Abstract
While the field of microbial biogeography has largely focused on the contributions of abiotic factors to community patterns, the potential influence of biotic interactions in structuring microbial communities, such as those mediated by the production of specialized metabolites, remains largely unknown. Here, we examined the relationship between microbial community structure and specialized metabolism at local spatial scales in marine sediment samples collected from the Long-Term Ecological Research (LTER) site in Moorea, French Polynesia. By employing a multi-omic approach to characterize the taxonomic, functional, and specialized metabolite composition within sediment communities, we find that biogeographic patterns were driven by local scale processes (e.g., biotic interactions) and largely independent of dispersal limitation. Specifically, we observed high variation in biosynthetic potential (based on Bray-Curtis dissimilarity) between samples, even within 1 m2 plots, that reflected uncharacterized chemical space associated with site-specific metabolomes. Ultimately, connecting biosynthetic potential to community metabolomes facilitated the in situ detection of natural products and revealed new insights into the complex metabolic dynamics associated with sediment microbial communities. Our study demonstrates the potential to integrate biosynthetic genes and metabolite production into assessments of microbial community dynamics.
Subject terms: Biogeography, Metabolomics, Microbial ecology, Environmental chemistry, Metagenomics
Introduction
The biogeographic processes that generate and maintain microbial diversity, such as environmental selection, dispersal, and species interactions, offer insights into the mechanisms that structure microbiomes [1]. At large (i.e., continental) spatial scales, community structure is primarily driven by dispersal limitation with environmental filters selecting for specific taxa at increasingly smaller spatial scales [2, 3]. At local spatial scales, there is an assumption that biotic interactions (e.g., predation, competition), including those mediated by chemical communication [4], largely account for the differences among communities (beta-diversity) [5]. The wide array of biologically active microbial specialized metabolites produced in response to both environmental cues and species interactions likely contribute to differences in community composition at small spatial scales [6, 7]. However, understanding how these complex chemical interactions contribute to community dynamics has largely been ignored.
While there is a growing appreciation for microbial chemical ecology [8], challenges associated with the in situ detection of specialized metabolites has hampered our understanding of their effects on community dynamics. For instance, marine sediment bacteria are prolific producers of small molecule natural products or specialized metabolites [9] that include potent antibiotics and cytotoxins. Yet, most of our knowledge of natural product production is inferred from lab cultures, providing limited insights into their ecological roles within a community context [10, 11]. Due to these challenges, molecules mediating such interactions are often inferred from genomic potential as estimated through the detection of natural product biosynthetic gene clusters (BGCs) or other biosynthetic enzymes [12, 13]. These environmental surveys have identified habitat-specific patterns of biosynthetic potential [14, 15], suggesting that specialized metabolites exhibit a biogeographic signal in concordance with taxonomic composition. Although these types of analyses are valuable, the environmental factors that elicit BGC expression remain largely unknown, as well as the concentrations at which compounds occur in nature and their roles in mediating community interactions. Given the recent advancements in mass spectrometry and other analytical tools, the in situ detection of microbial natural products [16, 17] and their influence on community structure remains an essential frontier for the field of microbial ecology.
Here, we sought to focus on both biosynthetic potential and realized natural product production in the context of marine sediment microbial community structure across varying spatial scales. To do so, we compared microbial community composition from 176 marine sediment samples collected from the Long-Term Ecological Research (LTER) site in Moorea, French Polynesia. While benthic cover, nutrient input, and microbial communities associated with coral reefs and the surrounding seawater have been tracked at the Moorea LTER, little work has been done on the sediment microbial communities. Given that shifts in sediment microbiomes can impact above-sediment community health and dynamics [18, 19], we aimed to broaden our understanding of this well-studied ecosystem.
To determine how microbial community structure and specialized metabolism are linked at local spatial scales, we employed a multi-omic approach to characterize the taxonomic composition, functional and biosynthetic potential, and community metabolomes within reef-associated sediments. We constrained our sampling to a single LTER site (LTER2) to minimize negative correlations between taxonomic community similarity and geographic distances (“distance-decay” effects), thereby limiting environmental variation and increasing our ability to determine if community metabolomes influence composition [3]. Furthermore, we employed a partially nested sampling design across eight replicate plots to provide sample comparisons ranging from cm to km scales within this relatively homogeneous environment (Fig. S1A). In doing so, we expected that variation in community composition (beta-diversity) would largely be independent of distance effects (i.e., high community similarity with low slope over the distance-decay curve). Rather, we hypothesized that local site differences could be linked to biosynthetic potential and community metabolomes, as evidenced by low compositional similarity in BGC composition and specialized metabolite production. Although most compounds detected using untargeted environmental metabolomics could not be identified, correlations between compounds and community structure were nonetheless established. Our paired metagenomic-metabolomic approach also facilitated linkages between unknown metabolites and their candidate biosynthetic machinery, which is useful for future natural product discovery efforts.
Results
Community patterns in marine sediments
Sediment samples were collected from eight replicate 1 m2 plots within and around the Long-Term Ecological site LTER2 in Moorea, French Polynesia (Fig. S1A). LTER2 is characterized by high nitrogen levels (all sites = 0.5 ± 0.06 %N based on algal tissue; Fig. S1B) and steady coral degradation over time (Fig. S1C). The eight collection sites (depth range 0.6–3.1 m) could be further defined as back (n = 4) and fringing (n = 4) reef, seven of which were characterized by light colored, relatively coarse sediments consistent with high calcium carbonate content. One fringing reef site (site 6) was dominated by algae and characterized by fine dark sediment containing small rocks. Our nested sampling design (16 subsamples × 8 sites + 4 transects per site = 176) allowed for 16 S rRNA gene community composition comparisons across a range of spatial scales. We observed similar community compositions across all sites at broad taxonomic levels with Proteobacteria and Cyanobacteria dominating the communities (Fig. S2A). At finer-taxonomic levels, as assessed using 16 S rRNA gene amplicon sequence variants (ASVs), we observed a distinct separation between the algae-dominated site (site 6) and the other reef sites (Fig. S2B), thus supporting previous findings that the transition between reef and algae-dominated habitats is reflected in microbial community composition [20].
To assess changes in community composition across cm to km scales within a more homogeneous environment, we excluded site 6 and narrowed our focus to the reef-dominated sediment samples (inset, Fig. S1A). We found that microbial community composition was largely structured at local scales (Fig. S3A; Bray-Curtis, p < 0.001 [PERMANOVA]), with site effects accounting for 18.2% of total variation and reef type (fringing vs. back reef) contributing an additional 7.2% (Table 1; p < 0.001 [PERMANOVA]). This was further supported by a distance-decay curve demonstrating a significant, negative slope with community similarity decreasing as a function of geographic distance (slope = −0.000013, p < 0.001; pink line in Fig. 1). While a small negative slope was observed, distance effects only explained 3.3% of the total variation in community composition (Table 1; reporting R2 [linear regression]), suggesting that factors other than dispersal limitation are responsible for community differences. Indeed, when we corrected for ASV phylogenetic relatedness (Weighted UniFrac), we observed a significantly shallower slope (Fig. S3C; slope = −0.000003, p < 0.001) suggesting that community variation over spatial distances was largely associated with differences among closely related taxa. Given the high correlation between beta-diversity metrics (Spearman’s ρ = 0.77, p < 0.001 [Mantel test]), the distance-decay patterns indicate that most compositional variation was due to local scale variation driven by site effects.
Table 1.
Results from permutational analysis of variance (PERMANOVA) for the effects of site, reef type (back vs. fringing), and geographic distance on microbial community metrics.
| Taxonomic composition | Functional composition | Biosynthetic potential | Realized potential | ||||
|---|---|---|---|---|---|---|---|
| ASV (Bray-Curtis) | ASV (UniFrac) | Metagenomes | Pfam | OBUs | GCFs | Metabolome | |
| Site (R2) | 0.18*** | 0.17*** | 0.25** | 0.22 | 0.19*** | 0.25*** | 0.28*** |
| Reef type (R2) | 0.07*** | 0.07*** | 0.07** | 0.07* | 0.04** | 0.06** | 0.09*** |
| Geographic distance (R2) | 0.03*** | 0.01*** | <0.0001 | <0.01 | 0.01** | 0.01** | 0.01*** |
Percent of variance explained depicted as R2 values for each effect with level of significance (*p < 0.05, **p < 0.01, ***p < 0.001).
ASV amplicon sequence variants of the 16 S rRNA gene, Pfam protein families, OBU operational biosynthetic unit, GCF gene cluster family.
Fig. 1. Biogeography of microbial community structure in reef-associated marine sediments.
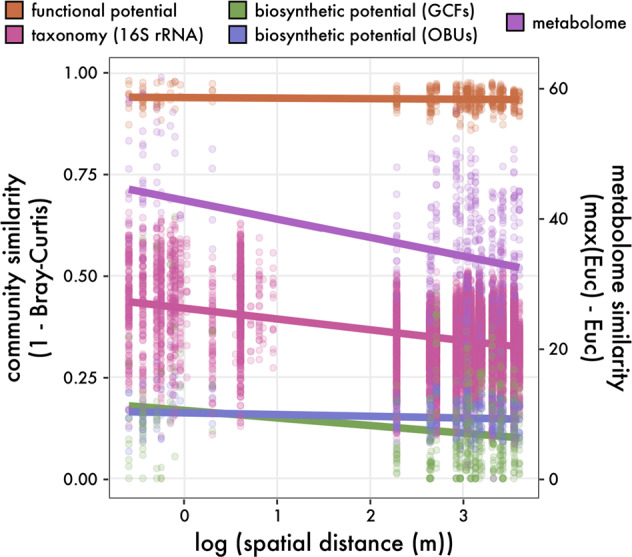
Solid lines denote the least squares linear regression across spatial scales for taxonomic, functional, biosynthetic potential (assessed based on both operational biosynthetic units (OBUs) of ketosynthase/condensation domains and gene cluster families (GCFs)), and community metabolomes. Each point represents a pairwise comparison between community samples with distances derived from Bray-Curtis metrics for all measurements except for metabolomes, which used Euclidean distances.
Functional conservation and biosynthetic variation across sites
Given the negative, shallow distance-decay slopes for taxonomic composition observed across small (<1 m) to large (ca. 1 km) spatial scales (Fig. S3C), we next asked whether functional potential exhibited similar patterns. To do so, we generated shotgun metagenomic data from 36 samples (meandepth = 205 M reads) across all eight sites to initially compare algal to reef samples. Due to possible sequencing biases, we first confirmed that the taxonomic patterns derived from the metagenomic data recapitulated the 16 S rRNA gene results (Table 1; Fig. S3D) and that site differences were consistent across technical replicates (Fig. S3E). Subsequently, we investigated functional potential across communities that may indicate environmental differences within LTER2. In terms of primary metabolic pathways, all sediment communities were similarly enriched in nitrogen cycling and methanogenesis genes (Fig. S4A) and common glycoside hydrolase genes related to the degradation of simple carbon substrates like oligosaccharides and starch (Fig. S4B). To compare functional potential with community composition, we again excluded the algae-dominated site 6 and found that 21.9% of the variation in total functional composition, based on protein family (Pfam) abundances, was explained by local site effects. This relationship did not significantly differ across sites (Table 1; p > 0.05 [PERMANOVA]), as we observed high functional redundancy across the reef sediment communities that was independent of distance effects: i.e., geographic distance accounted for only 0.3% of functional variation (orange line in Fig. 1; reporting R2 [linear regression]: slope = −0.0000011, p > 0.05). Similar to taxonomic composition, 7.4% of the total functional variation was significantly associated with reef type (back vs. fringing reef; p < 0.05 [PERMANOVA]), possibly owing to higher N input from nearshore storm runoff at the fringing reef sites [21].
Both taxonomic and functional variation analyses indicate that local (within) site processes are contributing to differences among microbial communities. Given that specialized metabolites can mediate biotic interactions [8], we further explored these functional differences across sediment communities by focusing on biosynthetic potential. To reduce biases introduced from assembly-based methods, we first extracted all ketosynthase (KS) and condensation (C) domains from the unassembled data, which are integral to the biosynthesis of polyketides (PKs) and nonribosomal peptides (NRPs), respectively. After clustering domains into operational biosynthetic units (OBUs) at 80% amino acid similarity, based on the prediction that domains in the same OBU will encode the biosynthesis of similar natural products [22, 23], we found high community dissimilarity in OBU composition across sites with a significant, but shallower distance effect compared to taxonomic composition (slope = −0.0000033, p < 0.01; blue line in Fig. 1). Distance effects only explained 1.1% of OBU compositional variation across samples while site effects explained 19.3% of variation and reef type (fringing vs. back reef) contributing an additional 4% (Table 1; both p < 0.01 [PERMANOVA]). Thus, broad community functions (i.e., primary metabolism) did not differ over geographic distances, but specialized microbial functions (i.e., specialized metabolism) exhibited fine-scale local differences among sediment communities.
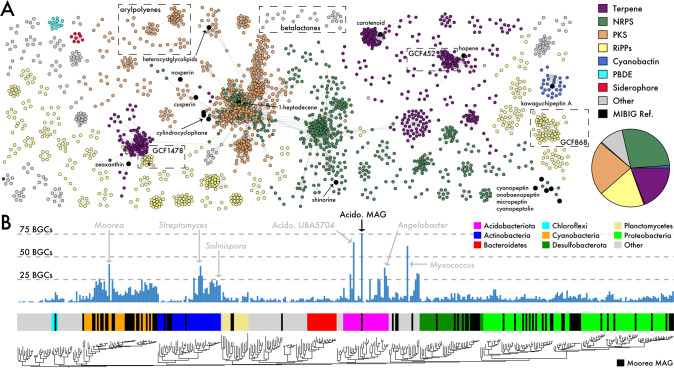
From the assembled metagenomes, we reconstructed 3,137 biosynthetic gene clusters (BGCs) across all samples (Fig. S5B). These were predominantly associated with NRP and terpene biosynthesis. After clustering these BGCs into 1147 gene cluster families (GCFs; Fig. 2A), we found that only 14 were related to experimentally characterized BGCs [24], the products of which include peptins, carotenoids, and heterocyst glycolipids (Fig. S5A). The remaining GCFs represent uncharacterized biosynthetic potential. We observed minimal GCF overlap among samples with only two shared across all eight sites. Both could be assigned to alphaproteobacterium based on metagenome-assembled genome (MAG) classifications (order: Kiloniellales; abundance16S = 0.38 ± 0.45%). These ubiquitous GCFs were predicted to encode an unknown thioaminidated ribosomal peptide (TfuA-related BGC) and a hopene-like terpene (GCF868 and GCF452 in Fig. 2A, respectively). Only one GCF was found in all reef-associated sites but absent in the algae-dominated samples (site 6). This GCF encoded a hybrid terpene-bacteriocin, with some similarity to the BGC associated with the common carotenoid zeaxanthin (Fig. 2A), and was observed in a group of Desulfobacterota MAGs (order: Desulfobacterales; abundance16S = 5.95 ± 2.6%). Most of the GCFs (accounting for 679 of the 3137 BGCs) shared no similarity with other GCFs within or across sites and thus were represented as unconnected nodes in the network. This lack of GCF overlap resulted in low compositional similarity with high GCF compositional differences across geographic distances (Table 1; slope = −0.0000097, p < 0.01; green line in Fig. 1), thus indicating unique biosynthetic gene assemblages within each sample.
Fig. 2. Biosynthetic potential across marine sediment communities.
A Similarity network of identified biosynthetic gene clusters (BGCs) clustered into gene cluster families (GCFs), colored by BGC classification. Inset pie chart depicts relative proportion of BGC classes. Black nodes indicate known reference BGCs from the MIBIG database. Singleton and doubleton GCFs not shown. B BGC distributions across the bacterial phylogeny colored by phyla. Moorea MAGs denoted in black. A subset of reference genomes/MAGs indicated in light gray.
To further explore primary and specialized taxonomic distributions, we examined the metabolic and biosynthetic potential of the 789 MAGs (Table S1), which accounted for 84% (2635 of 3137) of all assembled BGCs. These MAGs were predominantly identified as Cyanobacteria and Proteobacteria, mirroring our community composition results (Fig. S2A). We first analyzed our medium- (n = 80; completeness > 70%, contamination < 10%) and high-quality (n = 88; completeness > 85%, contamination < 10%) MAGs (dereplicated to n = 104) for primary metabolic potential. In terms of the dominant primary metabolic pathways across communities (Fig. S4A), Gammaproteobacteria MAGs encoded genes for the (de)nitrification and N fixation pathways while Desulfobacterota MAGs converted methylamine to ammonia as part of methanogenesis. In terms of biosynthetic potential, we compared our dereplicated medium- and high-quality MAGs to a variety of closely related genomes/MAGs from taxa that are known for high abundances of BGCs and specialized metabolite production (Fig. 2B). In particular, we assembled a high-quality Acidobacteriota MAG135.14 (91% complete, 4% contamination, genome size = 11.6 Mbp) from a single sample that encoded 76 BGCs (2.2 Mbp or 18.7% of the genome, average BGC length = 28.6 kbp), a number that surpasses any known MAG or genome including those documented across a global metagenomic dataset [25]. While the fragmentation of many BGCs likely skews BGC content, these approaches offer insights into unexplored chemical space and taxa. Together, both OBU and GCF metrics demonstrate high levels of variability in the biosynthetic potential of sediment microbial communities, even within 1 m2 sites. This likely reflects a wealth of uncharacterized biosynthetic potential that was not previously documented in global databases.
Connecting biosynthetic potential to community metabolomes
While gene-based surveys are indicative of biosynthetic potential, many BGCs are dependent on specific environmental triggers for expression [26] and thus remain silent under laboratory conditions [27]. Therefore, to investigate how biosynthetic potential translated to compound production, we utilized untargeted liquid chromatography–high-resolution mass spectrometry (LC-HRMS) to analyze extracts from the sediment samples (n = 36) used for the metagenomic analyses. In total, we identified 2164 unique molecular features and show that the major taxonomic differences between the reef and algae-dominated communities (Fig. S2B) translated to the metabolomes (Fig. S6A). After removing controls and features from the algae-dominated site 6, the reef-associated sediment metabolomes displayed clear site-specific signatures (Fig. 3A; Euclidean distance). Indeed, the community metabolome strongly reflected local site and reef type (fringing vs. back reef), accounting for 27.5% and 9.4% of the total variation, respectively (Table 1; both p < 0.001 [PERMANOVA]). The site-specific metabolome signatures were strongly reflected in our distance-decay curves (purple line in Fig. 1) where metabolite similarity was greatest within sites.
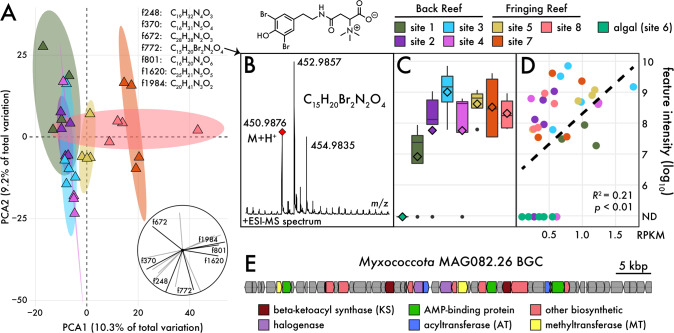
Fig. 3. Community metabolomes and candidate producers.
A Principal Component Analysis (PCA) showing differences in community metabolomes. Colored ellipses represent 75% confidence intervals around each site. Inset depicts contributions of the top molecular features to the principal dimensions with black lines denoting features with predicted molecular formulas shown above. B MS2 fragmentation of feature772 with diagnostic bromination signature, calculated mass, and probable molecular structure. C Abundance of feature772 across sites based on feature intensity in MS1 chromatograms. Black bars represent medians, diamonds represent means. D Linear regression of feature772 with the abundance of Myxococcota MAG082.26 (assessed by reads per kilobase mapped, RPKM). E Candidate BGC for feature772. Arrows represent individual genes and colored by predicted function.
While local site effects accounted for an increasing percent of variation explained when moving from taxonomy (16 S rRNA gene) to biosynthetic potential (GCF) to metabolomes (Table 1), we did not observe strong correlations across these metrics. For instance, metabolite composition did not correlate with either 16 S rRNA gene ASV (Spearman’s ρ = −0.02, p > 0.05 [Mantel test]) or OBU compositions (Spearman’s ρ = 0.16, p > 0.05 [Mantel test]). Rather, these compositional metrics may indicate that finer resolution is needed to correlate taxa, biosynthetic genes, and compounds. When we assessed the correlation between metabolomes and fine-scale taxonomic marker genes (i.e., diversity below the 16 S rRNA gene) derived from metagenomic data [28], we found higher congruence between metabolomes and taxa (Spearman’s ρ = 0.37, p < 0.01 [Mantel test]). These results suggest that finer levels of taxonomic resolution are needed to resolve relationships between specialized metabolites and their producers. Finally, these results indicate that deep metagenomic sequencing, as performed here, can promote connections between the inherently complex multi-omic datasets generated from environmental samples.
To determine how microbial community metabolomes are structured at local scales, we applied machine learning approaches to identify molecular features driving metabolomic variation across samples. Cheminformatic tools associated with the GNPS online platform facilitated the analysis of our large LC-MS datasets and the identification of molecules based on the comparison of MS/MS fragmentation spectra with over 580,000 library spectra [29]. We visualized the chemical space present in the sediment metabolomes using feature-based molecular networking [30] (Fig. S6B). Comparable with the biosynthetic potential encoded in the metagenomes, we observed a large proportion of singleton nodes in the network (1770, 81.8%) indicating the detection of large numbers of structurally unrelated compounds. In addition, 357 features (16.5% of the total) were observed at one site and 185 (8.5%) were shared between two sites, further highlighting the fine-scale spatial differences in community metabolomes. Further multivariate and random forest analyses revealed unidentified molecular features that were responsible for sample and site differences (inset in Fig. 3A).
While a majority of the compounds associated with the metabolomic differences among sites could not be identified, even after manual inspection, database searches [31], and other cheminformatic predictions [32], we were nonetheless successful in the identification of several natural products in the marine sediment extracts. The initial structural predictions based on MS/MS spectra comparisons with GNPS suggested 388 known features (Fig. S6B); however, many of these annotations proved spurious after manual inspection of their MS/MS mirror plots to reference spectra. Among the confidently identified features, most could be described as lipids, chlorophyll breakdown products (e.g., pheophorbide), and carotenoids (e.g., fucoxanthin). We also confidently identified several microbial specialized metabolites including coumarin derivatives and the cyanobacterial derived cyclic depsipeptides hantupeptins A-C [33, 34], which were only detected at site 8 (Fig. S6B). The latter of these features were initially annotated by GNPS as the closely related antanapeptins A-C [35], but manual inspection of the MS/MS spectra revealed peaks corresponding to m/z = 228.1033 and 246.1112 (Fig. S6C), which are diagnostic for the proline-3-phenyllactic acid fragments in the hantupeptins [33]. While the BGC for hantupeptins is unknown, we identified a highly fragmented NRPS BGC from the site 8 metagenome with homology to the distantly related anabaenopeptin BGC (Fig. S6D). This partial BGC included an LCL condensation domain and an adenylation domain selecting for valine (Fig. S6E), both of which are predicted for hantupeptin biosynthesis [33, 34]. Nevertheless, most molecular features could not be annotated and represent unexplored chemical space associated with sediment metabolomes.
Linking metabolites to producers
Pairing metagenomic and metabolomic datasets provides opportunities to link compounds to their cognate BGCs based on correlations in relative abundances and predicted biochemistry. While most molecular features provided only minimal structural information, we identified one with the diagnostic MS isotopic pattern of a dibrominated compound with the most intensive peak m/z = 452.9857 [M+H]+ and the lowest m/z = 450.9876 [M+H]+ (Fig. 3B). The calculated molecular formula (C15H20Br2N2O4) had no matches in the MarinLit database [31] and likely represents a new natural product. Extracts containing the highest amounts of this compound were pooled (Fig. S7A) and fractionated for LC-HRMS (Fig. S7B). The spectra suggested an alkaloid (Fig. S7C) consisting of a decarboxylated dibromotyrosine (high intensity peak m/z = 291.8975 [36], Fig. S7D) fused over an amide bond to an aspartic acid derived betaine, which corresponds to the observed mass loss of 159.089 Da and the high polarity (water soluble) of the compound (Fig. S7B). This new compound is structurally related to sponge-derived alkaloids [37]. However, due to the low compound yield, we were unable to explore its molecular structure further using nuclear magnetic resonance (NMR). This compound was observed at all sites except the algae-dominated site 6 with its highest abundance at site 3 (Fig. 3C). After correlating MAG abundances with the intensity of this dibrominated compound, we identified the Myxococcota MAG082.26 as a possible producing organism and a group of Desulfobacterota MAGs that have the potential to use this dibrominated molecule as an oxidant during respiration.
The high-quality Myxococcota MAG082.26 (93% complete) has a genome size of 7.7 Mbp and is distantly related to all reported genome or MAG sequences in this phylum, with its closest relative (77.4% average nucleotide identity [ANI]) belonging to the MAG-associated UBA9160 family. MAG082.26 was assembled from site 3, which contained the highest concentrations of the dibrominated aromatic metabolite, and was positively correlated with the mass of this compound across all samples (Fig. 3D; R2 = 0.21 [linear regression]). The biosynthesis of this molecule requires the halogenation of the aromatic phenyl ring (i.e., dibromotyrosine), which has been previously shown in marine myxobacterial metabolites such as salimabromide [38]. Myxococcota MAG082.26 harbors nine BGCs of which one contains two tetracycline 7-halogenase genes that share homology to brominases involved in the biosynthesis of the 2-arrylpyrrole moiety in pentabromopseudilins (MIBIG: BGC0000890/1) and bromopyrroles/bromophenols (MIBIG: BGC0001465). While the gene architecture of this BGC is unique (Table S2), it lacks the NRPS component (i.e., adenylation domain) expected for the biosynthesis of compound m/z = 452.9857 (Fig. 3E). While this remains the best candidate BGC assembled from our metagenomic data, more work is needed to confirm this connection.
While investigating linkages between MAGs, BGCs, and the dibrominated metabolite, we identified a group of five related MAGs (ANI ranges 77.2–98.5%) belonging to two bacterial families within the order Desulfobacterales that were highly correlated with the presence of this compound (R2 = 0.46 ± 0.01 [linear regression]). While these Desulfobacterota MAGs lacked the biosynthetic machinery to produce compound m/z = 452.9857, they encode a reductive dehalogenase (rdh) operon (Fig. S8A/B) along with genes for cobalamin biosynthesis, a key cofactor associated with reductive dehalogenation [39]. Moreover, the rdh operon is co-localized with several genes encoding electron transport proteins (i.e., ferredoxins and NADH-quinone reductase complexes; Fig. S8A), providing evidence that these microorganisms have the potential to use dibromophenol as a terminal electron acceptor during respiration. These findings add to the complex functional roles of natural products and support growing evidence that community metabolomes are shaped both by compound production and subsequent modification by other members of the community [26, 40].
Discussion
While the field of microbial biogeography has largely focused on the role of abiotic variables in structuring microbial distributions, there is growing appreciation for the role of microbial natural products in mediating biotic interactions within microbiomes [8]. The ability to assess environmental metabolomes is rapidly improving [16, 41], providing opportunities to transcend culture-based and synthetic community approaches or biosynthetic inferences derived from co-occurrence networks [42]. Here, we assessed the relationships between community metabolomes and composition at varying spatial scales by minimizing environmental variation through extensive sampling within a relatively homogeneous environment. We observed distinct biogeographic patterns associated with taxonomic composition, functional genes, biosynthetic potential, and the environmental detection of natural products. Finally, our multi-omics approach facilitated potential connections between metabolites, producing organisms, and those that may benefit from their production.
By reducing environmental variation through sampling within a single LTER site, we found evidence for biogeographic patterns that were largely independent of dispersal limitation. For example, geographic distance did not explain a large portion of community variance across all community measurements (<4% of total variance explained; Table 1), indicating that local, within site processes (e.g., biotic mechanisms) are driving community differences. This was evidenced from broad inter-habitat differences between the reef- and algae-dominated sites (Fig. S2C) and the high community similarity across spatial distances (Fig. 1), a pattern that was accentuated after correcting for phylogenetic relatedness (Fig. S3C). While ecological drift can account for local scale differences [43], our results suggest that biotic interactions, as measured by biosynthetic potential and specialized metabolite production, are linked to local biogeographic patterns. In both cases, we observed high community dissimilarity in biosynthetic potential (Fig. 1) and pronounced local site effects in community metabolome composition (Fig. 3A; Table 1). These patterns are largely congruent with observations of soil fungal communities, where local scale patterns were driven by extracellular enzyme production [44] that was not reflected in community composition.
Given the increasing awareness that natural products play important roles in structuring microbial communities, connecting chemical compounds with their producers (and associated biosynthetic machinery) and ultimately to ecological interactions remains a priority [45]. Despite major methodological advances, <5% of metabolites detected in marine systems can be identified [46] and fewer could be linked to ecological functions. Our results further demonstrate the limitations of cheminformatic tools for the identification of environmental specialized metabolites and the importance of stringent manual curation. In addition, untargeted metabolomics may miss low concentration compounds produced by members of the “rare biosphere” that are important contributors to the specialized metabolite pools in marine sediments [47]. Finally, correlation analyses built from co-occurrence networks of taxonomy and metabolites typically rely on coarse taxonomic markers, such as the 16 S rRNA gene. Given the lack of concordance between these parameters in our results, combined with evidence of high trait variation within 16S-defined taxa [48, 49], it appears that more highly resolved taxonomic marker are needed to assess relationships between metabolomes and the structure of complex microbial communities at fine spatial scales. A likely contributor to this observation is the fine-scale phylogenetic conservation in specialized metabolite production and BGC composition within taxa [50–53]. Nonetheless, at large spatial scales and across disparate environments, broad patterns of biosynthetic potential [14, 15, 54, 55] and the abundance of microbially-related metabolites have been shown to vary in a habitat-specific manner [56].
The integration of environmental metabolomes into microbiome research provides a new framework to address questions in microbial chemical ecology. The complex chemical landscapes associated with sediment microbial communities (Fig. 3A) coupled with the unprecedented biosynthetic potential of sediment MAGs (Fig. 2B) supports recent evidence that poorly studied phyla such as Acidobacteriota [57] and Myxococcota hold promise for future natural product discovery efforts. While this approach can facilitate the prioritization of BGCs for heterologous expression based on the predicted novelty of their biosynthetic products (Fig. 2A), it remains difficult to connect molecules with their cognate BGCs when dealing with complex microbial communities [58], as we have attempted to do with the hantupeptins (Fig. S6). Furthermore, the dynamics of specialized metabolism are complex, with molecules not only produced but also degraded or metabolized. Our detection of a dibrominated tyrosine metabolite (Fig. 3B) and a MAG containing the genetic potential to metabolize this compound provide examples of the interplay between these processes. Ultimately, a major goal remains to better understand the roles of environmental metabolomes in shaping microbial community composition.
Conclusion
Recent methodological advancements have enabled the incorporation of metabolomics into assessments of microbial diversity. Multi-omic studies of this type can begin to reveal the potential roles of specialized metabolites in structuring microbial communities in complex environmental biomes. These molecules likely mediate a myriad of largely undefined interactions among community members and the environment. A better understanding of environmental metabolomes and their functional roles will provide a new dimension to assessments of microbial diversity and distributions.
Methods and materials
Field site and sample collection
Sediment samples were collected from the barrier reef lagoon surrounding the island of Moorea, French Polynesia. We collected samples adjacent to the back and fringing reefs within eight replicate 1 m2 plots in and around the Long-Term Ecological Research (LTER) Site 2 (Fig. S1A). Plot locations were selected based on proximity to either the fringing reef (n = 4) or the back reef (n = 4). Sites 1–4 were designated as back reef sites with a depth range of 1.83–3.05 m. Site 5–8 were initially designated as fringing reef sites (<150 m from shore) that ranged in depth from 0.61–1.83 m, while site 6 was further designated as algae-dominated on the predominance of seaweeds and lack of living coral.
Within each 1 m2 plot, 16 subsamples were taken using a gridded design (Fig. S1A; n = 128). Additionally, sediments were sampled every meter along a 4 m transect in each cardinal direction away from the 1m2 plots (n = 256, 128 from plots, 128 from transects). All sediments samples (~80 g each) were collected in sterile Whirl-Pak bags and transferred on ice back to the field station for processing. Approximately 7 g of each sediment sample was transferred to a 15 mL Falcon tube with 7 mL of RNALater for subsequent community analyses. All tubes and the remaining sediment were stored at −20 °C until processing except when in transit to Scripps Institution of Oceanography when dry ice was used.
LTER data related to N content, heat stress, and coral degradation were sourced from publicly available data (see [21, 59]). Briefly, in R, public geospatial data from LTER monitoring was interpolated by converting geographic coordinates for our collection sites to geospatial vectors (st_as_sf in ‘sf’ package) and applying a kriging algorithm (kriging in ‘kriging’ package) to predict environmental values at our sites.
16S rRNA gene sequencing and analysis
All quadrat samples (n = 128) and transect tips (n = 32) were processed for DNA extraction. Mid-point transect samples (n = 4) were also processed for site 3, which was qualitatively identified as having the highest coral cover. All remaining transect samples from site 6 were also processed to increase spatial comparisons among the algae-dominated site (n = 12). In total, 176 samples were processed for 16 S rRNA gene sequencing. For DNA extraction, ~2 g of sediment in RNALater was processed by the Center for Microbiome Innovation (CMI) at UC San Diego using the MagAttract PowerSoil DNA KF kit (Qiagen). Samples were subsequently sequenced with the MiSeq System PE150 platform (Illumina) using the V4 region 515 forward primer (GTGYCAGCMGCCGCGGTAA) and the updated 806 reverse primer (GGACTACNVGGGTWTCTAAT) recommended by the Earth Microbiome Project [60]. For increased sequencing depth to capture the rare biosphere, two separate sequencing runs were performed for community analyses (see Supplemental Materials).
Metagenomic sequencing and analysis
For a subset of samples (n = 36) representing all eight sites, shotgun metagenomic libraries were constructed using a Nextera XT DNA library preparation kit (Illumina) at the CMI. To reduce sequencing biases, 20 of 36 samples were sequenced in triplicate on a HiSeq system (Illumina) with 150-bp paired end reads. Technical replicates were initially processed as independent samples and assessed for variance in taxonomic composition (see below). Upon validation, technical replicates were combined for all downstream analyses. The remaining samples were sequenced on a NovaSeq 6000 System (Illumina) with 150-bp paired end reads to achieve similar sequencing depth (meandepth = 205 M reads). Raw reads were quality trimmed with adapters removed using the BBMap toolkit (bbduk.sh) [61]. Processed reads were then analyzed using both a read-based (for taxonomic, functional, and KS/C biosynthetic potential) and assembly-based approaches (see Supplemental Materials).
Sediment metabolomics and mass spectrometry
For the same samples used to construct shotgun metagenomes, we partitioned sediments (wet weight 50 g) for chemical extractions with 100 mL 1:1 methanol dichloromethane mixture. The solvent was filtered and evaporated under reduced pressure. The extracts were resuspended in HPLC grade methanol with the supernatant removed and diluted to 1 mg/mL for LC-HRMS analysis. Process controls were prepared analogously using the same glassware. LC-HRMS analysis was performed on a 6530 Accurate-Mass QToF with ESI-source (Agilent) coupled with a 1260 Infinity HPLC (Agilent) equipped with a 150×4.6 mm Kinetex C18 5 µm column (Phenomenex, USA). HPLC was run starting at 20:80 acetonitrile: water with 0.1 % formic acid (FA) for 2 min followed by an 18 min gradient until 95 % acetonitrile (0.1 % FA), which we held for 2 min and increased to 100 % acetonitrile over 1 min and held for 2 min. We injected 5 µL of a 1 mg/mL sample solution (MeOH) at a flow of 1 mL/min. MS data were acquired over a range 135–1700 m/z in positive mode. All solvents were LCMS grade. Paired metagenomic and metabolomic datasets for each sample are available on the Paired Omics Data Platform [62]. Chemoinformatic analyses are detailed in Supplementary Materials.
Supplementary information
Acknowledgements
This work was supported by the National Institutes of Health (NIH) (R01GM085770) to PRJ, a University of California Academic Senate seed grant to PRJ/ABC/AB, a Scripps Postdoctoral Scholar Fellowship to ABC, a Marine Science Research Award from the University of California San Diego (UCSD) Academic Senate to PRJ/AMD, and a National Science Foundation Graduate Research Fellowship to AMD (DGE-1650112). We thank Mitchell Muskat for assistance in sample processing, April Lukowski, Bradley Moore, Douglas Sweeney, and Jennifer Martiny for helpful discussions and comments, Xiao Yang for assistance with geospatial data, members of the Jensen Lab for feedback on experimental design and assistance with field preparations, Rebecca Vega Thurber and Andrew Thurber for constructive discussions and field assistance, and all associated with the Richard B. Gump South Pacific Research Station for assistance with fieldwork. Finally, the authors thank the Center for Microbiome Innovation for sample processing and 16S rRNA gene sequencing (through a seed grant to AMD), the DNA Technologies and Expression Analysis Core at the UC Davis Genome Center (1S10OD010786-01) for shotgun metagenomic sequencing, and the San Diego Supercomputer Center at UCSD for computational resources. The authors declare that there is no conflict of interest regarding the publication of this article.
Author contributions
ABC, AMD, and PRJ designed and conceptualized the research project. AMD and PRJ performed fieldwork and sample collection. ABC and AB performed DNA extractions for metagenomes. AB performed metabolomic extractions, acquired and analyzed metabolomic data. ABC analyzed amplicon and metagenomic data and conducted statistical analyses with input from AB. ABC and PRJ wrote manuscript with input from all authors.
Funding
Open access funding provided by SCELC, Statewide California Electronic Library Consortium.
Data availability
Paired-end shotgun metagenomic and 16 S rRNA amplicon data were deposited in the NCBI Sequence Read Archive under the BioProject PRJNA611818. Public datasets for all metabolomic spectra files are available at massive.ucsd.edu (MSV000091150). All other data, including genomes, and relevant code used can be found at https://github.com/alex-b-chase/mooreaMS.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01410-3.
References
- 1.Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–12. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 2.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- 3.Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC. Drivers of bacterial β-diversity depend on spatial scale. Proc Natl Acad Sci USA. 2011;108:7850–4. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. J R Soc Interface. 2009;6:959–78. doi: 10.1098/rsif.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milke F, Wagner-Doebler I, Wienhausen G, Simon M. Selection, drift and community interactions shape microbial biogeographic patterns in the Pacific Ocean. ISME J. 2022;16:2653–65. doi: 10.1038/s41396-022-01318-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyc O, Song C, Dickschat JS, Vos M, Garbeva P. The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 2017;25:280–92. doi: 10.1016/j.tim.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Uchimiya M, Schroer W, Olofsson M, Edison AS, Moran MA. Diel investments in metabolite production and consumption in a model microbial system. ISME J. 2021;16:1306–17. doi: 10.1038/s41396-021-01172-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt R, Ulanova D, Wick LY, Bode HB, Garbeva P. Microbe-driven chemical ecology: past, present and future. ISME J. 2019;13:2656–63. doi: 10.1038/s41396-019-0469-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol. 2006;2:666–73. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 10.Palmer JD, Foster KR. Bacterial species rarely work together. Science. 2022;376:581–2. doi: 10.1126/science.abn5093. [DOI] [PubMed] [Google Scholar]
- 11.Rodríguez-Verdugo A, Ackermann M. Rapid evolution destabilizes species interactions in a fluctuating environment. ISME J. 2021;15:450–60. doi: 10.1038/s41396-020-00787-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharrar AM, Crits-Christoph A, Méheust R, Diamond S, Starr EP, Banfield JF. Bacterial secondary metabolite biosynthetic potential in soil varies with phylum, depth, and vegetation type. mBio. 2020;11:1–17. doi: 10.1128/mBio.00416-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy BVB, Kallifidas D, Kim JH, Charlop-Powers Z, Feng Z, Brady SF. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl Environ Microbiol. 2012;78:3744–52. doi: 10.1128/AEM.00102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlop-Powers Z, Owen JG, Reddy BVB, Ternei MA, Brady SF. Chemical-biogeographic survey of secondary metabolism in soil. Proc Natl Acad Sci USA. 2014;111:3757–62. doi: 10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh HW, Creamer KE, Chase AB, Klau LJ, Podell S, Jensen PR. Metagenomic data reveal type I polyketide synthase distributions across biomes. bioRxiv 2023.01.09.523365. 2023. 10.1101/2023.01.09.523365. [DOI] [PMC free article] [PubMed]
- 16.Tuttle RN, Demko AM, Patin N V., Kapono CA, Donia MS, Dorrestein P, et al. Detection of natural products and their producers in ocean sediments. Appl Environ Microbiol. 2019;85:e02830-18. [DOI] [PMC free article] [PubMed]
- 17.Roach TNF, Little M, Arts MGI, Huckeba J, Haas AF, George EE, et al. A multiomic analysis of in situ coral–turf algal interactions. Proc Natl Acad Sci USA. 2020;117:13588–95. doi: 10.1073/pnas.1915455117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gribben PE, Nielsen S, Seymour JR, Bradley DJ, West MN, Thomas T. Microbial communities in marine sediments modify success of an invasive macrophyte. Sci Rep. 2017;7:9845. doi: 10.1038/s41598-017-10231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Studivan MS, Rossin AM, Rubin E, Soderberg N, Holstein DM, Enochs IC. Reef sediments can act as a stony coral tissue loss disease vector. Front Mar Sci. 2022;8:2046. doi: 10.3389/fmars.2021.815698. [DOI] [Google Scholar]
- 20.Kelly LW, Williams GJ, Barott KL, Carlson CA, Dinsdale EA, Edwards RA, et al. Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc Natl Acad Sci USA. 2014;111:10227–32. doi: 10.1073/pnas.1403319111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donovan MK, Adam TC, Shantz AA, Speare KE, Munsterman KS, Rice MM, et al. Nitrogen pollution interacts with heat stress to increase coral bleaching across the seascape. Proc Natl Acad Sci USA. 2020;117:5351–7. doi: 10.1073/pnas.1915395117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klau LJ, Podell S, Creamer KE, Demko AM, Singh HW, Allen EE, et al. The Natural Product Domain Seeker version 2 (NaPDoS2) webtool relates ketosynthase phylogeny to biosynthetic function. J Biol Chem. 2022;298:102480. [DOI] [PMC free article] [PubMed]
- 23.Ziemert N, Lechner A, Wietz M, Millán-Aguiñaga N, Chavarria KL, Jensen PR. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci USA. 2014;111:E1130–9. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, van der Hooft JJJ, et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48:D454–8. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nayfach S, Roux S, Seshadri R, Udwary D, Varghese N, Schulz F, et al. A genomic catalog of Earth’s microbiomes. Nat Biotechnol. 2020;39:499–509. doi: 10.1038/s41587-020-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chevrette MG, Thomas CS, Hurley A, Rosario-Melendez N, Sankaran K, Tu Y, et al. Microbiome composition modulates secondary metabolism in a multispecies bacterial community. Proc Natl Acad Sci USA. 2022;119:e2212930119. doi: 10.1073/pnas.2212930119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chevrette MG, Gavrilidou A, Mantri S, Selem-Mojica N, Ziemert N, Barona-Gómez F. The confluence of big data and evolutionary genome mining for the discovery of natural products. Nat Prod Rep. 2021;38:2024–0. doi: 10.1039/D1NP00013F. [DOI] [PubMed] [Google Scholar]
- 28.Chase AB, Karaoz U, Brodie EL, Gomez-Lunar Z, Martiny AC, Martiny JBH. Microdiversity of an abundant terrestrial bacterium encompasses extensive variation in ecologically relevant traits. mBio. 2017;8:e01809–17. doi: 10.1128/mBio.01809-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–37. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nothias LF, Petras D, Schmid R, Dührkop K, Rainer J, Sarvepalli A, et al. Feature-based molecular networking in the GNPS analysis environment. Nat Methods. 2020;17:905–8. doi: 10.1038/s41592-020-0933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blunt JW, Copp BR, Keyzers RA, Munro MHG, Prinsep MR. Marine natural products. Nat Prod Rep. 2015;32:116–11. doi: 10.1039/C4NP00144C. [DOI] [PubMed] [Google Scholar]
- 32.Mohimani H, Gurevich A, Shlemov A, Mikheenko A, Korobeynikov A, Cao L, et al. Dereplication of microbial metabolites through database search of mass spectra. Nat Commun. 2018;9:4035. doi: 10.1038/s41467-018-06082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripathi A, Puddick J, Prinsep MR, Lee PPF, Tan LT. Hantupeptin A, a cytotoxic cyclic depsipeptide from a Singapore collection of Lyngbya majuscula. J Nat Prod. 2009;72:29–32. doi: 10.1021/np800448t. [DOI] [PubMed] [Google Scholar]
- 34.Tripathi A, Puddick J, Prinsep MR, Lee PPF, Tan LT. Hantupeptins B and C, cytotoxic cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. Phytochemistry. 2010;71:307–11. doi: 10.1016/j.phytochem.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Nogle LM, Gerwick WH. Isolation of four new cyclic depsipeptides, Antanapeptins A−D, and Dolastatin 16 from a Madagascan collection of Lyngbya majuscula. J Nat Prod. 2002;65:21–24. doi: 10.1021/np010348n. [DOI] [PubMed] [Google Scholar]
- 36.Mohanty I, Tapadar S, Moore SG, Biggs JS, Freeman CJ, Gaul DA, et al. Presence of bromotyrosine alkaloids in marine sponges is independent of metabolomic and microbiome architectures. mSystems. 2021;6:e01387–20. doi: 10.1128/mSystems.01387-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng J, Li J, Hamann MT. The marine bromotyrosine derivatives. Alkaloids Chem Biol. 2005;61:59–262. doi: 10.1016/S1099-4831(05)61002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Felder S, Dreisigacker S, Kehraus S, Neu E, Bierbaum G, Wright PR, et al. Salimabromide: unexpected chemistry from the obligate marine Myxobacterium Enhygromyxa salina. Chem - A Eur J. 2013;19:9319–24. doi: 10.1002/chem.201301379. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Lopez N, Ahn Y, Goldberg T, Bromberg Y, Kerkhof LJ, et al. Novel reductive dehalogenases from the marine sponge associated bacterium Desulfoluna spongiiphila. Environ Microbiol Rep. 2017;9:537–49. doi: 10.1111/1758-2229.12556. [DOI] [PubMed] [Google Scholar]
- 40.Stallforth P, Mittag M, Brakhage AA, Hertweck C, Hellmich UA. Functional modulation of chemical mediators in microbial communities. Trends Biochem Sci. 2023;48:71–81 [DOI] [PubMed]
- 41.Wegley Kelly L, Nelson CE, Petras D, Koester I, Quinlan ZA, Arts MGI, et al. Distinguishing the molecular diversity, nutrient content, and energetic potential of exometabolomes produced by macroalgae and reef-building corals. Proc Natl Acad Sci USA. 2022;119:e2110283119. doi: 10.1073/pnas.2110283119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, et al. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073. [DOI] [PubMed]
- 43.Ruokolainen L, Ranta E, Kaitala V, Fowler MS. When can we distinguish between neutral and non-neutral processes in community dynamics under ecological drift? Ecol Lett. 2009;12:909–19. doi: 10.1111/j.1461-0248.2009.01346.x. [DOI] [PubMed] [Google Scholar]
- 44.Talbot JM, Bruns TD, Taylor JW, Smith DP, Branco S, Glassman SI, et al. Endemism and functional convergence across the North American soil mycobiome. Proc Natl Acad Sci USA. 2014;111:6341–6. doi: 10.1073/pnas.1402584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran MA, Kujawinski EB, Schroer WF, Amin SA, Bates NR, Bertrand EM, et al. Microbial metabolites in the marine carbon cycle. Nat Microbiol. 2022;7:508–23. doi: 10.1038/s41564-022-01090-3. [DOI] [PubMed] [Google Scholar]
- 46.Bouslimani A, Sanchez LM, Garg N, Dorrestein PC. Mass spectrometry of natural products: current, emerging and future technologies. Nat Prod Rep. 2014;31:718–29. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–20. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chase AB, Martiny JBH. The importance of resolving biogeographic patterns of microbial microdiversity. Microbiol Aust. 2018;39:5–8. doi: 10.1071/MA18003. [DOI] [Google Scholar]
- 49.Larkin AA, Martiny AC. Microdiversity shapes the traits, niche space, and biogeography of microbial taxa. Environ Microbiol Rep. 2017;9:55–70. doi: 10.1111/1758-2229.12523. [DOI] [PubMed] [Google Scholar]
- 50.Chase AB, Sweeney D, Muskat MN, Guillén-Matus DG, Jensen PR. Vertical inheritance facilitates interspecies diversification in biosynthetic gene clusters and specialized metabolites. mBio. 2021;12:e02700–21. doi: 10.1128/mBio.02700-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chevrette MG, Currie CR. Emerging evolutionary paradigms in antibiotic discovery. J Ind Microbiol Biotechnol. 2019;46:257–71. doi: 10.1007/s10295-018-2085-6. [DOI] [PubMed] [Google Scholar]
- 52.Choudoir M, Rossabi S, Gebert M, Helmig D, Fierer N. A phylogenetic and functional perspective on volatile organic compound production by Actinobacteria. mSystems. 2019;4:e00295-18. [DOI] [PMC free article] [PubMed]
- 53.Chevrette MG, Carlos-Shanley C, Louie KB, Bowen BP, Northen TR, Currie CR. Taxonomic and metabolic incongruence in the ancient genus Streptomyces. Front Microbiol. 2019;10:2170. doi: 10.3389/fmicb.2019.02170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemetre C, Maniko J, Charlop-Powers Z, Sparrow B, Lowe AJ, Brady SF. Bacterial natural product biosynthetic domain composition in soil correlates with changes in latitude on a continent-wide scale. Proc Natl Acad Sci USA. 2017;114:11615–20. doi: 10.1073/pnas.1710262114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geers AU, Strube ML, Bentzon-Tilia M. Small spatial scale drivers of secondary metabolite biosynthetic diversity in environmental microbiomes. mSystems. 2023;e00724-22. 10.1128/msystems.00724-22. [DOI] [PMC free article] [PubMed]
- 56.Shaffer JP, Nothias L-F, Thompson LR, Sanders JG, Salido RA, Couvillion SP, et al. Standardized multi-omics of Earth’s microbiomes reveals microbial and metabolite diversity. Nat Microbiol. 2022;7:2128–50. doi: 10.1038/s41564-022-01266-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crits-Christoph A, Diamond S, Butterfield CN, Thomas BC, Banfield JF. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature. 2018;558:440–4. doi: 10.1038/s41586-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 58.Bogdanov A, Salib MN, Chase AB, Hammerlindl H, Muskat MN, Luedtke S, et al. Small molecule in situ resin capture-an organism independent strategy for natural product discovery. bioRxiv. 2023;2023:03.02.530684. 10.1101/2023.03.02.530684.
- 59.Adam TC, Burkepile DE, Holbrook SJ, Carpenter RC, Claudet J, Loiseau C, et al. Landscape-scale patterns of nutrient enrichment in a coral reef ecosystem: implications for coral to algae phase shifts. Ecol Appl. 2021;31:e2227. doi: 10.1002/eap.2227. [DOI] [PubMed] [Google Scholar]
- 60.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–63. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bushnell B. BBMap: a fast, accurate, splice-aware aligner. Berkeley, CA (United States): Lawrence Berkeley National Lab. (LBNL); 2014. [Google Scholar]
- 62.Schorn MA, Verhoeven S, Ridder L, Huber F, Acharya DD, Aksenov AA, et al. A community resource for paired genomic and metabolomic data mining. Nat Chem Biol. 2021;17:363–8. doi: 10.1038/s41589-020-00724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Paired-end shotgun metagenomic and 16 S rRNA amplicon data were deposited in the NCBI Sequence Read Archive under the BioProject PRJNA611818. Public datasets for all metabolomic spectra files are available at massive.ucsd.edu (MSV000091150). All other data, including genomes, and relevant code used can be found at https://github.com/alex-b-chase/mooreaMS.