Background:
Left ventricular noncompaction (LVNC) is a prevalent cardiomyopathy associated with excessive trabeculation and thin compact myocardium. Patients with LVNC are vulnerable to cardiac dysfunction and at high risk of sudden death. Although sporadic and inherited mutations in cardiac genes are implicated in LVNC, understanding of the mechanisms responsible for human LVNC is limited.
Methods:
We screened the complete exome sequence database of the Pediatrics Cardiac Genomics Consortium and identified a cohort with a de novo CHD4 (chromodomain helicase DNA-binding protein 4) proband, CHD4M202I, with congenital heart defects. We engineered a humanized mouse model of CHD4M202I (mouse CHD4M195I). Histological analysis, immunohistochemistry, flow cytometry, transmission electron microscopy, and echocardiography were used to analyze cardiac anatomy and function. Ex vivo culture, immunopurification coupled with mass spectrometry, transcriptional profiling, and chromatin immunoprecipitation were performed to deduce the mechanism of CHD4M195I-mediated ventricular wall defects.
Results:
CHD4M195I/M195I mice developed biventricular hypertrabeculation and noncompaction and died at birth. Proliferation of cardiomyocytes was significantly increased in CHD4M195I hearts, and the excessive trabeculation was associated with accumulation of ECM (extracellular matrix) proteins and a reduction of ADAMTS1 (ADAM metallopeptidase with thrombospondin type 1 motif 1), an ECM protease. We rescued the hyperproliferation and hypertrabeculation defects in CHD4M195I hearts by administration of ADAMTS1. Mechanistically, the CHD4M195I protein showed augmented affinity to endocardial BRG1 (SWI/SNF–related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4). This enhanced affinity resulted in the failure of derepression of Adamts1 transcription such that ADAMTS1-mediated trabeculation termination was impaired.
Conclusions:
Our study reveals how a single mutation in the chromatin remodeler CHD4, in mice or humans, modulates ventricular chamber maturation and that cardiac defects associated with the missense mutation CHD4M195I can be attenuated by the administration of ADAMTS1.
Keywords: cardiomyopathies; heart defects, congenital; mutation; myocardium; trabeculation
Novelty and Significance.
What Is Known?
Left ventricular noncompaction cardiomyopathy (LVNC) is one of the most prevalent congenital heart diseases, characterized by prominent left ventricular trabeculations, deep intertrabecular recesses, and a thin and compacted myocardium.
Mutations of CHD4 (chromodomain helicase DNA-binding protein 4) have been identified to be causative to congenital heart diseases that include LVNC.
ADAMTS1 (ADAM metallopeptidase with thrombospondin type 1 motif 1) is critical to terminate trabeculation during heart development.
What New Information Does This Article Contribute?
A patient-specific mouse model of CHD4M202I, mouse model CHD4M195I, develops ventricular trabecular noncompaction during embryogenesis and dies at birth.
Cardiomyocytes in CHD4M195I remain immature and continue to proliferate until birth.
The phenotype of CHD4M195I is associated with a dysregulation of the extracellular matrix composition.
CHD4M195I associates with BRG1 (SWI/SNF–related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4) to repress Adamts1, and restoration of ADAMTS1 rescues hypertrabeculation and cardiomyocyte over-proliferation in the CHD4M195I/M195I mice.
LVNC is a complex and heterogeneous cardiomyopathy associated with diverse clinical presentations and genetic causes. Despite extensive research, the underlying mechanisms that cause LVNC remain poorly understood. This study identified a de novo CHD4 mutation (CHD4M202I) in a cohort with congenital heart defects. We generated a mouse model (CHD4M195I) of the human CHD4 mutation and found that CHD4M195I/M195I mice developed biventricular hypertrabeculation and noncompaction and died at birth. Excessive trabeculation was associated with increased cardiomyocyte proliferation, extracellular matrix (ECM) protein accumulation, and reduced expression of ADAMTS1, an ECM metalloprotease that terminates trabeculation during heart development. Restoration of ADAMTS1 pharmacologically rescued the hypertrabeculation and cardiomyocyte over-proliferation defects. This study provides a unique mouse model of ventricular noncompaction cardiomyopathy that faithfully recapitulates the human genetic condition, highlighting the potential role of ADAMTS1 as a therapeutic target for treating LVNC.
Meet the First Author, see p 4
Editorial, see p 68
Left ventricular noncompaction (LVNC), the third most prevalent cardiomyopathy, is characterized by excessive ventricular trabeculae (noncompaction), a thin myocardium, and deep intertrabecular recesses among the trabeculae. Left ventricular noncompaction mainly affects the LV, but isolated right ventricular or biventricular noncompaction has also been reported.1–5 Left ventricular noncompaction can be asymptomatic, but patients with LVNC are at high risk of developing heart failure and sudden death.6 Genetic screening of individual patients with LVNC has identified mutations in genes encoding transcription factors, sarcomere, nuclear membrane, RNA-binding proteins, and ion channel proteins7–10; however, a significant portion of LVNC cases are of unknown etiology.11 Moreover, the identification of the molecular and cellular mechanisms that orchestrate the pathogenesis of LVNC is compromised by the absence of a genetic model that faithfully recapitulates patients’ conditions.
As a structural cardiac defect, LVNC involves ventricular chamber maturation, during which ventricular trabeculation and compaction are the most critical morphogenetic events.12 Trabeculation is initiated by myocardial sprouting that extends from the compact monolayer and protrudes into the extracellular matrix (ECM), also referred to as the cardiac jelly at approximately embryonic day (E) 8.5 in mice and approximately day 28 in humans.13 Trabecular myocardium grows by the proliferation of cardiomyocytes at early developmental stages. By E13.5 in mice and approximately day 40 in humans, the trabeculae undergo significant compaction and form a thickened, tightly packed ventricular wall with a relatively smooth inner surface.12 Persistent trabeculation and a reduced level of compaction6,14,15 typically characterize LVNC.
Abnormal development of the ventricular chambers of the heart is the basis of a significant number of congenital heart defects. Therefore, a mechanistic understanding of cardiac compaction is crucial for improving the treatment of structural heart disease. Large, multicomponent complexes that modify chromatin control cardiac gene expression. Prominent among these complexes is the NuRD (nucleosome remodeling and deacetylase) complex.16–18 The NuRD complex is essential for many developmental events, including ensuring proper timing of the switch from stem cell lineages to differentiated cell types, maintaining cell differentiation, and activating DNA damage response pathways.19–24 Mutations in CHD4 (chromodomain helicase DNA-binding protein 4), the core catalytic component of the NuRD, lead to congenital cardiac malformation, including atrial and ventricular septal defects.25 CHD4 is essential for cardiac development, and it represses inappropriate expression of the skeletal and smooth muscle programs in the developing heart.26–28 Nonetheless, neither CHD4 nor the NuRD complex can directly bind DNA. Instead, we have shown that CHD4 is recruited to target cardiac genes by interaction with the essential cardiac transcription factors GATA4 (GATA binding protein 4), NKX2-5 (NK2 homeobox 5) and TBX5 (T-box transcription factor 5).29
Although CHD4 is crucial for heart development, and its disease relevance is well established, many questions remain regarding the mechanism of CHD4’s function. Critically, we do not understand how a single missense mutation in CHD4, a broadly expressed catalytic unit of the NuRD complex, leads to cardiac-specific disease in humans. To determine the basis of CHD4 mutations in human congenital heart disease, we screened the complete exome sequence database of the Pediatrics Cardiac Genomics Consortium30,31 and identified a cohort with a de novo CHD4 proband, CHD4M202I, with congenital heart defects. Using CRISPR/CAS9, we generated a humanized mouse model of CHD4M202I (mouse CHD4M195I). We demonstrate that CHD4M195I/M195I mice displayed critical aspects of human LVNC. CHD4M195I/M195I hearts failed to undergo compaction, and cardiomyocytes were retained in an immature proliferating state through birth. The LVNC phenotype was associated with increased extracellular matrix proteins and a marked reduction in the ECM protease ADAMTS1 (ADAM metallopeptidase with thrombospondin type 1 motif 1). We demonstrate that Adamts1 is a direct target of CHD4 and that exogenous administration of ADAMTS1 or the introduction of a pharmacological compound that increases ADAMTS1 expression rescues key facets of LVNC ex vivo and in utero in CHD4M195I/M195I mice. Together, we have revealed a mechanism of how CHD4 may cause congenital heart diseases, and we present prospects for treatment.
Methods
Data Availability
An expanded Materials and Methods section is available in the Supplemental Material, and the sources of critical reagents are provided in the Major Resources Table. The raw RNA-sequencing data are available through the Gene Expression Omnibus (GEO) public functional genomics data repository with the accession number GSE229050. On reasonable request, the data, analytic methods, and study materials will be made available to other researchers to reproduce the results or replicate the procedures.
Ethics Statement
The Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill approved all animal experiments, which conformed to the Guide for the Care and Use of Laboratory Animals.
Results
Generation of a Patient-Specific Mouse Model of CHD4 M202I
To define the mechanisms by which mutations in CHD4 lead to cardiac disease, we screened the complete exome sequence database of the Pediatrics Cardiac Genomics Consortium31 and identified a cohort with a de novo CHD4 proband CHD4M202I. This proband was associated with ventricular septal defects and conotruncal abnormalities (Figure 1A). Surprisingly, CHD4M202I mapped to a highly evolutionarily conserved region (Figure S1A) of CHD4 of unknown biological function.32–34 To determine the mechanism by which CHD4M202I causes cardiac disease, we used a CRISPR/CAS9 gene-editing system to engineer a humanized mouse model of CHD4M202I (mouse CHD4M195I; Figure S1B). Founding males were bred to wild-type (WT) C57BL/6J females, and the genotypes of the founding males and all F2 offspring were confirmed by polymerase chain reaction genotyping and sequencing (Figure S1C and S1D). Immunoblot analysis and immunohistochemistry at E18.5 showed that intact CHD4M195I protein was present in homozygous CHD4M195I/M195I hearts at endogenous levels and localized to the nucleus (Figure S1E and S1F).
Figure 1.
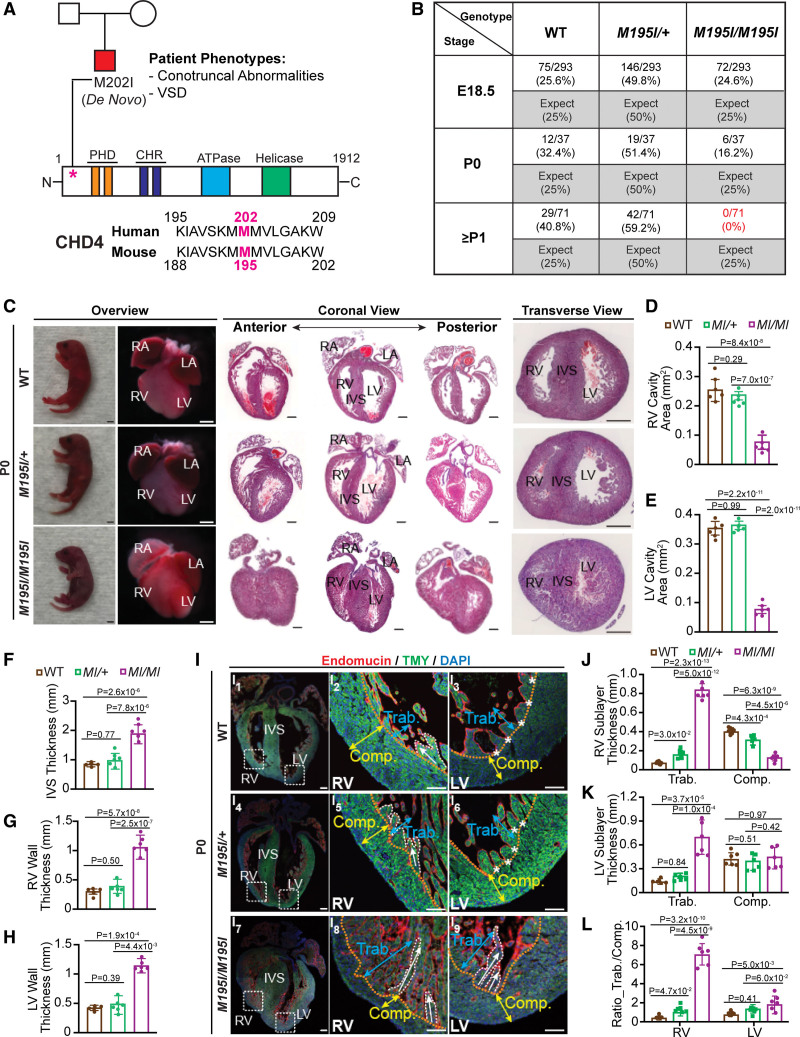
CHD4 (chromodomain helicase DNA-binding protein 4)M195I/M195I neonatal mice develop ventricular noncompaction and die at birth. A, Top, CHD4M202I pedigree (squares, male; circle, female; ventricular septal defect [VSD]) and schematic of CHD4 protein domains. Bottom, Protein sequence alignment of human and mouse CHD4 orthologs. B, Table of expected and observed Mendelian ratios of offspring at indicated developmental stages from the intercross of CHD4M195I/+ mice. Thirty-eight, 7, and 14 litters were recovered at embryonic day (E) 18.5, postnatal day (P) 0, and P1, respectively. C, Representative P0 neonates, whole mount and hemotoxylin and eosin (H&E)–stained paraffin coronal and transverse sections of P0 wild-type (WT), heterozygote (CHD4M195I/+), and homozygote (CHD4M195I/M195I) mouse hearts. Scale bars, 2 mm (overview of neonates) and 0.5 mm (overview of whole mount and H&E stains). D through H, Quantification of right ventricular (RV; D) and left ventricular (LV; E) ventricular cavity area, interventricular septum (IVS; F), RV wall (G), and LV wall (H) thickness on P0 WT, CHD4M195I/M195I, and CHD4M195I/M195I mouse heart sections. I, Representative images of immunofluorescent (endomucin, TMY [tropomyosin], and 4′,6-diamidino-2-phenylindole [DAPI]) stained sections from P0 mouse hearts. The approximate boundaries between compact myocardium (Comp.; indicated by yellow double-headed arrows) and trabecular myocardium (Trab.; indicated by blue double-headed arrows) are indicated by orange dashed lines. Trabeculae are delineated with the white dashed curve, and white asterisks indicate fused trabeculae. White arrows indicate the direction of trabecular projections. Scale bars, 0.5 mm (overview of the whole heart) and 0.2 mm (magnified views on RV and LV). J and K, RV (J) and LV (K) sublayer thickness measurements on P0 mouse hearts. L, Ratio of trabecular layer thickness to compact layer thickness in (J) and (K). Data in D–H and J–L are represented as mean±SEM. For all experiments in C–L, n=6 individual hearts per genotype, each point representing the mean value of 5 technical measurements from an individual biological replicate, so 30 measures were acquired per genotype. The Shapiro-Wilk test was applied to test the normality of variables. One-way ANOVA (D–H) or 2-way ANOVA (J–L) followed by Turkey post hoc test were used for comparisons. M195I/M195I or MI/MI represents CHD4M195I/M195I, same with all main and Supplemental Figures S1,S12, S14 through S16, S18 through S20, and S22. CHR, chromodomain; LA, left atrium; PHD, plant homeodomain; and RA, right atrium.
Neonatal Homozygous CHD4 M195I/M195I Develop Noncompaction Cardiomyopathy
Heterozygous CHD4M195I/+ mice were viable, fertile, recovered at expected Mendelian ratios, and exhibited no overt phenotypic abnormalities (Figure 1B; Figure S2). Conversely, homozygous CHD4M195I/M195I mice died at or shortly after birth, with none of the CHD4M195I/M195I mice surviving >2 days (Figure 1B). Anatomic analysis at postnatal day 0 (P0) revealed that CHD4M195I/M195I mouse hearts had a dramatically reduced ventricular cavity with a concomitant increase in the thickness of the ventricular walls and septa compared with WT or CHD4M195I/+ (Figure 1C through 1H). The histological examination further revealed that ≈20% of CHD4M195I/M195I hearts displayed ventricular septal defects (Figure S3). However, we did not detect alterations in the development of the aorta and pulmonary artery (Figure S4).
Studies have shown that conditional loss of Chd4 in skeletal muscle leads to myopathy.28 To determine if CHD4M195 is essential for skeletal muscle homeostasis, we conducted histological analyses of E18.5 hindlimbs. We observed that the CHD4M195I/M195I embryonic limbs have fewer but larger myofibers that are disorganized versus controls (Figure S5A through S5C). We have further observed that the distal hypertrophic chondrocyte layer of the tibia in the CHD4M195I/M195I embryonic limbs is thicker versus WT embryos (Figure S5D). Consistently, we find a more significant number of PAX7 (paired Box 7)+ cells (activated muscle satellite cells) and a concomitant increase in Ki67+/PAX7+ in the CHD4M195I/M195I tissue (Figure S6). However, we found no significant alteration in the expression of the skeletal muscle-specific contractile protein, eMHC (embryonic myosin heavy chain), between CHD4M195I/M195I and WT embryos (Figure S7). In accordance, the length of the tibial bone and the embryo are not significantly different (Figure S8A and S8B), and we find no significant difference in the body weight of CHD4M195I/M195I embryos versus controls (Figure S8C). In aggregate, these data suggest that CHD4M195 is essential for cardiac development, myofibril organization, and viability.
CHD4 M195I/M195I and Ventricular Noncompaction
As the CHD4 proband had no reported skeletal defects and, as skeletal defects associated with CHD4 skeletal conditional null mice28 are not lethal, the skeletal defects associated with CHD4M195I/M195I would not be predicted to be the cause of death in CHD4M195I/M195I mice. Therefore, we focused on the function of CHD4M195 in cardiac development. LVNC is a cardiac disease associated with increased cardiac failure in children and adults.35,36 In addition, LVNC is associated with hypertrabeculation of the myocardium of the left ventricle (LV), and occasionally the right ventricle (RV).1–5 Therefore, to assess the compact and trabecular layers of CHD4M195I/M195I embryos, we conducted immunofluorescence with Endomucin (red) and TMY (tropomyosin; green) antibodies to delineate chamber endocardium and myocardium, respectively (Figure 1I; Figure S9). In WT and CHD4M195I/+ hearts, the trabeculae were folded and oriented parallel to the free wall of the RV (white arrows, Figure 1I2 and 1I5). Moreover, the trabeculae assimilated into the compact zones clearing the ventricular lumen (asterisks, Figure 1I3 through 1I6). In contrast, consistent with LVNC phenotype,6,14,15 the trabeculae in the CHD4M195I/M195I hearts remained straight and protruded perpendicularly to the compact zones and extended across the lumen (white arrows, Figure 1I8 and 1I9). By examining the ratio of the layer thickness of the trabecular-to-compact, we found severe RV and modest LV noncompaction in CHD4M195I/M195I hearts. In contrast, CHD4M195I/+ hearts displayed RV but not LV noncompaction (Figure 1J through 1L). Together, these findings demonstrated that CHD4M195I is associated with critical aspects of the LVNC phenotype.
Figure 2.
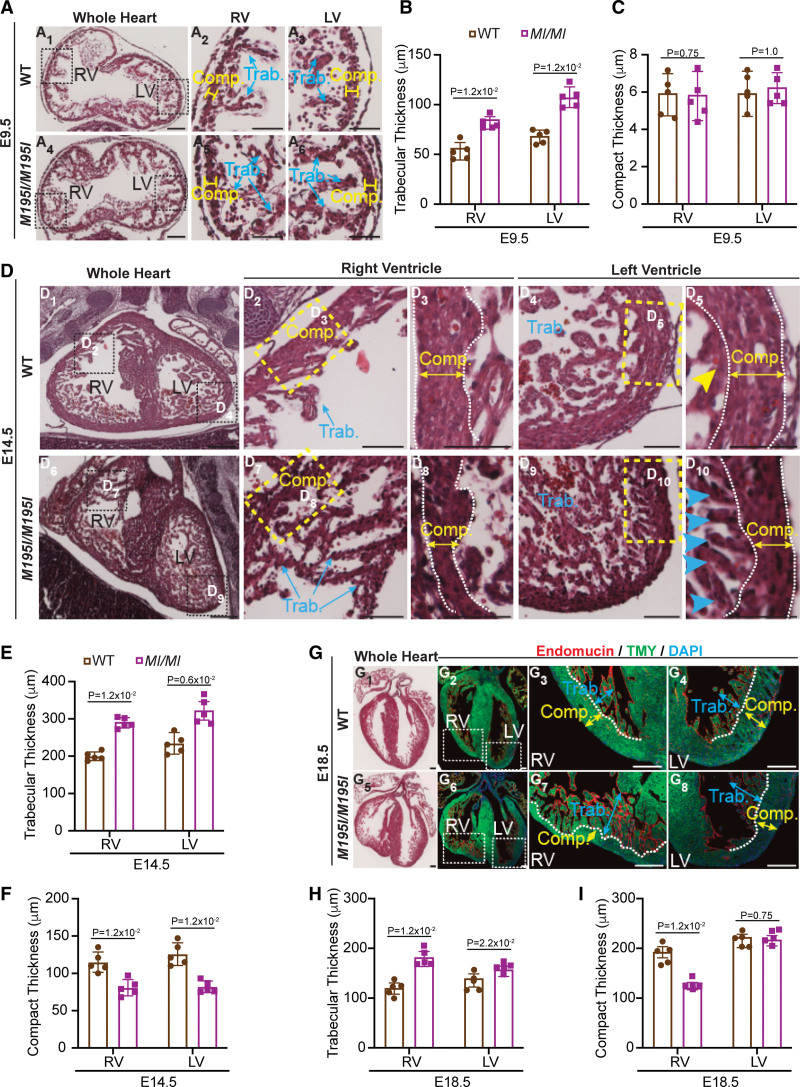
Ventricular noncompaction in CHD4M195I/M195I hearts dynamically proceeds over developmental stages. A and D, Representative images of hemotoxylin and eosin (H&E)–stained paraffin sections of wild-type (WT) and CHD4M195I/M195I hearts at embryonic day (E) 9.5 (A) and E14.5 (D), trabeculae assimilated into the compact myocardium are indicated by yellow arrowhead (D5) and nonassimilated trabeculae are indicated by blue arrowheads (D10). G, Representative images of H&E-stained (G1 and G5) and immunofluorescent (endomucin, TMY [tropomyosin], and 4′,6-diamidino-2-phenylindole [DAPI]; G2–G4 and G6–G8)-stained paraffin sections of WT and CHD4M195I/M195I hearts at E18.5; the approximal boundaries between compact myocardium (Comp.; indicated by yellow double-headed arrows) and trabecular myocardium (Trab.; indicated by blue double-headed arrows) are indicated by orange dashed lines. Scale bars, 50 (A), 250 (D), and 200 µm (G). B, C, E, F, H, and I, quantifications of sublayer thicknesses of WT and CHD4M195I/M195I heart sections from E9.5 (B and C), E14.5 (E and F), and E18.5 (H and I). For all experiments in A–I, n=5 individual hearts per genotype per stage, each point representing the mean value of 5 technical measurements from an individual biological replicate, so 25 measures were required per genotype per stage. Data in B, C, E, F, H, and I are represented as mean±SEM. Mann-Whitney U test was performed for comparisons. LV indicates left ventricle; and RV, right ventricle.
Figure 5.
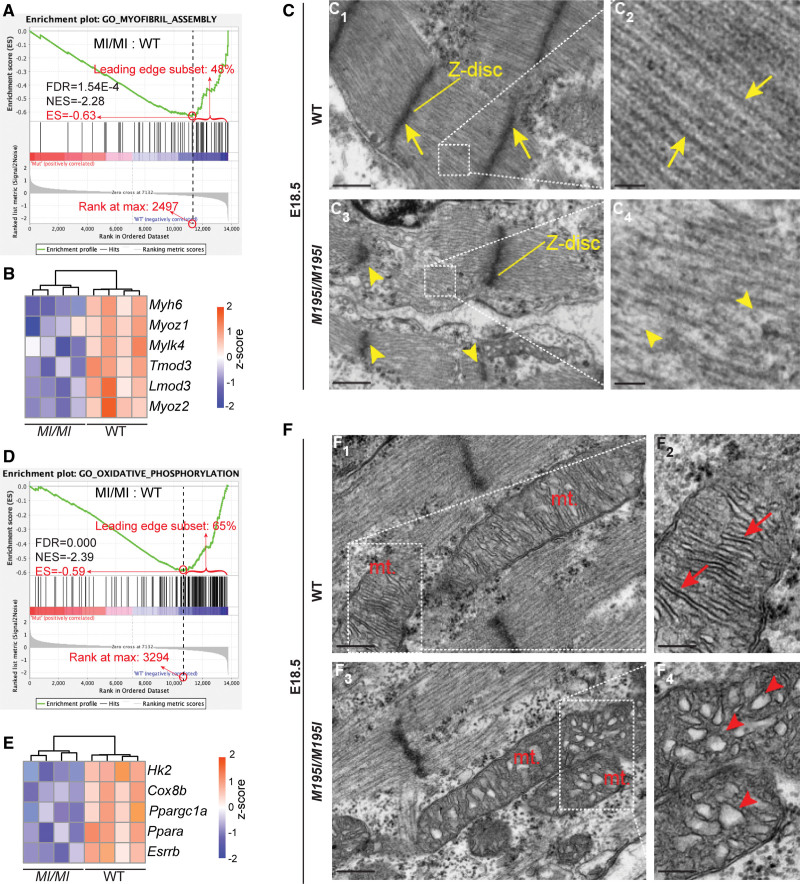
Cardiomyocytes in CHD4M195I/M195I are immature. A and D, Gene set enrichment analysis showing the downregulated gene sets of myofibril assembly (A) and oxidative phosphorylation (D) in CHD4M195I/M195I. B and E, Heatmaps of representative misregulated genes involved in A and D. Color bar: Z score. C, Representative transmission electron microscopy images of myofibrils in embryonic day (E) 18.5 hearts. Yellow arrows indicate well-formed Z-disc (C1) and sarcomeres (C2) in wild-type (WT) hearts and yellow arrowheads indicate weak, deficient Z-disc formation (C3) and poorly organized sarcomeres (C4) in CHD4M195I/M195I hearts. Scale bars, 500 nm (overview) and 100 nm (magnified). F, Representative transmission electron microscopy images of mitochondria (mt.) in E18.5 hearts. Red arrows indicate well-formed mitochondrial cristae (F2), and red arrowheads indicate poorly organized mitochondrial cristae (F4). Scale bars, 250 nm (overview) and 100 nm (magnified). n=3 individual hearts per genotype in C and F. ES indicates enrichment score; FDR, FDR, false discovery rate; and NES, normalized enrichment score.
Figure 3.
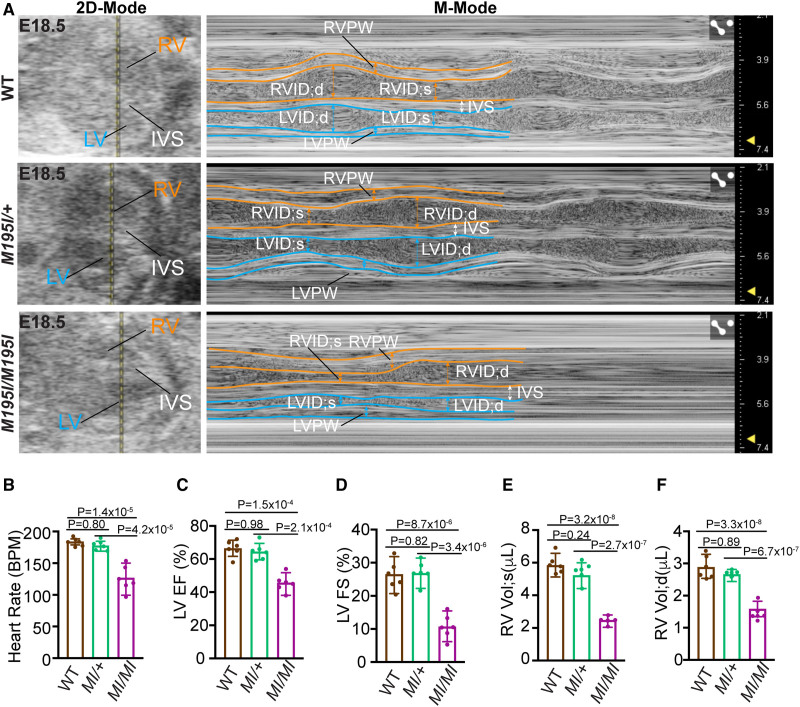
Impaired cardiac function in CHD4M195I/M195I mice. A, In utero echocardiography analysis of representative embryonic day (E) 18.5 wild-type (WT), CHD4M195I/+, and CHD4M195I/M195I embryonic hearts. Representative images showing 2-dimensional (2D)-Mode (left) and M-Mode (right) of the echocardiography. B–F, Quantification of heart rate (B), left ventricular (LV) ejection fraction (LV EF, C), LV fractional shortening (LV FS, D), end-systolic right ventricular (RV) volume (RV Vol;s, E), and end-diastolic right ventricular volume (RV Vol;d, F). n=6 individual embryos per genotype from 4 pregnant female CHD4M195I/+ mice, each point representing the mean value of 2 technical measurements from an individual biological replicate. Data in B–F are represented as mean±SEM. The Shapiro-Wilk test was applied to test the normality of variables. One-way ANOVA followed by Turkey post hoc test was used for comparisons. BPM indicates beat per minute; IVS, interventricular septum; LVID;d, end-diastolic LV internal diameter; LVID;s, end-systolic LV internal diameter; LVPW, LV posterior wall; RVID;d, end-diastolic RV internal diameter; RVID;s, end-systolic RV internal diameter; and RVPW, RV posterior wall.
Figure 6.
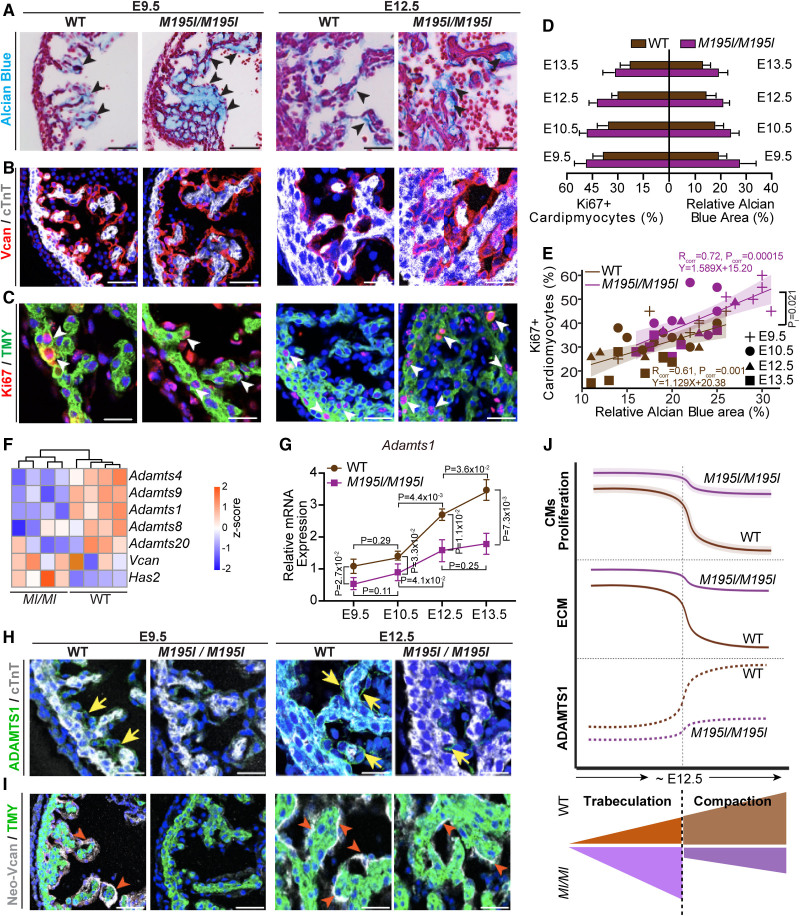
Cardiac extracellular matrix (ECM) dynamics in CHD4M195I/M195I are dysregulated. A, Representative images of Alcian Blue staining for ECM on embryonic day (E) 9.5 and E12.5 wild-type (WT) and CHD4M195I/M195I heart paraffin sections. ECM is indicated by black arrowheads. Scale bars, 50 µm (E9.5) and 100 µm (E12.5). B, Representative images of immunofluorescent (Vcan [versican], cTnT [cardiac Troponin T], and 4′,6-diamidino-2-phenylindole [DAPI]) stained paraffin sections from E9.5 and E12.5 mouse hearts. Scale bars, 50 µm. C, Representative images of immunofluorescent-(Ki67, TMY [tropomyosin], and DAPI) stained paraffin sections from E9.5 and E12.5 mouse hearts. Proliferating cardiomyocytes (Ki67+/TMY+) are indicated by white arrowheads. Scale bars, 25 µm (E9.5) and 50 µm (E12.5). D, Quantification of proliferating cardiomyocyte (Ki67+) ratio and relative Alcian Blue-positive area in indicated stages. E, Scatter plot showing Ki67+ cardiomyocyte (%) relative to Alcian Blue-positive area (%) in WT (brown symbols) or CHD4M195I/M195I (purple symbols) hearts. The Shapiro-Wilk test was applied to test the normality of variables. Pearson correlation coefficients were calculated to determine the correlation between Ki67+ cardiomyocytes (%) and Alcian Blue-positive area (%). Rcorr: Pearson correlation coefficient; Pcorr: significance level (P value) for determining correlation. Linear regression equations and confidence bands (95% confidence level) were indicated for each genotype. The linear regression analysis conducted an interaction test with a P value (PI)=0.021 between the Alcian Blue (%) and genotype (WT or CHD4M195I/M195I). In experiments in A–E, n=6 individual hearts per genotype per stage, and 2 technical replicates were performed for each biological replicate. Twenty cardiomyocytes were counted in each technical replicate, so 240 cardiomyocytes were counted per genotype per stage in C, D, and E. F, Heatmap of representative genes identified in E18.5 RNA-sequencing (RNA-seq) involved ECM dynamics. Color bar: Z score. G, Real-time quantitative polymerase chain reaction of Adamts1 expression in WT and CHD4M195I/M195I hearts at indicated stages. In both genotypes, 3 hearts were pooled as 1 biological replicate for E9.5 and E10.5, 2 hearts were pooled as 1 biological replicate for E12.5 and E13.5. Three biological replicates for each stage per genotype. The Pgk1 gene was used as the internal control, and Adamts1 expression in E9.5 WT hearts was normalized as 1.0; expression of all other stages and CHD4M195I/M195I hearts was normalized to E9.5 WT hearts. Data are represented as mean±SEM. Mann-Whitney U test was performed for comparisons. H and I, Representative images of immunofluorescent (ADAMTS1 [ADAM metallopeptidase with thrombospondin type 1 motif 1], cTnT, and DAPI, H; or Neo-Vcan, TMY, and DAPI, I) stained sections of E9.5 and E12.5 mouse hearts. ADAMTS1 and Neo-Vcan were indicated by yellow arrows or red arrowheads, respectively. Scale bars, 50 µm. n=6 per genotype per stage in H and I. J, A proposed schematic for dynamics of cardiomyocyte (CM) proliferation, ECM component, ADAMTS1 expression patterns, and sublayers development in WT and CHD4M195I/M195I hearts.
Figure 8.
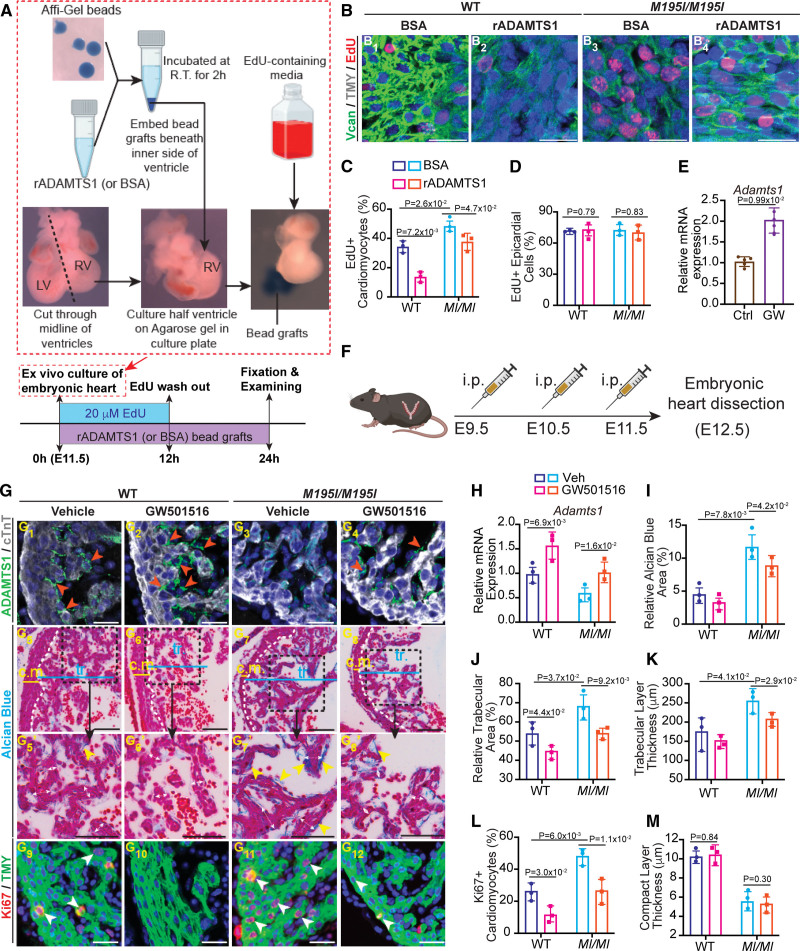
Restoration of ADAMTS1 (ADAM metallopeptidase with thrombospondin type 1 motif 1) rescues hypertrabeculation in CHD4M195I/M195I hearts. A, Workflow for treating the cardiac explants with recombinant ADAMTS1 protein. Hearts were dissected at embryonic day (E) 11.5 and cut through the middle line of the ventricles; the right ventricles (RVs) were cultured on low-melting point agarose gel. Recombinant ADAMTS1 protein (bovine serum albumin [BSA] was used as control) was incubated with the Affi-Gel bead at ambient temperature (R.T.) for 2 hours. Then the ADAMTS1 bead grafts were embedded beneath the inner side of the cultured ventricles for 24 hours. Explant ventricles were cultured in 4′,6-diamidino-2-phenylindole [DAPI]-containing media for the first 12 hours, and then cultured with EdU-free media for another 12 hours. B, Representative images of immunofluorescent (EdU, TMY [tropomyosin], and 4′,6-diamidino-2-phenylindole [DAPI])-stained sections of ventricle explants. Scale bars, 50 µm. C and D, Quantification of EdU+ cardiomyocytes (C) and epicardial cells (D) in the explants of 4 conditions in B (n=3 per condition). E, Real-time quantitative polymerase chain reaction (RT-qPCR) of Adamts1 expression in ADAMTS1 agonist (GW501516) or vehicle-treated (Control group [Ctrl]) wild-type (WT) heart explants. The Pgk1 gene was used as the internal control; n=5 explants per group. F, Workflow for treating pregnant CHD4M195I/+ dams with GW501516. CHD4M195I/+ mice were intercrossed for timed mating, and GW501516 was administered to the plugged CHD4M195I/+ mice by intraperitoneal injection at E9.5, E10.5, and E11.5. Embryonic hearts were dissected and examined at E12.5 (n=3 GW501516-treated CHD4M195I/+ dams). G, Representative images of ADAMTS1, cTnT immunofluorescence (G1–G4), Alcian Blue staining (G5–G8′), and Ki67, TMY immunofluorescence (G9–G12) performed on embryonic hearts that were dissected from GW501516-treated CHD4M195I/+ dams. ADAMTS1+ stains are indicated by orange arrowheads (G1–G4). White double-headed arrows indicate the thickness of trabeculae, and Alcian Blue-positive stains are indicated by yellow arrowheads (G5–G8′). EdU+ cardiomyocytes are indicated by white arrowheads (G9–G12). Scale bars, 50 µm (G1–G4, G9–G12) and 100 µm (G5–G8′). H, RT-qPCR of Adamts1 expression in the embryonic hearts of 4 conditions. The Pgk1 gene was used as the internal control. I through M, Quantification of relative Alcian Blue area (I), relative trabecular area (J), trabeculae thickness (K), Ki67+ cardiomyocyte ratio (L), and compact layer thickness (M). n=3 independent hearts for each condition in G–M, each point representing the mean value of 5 technical measurements from an individual biological replicate. Twenty cardiomyocytes (or 10 epicardial cells in D) were counted in each technical replicate, so 300 cardiomyocytes (or 150 epicardial cells) were counted in B, C, D, and L. All data in C–E, H–M are represented as mean±SEM. Mann-Whitney U test was performed for comparison in E, and a 2-way ANOVA followed by Tukey post hoc test was used for comparisons in C, D, and H–M. c.m. indicates compact myocardium; LV, left ventricular; rADAMTS1, recombinant ADAMTS1; tr., trabeculae; Vcan, versican; and Veh, vehicle.
Ventricular Noncompaction in CHD4 M195I/M195I Hearts
To define the onset of noncompaction in CHD4M195I/M195I embryos, we conducted histological analysis on CHD4M195I/M195I and WT embryonic hearts from E9.5 through E18.5. At E9.5, LV and RV in WT embryos displayed trabeculae as single sprouts with few interconnections (Figure 2A1 through 2A3). In contrast, trabeculae in the CHD4M195I/M195I hearts were longer (82.51±5.31 and 103.12±8.55 µm in the CHD4M195I/M195I hearts, compared with 60.24±7.53 and 72.50±4.62 µm in the WT hearts, in the RV and LV, respectively; Figure 2B), they protruded into the ventricular cavities and formed a ventricular mesh network (Figure 2A4 through 2A6). At this stage, alternations in the morphology of CHD4M195I/M195I hearts appeared to be confined to the trabecular layer. We did not observe a significant difference in thickness of the compact layer in either the RV or LV (5.88±1.46 and 6.07±1.08 µm in the CHD4M195I/M195I hearts, compared with 6.08±1.34 and 5.67±1.57 µm in the WT hearts, in the RV and LV, respectively; Figure 2C). However, by E12.5, in CHD4M195I/M195I hearts, we found a thinning of the compact layer (11.58±1.72 and 7.37±2.51 µm in the CHD4M195I/M195I hearts, compared with 16.84±2.45 and 12.54±3.73 µm in the WT hearts, in the RV and LV, respectively; Figure S10A and S10C) in addition to excess trabeculae (219.22±27.21 and 222.70±20.50 µm in the CHD4M195I/M195I hearts, compared with 126.42±30.86 and 175.18±18.79 µm in the WT hearts, in the RV and LV, respectively; Figure S10B).
Figure 4.
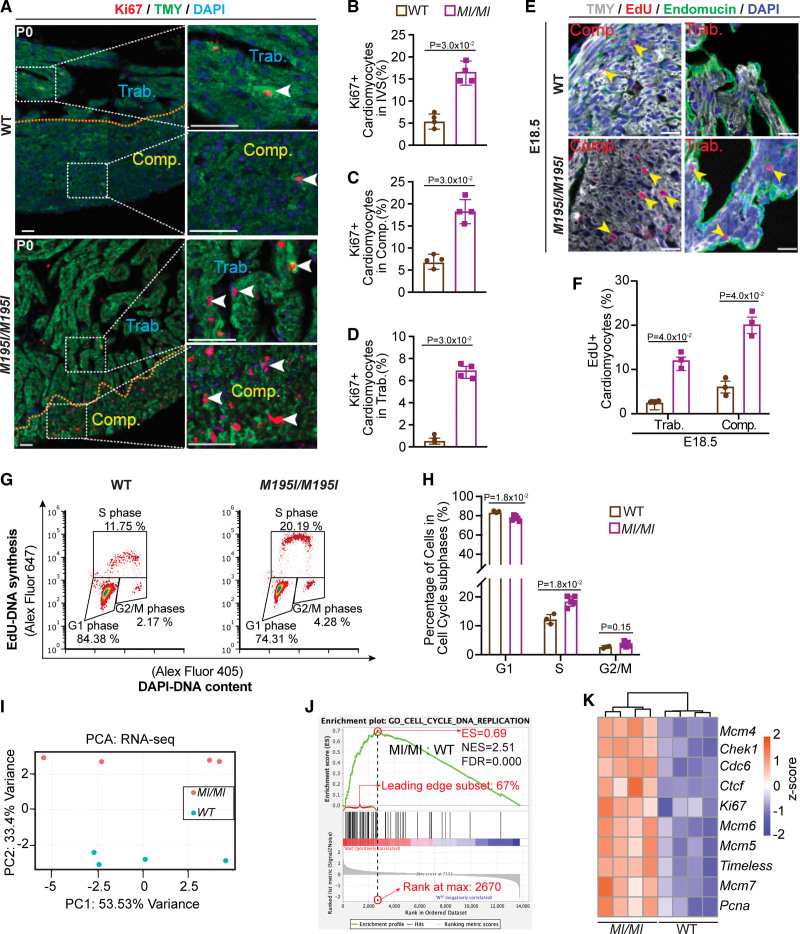
Cardiomyocyte proliferation is increased in CHD4M195I/M195I hearts. A, Representative images of immunofluorescent (Ki67, TMY [tropomyosin], and 4′,6-diamidino-2-phenylindole [DAPI]) stained paraffin sections from P0 wild-type (WT) and CHD4M195I/M195I mouse hearts. The approximate boundaries between compact myocardium (Comp.) and trabecular myocardium (Trab.) are indicated by orange dashed lines. Proliferating cardiomyocytes (Ki67+/TMY+) are indicated by white arrowheads. Scale bars: 50 µm. B through D, Quantification of proliferating cardiomyocyte ratios (Ki67+/total cardiomyocytes) in the interventricular septum (IVS, B), Comp. (C), and Trab. (D). In A–D, n=4 individual P0 hearts per genotype, each point representing the mean value of 3 technical replicates (30 cardiomyocytes were counted per technical replicate) from an individual biological replicate, so 360 cardiomyocytes were counted in total per genotype. E, Representative images of immunofluorescent (TMY, 5-Ethynyl-2′-deoxyuridine [EdU], endomucin, and DAPI) stained paraffin sections from embryonic day (E) 18.5 WT and CHD4M195I/M195I mouse hearts. Yellow arrowheads indicate proliferating cardiomyocytes. Scale bars, 20 µm. F, Quantification of proliferating cardiomyocytes ratio (EdU+/total cardiomyocytes) in E18.5 WT and CHD4M195I/M195I hearts (n=3 individual E18.5 hearts per genotype, each point representing the mean value of 3 technical replicates (30 cardiomyocytes were counted per technical replicate) from an individual biological replicate, so 270 cardiomyocytes were counted per genotype). G, Flow cytometry analysis for the proliferating cardiomyocytes derived from E18.5 hearts. H, Quantification for the percent of cardiomyocytes at each cell cycle phase (n=5 individual CHD4M195I/M195I hearts, n=3 WT individual hearts). Data in B–D, F, and H are represented as mean±SEM. Mann-Whitney U test was performed for comparisons. I, Principal component analysis (PCA) plot of gene expression data from RNA-sequencing (RNA-seq) of n=4 replicates for each genotype. J, Gene set enrichment analysis showing the upregulated gene set of cell cycle DNA replication in CHD4M195I/M195I. K, Heatmap of representative misregulated genes involved in J. Color bar: Z score. ES indicates enrichment score; FDR, false discovery rate; NES, normalized enrichment score; and PC, principal component.
By E14.5, in WT hearts, portions of the trabeculae were assimilated into an organized compact zone (Figure 2D1 through 2D5), whereas in CHD4M195I/M195I hearts, the trabeculae were thicker (269.13±22.25 and 307.09±40.52 µm in the CHD4M195I/M195I hearts, compared with 186.31±20.45 and 237.05±38.81 µm in the WT hearts, in the RV and LV, respectively; Figure 2E), while the compact layer was thinner (72.32±17.14 and 70.14±13.56 µm in the CHD4M195I/M195I hearts, compared with 113.31±12.82 and 125.23±21.35 µm in the WT hearts, in the RV and LV, respectively; Figure 2F). We observed a failure of the trabeculae to fold (Figure 2D6 through 2D10). These dysregulated trabecular phenotypes persisted in E16.5 (Figure S10D through S10F) and E18.5 (Figure 2G through 2I). However, the thickness of the compact zones in LV was not significantly different compared with WT hearts at these stages (E16.5: 172.08±23.86 and 165.57±22.83 µm for the CHD4M195I/M195I and WT LV compact layer, respectively, Figure S10F; E18.5: 217.09±18.71 and 205.24±10.82 µm for the CHD4M195I/M195I and WT LV compact layer, respectively, Figure 2I) and at P0 (386.31±105.25 and 389.41±122.04 µm for the CHD4M195I/M195I and WT LV compact layer, respectively, Figure 1K). These findings demonstrated that CHD4M195I/M195I hearts underwent dynamic remodeling leading to an LVNC phenotype and death at or shortly after birth.
Impaired Cardiac Function in CHD4 M195I/M195I Mice
To delineate the physiological consequences of CHD4M195I, we performed ultrasound pulsed wave Doppler on E18.5 embryos in utero without surgical manipulation of the dam or embryos.26,37,38 This approach enabled us to measure the effect of biventricular noncompaction on cardiac function in the developing heart. Short-axis M-mode echocardiograms showed little ventricular wall movement in the CHD4M195I/M195I (Figure 3A; Video S1 through S3), which confirmed severe cardiac contractile dysfunction and consequently reduced heart rate (Figure 3B), left ventricular ejection fraction (Figure 3C), left ventricular fractional shortening (Figure 3D), right ventricular end-systolic and diastolic volume (Figure 3E and 3F). No significant changes were identified in the CHD4M195I/+ embryos or adults (Figure 3A; Figure S11; Videos S4 and S5). These studies demonstrated that CHD4M195I/M195I physiologically models LVNC.
Hypertrabeculation Is Accompanied by the Over-Proliferation of Cardiomyocytes
We next queried whether hypertrabeculation in CHD4M195I/M195I hearts resulted from the inappropriate proliferation of cardiomyocytes. Results revealed a significant increase in the coexpression of Ki67, a marker of proliferation, and the cardiomyocyte marker TMY in CHD4M195I/M195I P0 mice (Figure 4A through 4D). Consistently, quantifying dissociated cardiomyocytes from P0 hearts confirmed an increase in the number of cardiomyocytes in CHD4M195I/M195I hearts compared with WT (Figure S12A). Correspondingly, we found an increase in cardiomyocyte proliferation in vivo as measured by an increase in 5-Ethynyl-2′-deoxyuridine (EdU) incorporation (Figure 4E and 4F) and Ki67/TMY costaining at E18.5 (Figure S12B and S12C).
To confirm and extend these findings, we conducted cell cycle profiling of cardiomyocytes derived from CHD4M195I/M195I and WT mice that had been labeled with EdU (Figure S13). We found a significant decrease in CHD4M195I/M195I (76.2%) compared with WT (84.3%) cardiomyocytes in the G1 phase and a concomitant increase of cardiomyocytes in the S phase (19.8% in CHD4M195I/M195I versus 12.3% in WT) and an increase in G2/M phases (3.8% in CHD4M195I/M195I versus 2.2% in WT; Figure 4G and 4H). In sum, CHD4M195I/M195I cardiomyocytes displayed an increased and prolonged state of proliferation.
We further found that cardiomyocyte proliferation and hypertrabeculation were not associated with cardiomyocyte hypertrophy. Wheat germ agglutinin and TMY double immunostaining (Figure S14A) on sections of P0 hearts did not show a significant difference in cardiomyocyte size between the WT and CHD4M195I/M195I hearts (Figure S14B). These results indicated that the hypertrabeculation was caused by the over-proliferation of cardiomyocytes and the overgrowth of the trabecular layer at the expense of the compact layer.
CHD4M195I Leads to Upregulation of Cell Cycle Pathways
Because the phenotype and timing of CHD4M195I/M195I are significantly different from cardiac Chd4 null mutations,26,28 we hypothesized that CHD4M195I regulates a distinct transcriptional network essential for cardiac development and function. To test this hypothesis, we performed RNA-sequencing analyses on E18.5 WT and CHD4M195I/M195I hearts and identified 323 genes that were differentially expressed (adjusted P<0.05, |log2[fold change]|>0.5), with 113 genes upregulated and 210 genes downregulated in CHD4M195I/M195I (Figure 4I; Figure S15A). Consistent with our in vivo analyses at P0, gene set enrichment analysis revealed the overrepresented classifications in the CHD4M195I/M195I hearts to be those of cell cycle-related biological processes, including DNA replication initiation, cell cycle checkpoint control, nucleosome assembly, and cytokinesis (Figure 4J; Figure S15B). Representative differential genes included the core subunits of the DNA helicases Mcm(4–7), essential for DNA replication39 (Figure 4K). Thus, CHD4M195I was associated with an increase in cell cycle components and a related increase in cardiomyocyte proliferation.
Cardiomyocytes in CHD4 M195I/M195I Mice Are in an Immature State
In mice at E18.5, cardiomyocytes initiate multiple adaptations for the heart to transit from fetal to adult states.40–42 In line with the higher cardiomyocyte proliferation activity of CHD4M195I/M195I hearts (Figure 4), our gene set enrichment analysis classification also revealed that the downregulated genes in CHD4M195I/M195I hearts were significantly enriched in differentiation functions, such as muscle cell development, sarcomere organization, and myofibril assembly (Figure 5A; Figure S15B). Myosin heavy chain 6 (Myh6), a dominant adult sarcomeric isoform in rodents,43,44 was among the representative downregulated genes in CHD4M195I/M195I hearts (Figure 5B).
In heart development, cardiomyocyte division is associated with disassembling organized contractile fibrils.45,46 The observation that CHD4M195I/M195I cardiomyocytes showed downregulation of gene sets related to mature cardiomyocytes led us to hypothesize that proliferating cardiomyocytes in CHD4M195I/M195I remained in an immature state and were unable to form functioning sarcomeres. To test this hypothesis, we used transmission electron microscopy to examine the morphology of myofibrils. By E18.5, control hearts were composed of actin filaments assembled into individual contractile units anchored by Z discs (Figure 5C1 and 5C2). In contrast, CHD4M195I/M195I hearts displayed a severe reduction in myosin filament density and a concomitant loss or deformation of Z discs (Figure 5C3 and 5C4). Congruently, in CHD4M195I/M195I hearts, we found a failure of TMY to localize into discrete sarcomeres (Figure S15C). These findings suggested that proliferating cardiomyocytes failed to incorporate into organized sarcomeres, which may explain the observation of deficient contractile function in the mutant mice as assessed from echocardiograms (Figure 3).
During maturation, cardiomyocytes undergo a metabolic transition from glycolysis to oxidative phosphorylation to produce sufficient ATP to sustain heart function.47,48 Consistent with our finding that CHD4M195I/M195I cardiomyocytes retain properties of immature cells, we found that genes associated with oxidative phosphorylation were underrepresented in the transcriptional profile of CHD4M195I/M195I hearts (Figure 5D). Prominent examples were the downregulation of hexokinase 2 (Hk2) and cytochrome C oxidase 8b (Cox8b; Figure 5E), 2 enzymes essential for metabolism in mature cardiomyocytes.49,50 In addition, metabolic transcriptional regulators of mature cardiomyocytes (Ppargc1a, Ppara, Esrrb) were also significantly downregulated in the CHD4M195I/M195I hearts (Figure 5E). The changes in gene expression in CHD4M195I/M195I cardiomyocytes correlated with alteration in the maturation of the mitochondria. Mitochondria in WT hearts contained densely organized cristae (Figure 5F1 and 5F2), the structure housing the electron transport chain and ATP synthase. In contrast, the mitochondrial cristae in the CHD4M195I/M195I were immature and poorly aligned (Figure 5F3 and 5F4). Collectively, our molecular and cellular data strongly implied that cardiomyocytes in CHD4M195I/M195I hearts continued to proliferate until birth and were maintained in an immature state.
CHD4 M195I Leads to a Downregulation of ADAMTS1 and a Concomitant Ectopic Accumulation of the Extracellular Matrix
A fine-tuned balance between the synthesis and degradation of ECM (extracellular matrix) components is required for proper trabeculation and compaction in developing hearts.13,51–55 The ECM environment provides essential extracellular signals for cell proliferation, migration, and differentiation, all processes altered in CHD4M195I/M195I cardiac tissue. These observations, coupled with results from our transcriptional profiling and histological analyses, led to the hypothesis that ECM dynamics were dysregulated in CHD4M195I/M195I hearts. To test this hypothesis, we analyzed the cardiac ECM at E9.5, E10.5, E12.5, and E13.5 with Alcian Blue to detect acidic mucopolysaccharides, including hyaluronan and proteoglycans. CHD4M195I/M195I hearts showed a robust increase in Alcian Blue in the ventricular cardiac jelly compared with WT at each stage (Figure 6A, Figure S16A, and S16B). We further found an accompanying increase in Vcan (Versican), a large proteoglycan that functions in the ventricular septal formation56 (Figure 6B through 6D; Figure S16A and S16B). Consistently, we found a significant positive correlation between the relative Alcian Blue-positive area and proliferating cardiomyocytes (Ki67+/TMY+) in CHD4M195I/M195I hearts (Figure 6E). Thus, cardiomyocyte over-proliferation is associated with ECM accumulation.
Interestingly, although we saw a marked increase in Vcan protein in CHD4M195I/M195I versus WT hearts, we did not observe an increase in Vcan mRNA (Figure 6F; Figure S16C), which suggested that accumulation of Vcan was posttranslationally controlled. In parallel, we observed that cardiac expression of ADAMTS1, a critical metalloproteinase for degrading ECM and terminating trabeculation,13,54 was significantly lower in the CHD4M195I/M195I at both gene and protein levels (Figure 6G and 6H; Figure S16A and S16B). These changes were accompanied by decreased expression of Neoversican (Figure 6I; Figure S16A and S16B), a readout of metalloprotease activity.54 Taken together, our data suggested that CHD4M195I represses Adamts1 transcription, at or after E12.5, leading to an accumulation of components of the cardiac jelly (ie, Vcan), elevated and sustained proliferation of immature cardiomyocytes, and a failure to terminate cardiac trabeculation (Figure 6J).
Augmented Interaction Between CHD4 and BRG1 in CHD4 M195I/M195I Hearts
The findings that Adamts1 was downregulated in CHD4M195I/M195I cardiomyocytes and the associated increase in the ADAMTS1 substrate Vcan led us to investigate whether Adamts1 is a direct target of CHD4. BRG1 (SWI/SNF–related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4), a component of the multiprotein chromatin-remodeling SWI/SNF (Switch/Sucrose Non-Fermentable) complex,57,58 represses Adamts1 in endocardial cells to produce an ECM-rich environment that supports trabeculation. By E12.5, BRG1-mediated repression of Adamts1 is relieved, dissipating the cardiac jelly and preventing excessive trabeculation.54
Endogenous BRG1 interacts with CHD459,60 by directly associating with the N-terminal fragment of CHD4.60,61 Coincidently, the CHD4M195I mutation occurs within the CHD4 N-terminus. Thus, we hypothesized that the CHD4M195I protein has a higher affinity than WT CHD4 for BRG1, thereby repressing Adamts1 in CHD4M195I/M195I.
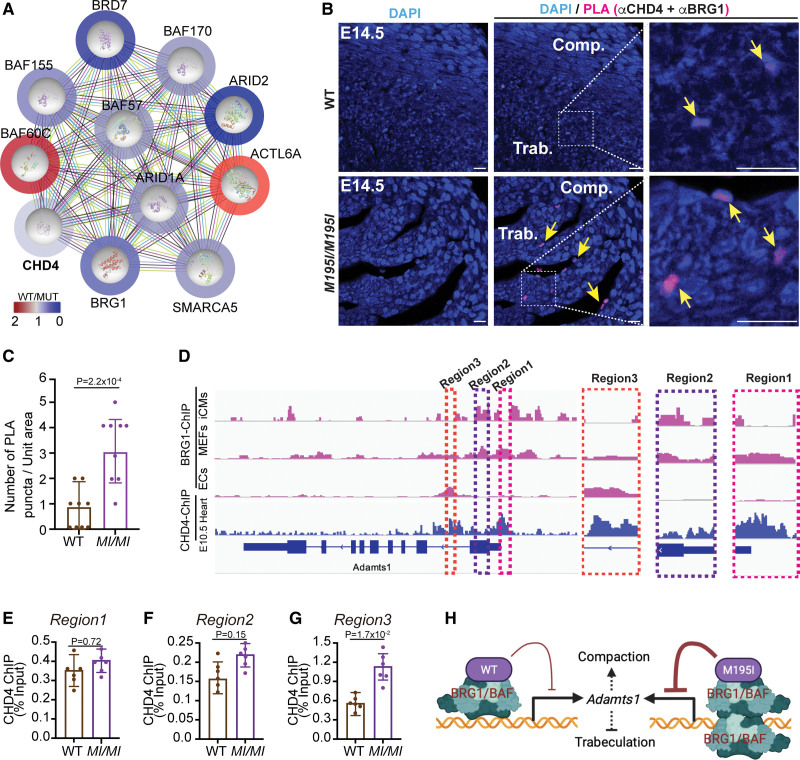
To test this model, in the presence of universal nuclease, we isolated the CHD4 cardiac endogenous interactome under physiological conditions from WT and CHD4M195I/M195I hearts and performed mass spectrometry analyses. The complexes were derived from E13.5 hearts, the time point when CHD4M195I/M195I cardiomyocytes show an increase in the ADAMTS1 substrate Vcan and have an increase in cardiomyocyte proliferation (Figure 6). We recovered CHD4 from E13.5 cardiac tissue at 57% coverage of the theoretical maximum of 86.6% with trypsin digest of all amino acids (Figure S17). We used an unbiased gene ontology-based bioinformatics classification to screen the functions of proteins associated with CHD4. This analysis showed CHD4 in association with ten components of the SWI/SNF complex, BRG1, SMARCA5 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A member 5), ACTL6A (actin-like 6A), ARID1A (AT-rich interaction domain 1A), BAF60C (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily D member 3), BAF155 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily C member 1), BRD7 (bromodomain containing 7), BAF57 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily E member 1), BAF170 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily C member 2), and ARID2 (AT-rich interaction domain 2). Thus, in E13.5 hearts, CHD4 associated with all the components required for a functional SWI/SNF complex. Moreover, after standardization, CHD4M195I/M195I was found to be associated with a greater number of spectra and a greater area under the curve for components of the SWI/SNF complex, including BRG1, compared with WT CHD4 (Figure 7A; Figure S18). In aggregate, these data indicated that, compared with the WT, CHD4M195I protein might interact with a higher affinity with the SWI/SNF complex in the developing heart.
To confirm the CHD4-BRG1 interaction, we performed an in situ proximity ligation assay with E14.5 heart sections. We observed interaction between BRG1 and CHD4 in the nuclei at the edge of the trabecular myocardium (Figure 7B) in both WT and mutant hearts (yellow arrows; Figure 7B). Consistent with the immunopurification–mass spectrometry results, the interaction between BRG1 and mutant CHD4 in the heart was more prominent than in WT hearts (Figure 7C). These results demonstrated that CHD4 and BRG1 interact in vivo in embryonic heart tissue.
Figure 7.
CHD4 (chromodomain helicase DNA-binding protein 4)M195I highly associates with BRG1 (SWI/SNF–related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4) to repress Adamts1. A, Protein network-based prediction with search tool for recurring instances of neighbouring genes (STING) showing CHD4 interactome in embryonic day (E) 13.5 hearts. The color bar and circles outside of each protein represent the ratio of M195I CHD4 spectra counts to wild-type (WT) CHD4. Two biological replicates per genotype for the mass spectrometry. Fifteen hearts were pooled for 1 replicate. B, Representative images of in situ Proximity Ligation Assay (PLA), counterstained with 4′,6-diamidino-2-phenylindole [DAPI], performed with anti-CHD4 and anti-BRG1 antibodies on E14.5 WT and CHD4M195I/M195I heart sections. Scale bars, 20 µm. Yellow arrows indicate positive PLA signals; n=3 individual hearts per genotype. C, Quantification of the PLA puncta. Data are represented as mean±SEM. Mann-Whitney U test was performed for comparisons. D, Visualization by integrative genomics viewer (IVG) browser of BRG1 or CHD4 chromatin immunoprecipitation sequencing (ChIP-seq) signals across the Adamts1 locus. Regions of interest are magnified in dashed frames. Data of BRG1 ChIP-seq and CHD4 ChIP-seq were retrieved from published studies: BRG1 ChIP-seq on iCMs (GSE116281),58 on MEFs (GSM2671190), on ECs (GSE152892),62 CHD4 ChIP-seq on E10.5.26 E through G, ChIP–quantitative polymerase chain reaction (qPCR) for CHD4-ChIP samples from E12.5 WT and CHD4M195I/M195I hearts with primers against the 3 indicated regions in D. Two biological replicates for the ChIP, 15 pooled hearts for each replicate. Data in E–G are represented as mean±SEM, and each biological replicate was analyzed 3 times in the qPCR assay. Mann-Whitney U test was performed for comparisons. H, Schematic for regulation of Adamts1 transcription by CHD4 and BRG1 association. ACTL6A indicates actin-like 6A; ARID1A, AT-rich interaction domain 1A; ARID2, AT-rich interaction domain 2; BAF57, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily E member 1; BAF60C, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily D member 3; BAF155, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily C member 1; BAF170, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily C member 2; BRD7, bromodomain containing 7; Comp., compact myocardium; ECs: endothelial cells; iCMs, iPSC-derived cardiomyocytes; MEFs: mouse embryonic fibroblasts; MUT, mutant; SMARCA5, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A member 5; and Trab., trabecular myocardium.
CHD4M195I-BRG1 Represses Adamts1
Chromatin immunoprecipitation–sequencing reveals that BRG1 and CHD4 cooccupy target genes genome-wide,61,63 and BRG1 and CHD4 dynamically occupy 3 cis-regulatory regions in the Adamts1 locus,54 that is, regions 1 to 3 (Figure 7D). To determine whether CHD4M195I affects the binding of CHD4 at Adamts1, we performed chromatin immunoprecipitation–quantitative polymerase chain reaction on WT and CHD4M195I/M195I hearts at E12.5. We found that CHD4M195I/M195I had a higher occupancy at Region 3 (the first intron of Adamts1) than WT CHD4, whereas there was no significant difference at region 1 or region 2 (Figure 7E through 7G). These results suggested that CHD4M195I/M195I associates more frequently or with a greater affinity with BRG1 at the first intron of the Adamts1 locus, thereby maintaining repression of Adamts1 (Figure 7H).
Restoration of ADAMTS1 Rescues Overgrowth of Trabeculae
Our data suggested that ADAMTS1 is essential in dissipating the cardiac jelly and preventing excessive trabeculation.54 To test this hypothesis in vivo, we examined the ability of rADAMTS1 (recombinant ADAMTS1) to restore the termination of trabeculation in hearts derived from CHD4M195I/M195I E11.5 embryos. For these studies, we first used heart explant cultures,64–68 in which the RVs were isolated and cultured in the presence of bead grafts that contained rADAMTS1 or BSA (Figure 8A). We observed that rADAMTS1 significantly inhibited the expression of Vcan and reduced the number of cycling cardiomyocytes (Figure 8B and 8C; Figure S19). Importantly, the changes in Vcan expression were not associated with changes in the proliferation of epicardial cells (Figure 8D), which suggested that rADAMTS1 acts in a cardiac cell type-specific manner. Thus, these data demonstrated that restoration of ADAMTS1 to the endocardial environment of CHD4M195I/M195I hearts inhibited ECM accumulation and cardiomyocyte proliferation.
Studies with breast cancer cell lines demonstrated that the pharmacological compound GW501516 acts by transcriptionally upregulating ADAMTS1.69 We tested the ability of the GW501516 to upregulate ADAMTS1 in heart tissue. Real-time polymerase chain reaction of Adamts1 derived from control and GW501516 treated heart explants confirmed that GW501516 significantly upregulated Adamts1 mRNA in cardiac tissue (Figure 8E). Next, we tested the ability of GW501516 to restore cessation of trabeculation in utero. To this end, we intercrossed CHD4M195I/+ mice and injected GW501516 intraperitoneally into pregnant CHD4M195I/+ females carrying litters between stages E9.5 to E11.5 (once a day; Figure 8F). At E12.5, we found that Adamts1 mRNA and protein were significantly induced in cardiomyocytes by GW501516 in the WT and CHD4M195I/M195I hearts (Figure 8G1 through 8G4 and 8H). Strikingly, cardiomyocyte proliferation decreased, and there was a concomitant decrease in the thickness of the trabeculae layer upon GW501516 treatment of CHD4M195I/M195I hearts (Figure 8G5 through 8G12, 8I through 8L). However, the GW501516 treatment did not affect the thickness of the compact myocardium (Figure 8M). Further treatment of pregnant females derived from the intercross of CHD4M195I heterozygous mice with GW501516 at E9.5, E11.5, E13.5, E15.5, and E17.5, and the cardiac phenotype was analyzed at P0 pups. We observed that GW501516 rescued the ventricular wall thickness in CHD4M195I/M195I hearts but did not rescue the compact myocardium, and hence, the pups were not brought to term (Figure S20). These studies indicated that CHD4M195 acting through ADAMTS1 regulates vital aspects of LVNC at mid-gestation by controlling the cessation of trabeculae growth.
Discussion
Mutations in CHD4, the catalytic component of NuRD complex, lead to congenital heart disease, including atrial and ventricular septal defects.26,28,32,70,71 However, CHD4 is expressed in most cell types; thus, it was not known how patients with CHD4 missense mutations display a restricted cardiac phenotype. In this study, we identified a proband for CHD4 (CHD4M202I) who had cardiac abnormalities, and we generated a humanized mouse model for this mutation (CHD4M195I). Using our model, we established the mechanisms by which CHD4M195I leads to impaired cardiac function, and we identified potential approaches to LVNC therapy. Critically, we showed that administration of ADAMTS1 in culture or in utero rescued aspects of the LVNC-associated phenotype. Mice homozygous for CHD4M195I displayed cardiac abnormalities significantly different than cardiac Chd4 heterozygous or homozygous null mutations.26,28 Most notably, CHD4M195I/M195I, unlike cardiac null CHD4 embryos, display an LVNC phenotype that includes cardiac noncompaction, hypertrabeculation, and cardiomyocyte over-proliferation. Collectively, these findings strongly imply that CHD4M202I activity regulates a transcriptional network distinct from the function of CHD4 during the early phases of cardiogenesis.
Left Ventricular Noncompaction
Whether and how cardiomyocyte proliferation leads to LVNC hypertrabeculation are a controversial issue. Recent fate mapping studies revealed that abolishing proliferation in the compact layer can lead to a hypertrabeculation phenotype in mice.72 Consistently, cardiac conditional mutations in Jag1, Jag2, Prdm16, and RBPMS all lead to decreased proliferation and hypertrabeculated hearts.10,14,73 Conversely, other studies suggest that trabeculation occurs at the expense of the compact layer. For example, Luxan et al. demonstrated that mutations in the E3 ubiquitin ligase MIB1 (mindbomb homolog 1) led to LVNC in mice and humans. Enlarged noncompacted trabeculae in MIB1 mice showed increased cardiomyocyte proliferation and a downregulation of Notch activity.15 Analogously, deletion of Nkx2–5 in trabecular myocardium74 and global knockout of Fkbp1a75 or Plxnd153 led to hypertrabeculation and reduced compaction that is associated with an increase in cardiomyocyte proliferation. One potential contradiction in these latter findings is that there was an increase in the proliferation of cardiomyocytes in the compact layer, yet, the compact layer was thinner than in controls. We favor the explanation that cardiomyocyte proliferation leads to noncompaction akin to a zebrafish model in which proliferation-induced cellular crowding at the tissue scale triggers tension heterogeneity among cardiomyocytes. The crowding, in turn, drives cardiomyocytes with higher contractility to delaminate and seed the trabecular layer.76 Thus, CHD4M195I leads to the over-proliferation and heterogenicity of cardiomyocytes, which leads to a greater number of cardiomyocytes that enter and proliferate within the trabecular layer. Interestingly, we observed p21 (Cdkn1a), p19 (Cdkn2d), and Cdkn3, potent cell cycle inhibitors, were significantly downregulated in the CHD4M195I/M195I hearts in our RNA-sequencing data (Figure S21A). We also identified that p21, p19, and Cdkn3 were direct targets of CHD426 (Figure S21B and S21C). These results may further demonstrate that the noncompaction phenotype is related to over-proliferation in the CHD4M195I/M195I hearts. In future studies, it will be informative to generate a cardiomyocyte conditional knockin CHD4M195I allele and determine if it phenocopies the global CHD4M195I/M195I allele.
ADMATS1 and LVNC
Recent genome-wide association studies implicated the family of ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) proteases in cardiovascular diseases.77–79 In the heart, the ADAMTS proteins are mainly secreted by cardiac fibroblasts (ie, ADAMTS5)80 or endothelial cells (ie, ADAMTS1,54 ADAMTS1381). Abundant extracellular matrix proteins, including the ADAMTs proteins, induce the differentiation and proliferation of cardiomyocytes. Previous studies demonstrated that Adamts1 is a direct target of BRG1 in the endocardium, and the suppression of Adamts1 by BRG1 is essential for the termination of trabeculation.54 Here we demonstrated that CHD4 (and CHD4M195I) interacts with BRG1 at the endocardium, and CHD4M195I/M195I led to a downregulation of ADAMTS1 and an accumulation of ECM components that might promote cardiomyocyte proliferation. These results strongly suggest that CHD4M195I robustly represses endocardial Adamts1 expression such that the CHD4M195I/M195 hearts fail to terminate trabeculation and display LVNC phenotype. Furthermore, administration of ADAMTS1 attenuated key LVNC properties of CHD4M195I/M195I hearts.
Disruption of Notch signaling in myocardium or endocardium leads to cardiac noncompaction.14,15,53,75,82–85 In the present study, transcriptional profiling of CHD4M195I/M195I hearts revealed only a modest decrease in a limited number of components of the Notch pathway, for example, Dll4, Jag2, and Hey2 (Figure S22). Knockout Dll4 in the endocardium is embryonic lethal14; in our CHD4M195I/M195I hearts, Dll4 is significantly downregulated (Figure S22A). However, no evidence showed that GW501516, the PPARδ (Peroxisome proliferator activated receptor delta)-selective agonist, can induce Dll4 expression in the heart. It has been demonstrated that there was no significant difference in the expression of Dll4 or other key components of Notch signaling between Pparb+/+ and Pparb−/− cells,86 suggesting that GW501516 is unlikely to restore the expression of Dll4 in the CHD4M195I/M195I hearts to ensure appropriate vascularization in the heart, which might explain why treatment with GW501516 cannot completely rescue the lethality. The CHD4M195I mutation occurs in a highly conserved N-terminal region of human CHD4 termed CHD4-N (residues 145–225). In vitro, CHD4-N binds poly (ADP-ribose) with higher affinity than it binds DNA, suggesting that CHD4-N recognizes the DNA backbone instead of making specific interactions with the nucleotides. Whether the M202I mutation affects CHD4’s affinity or recruitment to DNA remains to be determined. Our findings suggest that CHD4M195I acts either downstream or in parallel to Notch signaling. Thus, it will be interesting to determine whether the administration of ADAMTS1 rescues LVNC defects that result from alterations in Notch pathway components.
Conclusions
Our CHD4M195I/M195I mouse model recapitulated the ventricular noncompaction abnormality in human LVNC. Thus, CHD4M195I/M195I provided a comprehensive understanding of the mechanisms of hypertrabeculation and ventricular noncompaction, and the model defined a previously unknown function of CHD4 in heart development and diseases. Moreover, the observation that the administration of ADAMTS1 ameliorated aspects of LVNC suggests a possible ADAMTS1-based therapeutic approach for patients with LVNC.
Article Information
Acknowledgments
We greatly thank the generation of the CHD4M195I mouse line by the Animal Model Core at the University of North Carolina at Chapel Hill (UNC). The authors thank the UNC Microscopy Services Laboratory Core for the processing and imaging the transmission electron microscopy. The authors also thank the performance of proteomics analysis by the UNC Michael Hooker Proteomics Center.
Sources of Funding
This work was supported by grants R01HL156424 National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute (NHLBI) to F.L. Conlon and R35-GM141833 NIH/National Institute of General Medical Sciences (NIGMS) to J.G. Cook.
Disclosures
None.
Supplemental Material
Supplemental Methods
Figures S1–S22
Videos S1–S5
Uncut immunoblots
Major Resources Table
Supplementary Material
Nonstandard Abbreviations and Acronyms
- ADAMTS1
- ADAM metallopeptidase with thrombospondin type 1 motif 1
- BRG1
- SWI/SNF–related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4
- CHD4
- chromodomain helicase DNA-binding protein 4
- ECM
- extracellular matrix
- LV
- left ventricle
- LVNC
- left ventricular noncompaction
- NuRD
- nucleosome remodeling and deacetylase
- RV
- right ventricle
For Sources of Funding and Disclosures, see page 65.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.122.322223.
References
- 1.Finsterer J, Stollberger C, Towbin JA. Left ventricular noncompaction cardiomyopathy: cardiac, neuromuscular, and genetic factors. Nat Rev Cardiol. 2017;14:224–237. doi: 10.1038/nrcardio.2016.207 [DOI] [PubMed] [Google Scholar]
- 2.Ramakumar V, Sharma V, Mishra S. Left ventricular non-compaction. Eur Heart J. 2021;42:2398. doi: 10.1093/eurheartj/ehab099 [DOI] [PubMed] [Google Scholar]
- 3.Fazio G, Lunetta M, Grassedonio E, Gullotti A, Ferro G, Bacarella D, Lo Re G, Novo G, Massimo M, Maresi E, et al. Noncompaction of the right ventricle. Pediatr Cardiol. 2010;31:576–578. doi: 10.1007/s00246-010-9652-6 [DOI] [PubMed] [Google Scholar]
- 4.Ranganathan A, Ganesan G, Sangareddi V, Pillai AP, Ramasamy A. Isolated noncompaction of right ventricle--a case report. Echocardiogr. 2012;29:E169–E172. doi: 10.1111/j.1540-8175.2012.01671.x [DOI] [PubMed] [Google Scholar]
- 5.Syed MP, Doshi A, Pandey D, Kate Y, Harizi R. A rare case of biventricular non-compaction. BMJ Case Rep. 2020;13:e231154. doi: 10.1136/bcr-2019-231154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiford BC, Subbarao VD, Mulhern KM. Noncompaction of the ventricular myocardium. Circulation. 2004;109:2965–2971. doi: 10.1161/01.CIR.0000132478.60674.D0 [DOI] [PubMed] [Google Scholar]
- 7.Klaassen S, Probst S, Oechslin E, Gerull B, Krings G, Schuler P, Greutmann M, Hurlimann D, Yegitbasi M, Pons L, et al. Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation. 2008;117:2893–2901. doi: 10.1161/CIRCULATIONAHA.107.746164 [DOI] [PubMed] [Google Scholar]
- 8.Towbin JA. Ion channel dysfunction associated with arrhythmia, ventricular noncompaction, and mitral valve prolapse: a new overlapping phenotype. J Am Coll Cardiol. 2014;64:768–771. doi: 10.1016/j.jacc.2014.06.1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashraf H, Pradhan L, Chang EI, Terada R, Ryan NJ, Briggs LE, Chowdhury R, Zarate MA, Sugi Y, Nam HJ, et al. A mouse model of human congenital heart disease: high incidence of diverse cardiac anomalies and ventricular noncompaction produced by heterozygous Nkx2-5 homeodomain missense mutation. Circ Cardiovasc Genet. 2014;7:423–433. doi: 10.1161/CIRCGENETICS.113.000281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan P, Wang Z, Morales MG, Zhang Y, Bassel-Duby R, Liu N, Olson EN. RBPMS is an RNA-binding protein that mediates cardiomyocyte binucleation and cardiovascular development. Dev Cell. 2022;57:959–973.e7. doi: 10.1016/j.devcel.2022.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pantazis AA, Elliott PM. Left ventricular noncompaction. Curr Opin Cardiol. 2009;24:209–213. doi: 10.1097/HCO.0b013e32832a11e7 [DOI] [PubMed] [Google Scholar]
- 12.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat Rec. 2000;258:319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O [DOI] [PubMed] [Google Scholar]
- 13.Del Monte-Nieto G, Ramialison M, Adam AAS, Wu B, Aharonov A, D’Uva G, Bourke LM, Pitulescu ME, Chen H, de la Pompa JL, et al. Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature. 2018;557:439–445. doi: 10.1038/s41586-018-0110-6 [DOI] [PubMed] [Google Scholar]
- 14.D’Amato G, Luxan G, del Monte-Nieto G, Martinez-Poveda B, Torroja C, Walter W, Bochter MS, Benedito R, Cole S, Martinez F, et al. Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 2016;18:7–20. doi: 10.1038/ncb3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luxan G, Casanova JC, Martinez-Poveda B, Prados B, D’Amato G, MacGrogan D, Gonzalez-Rajal A, Dobarro D, Torroja C, Martinez F, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med. 2013;19:193–201. doi: 10.1038/nm.3046 [DOI] [PubMed] [Google Scholar]
- 16.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664 [DOI] [PubMed] [Google Scholar]
- 17.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4 [DOI] [PubMed] [Google Scholar]
- 19.O’Shaughnessy-Kirwan A, Signolet J, Costello I, Gharbi S, Hendrich B. Constraint of gene expression by the chromatin remodelling protein CHD4 facilitates lineage specification. Development. 2015;142:2586–2597. doi: 10.1242/dev.125450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung H, Kohnken R, Svaren J. The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J Neurosci. 2012;32:1517–1527. doi: 10.1523/JNEUROSCI.2895-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashiwagi M, Morgan BA, Georgopoulos K. The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development. 2007;134:1571–1582. doi: 10.1242/dev.001750 [DOI] [PubMed] [Google Scholar]
- 22.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005 [DOI] [PubMed] [Google Scholar]
- 23.Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida T, Hu Y, Zhang Z, Emmanuel AO, Galani K, Muhire B, Snippert HJ, Williams CJ, Tolstorukov MY, Gounari F, et al. Chromatin restriction by the nucleosome remodeler Mi-2beta and functional interplay with lineage-specific transcription regulators control B-cell differentiation. Genes Dev. 2019;33:763–781. doi: 10.1101/gad.321901.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilczewski CM, Hepperla AJ, Shimbo T, Wasson L, Robbe ZL, Davis IJ, Wade PA, Conlon FL. CHD4 and the NuRD complex directly control cardiac sarcomere formation. Proc Natl Acad Sci U S A. 2018;115:6727–6732. doi: 10.1073/pnas.1722219115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malecova B, Dall’Agnese A, Puri PL. Muscles cannot break a NuRDy heart. EMBO J. 2016;35:1600–1602. doi: 10.15252/embj.201694835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-Del Arco P, Perdiguero E, Yunes-Leites PS, Acin-Perez R, Zeini M, Garcia-Gomez A, Sreenivasan K, Jimenez-Alcazar M, Segales J, Lopez-Maderuelo D, et al. The chromatin remodeling complex Chd4/NuRD controls striated muscle identity and metabolic homeostasis. Cell Metab. 2016;23:881–892. doi: 10.1016/j.cmet.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 29.Robbe ZL, Shi W, Wasson LK, Scialdone AP, Wilczewski CM, Sheng X, Hepperla AJ, Akerberg BN, Pu WT, Cristea IM, et al. CHD4 is recruited by GATA4 and NKX2-5 to repress noncardiac gene programs in the developing heart. Genes Dev. 2022;36:468–482. doi: 10.1101/gad.349154.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gelb B, Brueckner M, Chung W, Goldmuntz E, Kaltman J, Kaski JP, Kim R, Kline J, Mercer-Rosa L, Porter G, et al. ; Pediatric Cardiac Genomics Consortium. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circ Res. 2013;112:698–706. doi: 10.1161/CIRCRESAHA.111.300297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss K, Lazar HP, Kurolap A, Martinez AF, Paperna T, Cohen L, Smeland MF, Whalen S, Heide S, Keren B, et al. The CHD4-related syndrome: a comprehensive investigation of the clinical spectrum, genotype-phenotype correlations, and molecular basis. Genet Med. 2020;22:389–397. doi: 10.1038/s41436-019-0612-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva AP, Ryan DP, Galanty Y, Low JK, Vandevenne M, Jackson SP, Mackay JP. The N-terminal region of chromodomain helicase DNA-binding protein 4 (CHD4) is essential for activity and contains a High Mobility Group (HMG) box-like-domain that can bind poly(ADP-ribose). J Biol Chem. 2016;291:924–938. doi: 10.1074/jbc.M115.683227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan MR, Hsieh HJ, Dai H, Hung WC, Li K, Peng G, Lin SY. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J Biol Chem. 2012;287:6764–6772. doi: 10.1074/jbc.M111.287037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stacey RB, Caine AJ, Jr, Hundley WG. Evaluation and management of left ventricular noncompaction cardiomyopathy. Curr Heart Fail Rep. 2015;12:61–67. doi: 10.1007/s11897-014-0237-1 [DOI] [PubMed] [Google Scholar]
- 36.Kawel N, Nacif M, Arai AE, Gomes AS, Hundley WG, Johnson WC, Prince MR, Stacey RB, Lima JA, Bluemke DA. Trabeculated (noncompacted) and compact myocardium in adults: the multi-ethnic study of atherosclerosis. Circ Cardiovasc Imaging. 2012;5:357–366. doi: 10.1161/CIRCIMAGING.111.971713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim W, Seidah NG, Prat A. In utero measurement of heart rate in mouse by noninvasive M-mode echocardiography. J Vis Exp. 2013;81:e50994. doi: 10.3791/50994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Touma M. Fetal mouse cardiovascular imaging using a high-frequency ultrasound (30/45MHZ) system. J Vis Exp. 2018;135:57210. doi: 10.3791/57210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mei L, Kedziora KM, Song EA, Purvis JE, Cook JG. The consequences of differential origin licensing dynamics in distinct chromatin environments. Nucleic Acids Res. 2022;50:9601–9620. doi: 10.1093/nar/gkac003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo Y, Pu WT. Cardiomyocyte maturation: new phase in development. Circ Res. 2020;126:1086–1106. doi: 10.1161/CIRCRESAHA.119.315862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, Grinsfelder D, Rothermel BA, Chen R, Garcia JA, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kannan S, Kwon C. Regulation of cardiomyocyte maturation during critical perinatal window. J Physiol. 2020;598:2941–2956. doi: 10.1113/JP276754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franco D, Lamers WH, Moorman AF. Patterns of expression in the developing myocardium: towards a morphologically integrated transcriptional model. Cardiovasc Res. 1998;38:25–53. doi: 10.1016/s0008-6363(97)00321-0 [DOI] [PubMed] [Google Scholar]
- 44.Lyons GE, Schiaffino S, Sassoon D, Barton P, Buckingham M. Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol. 1990;111:2427–2436. doi: 10.1083/jcb.111.6.2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060 [DOI] [PubMed] [Google Scholar]
- 46.Ahuja P, Perriard E, Perriard JC, Ehler E. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J Cell Sci. 2004;117:3295–3306. doi: 10.1242/jcs.01159 [DOI] [PubMed] [Google Scholar]
- 47.Neubauer S. The failing heart--an engine out of fuel. N Engl J Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052 [DOI] [PubMed] [Google Scholar]
- 48.Lopaschuk GD, Jaswal JS. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J Cardiovasc Pharmacol. 2010;56:130–140. doi: 10.1097/FJC.0b013e3181e74a14 [DOI] [PubMed] [Google Scholar]
- 49.Calmettes G, John SA, Weiss JN, Ribalet B. Hexokinase-mitochondrial interactions regulate glucose metabolism differentially in adult and neonatal cardiac myocytes. J Gen Physiol. 2013;142:425–436. doi: 10.1085/jgp.201310968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, et al. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016;39:480–490. doi: 10.1016/j.devcel.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lockhart M, Wirrig E, Phelps A, Wessels A. Extracellular matrix and heart development. Birth Defects Res A Clin Mol Teratol. 2011;91:535–550. doi: 10.1002/bdra.20810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Jr, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106:349–360. doi: 10.1172/JCI10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandireddy R, Cibi DM, Gupta P, Singh A, Tee N, Uemura A, Epstein JA, Singh MK. Semaphorin 3E/PlexinD1 signaling is required for cardiac ventricular compaction. JCI Insight. 2019;4:e125908. doi: 10.1172/jci.insight.125908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamura H, Zhang M, Markwald RR, Mjaatvedt CH. A heart segmental defect in the anterior-posterior axis of a transgenic mutant mouse. Dev Biol. 1997;186:58–72. doi: 10.1006/dbio.1997.8559 [DOI] [PubMed] [Google Scholar]
- 56.Hatano S, Kimata K, Hiraiwa N, Kusakabe M, Isogai Z, Adachi E, Shinomura T, Watanabe H. Versican/PG-M is essential for ventricular septal formation subsequent to cardiac atrioventricular cushion development. Glycobiology. 2012;22:1268–1277. doi: 10.1093/glycob/cws095 [DOI] [PubMed] [Google Scholar]
- 57.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hota SK, Johnson JR, Verschueren E, Thomas R, Blotnick AM, Zhu Y, Sun X, Pennacchio LA, Krogan NJ, Bruneau BG. Dynamic BAF chromatin remodeling complex subunit inclusion promotes temporally distinct gene expression programs in cardiogenesis. Development. 2019;146:dev174086. doi: 10.1242/dev.174086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh AP, Foley JF, Rubino M, Boyle MC, Tandon A, Shah R, Archer TK. Brg1 enables rapid growth of the early embryo by suppressing genes that regulate apoptosis and cell growth arrest. Mol Cell Biol. 2016;36:1990–2010. doi: 10.1128/MCB.01101-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimono Y, Murakami H, Kawai K, Wade PA, Shimokata K, Takahashi M. Mi-2 beta associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J Biol Chem. 2003;278:51638–51645. doi: 10.1074/jbc.M309198200 [DOI] [PubMed] [Google Scholar]
- 61.Sun X, Yu W, Li L, Sun Y. ADNP controls gene expression through local chromatin architecture by association with BRG1 and CHD4. Front Cell Dev Biol. 2020;8:553. doi: 10.3389/fcell.2020.00553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moonen J-R, Chappell J, Shi M, Shinohara T, Li D, Mumbach MR, Zhang F, Nair RV, Nasser J, Mai DH, et al. KLF4 recruits SWI/SNF to increase chromatin accessibility and reprogram the endothelial enhancer landscape under laminar shear stress. Nat Commun. 2022;13:4941. doi: 10.1038/s41467-022-32566-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morris SA, Baek S, Sung MH, John S, Wiench M, Johnson TA, Schiltz RL, Hager GL. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dyer LA, Patterson C. A novel ex vivo culture method for the embryonic mouse heart. J Vis Exp. 2013;75:e50359. doi: 10.3791/50359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao J, Poss KD. Explant culture of adult zebrafish hearts for epicardial regeneration studies. Nat Protoc. 2016;11:872–881. doi: 10.1038/nprot.2016.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rhee S, Chung JI, King DA, D’Amato G, Paik DT, Duan A, Chang A, Nagelberg D, Sharma B, Jeong Y, et al. Endothelial deletion of Ino80 disrupts coronary angiogenesis and causes congenital heart disease. Nat Commun. 2018;9:368. doi: 10.1038/s41467-017-02796-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Powers N, Huang GN. Visualization of regenerating and repairing hearts. Clin Sci (Lond). 2022;136:787–798. doi: 10.1042/CS20211116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rhee S, Paik DT, Yang JY, Nagelberg D, Williams I, Tian L, Roth R, Chandy M, Ban J, Belbachir N, et al. Endocardial/endothelial angiocrines regulate cardiomyocyte development and maturation and induce features of ventricular non-compaction. Eur Heart J. 2021;42:4264–4276. doi: 10.1093/eurheartj/ehab298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ham SA, Yoo T, Lee WJ, Hwang JS, Hur J, Paek KS, Lim DS, Han SG, Lee CH, Seo HG. ADAMTS1-mediated targeting of TSP-1 by PPARdelta suppresses migration and invasion of breast cancer cells. Oncotarget. 2017;8:94091–94103. doi: 10.18632/oncotarget.21584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pinard A, Guey S, Guo D, Cecchi AC, Kharas N, Wallace S, Regalado ES, Hostetler EM, Sharrief AZ, Bergametti F, et al. The pleiotropy associated with de novo variants in CHD4, CNOT3, and SETD5 extends to moyamoya angiopathy. Genet Med. 2020;22:427–431. doi: 10.1038/s41436-019-0639-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SH, Thienpont B, McRae J, Fitzgerald TW, Singh T, Swaminathan GJ, et al. ; INTERVAL Study. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–1065. doi: 10.1038/ng.3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tian X, Li Y, He L, Zhang H, Huang X, Liu Q, Pu W, Zhang L, Li Y, Zhao H, et al. Identification of a hybrid myocardial zone in the mammalian heart after birth. Nat Commun. 2017;8:87. doi: 10.1038/s41467-017-00118-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu T, Liang Z, Zhang Z, Liu C, Zhang L, Gu Y, Peterson KL, Evans SM, Fu XD, Chen J. PRDM16 is a compact myocardium-enriched transcription factor required to maintain compact myocardial cardiomyocyte identity in left ventricle. Circulation. 2022;145:586–602. doi: 10.1161/CIRCULATIONAHA.121.056666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choquet C, Nguyen THM, Sicard P, Buttigieg E, Tran TT, Kober F, Varlet I, Sturny R, Costa MW, Harvey RP, et al. Deletion of Nkx2-5 in trabecular myocardium reveals the developmental origins of pathological heterogeneity associated with ventricular non-compaction cardiomyopathy. PLoS Genet. 2018;14:e1007502. doi: 10.1371/journal.pgen.1007502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen H, Zhang W, Sun X, Yoshimoto M, Chen Z, Zhu W, Liu J, Shen Y, Yong W, Li D, et al. Fkbp1a controls ventricular myocardium trabeculation and compaction by regulating endocardial Notch1 activity. Development. 2013;140:1946–1957. doi: 10.1242/dev.089920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Priya R, Allanki S, Gentile A, Mansingh S, Uribe V, Maischein HM, Stainier DYR. Tension heterogeneity directs form and fate to pattern the myocardial wall. Nature. 2020;588:130–134. doi: 10.1038/s41586-020-2946-9 [DOI] [PubMed] [Google Scholar]
- 77.Saleheen D, Zhao W, Young R, Nelson CP, Ho W, Ferguson JF, Rasheed A, Ou K, Nurnberg ST, Bauer RC, et al. Loss of cardioprotective effects at the ADAMTS7 locus as a result of gene-smoking interactions. Circulation. 2017;135:2336–2353. doi: 10.1161/CIRCULATIONAHA.116.022069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, Burnett MS, Devaney JM, Knouff CW, Thompson JR, et al. ; Myocardial Infarction Genetics Consortium. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prins BP, Mead TJ, Brody JA, Sveinbjornsson G, Ntalla I, Bihlmeyer NA, van den Berg M, Bork-Jensen J, Cappellani S, Van Duijvenboden S, et al. Exome-chip meta-analysis identifies novel loci associated with cardiac conduction, including ADAMTS6. Genome Biol. 2018;19:87. doi: 10.1186/s13059-018-1457-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abonnenc M, Nabeebaccus AA, Mayr U, Barallobre-Barreiro J, Dong X, Cuello F, Sur S, Drozdov I, Langley SR, Lu R, et al. Extracellular matrix secretion by cardiac fibroblasts: role of microRNA-29b and microRNA-30c. Circ Res. 2013;113:1138–1147. doi: 10.1161/CIRCRESAHA.113.302400 [DOI] [PubMed] [Google Scholar]
- 81.Wang A, Duan Q, Wu J, Liu X, Sun Z. The expression of ADAMTS13 in human microvascular endothelial cells. Blood Coagul Fibrinolysis. 2016;27:464–466. doi: 10.1097/MBC.0000000000000405 [DOI] [PubMed] [Google Scholar]
- 82.Venkatesh DA, Park KS, Harrington A, Miceli-Libby L, Yoon JK, Liaw L. Cardiovascular and hematopoietic defects associated with Notch1 activation in embryonic Tie2-expressing populations. Circ Res. 2008;103:423–431. doi: 10.1161/CIRCRESAHA.108.177808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang J, Bucker S, Jungblut B, Bottger T, Cinnamon Y, Tchorz J, Muller M, Bettler B, Harvey R, Sun QY, et al. Inhibition of Notch2 by Numb/Numblike controls myocardial compaction in the heart. Cardiovasc Res. 2012;96:276–285. doi: 10.1093/cvr/cvs250 [DOI] [PubMed] [Google Scholar]
- 84.Watanabe Y, Kokubo H, Miyagawa-Tomita S, Endo M, Igarashi K, Aisaki K, Kanno J, Saga Y. Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development. 2006;133:1625–1634. doi: 10.1242/dev.02344 [DOI] [PubMed] [Google Scholar]
- 85.Mysliwiec MR, Bresnick EH, Lee Y. Endothelial Jarid2/Jumonji is required for normal cardiac development and proper Notch1 expression. J Biol Chem. 2011;286:17193–17204. doi: 10.1074/jbc.M110.205146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muller-Brusselbach S, Komhoff M, Rieck M, Meissner W, Kaddatz K, Adamkiewicz J, Keil B, Klose KJ, Moll R, Burdick AD, et al. Deregulation of tumor angiogenesis and blockade of tumor growth in PPARbeta-deficient mice. EMBO J. 2007;26:3686–3698. doi: 10.1038/sj.emboj.7601803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shi W, Xu C, Gong Y, Wang J, Ren Q, Yan Z, Mei L, Tang C, Ji X, Hu X, et al. RhoA/Rock activation represents a new mechanism for inactivating Wnt/beta-catenin signaling in the aging-associated bone loss. Cell Regen. 2021;10:8. doi: 10.1186/s13619-020-00071-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang JR, Wang CJ, Xu CY, Wu XK, Hong D, Shi W, Gong Y, Chen HX, Long F, Wu XM. Signaling cascades governing Cdc42-mediated chondrogenic differentiation and mensenchymal condensation. Genetics. 2016;202:1055–1069. doi: 10.1534/genetics.115.180109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dorr KM, Amin NM, Kuchenbrod LM, Labiner H, Charpentier MS, Pevny LH, Wessels A, Conlon FL. Casz1 is required for cardiomyocyte G1-to-S phase progression during mammalian cardiac development. Development. 2015;142:2037–2047. doi: 10.1242/dev.119107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ehler E, Moore-Morris T, Lange S. Isolation and culture of neonatal mouse cardiomyocytes. J Vis Exp. 2013;79:50154. doi: 10.3791/50154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Waldron L, Steimle JD, Greco TM, Gomez NC, Dorr KM, Kweon J, Temple B, Yang XH, Wilczewski CM, Davis IJ, et al. The cardiac TBX5 interactome reveals a chromatin remodeling network essential for cardiac septation. Dev Cell. 2016;36:262–275. doi: 10.1016/j.devcel.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shi W, Sheng X, Dorr KM, Hutton JE, Emerson JI, Davies HA, Andrade TD, Wasson LK, Greco TM, Hashimoto Y, et al. Cardiac proteomics reveals sex chromosome-dependent differences between males and females that arise prior to gonad formation. Dev Cell. 2021;56:3019–3034.e7. doi: 10.1016/j.devcel.2021.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim GH. Murine fetal echocardiography. J Vis Exp. 2013;72:4416. doi: 10.3791/4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 95.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
An expanded Materials and Methods section is available in the Supplemental Material, and the sources of critical reagents are provided in the Major Resources Table. The raw RNA-sequencing data are available through the Gene Expression Omnibus (GEO) public functional genomics data repository with the accession number GSE229050. On reasonable request, the data, analytic methods, and study materials will be made available to other researchers to reproduce the results or replicate the procedures.