusion
Deciphering the tumor immune microenvironment is key to improving the prognostication and treatment of colorectal adenocarcinoma. Spatial assessment technologies will continue to play a key role in delineating crucial architectural and cellular components of CRC tumor metastasis. Herein, the Digital Spatial Profiler was utilized to uncover spatial proteomics biomarkers of nodal and distant metastasis. This study identified proteomics markers that can provide additional predictive value for metastasis (by restricting to pT3 patients) when used in conjunction (eg, relative abundance, co-expression, and interaction). There is significant room for further investigation through unbiased and untargeted spatial RNA assays, as emergent themes appear contradictory to components of the previous literature. There are plans to identify independent risk factors for recurrence and survival using this spatial approach given adequate patient follow-up. Future plans involve leveraging high-dimensional spatial mRNA and single-cell assays to further contextualize metastasis etiology and pathogenesis and to operationalize significant results into informative tests that complement existing assessment methods for CRC prognosis.
Acknowledgments
We thank Gabriel Brooks and Linda Vahdat for thoughtful discussions of the subject matter.
Footnotes
Supported by NIH grants R01CA216265, R01CA253976, and P20GM104416 (B.C.C.); NIH subawards P20GM104416 and P20GM130454 (J.J.L.); Dartmouth College Neukom Institute for Computational Science CompX awards (B.C.C., J.J.L., and L.J.V.); and Dartmouth Cancer Center, Department of Pathology and Laboratory Medicine Clinical Genomics and Advanced Technologies EDIT program.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2023.02.020.
Contributor Information
Joshua J. Levy, Email: joshua.j.levy@dartmouth.edu.
Gregory J. Tsongalis, Email: gregory.j.tsongalis@hitchcock.org.
Supplemental Data
Supplemental Figure S1.
Volcano plots illustrating differentially expressed protein markers of any metastasis. Results stratified by tissue architecture: intratumoral (Intra; A, D, and G); peritumoral (Inter; B, E, and H); and away/stroma (Away; C, F, and I); results also stratified by mismatch repair (MMR) status: MMR proficient (pMMR; D–F) and MMR deficient (dMMR; G–I); statistical significance cutoff at α=0.05; x-axis indicates effect size and directionality (positive x-value indicates metastasis-related marker; negative indicates decreased metastasis risk); y-axis indicates effect significance (positive y-value indicates lower P value). Intra, Inter, and Away are described in Data Acquisition and Preprocessing.FOXP3, forkhead box P3; GZMB, granzyme B; PanCk, pancytokeratin; PD-L2, programmed death ligand 2.
Supplemental Figure S2.
Box plots and beeswarm scatterplots of select protein marker expression for biomarkers predictive of any metastasis, stratified by mismatch repair (MMR) status: programmed death ligand 2 (PD-L2) inside the tumor (Intra; A); CD56 at the interface (Inter; B); CD11c away from tumor (Away; C); and forkhead box P3 (FOXP3) away from tumor (D). Marker expression plotted in beeswarm plots was filtered on the basis of detection of outliers using a modified Tukey outlier test—after this initial filtering, only points between the 10% and 90% quantiles for each stratum were included. Intra, Inter, and Away are described in Data Acquisition and Preprocessing. dMMR, MMR deficient; pMMR, MMR proficient.
Supplemental Figure S3.
Volcano plots illustrating differentially expressed protein markers of distant metastasis. Results stratified by tissue architecture: intratumoral (Intra; A, D, and G); peritumoral (Inter; B, E, and H); and away/stroma (Away; C, F, and I); results also stratified by mismatch repair (MMR) status: MMR proficient (pMMR; D–F) and MMR deficient (dMMR; G–I); statistical significance cutoff at α=0.05;; x-axis indicates effect size and directionality (positive x-value indicates metastasis-related marker; negative indicates decreased metastasis risk); y-axis indicates effect significance (positive y-value indicates lower P value). Intra, Inter, and Away are described in Data Acquisition and Preprocessing. GZMB, granzyme B; PanCk: pancytokeratin; PD-L1, programmed death ligand 1.
Supplemental Figure S4.
Box plots and beeswarm scatterplots of select protein marker expression for biomarkers predictive of distant metastasis, stratified by mismatch repair (MMR) status: granzyme B (GZMB) inside the tumor (Intra; A); GZMB at the interface (Inter; B); CD11c away from tumor (Away; C); and Ki-67 away from tumor (D). Marker expression plotted in beeswarm plots was filtered on the basis of detection of outliers using a modified Tukey outlier test—after this initial filtering, only points between the 10% and 90% quantiles for each stratum were included. Intra, Inter, and Away are described in Data Acquisition and Preprocessing. dMMR, MMR deficient; pMMR, MMR proficient.
Supplemental Figure S5.
Box plots and beeswarm scatterplots of relative protein expression between two markers, predictive of any metastasis, stratified by mismatch repair (MMR) status: β-2-microglobulin/programmed death ligand 2 (PD-L2) inside the tumor (Intra; A); CD11c/granzyme B (GZMB) at the interface (Inter; B); CD11c/GZMB away from tumor (Away; C); and CD11c/CD14 away from tumor (D). Marker expression plotted in beeswarm plots was filtered on the basis of detection of outliers using a modified Tukey outlier test—after this initial filtering, only points between the 10% and 90% quantiles for each stratum were included. Intra, Inter, and Away are described in Data Acquisition and Preprocessing. dMMR, MMR deficient; pMMR, MMR proficient.
Supplemental Figure S6.
Box plots and beeswarm scatterplots of relative protein expression between two markers, predictive of distant metastasis, stratified by mismatch repair (MMR) status: CD8/programmed death ligand 1 (PD-L1) inside the tumor (Intra; A); CD4/PD-L1 inside the tumor (B); CD11c/granzyme B (GZMB) at the interface (Inter; C); and CD11c/CD34 away from tumor (Away; D). Marker expression plotted in beeswarm plots was filtered on the basis of detection of outliers using a modified Tukey outlier test—after this initial filtering, only points between the 10% and 90% quantiles for each stratum were included. Intra, Inter, and Away are described in Data Acquisition and Preprocessing. dMMR, MMR deficient; pMMR, MMR proficient.
Supplemental Figure S7.
Posterior interval estimates from Markov Chain Monte Carlo (MCMC) draw from bayesian generalized linear mixed effects models for prediction of any metastasis from standardized protein markers and their interactions, stratified by architecture; predictors were derived from mixed effects machine learning models and subselected using Horseshoe Least Absolute Shrinkage and Selection Operator (LASSO) before unpenalized statistical testing. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S8.
Box plots and beeswarm scatterplots of select protein marker expression, conditional on cell type (stratified by median expression), predictive of any metastasis: CD34 expression stratified by programmed death ligand 1 (PD-L1)–expressing cells inside the tumor (Intra; A), CD27 stratified by fibronectin at the interface (Inter; B), and CD27 stratified by CD40 away from the tumor (Away; C). Marker expression plotted in beeswarm plots was filtered on the basis of detection of outliers using a modified Tukey outlier test—after this initial filtering, only points between the 10% and 90% quantiles for each stratum were included. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S9.
Posterior interval estimates from Markov Chain Monte Carlo (MCMC) draw from bayesian generalized linear mixed effects models for prediction of nodal metastasis from standardized protein markers and their interactions, stratified by architecture; predictors were derived from mixed effects machine learning models and subselected using Horseshoe Least Absolute Shrinkage and Selection Operator (LASSO) before unpenalized statistical testing. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S10.
Posterior interval estimates from Markov Chain Monte Carlo (MCMC) draw from bayesian generalized linear mixed effects models for prediction of distant metastasis from standardized protein markers and their interactions, stratified by architecture; predictors were derived from mixed effects machine learning models and subselected using Horseshoe Least Absolute Shrinkage and Selection Operator (LASSO) before unpenalized statistical testing. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S11.
Differential co-expression networks predictive of any metastasis, stratified by three tissue architectures (Intra, Inter, and Away). C indicates whether co-expression was conserved between patients with and without metastasis; D indicates whether co-expression differed between patients with and without metastasis; and S indicates whether significant co-expression was specific to either patients with or without metastasis. Edges between markers indicate whether relationship could be characterized by C, D or S. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S12.
Rank-based summaries of proteins important to the differential co-expression networks predictive of any metastasis, stratified by three tissue architectures (Intra, Inter, and Away). C indicates whether co-expression was conserved between patients with and without metastasis; D indicates whether co-expression differed between patients with and without metastasis; and S indicates whether significant co-expression was specific to either patients with or without metastasis. Rank indicates eigenvector centrality of protein within each of the networks (lower rank indicates importance in network); proteins with top-10 overall rank were selected for viewing. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S13.
Differential co-expression scatterplots between select protein markers, stratified by any metastasis status within three tissue architectures (Intra, Inter, and Away). C indicates whether co-expression was conserved between patients with and without metastasis; D indicates whether co-expression differed between patients with and without metastasis; and S indicates whether significant co-expression was specific to either patients with or without metastasis. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S14.
Differential co-expression networks predictive of nodal metastasis, stratified by three tissue architectures (Intra, Inter, and Away). C indicates whether co-expression was conserved between patients with and without metastasis; D indicates whether co-expression differed between patients with and without metastasis; and S indicates whether significant co-expression was specific to either patients with or without metastasis. Edges between markers indicate whether relationship could be characterized by C, D or S. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S15.
Rank-based summaries of proteins important to the differential co-expression networks predictive of nodal metastasis, stratified by three tissue architectures (Intra, Inter, and Away). C indicates whether co-expression was conserved between patients with and without metastasis; D indicates whether co-expression differed between patients with and without metastasis; and S indicates whether significant co-expression was specific to either patients with or without metastasis. Rank indicates eigenvector centrality of protein within each of the networks (lower rank indicates importance in network); proteins with top-10 overall rank were selected for viewing. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S16.
Differential co-expression networks predictive of distant metastasis, stratified by three tissue architectures (Intra, Inter, and Away). C indicates whether co-expression was conserved between patients with and without metastasis; D indicates whether co-expression differed between patients with and without metastasis; and S indicates whether significant co-expression was specific to either patients with or without metastasis. Edges between markers indicate whether relationship could be characterized by C, D or S. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S17.
Rank-based summaries of proteins important to the differential co-expression networks predictive of distant metastasis, stratified by three tissue architectures (Intra, Inter, and Away). C indicates whether co-expression was conserved between patients with and without metastasis; D indicates whether co-expression differed between patients with and without metastasis; and S indicates whether significant co-expression was specific to either patients with or without metastasis. Rank indicates eigenvector centrality of protein within each of the networks (lower rank indicates importance in network); proteins with top-10 overall rank were selected for viewing. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S18.
Differential co-expression scatterplots between select protein markers, stratified by distant metastasis status within three tissue architectures (Intra, Inter, and Away). C indicates whether co-expression was conserved between patients with and without metastasis; D indicates whether co-expression differed between patients with and without metastasis; and S indicates whether significant co-expression was specific to either patients with or without metastasis. Intra, Inter, and Away are described in Data Acquisition and Preprocessing.
Supplemental Figure S19.
Hierarchical clustering of predictive markers of metastasis (METS) extracted from study, within three tissue architectures (Intra, Inter, and Away), stratified by mismatch repair (MMR) status; Fisher exact tests were used to compare cluster assignment with presence of metastasis to report P values at bottom of plot to indicate overall associations. Intra, Inter, and Away are described in Data Acquisition and Preprocessing. dMMR, MMR deficient; pMMR, MMR proficient.
References
- 1.Carethers J.M., Doubeni C.A. Causes of socioeconomic disparities in colorectal cancer and intervention framework and strategies. Gastroenterology. 2020;158:354–367. doi: 10.1053/j.gastro.2019.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasi P.M., Shahjehan F., Cochuyt J.J., Li Z., Colibaseanu D.T., Merchea A. Rising proportion of young individuals with rectal and colon cancer. Clin Colorectal Cancer. 2019;18:e87–e95. doi: 10.1016/j.clcc.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Slattery M.L. Diet, lifestyle, and colon cancer. Semin Gastrointest Dis. 2000;11:142–146. [PubMed] [Google Scholar]
- 4.Araghi M., Soerjomataram I., Bardot A., Ferlay J., Cabasag C.J., Morrison D.S., De P., Tervonen H., Walsh P.M., Bucher O. Changes in colorectal cancer incidence in seven high-income countries: a population-based study. Lancet Gastroenterol Hepatol. 2019;4:511–518. doi: 10.1016/S2468-1253(19)30147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 6.Qaderi S.M., Galjart B., Verhoef C., Slooter G.D., Koopman M., Verhoeven R.H., de Wilt J.H., van Erning F.N. Disease recurrence after colorectal cancer surgery in the modern era: a population-based study. Int J Colorectal Dis. 2021;36:2399–2410. doi: 10.1007/s00384-021-03914-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruni D., Angell H.K., Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20:662–680. doi: 10.1038/s41568-020-0285-7. [DOI] [PubMed] [Google Scholar]
- 8.Dalerba P., Sahoo D., Paik S., Guo X., Yothers G., Song N., Wilcox-Fogel N., Forgó E., Rajendran P.S., Miranda S.P., Hisamori S., Hutchison J., Kalisky T., Qian D., Wolmark N., Fisher G.A., van de Rijn M., Clarke M.F. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N Engl J Med. 2016;374:211–222. doi: 10.1056/NEJMoa1506597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarazona N., Gimeno-Valiente F., Gambardella V., Huerta M., Roselló S., Zuniga S., Calon A., Carbonell-Asins J.A., Fontana E., Martinez-Ciarpaglini C., Eason K., Rentero-Garrido P., Fleitas T., Papaccio F., Moro-Valdezate D., Nyamundanda G., Castillo J., Espí A., Sadanandam A., Roda D., Cervantes A. Detection of postoperative plasma circulating tumour DNA and lack of CDX2 expression as markers of recurrence in patients with mmuneed colon cancer. ESMO Open. 2020;5:e000847. doi: 10.1136/esmoopen-2020-000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee H., Sha D., Foster N.R., Shi Q., Alberts S.R., Smyrk T.C., Sinicrope F.A. Analysis of tumor microenvironmental features to refine prognosis by T, N risk group in patients with stage III colon cancer (NCCTG N0147) (Alliance) Ann Oncol. 2020;31:487–494. doi: 10.1016/j.annonc.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lizardo D.Y., Kuang C., Hao S., Yu J., Huang Y., Zhang L. Immunotherapy efficacy on mismatch repair-deficient colorectal cancer: from bench to bedside. Biochim Biophys Acta Rev Cancer. 2020;1874:188447. doi: 10.1016/j.bbcan.2020.188447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon H.H., Shi Q., Heying E.N., Muranyi A., Bredno J., Ough F., Djalilvand A., Clements J., Bowermaster R., Liu W.-W., Barnes M., Alberts S.R., Shanmugam K., Sinicrope F.A. Intertumoral heterogeneity of CD3+ and CD8+ T-cell densities in the microenvironment of DNA mismatch-repair–deficient colon cancers: implications for prognosis. Clin Cancer Res. 2019;25:125–133. doi: 10.1158/1078-0432.CCR-18-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senthil M., Trisal V., Paz I.B., Lai L.L. Prediction of the adequacy of lymph node retrieval in colon cancer by hospital type. Arch Surg. 2010;145:840–843. doi: 10.1001/archsurg.2010.182. [DOI] [PubMed] [Google Scholar]
- 14.Baxter N.N., Virnig D.J., Rothenberger D.A., Morris A.M., Jessurun J., Virnig B.A. Lymph node evaluation in colorectal cancer patients: a population-based study. J Natl Cancer Inst. 2005;97:219–225. doi: 10.1093/jnci/dji020. [DOI] [PubMed] [Google Scholar]
- 15.Ong M.L., Schofield J.B. Assessment of lymph node involvement in colorectal cancer. World J Gastrointest Surg. 2016;8:179. doi: 10.4240/wjgs.v8.i3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schofield J.B., Mounter N.A., Mallett R., Haboubi N.Y. The importance of accurate pathological assessment of lymph node involvement in colorectal cancer. Colorectal Dis. 2006;8:460–470. doi: 10.1111/j.1463-1318.2006.01044.x. [DOI] [PubMed] [Google Scholar]
- 17.Hartgrink H.H., van de Velde C.J.H., Putter H., Bonenkamp J.J., Klein Kranenbarg E., Songun I., Welvaart K., van Krieken J.H.J.M., Meijer S., Plukker J.T.M., van Elk P.J., Obertop H., Gouma D.J., van Lanschot JJB, Taat C.W., de Graaf P.W., von Meyenfeldt M.F., Tilanus H., Sasako M. Extended lymph node dissection for gastric cancer: who may benefit? final results of the randomized Dutch gastric cancer group trial. J Clin Oncol. 2004;22:2069–2077. doi: 10.1200/JCO.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 18.Pelka K., Hofree M., Chen J.H., Sarkizova S., Pirl J.D., Jorgji V., Bejnood A., Dionne D., William H.G., Xu K.H. Spatially organized multicellular immune hubs in human colorectal cancer. Cell. 2021;184:4734–4752. doi: 10.1016/j.cell.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norgaard Z., Zollinger D., Reeves J., Zhou Z., Kriner M., McKay-Fleisch J., Bahrami A., Warren S., Church S., Merritt C., Hoang M., Beechem J. Abstract 2825: high-plex, spatial RNA profiling of tumor infiltrating leukocytes and the tumor microenvironment of microsatellite instable colorectal cancer using GeoMxTM Digital Spatial Profiler. Cancer Res. 2020;80:2825. [Google Scholar]
- 20.Lazarus J., Maj T., Smith J.J., Perusina Lanfranca M., Rao A., D’Angelica M.I., Delrosario L., Girgis A., Schukow C., Shia J., Kryczek I., Shi J., Wasserman I., Crawford H., Nathan H., Pasca Di Magliano M., Zou W., Frankel T.L. Spatial and phenotypic immune profiling of metastatic colon cancer. JCI Insight. 2018;3:e121932. doi: 10.1172/jci.insight.121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uttam S., Stern A.M., Sevinsky C.J., Furman S., Pullara F., Spagnolo D., Nguyen L., Gough A., Ginty F., Lansing Taylor D., Chakra Chennubhotla S. Spatial domain analysis predicts risk of colorectal cancer recurrence and infers associated tumor microenvironment networks. Nat Commun. 2020;11:3515. doi: 10.1038/s41467-020-17083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dogan A., Demircioglu S. Assessment of the neutrophil-lymphocyte ratio in classic Hodgkin lymphoma patients. Pak J Med Sci. 2019;35:1270–1275. doi: 10.12669/pjms.35.5.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nearchou I.P., Gwyther B.M., Georgiakakis E.C.T., Gavriel C.G., Lillard K., Kajiwara Y., Ueno H., Harrison D.J., Caie P.D. Spatial immune profiling of the colorectal tumor microenvironment predicts good outcome in stage II patients. Npj Digit Med. 2020;3:1–10. doi: 10.1038/s41746-020-0275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nearchou I.P., Lillard K., Gavriel C.G., Ueno H., Harrison D.J. Caie PD: Automated analysis of lymphocytic infiltration, tumor budding, and their spatial relationship improves prognostic accuracy in colorectal cancer. Cancer Immunol Res. 2019;7:609–620. doi: 10.1158/2326-6066.CIR-18-0377. [DOI] [PubMed] [Google Scholar]
- 25.Peltomäki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 26.Kheirelseid E.A.H., Miller N., Chang K.H., Curran C., Hennessey E., Sheehan M., Kerin M.J. Mismatch repair protein expression in colorectal cancer. J Gastrointest Oncol. 2013;4:397–408. doi: 10.3978/j.issn.2078-6891.2013.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy J.J., Bobak C.A., Nasir-Moin M., Veziroglu E.M., Palisoul S.M., Barney R.E., Salas L.A., Christensen B.C., Tsongalis G.J., Vaickus L.J. Mixed effects machine learning models for colon cancer metastasis prediction using spatially localized mmune-oncology markers. Pac Symp Biocomput. 2022;27:175–186. [PMC free article] [PubMed] [Google Scholar]
- 28.Sinicrope F.A., Shi Q., Hermitte F., Zemla T.J., Mlecnik B., Benson A.B., Gill S., Goldberg R.M., Kahlenberg M.S., Nair S.G., Shields A.F., Smyrk T.C., Galon J., Alberts S.R. Contribution of immunoscore and molecular features to survival prediction in stage III colon cancer. JNCI Cancer Spectr. 2020;4:pkaa023. doi: 10.1093/jncics/pkaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giannini R., Zucchelli G., Giordano M., Ugolini C., Moretto R., Ambryszewska K., Leonardi M., Sensi E., Morano F., Pietrantonio F., Cremolini C., Falcone A., Fontanini G. Immune profiling of deficient mismatch repair colorectal cancer tumor microenvironment reveals different levels of immune system activation. J Mol Diagn. 2020;22:685–698. doi: 10.1016/j.jmoldx.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Hinerfeld D., Barker K., Merritt C., Beechem J. Validation of antibody panels for high-plex immunohistochemistry applications. J Biomol Tech JBT. 2019;30:S40–S41. [Google Scholar]
- 31.Rosenbloom A., Cronin J., Bonnett S. 34 Multi-step antibody validation for the geomx® digital spatial profiler. J Immunother Cancer. 2020;8(Suppl 3):AO2–A21. [Google Scholar]
- 32.Makowski D., Ben-Shachar M.S., Chen S.H.A., Lüdecke D. Indices of effect existence and significance in the Bayesian framework. Front Psychol. 2019;10:2767. doi: 10.3389/fpsyg.2019.02767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Searle S.R., Speed F.M., Milliken G.A. Population marginal means in the linear model: an alternative to least squares means. Am Stat Taylor Francis. 1980;34:216–221. [Google Scholar]
- 34.Gelman A., Tuerlinckx F. Type S error rates for classical and Bayesian single and multiple comparison procedures. Comput Stat. 2000;15:373–390. [Google Scholar]
- 35.McElreath R. CRC Press; Boca Raton, FL: 2020. Statistical Rethinking: A Bayesian Course With Examples in R and Stan. [Google Scholar]
- 36.Carpenter B., Gelman A., Hoffman M.D., Lee D., Goodrich B., Betancourt M., Brubaker M., Guo J., Li P., Riddell A. Stan: a probabilistic programming language. J Stat Softw. 2017;76:1–32. doi: 10.18637/jss.v076.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bürkner P.-C. Advanced Bayesian multilevel modeling with the R package brms. The R Journal. 2018;10:395–411. [Google Scholar]
- 38.Bürkner P.-C. brms: An R package for Bayesian multilevel models using stan. J Stat Softw. 2017;80:1–28. [Google Scholar]
- 39.Sigrist F. Latent gaussian model boosting. IEEE Trans Pattern Anal Mach Intell. 2022;45:1894–1905. doi: 10.1109/TPAMI.2022.3168152. [DOI] [PubMed] [Google Scholar]
- 40.Tan Y.V., Roy J. Bayesian additive regression trees and the general BART model. Stat Med. 2019;38:5048–5069. doi: 10.1002/sim.8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy J.J., O’Malley A.J. Don’t dismiss logistic regression: the case for sensible extraction of interactions in the era of machine learning. BMC Med Res Methodol. 2020;20:171. doi: 10.1186/s12874-020-01046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhadra A., Datta J., Polson N.G., Willard B. Lasso meets horseshoe. Stat Sci JSTOR. 2019;34:405–427. [Google Scholar]
- 43.Bartonicek A., Wickham S.R., Pat N., Conner T.S. The value of Bayesian predictive projection for variable selection: an example of selecting lifestyle predictors of young adult well-being. BMC Public Health. 2021;21:695. doi: 10.1186/s12889-021-10690-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savino A., Provero P., Poli V. Differential co-expression analyses allow the identification of critical signalling pathways altered during tumour transformation and progression. Int J Mol Sci. 2020;21:9461. doi: 10.3390/ijms21249461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen J.P., Almaas E. csdR, an R package for differential co-expression analysis. BMC Bioinformatics. 2022;23:79. doi: 10.1186/s12859-022-04605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia L., Yao W., Jiang Y., Li Y., Wang Z., Li H., Huang F., Li J., Chen T., Zhang H. Development of interactive biological web applications with R/Shiny. Brief Bioinform. 2022;23:bbab415. doi: 10.1093/bib/bbab415. [DOI] [PubMed] [Google Scholar]
- 47.Salti S.M., Hammelev E.M., Grewal J.L., Reddy S.T., Zemple S., Grossman W.J., Grayson M.H., Verbsky J.W. Granzyme B regulates antiviral CD8+ T cell responses. J Immunol. 2011;187:6301–6309. doi: 10.4049/jimmunol.1100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daemen A., Udyavar A.R., Sandmann T., Li C., Bosch L.J.W., O’Gorman W., Li Y., Au-Yeung A., Takahashi C., Kabbarah O., Bourgon R., Hegde P., Bais C., Das Thakur M. Transcriptomic profiling of adjuvant colorectal cancer identifies three key prognostic biological processes and a disease specific role for granzyme B. PloS One. 2021;16:e0262198. doi: 10.1371/journal.pone.0262198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buzza M.S., Zamurs L., Sun J., Bird C.H., Smith A.I., Trapani J.A., Froelich C.J., Nice E.C., Bird P.I. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin ∗. J Biol Chem. 2005;280:23549–23558. doi: 10.1074/jbc.M412001200. [DOI] [PubMed] [Google Scholar]
- 50.Wang H., Huang Y., He J., Zhong L., Zhao Y. Dual roles of granzyme B. Scand J Immunol. 2021;94:e13086. [Google Scholar]
- 51.Dermani F.K., Samadi P., Rahmani G., Kohlan A.K., Najafi R. PD-1/PD-L1 immune checkpoint: potential target for cancer therapy. J Cell Physiol. 2019;234:1313–1325. doi: 10.1002/jcp.27172. [DOI] [PubMed] [Google Scholar]
- 52.Miller T.J., Anyaegbu C.C., Lee-Pullen T.F., Spalding L.J., Platell C.F., McCoy M.J. PD-L1+ dendritic cells in the tumor microenvironment correlate with good prognosis and CD8+ T cell infiltration in colon cancer. Cancer Sci. 2021;112:1173–1183. doi: 10.1111/cas.14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y., Xu B., Hu W.-W., Chen L.-J., Wu C.-P., Lu B.-F., Shen Y.-P., Jiang J.-T. High expression of CD11c indicates favorable prognosis in patients with gastric cancer. World J Gastroenterol WJG. 2015;21:9403–9412. doi: 10.3748/wjg.v21.i31.9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee H., Lee H.J., Song I.H., Bang W.S., Heo S.-H., Gong G., Park I.A. CD11c-positive dendritic cells in triple-negative breast cancer. In Vivo. 2018;32:1561–1569. doi: 10.21873/invivo.11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun L.L., Ellerman D., Mathieu M., Hristopoulos M., Chen X., Li Y., Yan X., Clark R., Reyes A., Stefanich E., Mai E., Young J., Johnson C., Huseni M., Wang X., Chen Y., Wang P., Wang H., Dybdal N., Chu Y.-W., Chiorazzi N., Scheer J.M., Junttila T., Totpal K., Dennis M.S., Ebens A.J. Anti-CD20/CD3 T cell-dependent bispecific antibody for the treatment of B cell malignancies. Sci Transl Med. 2015;7:287ra70. doi: 10.1126/scitranslmed.aaa4802. [DOI] [PubMed] [Google Scholar]
- 56.Shibuya T.Y., Nugyen N., McLaren C.E., Li K.-T., Wei W.-Z., Kim S., Yoo G.H., Rogowski A., Ensley J., Sakr W. Clinical significance of poor CD3 response in head and neck cancer1. Clin Cancer Res. 2002;8:745–751. [PubMed] [Google Scholar]
- 57.Wang B., Li F., Guo L., Lu S., Ma J., Ma Y., Meng Y., Wang J., Zhou X., Fu W. Loss of survival advantage for deficient mismatch repair in patients with advanced colorectal cancer may be caused by changes in prognostic value of CD8+T cell. World J Surg Oncol. 2020;18:196. doi: 10.1186/s12957-020-01970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saleh R., Elkord E. FoxP3+ T regulatory cells in cancer: prognostic biomarkers and therapeutic targets. Cancer Lett. 2020;490:174–185. doi: 10.1016/j.canlet.2020.07.022. [DOI] [PubMed] [Google Scholar]
- 59.Karanikas V., Speletas M., Zamanakou M., Kalala F., Loules G., Kerenidi T., Barda A.K., Gourgoulianis K.I., Germenis A.E. Foxp3 expression in human cancer cells. J Transl Med. 2008;6:19. doi: 10.1186/1479-5876-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ling A., Edin S., Wikberg M.L., Öberg Å., Palmqvist R. The intratumoural subsite and relation of CD8+ and FOXP3+ T lymphocytes in colorectal cancer provide important prognostic clues. Br J Cancer. 2014;110:2551–2559. doi: 10.1038/bjc.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hua W., Yuan A., Zheng W., Li C., Cui J., Pang Z., Zhang L., Li Z., Goll R., Cui G. Accumulation of FoxP3+ T regulatory cells in the tumor microenvironment of human colorectal adenomas. Pathol Res Pract. 2016;212:106–112. doi: 10.1016/j.prp.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Fidelle M., Yonekura S., Picard M., Cogdill A., Hollebecque A., Roberti M.P., Zitvogel L. Resolving the paradox of colon cancer through the integration of genetics, immunology, and the microbiota. Front Immunol. 2020;11:600886. doi: 10.3389/fimmu.2020.600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang S., Liu Y., Li M.-Y., Ng C.S.H., Yang S.-L., Wang S., Zou C., Dong Y., Du J., Long X., Liu L.-Z., Wan I.Y.P., Mok T., Underwood M.J., Chen G.G. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16:124. doi: 10.1186/s12943-017-0700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuwahara T., Hazama S., Suzuki N., Yoshida S., Tomochika S., Nakagami Y., Matsui H., Shindo Y., Kanekiyo S., Tokumitsu Y., Iida M., Tsunedomi R., Takeda S., Yoshino S., Okayama N., Suehiro Y., Yamasaki T., Fujita T., Kawakami Y., Ueno T., Nagano H. Intratumoural-infiltrating CD4 + and FOXP3 + T cells as strong positive predictive markers for the prognosis of resectable colorectal cancer. Br J Cancer. 2019;121:659–665. doi: 10.1038/s41416-019-0559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ladoire S., Martin F., Ghiringhelli F. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: the paradox of colorectal cancer. Cancer Immunol Immunother. 2011;60:909–918. doi: 10.1007/s00262-011-1046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sconocchia G., Eppenberger S., Spagnoli G.C., Tornillo L., Droeser R., Caratelli S., Ferrelli F., Coppola A., Arriga R., Lauro D., Iezzi G., Terracciano L., Ferrone S. NK cells and T cells cooperate during the clinical course of colorectal cancer. Oncoimmunology. 2014;3:e952197. doi: 10.4161/21624011.2014.952197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rocca Y.S., Roberti M.P., Juliá E.P., Pampena M.B., Bruno L., Rivero S., Huertas E., Sánchez Loria F., Pairola A., Caignard A., Mordoh J., Levy E.M. Phenotypic and functional dysregulated blood NK cells in colorectal cancer patients can be activated by cetuximab plus IL-2 or IL-15. Front Immunol. 2016;7:413. doi: 10.3389/fimmu.2016.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horzum U., Yoyen-Ermis D., Taskiran E.Z., Yilmaz K.B., Hamaloglu E., Karakoc D., Esendagli G. CD66b+ monocytes represent a proinflammatory myeloid subpopulation in cancer. Cancer Immunol Immunother. 2021;70:75–87. doi: 10.1007/s00262-020-02656-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nair K.S., Zingde S.M. Adhesion of neutrophils to fibronectin: role of the CD66 antigens. Cell Immunol. 2001;208:96–106. doi: 10.1006/cimm.2001.1772. [DOI] [PubMed] [Google Scholar]
- 70.Lin C., Lin W., Yeh S., Li L., Chang C. Infiltrating neutrophils increase bladder cancer cell invasion via modulation of androgen receptor (AR)/MMP13 signals. Oncotarget. 2015;6:43081–43089. doi: 10.18632/oncotarget.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang N., Feng Y., Wang Q., Liu S., Xiang L., Sun M., Zhang X., Liu G., Qu X., Wei F. Neutrophils infiltration in the tongue squamous cell carcinoma and its correlation with CEACAM1 expression on tumor cells. PloS One. 2014;9:e89991. doi: 10.1371/journal.pone.0089991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Governa V., Trella E., Mele V., Tornillo L., Amicarella F., Cremonesi E., Muraro M.G., Xu H., Droeser R., Däster S.R., Bolli M., Rosso R., Oertli D., Eppenberger-Castori S., Terracciano L.M., Iezzi G., Spagnoli G.C. The interplay between neutrophils and CD8+ T cells improves survival in human colorectal cancer. Clin Cancer Res. 2017;23:3847–3858. doi: 10.1158/1078-0432.CCR-16-2047. [DOI] [PubMed] [Google Scholar]
- 73.Zheng W., Wu J., Peng Y., Sun J., Cheng P., Huang Q. Tumor-associated neutrophils in colorectal cancer development, progression and immunotherapy. Cancers. 2022;14:4755. doi: 10.3390/cancers14194755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rick J.W., Chandra A., Dalle Ore C., Nguyen A.T., Yagnik G., Aghi M.K. Fibronectin in malignancy: cancer-specific alterations, pro-tumoral effects, and therapeutic implications. Semin Oncol. 2019;46:284–290. doi: 10.1053/j.seminoncol.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan Z., Hu H., Zhu Y., Zhang W., Fang Q., Qiao T., Ma T., Wang M., Huang R., Tang Q., Gao F., Zou C., Gao X., Wang G., Wang X. Colorectal cancer cell intrinsic fibroblast activation protein alpha binds to Enolase1 and activates NF-κB pathway to promote metastasis. Cell Death Dis. 2021;12:543. doi: 10.1038/s41419-021-03823-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu J., Huang C., Peng C., Xu F., Li Y., Yutaka Y., Xiong B., Yang X. Stromal fibroblast activation protein alpha promotes gastric cancer progression via epithelial-mesenchymal transition through Wnt/β-catenin pathway. BMC Cancer. 2018;18:1099. doi: 10.1186/s12885-018-5035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y., Yu Z., Lv M., Cai Z., Zou Y., Lan P., Wu X., Gao F. Cancer-associated fibroblasts impact the clinical outcome and treatment response in colorectal cancer via immune system modulation: a comprehensive genome-wide analysis. Mol Med. 2021;27:139. doi: 10.1186/s10020-021-00402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coto-Llerena M., Ercan C., Kancherla V., Taha-Mehlitz S., Eppenberger-Castori S., Soysal S.D., Ng C.K.Y., Bolli M., von Flüe M., Nicolas G.P., Terracciano L.M., Fani M., Piscuoglio S. High expression of FAP in colorectal cancer is associated with angiogenesis and immunoregulation processes. Front Oncol. 2020;10:979. doi: 10.3389/fonc.2020.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Filip S., Vymetalkova V., Petera J., Vodickova L., Kubecek O., John S., Cecka F., Krupova M., Manethova M., Cervena K., Vodicka P. Distant metastasis in colorectal cancer patients—do we have new predicting clinicopathological and molecular biomarkers? A comprehensive review. Int J Mol Sci. 2020;21:5255. doi: 10.3390/ijms21155255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu C., Wang T., Yang J., Zhang J., Wei S., Guo Y., Yu R., Tan Z., Wang S., Dong W. Distant metastasis pattern and prognostic prediction model of colorectal cancer patients based on big data mining. Front Oncol. 2022;12:878805. doi: 10.3389/fonc.2022.878805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Naxerova K., Reiter J.G., Brachtel E., Lennerz J., Van de Wetering M., Rowan A., Cai T., Clevers H., Swanton C., Nowak M.A., Elledge S.J., Jain R.K. Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357:55–60. doi: 10.1126/science.aai8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuo Y.-T., Tsai W.-S., Hung H.-Y., Hsieh P.-S., Chiang S.-F., Lai C.-C., Chern Y.-J., Hsu Y.-J., You J.-F. Prognostic value of regional lymph node involvement in patients with metastatic colorectal cancer: palliative versus curative resection. World J Surg Oncol. 2021;19:150. doi: 10.1186/s12957-021-02260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moses L., Pachter L. Museum of spatial transcriptomics. Nat Methods. 2022;19:534–546. doi: 10.1038/s41592-022-01409-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Example analysis code used to generate article results can be found in the following GitHub repository (https://github.com/jlevy44/Colon_Protein_DSP_Study, last accessed November 15, 2022). Because of patient privacy restrictions and ongoing efforts to expand the study cohort for public access, the spatial proteomics data featured in this study can be made available on reasonable request. Raw expression data can also be explored using the RShiny web application (see Web Application for Result Viewing).