Abstract
Phages are prevalent in diverse environments and play major ecological roles attributed to their tremendous diversity and abundance. Among these viruses, transposable phages (TBPs) are exceptional in terms of their unique lifestyle, especially their replicative transposition. Although several TBPs have been isolated and the life cycle of the representative phage Mu has been extensively studied, the diversity distribution and ecological functions of TBPs on the global scale remain unknown. Here, by mining TBPs from enormous microbial genomes and viromes, we established a TBP genome dataset (TBPGD), that expands the number of accessible TBP genomes 384-fold. TBPs are prevalent in diverse biomes and show great genetic diversity. Based on taxonomic evaluations, we propose the categorization of TBPs into four viral groups, including 11 candidate subfamilies. TBPs infect multiple bacterial phyla, and seem to infect a wider range of hosts than non-TBPs. Diverse auxiliary metabolic genes (AMGs) are identified in the TBP genomes, and genes related to glycoside hydrolases and pyrimidine deoxyribonucleotide biosynthesis are highly enriched. Finally, the influences of TBPs on their hosts are experimentally examined by using the marine bacterium Shewanella psychrophila WP2 and its infecting transposable phage SP2. Collectively, our findings greatly expand the genetic diversity of TBPs, and comprehensively reveal their potential influences in various ecosystems.
Subject terms: Bacteriophages, Microbial ecology
Introduction
Bacteriophages, which are viruses that infect and parasitize bacteria, are the most abundant biological entities and critical ecological regulators in diverse natural and artificial environments [1, 2]. Among bacteriophages, temperate phages are capable of both lytic and lysogenic infections, and they typically integrate their own genomes into the host genome to form prophages during lysogenic infection [3]. Prophages are widespread in bacterial genomes, which is well supported by a previous analysis in which 46% of 2110 analyzed bacterial genomes were infected by a total of 2246 prophages [4]. In-depth studies on temperate phages that infect some model microorganisms, such as Escherichia coli and Salmonella enterica, have shown that they significantly influence a variety of physiological functions and basic cellular life activities of the bacterial host, including DNA replication, gene transcription, protein expression, growth, motility, biofilm formation, and environmental stress resistance [3, 5–7].
Among temperate phages, transposable bacteriophages (TBPs) are unique in that they replicate by transposition [8, 9]. Phage Mu, the representative TBP, was isolated nearly 60 years ago from E. coli strain K-12 [10]. Mu phage taxonomically classified into Myoviridae historically, has a genome size of ~38 kb, terminated by a 5′-TG-CA-3′ inverted repeat [9, 11]. After integration into the host genome and upon entering the lytic cycle, Mu initiates a “copy-and-paste” (replicative) transposition, a process that is conducted by a complex known as the transpososome, which mainly involves a DDE family transposase (MuA) and an AAA + ATPase (MuB) [12]. Replicative transposition results in the random integration of multiple copies of Mu into different loci in the host genome, thereby potentially causing a variety of host gene mutations, including inversions, duplications, deletions, and gene fusions [8, 9]. The headful mechanism is used for Mu phage DNA packaging. Specifically, the initial packaging site (pac) is located 50–150 bp upstream of the left end of the Mu DNA (attL), while the packaging terminates nonspecific locations after the phage head is filled with DNA, which leads to a host DNA sequence (normally 1–1.5 kb, up to 3 kb in some cases) that is downstream of the right end of the Mu DNA (attR) and is also packaged into the head [13, 14]. The extra packaging of host chromosomal and plasmid DNA into phage particles thus makes Mu to be a generalized transducing phage [15].
In addition to Mu, several other TBPs, such as D108, B3, BcepMu, RcapMu, SuMu, SfMu, and ФSHP3, which infect E. coli, Pseudomonas aeruginosa, Burkholderia cenocepacia, Rhodobacter capsulatus, Haemophilus parasuis, Shigella flexneri, and Stenotrophomonas maltophilia, respectively, have been successively isolated and characterized [16–22]. Although not yet isolated, some Mu-like prophages have been found in multiple bacterial genomes [23–25]. Moreover, the isolation niches of TBPs are diverse, including the human microbiome, marine water, hydrothermal vents and plateau wetlands [26–29]. Previously, two Mu-like phages encoding diguanylate cyclase and UDP-sulfoquinovose synthase were identified in viromes from the Cariaco Basin, indicating a potential impact of TBPs on microbial processes in oxygen-deficient marine seawater [30]. Altogether, these results suggest that TBPs are widespread in natural environments. Previously, based on the characteristic features of 26 sequenced TBP genomes, a new family, “Saltoviridae”, with two subfamilies, “Myosaltovirinae” and “Siphosaltovirinae”, was proposed [31]. Subsequently, four conserved proteins of TBPs (Mor, GemA, portal protein and transposase) were used as markers for the screening of TBPs in Enterobacteria, Pseudomonas, and Leptospiraceae genomes, leading to the discovery of hundreds of predicted transposable prophages [32, 33]. These pioneering works expand the TBP genome dataset and further support the existence of the newly proposed family “Saltoviridae”. Despite this progress, the distribution and diversity of TBPs in a wide range of prokaryotic genomes and environments remain unknown. The genes encoding transposases, which are conserved marker proteins of TBPs, have been shown to be the most abundant and widespread genes in microbial genomes and metagenomes [34]. Based on the above evidence and clues, it is reasonable to hypothesize that TBPs are widely distributed in various environments and microorganisms and have potentially important ecological functions.
In this study, to address this hypothesis, we first collected and curated genomic information of all isolated TBPs and identified six conserved proteins by comparative genome analysis. Subsequently, we mined TBPs from publicly available microbial genomes and established the first TBP genome dataset (TBPGD). The distribution, diversity, and gene content of TBPs were systematically evaluated, and the influences of TBPs on their hosts were experimentally examined by using the marine bacterium Shewanella psychrophila WP2 and its integrated transposable prophage SP2. The results of this study greatly contribute to a comprehensive understanding of the genetic and life-history traits of TBPs and their ecological importance.
Results
Transposable phages (TBPs) are prevalent globally
According to the designed bioinformatic workflow (Supplementary Figs. S1, S2), we first searched the literature for previously isolated TBPs. A total of 48 TBPs were retained after manual curation, and they were used as the reference dataset for subsequent TBP detection (Supplementary Table S1). We performed gene annotation and protein family analysis of these reference TBP genomes and found that 6 proteins (GemA, Mor, portal protein, head-tail connector protein, virion morphogenesis protein, and transposases) were encoded by all the reference TBPs, and they were therefore considered to be conserved proteins of TBPs (Supplementary Fig. S3). Specifically, although the gene encoding DDE_2-type transposase (PF02914.17) was absent in 9 TBPs, all of them encoded the IS240 transposase (PF13610.8, HHblits-probability >87.6%), which also belongs to the DDE family. Moreover, these IS240 transposases shared high sequence and structural similarity with transposases in other reference TBPs, especially the transposase encoded by phage B3 (pairwise identity >40%, Z score >27.3), indicating that the transposase (including two protein families, according to the Pfam categories) is one of the conserved proteins of TBPs (Supplementary Fig. S4). The six identified conserved proteins are critical in the TBP life cycle, including in replicative transposition, transcription, and assembly (Fig. 1a). To verify the reliability of these conserved proteins for TBP identification, we used them as queries to search the RefSeq virus database (n = 11,080), obtaining in a total of 40 viral genomes. Among them, 35 were already present in our reference TBP genomes, and the remaining five genomes exhibited high similarity to the reference TBPs (Supplementary Fig. S5), indicating that no false positives were produced in the search and that these six conserved proteins can be used for mining TBP genomes in large-scale datasets.
Fig. 1. The distribution of transposable bacteriophages (TBPs) across Earth’s biomes.
a Schematic life cycle of the TBPs. The six conserved marker proteins of TBPs are indicated in red. For clarity, the phage genomic DNA and virions are not to scale relative to the bacterial host cell and chromosome. b Geographic distribution of TBPs. Each point represents a geographic site, and only the TBP genomes (n = 5320) with available geographic coordinates are shown. The size of each point is proportional to the number of genomes found at that site, and the colors differentiate the derived environments: host-associated (orange), aquatic (blue), terrestrial/sediment (brown) or other (environmental information unavailable, green). c Distribution of TBPs across biomes and sub-biomes, based on environmental metadata of their derived host. The values in each pie chart represent the number of TBP genomes derived from the specific biomes and sub-biomes.
Given that all TBPs found thus far are integrated into the host genome and exist as prophages [8], we identified TBPs mainly from prokaryotic genomes (n = 216,709 for bacteria and n = 1156 for archaea) in the RefSeq database by performing a two round search (Supplementary Figs. S1, S2, and see Methods for further details). In addition, the currently largest virus database, IMG/VR v2.0 [35] (n = 760,445), and the largest marine virus dataset, GOV 2.0 [36] (n = 488,128), were combined as the query datasets for TBP mining. Overall, 18,449 TBP genomes were obtained, thereby expanding the quantity of TBP genomes 384-fold.
The TBPs identified to date are widely distributed in a variety of different geographic locations and biomes worldwide, including host-associated, aquatic, terrestrial and sediment biomes (Fig. 1b and Supplementary Table S3). In the TBPGD, host-associated TBPs accounted for the majority of the genomes, and most of them were derived from human-associated samples (Fig. 1c), which was probably due to the high enrichment of prokaryotic genomes from these environments in the database. Despite these preferences, our data unprecedentedly show that TBPs cover extremely diverse environments (Supplementary Table 3). In particular, we obtained 297 TBP genomes from oceans, which represent the largest ecosystem on Earth. The recruitment analysis showed that some of them were widespread in various marine areas, and multiple TBPs exhibited a high abundance in specific ocean zones (Supplementary Fig. S6), indicating that the oceans are important habitats for TBPs.
Vast genetic diversity of TBPs
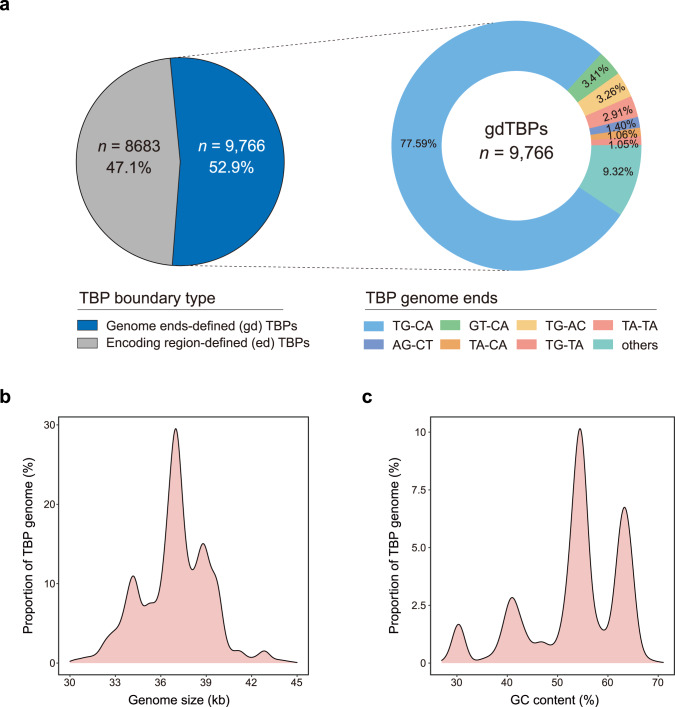
In the TBPGD, more than half of the TBPs (52.9%) were genome ends-defined (gdTBPs), representing TBPs with complete genome sequence, and the remaining TBPs (47.1%) were encoding region-defined (edTBPs) (Fig. 2a). Among gdTBPs, classical TG-CA genome ends accounted for the highest proportion (77.59%), followed by GT-CA (3.41%) and TG-AC (3.26%) ends. Furthermore, CheckV assessment revealed that the majority (74.51%) of the TBP genomes were of complete or high quality, and this proportion was higher for gdTBPs (87.75%) than that for edTBPs (59.59%) (Supplementary Fig. S7). Most TBPs were located in host chromosomes, although several of them (n = 24) were present in plasmids; this phenomenon has also been observed in Leptospira weilii strains [33]. Further analysis indicated that some of these TBP-plasmid host genomes were simultaneously integrated with diverse TBPs, corresponding to either different copies or different species (Supplementary Fig. S8), suggesting that these TBP plasmids originated from either replicative transposition of a chromosomal copy or independent infection events.
Fig. 2. Genome properties of the TBPs.
a Composition of the genome ends of TBPs. The left pie chart indicates the proportion of TBPs for which exact genome ends were defined (gdTBPs), and the right pie chart shows the percentages of different genome ends among the gdTBPs. b, c Density plots of the genome size (b) and GC content (c) distributions of TBPs.
The genome size of gdTBPs were concentrated between 33 and 40 kb (90.05%) (Fig. 2b), and the GC content of these TBPs ranged from 26 to 71%. The density plot of the GC content showed four significant peak intervals: low GC content (26–35%, n = 557), intermediate GC content (35–45%, n = 1420; 45–60%, n = 4968), and high GC content (60–71%, n = 2821), suggesting the presence of different clades within TBPs (Fig. 2c). Generally, the distribution of genome size and GC content among the newly identified TBPs was consistent with that of the reference TBPs (Supplementary Fig. S9), indicating their intrinsic association and the reliability of our TBP screening strategy.
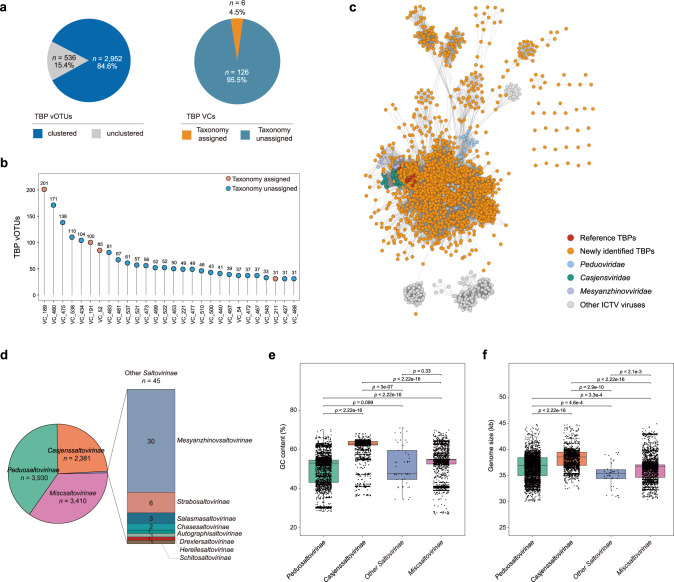
To assess the diversity of TBPs, we grouped all the gdTBPs into 3488 viral operational taxonomic units (vOTUs), equivalent to a species-level taxonomy. Furthermore, these vOTUs were clustered by vConTACT2 via protein-sharing networks (Fig. 3a, b and Supplementary Table S4), in which 2952 vOTUs formed 132 viral clusters (VCs). In addition, 536 vOTUs belonged to the overlap (n = 378), outlier (n = 153), or clustered/singleton (n = 5) types in the network and could thus not be assigned to VCs. Among the 132 VCs, only 6 contained viruses with assigned taxonomy, while the other 126 VCs could not be classified (Fig. 3a). Furthermore, among the 28 genome-enriched VCs (TBP genomes >30), only 4 (14.3%) harbored viruses with known taxonomy (Fig. 3b). Specifically, VC_189 contained the highest number of vOTUs (n = 201), 21 of which belonged to the genus Casadabanvirus, and the remaining 180 were newly identified herein. Three other vOTU-enriched VCs containing taxonomically classified viruses were VC_191 (Beetrevirus, n = 100), VC_52 (Muvirus, n = 85), and VC_211 (Bcepmuvirus, n = 31), and the numbers of known vOTUs they contained were expanded 32.3, 41.5,- and 14.5-fold, respectively, by the TBPGD. Collectively, these results strongly suggested a high unknown diversity of TBPs.
Fig. 3. Diversity and taxonomic classification of the TBP genome dataset (TBPGD).
a, b Taxonomic compositions of the TBPs at the viral cluster (VC) level. a The left pie chart indicates the percentage of TBP vOTUs that could be clustered into VCs, and the right pie graph shows the percentage of TBP VCs with assigned taxonomy. b TBP VCs with enriched genomes. The orange and blue circles indicate VCs with and without assigned taxonomy, respectively. For clarity, only TBP VCs with >30 genomes are shown. c, d Taxonomic compositions of the TBPs at the subfamily level. The number of genomes belonging to each family are indicated in different portions of the diagram. e, f Comparison of TBPs belonging to different subfamilies in terms of genome size (e) and GC content (f). The black horizontal lines in the box plots correspond to the median. The differences between different TBP families were assessed by the two-tailed Student’s t test, and p values are shown for each comparison.
We subsequently sought to explore the taxonomy of TBPs at the family level. The PhaGCN2 assessment showed that gdTBPs in the TBPGD belonged mainly to Peduoviridae (n = 3930) and Casjensviridae (n = 2381), followed by Mesyanzhinovviridae (n = 30) and other viral families (Fig. 3c and Supplementary Table S5). Previously, Hulo et al. proposed the establishment of the new family Saltoviridae of Caudovirales for transposable phages, with two subfamilies, Myosaltoviridae and Siphosaltoviridae, included in Saltoviridae [31]. However, the order Caudovirales has been abolished and reclassified as the class Caudoviricetes, and the existing subfamilies in Caudovirales have accordingly been tentatively elevated to families [37]. Based on the above evidence and the current situation, we propose the categorization of TBPs into different candidate subfamilies, named Peduosaltovirinae, Casjenssaltovirinae, and other Saltovirinae. Those TBPs (n = 3410) that could not be categorized into current viral taxa, were tentatively classified as Miscsaltovirinae (“Misc” for miscellaneous) (Fig. 3d). To further examine the differences among the four TBP groups, we compared their genome sizes and GC contents, and significant differences were observed (Fig. 3e, f), consistent with the aforementioned four distinct peak intervals of GC content (Fig. 2c). Moreover, the distribution of viral protein families among the four groups indicated that proteins related to integration and excision, the head and packaging, transcription regulation, the connector, the tail and lysis were broadly shared across these groups (Supplementary Fig. S10), indicating that they could be placed in a same higher taxon.
TBPs infect multiple bacterial phyla
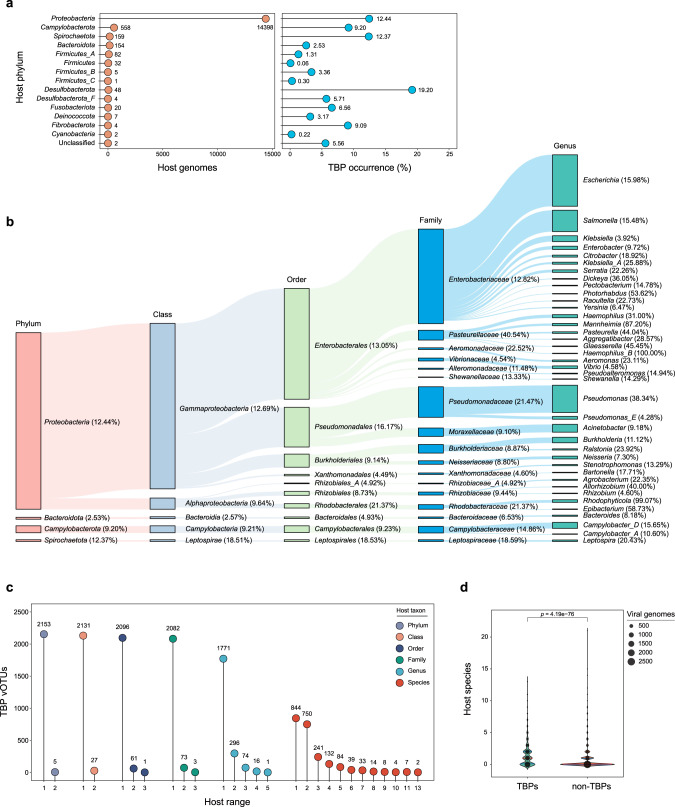
When we analyzed the phylogenetic distribution of TBP hosts (Supplementary Table S3), the host range of TBPs was found to cover 14 bacterial phyla (according to the Genome Taxonomy Database [38], GTDB), while no TBP-infecting archaea were found (Fig. 4a). The majority of TBPs infect Proteobacteria, and they maintained a high occurrence (12.42%) in this phylogenetic clade. In addition, TBPs also have a high incidence in Spirochaetota and Desulfobacterota, but due to the low number of genomes in these two phyla, only 215 and 53 TBPs infecting these phyla, respectively, were identified. Then, we examined the TBP host distribution at different taxonomic levels (Fig. 4b). The families Enterobacteriaceae, Pseudomonadaceae, Pasteurellaceae, and Burkholderiaceae contributed many TBP hosts. At the genus level, the TBP-infecting hosts were highly enriched in Haemophilus_B (100%), Rhodophyticola (99.07%), Manheimia (87.20%), and Epibacterium (58.73%). In terms of the number of genomes, Escherichia, Pseudomonas, Salmonella, Acinetobacter, and Campylobacter_D are the dominant host genera of TBPs.
Fig. 4. Host distribution of TBPs.
a Phylogenetic distribution of TBP hosts in different bacterial phyla. The values next to each circle represent the number of recovered TBP genomes (left panel) or TBP occurrence (right panel) for the specific bacterial phylum. b Phylogenetic distribution of TBP hosts at different taxonomic levels. The percentage in parentheses represents the proportion of TBP-containing genomes out of all genomes in each specific taxon. For clarity, only the genera with >30 TBP host genomes are shown. c Host range of gdTBPs. The number of TBP genomes and corresponding host ranges at different taxonomic levels are displayed. d Estimation of host ranges (at the species level) for TBPs (n = 3488) and non-TBPs (n = 3611). Host ranges were estimated by matching gdTBP genomes and prokaryotic CRISPR spacers.
We further noted that multiple TBPs were present in single host. Among the 15,476 TBP host bacteria, 1721 contained at least two TBPs (with a maximum of 12) in their genomes (Supplementary Fig. S11). In some cases, the TBPs existing in these polylysogens belonged to different phage species (up to 6). For example, for one of the polylysogens of TBPs (Klebsiella pneumoniae RHBSTW-00832) (Supplementary Fig. S12), the genomic analysis demonstrated that its chromosome was integrated with 4 TBP genomes (total length = 156.43 kb). Among these genomes, TBP_3382, TBP_3383, and TBP_3384 showed the same genome, while another TBP (TBP_3381) was significantly distinct from these TBPs.
Different TBP subfamilies were found to have different host profiles (Supplementary Fig. S13), among which, Peduosaltovirinae and Casjenssaltovirinae mainly infected Escherichia and Pseudomonas, respectively, while the dominant hosts of the Miscsaltovirinae were Escherichia, Salmonella, and Acinetobacter. We compared the host genome sizes and GC content of different TBP families and found significant differences among them (Supplementary Fig. S14), implying that both TBPs and their hosts may have differentiated during their coevolutionary history.
In addition to the original hosts in which the TBPs were integrated, the host range of the TBPs was further quantitatively assessed by clustered regularly interspaced short palindromic repeat (CRISPR) spacer matching. Although most TBPs show relatively narrow host ranges, we identified numerous multitaxon-infecting TBPs (Fig. 4c). For instance, we found a gdTBP (TBP_3566, genome length = 36,774 bp) belonging to Peduosaltovirinae, whose original host was affiliated with Morganella. CRISPR spacer matching indicated that its hosts also included three Escherichia albertii strains and the Salmonella enterica strain BCW_2822 (Supplementary Fig. S15), suggesting that this TBP is able to infect at least three genera. Since TBPs can infect a variety of bacterial hosts, we sought to examine whether TBPs have a wider host range than other phages. We selected 3488 vOTUs belonging to gdTBPs. As a control, we collected a total of 3611 phages belonging to Caudoviricetes from the ICTV database as the non-TBP group. The comparison results showed that TBPs had a significantly wider host range at the species level (mean = 1.38) than non-TBPs (mean = 0.68, p = 4.19e−76) (Fig. 4d), suggesting that TBPs show stronger infectivity across host species.
GSIEs and DGRs probably do not contribute to the wide host range of TBPs
Previous studies have indicated that some TBPs, such as Mu phage, possess tropism-switching genetic cassettes (i.e., G segment inversion elements, GSIEs) [39, 40]. GSIEs play a critical role in allowing TBPs to modify their tail fiber proteins, thus expanding host ranges [40]. To investigate whether GSIEs are related to the wide host range of TBPs, we detected GSIEs in the genomes of TBPs and non-TPBs and found that their occurrence in the former (18.8%) was considerably higher than that in the latter (0.6%) (Supplementary Fig. S16a). Furthermore, we compared the host ranges (at the species, genus, and family level) of TBPs with and without GSIEs. However, the number of hosts was significantly lower for the GSIE-encoding TBPs than for the GSIE-lacking TBPs, suggesting that GSIE do not contribute to the broad host range of TBPs (Supplementary Fig. S16b–d).
In addition, we analyzed the contribution of diversity-generating retroelements (DGRs) to the broad host range of TBPs. Although the occurrence of DGRs was higher (3.8%) among gdTBP genomes than among non-TBPs (0.78%), the host ranges of TBPs with DGRs were significantly narrower than those of TBPs without DGRs at the species, genus, and family levels (Supplementary Fig. S17), implying that DGRs are not a major contributor to the broad host range of TBPs.
Potential ecological influences of TBPs
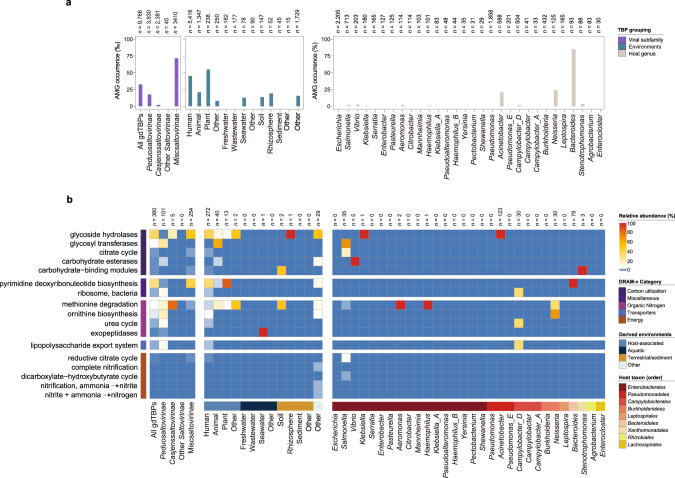
Although numerous TBPs have been identified, systematic assessment of their potential impacts in ecosystems is lacking. To this end, we identified auxiliary metabolic genes (AMGs) carried in gdTBP and edTBP genomes (Fig. 5 and Supplementary Fig. S18). In total, 413 AMGs were identified from the 9,766 gdTBP genomes, of which 339 AMGs could be assigned to functional modules (according to the DRAM-v category) [41], and they covered 24 KEGG Orthology (KO) pathways (Supplementary Table S6). In particular, Miscsaltovirinae showed the highest AMG coverage (7.12%) among the TBP subfamilies (Fig. 5a). In terms of the environmental distribution, TBPs from plant (5.46%), humans (4.51%), and animals (2.08%) showed a relatively high incidence of AMGs. Among different host taxa, AMGs were relatively enriched in TBPs infecting Bacteroides (84.95%), Neisseria (24%), and Acinetobacter (20.92%).
Fig. 5. Distribution of auxiliary metabolic genes (AMGs) encoded by TBPs.
a AMG coverage in different groups, divided by TBP families, derived environments and host taxa. The number of phage genomes contained in each group is shown at the top of the bar chart. b The heatmap shows the relative abundance and functional category of AMGs in each grouping. Only the AMG-encoding TBPs were included in the analysis. The number of TBP genomes contained in each group is shown at the top of the heatmap. The AMGs were identified and annotated by DRAM-v [41]. MISC, miscellaneous.
We analyzed the specific functions of AMGs encoded by TBPs (Fig. 5b). Overall, the dominant AMGs were related to carbon utilization and miscellaneous (MISC) functions. Specifically, the genes involved in glycoside hydrolases and pyrimidine deoxyribonucleotide biosynthesis were highly enriched. The types and abundances of AMGs enriched in different TBP families showed significant differences. For example, glycosyl transferase and methionine degradation-encoding genes were the most abundant AMGs in Peduosaltovirinae and Casjenssaltovirinae, respectively. Moreover, the AMGs encoded by TBPs derived from different environments and hosts were also significantly different. These data suggest that the occurrence of AMGs in TBPs is likely influenced by a combination of viral clades, environmental factors and host taxa.
Given that all TBPs can be integrated as prophages, we sought to identify “hotspot” sites for TBP integration (Supplementary Table S7). Most of the TBPs (68.5%) were located in noncoding regions of the host genome, and 26.1% and 5.0% of the TBPs were integrated into protein-encoding genes and tRNAs, respectively. TBPs were preferentially integrated into genes related to transporters, and the threonine tRNA seemed to be a hotspot tRNA integrated by transposable prophages (Supplementary Fig. S19).
TBPs influence the transcriptomes of marine bacterial hosts
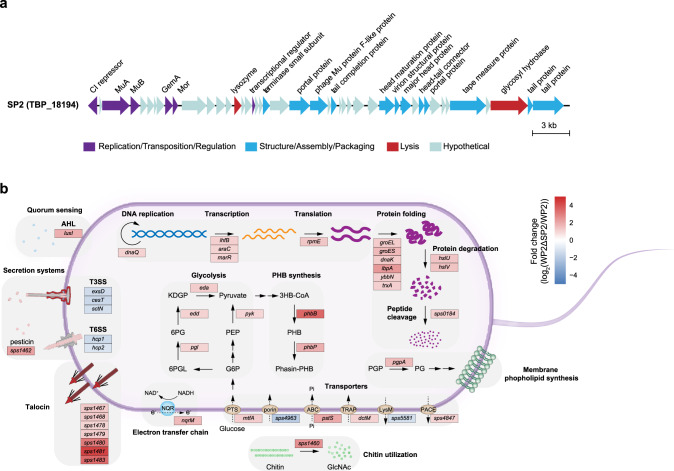
To further assess the impacts of TBPs on hosts, the marine bacterium S. psychrophila WP2 (hereafter referred to as WP2) [42, 43], which was isolated from deep-sea sediment in the western Pacific Ocean and harbors a transposable phage SP2 (TBP_18194, genome size = 38.9 kb), was used as a representative TBP-host system. The SP2 genome contains 45 open reading frames (ORFs), but the functions of 56% of them are unknown (Fig. 6a). The recruitment analysis showed that SP2-like TBPs are prevalent in the Pacific Ocean (Supplementary Fig. S20).
Fig. 6. Influences of the marine transposable phage SP2 on the host transcriptome.
a Genomic map of the prophage SP2 in the marine bacterium Shewanella psychrophila WP2. The arrows depict the location and direction of predicted proteins on the phage genomes, and the fill colors indicate different functional categories of genes, as indicated in the legend. b Graphic display of differentially expressed genes (DEGs) categorized by function in S. psychrophila WP2 after SP2 deletion. The transcriptome data represent three biologically independent samples for each strain (WP2 and WP2ΔSP2). Normalized differential expression levels (fold changes of WP2ΔSP2 versus WP2) are represented by heatmaps in boxes according to the scale bar (log2 scale) from most upregulated (red) to most downregulated (blue). The proteins encoded by the DEGs are shown in each box.
To investigate the impacts of SP2 on the host, we deleted SP2 from the WP2 genome to obtain the WP2ΔSP2 strain. Although SP2 did not significantly influence the growth of WP2 (Supplementary Fig. S21), it had a substantial effect on the transcriptome of WP2. The transcriptomic data were validated via RT-qPCR analysis, which revealed a high correlation coefficient (r2 = 0.92) (Supplementary Fig. S22), indicating that the transcriptomic data were reliable and could be used for follow-up analysis. Overall, 71 differentially expressed genes (DEGs) (false discovery rate [FDR] < 0.05 and fold change > 2) were identified (WP2ΔSP2 versus WP2) (Supplementary Table S8). The majority of the DEGs (n = 58, 81.7%) were upregulated. The transcriptional levels of 4 genes (pgl, edd, eda, and pyk) and 2 genes (phbB and phbP) involved in glycolysis and polyhydroxybutyrate (PHB) synthesis, respectively, were significantly increased in WP2ΔSP2 compared with WP2(Fig. 6b). Moreover, the genes participating in central dogma processes (DNA replication, transcription, and translation), including dnaQ, infB, araC, marR, and rpmE were upregulated, and the transcription levels of multiple genes involved in protein folding and degradation were significantly higher in WP2ΔSP2 than in WP2. DEGs responsible for the electron transfer chain (nqrM), membrane phospholipid synthesis (pgpA), chitin utilization (sps1460), and talocin production (sps1467–1483) were also discovered by transcriptome analysis. In addition, we identified five downregulated DEGs associated with secretion systems (type III and VI) and transporters (porin and LysM). These results suggested that TBPs can significantly alter a variety of host physiological functions, especially central carbon metabolism and several basic cellular activities.
Discussion
Since Mu phage, a representative transposable phage, was discovered in the type strain of E. coli [10], in-depth investigations have revealed its life cycle and the underlying molecular mechanisms, including replicative transposition and transpososome structures [12, 44–47]. Subsequently, a variety of TBPs have been isolated and characterized (Supplementary Table S1) [16–22, 48–53]. In recent years, with the rapidly expanding number of prokaryotic genomes available, by means of marker protein and genomic analyses, the existence of TBPs in some microbial taxa, such as Firmicutes, P. aeruginosa and Leptospira, has been surveyed [33, 54, 55]. Despite these advances, we still lack an understanding of the global-scale distribution of TBPs across a broad range of prokaryotic taxa. In this study, we performed comprehensive and systematic mining of TBPs by using massive microbial genome sequences and their environmental information and constructed a TBPGD, containing 18,449 TBP genomes and their host and environmental information. These TBPs cover a wide range of genome sizes, and they are not only present in bacterial chromosomes but also integrated into plasmids, as previously reported [33]. Building on the seminal works of the Mu research community and the recent proposed taxonomy for transposable phages [8, 31], we now provide evidence to support the expansion of TBP taxa to several candidate subfamilies. In addition, our TBPGD contains a large number of taxonomically unclassified TBPs that shared relatively low similarity with known viruses. We tentatively classified them as Miscsaltovirinae, allowing further amendments and refinements in the future when more evidence is available. Considering that the TBPs isolated to date account for only 0.29% of the total number of genomes in the TBPGD, undoubtedly, these discovered TBPs, especially the representative TBPs, still need to be isolated and characterized in the future to study the life-history traits of these new TBP taxa in depth. These findings notwithstanding, accumulative analysis showed that the TBPGD was not saturated at the protein cluster (PC), species, and viral cluster (VC) levels (Supplementary Fig. S23), suggesting that there is still an enormous unknown diversity awaiting discovery. It is worth noting that all the TBPs isolated to date were found to exclusively infect bacteria. However, considering that the number of archaeal genomes in the RefSeq dataset used in this study was only equivalent to 0.5% of the number of bacterial genomes, the existence of TBPs infecting archaea remains to be explored.
It is well known that bacteriophages are critical genetic information carriers and transporters in horizontal gene transfer (HGT) [56]. Among the diverse phage taxa, the unique features of TBPs likely make prominent players in HGT. First, TBPs exhibit a significantly wider host range than other phages (non-TBPs) belonging to Caudoviricetes (Fig. 4). Second, replicative transposition allows them to randomly insert themselves at multiple sites on the host genome [8, 9] so that the host DNA sequences flanking both ends of the TBP genomes are more diverse than those of other phages with relatively fixed insertion sites. Furthermore, TBPs carry multiple functional AMGs (Fig. 5), and these metabolically important AMGs undoubtedly confer significant effects on the physiological functions of the host. Based on the above evidence, we strongly believe that TBPs function as “super messengers” in HGT, thus representing an important driving force for microbial gene exchange, genetic diversification and the formation of new species in various ecosystems.
Previously, the Mu lysogens of E. coli have been shown to have a higher growth rate than nonlysogens in glucose-limited chemostats, and this phenomenon was believed to be correlated with the higher metabolic activity of the lysogens [57]. Nevertheless, the effects of TBPs on host physiology have not been explored in environmental microorganisms, especially under natural conditions. In this study, we addressed this issue by using a marine TBP and its host S. psychrophila WP2 as representatives, and transcriptome analysis was performed under simulated in situ environmental conditions for a deep-sea bacterium (20 MPa and 4 °C). Previously, several marine TBPs have been isolated and characterized [29, 31, 58], and two Mu-like phages were identified in viromes from marine seawater from the Cariaco Basin [30]. Moreover, Shewanella is a proteobacterial genus that is prevalent in diverse marine environments [59, 60]. In addition, recruitment analysis indicated that SP2-like TBPs are prevalent in the Pacific Ocean virome (POV). Therefore, the transcriptome analysis executed in this study very likely reflects the potential effects of TBPs on their hosts in natural environments. SP2 significantly affected the expression of multiple important functional genes in the host (Fig. 6b), albeit it does not harbor identifiable AMGs in the genome (Fig. 6a). This result suggests that AMG-lacking TBPs, which account for a majority of genomes in the TBPGD (97.92%), can probably have profound effects on their corresponding hosts.
Compared with other phages, TBPs encode a variety of typical conserved proteins, such as the DDE family transposase, GemA, and Mor, and feature unique life cycle processes, such as replicative transposition [8, 9, 40]. The origin and evolutionary history of TBPs are intriguing and worth further exploration. This issue has been addressed for Leptospiraceae TBPs but remains challenging owing to the frequent horizontal transfer and recombination of TBP genes [33]. In terms of viral and host taxonomy, the TBPs discovered thus far all belong to Caudoviricetes and mainly infect Proteobacteria, suggesting a putative evolutionary scenario in which an ancient tailed phage occasionally acquired transposition elements (TnpA and TnpB), thereby fostering the origination of TBPs. It remains unknown whether TBPs originated in Proteobacteria and then gradually spread to other prokaryotes. Owing to the obvious bias of the genome composition in public databases, we cannot rule out the possibility that TBPs originally originated from other phyla (particularly those with high TBP occurrence) and then spread to different microbial clades within the proteobacteria. Previously, the Mu-like head phage group, which was represented by two deep seawater-derived phages (vB_ThpS-P1 and vB_PeaS-P1), was proposed and found to be common in the marine environment [26]. This phage group displayed distinct features from the Mu and Mu-like phages, including a lack of the Gam (host nuclease inhibitor)- and Mor (transcription activator)- encoding genes and the absence of random host-derived sequences at its genomic DNA termini [26]. The Mu-like head phage group seems to have originated via recombination between TBPs and other phages. However, whether these phages are capable of replicative transposition, thereby belonging to the TBP family, is currently unknown. Transposases are among the most abundant genes in microbial genomes and harbor great diversity [34, 61]. Nonetheless, all the identified transposases belong exclusively to the DDE family. Are there TBPs that contain other types of transposases? In fact, abundant transposases have been found to exist in the genomes of other viral groups, such as Inovirus [62]. Can they perform replicative transposition? If so, the definition and membership of TBPs is expected to be further expanded. If not, why did TBPs specifically select the DDE family of transposases? We believe that these issues are worth exploring in future studies.
Methods
Identification of the conserved protein families of TBPs
The genomic sequences of isolated TBPs were retrieved from the NCBI GenBank database [63]. To avoid missing putative TBPs in our screen, we searched the literature and the ICTV database [64]. The literature for isolated TBPs was searched using databases, including Google Scholar, PubMed, and Web of Science. The searches were limited to peer-reviewed publications written in English. All the putative TBPs were carefully curated to ensure that they met the criteria for TBPs previously defined by experts in the research field [8, 9], according to their genomic features and annotation information. The protein families of the 48 reference TBPs were identified by HHblits from HH-Suite3 [65] using the Pfam database [66], and the best hit with a probability >95% was considered the homologue of the query protein, according to the instructions of HH-Suite [65]. The Sequence Demarcation Tool (SDT, v1.2) [67] was used to calculate the AAI matrix of all TBP transposases. AlphaFold (v2.1) [68] was used to predict the structures of these transposases, and then DALI (online version, http://ekhidna2.biocenter.helsinki.fi/dali/) [69] was used to calculate the similarity matrix of these structures.
Identification of TBPs in prokaryotic and viral databases
The prokaryotic genome sequences were downloaded from the NCBI RefSeq Bacteria (n = 216,709) and Archaea (n = 1156) databases (Release 206) [70]. The viral genome sequences in the RefSeq Viral Database (Release 206, n = 11,080) [70], IMG/VR (v.2.0, n = 760,445) [35], and Global Ocean Viromes 2.0 (GOV 2.0, n = 488,128) [36] were downloaded from the NCBI database [71], the Joint Genome Institute’s (JGI) Genome Portal [72, 73], and iVirus [74], respectively.
The ORFs of genomes from the four datasets mentioned above were predicted by Prodigal (v2.6.3) [75] using the parameter “-p meta”. All the predicted proteins were compared with the conserved protein families of TBPs using hmmsearch (v3.3.2) from HMMER [76] with the threshold “score ≥30 and e ≤0.001”. For viruses from the RefSeq Viral Database, IMG/VR v2.0, and GOV 2.0, only the viral genomes containing all six conserved protein families were considered as putative TBPs. For genomes from the NCBI RefSeq Bacteria and Archaea databases, we set the following criterion: if there were six conserved TBP proteins in a genomic region of 40 kb (the length of isolated TBPs is generally between 35 and 40 kb), then that region was considered to contain a potential TBP. The smallest fragment containing six TBP marker proteins was referred to as the core TBP region. To determine the TBP boundaries, the core TBP region was extended to the left and right to obtain an 80 kb genomic region containing potential TBPs, referred to as the candidate TBP region. This region contained both potential TBP and host sequences at both ends. Next, we delimited the boundaries of transposable prophages by searching for its attachment sites (att) in the host genome. Specifically, BLASTn (v2.5.0 + ) [77] was used to align the candidate TBP region with all the other genomes in the same bacterial genus. If two fragments (queries) flanking the core TBP region could be aligned with two fragments (subjects) in another bacterial genome and they were identified as homologous sequences (cut-off e-value of 1e−5), and showed a 5 bp or 6 bp overlap at their ends (subjects), then they were identified as attB (in the subject genome), attL, or attR (in the query genome). Through this comparison, genome ends-defined (gd) TBPs were obtained (supplementary Fig. S2). Based on these obtained gdTBPs, we used BLASTn (cut-off e-value of 1e−5) to determine the genome boundaries in other candidate TBP regions. We compared gdTBPs with candidate TBP regions whose genome boundaries could not be determined by this method using BLASTp (v2.5.0) [77] (cut-off value of 1e−5) to obtain encoding region-defined (ed) TBPs. To further exclude possible false positives in edTBPs, all genomes with lengths >45 kb (n = 77) and <30 kb (n = 1031) as well as those harboring abnormal TBP regions (n = 316) were carefully manually curated.
To explore more diverse TBPs, we further searched for TBPs that were more distantly related to gdTBPs. Firstly, we extracted the sequences of transposases within gdTBPs, and we then compared them with the bacterial and archeal proteins in Refseq by BLASTp searches using DIAMOND (v2.0.2.140) [78] in ultra-sensitive mode with cut-offs of a coverage >50% and e-value <1e−3. Then the candidate TBP regions were obtained by using these homologous transposases in the bacterial genome as anchor points. The proteins in these regions were annotated using the PHROG database [79], and only the regions containing viral structural proteins were used for further analysis. We used the same procedure described above (Supplementary Fig. S2) to identify the gdTBPs contained in these candidate TBP regions. All TBPs from the RefSeq Viral Database were considered as genome boundary defined TBPs (gbTBP), as they have complete genome sequences.
Viral taxonomic assignment and network analysis
A total of 9766 gdTBP genomes were compiled and clustered by CD-HIT (v4.8.1) [80] using the widely recognized cut-off value of 95% average nucleotide identity (ANI) over an 85% alignment fraction [81], and the produced 3488 vOTUs were approximately at the species level. Protein-sharing network analysis of viral populations was performed by vConTACT (v2.0) [82]. In brief, the protein sequences of the vOTUs were grouped into PCs via all-to-all BLASTp by DIAMOND (v0.9.14.115) [78] with the default parameters of vConTACT (v2.0) [82]. The degree of similarity between the vOTUs was calculated based on the number of shared PCs. Then, pairs of closely related vOTUs with a similarity score of ≥1 were grouped into viral clusters (VCs), which were approximately at the genus level. The viral genomes from the class Caudoviricetes in ICTV (VMR_21-221122_MSL37) [64] were included in the networks used as references. The networks were visualized by Cytoscape (v3.8.2) [83] using a prefuse force-directed model. For cumulative analysis, 100 random TBP genome sets were generated, and the total number of PCs, vOTUs, and VCs identified from this set was calculated.
The family-level taxonomy of TBPs was assigned by PhaGCN (v2.0) [84] using a recommended cut-off score >0.5. Subsequently, all taxonomic assignments were subjected to manual inspection. To examine the gene contents of the TBPs, all protein sequences were aligned to the PHROG database [79] by BLASTp (v2.5.0) with an e-value cut-off of 1e−5.
Determining the distribution of TBPs in the biome and host
The taxonomic classification of all bacterial genomes was performed by GTDB-Tk (v1.5.0) [85] using the “classify_wf” pipeline. Briefly, 120 marker proteins in the bacterial genomes were identified, concatenated and aligned [75, 76]. Then, the maximum likelihood placement of each genome in the GTDB-Tk reference tree was determined [86]. The placement in the reference tree, relative evolutionary divergence and/or ANI to reference genomes were combined to classify each given bacterial genome [85]. For TBPs derived from prokaryotic genomes, their biomes were determined by extracting the biosample information of the host genomes from the NCBI database [71]. For TBPs derived from IMG/VR, this information was retrieved from the JGI Genome Portal [72, 73]. As previously reported [36], all the biomes of GOV 2.0-derived TBPs were categorized as seawater biomes.
Detection of GSIEs and DGRs
To identify functional GSIEs, we first retrieved the UniProt database [87] for the experimentally verified serine recombinase gin (Gin), and then the Gin protein sequences of the Escherichia phage Mu and D108 were used as references for subsequent detection. In addition, the tail fibre protein sequences were retrieved from the PHROG database [79]. The Gin and tail fibre proteins in viral genomes were identified by BLASTp using DIAMOND (v2.0.2.140) [78] with cut-off e-value of 1e−5. Only a Gin protein with an adjacent tail fibre protein (within two proteins downstream or upstream of Gin) was considered a functional GSIE. The virus-encoded DGRs, including reverse transcriptases (RTs), known template sequences (TRs), and variable repeats (VRs), were predicted by MetaCSST (v1.0) based on the Generalized Hidden Markov Model (GHMM) with default parameters [88].
Estimation of host range for bacteriophages
The putative phage hosts were predicted based on sequence similarity between spacers in prokaryotic CRISPR regions and protospacers in the phage genomes, as previously described [89]. Specifically, the CRISPR regions of all the prokaryotic genomes in the RefSeq database (Release 206) [70] were identified using the CRISPR Recognition Tool (CRT, v1.2) [90] with optimized parameters of “-minRL 20, -maxRL 50, -minSL 20, -maxSL 60, -searchWL 7”. As previously described [91], the ratio of the spacer lengths to the repeat lengths was limited to between 0.6 and 2.5, and CRISPR regions with <3 spacers were ignored. The retained CRISPR spacers were aligned with the phage genomes using BLASTn (v2.10.1) to identify protospacers in the phage genomes, and only matches satisfying the thresholds of ≥95% identity and ≤2 SNPs were selected. The CRISPR spacer matches were then used to estimate the species-level host range for each phage. Host range (number of host species) was compared between different groupings. Only gbTBPs were included in the analysis. Accordingly, we collected phages belonging to Caudoviricetes in the ICTV database [64], and they were regarded as the non-TBP group (n = 3611).
Identification of AMGs
The 18,449 TBPs were first run through the “--prep-for-dramv” function in VirSorter2 (v2.2.2) [92] to produce affi-contigs.tab files. DRAM-v, the viral mode of DRAM (v1.2.0) [41], was used to annotate all VirSorter 2-produced files and identify AMGs encoded by TBP genomes. The putative AMGs were distilled from the annotations based on their metabolic flag and their AMG score. All the qualified AMGs from TBPs had an AMG score of 1 or 2, which meant that the AMGs were flanked by two viral hallmark genes or flanked by one viral hallmark gene and one viral-like gene, respectively [41]. Only the AMGs assigned functional modules were used for further analysis.
Identification of integration sites
The attachment sites attL and attR, which are utilized for TBP integration into the host genome, normally displaying 5 bp or 6 bp direct repeats (DRs) close to the prophage boundaries [8, 9], were identified as described above. Hence, the sequences of noncoding regions flanking both ends of TBPs were extracted from host genomes, and the putative integration sites of TBPs were identified by inhouse scripts. The sequences were subjected to tBLASTx alignment against the KEGG database [93] online with an e-value cut-off of 1e−5 to predict protein-coding genes. In addition, tRNAscan-SE (v2.0.9) [94] in the “B” model was used to predict tRNAs at the integration loci.
Statistical analysis
Student’s t test was adopted to test the significance of all differences between groupings, using the R package ggsignif (v0.6.3) [95] and the Python function “ttest_ind” from Scipy (v1.6.2) [96]. For the DEGs in the transcriptomic analysis, the averaged fold changes (FCs) of transcription levels were based on three biologically independent samples. p values corresponding to differential gene expression tests were calculated by edgeR based on an overdispersed Poisson model [97]. The false discovery rate (FDR) was used in the multiple hypothesis testing to correct p value by the Benjamini-Hochberg (BH) method [98].
Recruitment analysis
The recruitment analysis of SP2 was performed as previously described [99]. Briefly, the raw reads of the POV [100] were downloaded from iVirus [74], and then the SP2 genome was compared to raw reads of POV by BLASTn (v2.10.1) with a cut-off e-value of 1e−3. Only reads that had ≥50% identity (nucleic acid) were considered in the recruitment plots.
Culture conditions and growth assays
The Shewanella strains (Supplementary Table S9) were cultured in modified 2216E marine (2216E) medium (5 g/L tryptone, 1 g/L yeast extract, 0.1 g/L FePO4, 34 g/L NaCl) with shaking at 220 rpm at different temperatures (15 °C or 4 °C) or using stainless steel pressure vessels for cultivation at high hydrostatic pressure (20 MPa). The E. coli strain was incubated in lysogeny broth (LB) medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) supplemented with 50 μg/mL DL-α, ε-diaminopimelic acid (DAP) (Sigma, St. Louis, USA) at 37 °C. For solid media, agar-A (Bio Basic Inc., Ontario, Canada) was added at a concentration of 1.5% (w/v). When needed, the antibiotic chloramphenicol (Cm) (Sigma, St. Louis, USA) was added to the media at final concentrations of 25 μg/mL and 12.5 μg/mL for E. coli and Shewanella, respectively. Growth assays of the Shewanella strains were performed using turbidity measurements at 600 nm with a spectrophotometer (UV-2550, Shimadzu, Kyoto, Japan).
Construction of the prophage SP2 deletion strain
The SP2 prophage deletion mutant was constructed by a recombination knock-out method as described previously [101]. Briefly, the upstream and downstream fragments flanking both ends of SP2 were amplified with PCR primer pairs (Supplementary Table S110). These two fragments were used as templates in a second round of fusion PCR, resulting in a fusion fragment containing the flanking sequence of SP2. Then, the PCR product was cloned into the suicide plasmid pRE112, yielding pRE112-SP2. This plasmid was transformed into E. coli WM3064 and then into WP2 by two-parent conjugation. The transconjugant was selected by chloramphenicol resistance and verified by PCR. The WP2 strain with pRE112-SP2 inserted into the chromosome was plated on 2216E agar medium supplemented with 10% sucrose. Finally, the successful prophage deletion mutant WP2ΔSP2 was screened and confirmed by PCR and subsequent DNA sequencing.
RNA isolation and RT-qPCR
The cultures of S. psychrophila WP2 strains were collected and frozen in liquid nitrogen immediately when the cells reached late exponential phase. Total RNA was isolated with a TRI reagent-RNA isolation kit (Molecular Research Center, Cincinnati, USA) and treated with DNase I at 37 °C for 1 h to remove DNA contamination. The purified RNA was reverse transcribed to cDNA by a RevertAid First Strand cDNA Synthesis Kit (Fermentas, Maryland, USA). The primer pairs used to amplify the selected genes for RT-qPCR were designed using Primer-BLAST [102], and PCR cycling was conducted using a StepOnePlus real-time PCR system (Thermo Fisher Scientific) in 20 μl reaction mixtures that included 1× SYBR Green I Universal PCR Master Mix (Thermo Fisher Scientific), 0.5 μM each primer, and 1 μl of cDNA template.
Transcriptomic analysis
Strand-specific transcriptome sequencing was performed at Magigene Biotechnology Co., Ltd. (Guangdong, China) as described previously [103]. First, rRNA was removed using an Epicentre Ribo-Zero rRNA Removal Kit (Epicentre, Madison, WI, USA), and a cDNA library was prepared with a NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) according to the manufacturer’s instructions. The initial quantification of the library was carried out using a Qubit Fluorometer (Life Technologies, Carlsbad, CA, USA), and the insertion fragment size of the library was determined with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The effective concentration of the library was quantified accurately via qPCR (effective concentration > 2 nM). The different libraries were pooled together in a flow cell according to the effective concentration and the target offline data volume. After clustering, the HiSeq System (Illumina, San Diego, USA) was used for paired-end sequencing. The raw data were filtered and evaluated by FASTp software (v0.19.7) [104], after which the clean reads were mapped to the S. psychrophila WP2 genome (NZ_CP014782.1) by HISAT software (v2.1.0) [105]. RSEM (v1.3.1) [106] was used to calculate the read counts per sample, and the sequencing results were evaluated in terms of quality, alignment, saturation, and distribution of reads on the reference genome by DEGseq (v1.36.0) [107]. Gene expression was calculated on the basis of the number of reads mapped to each gene using the fragments per kilobase per million mapped reads (FPKM) method [108] and analyzed by edgeR (v3.20.2) [97]. The DEGs were identified according to the following standards: FDR < 0.05 and FPKM fold change (FC) ≥ 2 between two samples. For each strain, three biologically independent samples were used for the RNA-seq analysis.
Supplementary information
Acknowledgements
This work was financially supported by the Hainan Provincial Joint Project of Sanya Yazhou Bay Science and Technology City (grant no. 2021JJLH0057), the National Natural Science Foundation of China (grant nos. 42176095, 91851113, 41921006), the National Key R&D Program of China (grant no. 2021YFF0501300), and the Oceanic Interdisciplinary Program of Shanghai Jiao Tong University (project no. SL2021PT201). We would like to thank Prof. Ariane Toussaint for helpful suggestions on TBP identification. We are grateful to the editor and two anonymous reviewers for their comments that were instrumental in improving the paper.
Author contributions
HJ conceived and designed the research; MZ, QS, and ST collected and curated genomic and metadata; MZ performed the bioinformatic and statistical analysis; MZ and SL conducted the microbiological experiments; YH and XT helped in RNA isolation; MZ and HJ analyzed and interpreted the data; HJ and MZ wrote the paper; XX and YY provided useful comments to improve the paper; HJ supervised the project. All the authors reviewed the results and approved the paper.
Data availability
All the identified TBP genomic sequences (n = 18,449) have been deposited in CyVerse (available at https://data.cyverse.org/dav-anon/iplant/home/zhangmujie/TBPGD/TBPGD.zip) and in the National Omics Data Encyclopedia (NODE) under project ID OEP003495. The transcriptomic data from the current study have been deposited in NODE under project ID OEP002984.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01414-z.
References
- 1.Dion MB, Oechslin F, Moineau S. Phage diversity, genomics and phylogeny. Nat Rev Microbiol. 2020;18:125–38.. doi: 10.1038/s41579-019-0311-5. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerman AE, Howard-Varona C, Needham DM, John SG, Worden AZ, Sullivan MB, et al. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat Rev Microbiol. 2020;18:21–34. doi: 10.1038/s41579-019-0270-x. [DOI] [PubMed] [Google Scholar]
- 3.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol. 2015;13:641–50. doi: 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 4.Touchon M, Bernheim A, Rocha EP. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016;10:2744–54.. doi: 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahl A, Battesti A, Ansaldi M. Prophages in Salmonella enterica: a driving force in reshaping the genome and physiology of their bacterial host? Mol Microbiol. 2019;111:303–16.. doi: 10.1111/mmi.14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 2017;11:1511–20.. doi: 10.1038/ismej.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argov T, Azulay G, Pasechnek A, Stadnyuk O, Ran-Sapir S, Borovok I, et al. Temperate bacteriophages as regulators of host behavior. Curr Opin Microbiol. 2017;38:81–7. doi: 10.1016/j.mib.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Toussaint A, Rice PA. Transposable phages, DNA reorganization and transfer. Curr Opin Microbiol. 2017;38:88–94. doi: 10.1016/j.mib.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Harshey RM. Transposable Phage Mu. Microbiology spectrum. 2014;2. 10.1128/microbiolspec.MDNA3-0007-2014. [DOI] [PMC free article] [PubMed]
- 10.Taylor AL. Bacteriophage-induced mutation in Escherichia coli. PNAS. 1963;50:1043–51. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harshey RM. The Mu story: how a maverick phage moved the field forward. Mobile. DNA. 2012;3:21. doi: 10.1186/1759-8753-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuno N, Dramićanin M, Mizuuchi M, Adam J, Wang Y, Han Y-W, et al. MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition. PNAS. 2013;110:E2441–50. doi: 10.1073/pnas.1309499110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.George M, Bukhari AI. Heterogeneous host DNA attached to the left end of mature bacteriophage Mu DNA. Nature. 1981;292:175–6. doi: 10.1038/292175a0. [DOI] [PubMed] [Google Scholar]
- 14.Groenen MAM, Putte PVD. Mapping of a site for packaging of bacteriophage Mu DNA. Virology. 1985;144:520–2. doi: 10.1016/0042-6822(85)90292-2. [DOI] [PubMed] [Google Scholar]
- 15.Howe MM. Transduction by Bacteriophage MU-l. Virology. 1973;55:103–17. [DOI] [PubMed]
- 16.Gill GS, Hull RC. Mutator bacteriophage D108 and its DNA: an electron microscopic characterization. J Virol. 1981;37:420–30. doi: 10.1128/jvi.37.1.420-430.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J Bacteriol. 2004;186:6560–74. doi: 10.1128/JB.186.19.6560-6574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Summer EJ, Gonzalez CF, Carlisle T, Mebane LM, Cass AM, Savva CG, et al. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. J Mol Biol. 2004;340:49–65. doi: 10.1016/j.jmb.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 19.Fogg PCM, Hynes AP, Digby E, Lang AS, Beatty JT. Characterization of a newly discovered Mu-like bacteriophage, RcapMu, in Rhodobacter capsulatus strain SB1003. Virology. 2011;421:211–21. doi: 10.1016/j.virol.2011.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Zehr ES, Tabatabai LB, Bayles DO. Genomic and proteomic characterization of SuMu, a Mu-like bacteriophage infecting Haemophilus parasuis. BMC Genom. 2012;13:331. doi: 10.1186/1471-2164-13-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakhetia R, Verma NK. Identification and Molecular Characterisation of a Novel Mu-Like Bacteriophage, SfMu, of Shigella flexneri. PLoS ONE. 2015;10:e0124053. doi: 10.1371/journal.pone.0124053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Zhang Y, Jiang Y, Wu H, Sun W, Huang Y-P. Characterization and Genomic Analysis of ΦSHP3, a New Transposable Bacteriophage Infecting Stenotrophomonas maltophilia. J Virol. 2021;95:e00019–21.. doi: 10.1128/JVI.00019-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masignani V, Giuliani MM, Tettelin H, Comanducci M, Rappuoli R, Scarlato V. Mu-like Prophage in serogroup B Neisseria meningitidis coding for surface-exposed antigens. Infect Immun. 2001;69:2580–8. doi: 10.1128/IAI.69.4.2580-2588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan GJ, Hatfull GF, Casjens S, Hendrix RW. Bacteriophage Mu Genome Sequence Analysis and comparision with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J Mol Microbiol. 2002;317:337–59. doi: 10.1006/jmbi.2002.5437. [DOI] [PubMed] [Google Scholar]
- 25.Guo Q, Chen B, Tu Y, Du S, Chen X. Prophage LambdaSo uses replication interference to suppress reproduction of coexisting temperate phage MuSo2 in Shewanella oneidensis MR-1. Environ Microbiol. 2019;21:2079–94.. doi: 10.1111/1462-2920.14592. [DOI] [PubMed] [Google Scholar]
- 26.Tang K, Lin D, Zheng Q, Liu K, Yang Y, Han Y, et al. Genomic, proteomic and bioinformatic analysis of two temperate phages in Roseobacter clade bacteria isolated from the deep-sea water. BMC Genom. 2017;18:485. doi: 10.1186/s12864-017-3886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szafrański SP, Kilian M, Yang I. Wieden GBd, Winkel A, Hegermann J, et al. Diversity patterns of bacteriophages infecting Aggregatibacter and Haemophilus species across clades and niches. ISME J. 2019;13:2500–22.. doi: 10.1038/s41396-019-0450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui Z, Xu Z, Wei Y, Zhang Q, Qin K, Ji X. Characterization and Genome Analysis of a Novel Mu-like Phage VW-6B Isolated from the Napahai Plateau Wetland of China. Curr Microbiol. 2021;78:150–8. doi: 10.1007/s00284-020-02277-9. [DOI] [PubMed] [Google Scholar]
- 29.Lin D, Tang K, Han Y, Li C, Chen X. Genome sequence of an inducible phage in Rhodovulum sp. P5 isolated from the shallow-sea hydrothermal system. Mar Genom. 2016;30:93–5. doi: 10.1016/j.margen.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Mara P, Vik D, Pachiadaki MG, Suter EA, Poulos B, Taylor GT, et al. Viral elements and their potential influence on microbial processes along the permanently stratified Cariaco Basin redoxcline. ISME J. 2020;14:3079–92.. doi: 10.1038/s41396-020-00739-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hulo C, Masson P, Mercier PL, Toussaint A. A structured annotation frame for the transposable phages: a new proposed family “Saltoviridae” within the Caudovirales. Virology. 2015;477:155–63.. doi: 10.1016/j.virol.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Toussaint A, Gijsegem FV. Extension of the transposable bacterial virus family: two genomic organisations among phages and prophages with a Tn552-related transposase. Res Microbiol. 2018;169:495–9. doi: 10.1016/j.resmic.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Ndela EO, Enault F, Toussaint A. Transposable Prophages in Leptospira: An Ancient, Now Diverse, Group Predominant in Causative Agents of Weil’s Disease. Int J Mol Sci. 2021;22:13434. doi: 10.3390/ijms222413434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–17. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paez-Espino D, Roux S, Chen I-MA, Palaniappan K, Ratner A, Chu K, et al. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 2019;47:D678–D86. doi: 10.1093/nar/gky1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregory AC, Zayed AA, Conceição-Neto N, Temperton B, Bolduc B, Alberti A, et al. Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell. 2019;177:1109–23.. doi: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner D, Kropinski AM, Adriaenssens EM. A Roadmap for Genome-Based Phage Taxonomy. Viruses. 2021;13:506. doi: 10.3390/v13030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, Hugenholtz P. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50:D785–D94.. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritacco CJ, Kamtekar S, Wang J, Steitz TA. Crystal structure of an intermediate of rotating dimers within the synaptic tetramer of the G-segment invertase. Nucleic Acids Res. 2013;41:2673–82. doi: 10.1093/nar/gks1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howe MM, Pato ML. Phage Mu. Reference Module in Life Sciences. 2017:1–6. 10.1016/B978-0-12-809633-8.06883-7.
- 41.Shaffer M, Borton MA, McGivern BB, Zayed AA, Rosa SLL, Solden LM, et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020;48:8883–900.. doi: 10.1093/nar/gkaa621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao X, Wang P, Zeng X, Bartlett DH, Wang F. Shewanella psychrophila sp. nov. and Shewanella piezotolerans sp. nov., isolated from west Pacific deep-sea sediment. Int J Syst Evolut Microbiol. 2007;57:60–5. doi: 10.1099/ijs.0.64500-0. [DOI] [PubMed] [Google Scholar]
- 43.Xu G, Jian H, Xiao X, Wang F. Complete genome sequence of Shewanella psychrophila WP2, a deep-sea bacterium isolated from west Pacific sediment. Mar Genom. 2017;35:19–21. doi: 10.1016/j.margen.2017.03.004. [DOI] [Google Scholar]
- 44.Namgoong S-Y, M.Harshey R. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 1988;17:3775–85. doi: 10.1093/emboj/17.13.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuuchi M, Mizuuchi K. Conformational isomerization in phage Mu transpososome assembly effects of the transpositional enhancer and of MuB. EMBO J. 2001;20:6927–35. doi: 10.1093/emboj/20.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han Y-W, Mizuuchi K. Phage Mu transposition immunity: protein pattern formation along DNA by a diffusion-ratchet mechanism. Mol Cell. 2010;39:48–58. doi: 10.1016/j.molcel.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi W, Jang S, Harshey RM. Mu transpososome and RecBCD nuclease collaborate in the repair of simple Mu insertions. PNAS. 2014;111:14112–7. doi: 10.1073/pnas.1407562111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang PW, Chu L, Guttman DS. Complete sequence and evolutionary genomic analysis of the Pseudomonas aeruginosa transposable bacteriophage D3112. J Bacteriol. 2004;186:400–10. doi: 10.1128/JB.186.2.400-410.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goudie AD, Lynch KH, Seed KD, Stothard P, Shrivastava S, Wishart DS, et al. Genomic sequence and activity of KS10, a transposable phage of the Burkholderia cepacia complex. BMC Genom. 2008;9:615. doi: 10.1186/1471-2164-9-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung I-Y, Cho Y-H. Complete genome sequences of two Pseudomonas aeruginosa temperate phages, MP29 and MP42, which lack the phage-host CRISPR interaction. J Virol. 2012;86:8336. doi: 10.1128/JVI.01127-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J, Kong Y, Li X, Yang S. A novel transposable Mu-like prophage in Bacillus alcalophilus CGMCC 1.3604 (ATCC 27647) Virol Sin. 2015;30:63–5. doi: 10.1007/s12250-014-3497-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thi BVT, Khanh NHP, Namikawa R, Miki K, Kondo A, Thi PTD, et al. Genomic characterization of Ralstonia solanacearum phage ΦRS138 of the family Siphoviridae. Arch Virol. 2016;161:483–6. doi: 10.1007/s00705-015-2654-1. [DOI] [PubMed] [Google Scholar]
- 53.Cornuault JK, Petit M-A, Mariadassou M, Benevides L, Moncaut E, Langella P, et al. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome. 2018;6:65. doi: 10.1186/s40168-018-0452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toussaint A. Transposable Mu-like phages in Firmicutes: new instances of divergence generating retroelements. Res Microbiol. 2013;164:281–7. doi: 10.1016/j.resmic.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 55.Cazares A, Mendoza-Hernández G, Guarneros G. Core and accessory genome architecture in a group of Pseudomonas aeruginosa Mu-like phages. BMC Genom. 2014;15:1146. [DOI] [PMC free article] [PubMed]
- 56.Touchon M, Sousa JAMD, Rocha EP. Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr Opin Microbiol. 2017;38:66–73. doi: 10.1016/j.mib.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 57.Edlin G, Lin L, Bitner R. Reproductive fitness of P1, P2, and Mu lysogens of Escherichia coli. J Virol. 1977;21:560–4. doi: 10.1128/jvi.21.2.560-564.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engelhardt T, Sahlberg M, Cypionka H, Engelen B. Biogeography of Rhizobium radiobacter and distribution of associated temperate phages in deep subseafloor sediments. ISME J. 2013;7:199–209. doi: 10.1038/ismej.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, et al. Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 60.Lemaire ON, Méjean V, Iobbi-Nivol C. The Shewanella genus: ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol Rev. 2020;44:155–70.. doi: 10.1093/femsre/fuz031. [DOI] [PubMed] [Google Scholar]
- 61.Hickman AB, Dyda F. DNA Transposition at Work. Chem Rev. 2016;116:12758–84.. doi: 10.1021/acs.chemrev.6b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roux S, Krupovic M, Daly RA, Borges AL, Nayfach S, Schulz F, et al. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat Microbiol. 2019;4:1895–906.. doi: 10.1038/s41564-019-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Schoch CL, Sherry ST, et al. GenBank. Nucleic Acids Res. 2021;49:D92–D6.. doi: 10.1093/nar/gkaa1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018;46:D708–D17.. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steinegger M, Meier M, Mirdita M, Vöhringer H, Haunsberger SJ, Söding J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinform. 2019;20:473. doi: 10.1186/s12859-019-3019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D9.. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE. 2014;9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holm L. Using Dali for Protein Structure Comparison. In: Gάspάri Z, editor. Structural Bioinformatics: Methods and Protocols. Methods in Molecular Biology. Springer Science+Business Media; 2020;2112:29–42. [DOI] [PubMed]
- 70.Li W, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, et al. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49:D1020–D8.. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–45. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014;42:D26–31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bolduc B, Zablocki O, Guo J, Zayed AA, Vik D, Dehal P, et al. iVirus 2.0: Cyberinfrastructure-supported tools and data to power DNA virus ecology. ISME Commun. 2021;1:77. doi: 10.1038/s43705-021-00083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W4.. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 79.Terzian P, Ndela EO, Galiez C, Lossouarn J, Bucio REP, Mom R, et al. PHROG: families of prokaryotic virus proteins clustered using remote homology. NAR Genom Bioinforma. 2021;3:lqab067. doi: 10.1093/nargab/lqab067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, et al. Minimum Information about an Uncultivated Virus Genome (MIUViG) Nat Biotechnol. 2019;37:29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jang HB, Bolduc B, Zablocki O, Kuhn JH, Roux S, Adriaenssens EM, et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol. 2019;37:632–9. doi: 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
- 83.Shannon P, Andrew M, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang J-Z, Yuan W-G, Shang J, Shi Y-H, Yang L-L, Liu M, et al. Virus classification for viral genomic fragments using PhaGCN2. Brief Bioinforma. 2023;24:1–9. doi: 10.1093/bib/bbac505. [DOI] [PubMed] [Google Scholar]
- 85.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsen FA, Kodner RB, Armbrust EV. pplacer: linear time maximum-likelihood and bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinform. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Consortium TU. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D9.. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yan F, Yu X, Duan Z, Lu J, Jia B, Qiao Y, et al. Discovery and characterization of the evolution, variation and functions of diversity-generating retroelements using thousands of genomes and metagenomes. BMC Genom. 2019;20:595. doi: 10.1186/s12864-019-5951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jian H, Yi Y, Wang J, Hao Y, Zhang M, Wang S, et al. Diversity and distribution of viruses inhabiting the deepest ocean on Earth. ISME J. 2021;15:3094–110.. doi: 10.1038/s41396-021-00994-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, et al. Uncovering Earth’s virome. Nature. 2016;536:425–30. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 92.Guo J, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9:37. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D51.. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan PP, Lin BY, Mak AJ, Lowe TM. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49:9077–96.. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ahlmann-Eltze C, Patil I. Ggsignif: R package for displaying significance brackets for ‘ggplot2’. PsyArXiv. 2021. 10.31234/osf.io/7awm6.
- 96.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–72.. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Benjamini Y, Hochberg Y. Controlling the False Discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol) 1995;1:289–300. [Google Scholar]
- 99.Meng C, Li S, Fan Q, Chen R, Hu Y, Xiao X, et al. The thermo-regulated genetic switch of deep-sea filamentous phage SW1 and its distribution in the Pacific Ocean. FEMS Microbiol Lett. 2020;367:fnaa094. doi: 10.1093/femsle/fnaa094. [DOI] [PubMed] [Google Scholar]
- 100.Hurwitz BL, Sullivan MB. The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE. 2013;8:e57355. doi: 10.1371/journal.pone.0057355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jian H, Xiao X, Wang F. Role of filamentous phage SW1 in regulating the lateral flagella of Shewanella piezotolerans strain WP3 at low temperatures. Appl Environ Microbiol. 2013;79:7101–9. doi: 10.1128/AEM.01675-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jian H, Xu G, Yi Y, Hao Y, Wang Y, Xiong L, et al. The origin and impeded dissemination of the DNA phosphorothioation system in prokaryotes. Nat Commun. 2021;12:6382. doi: 10.1038/s41467-021-26636-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i90.. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–8. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 108.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Baren MJV, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the identified TBP genomic sequences (n = 18,449) have been deposited in CyVerse (available at https://data.cyverse.org/dav-anon/iplant/home/zhangmujie/TBPGD/TBPGD.zip) and in the National Omics Data Encyclopedia (NODE) under project ID OEP003495. The transcriptomic data from the current study have been deposited in NODE under project ID OEP002984.