Abstract
Polycomb repressive complexes are a family of chromatin modifier enzymes which are critical for regulating gene expression and maintaining cell-type identity. The reversible chemical modifications of histone H3 and H2A by the Polycomb proteins are central to its ability to function as a gene silencer. PRC2 is both a reader and writer of the trimethylation of histone H3 lysine 27 (H3K27me3) which serves as a marker for transcription repression, and heterochromatin boundaries. Over the last few years, several studies have provided key insights into the mechanisms regulating the recruitment and activation of PRC2 at Polycomb target genes. In this review, we highlight the recent structural studies which have elucidated the roles played by Polycomb cofactor proteins in mediating crosstalk between histone post-translational modifications and the recruitment of PRC2 and the stimulation of PRC2 methyltransferase activity.
Introduction
Eukaryotic cells pack their DNA (~6 billion bases) into the small volume of the nucleus (1 × 102−1 × 105 μm3) by organizing their DNA into complex but defined structures, with the fundamental unit of such a structure being the nucleosome. The nucleosome is a hetero octamer consisting of two copies of four different histone proteins (H2A, H2B, H3 and H4) with a 147 bases of DNA wrapped around the histone protein octamer core [1,2]. One hallmark of the nucleosome is that its higher-order organization as well as access to the DNA can be modulated by chemical modifications of the histone proteins, termed histone post-translational modifications. Some histone post-translational modifications such as tri-methylation of histone H3 at lysine 4 or 36 (H3K4me3 or H3K36me3) mark them as regions with active transcription, whereas others such as H3K27me3 or H3K9me3 mark them for transcription repression where the DNA and histones are in a compact configuration which does not allow access to transcription machinery [3,4]. The chromatin architecture changes mediated by these epigenetic histone post-translational modifications are hence crucial for the transcriptional regulation underlying key cellular events such as cell differentiation and maintenance of cell-type identity [5,6]. The enzymes that read, write, or erase histone post-translational modifications are collectively known as chromatin modifiers [4]. The Polycomb group of proteins (PcG) are evolutionarily conserved chromatin modifier enzymes which play critical roles in cell differentiation, pluripotency, maintenance of cell type identity, epigenetic memory during imprinting, and are also one of the most frequently mutated enzymes in cancers [7,8]. The first Polycomb gene was discovered almost a century ago in D. melanogaster [9]. However, it wasn’t until two decades later that both the Polycomb (PcG and Trithorax (Trx)) group of proteins were discovered to play vital roles in gene-body patterning and development in D. melanogaster through the regulation of Homeotic (Hox) genes [9–14]. PcG and Trx homologs have since been discovered in various other eukaryotic species from fungi to mammals hinting at an evolutionarily conserved role for these enzymes [7]. The balanced activity of PcG and Trx enzymes is important for a plethora of regulatory processes such as X chromosome inactivation, epigenetic inheritance of memory or imprinting, stem cell differentiation, and cell cycle control [15–17]. The PcG family consists of three major sub-family of complexes: Polycomb Repressive Complexes 1 (PRC1), Polycomb Repressive Complex 2 (PRC2) and Polycomb Repressive De-ubiquitinase (PR-DUB) [7,18,19]. PRC2 is a methyltransferase (HMTase) that catalyzes the mono-, di- and tri-methylation of histone H3 at lysine 27 (H3K27me1/2/3) marking them for transcription repression [20–22]. PRC2 is also responsible for the spreading of the transcriptionally repressive H3K27me3 mark to neighboring nucleosomes resulting in heterochromatin boundaries [23]. Recent studies have shown that all three members of the PRC family of proteins work together in a hierarchical manner to regulate the transcriptional landscape during cellular differentiation [24,25].
The composition of the functional core-PRC2 enzyme is conversed across species and consists of four proteins: Enhancer of Zester Homolog 1 or 2 (EZH1 or EZH2), Embryonic Ectoderm Development (EED), Suppressor of Zeste 12 (SUZ12), and Retinoblastoma Binding Protein 46 or 48 (RBAP46 or RBAP48) [7]. Several biochemical and functional studies have identified EZH1 or EZH2 to be the catalytic subunit which contains the SET (Su(var)3–9, Enhancer-of-zeste and Trithorax) domain responsible for HMTase activity [20,26–28]. It was further shown that the EZH1 or EZH2 SET domain in isolation had no activity with the minimal functional complex for the methyltransferase activity required EED and the VEFS-domain of SUZ12 [26]. In vitro studies have shown that PRC2–EZH1 has relatively weak methyltransferase activity compared with PRC2–EZH2 on mono- and oligo-nucleosome substrates [29,30]. However, negative-stain electron microscopy studies with oligo-nucleosomes have shown that PRC2–EZH1 rather than PRC2–EZH2 could play a role in chromatin compaction [29,31]. In vivo studies together with mass spectrometry based proteomics analyses have identified that core-PRC2 associates with several accessory factors such as Adipocyte Enhancer-Binding Protein 2 (AEBP2), Jumonji and AT-Rich Interaction Domain 2 (JARID2), Metal Response Element Binding Transcription Factor 2 (MTF2), Plant Homeodomain Finger Protein 1 or 19 (PHF1/PHF19), Elongin BC and Polycomb repressive complex 2-associated protein (EPOP), and PRC2-associated LCOR isoform 1 or 2 (PALI1/2) [32,33]. These studies have categorized PRC2 to form distinct sub-complexes named PRC2.1 and PRC2.2, each containing different set of cofactors [32]. PRC2.1 complexes contain cofactors such as Polycomb-like proteins (PHF1/PHF19/MTF2) or EPOP or PALI1/2 [34–39], whereas PRC2.2 contains cofactors JARID2 and AEBP2 [31,32]. The cofactors of PRC2 have been shown to be crucial for the regulation of the HMTase activity by stabilizing the PRC2 complex and assembly, and together with long non-coding RNAs (lncRNA) also play a vital role in the recruitment of PRC2 to specific gene loci [40–48]. While the recruitment of PRC2 to polycomb targets in D. melanogaster are aided by the presence of specific DNA sequences termed Polycomb Recognition Elements (PRE), PRC2 recruitment in humans seems to be more complex [49–51]. However, in both D. melanogaster and humans, PRC2 is enriched in gene loci with CpG islands [51]. In this review, we focus exclusively on the recent structural studies that have advanced our understanding of how both PRC2.1 and PRC2.2 activities and recruitment to chromatin are regulated by different cofactors (Figure 1 and Table 1). For other aspects of Polycomb regulation, we would like to refer the readers to these recent reviews [7,19,37–39,42,43,52–54].
Figure 1.
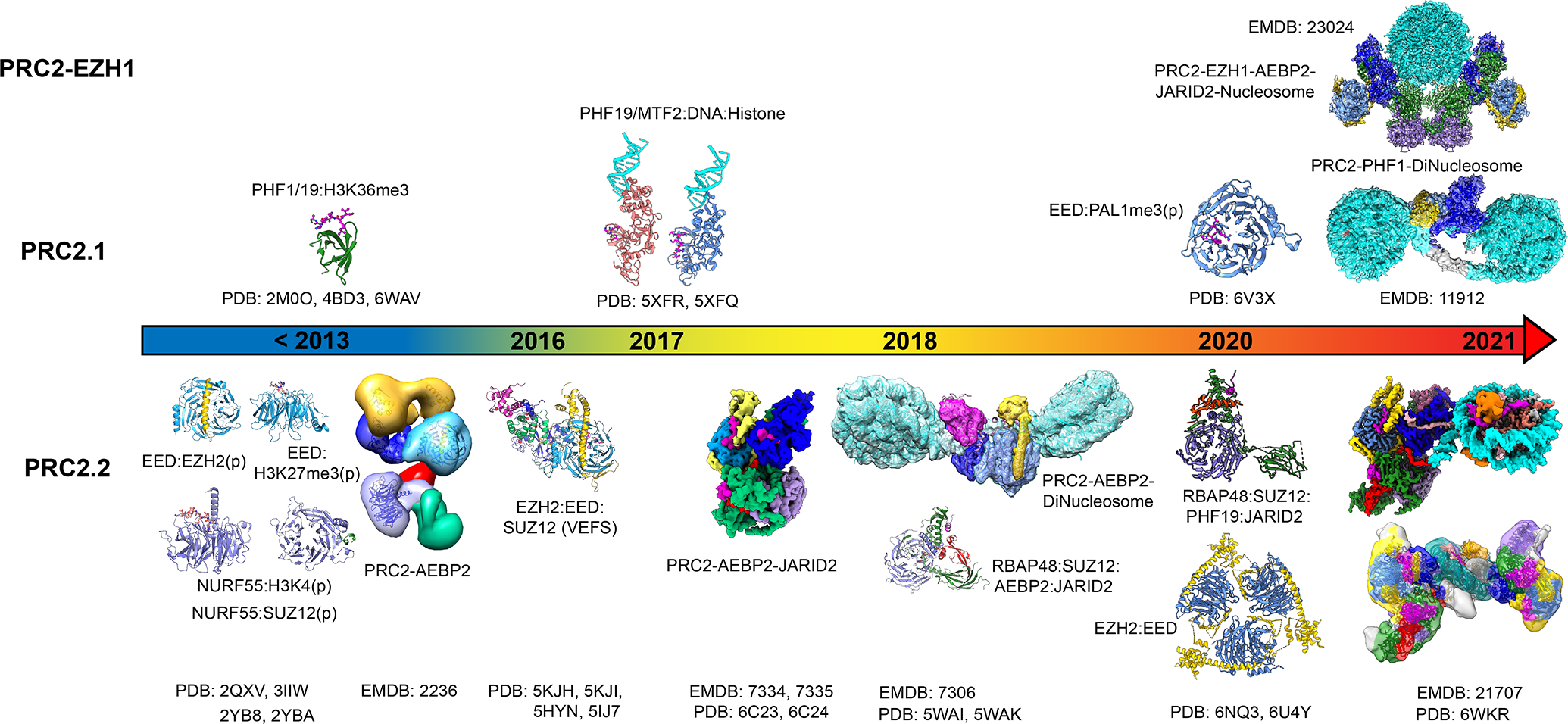
Schematic overview of the various structural studies of PRC2–EZH1, PRC2.1 and PRC2.2.
Table 1.
Structural studies on Polycomb Repressive Complexes
| Year | Citation # | Title | PDB | EMDB | Method | Organism |
|---|---|---|---|---|---|---|
|
| ||||||
| 2007 | [55] | Structural Basis of EZH2 Recognition by EED | 2QXV | X-ray | Mus musculus | |
| 2009 | [23] | Role of the polycomb protein EED in the propagation of repressive histone marks | 3IIW | X-ray | Homo sapiens | |
| 2011 | [59] | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks. | 2YBA 2YB8 |
X-ray X-ray |
Drosophila melanogaster Drosophila melanogaster |
|
| 2012 | [36] | Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity | 4BD3 | NMR | Homo sapiens | |
| 2012 | [47] | Molecular architecture of human polycomb repressive complex 2 | 2236 | EM | Homo sapiens | |
| 2013 | [63] | An H3K36 Methylation-Engaging Tudor Motif of Polycomb-like Proteins Mediates PRC2 Complex Targeting. | 2M0O | X-ray | Homo sapiens | |
| 2015 | [58] | Structural basis of histone H3K27 tri-methylation by an active polycomb repressive complex 2 | 5KJH 5KJI |
X-ray X-ray |
Chaetomium thermophilum Chaetomium thermophilum |
|
| 2016 | [73] | Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2 | 5HYN | X-ray | Homo sapiens | |
| 2016 | [74] | Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. | 5IJ7 | X-ray | Anolis carolinensis, Homo sapiens | |
| 2017 | [72] | Polycomb-like proteins link the PRC2 complex to CpG islands |
5XFR 5XFQ |
X-ray X-ray |
Homo sapiens Homo sapiens |
|
| 2018 | [48] | Structures of human PRC2 with its cofactors AEBP2 and JARID2. | 6C23 6C24 |
7334 7335 |
cryo-EM cryo-EM |
Homo sapiens Homo sapiens |
| 2018 | [77] | Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. | 7306 | Cryo-EM | Homo sapiens | |
| 2018 | [75] | Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding |
5WAI 5WAK |
X-ray X-ray |
Homo sapiens, Neovison vison Homo sapiens |
|
| 2020 | [103] | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19. | 6WAV | X-ray | Homo sapiens | |
| 2020 | [89] | Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3. | 11912 | Cryo-EM | Drosophila | |
| 2020 | [94] | A Dimeric Structural Scaffold for PRC2-PCL Targeting to CpG Island Chromatin. | 6NQ3 | X-ray | Homo sapiens | |
| 2020 | [96] | A partially disordered region connects gene repression and activation functions of EZH2. | 6U4Y | X-ray | Homo sapiens | |
| 2021 | [56] | Crystal structure of EED in complex with PALI1-K1241me3 peptide | 6V3X | X-ray | Homo sapiens | |
| 2021 | [76] | JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. | 6WKR | 21707 | cryo-EM | Homo sapiens |
| 2021 | [82] | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction. | 23024 | cryo-EM | Homo sapiens | |
Early insights into Polycomb structure and function (before 2020)
Architecture and interaction landscape of PRC2 subunits
The earliest structural studies of PRC2 focused on individual domains or subunits. These studies highlighted for the first time how the WD40 repeat within EED recognizes specific regions within EZH2 [55]. In addition, EED also binds to specific histone H3 peptides and is shown to bind transcriptionally repressive H3K9me3 or H3K27me3 peptides with higher affinity than H3K4me3 or H3K36me3 peptide which mark regions of active transcription [23]. JARID2 aa K116 and more recently PALI1 aa 1241 have both been shown to be trimethylated by PRC2 and in its trimethylated state (JARID2 K116me3 or PALI1 K1241me3) mimic H3K27me3 in their binding to EED [48,56,57]. This preferential binding of EED to select trimethylated lysine residues is important and necessary for the allosteric activation of EZH2 (SET) HMTase activity [23,42,58]. Besides EED, the Nurf55 subunit (RBAP46/48 in human) which also contains WD40 repeats is found to bind both the N-terminal tail of the unmodified H3 as well as a segment of SUZ12 (aa 79–91) [59]. Structural studies complemented biochemical studies on the interaction landscape of PRC2 with the identification of a C-terminal segment of SUZ12, containing the VEFS domain, as necessary for the EZH2 (SET) HMTase activity [26]. Such studies also showed that cofactors AEBP2, JARID2, PHF1/19 co-localize with EZH2, EED and SUZ12 subunits in H3K27me3 rich regions in vivo in embryonic stem cells [26,47,57,60–64]. Moreover, AEBP2 and JARID2 were also observed to co-localize with PRC2 components in H3K4me3 enriched promoter regions [40,65–67]. A recent study found that JARID2 and MTF2 regulate a distinct set of Polycomb target genes hinting at the distinct functional role of PRC2.1 and PRC2.2 [68]. Biochemical studies have also identified EZH2, EED and SUZ12 together with these cofactors to be important for interaction with CpG DNA and lncRNA [38,39,44–46,69–71]. JARID2 binding to EED is also thought to be important for the allosteric stimulation of PRC2 activity [48,57]. PHF1 or PHF19 recruits both PRC2 and NO66 (H3K36me3 demethylase) to H3K36me3 containing regions in stem cells [35]. All three of PHF1, PHF19 and MTF2 are known to stimulate the H3K27me3 activity of core-PRC2, yet the precise mechanism for how these cofactors stimulate PRC2 activity remains unknown [34,36,39]. Crystal structures of PHF1 and MTF2 with H3K36me3 containing H3 peptide (aa 34–41) and unmethylated CpG DNA hinted at the role played by histone post-translational modifications together with these cofactors in mediating PRC2 recruitment [72].
Structural organization and allosteric activation of PRC2 activity
The first complete structure of the human PRC2 containing cofactor AEBP2 came from a negative-stain electron microscopy study which showed that PRC2–AEBP2 can be divided into two regions: top and bottom lobes [47]. EZH2, EED and parts of the C-terminus of SUZ12 was shown to localize to the top lobe with AEBP2 linking the top lobe with RBAP46/48 and N-terminus of SUZ12 which were localized to the bottom lobe [47]. This structure provided the first hint that besides the role played by AEBP2 in the recruitment of PRC2, AEBP2 is required for the structural stability of PRC2.2, agreeing with biochemical studies which have shown that PRC2 containing AEBP2 was a better methyltransferase than core-PRC2 [31]. Following this study, high-resolution structures of the catalytic lobe of PRC2 containing EZH2, EED and the C-terminus of SUZ12 containing the VEFS domain from different organisms provided a detailed look into how the EZH2 (SET) active site formation required the co-assembly of EZH2 with both EED and the SUZ12 (VEFS) domain [58,73,74]. These structures also showed for the first time how EED binding to H3K27me3, the catalytic product of PRC2 HMTase activity, initiates a feed-forward mechanism of H3K27 methylation through the allosteric stimulation of EZH2 (SET) domain. The cryo-EM structure of the human PRC2 with cofactors AEBP2 and JARID2 provided the first high-resolution snapshots of PRC2, both top and bottom lobes. This study identified two active confirmations of PRC2 and discovered how both cofactors, AEBP2 and JARID2, simultaneously mimic histone tails in their interaction with PRC2 [48]. The structures showed that JARID2 is both a substrate (binds EZH2 (SET) active site) and mimics H3K27me3 when trimethylated at aa 116 resulting in its interaction with EED, which in turn allosterically stimulates PRC2 activity. The structural visualization in this study agreed excellently with previous biochemical studies which identified JARID2 as a substrate for PRC2 [57]. Unexpectedly, AEBP2 is found to mimic unmodified histone H3 tail in its interaction with RBAP46/48 and this was also subsequently observed in a structural study of the bottom lobe of PRC2 [48,75]. The observation that both AEBP2 and JARID2, and more recently PALI1 mimic histone tails in their interaction with PRC2 prompts the question as to whether other cofactors such as PHF1/PHF19/MTF2 could also employ a yet unknown but similar mechanism as part of the PRC2.1 complex. The cryo-EM structure of PRC2–AEBP2–JARID2 provided a structural visualization of why SUZ12 was known to be critical for PRC2 activity [76]. SUZ12 is required for the assembly of the full PRC2 complex and does so by interacting with all the core-PRC2 subunits namely EZH2, EED and RBAP46/48, and interacts with both cofactors AEBP2 and JARID2. In addition, the SUZ12 N-terminus (aa 100–400) folds into an RRM-like fold, rich in beta-sheet and is shown to be relevant for the interaction with XIST lncRNA [48]. This structure provided the first clues to how PRC2 can be bound to both a substrate and an allosteric activator at the same time, a mechanism which could be important for the spreading of H3K27me3 to neighboring nucleosomes. This feed-forward mechanism of H3K37 methylation was directly visualized in the cryo-EM structure of PRC2 containing AEBP2 bound to di-nucleosome [77]. This structure showed for the first time that both EED and EZH2 (SET, CXC) formed the primary interface for interaction with nucleosome [77]. Surprisingly, PRC2 interacts predominantly with the nucleosome DNA unlike most other chromatin modifiers which interact with both the DNA and the conserved histone acidic patch surface [78]. The structure showed that the nucleosome containing H3K27me3 bound to EED such that the H3K27me3 is threaded into the EED aromatic-cage, while the substrate nucleosome containing unmodified H3 tail binds to EZH2 (SET, CXC) in a configuration that allows the H3 tail to bind the active site. This simultaneous interaction of H3K27me3 with EED and unmodified H3 tail with EZH2 (SET), respectively, provided a mechanistic explanation for biochemical studies which showed that PRC2 displayed higher activity for di-nucleosomes compared with mono-nucleosome substrates [77,79,80]. Besides the EED and EZH2 (SET, CXC) interface for nucleosome interaction, a crystal structure of the bottom lobe of PRC2 containing RBAP46/48, SUZ12 (C2), AEBP2 and JARID2 suggested that the bottom lobe of PRC2 could also potentially be a site for nucleosome interaction [75]. Negative-stain 2D class average analysis from the cryo-EM study of PRC2–AEBP2 bound to di-nucleosome had previously hinted at a di-nucleosome arrangement such that one nucleosome may interact with the bottom lobe [77]. These studies together with previous crystal structure of Nurf55 bound to unmodified H3 tail suggest that there may be more complex PRC2-nucleosome interactions possible which play a role in the spreading of the transcriptionally repressive H3K27me3 mark. The structural studies of PRC2 with cofactors together with biochemical studies have provided insight into the assembly and regulation of PRC2, piece-by-piece [43,54]. Insights into the regulation of PRC2 activity at chromatin have so far come from biochemical studies which demonstrate how different histone post-translational marks, particularly H3K4me3 and H3K36me3, affect PRC2 activity [59,80,81]. However, structural, and mechanistic insights into how histone post-translational marks together with cofactors both recruit and regulate PRC2 HMTase activity on chromatin remains to be studied in more detail. In the next section, we highlight recent structural studies which have begun to address some of these questions.
Recent structural insights into Polycomb Repressive Complexes (2020 – present)
JARID2, AEBP2 recognize H2AK119ub1 and CpG DNA to recruit PRC2
Despite several studies identifying the roles played by RNA, histone post-translational marks, CpG DNA, and cofactors in the recruitment of PRC2, specific mechanistic insight into how these factors contribute to the recruitment and activation of PRC2 on chromatin has remained elusive. Recent studies have provided the first structural snapshots of different PRC2 complexes interacting with complex chromatin substrates which has enabled the identification of specific role played by the different factors [76,77,82]. Besides the PRC2.2-specific cofactors AEBP2 and JARID2, biochemical studies have shown that histone post-translational marks such as PRC1-mediated H2AK119ub1, are necessary for the proper recruitment of PRC2 [24,83,84]. In addition, JARID2 has also been identified recently to harbor a ubiquitin-interaction like motif (UIM) which interacts with H2AK119ub1 and is hence thought to be important for the H2AK119ub1-mediated recruitment of PRC2 to chromatin [85]. The recent cryo-EM structure of PRC2–AEBP2–JARID2 bound to a H2AK119ub1 and CpG DNA-containing nucleosome revealed that JARID2 not only allosterically activates PRC2 but also specifically interacts with H2AK119ub1-containing nucleosome (Figure 2) [76]. Cryo-EM structures of PRC2–AEBP2–JARID2 with or without the UIM region in complex with nucleosome with or without H2AK119ub1 demonstrated that the presence of both the JARID2 UIM as well as the H2AK119ub1 is necessary for the stable interaction of JARID2 with nucleosome [76]. In addition to JARID2, AEBP2 was previously uncovered to be important for interaction with CpG DNA through its lysine-arginine rich motif [70,86]. The recent cryo-EM study directly observed and verified this interaction between AEBP2 and CpG DNA [76]. Interestingly, this study also identified a previously unrecognized role of the tandem zinc-fingers of AEBP2 in its interaction with H2AK119ub1. While the tandem zinc-fingers of AEBP2 was previously observed to bind CpG DNA [70], this role of AEBP2’s tandem zinc-fingers in recognizing H2AK119ub1 has previously not been shown. The results from this cryo-EM structural study will guide further biochemical and in vivo studies on the precise role played by cofactors JARID2 and AEBP2 in the recruitment and activation of PRC2.
Figure 2. JARID2, AEBP2 regulate PRC2.2 activity in the presence of histone post-translational modification.
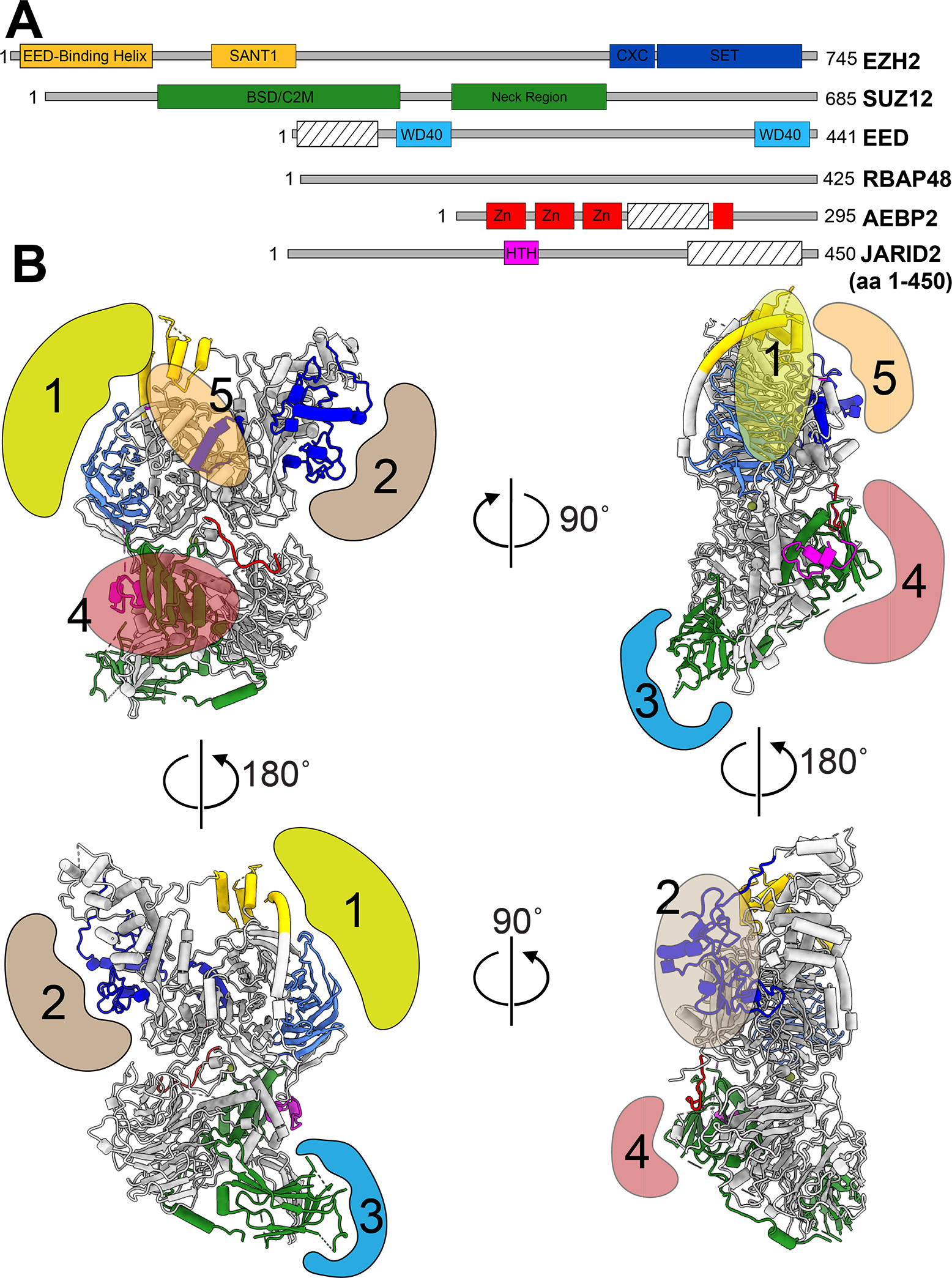
(A) Atomic model of PRC2.2 bound to nucleosome containing H2AK119ub1 (PDB: 6WKR) showing cryo-EM density for JARID2 UIM region (magenta), ubiquitin (orange) and AEBP2 tandem zinc fingers (red). (Left) Close-up of the JARID2 UIM interaction with ubiquitin (indicated by black box) is shown with arrow representing the hydrophobic interface of interaction. (Right) Close-up of the interaction between AEBP2 zinc-fingers and ubiquitin. The interaction interface is a mix of hydrophobic and electrostatic interactions. (B) Cryo-EM reconstruction of PRC2.2 bound to nucleosome containing H3K4me3 showing the absence of H3 tail engagement within the EZH2 (SET) active site. The canonical H3 tail binding groove within EZH2 (SET) is shown in green. In addition to the absence of H3 tail engagement, changes in the EZH2 (SANT1) (indicated in gold) is also shown. (C) Overlay of the cryo-EM reconstructions of the two states observed for PRC2.2 interact with nucleosome containing H3K4me3. The close-up view of the changes between state 1 (white) and state 2 (pink) are shown on the right. These changes are primarily localized to regions 1 (EZH2 (SANT1) and EED allosteric binding site) and region 2 (H3 tail binding groove within EZH2 (SET).
Regulation of PRC2 HMTase activity by histone post-translational modifications and RNA
As important as it is to recruit PRC2 to specific genomic loci, it is equally critical to restrict PRC2 activity to these specific regions to avoid aberrant gene repression. Several studies have shown that both nascent RNA as well as active histone post-translational marks such as H3K4me3 and H3K36me3 play key roles in this process [59,69,70,87–89]. While structural insight into how RNA regulates PRC2 activity remains elusive, RNA has been shown to bind several different regions within PRC2 including the catalytic EZH2 (SET) domain [76,90]. Since EZH2(SET) is one of the primary site of interaction with nucleosome substrates, the current hypothesis is that that RNA likely competes with PRC2’s natural substrate to inhibit PRC2 activity (Figure 3) [45,69,76,77,91]. Besides RNA, the inhibitory effect of active transcription marks namely H3K4me3 and H3K36me3, on core-PRC2 complex (lacking cofactors) has been well studied [59,92]. However, the precise molecular mechanism through which H3K4me3 and H3K36me3 inhibit PRC2 activity has remained unclear. While H3K4me3 and H3K36me3 do indeed inhibit core-PRC2 activity, the HMTase activity of PRC2 containing cofactors JARID2 and AEBP2 seems to only be partially inhibited [76]. Cryo-EM structures of PRC2–AEBP2–JARID2 bound to H3K4me3 containing nucleosome show the presence of multiple conformations, one with H3 tail engaged in the EZH2(SET) catalytic domain and one without (Figure 2) [76]. The partial inhibition of PRC2–AEBP2–JARID2 HMTase activity on H3K4me3-containing nucleosomes could hence likely be due to the increased conformational flexibility of the H3 tail (Figure 2) [93]. The in vitro HMTase activity assays carried out with nucleosome substrates containing H3K4me3 or H3K36me3 on both H3 tails detected the co-presence of H3K27me3 [76]. However, mass spectrometry analysis had previously failed to detect nucleosomes in vivo containing both H3K4me3 or H3K36me3 and H3K27me3 on the same histone H3 tail [81,92]. While the lack of detection does not necessarily imply absence, it does pose the question of the in vivo functional relevance of JARID2 and AEBP2 mediated partial activity on H3K4me3 or H3K36me3 containing nucleosomes. This is an interesting avenue for future studies since JARID2 and AEBP2 have previously been shown to co-localize to H3K4me3-enriched gene promoter regions [40,65]. Perhaps other mechanisms such as the RNA-mediated inhibition and/or eviction of PRC2 by chromatin remodelers and RNA Polymerase are primarily responsible for the inhibition of PRC2 activity on actively transcribed genes.
Figure 3. RNA binding regions with PRC2.
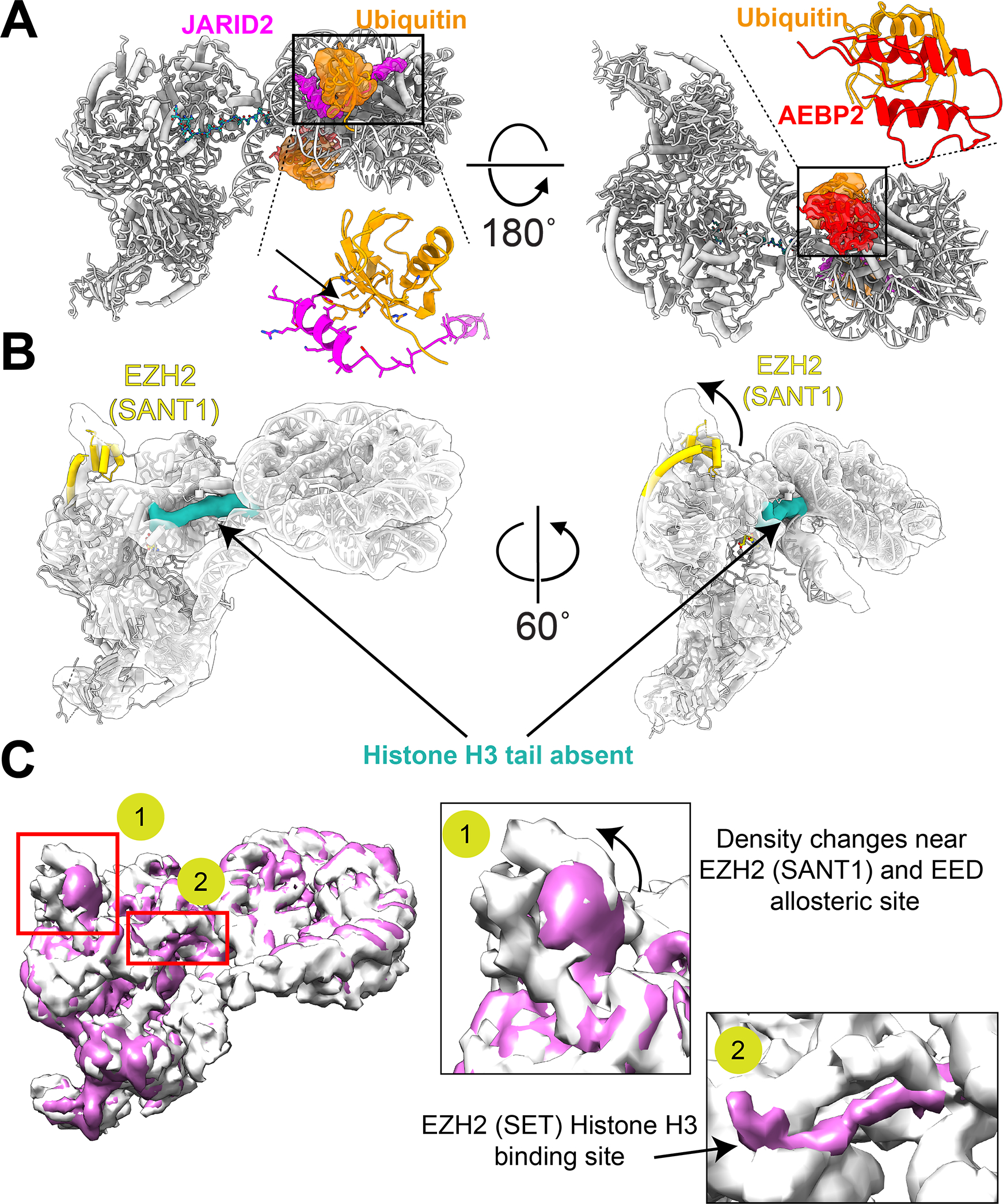
(A) Schematic representation of the RNA-binding regions within PRC2.2. The color schemes are kept the same as in [48,76]. Regions marked with striped boxes are known RNA-binding regions but have so far not been visualized in structural studies. (B) Spatial mapping of the known RNA-binding regions, 1 through 5, to the atomic model of PRC2.2 (PDB: 6WKR). The regions are colored to distinguish their known roles in the binding to different nucleic acid substrates such as nucleosomes, and RNA: (gold, #1) EED:EZH2 (SANT1) region known to interact with RNA, and nucleosome containing H3K27me3, (brown, #2): EZH2 (SET, CXC) and region within AEBP2 (not observed so far) known to interact with RNA and substrate nucleosome, (blue, #3): SUZ12 (BSD (β-sheet domain) or C2 known to interact with lncRNA (XIST), (salmon, #4): SUZ12 (zinc-finger), SUZ12 (neck region), JARID2 (HTH), AEBP2 (lysine-arginine rich segment) known to interact with RNA and CpG DNA, (yellow, #5): EZH2 (SRM, SET) regions known to interact with RNA. The nucleic acid (DNA/RNA) binding for regions #1,#2,#5 [77,90], and regions #3, #4 [48,71], respectively are inferred from the indicated references.
Insights from structural studies of PRC2.1 and PRC2–EZH1
Besides PRC2.2, there have been notable structural studies including those on PRC2.1, PRC2–EZH1 and a subcomplex comprised of SUZ12–RBAP48–PHF19–JARID2 [82,89,94]. EZH1 is a paralog of EZH2 and shares 63% overall sequence identity with 94% identity in the catalytic SET domain [29]. PRC2–EZH1 and PRC2–EZH2 share the other three core components namely, EED, SUZ12, and RBAP46/48. While PRC2–EZH1 is a catalytically weak methyltransferase compared with PRC2–EZH2 [31], PRC2–EZH1 is thought to be important for gene repression through its methyltransferase-independent chromatic compaction activity [29]. However, a critical distinction between PRC2–EZH1 mediated chromatin compaction versus the canonical PRC1-mediated chromatin compaction is that PRC2–EZH1 requires nucleosomes with histone tails whereas canonical PRC1 can compact tailless nucleosome arrays [29]. However, the structural description of the chromatin compaction by PRC2–EZH1 or canonical PRC1 remains to be elucidated. The recent cryo-EM study of PRC2–EZH1 together with cofactors AEBP2 and JARID2 and mono-nucleosome provided the first visualization of PRC2–EZH1 bound to nucleosome (Figure 4) [82]. PRC2–EZH1 is seen to undergo a major conformational change upon interaction with nucleosome, with the bottom lobe consisting of RBAP46, SUZ12, AEBP2, and JARID2 rotated ~170° such that this lobe interacts with another copy from a second PRC2–EZH1 which presumably is bound to the other H3 histone tail of the nucleosome (Figure 4) [82]. This bottom lobe-bottom lobe interaction is thought to be further stabilized by the domain swapped conformation seen previously in the crystal structure of the bottom lobe sub-complex consisting of parts of SUZ12, RBAP46, PHF19 and JARID2 [94]. Similar observations with two copies of PRC2–AEBP2–JARID2 bound to the same nucleosome has been seen before, however, the two PRC2.2 complexes were not observed to directly interact with each other [76]. It is currently unknown why two different interaction geometries, one with domain swapping and one without is observed for PRC2–EZH1 and PRC2–EZH2, respectively. Different PRC2–EZH2 dimer geometries without nucleosomes, including the currently known SUZ12 domain-swapped dimer, have been observed using negative-stain electron microscopy (unpublished-) and small angle x-ray scattering [95]. In addition, a minimal EZH2–EED complex has also been recently observed to form higher-order structures which is thought to be relevant for the gene activation function of EZH2 in cancers independent of its canonical role within PRC2 [96]. The higher-order oligomeric structures of PRC2 and their in vivo relevance for gene expression regulation remains to be explored further.
Figure 4. Oligomeric forms of PRC2–EZH1 and PRC2.2.
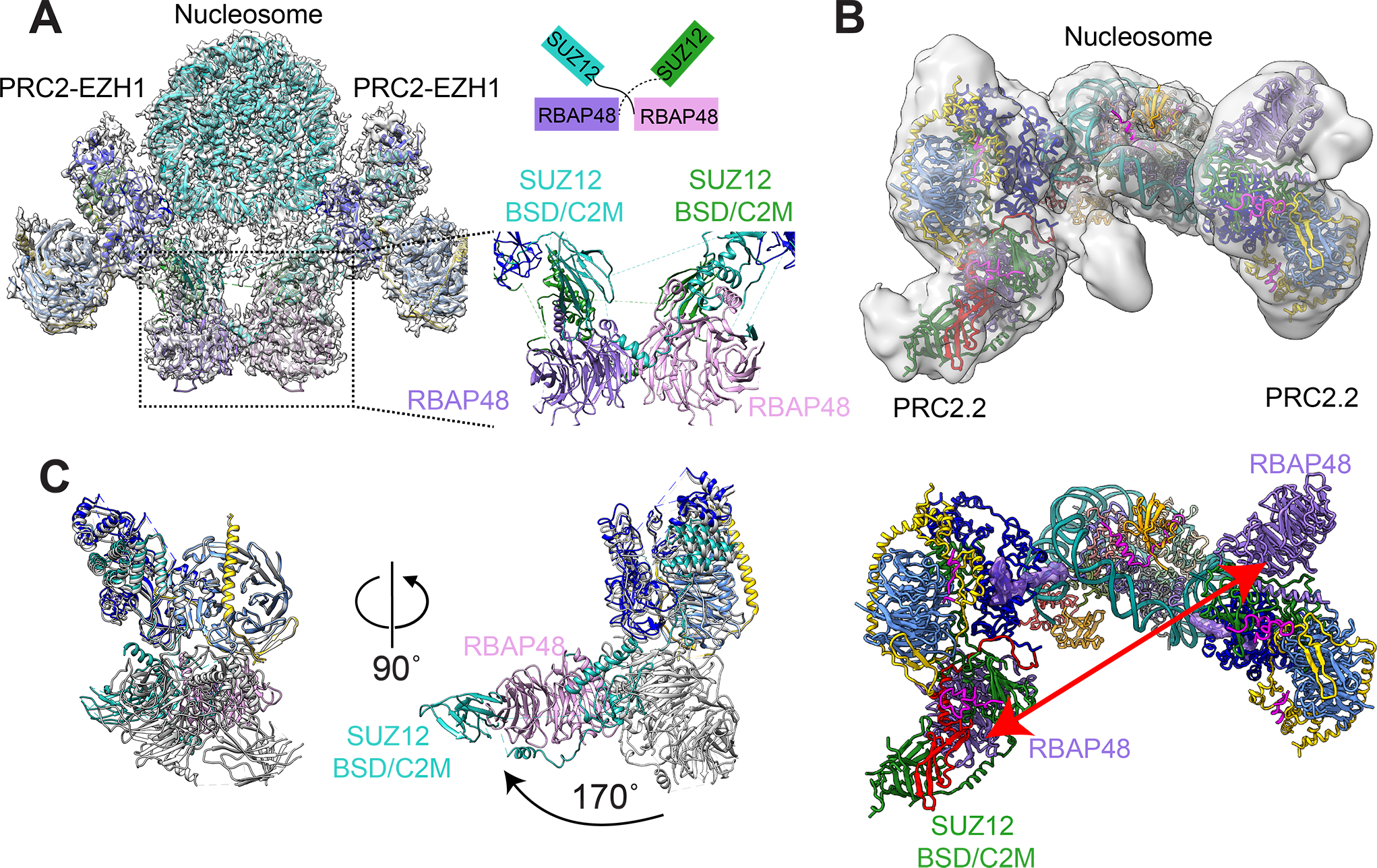
(A) Cryo-EM reconstruction of PRC2–EZH1 dimer bound to nucleosome is shown with the model of PRC2–EZH1 and nucleosome represented in cartoon format (EMDB: 23024). Close-up of the RBAP48–SUZ12 domain swapped dimer interaction (black dashed box) observed in the cryo-EM reconstruction is shown. Schematic representation of the domain swapped dimer is shown above for clarity. (B) Cryo-EM reconstruction of two PRC2–AEBP2–JARID2 (PRC2.2) complexes interacting with one nucleosome containing H2AK119ub1 is shown with the model represented in cartoon format (EMDB: 21707). The interaction geometry of PRC2.2 is different from that observed for PRC2–EZH1 in (A). (C) (Left) Overlay of PRC2–EZH1 in apo state (white) and PRC2–EZH1 in nucleosome bound state (colored the same as other figures) is shown to highlight the 170° rotation of the RBAP48–SUZ12 (BSD or C2) in the nucleosome bound state. (Right) Model of the two PRC2.2 bound to nucleosome containing H2AK119ub1 shows that the RBAP48–SUZ12 (BSD) are far apart (red arrow) and do not interact with each other.
Interaction of PRC2.1 with chromatin
While the last few years have seen several structural studies on PRC2.2 containing AEBP2 and JARID2 cofactors, there have been no complete high-resolution structural information for PRC2.1 complexes involving cofactors (PHF1, PHF19, MTF2, EPOP, PALI1). Structural information for individual domains of PHF1, PHF19, MTF2 bound to either modified histone tails or histone tails and DNA have alluded to their functional role in mediating crosstalk with histone post-translational modifications and CpG DNA [72]. Recent biochemical studies have identified key residues in SUZ12 that can dictate the formation of PRC2.1 versus PRC2.2 in vivo [97]. This is particularly relevant since structural studies have shown that SUZ12 is critical for the assembly of full PRC2 [48]. However, a functional description of core-PRC2 interaction with PHF/19, MTF2, and other cofactors within PRC2.1 in both free and nucleosome bound states as well as a mechanistic description of how these specific cofactors regulate PRC2 activity remains to be determined. This is particularly important for understanding how these cofactors such as PHF1/19 regulate PRC2 activity in the presence of active transcription marks such as H3K4me3 or H3K36me3 as well as how mutations in these cofactors or histone tails affect PRC2 activity. To address these questions, a recent cryo-EM study aimed to visualize the structure of PRC2–PHF1 bound to di-nucleosomes [89]. The overall structure agreed excellently with the previous cryo-EM study of PRC2–AEBP2 bound to di-nucleosomes [77]. The structural and biochemical analysis from this study showed that H3K36 is important for the proper channeling of the H3 tail into the catalytic site of the EZH2 (SET) domain. However, this structural study suffered from the same technical bottleneck as the previous cryo-EM study by Poepsel et al. [77] where the bottom lobe of PRC2 was not visible. This perhaps could be one of the reasons for the lack of visualization of the PHF1 cofactor and its potential role in the interaction of PRC2 with nucleosome. However, this study and the recent cryo-EM study of PRC2–AEBP2–JARID2 bound to H2AK119ub1 containing nucleosome both visualized the histone H3 tail bound to the EZH2(SET) domain at high resolution [76]. The H3K36 residue is found to be well positioned at the interface of the EHZ2(SET) domain and the nucleosome DNA. It remains to be elucidated whether the presence of H3K36me3 disrupts the positioning of H3 tail or results in an increase in the flexibility of the H3 tail, similar to H3K4me3, resulting in reduced PRC2 HMTase activity.
Outlook
Despite the advances in structural and biochemical studies on understanding PRC2 regulation, there remains several interesting questions unanswered. Here, we highlight three that are likely to be the subject of future structural studies: (i) the structural assembly and characterization of PRC2.1, (ii) structural basis for RNA-mediated regulation of PRC2, and (iii) the cofactor mediated crosstalk between histone post-translational modifications (H3K4me3/K36me3) and PRC2.1/PRC2.2. In contrast with PRC2.2, visualization of the interactions of cofactors PHF1/19, MTF2 and others with the complete core-PRC2 assembly remains to be elucidated. Structural studies aimed at PRC2.1 will also serve as a platform for understanding the role played by these cofactors in regulating both the activity and interactions of PRC2.1 with nucleosome. Despite extensive biochemical studies highlighting the role played by RNA in the recruitment and regulation of PRC2 activity, there have been no direct visualization or structural model of the interaction of RNA with PRC2. It remains to be elucidated how RNA can orchestrate dual functions as a recruitment factor and an inhibitor of PRC2 activity. In addition, the mechanistic insights into the alleviation of RNA-mediated inhibition post-recruitment are also not clearly understood. Recent cryo-EM based structural studies have addressed some of the technical bottlenecks such as air-water interface mediated damage which had hindered the characterization of PRC2-nucleosome complexes [76,98–100]. These advances have not only provided the capability to assemble intact PRC2-nucleosome complexes but have also allowed for the direct visualization of the role played by cofactors. The application of these new methods for the structural studies of PRC2.1, and PRC2 with RNA, both of which have so far eluded detailed mechanistic descriptions will open new avenues for biochemical and in vivo studies. Lastly, structural studies aimed at deciphering the functional relevance of the recently observed higher-order PRC2 assemblies would provide a more complete picture of PRC2 function in vivo. A cross-disciplinary approach involving in vivo cell biology, in vitro biochemistry, structural biology, and going forward in situ structural cell biology approaches using cryo-electron tomography will be necessary to provide new insights into the mechanisms underlying PRC2 function [101,102].
Perspectives.
Polycomb Repressive Complexes are key epigenetic regulators of cell differentiation and cell-type identity. Polycomb Repressive Complexes are some of the most mutated enzymes in various cancers. Structural and functional insights into Polycomb Repressive Complexes are crucial for understanding how the activity of these enzymes are spatially and temporally regulated.
The recruitment and activity of various Polycomb Repressive Complexes are tightly regulated by cofactor proteins, histone post-translational modifications and RNA.
The crosstalk between cofactor proteins, histone post-translational modifications and RNA remains unclear, and will be the subject of future studies. Structural insights into several classes of Polycomb Repressive Complexes including PRC2.1 will likely be addressed in future studies.
Acknowledgements
We would like to thank Dr. Avinash Patel, members of the Nogales and Cech labs for valuable feedback.
Funding
This work was funded in part by V.K’s start-up funds from University of Colorado, Boulder, and by the National Institute of General Medical Sciences (NIH K99/R00 1K99GM132544).
Abbreviations
- AEBP2
Adipocyte Enhancer-Binding Protein 2
- EED
Embryonic Ectoderm Development
- JARID2
Jumonji and AT-Rich Interaction Domain 2
- MTF2
Metal Response Element Binding Transcription Factor 2
- PRC1
Polycomb Repressive Complexes 1
- SUZ12
Suppressor of Zeste 12
- UIM
ubiquitin-interaction like motif
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF and Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD (1974) Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871 10.1126/science.184.4139.868 [DOI] [PubMed] [Google Scholar]
- 3.Ptashne M (2009) Binding reactions: epigenetic switches, signal transduction and cancer. Curr. Biol. 19, R234–R241 10.1016/j.cub.2009.02.015 [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T (2007) Chromatin modifications and their function. Cell 128, 693–705 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 5.Reik W (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 10.1038/nature05918 [DOI] [PubMed] [Google Scholar]
- 6.Sexton T and Cavalli G (2015) The role of chromosome domains in shaping the functional genome. Cell 160, 1049–1059 10.1016/j.cell.2015.02.040 [DOI] [PubMed] [Google Scholar]
- 7.Margueron R and Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laugesen A, Hojfeldt JW and Helin K (2016) Role of the polycomb repressive complex 2 (PRC2) in transcriptional regulation and cancer. Cold Spring Harb. Perspect. Med 6, a026575 10.1101/cshperspect.a026575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis EB (1978) A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- 10.Paro R and Hogness DS (1991) The polycomb protein shares a homologous domain with a heterochromatin-associated protein of drosophila. Proc. Natl Acad Sci. U.S.A. 88, 263–267 10.1073/pnas.88.1.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ringrose L and Paro R (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 38, 413–443 10.1146/annurev.genet.38.072902.091907 [DOI] [PubMed] [Google Scholar]
- 12.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B and Cavalli G (2007) Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745 10.1016/j.cell.2007.02.009 [DOI] [PubMed] [Google Scholar]
- 13.Schuettengruber B, Martinez AM, lovino N and Cavalli G (2011) Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell. Biol. 12, 799–814 10.1038/nrm3230 [DOI] [PubMed] [Google Scholar]
- 14.Jürgens G (1985) A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316, 153–155 10.1038/316153a0 [DOI] [Google Scholar]
- 15.Sparmann A and van Lohuizen M (2006) Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6, 846–856 10.1038/nrc1991 [DOI] [PubMed] [Google Scholar]
- 16.Bracken AP and Helin K (2009) Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer 9, 773–784 10.1038/nrc2736 [DOI] [PubMed] [Google Scholar]
- 17.Pietersen AM and van Lohuizen M (2008) Stem cell regulation by polycomb repressors: postponing commitment. Curr. Opin. Cell Biol 20, 201–207 10.1016/j.ceb.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 18.Di Croce L and Helin K (2013) Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- 19.Chittock EC, Latwiel S, Miller TC and Muller CW (2017) Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 45, 193–205 10.1042/BST20160173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A and Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196 10.1016/S0092-8674(02)00975-3 [DOI] [PubMed] [Google Scholar]
- 21.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B et al. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 10.1016/S0092-8674(02)00976-5 [DOI] [PubMed] [Google Scholar]
- 22.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P and Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 16, 2893–2905 10.1101/gad.1035902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, et al. (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 10.1038/nature08398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blackledge NP, Fursova NA, Kelley JR, Huseyin MK, Feldmann A and Klose RJ (2020) PRC1 catalytic activity is central to polycomb system function. Mol. Cell 77, 857–874 e9 10.1016/j.molcel.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aranda S, Mas G and Di Croce L (2015) Regulation of gene transcription by polycomb proteins. Sci. Adv. 1, e1500737 10.1126/sciadv.1500737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao R and Zhang Y (2004) SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15, 57–67 10.1016/j.molcel.2004.06.020 [DOI] [PubMed] [Google Scholar]
- 27.Lhuillier-Akakpo M, Frapporti A, Denby Wilkes C, Matelot M, Vervoort M, Sperling L et al. (2014) Local effect of enhancer of zeste-like reveals cooperation of epigenetic and cis-acting determinants for zygotic genome rearrangements. PLoS Genet. 10, e1004665 10.1371/journal.pgen.1004665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P et al. (2002) Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298, 1039–1043 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- 29.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL et al. (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32, 503–518 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee CH, Holder M, Grau D, Saldana-Meyer R, Yu JR, Ganai RA et al. (2018) Distinct stimulatory mechanisms regulate the catalytic activity of polycomb repressive complex 2. Mol Cell 70, 435–448 e5 10.1016/j.molcel.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Son J, Shen SS, Margueron R and Reinberg D (2013) Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 27, 2663–2677 10.1101/gad.225888.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauri S, Comoglio F, Seimiya M, Gerstung M, Glatter T, Hansen K et al. (2016) A high-density map for navigating the human polycomb complexome. Cell Rep. 17, 583–595 10.1016/j.celrep.2016.08.096 [DOI] [PubMed] [Google Scholar]
- 33.Conway E, Jerman E, Healy E, Ito S, Holoch D, Oliviero G et al. (2018) A family of vertebrate-specific polycombs encoded by the LCOR/LCORL genes balance PRC2 subtype activities. Mol. Cell 70, 408–421 e8 10.1016/j.molcel.2018.03.005 [DOI] [PubMed] [Google Scholar]
- 34.Sarma K, Margueron R, Ivanov A, Pirrotta V and Reinberg D (2008) Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol. Cell Biol 28, 2718–2731 10.1128/MCB.02017-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brien GL, Gambero G, O’Connell DJ, Jerman E, Turner SA, Egan CM et al. (2012) Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase N066 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 19, 1273–1281 10.1038/nsmb.2449 [DOI] [PubMed] [Google Scholar]
- 36.Ballare C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G et al. (2012) Phf19 links methylated Lys36 of histone H3 to regulation of polycomb activity. Nat. Struct. Mol. Biol. 19, 1257–1265 10.1038/nsmb.2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunkapiller J, Shen Y, Diaz A, Cagney G, McCleary D, Ramalho-Santos M et al. (2012) Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal. PLoS Genet. 8, e1002576 10.1371/journal.pgen.1002576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casanova M, Preissner T, Cerase A, Poof R, Yamada D, Li X et al. (2011) Polycomblike 2 facilitates the recruitment of PRC2 polycomb group complexes to the inactive X chromosome and to target loci in embryonic stem cells. Development 138, 1471–1482 10.1242/dev.053652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J et al. (2010) Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell 6, 153–166 10.1016/j.stem.2009.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G, Margueron R, Ku M, Chambon P, Bernstein BE and Reinberg D (2010) Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24, 368–380 10.1101/gad.1886410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K et al. (2014) Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 53, 290–300 10.1016/j.molcel.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasinath V, Poepsel S and Nogales E (2019) Recent structural insights into polycomb repressive complex 2 regulation and substrate binding. Biochemistry 58, 346–354 10.1021/acs.biochem.8b01064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X (2021) A structural perspective on gene repression by polycomb repressive complex 2. Subcell. Biochem. 96, 519–562 10.1007/978-3-030-58971-4_17 [DOI] [PubMed] [Google Scholar]
- 44.Davidovich C and Cech TR (2015) The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA 21, 2007–2022 10.1261/rna.053918.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davidovich C, Zheng L, Goodrich KJ and Cech TR (2013) Promiscuous RNA binding by polycomb repressive complex 2. Nat. Struct. Mol. Biol. 20, 1250–1257 10.1038/nsmb.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan J, Dutta B, Hee YT and Chng WJ (2019) Towards understanding of PRC2 binding to RNA. RNA Biol. 16, 176–184 10.1080/15476286.2019.1565283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R and Nogales E (2012) Molecular architecture of human polycomb repressive complex 2. eLife 1, e00005 10.7554/eLife.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasinath V, Faini M, Poepsel S, Reif D, Feng XA, Stjepanovic G et al. (2018) Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359, 940–944 10.1126/science.aar5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan CS, Rastelli L and Pirrotta V (1994) A polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMB0 J. 13, 2553–2564 10.1002/j.1460-2075.1994.tb06545.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muller J and Kassis JA (2006) Polycomb response elements and targeting of polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16, 476–484 10.1016/j.gde.2006.08.005 [DOI] [PubMed] [Google Scholar]
- 51.Laugesen A, Hojfeldt JW and Helin K (2019) Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell 74, 8–18 10.1016/j.molcel.2019.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz YB and Pirrotta V (2013) A new world of polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genet. 14, 853–864 10.1038/nrg3603 [DOI] [PubMed] [Google Scholar]
- 53.van Mierlo G, Veenstra GJC, Vermeulen M and Marks H (2019) The complexity of PRC2 subcomplexes. Trends Cell Biol. 29, 660–671 10.1016/j.tcb.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 54.Uckelmann M and Davidovich C (2021) Not just a writer: PRC2 as a chromatin reader. Biochem. Soc. Trans. 49, 1159–1170 10.1042/BST20200728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han Z, Xing X, Hu M, Zhang Y, Liu P and Chai J (2007) Structural basis of EZH2 recognition by EED. Structure 15, 1306–1315 10.1016/j.str.2007.08.007 [DOI] [PubMed] [Google Scholar]
- 56.Zhang Q, Agius SC, Flanigan SF, Uckelmann M, Levina V, Owen BM et al. (2021) PALI1 facilitates DNA and nucleosome binding by PRC2 and triggers an allosteric activation of catalysis. Nat. Commun. 12, 4592 10.1038/s41467-021-24866-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanulli S, Justin N, Teissandier A, Ancelin K, Portoso M, Caron M et al. (2015) Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell 57, 769–783 10.1016/j.molcel.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiao L and Liu X (2015) Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383 10.1126/science.aac4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- 60.Vizan P, Beringer M, Ballare C and Di Croce L (2015) Role of PRC2-associated factors in stem cells and disease. FEBS J. 282, 1723–1735 10.1111/febs.13083 [DOI] [PubMed] [Google Scholar]
- 61.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV et al. (2010) JARID2 regulates binding of the polycomb repressive complex 2 to target genes in ES cells. Nature 464, 306–310 10.1038/nature08788 [DOI] [PubMed] [Google Scholar]
- 62.Cao R and Zhang Y (2004) The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14, 155–164 10.1016/j.gde.2004.02.001 [DOI] [PubMed] [Google Scholar]
- 63.Cai L, Rothbart SB, Lu R, Xu B, Chen WY, Tripathy A et al. (2013) An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol. Cell 49, 571–582 10.1016/j.molcel.2012.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao Q, Wang X, Zhao M, Yang R, Malik R, Qiao Y et al. (2014) The central role of EED in the orchestration of polycomb group complexes. Nat. Commun. 5, 3127 10.1038/ncomms4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS et al. (2010) GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 6, e1001244 10.1371/journal.pgen.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grijzenhout A, Godwin J, Koseki H, Gdula MR, Szumska D, McGouran JF et al. (2016) Functional analysis of AEBP2, a PRC2 polycomb protein, reveals a trithorax phenotype in embryonic development and in ESCs. Development 143, 2716–2723 10.1242/dev.123935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petracovici A and Bonasio R (2021) Distinct PRC2 subunits regulate maintenance and establishment of polycomb repression during differentiation. Mol. Cell 81, 2625–2639 e5 10.1016/j.molcel.2021.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaneko S, Son J, Shen SS, Reinberg D and Bonasio R (2013) PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1258–1264 10.1038/nsmb.2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, Paucek RD, Gooding AR, Brown ZZ, Ge EJ, Muir TW et al. (2017) Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 24, 1028–1038 10.1038/nsmb.3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Betancur JG and Tomari Y (2015) Cryptic RNA-binding by PRC2 components EZH2 and SUZ12. RNA Biol. 12, 959–965 10.1080/15476286.2015.1069463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P et al. (2017) Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549, 287–291 10.1038/nature23881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E et al. (2016) Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 7, 11316 10.1038/ncomms11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brooun A, Gajiwala KS, Deng YL, Liu W, Bolanos B, Bingham P et al. (2016) Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat. Commun. 7, 11384 10.1038/ncomms11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen S, Jiao L, Shubbar M, Yang X and Liu X (2018) Unique structural platforms of Suz12 dictate distinct classes of PRC2 for chromatin binding. Mol. Cell 69, 840–852 e5 10.1016/j.molcel.2018.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kasinath V, Beck C, Sauer P, Poepsel S, Kosmatka J, Faini M et al. (2021) JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 371, eabc3393 10.1126/science.abc3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poepsel S, Kasinath V and Nogales E (2018) Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 25, 154–162 10.1038/s41594-018-0023-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K and Hansen JC (2013) The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface 10, 20121022 10.1098/rsif.2012.1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N et al. (2012) Dense chromatin activates polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337, 971–975 10.1126/science.1225237 [DOI] [PubMed] [Google Scholar]
- 80.Ge EJ, Jani KS, Diehl KL, Muller MM and Muir TW (2019) Nucleation and propagation of heterochromatin by the histone methyltransferase prc2: geometric constraints and impact of the regulatory subunit JARID2. J. Am. Chem. Soc. 141, 15029–15039 10.1021/jacs.9b02321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voigt P, LeRoy G, Drury WJ III, Zee BM, Son J, Beck DB et al. (2012) Asymmetrically modified nucleosomes. Cell 151, 181–193 10.1016/j.cell.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grau D, Zhang Y, Lee CH, Valencia-Sanchez M, Zhang J, Wang M et al. (2021) Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction. Nat. Commun. 12, 714 10.1038/s41467-020-20775-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tamburri S, Lavarone E, Fernandez-Perez D, Conway E, Zanotti M, Manganaro D et al. (2020) Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol. Cell 77, 840–856 e5 10.1016/j.molcel.2019.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M et al. (2014) Histone H2A monoubiquitination promotes histone H3 methylation in polycomb repression. Nat. Struct. Mol. Biol. 21, 569–571 10.1038/nsmb.2833 [DOI] [PubMed] [Google Scholar]
- 85.Cooper S, Grijzenhout A, Underwood E, Ancelin K, Zhang T, Nesterova TB et al. (2016) Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between polycomb complexes PRC1 and PRC2. Nat. Commun. 7, 13661 10.1038/ncomms13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun A, Li F, Liu Z, Jiang Y, Zhang J, Wu J et al. (2018) Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4. Protein Cell 9, 738–742 10.1007/s13238-017-0483-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Q, McKenzie NJ, Warneford-Thomson R, Gail EH, Flanigan SF, Owen BM et al. (2019) RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nat. Struct. Mol. Biol. 26, 237–247 10.1038/s41594-019-0197-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaneko S, Son J, Bonasio R, Shen SS and Reinberg D (2014) Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 28, 1983–1988 10.1101/gad.247940.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finogenova K, Bonnet J, Poepsel S, Schafer IB, Finkl K, Schmid K et al. (2020) Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3. eLife 9, e61964 10.7554/eLife.61964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Long Y, Bolanos B, Gong L, Liu W, Goodrich KJ, Yang X et al. (2017) Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. eLife 6, e31558 10.7554/eLife.31558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yuan W, Xu M, Huang C, Liu N, Chen S and Zhu B (2011) H3k36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 10.1074/jbc.M110.194027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bowman GD and Poirier MG (2015) Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 115, 2274–2295 10.1021/cr500350x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen S, Jiao L, Liu X, Yang X and Liu X (2020) A dimeric structural scaffold for PRC2-PCL targeting to CpG island chromatin. Mol. Cell 77, 1265–1278 e7 10.1016/j.molcel.2019.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davidovich C, Goodrich KJ, Gooding AR and Cech TR (2014) A dimeric state for PRC2. Nucleic Acids Res. 42, 9236–9248 10.1093/nar/gku540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiao L, Shubbar M, Yang X, Zhang Q, Chen S, Wu Q et al. (2020) A partially disordered region connects gene repression and activation functions of EZH2. Proc. Natl Acad. Sci. U.S.A. 117, 16992–17002 10.1073/pnas.1914866117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Youmans DT, Gooding AR, Dowell RD and Cech TR (2021) Competition between PRC2.1 and 2.2 subcomplexes regulates PRC2 chromatin occupancy in human stem cells. Mol. Cell 81, 488–501 e9 10.1016/j.molcel.2020.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang F, Liu Y, Yu Z, Li S, Feng S, Cheng Y et al. (2020) General and robust covalently linked graphene oxide affinity grids for high-resolution cryo-EM. Proc. Natl Acad. Sci. U.S.A. 117, 24269–24273 10.1073/pnas.2009707117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu G, Li K and Jiang W (2016) Antibody-based affinity cryo-EM grid. Methods 100, 16–24 10.1016/j.ymeth.2016.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Han BG, Watson Z, Kang H, Pulk A, Downing KH, Cate J et al. (2016) Long shelf-life streptavidin support-films suitable for electron microscopy of biological macromolecules. J. Struct. Biol. 195, 238–244 10.1016/j.jsb.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK et al. (2016) Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972 10.1126/science.aad8857 [DOI] [PubMed] [Google Scholar]
- 102.Turk M and Baumeister W (2020) The promise and the challenges of cryo-electron tomography. FEBS Lett. 594, 3243–3261 10.1002/1873-3468.13948 [DOI] [PubMed] [Google Scholar]
- 103.Dong C, Nakagawa R, Oyama K, Yamamoto Y, Zhang W, Dong A et al. (2020) Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19. eLife 9, e58675 10.7554/eLife.58675 [DOI] [PMC free article] [PubMed] [Google Scholar]


