Abstract
Virus host shifts, where a virus transmits to and infects a novel host species, are a major source of emerging infectious disease. Genetic similarity between eukaryotic host species has been shown to be an important determinant of the outcome of virus host shifts, but it is unclear if this is the case for prokaryotes where anti-virus defences can be transmitted by horizontal gene transfer and evolve rapidly. Here, we measure the susceptibility of 64 strains of Staphylococcaceae bacteria (48 strains of Staphylococcus aureus and 16 non-S. aureus species spanning 2 genera) to the bacteriophage ISP, which is currently under investigation for use in phage therapy. Using three methods–plaque assays, optical density (OD) assays, and quantitative (q)PCR–we find that the host phylogeny explains a large proportion of the variation in susceptibility to ISP across the host panel. These patterns were consistent in models of only S. aureus strains and models with a single representative from each Staphylococcaceae species, suggesting that these phylogenetic effects are conserved both within and among host species. We find positive correlations between susceptibility assessed using OD and qPCR and variable correlations between plaque assays and either OD or qPCR, suggesting that plaque assays alone may be inadequate to assess host range. Furthermore, we demonstrate that the phylogenetic relationships between bacterial hosts can generally be used to predict the susceptibility of bacterial strains to phage infection when the susceptibility of closely related hosts is known, although this approach produced large prediction errors in multiple strains where phylogeny was uninformative. Together, our results demonstrate the ability of bacterial host evolutionary relatedness to explain differences in susceptibility to phage infection, with implications for the development of ISP both as a phage therapy treatment and as an experimental system for the study of virus host shifts.
Author summary
Virus host shifts, where a virus jumps into a new host from another species, are a major source of emerging infectious diseases. Virus host shifts have been shown to be more likely between animals and plant species that are closely related. However, there is much to learn about how relatedness predicts susceptibility in bacterial hosts. Here, we used a panel of 64 Staphylococcus bacteria and a virus that infects them to investigate how the relationship between host species influences their susceptibility to infection. We find high variation in susceptibility to infection across the host panel and that most of that variation can be explained by the relationship between hosts. This effect is seen consistently using three methods for assessing susceptibility and can be seen both within and between species. Additionally, we find that the relationship between hosts allows us to predict the susceptibility of an unknown host with some accuracy, although there are instances where susceptibility cannot be predicted. Overall, this suggests that closely related hosts will show similar susceptibility to infection and that the relationship between bacterial hosts can be used to predict the susceptibility of an unknown host with limited success.
Introduction
Host shifts, where a pathogen jumps into a novel host species and establishes onward transmission, are a major source of emerging infectious diseases. While host shifts can occur with many types of pathogen, viruses are the most prolific [1, 2]. Accordingly, viruses originating in non-human animals make up the majority of recently emerged human infections [3–5], including several human pandemics: HIV-1, which jumped into humans from chimpanzees [3, 6]; influenza A viruses, which commonly emerge from wild aquatic birds [7–10]; and most recently SARS-CoV-2, which likely transmitted into humans from a bat reservoir [11–13]. Given the scale and speed at which emerging viruses can impact host populations, understanding the underlying causes of virus host shifts has become a major goal of infectious disease research.
The evolutionary relationships between hosts are a key factor in determining the success of a pathogen following transmission to a novel host species, and several studies have investigated the ability of host evolutionary relatedness to explain variation in infection traits. These studies have shown that virulence tends to increase [14–18], and transmission rate [14, 19] and pathogen load [18, 20, 21] decrease with greater evolutionary distance between donor and recipient host species. These ‘distance effects’ have been seen in viruses [19–21], bacterial pathogens [22–24], fungi [25, 26], and nematodes [18], as well as in reconstructions of virus host shifts and cross species transmissions in nature [19, 27]. Additionally, closely related species may share similar levels of susceptibility independent of evolutionary distance to the natural host. These ‘clade effects’ create a patchwork of host clades across a phylogeny that vary in their susceptibility to a pathogen, and have been demonstrated in experimental infections of fruit flies [17, 21] and in pathogens that repeatedly jump between distantly related hosts in nature [28–32]. These effects can act concurrently to influence host shifts, with distance effects making successful pathogen shifts into closely related hosts more likely, and clade effects allowing for host shifts into more distantly related clades of susceptible hosts.
While most studies investigate the role of eukaryotic host phylogenies, few have examined the influence of the host phylogeny on susceptibility to infection in a prokaryotic system. Host phylogenetic effects follow the conventional wisdom that more closely related species share more similar phenotypes, and so present similar environments to invading pathogens. In bacteria, there are several mechanisms by which bacteriophage (viruses that infect bacteria; hereafter ‘phage’) resistance may be acquired in a phylogenetically intractable way, including horizontal gene transfer (HGT) [33], recombination [34], and the acquisition of prophages conferring resistance to further phage infection [35]. It was previously demonstrated that the transfer of plasmids between bacterial hosts is more likely between recipients with near identical genomes to the plasmid donor, but the likelihood of plasmid transfer was not correlated with genetic distance between donors and recipients at larger evolutionary distances [36]. As such, mobile genetic elements containing phage resistance genes may transmit between bacterial hosts in a way that does not segregate phylogenetically. Additionally, it has been demonstrated that evolution of the bacterial core genome is almost entirely driven by recombination, and thus more determined by the spatial structure of microbes rather than clonal inheritance [34]. Finally, the presence of prophage in bacterial genomes providing superinfection immunity to lytic phage has been characterised [37–41], including in Staphylococcus [42], meaning that prior infection with temperate bacteriophage could influence the pattern of susceptibility to lytic phage across the phylogeny [35, 43]. Together, we may expect these mechanisms to lead to weaker phylogenetic signal in virus susceptibility in bacterial hosts than that seen in animals where immunity is largely vertically transmitted.
Recent years have seen a resurgence in interest in the use of phage to treat bacterial infections due to the continuing emergence of antimicrobial resistance [44–47]. Phage present a promising alternative to traditional antimicrobials in that they are self-amplifying, self-limiting, and have proved effective in the treatment of drug-resistant bacterial infections such as methicillin-resistant Staphylococcus aureus (MRSA) [48–50] and Pseudomonas aeruginosa [50]. Two key considerations in the design of phage therapies are the host range of the phage–and so the range of bacterial strains or species it can be used to treat–and the efficiency of the phage in replicating and killing its bacterial host, which is linked to host susceptibility. Studies have shown that phage host range can vary from a single host species [51–55] to broad host range generalists [51, 53–59], although a consensus has yet to be reached on the most effective method for quantifying phage host range [60–62]. Additionally, studies investigating susceptibility to phage across bacterial isolates have shown variation in susceptibility at both the host strain and species level [56, 57, 59].
An ability to explain and predict variation in phage host range and susceptibility would be beneficial in the design of future therapies, allowing for the more efficient design of broad range phage cocktails with high efficacy against multiple pathogens. Recently, the evolutionary relationships between bacterial hosts were shown to explain some of the variation in susceptibility of a panel of S. aureus hosts to several staphylococcal phages when measured using plaque assays [63], suggesting that the structure of the host phylogeny may be a useful tool in predicting bacterial susceptibility to phage. It remains to be seen whether the host phylogeny can be a useful tool in explaining variation in bacterial susceptibility across broader phylogenetic scales (i.e., across bacterial species or genera), or when applied to different measures or components of bacterial susceptibility.
Here, we use a broad host range bacteriophage (Intravenous Staphylococcal Phage; ISP) and a panel of 64 Staphylococcaceae isolates (encompassing 2 genera and 17 Staphylococcus species) to investigate how patterns of bacterial susceptibility are influenced by the evolutionary relationships between hosts. ISP, a double stranded DNA virus in the family Myoviridae, is closely related to Staphylococcus phage G1 [64, 65] and has shown success in the treatment of antimicrobial resistant infections clinically [66]. ISP was isolated in the 1920s from an unknown source [65] and has a broad experimental host range within S. aureus, infecting 86% isolates tested in a previous study [67]. However, in the same study, ISP was unable to experimentally infect nine S. haemolyticus isolates, suggesting that this host range may be species-specific. Staphylococci are a well-established model for bacteria-phage interactions, with investigations into staphylococcal phages occurring since the 1910s. Lytic staphylococcal phages have demonstrated broad host ranges [52, 56, 68, 69], antibiofilm activity [68, 70], and vary in their efficacies against bacterial infection [71–73]. Several mechanisms that are important for the interaction between staphylococci and their phage have been characterised, including common cell surface receptors used for attachment [74–79], and host resistance mechanisms such as the overproduction of surface proteins to block adsorption [80–82], restriction modification systems [83–85], and CRISPR targeted degradation of phage DNA [86–89]. Despite this, much remains to be understood about the interactions between staphylococcal phages and their hosts. An improved understanding of the mechanisms that underpin bacteria-phage interactions may provide more general insights into virus-host interactions and their implications in the emergence of viral pathogens into novel populations.
In this study we demonstrate that ISP is a broad host range bacteriophage, able to infect both within and among species of Staphylococcaceae. Given that different methods of quantifying host range can give different estimations, we assessed the susceptibility of our bacterial host panel to ISP using three methods: plaque assays, optical density assays, and quantitative PCR. We observed considerable variation in susceptibility to ISP across the host panel and showed that a high proportion of the observed variation in susceptibility was attributable to the relationship between host species.
Methods
Staphylococcaceae isolates
This study made use of 64 strains of Staphylococcaceae, representing a broad phylogenetic and geographic range of hosts. These strains spanned 2 genera and 17 species that were estimated to have last shared a common ancestor ~122mya [90]. Multi-Locus Sequencing Typing (MLST) revealed that 5 Clonal Complexes (CC) of S. aureus were represented in this panel, including major complexes CC1 and CC8 (S1 Table).
Each Staphylococcaceae sample was streaked on a LB-agar Miller (Formedium) plate (1.5% agar) and incubated for 24 hours at 37°C. A single colony was isolated and used to inoculate 5mL of LB broth, which was incubated at 37°C, 180rpm for 24 hours. All isolates were stored in 25% glycerol at -80°C. When required, isolates were grown up by inoculating 5mL of high-salt LB Miller broth (Formedium) in a sterile 30mL glass universal with a scraping of the frozen culture and incubating at 37°C, 180rpm overnight. To ensure that any observed differences in susceptibility to ISP were not a function of host availability (i.e., differences in the concentration of bacterial hosts in solution), calibration curves using optical density (OD) and colony forming units (CFU)/mL were generated for each strain. Overnight cultures were then normalised by dilution in LB Miller broth to give similar host densities prior to their use in susceptibility assays.
Phage preparation
An isolate of ISP was kindly provided by Jean-Paul Pirnay and Maya Merabishvili at the Queen Astrid Military Hospital (Brussels, Belgium). ISP was propagated on the S. aureus strain 13S44S9 (chosen for consistency with previous studies [65]; hereafter referred to as the ‘propagation host’) and extracted using chloroform: 1mL aliquots of the suspension were treated with 10% chloroform, vortexed for 1.5 minutes, then centrifuged for 1 minute at 18 x g. The supernatant containing the phage was aliquoted into sterile 2mL Eppendorf tubes, and the process repeated a second time to ensure the complete removal of bacterial hosts.
The number of infectious phage present was quantified using plaque assays. ISP was 10-fold serially diluted in 1X M9 buffer (Merck Life Sciences) from 100 to 10−8. 100μL of overnight culture of 13S44S9 and 50μL of diluted phage were added to 5mL of LB-agar (0.5% agar), gently mixed, and poured over a 20mL LB-agar Miller plate. Plates were left to dry before being inverted and incubated at 37°C for 24 hours. Plaques were counted and plaque forming units (PFU)/mL determined based on the dilution with the highest number of discernible plaques. Each plaque assay was repeated at least 3 times to account for plating error.
Assessing susceptibility using plaque assays
10-fold serial dilutions of ISP (initial concentration 1.6x108 PFU/mL, determined by plaquing on 13S44S9) were prepared in 1X M9 buffer, ranging from 100 to 10−8. Bacterial isolates were diluted in LB Miller broth, plated out to a final density of 2x1010 bacteria/mL, and left to dry before 5μL of each ISP dilution was spotted on each LB-agar Miller plate. The plates were then inverted and incubated at 37°C. Plaques were counted after 24 hours and converted to PFU/μL, based on the dilution with the highest number of discernible plaques. Six biological replicates of the plaque assay were performed for each bacteria-phage pairing. As the range of PFU/mL across technical replicates in preliminary plaque assays was small (1.4–1.7x108 PFU/mL), only a single technical replicate per biological replicate was performed. Of the 64 bacterial hosts tested, 6 biological replicates were obtained for 42 strains, 5 replicates for 12 strains, and 4 replicates for 10 strains.
Assessing susceptibility using optical density assays
In each well of a 96-well plate, 180μL of LB Miller broth was added alongside 10μL of each Staphylococcaceae isolate diluted to an initial concentration of 1x106 CFU/mL. For infected plates, 10μL of ISP at a concentration of 5x104 PFU/mL was added to each well to achieve an MOI of 0.05 (final concentration of 5x104 CFU/mL and 2.5x103 PFU/mL). A low MOI was chosen to ensure any large changes in OD were due to multiple rounds of phage infection within the sample, requiring the production of viable phage progeny. For control plates, 10μL of 1X M9 buffer was added in place of ISP to maintain a final concentration of 5x104 CFU/mL. Samples with and without phage were kept on separate plates to minimise the chance of phage contaminating uninfected samples. Within-plate position was kept constant between infected and control plates for each biological replicate but randomised between replicates to minimise the effect of within-plate position effects. The effects of between-plate variation were estimated by comparing OD readings between biological replicates of the same infection conditions (standard deviation of 0.14) and were found to account for a small amount of variation compared with the effect of infection (standard deviation of 0.67). Three LB Miller broth and three 1X M9 buffer controls were added to each plate to check for contamination. The plates were sealed with an adhesive PCR plate seal (Thermo Scientific) and incubated for 24 hours at 37°C, 180rpm. Following the incubation, OD was read at 600nm on a MultiSkan Sky Microplate Spectrophotometer (Thermo Fisher). To standardise for any differences in bacterial growth rate, susceptibility was calculated as a proportion change in OD due to infection, as follows:
Several collected data points reported a proportion change in OD considerably less than zero. As this was likely the result of anomalous OD readings, outlier analysis was performed. Datapoints more than 1.5 times the interquartile range outside the upper and lower quartile of the data were considered minor outliers, and those more than 3 times the interquartile range outside the upper and lower quartiles were considered major outliers. All analyses included here were robust to the removal of both major and minor outliers (S2–S5 Tables), and so only data with all major and minor outliers removed is presented in the main text. Following outlier removal, 6 biological replicates were obtained for 56 of the bacterial strains tested, 5 replicates were obtained for 7 of the strains, and 3 replicates were obtained for 1 strain.
Assessing susceptibility using quantitative PCR
Infections used to measure susceptibility using quantitative (q)PCR were set up identically to those in the OD assays described above. Again, a low MOI of 0.05 was used to ensure that any large changes in viral load were due to multiple rounds of phage infection, requiring the production of viable phage progeny. Following the 24 hour incubation, 100μL of each sample was transferred to a sterile 1.5mL screw cap microcentrifuge tube and heat treated at 90°C for 10-minutes to inactivate the bacteria. Additionally, 12 samples of the aliquot of ISP used to infect the Staphylococcaceae strains were diluted 1:20 in LB Miller broth and extracted to provide an initial viral load at infection timepoint zero. To extract the viral DNA, 100μL of 10% w/v Chelex 100 (Merck Life Sciences), 2ul of 20ng/μL Proteinase K (Merck Life Sciences), and ~10 1mm zirconia beads (Thistle Scientific) were added to each sample before mechanically lysing the bacteria using an Omni Bead Ruptor 24 (Camlab). Samples were centrifuged briefly, and heat treated for 10 minutes at 95°C to inactivate the Proteinase K before being centrifuged for a further 5 minutes at 18 x g to sediment out the Chelex. For each sample, 50μL of the supernatant containing bacterial and viral DNA was transferred and stored at -20°C.
qPCR was performed on each of the samples using an Applied Biosystems StepOnePlus system with a Sensifast Hi-Rox Sybr kit (Bioline). Cycle conditions were as follows: initial denaturation at 95°C for 120 seconds, then 40 cycles of 95°C for 5 seconds, and 60°C for 30 seconds. ISP was measured using the following primers: forward, 5’- CCTGTACCGGCTTGACTCTC -3’; reverse, 5’- AGCTACAACCGAGCAGTTAGA -3’, which were confirmed to have a near 100% efficiency when tested in the presence of each bacterial isolate. Pilot experiments showed that normalisation to either staphylococcal genomic DNA or an exogenous DNA spike made little difference to the between sample variation in ISP viral load and, therefore, no normalisation was used.
For each sample, two technical replicates of the qPCR reaction were performed. Amplification of the correct product was confirmed by melt curve analysis: samples that had failed to amplify the product, showed evidence of melt curve contaminants, or departed from the melt curve peak of positive samples by ±1.5°C were excluded. Correction for plate effects between technical replicates was performed using a linear model as previously described [91, 92]. Mean viral Ct values from technical replicate pairs (Ct:24) were normalised to an initial dose of ISP (Ct:0) and converted to fold change in viral load using the 2–ΔCt method, where ΔCt = Ct:24 –Ct:0. Prior to analysis, these values were converted to a log10 fold change in viral load over 24 hours. Of the 64 bacterial strains tested, 6 biological replicates were obtained for 58 strains, 5 replicates for 5 strains, and 4 replicates for 1 strain.
Inferring the host phylogeny
To infer the evolutionary relationships between Staphylococcaceae isolates, a core genome phylogeny was constructed using BEAST v1.10 [93]. Briefly (see S1 Text, S1–S4 Figs, and S6–S7 Tables for full details), whole genome sequences were collected for 8 previously sequenced strains from NCBI (S1 Table), and the remaining 56 sequences were obtained through whole genome sequencing. Library preparations and sequencing were performed by MicrobesNG (Birmingham, UK). Genomic DNA libraries were prepared using the Nextera XT Library Prep Kit (Illumina) with twice the stated amount of input DNA and PCR elongation increased to 45-seconds. Pooled libraries were quantified using the Kapa Biosystems Library Quantification Kit for Illumina and sequenced using Illumina sequencers (HiSeq/NovaSeq) with a 250-bp paired end protocol. Sequence reads were deposited on NCBI under the BioProject ID: PRJNA894984 (see S1 Table). Genomes were assembled and quality controlled as described in S1 Text.
De novo assemblies were annotated using Prokka (v2.8.2) [94] and orthologous genes identified with Panaroo [95] using a sequence identity threshold of 0.7. Orthologous genes were then used to generate a core genome alignment of the 102 genes shared between each of the 64 Staphylococcaceae isolates. Phylogenetic trees were constructed using BEAST v1.10 [93]. Sequence alignments were fitted to a HKY substitution model using relaxed uncorrelated molecular clock models, gamma distributions of rate variation, and constant population size coalescent priors [93]. Separate substitution models and molecular clocks were fitted to 1st/2nd and 3rd codon positions, to reflect differences in selective constraint [96]. Two independent MCMC chains were run for each model until both convergence and a <10% burn-in was achieved. Convergence of all parameters was checked using Tracer v1.6 [97].
The within-S. aureus and among-species phylogenies were constructed by dropping the relevant tips from the 64-strain tree using the R package ape [98]. To ensure that the phylogenetic relationship between species and strains were accurately resolved in the 64-strain tree, separate phylogenetic trees were constructed using only the S. aureus samples or species isolates with 13S44S9 as a representative S. aureus strain (see S1 Text). As the topology of the smaller phylogenies matched that of the phylogenies made by dropping tips from the whole phylogeny, the dropped tip phylogenies were used for our analyses as they had comparable branch lengths to the 64-strain phylogeny.
Phylogenetic mixed models
Phylogenetic generalised linear mixed models were used to investigate the effect of host relatedness on susceptibility to infection with ISP, and to examine correlations between the methods used to assess susceptibility. Multivariate models were fitted using the R package MCMCglmm [99] with susceptibility, as determined by each method (plaque assay, OD, and qPCR), as the response variable. Unscaled trees were used to correct for differences in the evolutionary divergence of our within- and among-species trees.
The structures of the model were as follows:
| (1) |
| (2) |
In these models, yhim is the susceptibility measured by method m in the ith biological replicate of host h. The fixed effect β1 represents the intercepts for each method, the random effect μp represents the effects of the host phylogeny assuming a Brownian model of evolution, and e represents the model residuals. Model (1) also includes a strain-specific random effect that is independent of the host phylogeny (μs:hm). This explicitly estimates the non-phylogenetic component of between-strain variance and allows for the calculation of phylogenetic heritability (described below). μs:hm was removed from model (2) as model (1) failed to separate the phylogenetic and strain-specific effects. An additional version of model (1) was run with evolutionary distance from the propagation host added as a fixed effect (βd:hm). This was done to examine if the evolutionary distance from the propagation host explained any observed differences in susceptibility between strains, but was found to be non-significant and thus excluded from further models.
Susceptibilities measured by OD and qPCR were treated as normally distributed. However, to account for the zero-inflated nature of the plaque assay data, plaque assay susceptibility was divided into two separate variables–equivalent to a hurdle modelling approach–with a binary variable indicating whether the bacteria was permissive (1) or non-permissive (0) to infection, modelled using probit link function [100], and (conditional on being permissive) a continuous variable containing PFU/μL which was treated as normally distributed.
Within each of these models, the random effects and residuals were assumed to follow a multivariate normal distribution with a mean of 0 and covariance structure Vp⊗A for the phylogenetic effects, Vs⊗I for strain-specific effects, and Ve⊗I for residuals, where ⊗ represents the Kronecker product. A represents the host phylogenetic relatedness matrix, I an identity matrix, and V represents 4 × 4 covariance matrices describing the variances and covariances in susceptibility for the different methods. Specifically, the matrices Vp and Vs describe the phylogenetic and non-phylogenetic between-strain variances in susceptibility for each method and the covariances between them, whereas the residual covariance matrix Ve describes the within-strain variance that includes both true within-strain effects and measurement errors. Here, between strain variation refers to the variation in susceptibility between bacterial strains, whereas within-strain variation refers to variation in susceptibility between biological replicates of the same strain (i.e., variation within the same strain), including both within-strain genetic variance and measurement error. Because each biological replicate consists of a measurement from a single method, the covariances of Ve cannot be estimated and were set to 0. Additionally, the residual variance for the binary variable cannot be estimated and was fixed at 1.
Models were run for 13 million MCMC generations, sampled every 5,000 iterations with a burn-in of 3 million generations. Parameter expanded priors were placed on the covariance matrices, resulting in multivariate F distributions with marginal variances being scaled by 1000. Inverse-gamma priors were placed on the residual variances, with a shape and scale equal to 0.002. To ensure the model outputs were robust to changes in prior distribution, models were also fitted with inverse-Wishart priors, which gave quantitatively similar results.
To estimate an effect of phylogeny on the susceptibility of Staphylococcaceae to ISP, we calculated several metrics. First, repeatability, used in the study of quantitative traits and defined as how ‘repeatable’ a measurement is within a group compared with measurements made across groups, was calculated from model (2) as Vp/Vp+Ve, where Vp represents the phylogenetic (i.e., ‘across group’) variation, and Ve the within strain variation [101–103]. To estimate the proportion of between-strain variation that can be explained by phylogeny, we calculated phylogenetic heritability (h2) from model (1). Phylogenetic heritability is defined in two ways in the literature: either as the variance in average phenotype across strains that can be explained by phylogeny or as the variance in phenotype that can be explained by phylogeny [104, 105]. The former averages over the within-strain variation (Vs) such that (i.e., the proportion of the phylogenetic and non-phylogenetic strain variance explained by phylogeny, referred to as “phylogenetic heritability” in this paper), whereas the latter does not and (i.e., the proportion of the total variance explained by phylogeny, defined as the “phylogenetic heritability of total variance” in this paper). Inter-strain correlations in viral load between each method were calculated from model (2) Vp matrix as and the slopes (β) of each relationship as . Parameter estimates stated below are means of the posterior density, and 95% credible intervals (CIs) were taken to be the 95% highest posterior density intervals.
To visualise the outputs of our phylogenetic GLMMs as patterns of bacterial susceptibility across the host phylogeny, we plotted ancestral state reconstructions using methods described elsewhere (https://doi.org/10.6084/m9.figshare.7756526.v1). These plots take the trait values for each terminal node (i.e., the experimentally measured susceptibilities of extant bacterial hosts) and the inferred trait values of ancestral nodes from model (2) and plot them as colour gradients across the phylogeny, with changes in trait values along branches assumed to be smooth, fixed rate transitions as expected when following a Brownian model of trait evolution.
Leave-one-out cross-validation
To investigate the ability of the bacterial host phylogeny to predict the susceptibility of a novel host, leave-one-out cross-validation was used [106], whereby multiple versions of model (1) were fitted, each with the data from a single bacterial strain removed, and the model challenged to predict the susceptibility of the “unknown” host given only its evolutionary relationships to other Staphylococcaceae strains and their measured susceptibilities. For comparison, a null model containing no effect of host phylogeny was fitted. Prediction errors from the leave-one-out cross-validation and the null model were compared using Wilcoxon rank sum tests [107] to determine whether information on the relationship between host species significantly improved the ability of the model to predict the susceptibility of an unknown host.
Results
ISP is a broad host range phage with varying infectivity across Staphylococcaceae
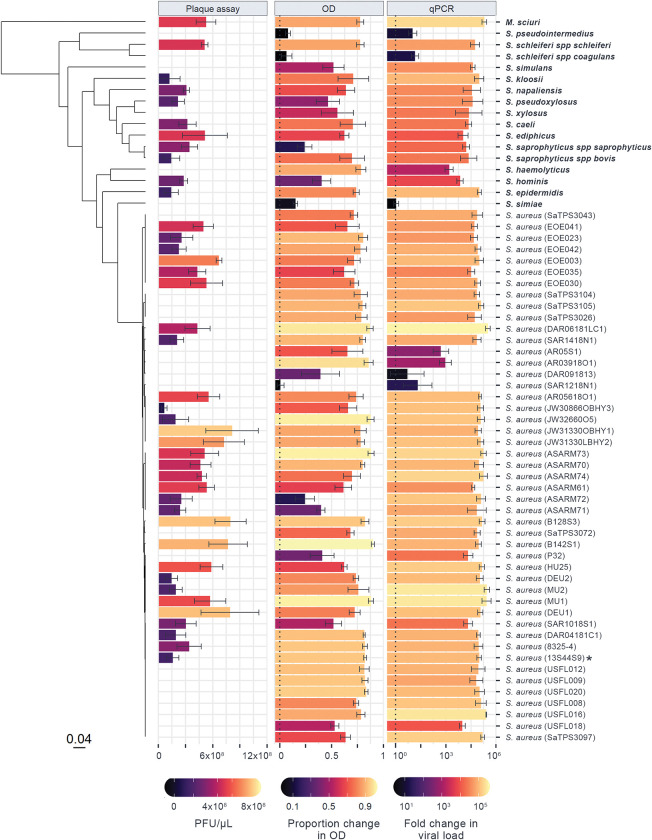
To investigate the ability of the host phylogeny to explain variation in susceptibility to virus infection in bacteria–and how different methods may vary in their quantification of phage host range–we experimentally infected 64 strains of Staphylococcaceae with the bacteriophage ISP and assessed susceptibility using three distinct methods: plaque assays, OD assays, and qPCR. When assessed using plaque assays, 64% of host strains were seen to be permissive to infection with ISP. However, both OD and qPCR measures of susceptibility showed that ISP was capable of infecting 97% of the host panel (Fig 1). Variation in susceptibility between Staphylococcaceae isolates was seen in every method: the mean PFU/μL of permissive hosts ranged from 2.2x103 in S. simulans to 8.1x105 in S. aureus strain JW31330OBHY1; the mean proportional decrease in OD ranged from 0.01 in S. aureus strain SAR1218N1 to 0.90 in S. aureus strain B142S1; and the mean change in qPCR viral load ranged from a 1.2-fold increase in S. simiae to a ~300,000-fold increase in S. aureus strain DAR06181LC1 (Fig 1). Together, these results show that ISP is both able to infect a broad range of Staphylococcaceae strains and species, and that susceptibility to ISP varies widely across the Staphylococcaceae family.
Fig 1. Susceptibility to ISP across a panel of 64 Staphylococcaceae isolates, measured using three distinct methods.
Bar lengths and colour show the mean change in ISP assessed by plaque assay (PFU/μL), optical density (proportion decrease in OD with infection after 24 hours), and qPCR (log10 fold change in viral load after 24 hours), with error bars representing the standard error of the mean across at least four biological replicates. The phylogeny of Staphylococcaceae hosts is presented on the left, with the scale bar representing the number of nucleotide substitutions per site. Strain names are presented on the right, with non-aureus species in bold and the propagation host labelled with an asterisk (a full version of the tree is available at https://doi.org/10.6084/m9.figshare.21642209.v1).
Susceptibility to ISP across Staphylococcaceae is explained by the host phylogeny
The phylogeny of Staphylococcaceae host species inferred here is broadly consistent with previous studies of these taxa [84], with the close phylogenetic relationships between species being generally well supported and hosts falling into two main clades (Fig 1). The Staphylococcaceae phylogeny is characterised by high susceptibility to ISP across a large number of isolates, with smaller clades showing reduced susceptibility to infection (i.e., the clade containing S. aureus strains AR05S1, AR03918O1, and DAR091813). Some clades show intermediate levels of susceptibility (e.g., the clade containing S. haemolyticus and S. hominis) while some contain both permissive and non-permissive strains (e.g., the clade containing S. aureus strains SAR1218N1 and AR05618O1).
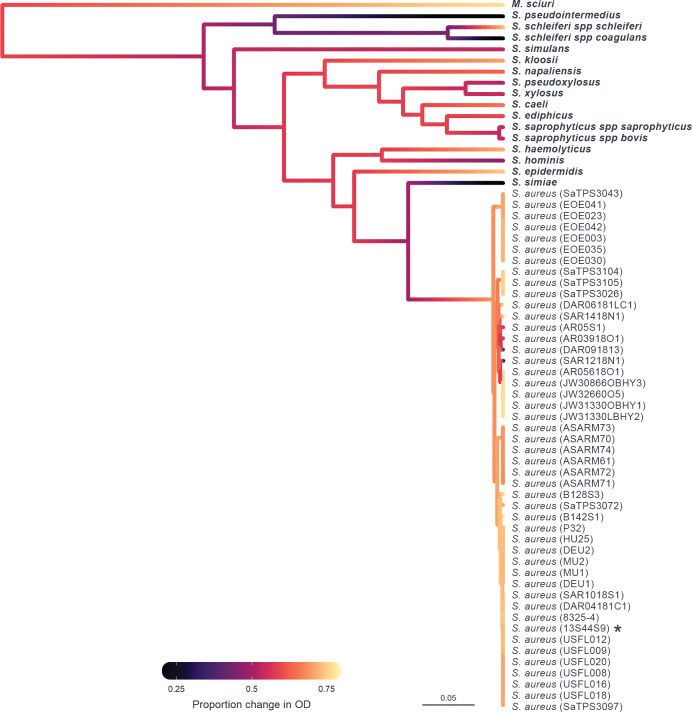
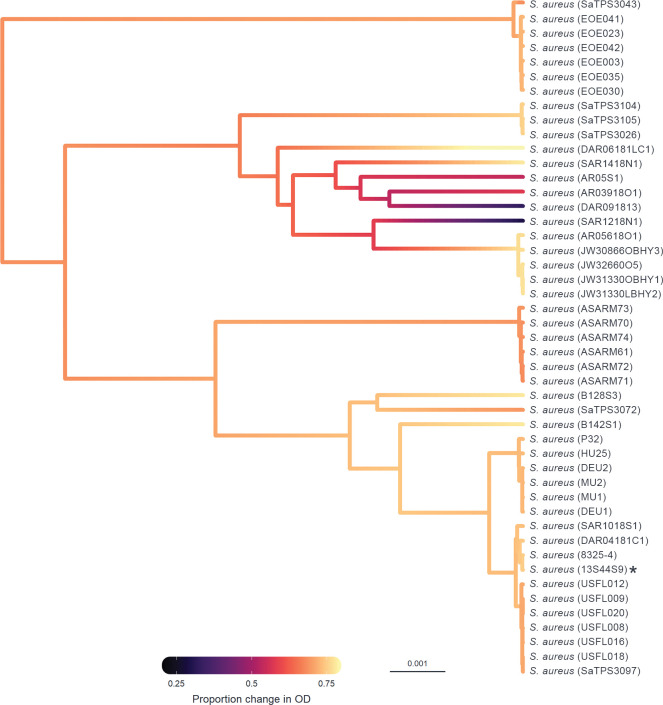
Phylogenetic generalised linear mixed models were fitted to the data to determine the proportion of variation in susceptibility explained by the host phylogeny (Table 1). Estimates of repeatability, phylogenetic heritability, and phylogenetic heritability of total variance for the binary plaque assay, OD, and qPCR data were close to 1 with narrow credible intervals. The convergence of phylogenetic heritability and repeatability estimates for the binary plaque assay, OD, and qPCR data at 1 suggests that the between-strain phylogenetic component explains a high proportion of the variation in susceptibility with little within-strain variation or measurement error (Table 1). Estimates of repeatability and phylogenetic heritability for the continuous (PFU/μL) component of the plaque assay had wide credible intervals spanning 0–1, suggesting that there is no effect of phylogeny when susceptibility is assessed using a continuous component of plaque assay. The effect of phylogeny on the susceptibility of Staphylococcaceae hosts to ISP can be further seen in ancestral state reconstructions (Figs 2 and 3), where different clades are seen to have similar susceptibilities to infection. This is particularly apparent within-S. aureus samples, where clades show either high, intermediate, or low susceptibility to ISP (Fig 3).
Table 1. Estimates for the repeatability and phylogenetic heritability across 64 Staphylococcaceae isolates.
Estimates of repeatability are taken from model (2) and estimates of phylogenetic heritability (the proportion of phylogenetic and non-phylogenetic strain variation explained by the host phylogeny) and phylogenetic heritability of the total variance (the proportion of total variation explained by the host phylogeny) are taken from model (1). PA = plaque assay, CI = credible interval.
| Repeatability | Phylogenetic heritability | Phylogenetic heritability of total variance | ||||
|---|---|---|---|---|---|---|
| Method | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI |
| Binary PA | 1.00 | 1.00, 1.00 | 1.00 | 1.00, 1.00 | 1.00 | 1.00, 1.00 |
| Continuous PA | 0.00 | 0.00, 0.00 | 0.68 | 0.01, 1.00 | 0.00 | 0.00, 0.00 |
| OD | 0.98 | 0.96, 0.99 | 0.97 | 0.94, 1.00 | 0.94 | 0.88, 0.98 |
| qPCR | 0.99 | 0.99, 1.00 | 1.00 | 1.00, 1.00 | 0.99 | 0.98, 1.00 |
Fig 2. Ancestral state reconstruction of susceptibility to ISP measured by OD.
Ancestral states were estimated from model (2) for each node and plotted in colour across the Staphylococcaceae host phylogeny, with the scale bar representing nucleotide substitutions per site. Colours represent susceptibility of a host to infection with ISP measured by OD (proportion change in optical density in infected compared to non-infected cultures), with black representing the lowest level of susceptibility and yellow the highest. Strain IDs are presented on the right, with non-aureus species in bold and the propagation host labelled with an asterisk (*).
Fig 3. Ancestral state reconstruction of the S. aureus host strains susceptibility to ISP measured by OD.
Ancestral states were estimated from model (2) for each node and plotted in colour across the Staphylococcus aureus host phylogeny, with the scale bar representing nucleotide substitutions per site. Colours represent susceptibility of a host to infection with ISP measured by OD (proportion change in optical density in infected compared to non-infected cultures), with black representing the lowest level of susceptibility and yellow the highest. Strain IDs are presented on the right and the propagation host labelled with an asterisk (*).
To determine if the observed phylogenetic signal is consistent across evolutionary scales, we reduced the phylogeny to one containing only the S. aureus samples (S8 Table) and one containing each of the Staphylococcaceae species and a single representative S. aureus strain (13S44S9) (S9 Table). Similar estimates of repeatability were observed for both the within-aureus and among-species phylogeny models. However, both models showed a reduced ability to estimate phylogenetic effect, with wide credible intervals around estimates (apart from estimates for binary plaque assay and qPCR in the within-S. aureus model). It is likely that the observed difference in ability to estimate heritability between the whole phylogeny model compared to the reduced phylogeny models is down to reduced statistical power, causing the latter models to struggle to separate the phylogenetic and strain-specific effects.
Finally, to ensure that ISP had not adapted to its propagation host prior to the experiment, we looked for an effect of distance from the propagation host (S. aureus strain, 13S44S9). No effect of distance from the propagation host on the susceptibility of Staphylococcaceae to ISP was found (β = 0.03, 95% credible interval: -1.46, 1.56), suggesting that the observed phylogenetic signal was not being driven by adaptation of ISP to the propagation host.
Measures of susceptibility from plaque assays, OD assays, and qPCR are positively correlated across hosts
Correlations between plaque assay, OD, and qPCR measures of susceptibility to ISP across bacterial host strains were estimated from the variance-covariance matrices of model (2) (Table 2). A strong positive inter-strain correlation was observed between susceptibility measured by OD and qPCR (r = 0.97, 95% CI: 0.92, 1:00), and a strong positive correlation was observed between the binary measure of plaque assay and OD (r = 0.94, 95% CI: 0.86, 1.00) and the binary measure of plaque assay and qPCR (r = 0.98, 95% CI: 0.94, 1.00). However, no evidence of a correlation was observed between the continuous plaque assay data and either OD or qPCR, with correlation coefficients approximately zero and credible intervals spanning -1 to 1 (Table 2). Similar correlations were observed between methods for the within S. aureus (S10 Table) and among-species (S11 Table) phylogenies, with the qPCR:OD correlation being the only one consistently and significantly positive. A correlation between the binary measure of PA and the other measures of susceptibility was observed in the among-species phylogeny but not the within-S. aureus phylogeny, suggesting that the correlation observed between these measures is being driven by strong correlations seen at the species level rather than the within-species level.
Table 2. Inter-strain correlations in susceptibility measures between methods.
Numbers show the mean estimates for the correlation strength (r, white cells) and slope (β, grey cells) between pairs of methods, with 95% credible intervals (CIs) indicated in brackets. The slopes were calculated with the variables in columns as x and variables in rows as y. Estimates with CIs that do not span zero are highlighted in bold. PA = plaque assay, *value on a probit scale.
| Binary PA | Continuous PA | OD | qPCR | |
|---|---|---|---|---|
| Binary PA | - | - |
0.94
(0.86, 1.00) |
0.98
(0.94, 1.00) |
| Continuous PA | - | - | -0.01 (-0.99, 0.99) |
-0.01 (-1.00, 1.00) |
| OD |
0.50*
(0.50, 0.50) |
-0.00 (-0.12, 0.17) |
- |
0.97
(0.92, 1.00) |
| qPCR |
0.51*
(0.50, 0.52) |
0.01 (-1.30, 0.97) |
7.45
(5.53, 9.59) |
- |
Mixed evidence for host phylogeny improving the accuracy of susceptibility predictions
As the host phylogeny explains a large proportion of the variation in ISP susceptibility, it may allow for the susceptibility of untested Staphylococcaceae strains and species to be predicted based on their evolutionary relationships to staphylococci with known susceptibilities. To investigate the ability of the bacterial host phylogeny to predict susceptibility, leave-one-out cross-validation was used, whereby multiple versions of model (2) were fitted, each with the data from a single bacterial strain removed, and the model challenged to predict the susceptibility of the “unknown” host given only its evolutionary relationships to other staphylococci strains and their measured susceptibilities (Fig 4).
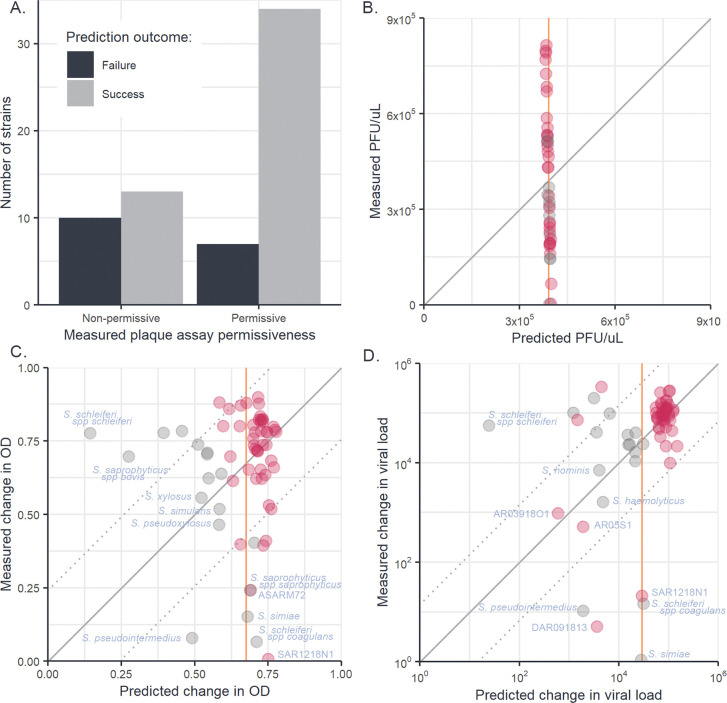
Fig 4. Leave-one-out cross-validation of phylogenetic mixed models fitted to plaque assay, OD, and qPCR data.
Predictions of “unknown” trait values from a leave-one-out cross-validation of phylogenetic model (2) for binary plaque assay (A), continuous (PFU/μL) plaque assay (B), change in OD with infection (C) and fold changes in viral load measured by qPCR (D). Each datapoint represents an individual strain of Staphylococcaceae whose measured trait value has been removed from model (2) and predicted from its evolutionary relationships to other Staphylococcaceae isolates and their trait values. Solid diagonal lines illustrate the location of 1:1 predictions and dotted lines indicate the root-mean-squared errors around these lines. Orange vertical lines represent the predicted trait values of all strains from a null (intercept only) model. For panels B, C, and D, points plotted in pink show the S. aureus strains whereas points plotted in grey show the non-S. aureus strains.
For the binary component of plaque assay, the phylogenetic model was able to correctly predict a strain’s permissiveness to ISP infection in 73% of cases (Fig 4A), whereas a null model without phylogeny predicted all hosts to be permissive, achieving an accuracy of 63%. When asked to predict the continuous (PFU/μL) component of plaque assays, errors from the phylogenetic and null models were indistinguishable (Wilcoxon rank sum test: W43,43 = 886, p = 0.74), as in the phylogenetic model most of the variation in PFU/μL was partitioned into the model residuals, causing trait values predicted from the phylogenetic component to cluster around the across-strain mean (Fig 4B). When asked to predict susceptibility measured by OD assay (Fig 4C), the errors produced by the phylogenetic model were also not significantly different from those produced by a null model (Wilcoxon rank sum test: W64,64 = 2111, p = 0.77). However, when predicting viral loads from qPCR data (Fig 4D), the phylogenetic model produced a small but significant decrease in error compared to the null model (Wilcoxon rank sum test: W64,64 = 2726, p < 0.01).
In the OD and qPCR assays most strains were both similar in susceptibility to their close relatives and showed susceptibility close to the across-strain mean (Figs 1, 4C, and 4D), causing the phylogenetic and null models to produce similar predictions for the majority of strains. In cases where hosts existed in clades with susceptibilities different from the across-strain mean, prediction accuracy increased when phylogeny was included (e.g., qPCR prediction for S. haemolyticus and S. hominis, OD prediction for S. xylosus and S. pseudoxylosus). However, when hosts had susceptibility near the across-strain mean and had close relatives that differed from this mean, the phylogenetic model produced larger errors in prediction than the null model (e.g., both OD and qPCR predictions for S. schleiferi spp schleiferi). Lastly, where hosts differed from the across-strain mean but had close relatives that conformed to the mean, both phylogenetic and null models were poor predictors of susceptibility (e.g., both OD and qPCR predictions for S. schleiferi spp coagulans and S. aureus SAR1218N1). Together, these results suggest that the host phylogeny may allow for the limited prediction of host susceptibility, but may mislead predictions in host strains that differ strongly from the susceptibilities of their close relatives.
Discussion
Closely related host species present similar environments to novel viruses [103, 108], and so tend to share similar levels of susceptibility [17, 19–21, 26]. Here, we have examined how susceptibility to a broad host range phage varies across a diverse panel of Staphylococcaceae bacteria and determined what proportion of the variation in phage susceptibility is explained by the relationships between bacterial hosts. ISP was capable of infecting 97% of the host strains investigated here when measured by OD and qPCR assays, but only 64% of these strains appeared permissive when tested by plaque assay. We found that variation in susceptibility across our Staphylococcaceae panel–measured using OD, qPCR, and the binary component of plaque assays–was largely explained by the host phylogeny, and these effects were seen at both within-species, and among-species phylogenetic scales. No effects of evolutionary distance from the propagation host were seen, with the patterns instead being driven by the existence of host clades sharing similar levels of susceptibility to ISP. Strong positive correlations were seen between the susceptibility measures from the binary plaque assay, OD, and qPCR assays, but not between these methods and the PFU/μL measures taken from plaque assays. Together, these results suggest that ISP has a broad ability to infect bacterial hosts within the Staphylococcaceae family, and that a large proportion of the variation in susceptibility to ISP across Staphylococcaceae hosts can be explained by the evolutionary relationship between bacterial hosts.
Recent studies investigating the host range of phages have employed host panels of varying phylogenetic scales, including analyses across diverse panels of bacterial species [51, 53, 57, 58, 109–111], and focused investigations in a high number of isolates within a single species [54, 55, 59]. This variable diversity of hosts selected for host range screening has often affected our interpretation of host range, with “broad host range” used to refer to both phages able to infect many strains within a single species and phage able to infect a large number of species within a genus. Here, we have demonstrated that ISP is capable of infecting two genera and 17 species within the Staphylococcaceae family. This expands upon the findings of a previous study which demonstrated that ISP was able to infect many strains of S. aureus [67] and extends the number of hosts that the phage can infect. The identification of phages with a broad host range is advantageous for phage therapy, where broad range phage can be used to treat a larger number of clinical infections. Having such phages available for study would also be beneficial to our growing understanding of virus host shifts, as it has been demonstrated that viruses with a broad host range are more likely to shift between species [112].
Both permissiveness and susceptibility of bacterial hosts to phage are key considerations when designing phage therapies. In our results, variation in permissiveness and susceptibility to ISP was apparent, both within S. aureus and among Staphylococcaceae species. This among-strain and among-species variation is likely due to differences in the molecular pathways influencing host range. For example, previous studies on phages have demonstrated that the highly conserved wall teichoic acid (WTA) serves as the primary receptor for staphylococcal phages [79], contributing to their broad host range. Further studies have characterised the anti-phage defence systems present in S. aureus and found that a combination of type I and type III restriction modification systems prevent the uptake of DNA originating from other species [113] and between S. aureus lineages [114], respectively, meaning that patterns of restriction can be clonal complex specific and lead to independent lineage evolution [115]. A Genome-wide association study (GWAS) aiming to identify genes associated with susceptibility measured by plaque assays identified putative loci affecting the host range of S. aureus bacteriophages, including: the TarJ, TagH, and TarP proteins which are involved in the production and transport of WTA to the bacterial cell surface; HsdS which determines the sequence specificity of the SauI restriction modification system; and several prophage associated genes, suggesting that superinfection immunity is playing a role in the susceptibility of S. aureus to bacteriophage [63]. Given the increase in phage host range observed in our study when susceptibility was assessed by either OD or qPCR, it is likely that future GWAS approaches based on susceptibility assessed by different methods may reveal further molecular pathways involved in bacteriophage host range. Further investigation of ISP to identify the specific molecular pathways by which it interacts with its host will improve our understanding of the factors that contribute to variation in susceptibility across bacterial hosts, allowing us to utilise ISP more efficiently as a therapeutic.
Bacterial evolution is known to involve several processes that could disrupt the phylogenetic heritability of susceptibility to phage. Notably, cell surface receptors can be easily altered by point mutations and components of phage resistance may be frequently gained and lost through horizontal gene transfer, which can lead to high levels of variation in the phage defence systems present in bacterial genomes [33, 116–120]. Previous investigations have shown that the carriage of prophage and the transfer of restriction modification systems can be lineage specific in bacteria [36, 115]. However, the extent to which these mechanisms are phylogenetically constrained is unknown. Despite the potential for horizontal gene transfer and large effect point mutations to disrupt the phylogenetic signal in phage susceptibility, our models suggest that the evolutionary relationships between a diverse panel of bacterial hosts can capture a large proportion of the variation in susceptibility to ISP. If horizontal gene transfer is occurring more frequently between closely related strains of Staphylococcaceae, then their influence on phage susceptibility may be phylogenetically conserved and this variation may have been captured in the core genome phylogeny. In any case, our results indicate that more closely related Staphylococcaceae strains and species are more likely to share similar susceptibilities to phage infection, consistent with patterns seen in previous studies of animal hosts and viruses [17, 19–21, 26]. Additionally, while the evolutionary relationship between hosts had been shown to influence susceptibility within a single species of Staphylococcus [63], we have found that the relationship between hosts can explain susceptibility over a much broader phylogenetic scale, across genera. Further work should aim to characterise the mechanisms underpinning the differences in susceptibility reported here. In particular, the role of horizontal gene transfer [33], superinfection immunity [35], and recombination [34] in the resistance of Staphylococcaceae to phage infection, as well as the evolutionary scale at which the relationship between host species is unable to explain variation to susceptibility.
In this study, we used three methods to assess the susceptibility of Staphylococcaceae to a bacteriophage: plaque assays, optical density assays, and qPCR. Susceptibility measured by binary plaque assay, OD, and qPCR were strongly positively correlated. However, no correlation was seen between the continuous component of plaque assay and either other method. While OD and qPCR suggest a broad host range for ISP, plaque assays underestimated the susceptibility of the host panel, with only 63% of strains appearing permissive to infection. This discrepancy may in part be due to differences in the environmental conditions imposed by each assay on the bacteria and phage. Bacteria in the plaque assay are sessile and aerobic, whereas bacteria are planktonic and anaerobic in the OD and qPCR assays. For facultative anaerobes, such as Staphylococcus, anoxia has been shown to lead to the differential expression of over 200 genes influencing a multitude of biological processes [121–126]. While the ability of phage to infect Staphylococcaceae species under varying oxygen concentrations is understudied, it is likely that differences in oxygen availability would influence bacterial physiology in a way that may affect phage infection [125, 127]. Further investigation into the influence of oxygen availability on phage infection, and the physiological relevance of this to various clinical infections may improve our ability to effectively utilise ISP as a therapeutic and explain the lack of correlation between the plaque assays and OD and qPCR used here. Interestingly, we observe a strong positive correlation between OD and qPCR even though both assays are measuring different aspects of infection, with OD providing a measure of damage done to the host and qPCR a measure of virus replication and persistence. That these two infection traits are strongly correlated suggests that staphylococcal phage immunity is strongly tied to their ability to disrupt and prevent phage replication, as opposed to mitigating damage while tolerating phage replication.
Being able to predict and prevent emerging infectious diseases is a major goal of scientific research, and one of the first steps of that process is being able to predict whether a host will be susceptible to a novel pathogen [128]. While our phylogenetic models suggest that a high proportion of the variation in susceptibility between host species can be attributed to the relationship between hosts, they showed a limited ability to predict the susceptibility of an unknown host given only its relationship to other hosts and their susceptibilities. For example, S. simiae is isolated within the phylogeny, with few close relatives, and had a poorly estimated susceptibility due to the distance between it and its nearest neighbour. Several additions may improve the accuracy of predictions of susceptibility using these models. Firstly, improving the depth of our phylogeny–i.e., adding more strains and species that reduce the number of isolated strains on long evolutionary branches–would increase the number of observations that the model can use to predict susceptibility. Secondly, furthering our understanding of the mechanisms by which susceptibility can change in a non-phylogenetically tractable way (i.e., via HGT, superinfection immunity, or recombination) may improve our ability to predict susceptibility when phylogeny is not informative. Predictive models that incorporate the evolutionary relationships between hosts (core genome phylogeny) alongside additional information about the presence of anti-phage defence systems and prophages may be better able to predict susceptibility in instances where phylogeny alone is not informative. Alternative metrics, such as genome composition bias, have been shown to outperform phylogeny in the prediction of infection traits [129], and may make useful additions to future predictive models [128–130]. Understanding the complexity of factors that contribute to viral susceptibility, and how they interact with one another, is an important step towards the prediction of susceptibility in unknown hosts. An ability to predict the susceptibility of a host to novel pathogens, particularly how susceptible a host will be to infection, would allow us to better prioritise resources to the prevention and control of emerging infectious diseases in humans and wildlife.
Together, our results demonstrate that the bacterial host phylogeny is an important determinant of phage susceptibility and replication across novel hosts, and that the relationship between host species may be a useful addition to models aiming to predict virus host range both in the context of emerging infectious disease and phage therapy. Further work is required to understand the specific interactions underlying variation in ISP susceptibility across bacterial hosts; the relationship between host damage, virus replication, and virus persistence in this system; and how the patterns of phage susceptibility across bacterial phylogenies may vary under different infection conditions and contexts.
Supporting information
(DOCX)
Clonal complex and sequence type, where available, were determined using PubMLST [23]. S. aureus strains missing a ST are coagulase negative and therefore do not have an MLST scheme and the S. aureus strains with STs but no CCs are not similar enough to other strains to be assigned a CC.
(DOCX)
Estimates of repeatability are taken from model (2) and estimates of phylogenetic heritability (the proportion of phylogenetic and non-phylogenetic strain variation explained by the host phylogeny) and phylogenetic heritability of the total variance (the proportion of total variation explained by the host phylogeny) are taken from model (1). PA = plaque assay, CI = credible interval.
(DOCX)
Estimates of repeatability are taken from model (2) and estimates of phylogenetic heritability (the proportion of phylogenetic and non-phylogenetic strain variation explained by the host phylogeny) and phylogenetic heritability of the total variance (the proportion of total variation explained by the host phylogeny) are taken from model (1). PA = plaque assay, CI = credible interval.
(DOCX)
Numbers show the mean estimates for the correlation strength (r, white cells) and slope (β, grey cells) between pairs of methods, with 95% credible intervals (CIs) indicated in brackets. The slopes were calculated with columns as x and rows as y. Estimates with CIs that do not span zero are highlighted in bold. PA = plaque assay, *value on a probit scale.
(DOCX)
Numbers show the mean estimates for the correlation strength (r, white cells) and slope (β, grey cells) between pairs of methods, with 95% credible intervals (CIs) indicated in brackets. The slopes were calculated with columns as x and rows as y. Estimates with CIs that do not span zero are highlighted in bold. PA = plaque assay, *value on a probit scale.
(DOCX)
(DOCX)
(DOCX)
Estimates of repeatability are taken from model (2) and estimates of phylogenetic heritability (the proportion of phylogenetic and non-phylogenetic strain variation explained by the host phylogeny) and phylogenetic heritability of the total variance (the proportion of total variation explained by the host phylogeny) are taken from model (1). PA = plaque assay, CI = credible interval.
(DOCX)
Estimates of repeatability are taken from model (2) and estimates of phylogenetic heritability (the proportion of phylogenetic and non-phylogenetic strain variation explained by the host phylogeny) and phylogenetic heritability of the total variance (the proportion of total variation explained by the host phylogeny) are taken from model (1). PA = plaque assay, CI = credible interval.
(DOCX)
Numbers show the mean estimates for the correlation strength (r, white cells) and slope (β, grey cells) between pairs of methods, with 95% credible intervals (CIs) indicated in brackets. The slopes were calculated with columns as x and rows as y. Estimates with CIs that do not span zero are highlighted in bold. PA = plaque assay, *value on a probit scale.
(DOCX)
Numbers show the mean estimates for the correlation strength (r, white cells) and slope (β, grey cells) between pairs of methods, with 95% credible intervals (CIs) indicated in brackets. The slopes were calculated with columns as x and rows as y. Estimates with CIs that do not span zero are highlighted in bold. PA = plaque assay, *value on a probit scale.
(DOCX)
MDS visualization of tree distances when λ = 0. As the projection often requires that multiple trees are plotted at the same co-ordinates, contour lines are used to indicate the density of points.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
Staphylococcus isolates were kindly provided by Edward Feil and Kay Fountain from the University of Bath and Gavin Paterson at the University of Edinburgh. ISP was originally isolated by the Eliava Institute (Tbilisi, Georgia) and kindly provided by Jean-Paul Pirnay and Maya Merabishvili at the Queen Astrid Military Hospital (Brussels, Belgium). Genome sequencing was provided by MicrobesNG (http://www.microbesng.com).
Data Availability
All data files and R scripts used in this study are available in an online repository: https://doi.org/10.6084/m9.figshare.21642209.v1.
Funding Statement
The work described in our manuscript was funded by support from the following sources: S.K.W. is supported by a studentship funded by the Biotechnology and Biological Sciences Research Council (BBSRC) South West Biosciences Doctoral Training Partnership (BB/M009122/1); M.M. was supported in part by both a PhD from the Medical Research Council and the Raymond and Beverly Sackler Fund; L.A.W. is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (109385/Z/15/Z); J.D.H. is supported by NERC (NE/P000924/1); A.B. is supported by NERC (NE/S000771/1 and NE/V012347/1); and B.L. and R.M.I are supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (109356/Z/15/Z). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008. Feb;451(7181):990–3. doi: 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoelzer K, Shackelton LA, Holmes EC, Parrish CR. Within-Host Genetic Diversity of Endemic and Emerging Parvoviruses of Dogs and Cats. J Virol. 2008;82(22):11096–105. doi: 10.1128/JVI.01003-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp PM, Hahn BH. The evolution of HIV-1 and the origin of AIDS. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365(1552):2487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolhouse MEJ, Adair K, Brierley L. RNA viruses: a case study of the biology of emerging infectious diseases. 2018;1(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woolhouse M, Scott F, Hudson Z, Howey R, Chase-Topping M. Human viruses: discovery and emergence. Philos Trans R Soc Lond B Biol Sci. 2012. Oct;367(1604):2864–71. doi: 10.1098/rstb.2011.0354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. HIV/AIDS. Global Health Observatory Data. 2018. [Google Scholar]
- 7.Johnson NPAS, Mueller J. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med. 2002;76(1):105–15. doi: 10.1353/bhm.2002.0022 [DOI] [PubMed] [Google Scholar]
- 8.Murray CJ, Lopez AD, Chin B, Feehan D, Hill KH. Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918–20 pandemic: a quantitative analysis. Lancet. 2006;368(9554):2211–8. doi: 10.1016/S0140-6736(06)69895-4 [DOI] [PubMed] [Google Scholar]
- 9.Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza a epidemic. Nature. 2009;459(7250):1122–5. doi: 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- 10.Worobey M, Han GZ, Rambaut A. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature. 2014;508(7495):254–7. doi: 10.1038/nature13016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lytras S, Xia W, Hughes J, Jiang X, Robertson DL. The animal origin of SARS-CoV-2. Science (1979). 2021;373(6558):968–70. doi: 10.1126/science.abh0117 [DOI] [PubMed] [Google Scholar]
- 12.Holmes EC, Goldstein SA, Rasmussen AL, Robertson DL, Crits-Christoph A, Wertheim JO, et al. The origins of SARS-CoV-2: A critical review. Cell. 2021;184(19):4848–56. doi: 10.1016/j.cell.2021.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Vol. 26, Nature medicine. 2020. p. 450–2. doi: 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guth S, Visher E, Boots M, Brook CE. Host phylogenetic distance drives trends in virus virulence and transmissibility across the animal-human interface. Philosophical Transactions of the Royal Society B: Biological Sciences. 2019;374(1782). doi: 10.1098/rstb.2019.0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrell MJ, Davies TJ. Disease mortality in domesticated animals is predicted by host evolutionary relationships. Proc Natl Acad Sci U S A. 2019;116(16):7911–5. doi: 10.1073/pnas.1817323116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mollentze N, Streicker DG, Murcia PR, Hampson K, Biek R. Virulence mismatches in index hosts shape the outcomes of cross-species transmission. Proc Natl Acad Sci U S A. 2020;117(46):28859–66. doi: 10.1073/pnas.2006778117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longdon B, Hadfield JD, Day JP, Smith SCL, McGonigle JE, Cogni R, et al. The Causes and Consequences of Changes in Virulence following Pathogen Host Shifts. PLoS Pathog. 2015;11(3):1–18. doi: 10.1371/journal.ppat.1004728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perlman SJ, Jaenike J. Infection Success in Novel Hosts: An Experimental and Phylogenetic Study of Drosophila-parasitic Nematodes. Evolution (N Y). 2003;57(3):544–57. doi: 10.1111/j.0014-3820.2003.tb01546.x [DOI] [PubMed] [Google Scholar]
- 19.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin I V, McCracken GF, Rupprecht CE. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010. Aug;329(5992):676–9. doi: 10.1126/science.1188836 [DOI] [PubMed] [Google Scholar]
- 20.Imrie RM, Roberts KE, Longdon B. Between virus correlations in the outcome of infection across host species: Evidence of virus by host species interactions. Evol Lett. 2021;5(5):472–83. doi: 10.1002/evl3.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longdon B, Hadfield JD, Webster CL, Obbard DJ, Jiggins FM. Host phylogeny determines viral persistence and replication in novel hosts. PLoS Pathog. 2011;7(9). doi: 10.1371/journal.ppat.1002260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tinsley MC, Majerus MEN. Small steps or giant leaps for male-killers? Phylogenetic constraints to male-killer host shifts. BMC Evol Biol. 2007;7(1):1–10. doi: 10.1186/1471-2148-7-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell JA, Goldman-Huertas B, Moreau CS, Baldo L, Stahlhut JK, Werren JH, et al. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution. 2008/11/19. 2009. Mar;63(3):624–40. doi: 10.1111/j.1558-5646.2008.00579.x [DOI] [PubMed] [Google Scholar]
- 24.Engelstädter J, Hurst GDD. The dynamics of parasite incidence across host species. Evol Ecol. 2006;20(6):603–16. [Google Scholar]
- 25.De Vienne DM, Hood ME, Giraud T. Phylogenetic determinants of potential host shifts in fungal pathogens. J Evol Biol. 2009;22(12):2532–41. doi: 10.1111/j.1420-9101.2009.01878.x [DOI] [PubMed] [Google Scholar]
- 26.Gilbert GS, Webb CO. Phylogenetic signal in plant pathogen-host range. Proc Natl Acad Sci U S A. 2007;104(12):4979–83. doi: 10.1073/pnas.0607968104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faria NR, Suchard MA, Rambaut A, Streicker DG, Lemey P. Simultaneously reconstructing viral crossspecies transmission history and identifying the underlying constraints. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368(1614). doi: 10.1098/rstb.2012.0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinert LA, Welch JJ, Suchard MA, Lemey P, Rambaut A, Fitzgerald JR. Molecular dating of human-to-bovid host jumps by Staphylococcus aureus reveals an association with the spread of domestication. Biol Lett. 2012;8(5):829–32. doi: 10.1098/rsbl.2012.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webby RJ, Webster RG. Emergence of influenza A viruses. Philosophical Transactions of the Royal Society B: Biological Sciences. 2001;356(1416):1817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wille M, Lisovski S, Roshier D, Ferenczi M, Hoye BJ, Leen T, et al. Strong phylogenetic and ecological effects on host competency for avian influenza in Australian wild birds. bioRxiv. 2022. Jan 1;2022.02.14.480463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrow LN, McNew SM, Mitchell N, Galen SC, Lutz HL, Skeen H, et al. Deeply conserved susceptibility in a multi-host, multi-parasite system. Ecol Lett. 2019. Jun 1;22(6):987–98. doi: 10.1111/ele.13263 [DOI] [PubMed] [Google Scholar]
- 32.Martel A, Blooi M, Adriaensen C, Van Rooij P, Beukema W, Fisher MC, et al. Wildlife disease. Recent introduction of a chytrid fungus endangers Western Palearctic salamanders. Science. 2014. Oct;346(6209):630–1. doi: 10.1126/science.1258268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernheim A, Sorek R. The pan-immune system of bacteria: antiviral defence as a community resource. Nat Rev Microbiol. 2020;18(2):113–9. doi: 10.1038/s41579-019-0278-2 [DOI] [PubMed] [Google Scholar]
- 34.Sakoparnig T, Field C, van Nimwegen E. Whole genome phylogenies reflect the distributions of recombination rates for many bacterial species. Nourmohammad A, Walczak AM, editors. Elife. 2021;10:e65366. doi: 10.7554/eLife.65366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, et al. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016. Dec;10(12):2854–66. doi: 10.1038/ismej.2016.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dimitriu T, Marchant L, Buckling A, Raymond B. Bacteria from natural populations transfer plasmids mostly towards their kin. Proc Biol Sci. 2019. Jun;286(1905):20191110. doi: 10.1098/rspb.2019.1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung IY, Jang HJ, Bae HW, Cho YH. A phage protein that inhibits the bacterial ATPase required for type IV pilus assembly. Proc Natl Acad Sci U S A. 2014. Aug;111(31):11503–8. doi: 10.1073/pnas.1403537111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cumby N, Edwards AM, Davidson AR, Maxwell KL. The bacteriophage HK97 gp15 moron element encodes a novel superinfection exclusion protein. J Bacteriol. 2012. Sep;194(18):5012–9. doi: 10.1128/JB.00843-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uc-Mass A, Loeza EJ, de la Garza M, Guarneros G, Hernández-Sánchez J, Kameyama L. An orthologue of the cor gene is involved in the exclusion of temperate lambdoid phages. Evidence that Cor inactivates FhuA receptor functions. Virology. 2004. Nov;329(2):425–33. doi: 10.1016/j.virol.2004.09.005 [DOI] [PubMed] [Google Scholar]
- 40.Susskind MM, Wright A, Botstein D. Superinfection exclusion by P22 prophage in lysogens of Salmonella typhimurium. IV. Genetics and physiology of sieB exclusion. Virology. 1974. Dec;62(2):367–84. doi: 10.1016/0042-6822(74)90399-7 [DOI] [PubMed] [Google Scholar]
- 41.Sun X, Göhler A, Heller KJ, Neve H. The ltp gene of temperate Streptococcus thermophilus phage TP-J34 confers superinfection exclusion to Streptococcus thermophilus and Lactococcus lactis. Virology. 2006. Jun;350(1):146–57. doi: 10.1016/j.virol.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 42.Kuntová L, Mašlaňová I, Obořilová R, Šimečková H, Finstrlová A, Bárdy P, et al. Staphylococcus aureus Prophage-Encoded Protein Causes Abortive Infection and Provides Population Immunity against Kayviruses. mBio. 2023. Feb;e0249022. doi: 10.1128/mbio.02490-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samson JE, Magadán AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. 2013. Oct;11(10):675–87. doi: 10.1038/nrmicro3096 [DOI] [PubMed] [Google Scholar]
- 44.World Health Organization. New report calls for urgent action to avert antimicrobial resistance crisis. 2019. [Google Scholar]
- 45.AMR Industry Alliance. 2020 progress report. 2020. [Google Scholar]
- 46.Perros M. A sustainable model for antibiotics. Science (1979). 2015;347(6226):1062–4. [DOI] [PubMed] [Google Scholar]
- 47.Bhargava K, Nath G, Bhargava A, Aseri GK, Jain N. Phage therapeutics: from promises to practices and prospectives. Appl Microbiol Biotechnol. 2021;9047–67. doi: 10.1007/s00253-021-11695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fish R, Kutter E, Wheat G, Blasdel B, Kutateladze M, Kuhl S. Bacteriophage treatment of intransigent Diabetic toe ulcers: A case series. J Wound Care. 2016;25:S27–33.26949862 [Google Scholar]
- 49.Lin DM, Koskella B, Lin HC. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther. 2017;8(3):162. doi: 10.4292/wjgpt.v8.i3.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Nieuwenhuyse B, Van der Linden D, Chatzis O, Lood C, Wagemans J, Lavigne R, et al. Bacteriophage-antibiotic combination therapy against extensively drug-resistant Pseudomonas aeruginosa infection to allow liver transplantation in a toddler. Nat Commun. 2022;13(1):5725. doi: 10.1038/s41467-022-33294-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kauffman KM, Chang WK, Brown JM, Hussain FA, Yang J, Polz MF, et al. Resolving the structure of phage-bacteria interactions in the context of natural diversity. Nat Commun [Internet]. 2022. Dec 1 [cited 2023 Mar 20];13(1). Available from: https://pubmed.ncbi.nlm.nih.gov/35042853/ doi: 10.1038/s41467-021-27583-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsieh SE, Lo HH, Chen ST, Lee MC, Tseng YH. Wide host range and strong lytic activity of Staphylococcus aureus lytic phage Stau2. Appl Environ Microbiol. 2011. Feb;77(3):756–61. doi: 10.1128/AEM.01848-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Göller PC, Elsener T, Lorgé D, Radulovic N, Bernardi V, Naumann A, et al. Multi-species host range of staphylococcal phages isolated from wastewater. Nat Commun. 2021. Dec 1;12(1). doi: 10.1038/s41467-021-27037-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piel D, Bruto M, Labreuche Y, Blanquart F, Goudenège D, Barcia-Cruz R, et al. Phage-host coevolution in natural populations. Nat Microbiol [Internet]. 2022;2022(7):1075–86. Available from: https://hal.science/hal-03738913 doi: 10.1038/s41564-022-01157-1 [DOI] [PubMed] [Google Scholar]
- 55.Wendling CC, Goehlich H, Roth O. The structure of temperate phage–bacteria infection networks changes with the phylogenetic distance of the host bacteria. Biol Lett [Internet]. 2018;14(11). Available from: doi: 10.1098/rsbl.2018.0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Flaherty S, Ross RP, Meaney W, Fitzgerald GF, Elbreki MF, Coffey A. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Appl Environ Microbiol. 2005. Apr;71(4):1836–42. doi: 10.1128/AEM.71.4.1836-1842.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis CM, Ruest MK, Cole JH, Dennis JJ. The Isolation and Characterization of a Broad Host Range Bcep22-like Podovirus JC1. Viruses [Internet]. 2022. May 1 [cited 2023 Mar 20];14(5). Available from: /pmc/articles/PMC9144972/ doi: 10.3390/v14050938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao C, Yi Z, Zhang Y, Wang Y, Zhu H, Afayibo DJA, et al. Characterization of a Broad-Host-Range Lytic Phage SHWT1 Against Multidrug-Resistant Salmonella and Evaluation of Its Therapeutic Efficacy in vitro and in vivo. Front Vet Sci [Internet]. 2021. Jun 10 [cited 2023 Mar 20];8:683853. Available from: /pmc/articles/PMC8222671/ doi: 10.3389/fvets.2021.683853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alves DR, Gaudion A, Bean JE, Perez Esteban P, Arnot TC, Harper DR, et al. Combined Use of Bacteriophage K and a Novel Bacteriophage To Reduce Staphylococcus aureus Biofilm Formation. Appl Environ Microbiol [Internet]. 2014. [cited 2023 Mar 20];80(21):6694. Available from: /pmc/articles/PMC4249044/ doi: 10.1128/AEM.01789-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duyvejonck H, Merabishvili M, Pirnay JP, De Vos D, Verbeken G, Van Belleghem J, et al. Development of a qPCR platform for quantification of the five bacteriophages within bacteriophage cocktail 2 (BFC2). Sci Rep [Internet]. 2019;9(1):1–10. Available from: 10.1038/s41598-019-50461-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson B, Rashid MH, Carter C, Pasternack G, Rajanna C, Revazishvili T, et al. Enumeration of bacteriophage particles. Bacteriophage [Internet]. 2011;1(2):86–93. Available from: 10.4161/bact.1.2.15456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan Mirzaei M, Nilsson AS. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One. 2015;10(3):e0118557. doi: 10.1371/journal.pone.0118557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moller AG, Winston K, Ji S, Wang J, Hargita Davis MN, Solís-Lemus CR, et al. Genes Influencing Phage Host Range in Staphylococcus aureus on a Species-Wide Scale. mSphere [Internet]. 2021. Feb 24 [cited 2023 Mar 20];6(1). Available from: https://pubmed.ncbi.nlm.nih.gov/33441407/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A. 2005/03/23. 2005. Apr 5;102(14):5174–9. doi: 10.1073/pnas.0501140102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, et al. Quality-Controlled Small-Scale Production of a Well-Defined Bacteriophage Cocktail for Use in Human Clinical Trials. PLoS One [Internet]. 2009. Mar 20 [cited 2023 Mar 21];4(3):4944. Available from: /pmc/articles/PMC2654153/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Nieuwenhuyse B, Van der Linden D, Chatzis O, Lood C, Wagemans J, Lavigne R, et al. Bacteriophage-antibiotic combination therapy against extensively drug-resistant Pseudomonas aeruginosa infection to allow liver transplantation in a toddler. Nat Commun [Internet]. 2022. Dec 1 [cited 2023 Apr 4];13(1). Available from: /pmc/articles/PMC9523064/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandersteegen K, Mattheus W, Ceyssens PJ, Bilocq F, de Vos D, Pirnay JP, et al. Microbiological and molecular assessment of bacteriophage ISP for the control of Staphylococcus aureus. PLoS One. 2011;6(9). doi: 10.1371/journal.pone.0024418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Synnott AJ, Kuang Y, Kurimoto M, Yamamichi K, Iwano H, Tanji Y. Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl Environ Microbiol. 2009. Jul;75(13):4483–90. doi: 10.1128/AEM.02641-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alves DR, Gaudion A, Bean JE, Perez Esteban P, Arnot TC, Harper DR, et al. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl Environ Microbiol. 2014. Nov;80(21):6694–703. doi: 10.1128/AEM.01789-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gutiérrez D, Briers Y, Rodríguez-Rubio L, Martínez B, Rodríguez A, Lavigne R, et al. Role of the Pre-neck Appendage Protein (Dpo7) from Phage vB_SepiS-phiIPLA7 as an Anti-biofilm Agent in Staphylococcal Species. Front Microbiol. 2015;6:1315. doi: 10.3389/fmicb.2015.01315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verstappen KM, Tulinski P, Duim B, Fluit AC, Carney J, van Nes A, et al. The Effectiveness of Bacteriophages against Methicillin-Resistant Staphylococcus aureus ST398 Nasal Colonization in Pigs. PLoS One. 2016;11(8):e0160242. doi: 10.1371/journal.pone.0160242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wills QF, Kerrigan C, Soothill JS. Experimental bacteriophage protection against Staphylococcus aureus abscesses in a rabbit model. Antimicrob Agents Chemother. 2005. Mar;49(3):1220–1. doi: 10.1128/AAC.49.3.1220-1221.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsuzaki S, Yasuda M, Nishikawa H, Kuroda M, Ujihara T, Shuin T, et al. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J Infect Dis. 2003. Feb;187(4):613–24. doi: 10.1086/374001 [DOI] [PubMed] [Google Scholar]
- 74.Chatterjee AN, Mirelman D, Singer HJ, Park JT. Properties of a novel pleiotropic bacteriophage-resistant mutant of Staphylococcus aureus H. J Bacteriol. 1969. Nov;100(2):846–53. doi: 10.1128/jb.100.2.846-853.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chatterjee AN. Use of bacteriophage-resistant mutants to study the nature of the bacteriophage receptor site of Staphylococcus aureus. J Bacteriol. 1969. May;98(2):519–27. doi: 10.1128/jb.98.2.519-527.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shaw DR, Chatterjee AN. O-Acetyl groups as a component of the bacteriophage receptor on Staphylococcus aureus cell walls. J Bacteriol. 1971. Oct;108(1):584–5. doi: 10.1128/jb.108.1.584-585.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shaw DR, Mirelman D, Chatterjee AN, Park JT. Ribitol teichoic acid synthesis in bacteriophage-resistant mutants of Staphylococcus aureus H. J Biol Chem. 1970. Oct;245(19):5101–6. [PubMed] [Google Scholar]
- 78.Brown S, Santa Maria JPJ, Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–36. doi: 10.1146/annurev-micro-092412-155620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xia G, Corrigan RM, Winstel V, Goerke C, Gründling A, Peschel A. Wall teichoic Acid-dependent adsorption of staphylococcal siphovirus and myovirus. J Bacteriol. 2011. Aug;193(15):4006–9. doi: 10.1128/JB.01412-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilkinson BJ, Holmes KM. Staphylococcus aureus cell surface: capsule as a barrier to bacteriophage adsorption. Infect Immun. 1979. Feb;23(2):549–52. doi: 10.1128/iai.23.2.549-552.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ohshima Y, Schumacher-Perdreau F, Peters G, Pulverer G. The role of capsule as a barrier to bacteriophage adsorption in an encapsulated Staphylococcus simulans strain. Med Microbiol Immunol. 1988;177(4):229–33. doi: 10.1007/BF00211222 [DOI] [PubMed] [Google Scholar]
- 82.Nordström K, Forsgren A. Effect of protein A on adsorption of bacteriophages to Staphylococcus aureus. J Virol. 1974;14(2):198–202. doi: 10.1128/JVI.14.2.198-202.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sadykov MR. Restriction–modification systems as a barrier for genetic manipulation of Staphylococcus aureus. In: The genetic manipulation of staphylococci. Springer; 2014. p. 9–23. [DOI] [PubMed] [Google Scholar]
- 84.Monk IR, Foster TJ. Genetic manipulation of Staphylococci-breaking through the barrier. Front Cell Infect Microbiol. 2012;2:49. doi: 10.3389/fcimb.2012.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Costa SK, Donegan NP, Corvaglia AR, François P, Cheung AL. Bypassing the Restriction System To Improve Transformation of Staphylococcus epidermidis. J Bacteriol. 2017. Aug;199(16). doi: 10.1128/JB.00271-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang S, Liu J, Shao F, Wang P, Duan G, Yang H. Analysis of the features of 45 identified CRISPR loci in 32 Staphylococcus aureus. Biochem Biophys Res Commun. 2015. Aug;464(3):894–900. doi: 10.1016/j.bbrc.2015.07.062 [DOI] [PubMed] [Google Scholar]
- 87.Li Q, Xie X, Yin K, Tang Y, Zhou X, Chen Y, et al. Characterization of CRISPR-Cas system in clinical Staphylococcus epidermidis strains revealed its potential association with bacterial infection sites. Microbiol Res. 2016. Dec;193:103–10. doi: 10.1016/j.micres.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 88.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007. Mar;315(5819):1709–12. doi: 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 89.Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014. Apr;54(2):234–44. doi: 10.1016/j.molcel.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, et al. TimeTree 5: An Expanded Resource for Species Divergence Times. Mol Biol Evol. 2022. Aug 1;39(8):msac174. doi: 10.1093/molbev/msac174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruijter JM, Thygesen HH, Schoneveld OJLM, Das AT, Berkhout B, Lamers WH. Factor correction as a tool to eliminate between-session variation in replicate experiments: Application to molecular biology and retrovirology. Retrovirology. 2006;3(May). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ruijter JM, Ruiz Villalba A, Hellemans J, Untergasser A, van den Hoff MJB. Removal of between-run variation in a multi-plate qPCR experiment. Biomol Detect Quantif. 2015;5:10–4. doi: 10.1016/j.bdq.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012. Aug;29(8):1969–73. doi: 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014. Jul;30(14):2068–9. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 95.Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020. Jul;21(1):1–21. doi: 10.1186/s13059-020-02090-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Vol. 23, Molecular biology and evolution. United States; 2006. p. 7–9. doi: 10.1093/molbev/msj021 [DOI] [PubMed] [Google Scholar]
- 97.Rambaut A, Suchard M, Xie D, Drummond A. Tracer v1.6. 2014. [Google Scholar]
- 98.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019. Feb 1;35(3):526–8. doi: 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
- 99.Hadfield JD. MCMC Methods for Multi-Response Generalized Linear Mixed Models: The MCMCglmm R Package. J Stat Softw. 2010. Feb 2;33(2 SE-Articles):1–22.20808728 [Google Scholar]
- 100.Razzaghi M. The Probit Link Function in Generalized Linear Models for Data Mining Applications. Journal of Modern Applied Statistical Methods [Internet]. 2013. [cited 2023 Apr 3];12(1):164–9. Available from: http://digitalcommons.wayne.edu/jmasmhttp://digitalcommons.wayne.edu/jmasm/vol12/iss1/19. [Google Scholar]
- 101.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits [Internet]. Genetics and Analysis of Quantitative Traits. Sinauer Associates; 1998. [cited 2023 Mar 23]. Available from: papers2://publication/uuid/6E5E388C-7C1E-49F6-937F-13FB4B8E5A68. [Google Scholar]
- 102.Falconer DS. Introduction to quantitative genetics. Pearson Education India; 1996. [Google Scholar]
- 103.Freckleton RP, Harvey PH, Pagel M. Phylogenetic Analysis and Comparative Data: A Test and Review of Evidence. Am Nat [Internet]. 2002. Dec 1;160(6):712–26. Available from: doi: 10.1086/343873 [DOI] [PubMed] [Google Scholar]
- 104.Housworth EA, Martins EP, Lynch M. The Phylogenetic Mixed Model. American Naturalist. 2004;163(1):84–96. [DOI] [PubMed] [Google Scholar]
- 105.Pagel M. Inferring the historical patterns of biological evolution. Nature [Internet]. 1999. Oct 28 [cited 2023 Mar 23];401(6756):877–84. Available from: https://pubmed.ncbi.nlm.nih.gov/10553904/. doi: 10.1038/44766 [DOI] [PubMed] [Google Scholar]
- 106.Hastie T, Tibshirani R, Friedman J. The Elements Of Statistical Learning. Aug, Springer. 2001. Jan 1;1. [Google Scholar]
- 107.Crawley M. Statistics: An Introduction Using R. 2005. Jan 1; [Google Scholar]
- 108.Poulin R, Krasnov BR, Mouillot D, Thieltges DW. The comparative ecology and biogeography of parasites. Philosophical Transactions of the Royal Society B: Biological Sciences. 2011. Aug 27;366(1576):2379–90. doi: 10.1098/rstb.2011.0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hussain FA, Dubert J, Elsherbini J, Murphy M, VanInsberghe D, Arevalo P, et al. Rapid evolutionary turnover of mobile genetic elements drives bacterial resistance to phages. Science [Internet]. 2021. Oct 22 [cited 2023 Mar 20];374(6566):488–92. Available from: https://pubmed.ncbi.nlm.nih.gov/34672730/. doi: 10.1126/science.abb1083 [DOI] [PubMed] [Google Scholar]
- 110.O’Flaherty S, Ross RP, Meaney W, Fitzgerald GF, Elbreki MF, Coffey A. Potential of the Polyvalent Anti-Staphylococcus Bacteriophage K for Control of Antibiotic-Resistant Staphylococci from Hospitals. Appl Environ Microbiol [Internet]. 2005. Apr [cited 2023 Mar 20];71(4):1836. Available from: /pmc/articles/PMC1082512/. doi: 10.1128/AEM.71.4.1836-1842.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hsieh SE, Lo HH, Chen ST, Lee MC, Tseng YH. Wide Host Range and Strong Lytic Activity of Staphylococcus aureus Lytic Phage Stau2. Appl Environ Microbiol [Internet]. 2011. Feb [cited 2023 Mar 20];77(3):756. Available from: /pmc/articles/PMC3028737/. doi: 10.1128/AEM.01848-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg Infect Dis. 2005;11(12):1842–7. doi: 10.3201/eid1112.050997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Corvaglia AR, François P, Hernandez D, Perron K, Linder P, Schrenzel J. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc Natl Acad Sci U S A [Internet]. 2010. Jun 29 [cited 2023 Mar 23];107(26):11954–8. Available from: /pmc/articles/PMC2900679/. doi: 10.1073/pnas.1000489107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Waldron DE, Lindsay JA. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol [Internet]. 2006. Aug [cited 2023 Mar 23];188(15):5578–85. Available from: https://pubmed.ncbi.nlm.nih.gov/16855248/. doi: 10.1128/JB.00418-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McCarthy AJ, Witney AA, Lindsay JA. Staphylococcus aureus Temperate Bacteriophage: Carriage and Horizontal Gene Transfer is Lineage Associated. Front Cell Infect Microbiol [Internet]. 2012. [cited 2023 Mar 23];2:6. Available from: /pmc/articles/PMC3417521/. doi: 10.3389/fcimb.2012.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Koonin E V, Makarova KS, Wolf YI. Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu Rev Microbiol. 2017. Sep;71:233–61. doi: 10.1146/annurev-micro-090816-093830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oliveira PH, Touchon M, Rocha EPC. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42(16):10618–31. doi: 10.1093/nar/gku734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015;13(11):722–36. doi: 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006. Jun;62(6):718–29. doi: 10.1007/s00239-005-0223-z [DOI] [PubMed] [Google Scholar]
- 120.Makarova KS, Wolf YI, Koonin E V. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013. Apr;41(8):4360–77. doi: 10.1093/nar/gkt157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chan PF, Foster SJ. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325–4. Microbiology (Reading). 1998. Sep;144 (Pt 9):2469–79. doi: 10.1099/00221287-144-9-2469 [DOI] [PubMed] [Google Scholar]
- 122.Kass EH, Kendrick MI, Tsai YC, Parsonnet J. Interaction of magnesium ion, oxygen tension, and temperature in the production of toxic-shock-syndrome toxin-1 by Staphylococcus aureus. J Infect Dis. 1987. Apr;155(4):812–5. doi: 10.1093/infdis/155.4.812 [DOI] [PubMed] [Google Scholar]
- 123.Ross RA, Onderdonk AB. Production of toxic shock syndrome toxin 1 by Staphylococcus aureus requires both oxygen and carbon dioxide. Infect Immun. 2000. Sep;68(9):5205–9. doi: 10.1128/IAI.68.9.5205-5209.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J Bacteriol. 2004. Apr;186(8):2430–8. doi: 10.1128/JB.186.8.2430-2438.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fuchs S, Pane J, Kohler C, Hecker M, Engelmann S. Anaerobic Gene Expression in Staphylococcus aureus. 2007;189(11):4275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999. Oct;67(10):5427–33. doi: 10.1128/IAI.67.10.5427-5433.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hern S, Vives MJ. Phages in Anaerobic Systems. 2020;1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Carlson CJ, Farrell MJ, Grange Z, Han BA, Mollentze N, Phelan AL, et al. The future of zoonotic risk prediction. Philos Trans R Soc Lond B Biol Sci. 2021. Nov;376(1837):20200358. doi: 10.1098/rstb.2020.0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mollentze N, Babayan SA, Streicker DG. Identifying and prioritizing potential human-infecting viruses from their genome sequences. PLoS Biol. 2021. Sep;19(9):e3001390. doi: 10.1371/journal.pbio.3001390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Babayan SA, Orton RJ, Streicker DG. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science. 2018. Nov;362(6414):577–80. doi: 10.1126/science.aap9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Clonal complex and sequence type, where available, were determined using PubMLST [23]. S. aureus strains missing a ST are coagulase negative and therefore do not have an MLST scheme and the S. aureus strains with STs but no CCs are not similar enough to other strains to be assigned a CC.
(DOCX)
Estimates of repeatability are taken from model (2) and estimates of phylogenetic heritability (the proportion of phylogenetic and non-phylogenetic strain variation explained by the host phylogeny) and phylogenetic heritability of the total variance (the proportion of total variation explained by the host phylogeny) are taken from model (1). PA = plaque assay, CI = credible interval.
(DOCX)
Estimates of repeatability are taken from model (2) and estimates of phylogenetic heritability (the proportion of phylogenetic and non-phylogenetic strain variation explained by the host phylogeny) and phylogenetic heritability of the total variance (the proportion of total variation explained by the host phylogeny) are taken from model (1). PA = plaque assay, CI = credible interval.
(DOCX)
Numbers show the mean estimates for the correlation strength (r, white cells) and slope (β, grey cells) between pairs of methods, with 95% credible intervals (CIs) indicated in brackets. The slopes were calculated with columns as x and rows as y. Estimates with CIs that do not span zero are highlighted in bold. PA = plaque assay, *value on a probit scale.
(DOCX)
Numbers show the mean estimates for the correlation strength (r, white cells) and slope (β, grey cells) between pairs of methods, with 95% credible intervals (CIs) indicated in brackets. The slopes were calculated with columns as x and rows as y. Estimates with CIs that do not span zero are highlighted in bold. PA = plaque assay, *value on a probit scale.
(DOCX)
(DOCX)
(DOCX)
Estimates of repeatability are taken from model (2) and estimates of phylogenetic heritability (the proportion of phylogenetic and non-phylogenetic strain variation explained by the host phylogeny) and phylogenetic heritability of the total variance (the proportion of total variation explained by the host phylogeny) are taken from model (1). PA = plaque assay, CI = credible interval.
(DOCX)
Estimates of repeatability are taken from model (2) and estimates of phylogenetic heritability (the proportion of phylogenetic and non-phylogenetic strain variation explained by the host phylogeny) and phylogenetic heritability of the total variance (the proportion of total variation explained by the host phylogeny) are taken from model (1). PA = plaque assay, CI = credible interval.
(DOCX)
Numbers show the mean estimates for the correlation strength (r, white cells) and slope (β, grey cells) between pairs of methods, with 95% credible intervals (CIs) indicated in brackets. The slopes were calculated with columns as x and rows as y. Estimates with CIs that do not span zero are highlighted in bold. PA = plaque assay, *value on a probit scale.
(DOCX)
Numbers show the mean estimates for the correlation strength (r, white cells) and slope (β, grey cells) between pairs of methods, with 95% credible intervals (CIs) indicated in brackets. The slopes were calculated with columns as x and rows as y. Estimates with CIs that do not span zero are highlighted in bold. PA = plaque assay, *value on a probit scale.
(DOCX)
MDS visualization of tree distances when λ = 0. As the projection often requires that multiple trees are plotted at the same co-ordinates, contour lines are used to indicate the density of points.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All data files and R scripts used in this study are available in an online repository: https://doi.org/10.6084/m9.figshare.21642209.v1.