Abstract
Plants’ requirement of Phosphorus (P) as an essential macronutrient is obligatory for their normal growth and metabolism. Besides restricting plants’ primary growth, P depletion affects both primary and secondary metabolism and leads to altered levels of sugars, metabolites, amino acids, and other secondary compounds. Such metabolic shifts help plants optimize their metabolism and growth under P limited conditions. Under P deprivation, both sugar levels and their mobilization change that influences the expression of Pi starvation-inducible genes. Increased sugar repartitioning from shoot to root help root growth and organic acids secretion that in turn promotes phosphate (Pi) uptake from the soil. Other metabolic changes such as lipid remodeling or P reallocation from older to younger leaves release the P from its bound forms in the cell. In this review, we summarize the metabolic footprinting of Pi-starved plants with respect to the benefits offered by such metabolic changes to intracellular Pi homeostasis.
Keywords: Acid phosphatases, Metabolic adaptations, Mineral nutrition, Organic acids, Phosphorus starvation, Root system architecture, Sucrose
Introduction
Phosphorus (P) is an elementary macronutrient for normal plant growth and development. Roots absorb P in its inorganic phosphate (Pi) or orthophosphate form. P participates in virtually all metabolic processes and contributes to about 0.2% of the dry weight of plants (Sulieman and Tran 2017). In the cellular context, its unimpeded availability is crucial to structural integrity (phospholipids in the cell membrane), nucleic acids (DNA and RNA) conservation, energy production (ATP), phosphorylated metabolites production, and nitrogen metabolism (Lambers and Plaxton 2015). Due to its widespread involvement in cellular activities, Pi starvation often compromises plant growth and hampers crop production, yield, and quality of products (Srivastava et al. 2021a). For example, Li et al. (2021) recently demonstrated the impact of P starvation on tomato fruit quality. They reported enhanced lycopene levels but lower starch and glucose levels in the fruits of Pi-starved plants than the Pi-sufficient counterparts. Application of Pi fertilizers in farmlands is a popular approach to replenish soil P levels. Unfortunately, the rock phosphate reserves, mined to produce chemical Pi fertilizers, are a finite resource and are expected to persist for only a few hundred years (Cordell et al. 2009; Walan et al. 2014). Although the several decades-long high fertilization practices in agriculture have yielded enhanced crop yields globally, the prolonged use of Pi-fertilizers has its side effects as it led to the development of high-yielding crop varieties with poor nutrient acquisition and use efficiency.
Due to its highly reactive nature, Pi remains unstable in the rhizosphere and is rapidly converted into its inorganic mineral complexes (mineral-P) with metal ions such as aluminium (Al), iron (Fe), or calcium (Ca) in a pH-dependent manner. Additionally, Pi is taken up by soil microorganisms and converted into organic Pi-monoesters (Po) compounds which may constitute up to 80% of total soil P (Feder et al. 2020). Only 20–30% of the externally applied P, in the form of chemical Pi fertilizers, is estimated to remain in bioavailable form. The remaining P is precipitated in the soils in its inorganic forms and Po or runs off into aquatic ecosystems, creating a perpetual Pi starvation situation for plants. Enzymes such as acid phosphatases, secreted by plants or soil microbes, are known to mineralize mineral-P complexes or Po to release Pi in the soils, followed by its uptake by the root epidermal cells through their plasma-membrane localized Pi transporters (Richardson et al. 2011). To enhance plant efficiency, plants can either increase PAE (phosphorus-acquisition-efficiency) or PUE (phosphorus-use-efficiency). PAE is defined as the capability of plant roots to obtain the freely available Pi from the soil, while PUE is the internal utilization and distribution inside the plant. Experts have recommended improving these traits to reduce Pi fertilizers usage in farming systems.
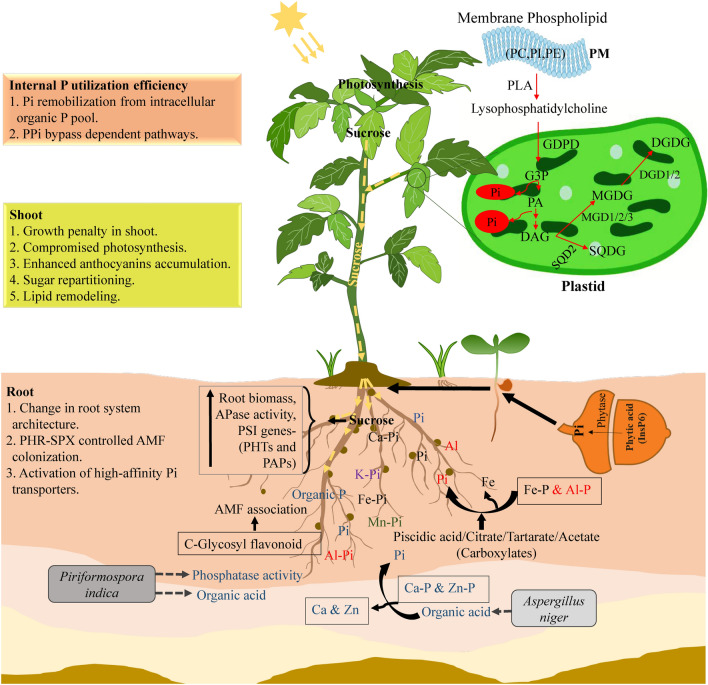
The adaptive strategy plants adopt to mitigate Pi starvation is called Pi-starvation response (PSR). It comprises morphological, physiological, biochemical, and molecular alterations to adjust their growth and survival under Pi-deficient environments. At the morphological levels, stunted shoot growth, reprogrammed root system architecture (RSA), increased root-to-shoot ratio, and longer and denser root hairs are the characteristic features of Pi-deficient plants (Fig. 1). These Pi-starved plants also develop denser and shorter lateral roots with shallower root growth angles to improve PAE (Strock et al. 2018). Cluster root (CR) formation in the members of several different families (e.g., Proteaceae, Casuarinaceae, Fabaceae, and Myricaceae) is another magnificent strategy to mobilize Pi from its enriched patches in the soil (Peret et al. 2014). Overall, the reprogrammed RSA is meant to increase the nutrient-scavenging ability of roots from topsoil layers, which is a significant source of Po due to the contribution of the decaying organic matter.
Fig. 1.
Schematic representation of the adaptive response of plants under P-limited conditions. Plants undergo morpho-physiological, biochemical, and molecular changes to cope with the depleted Pi levels. Altered root system architecture and its interaction with different microbial species, such as fungi (including mycorrhizal association) and plant growth-promoting rhizobacteria, help in increasing Pi accessibility for root uptake in the soil. The release of organic acids, phosphatases, and ribonucleases also facilitates the hydrolysis of organic P to release Pi in the rhizosphere. Membrane phospholipids are converted into glycolipids to free Pi from organic P compounds. Enhanced sucrose mobilization from shoots to roots supporting the reprogramming of root system architecture, the secretory acid phosphatase activity, and the robust activation of sucrose-dependent Pi starvation-induced genes such as high-affinity Pi transporters and purple acid phosphatases jointly contribute to the plant adaptation to low P availability. AMF Arbuscular mycorrhizal fungi, APase acidic phosphatase, DAG Diacylglycerol, GDPD Glycerophosphodiester phosphodiesterase, G3P Glycerol-3-phosphate, GLs Glycerolipids, P Phosphorus, PA Phosphatidic acid, PAPs Purple acid phosphatases, PC Phosphatidylcholine, PE Phosphatidylethanolamine, PHR Phosphate starvation response, PHTs Phosphate transporters, Pi Inorganic phosphate, PI Phosphatidylinositol, PLA Phospholipase A, PLs Phospholipids, Po Organic P, PM Plasma membrane, PPi Pyrophosphate, PSI Phosphate starvation-induced, MGDG Monogalactosyl-diacylglycerol, SPX; SYG1, PHO81, and Xpr1, SQDG Sulfoquinovosyl-diacylglycerol
Regarding cellular Po pools, ribosomal RNAs have the largest share (~ 50%), followed by P-lipids, P-esters, DNA, RNA, and phosphoproteins (Wang et al. 2021). Plant metabolism is greatly affected by depleting Pi levels in the cell. When Pi is scarce, intracellular ATP, ADP, and associated nucleoside levels decline, and Pi allocation between cytosol and organelles changes (Wang et al. 2021). Vacuole is the primary reservoir of the Pi pool and can store up to 95% of the total cellular Pi of a Pi-replete plant cell (Yang et al. 2017). The severe depletion of cytoplasmic Pi levels causes vacuolar Pi to be mobilized to the cytosol (Liu et al. 2015). Such mobilization is mediated by vacuolar phosphate transporters (VPTs) activation. In rice, OsSPX-MSF3 (SYG1, PHO81, and Xpr1-Major Facilitator Superfamily 3) seems to facilitate Pi efflux from the vacuole to the cytoplasm (Liu et al. 2016). Overexpression of this gene led to lower Pi retention in the vacuole, causing a low vacuolar-to-cytosolic Pi ratio in transgenic rice plants. Luan et al. (2019) recently reported a more dominant role of VPT1 over VPT3 in maintaining cytosol-to-vacuole Pi partitioning in Arabidopsis. The loss of function of these genes in the leaves disturbed the Pi partitioning to different organs as excessive Pi moves to flowers, most likely at the expense of leaf vacuolar Pi, causing a significant decline in the leaf’s Pi levels in the mutant. Besides VPTs, PHT members involved in Pi transport across the membranes of other organelles have also been identified. For example, PHT2;1 facilitates Pi import in chloroplasts. Mutation in this low-affinity transporter caused reduced intracellular Pi levels, disturbing plant growth and photosynthetic rates in the ospht2;1 plants. Interestingly, the mutant plants accumulated lower levels of flavonoids and their precursor, phenylalanine (Liu et al. 2020). AtPHT4;1, a thylakoid Pi transporter, is vital for ATP synthesis and plant growth in Arabidopsis seedlings (Karlsson et al. 2015). Another PHT, AtPHT4;6, is localized to the Golgi apparatus and facilitates Pi efflux for sugar-nucleotide metabolism (Cubero et al. 2009). Similarly, PHT3 members facilitate Pi transport across inner mitochondrial membranes for ATP synthesis (Zhu and Miao 2012). Jia et al. (2015) reported that mitochondrial membrane-localized phosphate transporter (AtMPT3) plays a critical role in ATP synthesis, ROS accumulation, and programmed cell death (PCD) and thus regulates the growth and development of Arabidopsis plants.
Under chronic Pi starvation conditions, when vacuolar and cytoplasmic Pi pools deplete considerably, the cellular Po pool is dramatically reduced (Del-Saz et al. 2018; Luo et al. 2020). It is anticipated that under such extreme conditions, Pi homeostasis becomes incompatible with plant PSR, and a more pronounced response to free Pi from additional Po sources, such as membrane PLs, is initiated (Lambers and Plaxton 2015). Thus, biochemical events such as the conversion of phospholipids to galactolipids in cell membranes, Pi-scavenging from nucleic acids, remobilization of Pi from older to younger organs, differential allocation of the available Pi to different organelles, ATP-dependent bypass enzymes and repartitioning of photosynthesis-derived sugars from shoots to roots are central to the metabolic adaptations in Pi-deficient plants. All these changes are well coordinated in plants and help them adopt an energy-saving strategy to cope with Pi starvation. This review summarizes these well-coordinated metabolic events underlying the plants’ response to Pi starvation.
During Pi starvation, the primary metabolic pathways undergo a transition toward Pi-independent processes
Primary metabolism is necessary for plant survival and growth, including photosynthesis, respiration, and synthesizing essential molecules such as AAs, sugars, and lipids. Conversely, secondary metabolism involves specialized pathways that allow plants to produce various bioactive compounds that contribute to their adaptation to different environments. One notable effect of Pi starvation is the re-prioritization of Pi from its conjugates, resulting in a change in phosphorylated metabolites. Pi starvation also impacts numerous energy-intensive activities, such as carbon metabolism, by decreasing the proportion of phosphorylated to non-phosphorylated polysaccharides (Ashihara et al. 1988). Substantial decreases in cellular Pi and adenylate concentrations activate alternative glycolytic carbon flow and mitochondrial electron transport mechanisms (Dissanayaka et al. 2021). The most striking evidence of the activation of these alternate metabolic routes has come from transcriptome and proteome studies where several genes encoding crucial enzymes of the alternate pathway, such as Phosphoenolpyruvate carboxylase (PEPC) and PEPC kinase (PEPCK) based glycolytic bypass in Arabidopsis, rice, white lupin, orange (Citrus sinensis L.) and melon (Cucumis melo L.) have been reported (Liang et al. 2014). Among the upregulated ones, triose phosphate isomerase (TPI), glucose-6-phosphate 1-epimerase (PGEM), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are known to favor neoglucogenesis in Lotus corniculatus (Zhao et al. 2023).
Similarly, the improved glutathione metabolism through higher protein abundance of glutathione S-transferase in wheat also supports the activation of alternate metabolic routes in Pi-starved plants (Zheng et al. 2023). Interestingly, while cellular ATP pools are tightly regulated with the Pi levels, the pyrophosphate (PPi) level is relatively insensitive to Pi starvation (Ferjani et al. 2012). Most of the enzymes of the alternate routes can utilize PPi in place of ATP to catalyze the reaction. Thus the depleted cellular Pi levels incite the activation of several bypass enzymes, such as pyrophosphate-dependent phosphofructokinase in white lupin (Uhde-Stone et al. 2003), pyruvate phosphate dikinase in Zea mays (Li et al. 2007), UDP-sugar pyrophosphorylase in Arabidopsis (Okazaki et al. 2009), and mitochondrial alternative oxidases in white lupin (Florez-Sarasa et al. 2014) that depend on inorganic pyrophosphate (PPi) for their activity. PPi-dependent glycolysis helps plants maintain the carbon flow down to the citric acid cycle. In this scheme, activation of mitochondrial alternative oxidases is critical as their enhanced activity helps sustain the boosted production of organic acids with minimal ATP consumption, which is critical for respiration to maintain cellular activity, especially under depleted Pi levels.
These Pi and adenylate reductions disturb the interchange of Pi and phosphorylated sugars between the cytosol and the chloroplast stroma, resulting in starch deposition and a drop in photosynthesis rate. During Pi starvation, significant changes in the allocation of photosynthates to phosphorylated and non-phosphorylated carbon metabolites and decreased hexokinase activity have been observed (Karthikeyan et al. 2007). Over-accumulation of starch in HXK-antisense plants (Veramendi et al. 1999) and the onset of Pi toxicity symptoms in HXK over-expressing plants (Dai et al. 1999) endorse the link between Pi homeostasis and hexokinase activity (Briat et al. 2015). This linkage is further supported by the hexokinase I gene knockout (gin2) mutant of Arabidopsis, where a significant reduction of glucose-6-phosphate is coupled with the decrease in the transcripts of several PSI genes, namely Pht1;1, Pht1;4, INDUCED BY PHOSPHATE STARVATION (At4, AtIPS1), and RNase2. Earlier, Müller et al. (2015) also discovered a significant drop in sugar P conjugates in white lupin due to cellular Pi deficit. In both shoots and roots, phosphorylated metabolites such as glucose-6-phosphate, fructose-6-phosphate, myo-inositol-phosphate, and glycerol-3-phosphate were decreased from 14 to 22 days post-Pi starvation. This decrement can be justified as an early reaction to Pi starvation, where the altered carbohydrate allocation between the shoot and root supports the reprogramming and expansion of the root system. Santosh et al. (2018) reported directional changes in sugar conjugates, i.e., drastically increased fructose and glucose in leaves, and glucose, D-xylose, and gluconate in the root. In a recent study by Iqbal et al. (2023), the authors found differential behavior of Pi-inexpensive pathways in tolerant (Jimian169) and sensitive (DES926) cotton varieties. This study reported an enhanced TCA cycle in Jimian169, especially in roots, to support higher root biomass production upon Pi starvation. Contrarily, glucose, fructose, and ribose levels declined more severely in roots of DES926 compared to Jimian169, suggesting better carbohydrate metabolism in the latter variety for improving low Pi tolerance.
Pi remobilization from external and internal nucleic acids sources contributes to the PAE and PUE of Pi-starved plants
The Po pools are the predominant form of P in the soils (Anderson 2015). Two major forms where P is fixed in soil Po pools are nucleotides (contributed by decaying organic matter) and Pi-monoesters such as phytic acid (Bünemann et al. 2008). P-deficient plants secrete ribonuclease and acid phosphatase enzymes to mineralize Po compounds and mobilize Pi in the soils. The plants’ ability to survive on nucleotides as the sole P source has been elegantly demonstrated in Arabidopsis seedlings (Robinson et al. 2012). A recent report demonstrating rapid Pi uptake by roots of the sedge Carex flacca from 33P-labeled DNA than other species in a mixed plant community highlights the importance of species-specific preference to different P sources, including nucleotides sources, for Pi acquisition in the same habitat (Phoenix et al. 2020). Metabolic studies have shown increased levels of adenosine, a constituent of ATP, NADPH, and RNA, in P-deficient tissues in Lupinus albus L. (Müller et al. 2015). CRs are also found to over-accumulate three metabolites, adenosine, adenine, and ribose, under Pi starvation. Recently, Luo et al. (2020) reported a decrease in the concentrations of 11 out of 14P-containing nucleotides in Stylosanthes roots exposed to Pi starvation. The authors reported sharply declined levels of nicotinic acid mononucleotide, cytidylic acid, uridine 5′-diphospho-D-glucose, uridine 5′-monophosphate, nicotinic acid adenine dinucleotide and adenosine monophosphate in Pi-deficient Stylosanthes roots, indicating that nucleotides level inside plant also changes under Pi starvation. For example, degradation of organellar DNA, such as that of mitochondria by exonuclease DEFECTIVE IN POLLEN ORGANELLE DNA DEGRADATION1 (DPD1) in senescent leaves, improve the PUE and fitness of Arabidopsis seedlings (Takami et al. 2018). As ~ 80% of cellular nucleic acids Po pool exists as rRNA, autophagy-mediated delivery of rRNA to the vacuole for recycling of Pi plays a profound role in plant growth under Pi-starvation (Stigter and Plaxton 2015; Bassham and MacIntosh 2017). Vacuolar PSI RNases and phosphodiesterases first metabolize rRNA to nucleotide monophosphates, which release Pi upon the subsequent action of PSI purple acid phosphatases (PAPs). Then VPTs transport Pi to the cytosol to synthesize organic compounds such as ATP (Floyd et al. 2015, 2017; Bassham and MacIntosh 2017). A similar mechanism operates in senescing organs where Pi-mobilizing enzymes such as RNases and PAPs are activated, linking senescence with Pi stress signaling cascades (Shane et al. 2014; Yang et al. 2017). One of the best examples of highly efficient Pi remobilization from senescing organs has been observed in Harsh hakea, a Proteaceae species that is well adapted to the highly P-impoverished habitats of Western Australia. These species can remobilize up to 90% of total Pi from the senescent leaf to younger organs by activating vacuolar and cell-wall-localized RNases and PAPs (Lambers and Plaxton 2015; Shane et al. 2014).
Phosphatases, majorly PAPs, play a vital role in enabling plants to adapt to Pi starvation as these enzymes facilitate the dephosphorylation of Po compounds into inorganic orthophosphate in the rhizosphere and intracellular compartments to release Pi (Feder et al. 2020; Srivastava et al. 2020). This process is particularly crucial for plants facing short-term Pi starvation, as observed in species such as Arabidopsis (AtPAP26) (Veljanovski et al. 2006), rice (OsACP1) (Deng et al. 2022), (OsPAP26) (Gao et al. 2017). AtPAP26, on the other hand, possesses a dual-targeted property, being present in both the vacuole and apoplast. As a result, it is considered a key player in both intracellular and extracellular phosphatase activity, thereby aiding in Pi remobilization within plants (Robinson et al. 2012; Shane et al. 2014). Nevertheless, extracellular acid phosphatases, such as PAP3b, OsPAP10c, and OsPAP10a, are secreted by roots into the rhizosphere, primarily aiding nutrient acquisition. These enzymes have been shown to enhance plant growth by increasing P availability from organic P compounds under specific environmental stimuli (Tian et al. 2012). In soybean, GmPAP4, GmPAP14, GmPAP7a, and GmPAP7b have been demonstrated to enhance the utilization of extracellular phytate and ATP. As GmPAP4 and GmPAP14 are induced upon organic P treatment, overexpression of these enzymes promotes the breakdown of organic P through increased acid phosphatase activity (Kong et al. 2014, 2018; Zhu et al. 2020).
Carbon repartitioning is crucial for differential growth and biochemical responses of Pi-starved plants
Sugars are ambidextrous as they act as a source of energy and signaling molecules under stress. As sucrose is the most abundant plant storage product with limited chemical activity, it is the primary carbohydrate transported through the phloem (Gangola and Ramadoss 2018). Due to the differential growth of roots under P-starved conditions, the sink tissues’ equilibrium shifts towards the underground parts. Sucrose plays a vital role in P starvation as its enhanced shoot-to-root mobilization is critical to support the reprogramming of RSA in Pi-deficient plants (Hammond and White 2008) (Fig. 1). Lei et al. (2011), in an elegant study, demonstrated the dynamic role of a SUCROSE TRANSPORTER 2 (SUC2) gene in bidirectional sucrose transport between root and shoot in Arabidopsis seedlings. The authors reported that overexpression of SUC2 in a hypersensitive to phosphate starvation 1 (hps1) mutant conferred greater sensitivity to mutant plants in almost all aspects of PSR. It was suggested that enhanced sucrose levels are responsible for such an exacerbated response. Consequent microarray analysis showed that 73% of the PSI genes are transcriptionally overactive under Pi-sufficient conditions in SUC2 overexpressing hps1 mutant plants. Sucrose levels influence PSR in the root, such as increased root-associated acid phosphatase activity (Srivastava et al. 2020), denser root hairs, and an increased number of lateral roots. However, at the molecular level, it has a role in the induction of PSI genes like those encoding PHTs, PAPs, and ROOT HAIR DEFECTIVE 6-LIKE (RSLs). Enhanced conversion of starch to sucrose and other carbohydrates also induces a myriad of PSI genes (Karthikeyan et al. 2007). Sucrose-induced expression of PSI genes in Arabidopsis has found support in several recently published studies in tomato and barley, where an external supply of sucrose has been found to activate PHT1 transporters in barley (Srivastava et al. 2021b), as well as several Snf1-Related protein kinases (Khurana and Akash 2021), and F-box family members (Akash et al. 2021) in tomato. The enhanced sucrose level has also been found to activate the AtMYB75 transcription factor to influence higher anthocyanin accumulation, one of the characteristic PSR, in Pi-starved Arabidopsis seedlings (Li et al. 2014).
In a recent study by Xiao et al. (2022), new insights have emerged regarding an additional function of SHR, a GRAS transcription factor known to play a vital role in various aspects of plant development, such as determining root patterning and cell fate. Notably, mutants like shr and scarecrow (scr) have already been recognized for their impact on sugar metabolism (Cui et al. 2012). The mutants exhibit increased sugar accumulation, mainly sucrose, glucose, and fructose, suggesting that shr and scr have distinct, independent functions in regulating sugar responses. This study also suggests that SHR may also be involved in regulating the allocation of Pi in plants. The findings upon Pi starvation have unveiled that inhibiting SHR in roots impedes phosphate redistribution to shoots in Arabidopsis. Another recent study by Agrawal et al. (2023) highlights the crucial role of a sucrose-inducible MEDIATOR SUBUNIT17 (MED17) as a node, connecting sucrose and auxin signaling. MED17 plays a critical role in conveying the transcriptional signal from sucrose to auxin-responsive and cell-cycle genes to regulate RSA, emphasizing the role of MED proteins as the transcriptional processor for optimum root system design in Arabidopsis. Another MEDIATOR protein MED16 has been found to control RSA remodeling in Arabidopsis Pi-deficient roots (Raya-Gonzalez et al. 2021). Transcriptome analysis of root tip showed that MED16 influences transcript levels of a large set of low-phosphate-induced genes involved in local and systemic signaling. Considering the conserved function of MED proteins in regulating gene expression and the involvement of MED16 in root PSR, a prominent role of MED17 in the reprogramming of RSA under Pi starvation cannot be ruled out. Overall, the evidence suggests that carbon remobilization under Pi starvation plays a critical role in determining PSR in plants.
Organic acids are critical in enhancing PAE under Pi-depleted conditions
Roots of Pi-deficient plants secrete organic acids (OAs) and protons to liberate Pi from its bound forms in soils (Srivastava et al. 2021a). Radioactive labeling of CO2 indicates that plants make a considerable investment in translocating their photosynthetically fixed carbon (approximately 20–40%), generally in the form of sucrose, to roots for supporting the exudation of OAs and small molecular compounds. The concentration of OAs in soil ranges from 0 to 50 µM for di/tricarboxylic acid, while for monocarboxylic acids, its concentration remains between 0 and 1 mM (Adeleke et al. 2017). Two main OAs secreted by Pi-deficient roots are malate and citrate, often called carboxylates. Citrate is the most effective OA at mobilizing Pi from the soil in Alfisols, followed by piscidic acid and malate (Zhu et al. 2022).
In addition to these two main OAs, isocitrate, oxoglutarate, citramalic acid, and polyhydroxy acids have also been reported to accumulate differentially in Pi-deficient roots and shoot tissues in tomato (Sung et al. 2015). This study reported gradually declined α-ketoglutarate, fumarate, and malate levels in Pi-deficient plants’ shoot and root tissues. In contrast, citrate accumulation was enhanced in both tissues. In another study, the amount of carboxylates in the cucumber xylem and phloem sap has been reported to increase under Pi starvation (Sun et al. 2022). Several studies that assessed the level of OAs secretion and its impact on PAE discovered it to be a highly variable species-specific trait, most likely due to varying ‘carbon’ cost versus the altered enhanced Pi uptake. The published literature shows that some species/cultivars have evolved to produce more OAs than others. For example, soybean genotypes and white lupin release more OAs than monocots with fibrous root systems, such as maize and wheat (Lyu et al. 2016). Similarly, Pi-tolerant varieties of maize secrete a more considerable amount of OAs than the sensitive ones (Luo et al. 2019). A positive correlation between root width and OAs exudation has also been reported, as species with thicker roots tend to release a higher amount of OAs than the plant species with thinner roots (Wen et al. 2020). Recent evidence also indicates the beneficial roles of higher OAs-secreting species in helping the growth of other non-Pi-mobilizing cohabitating species in Pi-depleted soils (Yu et al. 2020). Exudation of carboxylate secretion from proteoid roots by white lupin under P-deficient conditions enhanced the release of inorganic Pi from phosphorylated ferric hydroxide and from soil P (Uhde-stone et al. 2003).
The exudation of OAs is mainly associated with the increased activity of three main enzymes: malate dehydrogenase, citrate synthase, and PEP carboxylase (Ligaba et al. 2004). As anticipated, overexpression of these genes has been shown to improve both Pi acquisition and plant biomass in different plant species under P-deficient conditions (Koyama et al. 2000; Vance et al. 2003). Cytoplasmic PEP carboxylase is a peculiar enzyme in this scheme due to its central position in plant metabolism. Evidence suggests that PEPC-catalyzed anaplerotic CO2 fixation can contribute almost a third of the released carboxylates by white lupin CRs (Shane et al. 2016). The described experimental procedures in the published literature indicate that most studies have focused on their short-term effects in the rhizosphere, and the long-term advantages of the secreted OAs remain obscure, which warrants further investigations. Besides their direct roles in releasing Pi from its bound forms in the rhizosphere, the excretion of OAs also facilitates the recruitment of beneficial rhizobacteria, such as Pseudomonas fluorescens, to the root surface, thus activating the integrated stress response including adaptive changes in the root system architecture for enhanced Pi uptake (Fig. 1) (Wu et al. 2018; Srivastava et al. 2021a).
Differential regulation of amino acids is vital for altered metabolic fluxes and growth response under Pi starvation
While AAs are best recognized as protein building blocks, their potential for stress tolerance at cellular and physiological levels is sometimes underestimated. In plants, amino acid metabolism is connected to carbohydrate and secondary metabolism. Energy depletion in a cell is a serious concern during Pi starvation because of the decreased biosynthesis of the primary energy source ATP. The cellular pool of AAs is prone to change because of the need for a paradigm shift from growth to defense in various biotic and abiotic stressful situations. Hildebrandt (2018) showed that the proteome shift produces a free amino acid pool from which specific AAs are directed to be the precursor of defensive mechanisms depending on the needs. Stress may also necessitate favoring the growth of specific organs over others, resulting in a local energy shortage. For example, the reprogrammed root development for a larger surface area, at the expense of flowers or leaves in plants’ aerial parts, may enhance plants’ Pi uptake ability to survive short Pi starvation exposure (Batista-Silva et al. 2019). The osmoprotectant role of one of the best-studied AAs, proline (Pro), that stabilizes sub-cellular structures under drought, salinity, low temperature, heavy metal exposure, and UV radiations is well known (Hayat et al. 2012). A recent report by Zhang et al. (2021) revealed that Pro metabolism genes are activated under exogenous silicon application to revive low Pi-deprived tomato plants’ fitness.
The synthesis of 4-hydroxyproline from Pro is well recognized for its function during abiotic stress tolerance (Santosh et al. 2018). Pro hydroxylation by prolyl 4-hydroxylase (P4H) might be a central metabolic pathway in P-deficient plants. Under Pi starvation in tea (Camellia sinensis) plants, hydroxyproline levels build up through P4H expression stimulation (Santosh et al. 2018). The authors also found elevated amounts of other AAs, such as alanine (Ala), tryptophan (Trp), and tyrosine (Tyr), in the roots and shoots of P-deficient tea plants. In another study on white lupin, Müller et al. (2015) reported a higher abundance of Trp, followed by asparagine (Asn) and leucine (Leu), Phe, and Tyr in CRs compared to regular roots. Thus elevated production of free Trp, a precursor of auxins (Mano and Nemoto 2012), under Pi starvation may be directly correlated with the enhanced localized production of auxin in roots to reprogram RSA in rice (Shen et al. 2013; Wang et al. 2015). Monogalactosyldiacylglycerol, found in chloroplast membranes, is a molecule that plays a vital role in connecting and maintaining the activity of these membranes. It contains specific Trp residues that are essential for its function. The enhanced Trp levels may be required in Pi-starved roots to support glycolipids (GLs) biosynthesis (Ge et al. 2011). The cellular buildup of free Phe and Tyr, precursors of the flavanone naringenin, a well-known raw material for anthocyanin biosynthesis, also supports the significance of the changes in AAs levels for plant PSR in Pi-deficient plants (Nezamivand-Chegini et al. 2023).
In contrast to the commonly upregulated Ala, Trp, and Tyr AAs in roots, Hernández et al. (2009) reported an opposite pattern of amino acid buildup in common bean root nodules under Pi starvation. Non-targeted metabolic profiles indicated a considerable decrease in AAs and other nitrogen metabolites such as glycine (Gly), serine (Ser), Thr, and glutamate (Glu). While, numerous other AAs such as β-alanine, Asn, and Phe increased significantly. This drop in AAs levels was linked to the reduced expression of three aminoacyl-tRNA enzymes causing significant inhibition of this biosynthetic pathway. These findings highlight the variations in the metabolic response of non-colonized and colonized P-stressed bean roots (Hernández et al. 2007), which showed a considerable increase in amino acid content and suggested that metabolic response in roots subjected to Pi-starvation is dynamic. Additionally, nucleotide metabolism was overrepresented among the inhibited cellular activities of Pi-starved nodules, which might be exploited to prioritize the expression of specific PSI genes.
Chronic Pi starvation alters glycerolipid metabolism in Pi-deficient plants
The significant components of plant lipidome are glycerolipids (GLs) which can be further categorized into phospholipids (PLs) and GLs, the central polar lipids, and triacylglycerol (TAG), a neutral lipid. Concerning lipids, membranes in plants are unique since plasma membranes are rich in PLs, but the chloroplast envelope and thylakoid membranes are rich in GLs. PLs, are one of the most central targets for remodeling under chronic Pi starvation to facilitate Pi solubilization, as they contribute about 1/3 of plants’ total organic P content (Nakamura 2013). The conversion of PLs to GLs for producing free Pi is considered a prominent adaptation under PSR (Okazaki et al. 2017). The reduction in PLs allows Pi to be consumed for other prioritized cell functions and provides lipid moiety diacylglycerol (DAG) accessible for GL biosynthesis under Pi starvation (Li et al. 2006). Multiple PLs-hydrolysing enzymes, such as phospholipase D (PLD) and phospholipase C (PLC), mediate their conversion to GLs, predominantly monogalactosyl-diacylglycerol (MGDG), a dihexosylglycerolipid, digalactosyl-diacylglycerol (DGDG) and a sulfolipid, sulfoquinovosyl-diacylglycerol (SQDG) (Benning and Ohta 2005). MGDG and DGDG are nonionic lipids and are assumed to operate mainly as structural lipids for the creation of the lipid bilayer, whereas SQDG and PG are acidic lipids and are presumably necessary to maintain the balance of negative charges in the thylakoid membranes (Nakajima et al. 2018). Declined PLs intensities in Pi-deprived conditions were first observed in a non-photosynthetic bacterium, Pseudomonas diminuta (Minnikin et al. 1974). Due to a lengthy fatty acid (FA) moiety, GLs may be categorized based on their carbon numbers. Whereas 34-C DGDGs can be prokaryotic or eukaryotic in origin, 36-carbon DGDGs have been demonstrated to be only eukaryotic in origin (Härtel et al. 2000). Pi starvation also changes the cell membranes’ FA composition. Härtel et al. (2000) reported the unique nature of Pi starvation -induced DGDG by demonstrating an abundance of 16-carbon FAs in the C-1 position of the DGDG glycerol backbone. The FA content and position are assumed to be diagnostic for DGDG genesis via the plastid or ER pathways under Pi starvation in Arabidopsis. The spatial development, whether plants are grown in a controlled laboratory environment or in nature, can also impact lipid remodeling. Recently, Li et al. (2023b) revealed common and unique patterns in leaf lipid remodeling in a natural environment compared to laboratory conditions. In field-grown plants, high levels of plastidic lipids were observed, which might result from high levels of sunlight.
Furthermore, Pi starvation altered the composition of FAs in the seed of Camelina, a popular oil-seed crop. It increased monounsaturated FAs (MUFAs), namely oleic (18:1) and eicosenoic (20:1) acids, and decreased saturated FAs levels. Polyunsaturated fatty acids (PUFAs), especially (18:2) and (18:3) species, rise proportionately with the degree of Pi depletion, subsequently penalising overall seed yield to 10–15 times lower, indicating the adverse effect of Pi starvation on seed production in Camelina. While a temporary increase in PC (phosphatidylcholine) upon Pi starvation has also been reported in Arabidopsis (Jouhet et al. 2003), it was compensated by the decrease in the levels of minor lipids, such as phosphatidic acid (PA), phosphatidyl-Ser (PS), lysophosphatidylcholine (lysoPC), lysophosphatidylethanolamine (lysoPE), and lysophosphatidylglycerol (lysoPG) in rosettes and roots. The lysoPC levels are significantly reduced to 37% in rosettes and 64% in roots in Arabidopsis seedlings under Pi starvation (Li et al. 2006). Recent evidence also suggests that the changes associated with lipidomic amendment under Pi starvation may differ in the shoot and root tissues of the same plant. Pfaff et al. (2020), showed mutually exclusive changes in membrane lipids in shoot and root tissues of Pi-deficient tomato seedlings. While leaves majorly accumulate polyunsaturated TAG, roots predominantly accumulate GLs. The detailed analysis of FA profiles revealed that the molecular species conversion was more prominent in Pi-deprived roots than leaves, as evident from a considerable increase in 34:2, 34:3, and 36:4 GLs species. The authors also demonstrated that the differential degradation of PLs into subsequent TAGs and GLs is not regulated at the transcriptional level. Supported by the unaltered transcriptional induction of fatty acid biosynthesis, PLs biosynthesis, or GLs biosynthesis genes from plastidic pathways in this study confirms the lipid remodeling, rather than de-novo biosynthesis, as the primary mechanism for plants to adapt under Pi starvation.
The physiological necessity of lipid remodeling under Pi starvation has been addressed using several loss-of-function mutants. A particular emphasis has been given to the genes that engage in the process of SQDG biosynthesis pathway, namely SQD-B in Thermosynechococcus elongatus, SQD1 and UDP-glucose pyrophosphorylase3 (UGP3) in A. thaliana (Nakajima et al. 2018), genes responsible for enhanced GL biosynthesis, namely MGD synthase 1 (MGD1), and Phospholipase C5 (NPC5) in Arabidopsis (Gaude et al. 2008), MGD synthase 3 (MGD3) in rice (Verma et al. 2022) and also the genes liable for the degradation of PLs especially Glycerophosphodiester phosphodiesterases (GDPDs), in Arabidopsis (Cheng et al. 2011). Evidence suggests that lipid remodeling in Arabidopsis sqd2 mutants is spatially controlled, with increased chlorophyll breakdown in mature older leaves. In soybean, glucuronosyldiacyl-glycerol (GlcADG) accumulation dominates SQDG upon Pi starvation (Okazaki et al. 2017). GlcADG is considered one of the products of the same SQDG synthase. It does not seem to be restricted to only soybean and may function as a preferential alternative over SQDG plausibly in Pi-deficient plants of other dicot species (Okazaki et al. 2017).
The contribution of a less-traveled route, mediated by lipid acyl hydrolase (LAH) and GDPD enzymes during GLs remodeling under Pi starvation, remains far lesser known and is yet to be adequately scrutinized. This pathway works upon the hydrolyzation of PLs into free fatty acids and glycerophosphodiester (GPD) by LAH and subsequent hydrolyzation to glycerol-3-phosphate (G3P) and corresponding alcohols (Pfaff et al. 2020). However, this metabolic pathway is economically expensive and does not yield free Pi (Mehra et al. 2019). Recent investigations have yielded fresh perspectives and understanding regarding the role of OsGDPD2 and AtGDPD6 in plant growth and development. In a study by Mehra et al. (2019), it was found that rice PHOSPHATE STARVATION RESPONSE2 (OsPHR2) directly targets OsGDPD2 for transcriptional activation, and overexpression of this gene in rice plants led to several beneficial effects. The transgenic lines exhibited increased GDPD activity, Pi content, root development, and biomass accumulation compared to the wild-type plants. Additionally, these lines showed contrasting morpho-physiological and biochemical characteristics. Interestingly, the overexpression lines had elevated levels of various phosphorus-containing metabolites, including fatty acids (FAs), suggesting a potential role of OsGDPD2 in the de novo biosynthesis of GLs. Similarly, Ngo and Nakamura (2022) characterized AtGDPD6 and found that its overexpression in Arabidopsis plants resulted in significantly longer roots than the wild type and Atgdpd mutants. Notably, the cell morphology in the roots’ maturation, elongation, and meristem zones remained unaffected, indicating that the increased root length did not cause any abnormalities in root cell architecture. Taken together, these results clearly indicate the beneficial impact of GDPDs on root growth under differential Pi availability. These studies shed light on plant growth regulation’s molecular mechanisms and provide potential targets for improving crop productivity and nutrient utilization.
Altered flavonoid levels are an essential aspect of PSR
Flavonoids play diverse roles in plant protection against biotic and abiotic stresses. Flavonoids reduce reactive oxygen species (ROS) production by inhibiting enzymes that generate ROS and suppressing singlet oxygen. Around 10,000 structural variations of flavonoids have been found in plants so far. Based on their fundamental structures, flavonoids are classified as flavones, flavanones, flavonols, isoflavones, anthocyanins, and chalcones (Panche et al. 2016). In Arabidopsis, the concentration of many phenylpropanoids, flavonoids, and their derivatives increased in both roots and shoots during P-starvation (Pant et al. 2015). Flavonoids such as benzoxazinoids were strongly decreased in the roots of Pi-resistant accessions in maize. In contrast, the flavonoid quercetin-3,4-O-di-beta-glucopyranoside was preferentially accumulated in the leaves of Pi-sensitive genotypes (Luo et al. 2019). Similarly, the concentration of C-glycosylflavones was significantly high under P-limited conditions in melon roots (Akiyama et al. 2002). Accumulation of quercetin in melon roots also promoted mycorrhizal colonization and helped plants’ fitness by indirectly increasing the availability of micronutrients. Similarly, flavonoid exudates such as phenylamide, phenolic acid, and piscidic acid from white lupin, Stylosanthes, and Cajanus cajan roots was also reported causing a rise in P acquisition via facilitating Pi remobilization and roots interaction with microorganisms (Fig. 1) (Luo et al. 2020).
Flavonols have been observed to accumulate in Stylosanthes during Pi starvation (Luo et al. 2020). In this work, Pi starvation increased the amount of 41 out of 54 differently accumulated flavonoids in Stylosanthes roots. The quantities of kaempferol and its flavonol glycoside derivatives were considerably increased at low Pi conditions. Mo et al. (2021) recently reported that Pi starvation increased transcripts of COP9 signalosome subunit (GmCSN5A/B) in both young and old soybean leaves. Additionally, overexpression of these genes results in the activation of anthocyanin biosynthesis pathway genes, resulting in more significant accumulation in shoots under Pi starvation. Apart from their role as negative regulators of a subset of PSI genes in Arabidopsis and tomato (Osorio et al. 2019; Singh et al. 2023), SPX4 and SPX1/2 also interact with certain genes involved in the biosynthesis of flavonoids. For example, He et al. (2021) recently revealed that SPX4 directly interacts with the dihydroflavonol 4-reductase (DFR) gene, which is the rate-limiting step in anthocyanin biosynthesis, as demonstrated by significantly greater transcript levels in spx4 mutants and a 40% decrease in anthocyanin accumulation in SPX4-OE lines. Besides, Wang et al. (2022) discovered that chalcone synthase (PtCHS), chalcone isomerase (PtCHI), and flavonoid 3-hydroxylase (PtF3H) genes are overexpressed in Pinellia ternata under Pi starvation, leading to increased accumulation of total flavonoids. A recent study from Li et al. (2023a) revalidated the vital role of MYB-bHLH-WD40 (MBW) complex coupled with the transcription factor (TF) MYB75 and Production of Anthocyanin Pigments1 (PAP1) in the regulation of anthocyanin synthesis in Arabidopsis (He et al. 2021; Meng et al. 2021). Besides that, PHR1 is physically coupled with transcription factors engaged in anthocyanidin biosynthesis, such as PAP1/MYB75, MYB DOMAIN PROTEIN 113 (MYB113), and TRANSPARENT TESTA 8 (TT8) (Li et al. 2023a). These findings allow a novel mechanistic understanding of how P-deficient signaling depends on endogenous anthocyanins synthesis pathway, mediated by TFs, to enhance anthocyanins accumulation under Pi starvation.
Conclusions and perspectives
Plant acclimation to P starvation is a multifaceted process that comprises intertwined local and systemic signaling pathways (Li et al. 2022). Several efforts have been made to adapt the model systems for plant growth studies under a controlled environment to address the crosstalk between two or more stress factors. For instance, specific metabolic adaptations have been observed in conditions of varying light intensity and P supply (KC et al. 2021) or combined iron and Pi starvation (Kaur et al. 2021). It should be noted that the advances in plant systems biology already allow the concomitant analysis of the transcriptome, proteome, and metabolome in the same plant sample, which provides a comprehensive overview of the affected cellular processes upon Pi starvation. However, as outlined by (Dissanayaka et al. 2021), the protein interactome, the post-translational modifications, and the intracellular compartmentalization of key PSR-related players are other essential aspects of the plant response to P stress that remain largely unexplored to date. Still, identifying P stress-specific metabolomic signatures per se could bring valuable information about the Pi use efficiency of a genotype of interest, allowing the prediction of its adaptive potential (Watanabe et al. 2020). Here, discrimination between genotype-specific adaptive mechanisms and general stress responses to Pi starvation is a significant challenge. A paramount goal of such studies is identifying metabolic markers for selecting low Pi-tolerant crops. In most cases, the reproducibility of experimental results in search of novel metabolite markers is hampered by reported differences in the model plant growth conditions, dose and duration of the applied stress factor. Nevertheless, the thorough metabolome dissection emerges as a powerful source of knowledge that fuels novel strategies for improving crop resilience to low Pi availability.
Acknowledgements
This study was funded by the Indo-Bulgarian Bilateral Research Cooperation (projects INT/BLG/P-06/2019 and KP-06-India-9). In addition, R.K. acknowledges the financial support from the Science and Engineering Research Board (SERB), Government of India (CRG/2018/001033) and to the University of Hyderabad-IoE by MHRD (F11/9/2019-U3(A) and DBT-support to the School of Life Sciences under BUILDER program. A.R. thanks SERB, Government of India (CRG/2018/001033), and UoH BBL for his fellowships. R.S. and Akash thank the Council of Scientific and Industrial Research, Govt. of India, for the JRF and SRF fellowships and UoH BBL for the research fellowship.
Author contributions
The idea was conceived by RK, KM and GZ. The manuscript was written by AR, RS, and GS. Akash made the figure. Final drafts of the manuscript were read and approved by all authors.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kiril Mishev, Email: mishev@bio21.bas.bg.
Rahul Kumar, Email: rksl@uohyd.ac.in.
References
- Adeleke R, Nwangburuka C, Oboirien B. Origins, roles and fate of organic acids in soils: a review. S Afr J Bot. 2017;108:393–406. doi: 10.1016/j.sajb.2016.09.002. [DOI] [Google Scholar]
- Agrawal R, Singh A, et al. MEDIATOR SUBUNIT17 is required for transcriptional optimization of root system architecture in Arabidopsis. Plant Physiol. 2023;28:kiad129. doi: 10.1093/plphys/kiad129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akash PAP, et al. Identification, evolutionary profiling, and expression analysis of F-box superfamily genes under phosphate deficiency in tomato. Plant Physiol Biochem. 2021;162:349–362. doi: 10.1016/j.plaphy.2021.03.002. [DOI] [PubMed] [Google Scholar]
- Akiyama K, Matsuoka H, Hayashi H. Isolation and identification of a phosphate deficiency-induced C-glycosylflavonoid that stimulates arbuscular mycorrhiza formation in melon roots. Mol Plant Microbe Interact. 2002;15:334–340. doi: 10.1094/MPMI.2002.15.4.334. [DOI] [PubMed] [Google Scholar]
- Anderson G. Assessing organic phosphorus in soils. In: Khasawneh FE, Sample EC, Kamprath EJ, editors. The role of phosphorus in agriculture. Madison: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; 2015. pp. 411–431. [Google Scholar]
- Ashihara H, Li XN, Ukaji T. Effect of inorganic phosphate on the biosynthesis of purine and pyrimidine nucleotides in suspension-cultured cells of Catharanthus roseus. Ann Bot. 1988;61:225–232. doi: 10.1093/oxfordjournals.aob.a087547. [DOI] [Google Scholar]
- Bassham DC, MacIntosh GC. Degradation of cytosolic ribosomes by autophagy-related pathways. Plant Sci. 2017;262:169–174. doi: 10.1016/j.plantsci.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Batista-Silva W, Heinemann B, et al. The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 2019;42:1630–1644. doi: 10.1111/pce.13518. [DOI] [PubMed] [Google Scholar]
- Benning C, Ohta H. Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem. 2005;280:2397–2400. doi: 10.1074/jbc.R400032200. [DOI] [PubMed] [Google Scholar]
- Bozzo GG, Dunn EL, Plaxton WC. Differential synthesis of phosphate-starvation inducible purple acid phosphatase isozymes in tomato (Lycopersicon esculentum) suspension cells and seedlings. Plant Cell Environ. 2006;29(2):303–313. doi: 10.1111/j.1365-3040.2005.01422.x. [DOI] [PubMed] [Google Scholar]
- Briat JF, Rouached H, et al. Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1) Front Plant Sci. 2015;6:290. doi: 10.3389/fpls.2015.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünemann EK, Marschner P, et al. Soil organic phosphorus and microbial community composition as affected by 26 years of different management strategies. Biol Fertil Soils. 2008;44:717–726. doi: 10.1007/s00374-007-0254-2. [DOI] [Google Scholar]
- Cheng Y, Zhou W, et al. Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 2011;66:781–795. doi: 10.1111/j.1365-313X.2011.04538.x. [DOI] [PubMed] [Google Scholar]
- Cordell D, Drangert JO, White S. The story of phosphorus: global food security and food for thought. Glob Environ Chang. 2009;19:292–305. doi: 10.1016/j.gloenvcha.2008.10.009. [DOI] [Google Scholar]
- Cubero B, Nakagawa Y, et al. The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Mol Plant. 2009;2(3):535–552. doi: 10.1093/mp/ssp013. [DOI] [PubMed] [Google Scholar]
- Cui H, Hao Y, Kong D. SCARECROW has a SHORT-ROOT-independent role in modulating the sugar response. Plant Physiol. 2012;158(4):1769–1778. doi: 10.1104/pp.111.191502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai N, Schaffer A, et al. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell. 1999;11:1253–1266. doi: 10.1105/tpc.11.7.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del-Saz NF, Romero-Munar A, et al. (2018) Phosphorus concentration coordinates a respiratory bypass, synthesis and exudation of citrate, and the expression of high-affinity phosphorus transporters in Solanum lycopersicum. Plant Cell Environ. 2018;41(4):865–875. doi: 10.1111/pce.13155. [DOI] [PubMed] [Google Scholar]
- Deng S, Li J, et al. Rice ACID PHOSPHATASE 1 regulates Pi stress adaptation by maintaining intracellular Pi homeostasis. Plant, Cell Environ. 2022;45(1):191–205. doi: 10.1111/pce.14191. [DOI] [PubMed] [Google Scholar]
- Dissanayaka DMSB, Ghahremani M, et al. Recent insights into the metabolic adaptations of phosphorus-deprived plants. J Exp Bot. 2021;72(2):199–223. doi: 10.1093/jxb/eraa482. [DOI] [PubMed] [Google Scholar]
- Feder D, McGeary RP, et al. Structural elements that modulate the substrate specificity of plant purple acid phosphatases: avenues for improved phosphorus acquisition in crops. Plant Sci. 2020;294:110445. doi: 10.1016/j.plantsci.2020.110445. [DOI] [PubMed] [Google Scholar]
- Ferjani A, Segami S, et al. Regulation of pyrophosphate levels by H+-PPase is central for proper resumption of early plant development. Plant Signal Behav. 2012;7(1):38–42. doi: 10.4161/psb.7.1.18573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez-Sarasa IG, Lambers H, et al. The alternative respiratory pathway mediates carboxylate synthesis in white lupin cluster roots under phosphorus deprivation. Plant Cell Environ. 2014;37(4):922–928. doi: 10.1111/pce.12208. [DOI] [PubMed] [Google Scholar]
- Floyd BE, Morriss SC, et al. Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy. 2015;11:2199–2212. doi: 10.1080/15548627.2015.1106664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd BE, Mugume Y, et al. Localization of RNS2 ribonuclease to the vacuole is required for its role in cellular homeostasis. Planta. 2017;245:779–792. doi: 10.1007/s00425-016-2644-x. [DOI] [PubMed] [Google Scholar]
- Gangola MP, Ramadoss BR. Sugars play a critical role in abiotic stress tolerance in plants. In: Wani SH, editor. Biochemical, physiological and molecular avenues for combating abiotic stress tolerance in plants. London: Academic Press; 2018. pp. 17–38. [Google Scholar]
- Gao W, Lu L, et al. OsPAP26 encodes a major purple acid phosphatase and regulates phosphate remobilization in rice. Plant Cell Physiol. 2017;58(5):885–892. doi: 10.1093/pcp/pcx041. [DOI] [PubMed] [Google Scholar]
- Gaude N, Nakamura Y, et al. Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 2008;56:28–39. doi: 10.1111/j.1365-313X.2008.03582.x. [DOI] [PubMed] [Google Scholar]
- Ge C, Georgiev A, et al. Tryptophan residues promote membrane association for a plant lipid glycosyltransferase involved in phosphate stress. J Biol Chem. 2011;286:6669–6684. doi: 10.1074/jbc.M110.138495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, White PJ. Sucrose transport in the phloem: integrating root responses to phosphorus starvation. Journal of experimental botany. 2008;59(1):93–109. doi: 10.1093/jxb/erm221. [DOI] [PubMed] [Google Scholar]
- Härtel H, Dörmann P, Benning C. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci U S A. 2000;97:10649–10654. doi: 10.1073/pnas.180320497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat S, Hayat Q, et al. Role of proline under changing environments. Plant Signal Behav. 2012;7(11):1456–1466. doi: 10.4161/psb.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang X, et al. SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytol. 2021;230:205–217. doi: 10.1111/nph.17139. [DOI] [PubMed] [Google Scholar]
- Hernández G, Ramírez M, et al. Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol. 2007;144:752–767. doi: 10.1104/pp.107.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G, Valdés-López O, et al. Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 2009;151:1221–1238. doi: 10.1104/pp.109.143842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt TM. Synthesis versus degradation: directions of amino acid metabolism during Arabidopsis abiotic stress response. Plant Mol Biol. 2018;98:121–135. doi: 10.1007/s11103-018-0767-0. [DOI] [PubMed] [Google Scholar]
- Iqbal A, Qiang D, et al. Integrative physiological, transcriptome and metabolome analysis reveals the involvement of carbon and flavonoid biosynthesis in low phosphorus tolerance in cotton. Plant Physiol Biochem. 2023;196:302–317. doi: 10.1016/j.plaphy.2023.01.042. [DOI] [PubMed] [Google Scholar]
- Jia F, Wan X, et al. Overexpression of mitochondrial phosphate transporter 3 severely hampers plant development through regulating mitochondrial function in Arabidopsis. PLoS ONE. 2015;10(6):e0129717. doi: 10.1371/journal.pone.0129717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J, Maréchal E, et al. Transient increase of phosphatidylcholine in plant cells in response to phosphate deprivation. FEBS Lett. 2003;544:63–68. doi: 10.1016/S0014-5793(03)00477-0. [DOI] [PubMed] [Google Scholar]
- Karlsson PM, Herdean A, et al. The Arabidopsis thylakoid transporter PHT4;1 influences phosphate availability for ATP synthesis and plant growth. Plant J. 2015;84:99–110. doi: 10.1111/tpj.12962. [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, et al. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta. 2007;225:907–918. doi: 10.1007/s00425-006-0408-8. [DOI] [PubMed] [Google Scholar]
- Kaur G, Shukla V, et al. Physiological and molecular responses to combinatorial iron and phosphate deficiencies in hexaploid wheat seedlings. Genomics. 2021;113:3935–3950. doi: 10.1016/j.ygeno.2021.09.019. [DOI] [PubMed] [Google Scholar]
- Kc S, Long L, et al. Light intensity modulates the effect of phosphate limitation on carbohydrates, amino acids, and catechins in tea plants (Camellia sinensis L.) Front Plant Sci. 2021;12:2107. doi: 10.3389/fpls.2021.743781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana A, Akash RA. Identification of phosphorus starvation inducible SnRK genes in tomato (Solanum lycopersicum L.) J Plant Biochem Biotechnol. 2021;2021:1–12. [Google Scholar]
- Kong Y, Li X, et al. GmPAP4, a novel purple acid phosphatase gene isolated from soybean (Glycine max), enhanced extracellular phytate utilization in Arabidopsis thaliana. Plant Cell Rep. 2014;33:655–667. doi: 10.1007/s00299-014-1588-5. [DOI] [PubMed] [Google Scholar]
- Kong Y, Li X, et al. The soybean purple acid phosphatase GmPAP14 predominantly enhances external phytate utilization in plants. Front Plant Sci. 2018;9:292. doi: 10.3389/fpls.2018.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Kawamura A, et al. Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol. 2000;41(9):1030–1037. doi: 10.1093/pcp/pcd029. [DOI] [PubMed] [Google Scholar]
- Lambers H, Plaxton WC. Phosphorus: back to the roots. Annu Plant Rev. 2015;48:1–22. [Google Scholar]
- Lei M, Liu Y, et al. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 2011;156:1116–1130. doi: 10.1104/pp.110.171736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Welti R, Wang X. Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 2006;142:750–761. doi: 10.1104/pp.106.085647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Xu C, et al. Proteomic analysis of roots growth and metabolic changes under phosphorus deficit in maize (Zea mays L.) plants. Proteomics. 2007;7(9):1501–12. doi: 10.1002/pmic.200600960. [DOI] [PubMed] [Google Scholar]
- Li Y, Van Den Ende W, Rolland F. Sucrose induction of anthocyanin biosynthesis is mediated by DELLA. Mol Plant. 2014;7:570–572. doi: 10.1093/mp/sst161. [DOI] [PubMed] [Google Scholar]
- Li Z, Qiu Q, et al. Metabolite alteration in response to low phosphorus stress in developing tomato fruits. Plant Physiol Biochem. 2021;159:234–243. doi: 10.1016/j.plaphy.2020.12.023. [DOI] [PubMed] [Google Scholar]
- Li Z, Hu J, et al. Integrative analysis of the metabolome and transcriptome reveal the phosphate deficiency response pathways of alfalfa. Plant Physiol Biochem. 2022;170:49–63. doi: 10.1016/j.plaphy.2021.11.039. [DOI] [PubMed] [Google Scholar]
- Li H, He K, et al. Molecular mechanism of phosphorous signaling inducing anthocyanin accumulation in Arabidopsis. Plant Physiol Biochem. 2023;196:121–129. doi: 10.1016/j.plaphy.2023.01.029. [DOI] [PubMed] [Google Scholar]
- Li J, Su Y, et al. Phosphate deficiency modifies lipid composition and seed oil production in Camelina. Plant Sci. 2023;330:111636. doi: 10.1016/j.plantsci.2023.111636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang J, et al. Control of phosphate homeostasis through gene regulation in crops. Curr Opin Plant Biol. 2014;21:59–66. doi: 10.1016/j.pbi.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Ligaba A, Shen H, et al. The role of phosphorus in aluminium-induced citrate and malate exudation from rape (Brassica napus) Physiol Plant. 2004;120:575–584. doi: 10.1111/j.0031-9317.2004.0290.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Yang L, et al. A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc Natl Acad Sci U S A. 2015;112:E6571–E6578. doi: 10.1073/pnas.1514598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Huang TK, et al. (2016) Identification of plant vacuolar transporters mediating phosphate storage. Nat Commun. 2016;71(7):1–11. doi: 10.1038/ncomms11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Wang L, et al. Mutation of the chloroplast-localized phosphate transporter OsPHT2;1 reduces flavonoid accumulation and UV tolerance in rice. Plant J. 2020;102:53–67. doi: 10.1111/tpj.14611. [DOI] [PubMed] [Google Scholar]
- Luan M, Zhao F, et al. Vacuolar phosphate transporters contribute to systemic phosphate homeostasis vital for reproductive development in arabidopsis. Plant Physiol. 2019;179:640–655. doi: 10.1104/pp.18.01424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Ma P, et al. Metabolite profiling and genome-wide association studies reveal response mechanisms of phosphorus deficiency in maize seedling. Plant J. 2019;97:947–969. doi: 10.1111/tpj.14160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Liu Y, et al. Metabolic alterations provide insights into Stylosanthes roots responding to phosphorus deficiency. BMC Plant Biol. 2020;20:1–16. doi: 10.1186/s12870-020-2283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu Y, Tang H, et al. Major crop species show differential balance between root morphological and physiological responses to variable phosphorus supply. Front Plant Sci. 2016;7:1939. doi: 10.3389/fpls.2016.01939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63:2853–2872. doi: 10.1093/jxb/ers091. [DOI] [PubMed] [Google Scholar]
- Mehra P, Pandey BK, et al. A novel glycerophosphodiester phosphodiesterase improves phosphate deficiency tolerance in rice. Plant Cell Environ. 2019;42:1167–1179. doi: 10.1111/pce.13459. [DOI] [PubMed] [Google Scholar]
- Meng L, Qi C. Determinant factors and regulatory systems for anthocyanin biosynthesis in rice Apiculi and Stigmas. Rice. 2021;14:1–18. doi: 10.1186/s12284-021-00480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin DE, Abdolrahimzadeh H, Baddiley J. Replacement of acidic phosphates by acidic glycolipids in Pseudomonas diminuta. Nature. 1974;249:268–269. doi: 10.1038/249268a0. [DOI] [PubMed] [Google Scholar]
- Mo X, Zhang M, et al. Phosphate (Pi) starvation upregulated GMCSN5A/B participates in anthocyanin synthesis in soybean (Glycine max) dependent on pi availability. Int J Mol Sci. 2021;22:12348. doi: 10.3390/ijms222212348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Gödde V, et al. Metabolic adaptations of white lupin roots and shoots under phosphorus deficiency. Front Plant Sci. 2015;6:1–10. doi: 10.3389/fpls.2015.01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y, Umena Y, et al. Thylakoid membrane lipid sulfoquinovosyl-diacylglycerol (SQDG) is required for full functioning of photosystem II in Thermosynechococcus elongatus. J Biol Chem. 2018;293:14786. doi: 10.1074/jbc.RA118.004304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. Phosphate starvation and membrane lipid remodeling in seed plants. Prog Lipid Res. 2013;52:43–50. doi: 10.1016/j.plipres.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Nezamivand-Chegini M, Metzger S, et al. Integration of transcriptomic and metabolomic analyses provides insights into response mechanisms to nitrogen and phosphorus deficiencies in soybean. Plant Sci. 2023;326:111498. doi: 10.1016/j.plantsci.2022.111498. [DOI] [PubMed] [Google Scholar]
- Ngo AH, Nakamura Y. Phosphate starvation-inducible GLYCEROPHOSPHODIESTER PHOSPHODIESTERASE6 is involved in Arabidopsis root growth. J Exp Bot. 2022;73(9):2995–3003. doi: 10.1093/jxb/erac064. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Shimojima M, et al. A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis Is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell. 2009;21:892–909. doi: 10.1105/tpc.108.063925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Takano K, Saito K. Lipidomic analysis of soybean leaves revealed tissue-dependent difference in lipid remodeling under phosphorus-limited growth conditions. Plant Biotechnol. 2017;34:57–63. doi: 10.5511/plantbiotechnology.17.0113a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio MB, Ng S, et al. SPX4 Acts on PHR1-dependent and -independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol. 2019;181:332–352. doi: 10.1104/pp.18.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panche AN, Diwan AD, Chandra SR. Flavonoids: an overview. J Nutr Sci. 2016;5:e47. doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Pant P, et al. Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ. 2015;38:172–187. doi: 10.1111/pce.12378. [DOI] [PubMed] [Google Scholar]
- Peret B, Desnos T, et al. Root architecture responses. Search Phosphate Plant Physiol. 2014;166:1713–1723. doi: 10.1104/pp.114.244541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff J, Denton AK, et al. Phosphate starvation causes different stress responses in the lipid metabolism of tomato leaves and roots. Biochem Biophys Acta Mol Cell Biol Lipids. 2020;1865:158763. doi: 10.1016/j.bbalip.2020.158763. [DOI] [PubMed] [Google Scholar]
- Phoenix GK, Johnson DA, et al. Niche differentiation and plasticity in soil phosphorus acquisition among co-occurring plants. Nat Plants. 2020;64(6):349–354. doi: 10.1038/s41477-020-0624-4. [DOI] [PubMed] [Google Scholar]
- Raya-González J, Ojeda-Rivera JO, et al. (2021) MEDIATOR16 orchestrates local and systemic responses to phosphate scarcity in Arabidopsis roots. New Phytol. 2021;229(3):1278–1288. doi: 10.1111/nph.16989. [DOI] [PubMed] [Google Scholar]
- Richardson AE, Lynch JP, et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil. 2011;349:121–156. doi: 10.1007/s11104-011-0950-4. [DOI] [Google Scholar]
- Robinson WD, Park J, et al. The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana. J Exp Bot. 2012;63:6531–6542. doi: 10.1093/jxb/ers309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santosh KC, Liu M, et al. Metabolic changes of amino acids and flavonoids in tea plants in response to inorganic phosphate limitation. Int J Mol Sci. 2018;19:3683. doi: 10.3390/ijms19113683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane MW, Stigter K, et al. Senescence-inducible cell wall and intracellular purple acid phosphatases: Implications for phosphorus remobilization in Hakea prostrata (Proteaceae) and Arabidopsis thaliana (Brassicaceae) J Exp Bot. 2014;65:6097–6106. doi: 10.1093/jxb/eru348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane MW, Feil R, et al. Light-dependent activation of phosphoenolpyruvate carboxylase by reversible phosphorylation in cluster roots of white lupin plants: diurnal control in response to photosynthate supply. Ann Bot. 2016;118:637–643. doi: 10.1093/aob/mcw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Wang S, et al. OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.) Plant Cell Environ. 2013;36:607–620. doi: 10.1111/pce.12001. [DOI] [PubMed] [Google Scholar]
- Singh NRR, Roychowdhury A, et al. Silencing of SlSPX1 and SlSPX2 promote growth and root mycorrhization in tomato (Solanum lycopersicum L.) seedlings. Plant Sci. 2023;333:111723. doi: 10.1016/j.plantsci.2023.111723. [DOI] [PubMed] [Google Scholar]
- Srivastava R, Parida AP, et al. Identification, structure analysis, and transcript profiling of purple acid phosphatases under Pi deficiency in tomato (Solanum lycopersicum L.) and its wild relatives. Int J Biol Macromol. 2020;165:2253–66. doi: 10.1016/j.ijbiomac.2020.10.080. [DOI] [PubMed] [Google Scholar]
- Srivastava R, Basu S, Kumar R. Phosphorus starvation response dynamics and management in plants for sustainable agriculture. J Plant Biochem Biotechnol. 2021;30:829–847. doi: 10.1007/s13562-021-00715-8. [DOI] [Google Scholar]
- Srivastava R, Sirohi P, et al. The enhanced phosphorus use efficiency in phosphate-deficient and mycorrhiza-inoculated barley seedlings involves activation of different sets of PHT1 transporters in roots. Planta. 2021;254:1–17. doi: 10.1007/s00425-021-03687-0. [DOI] [PubMed] [Google Scholar]
- Stigter KA, Plaxton WC. Molecular mechanisms of phosphorus metabolism and transport during leaf senescence. Plants. 2015;4:773–798. doi: 10.3390/plants4040773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strock CF, De La Riva LM, Lynch JP. Reduction in root secondary growth as a strategy for phosphorus acquisition. Plant Physiol. 2018;176:691–703. doi: 10.1104/pp.17.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulieman S, Tran LSP. Legume nitrogen fixation in soils with low phosphorus availability: adaptation and regulatory implication. Berlin: Springer; 2017. pp. 1–287. [Google Scholar]
- Sun J, Li Q, et al. Analysis of metabolomic changes in xylem and phloem sap of cucumber under phosphorus stresses. Metabolites. 2022;12(4):361. doi: 10.3390/metabo12040361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J, Lee S, et al. Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 2015;241:55–64. doi: 10.1016/j.plantsci.2015.09.027. [DOI] [PubMed] [Google Scholar]
- Takami T, Ohnishi N, et al. Organelle DNA degradation contributes to the efficient use of phosphate in seed plants. Nat Plants. 2018;412(4):1044–1055. doi: 10.1038/s41477-018-0291-x. [DOI] [PubMed] [Google Scholar]
- Tian J, Wang C, et al. Overexpression of OsPAP10a, a root-associated acid phosphatase, increased extracellular organic phosphorus utilization in rice. J Int Plant Biol. 2012;54(9):631–639. doi: 10.1111/j.1744-7909.2012.01143.x. [DOI] [PubMed] [Google Scholar]
- Uhde-Stone C, Gilbert G, et al. Acclimation of white lupin to phosphorus deficiency involves enhanced expression of genes related to organic acid metabolism. Plant Soil. 2003;248:99–116. doi: 10.1023/A:1022335519879. [DOI] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003;157(3):423–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- Veljanovski V, Vanderbeld B, et al. Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived arabidopsis suspension cells and seedlings. Plant Physiol. 2006;142:1282. doi: 10.1104/pp.106.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veramendi J, Willmitzer L, Trethewey RN. In vitro grown potato microtubers are a suitable system for the study of primary carbohydrate metabolism. Plant Physiol Biochem. 1999;37:693–697. doi: 10.1016/S0981-9428(00)80100-X. [DOI] [Google Scholar]
- Verma L, Bhadouria J, et al. Monogalactosyl diacylglycerol synthase 3 affects phosphate utilization and acquisition in rice. J Exp Bot. 2022;73(14):5033–5051. doi: 10.1093/jxb/erac192. [DOI] [PubMed] [Google Scholar]
- Walan P, Davidsson S, et al. Phosphate rock production and depletion: regional disaggregated modeling and global implications. Resour Conserv Recycl. 2014;93:178–187. doi: 10.1016/j.resconrec.2014.10.011. [DOI] [Google Scholar]
- Wang C, Yue W, et al. Rice SPX-major facility superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. Plant Physiol. 2015;169:01005. doi: 10.1104/pp.15.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kuo HF, Chiou TJ. Intracellular phosphate sensing and regulation of phosphate transport systems in plants. Plant Physiol. 2021;187:2043–2055. doi: 10.1093/plphys/kiab343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu J, et al. Involvement of PtPHR1 in phosphates starvation-induced alkaloid biosynthesis in Pinellia ternata (Thunb.) Breit. Front Plant Sci. 2022;13:2709. doi: 10.3389/fpls.2022.914648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Walther D, et al. Metabolomic markers and physiological adaptations for high phosphate utilization efficiency in rice. Plant Cell Environ. 2020;43:2066–2079. doi: 10.1111/pce.13777. [DOI] [PubMed] [Google Scholar]
- Wen T, Yuan J, et al. Enrichment of beneficial cucumber rhizosphere microbes mediated by organic acid secretion. Hortic Res. 2020;1:7. doi: 10.1038/s41438-020-00380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Kobayashi Y, et al. Organic acid excretion from roots: a plant mechanism for enhancing phosphorus acquisition, enhancing aluminum tolerance, and recruiting beneficial rhizobacteria. Soil Sci Plant Nutr. 2018;64:697–704. doi: 10.1080/00380768.2018.1537093. [DOI] [Google Scholar]
- Xiao X, Zhang J, et al. SHORT-ROOT stabilizes PHOSPHATE1 to regulate phosphate allocation in Arabidopsis. Nat Plants. 2022;9:1074–1081. doi: 10.1038/s41477-022-01231-w. [DOI] [PubMed] [Google Scholar]
- Yang SY, Huang TK, et al. Role of vacuoles in phosphorus storage and remobilization. J Exp Bot. 2017;68:3045–3055. doi: 10.1093/jxb/erw481. [DOI] [PubMed] [Google Scholar]
- Yu RP, Li XX, et al. Phosphorus facilitation and covariation of root traits in steppe species. New Phytol. 2020;226(5):1285–1298. doi: 10.1111/nph.16499. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen H, et al. Comparative transcriptomic and metabolomic analyses reveal the protective effects of silicon against low phosphorus stress in tomato plants. Plant Physiol Biochem. 2021;166:78–87. doi: 10.1016/j.plaphy.2021.05.043. [DOI] [PubMed] [Google Scholar]
- Zhao X, Chen KK, et al. Transcriptome analysis provides insights into the response of Lotus corniculatus roots to low-phosphorus stress. Front Plant Sci. 2023;14:1089380. doi: 10.3389/fpls.2023.1089380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Wang R, et al. Comparative physiological and proteomic response to phosphate deficiency between two wheat genotypes differing in phosphorus utilization efficiency. J Proteomics. 2023;280:104894. doi: 10.1016/j.jprot.2023.104894. [DOI] [PubMed] [Google Scholar]
- Zhu W, Miao Q, et al. The mitochondrial phosphate transporters modulate plant responses to salt stress via affecting ATP and gibberellin metabolism in Arabidopsis thaliana. PLoS One. 2012 doi: 10.1371/journal.pone.0043530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Chen M, et al. Characterization of purple acid phosphatase family and functional analysis of GmPAP7a/7b involved in extracellular ATP utilization in soybean. Front Plant Sci. 2020;11:661. doi: 10.3389/fpls.2020.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Bing H, Wu Y. Citric acid promotes the mobilization of phosphorus under the lower concentration of low molecular weight organic acids in acidic forest soil. Adsorpt Sci Technol. 2022 doi: 10.1155/2022/5071907. [DOI] [Google Scholar]