Abstract
Post-translational modifications (PTM) of proteins increase the functional diversity of the proteome and have been implicated in the pathogenesis of numerous diseases. The most widely understood modifications include phosphorylation, methylation, acetylation, O-linked/N-linked glycosylation, and ubiquitination, all of which have been extensively studied and documented. Citrullination is a historically less explored, yet increasingly studied, protein PTM which has profound effects on protein conformation and protein-protein interactions. Dysregulation of protein citrullination has been associated with disease development and progression. Identification and characterization of citrullinated proteins is highly challenging, complicated by the low cellular abundance of citrullinated proteins, making it difficult to identify and quantify the extent of citrullination in samples, coupled with challenges associated with development of mass spectrometry (MS)-based methods, as the corresponding mass shift is relatively small, +0.984 Da, and identical to the mass shift of deamidation. The focus of this review is to discuss recent advancements of citrullination-specific MS approaches and integration of the potential methodology for improved citrullination identification and characterization. In addition, the association of citrullination in disease networks is also highlighted.
Keywords: Citrullination, mass spectrometry, PTM, methodology, quantitation, proteomics, disease network
1. Introduction
Citrullination is a post-translational modification (PTM) of great interest, characterized by the conversion of an arginine residue to a citrulline residue. Mechanistically this modification occurs through the transformation of a guanidium group, located on an arginine residue, to an ureido group, facilitated by peptidylarginine deiminase (PAD) enzymes [1–4]. In humans, PADs are a family of calcium-dependent enzymes composed of five isozymes (PAD1–4, and PAD6) which are encoded by five genes arrayed in tandem on chromosome 1 (Figure 1). These enzymes have been noted to be highly homologous, sharing 50% or higher sequence identity, with the greatest regions of similarity found toward the C-termini of the protein [4]. PADs are widely distributed in various tissues and each PAD has its specific location and preferred substrates. PAD1 is known to be primarily expressed in the epidermis and uterus, with keratins and filaggrins serving as the primary citrullination substrates within the epidermis [5,6]. PAD2 is most frequently associated with the secretory glands, uterus, spleen, and pancreas, though it is especially abundant in skeletal muscle and the brain [7–10]. PAD3 is primarily localized in hair follicles and the epidermis, where colocalization and citrullination of trichohyalin, a structural protein of the inner root sheath of hair follicles, occurs during routine hair follicle hardening [11,12]. PAD4 is found mainly in white blood cells and tumors, broadly distributed and associated with various tissue origins [13–15], PAD4 is the only isozyme with confirmed presence within the cellular nucleus, where it plays a role in histone deimination, though recent studies have reported that PAD2 may also exist in the nucleus [7]. PAD6 is essential for oocyte cytoskeletal sheet formation and female fertility, and is mainly found in mouse eggs, embryos, and oocytes [16–18]. Different from PADs 1−4, which are catalytically active, PAD6 has lost some conserved Ca2+ binding residues and conserved cysteine residues, thus it cannot be considered as an active deiminase [19]. A comprehensive summary of distribution of each type of PAD is illustrated in Figure 1. PAD-knockout models have served as a valuable resource to gain an understanding of the PAD-mediated roles of citrullination in various pathological pathways and diseases. In one instance, a PAD2-knockout model displayed improved survival in instances of hemorrhagic shock, a form of hypovolemic shock in which severe blood loss leads to inadequate oxygen delivery[20]. In another study, a PAD2-knockout mouse had reportedly lower levels of citrullination in the central nervous system (CNS)[21]. It has also been reported in one instance that the knockout of PAD2 did not produce any significant phenotype in the nervous system, bolstering the importance of myelin basic protein (MBP) citrullination in axonal electrical signal transmission[13], and PAD2-catalyzed citrullination was deemed non-essential for the development of autoimmune encephalomyelitis, a condition associated with inflammation[21]. Alternatively, it was determined that PAD4-knockout mice are more susceptible to bacterial infections than wild-type mice, due to a lack of neutrophil extracellular trap (NETs) formation after stimulation with chemokines or incubation with bacteria[22]. Evidence of decreased severity of autoimmune arthritis was also observed in PAD4-knockout mice[23]. Under normal physiological conditions, PADs usually maintain an inactive status, given the low internal calcium concentration. PADs can be activated and have normal functionality during certain events, such as apoptosis and terminal differentiation of cells, where calcium levels are above the physiological concentration (10−8 to 10−6 M)[24]. Given the widespread distribution of PADs, citrullination has been also reported to play other important roles as part of normal physiological functions, including normal function of the immune system, gene regulation, apoptosis, skin keratinization, hair growth, myelin formation, NETs formation, the insulation of neurons, and the plasticity of the central nervous system[25].
Figure 1.
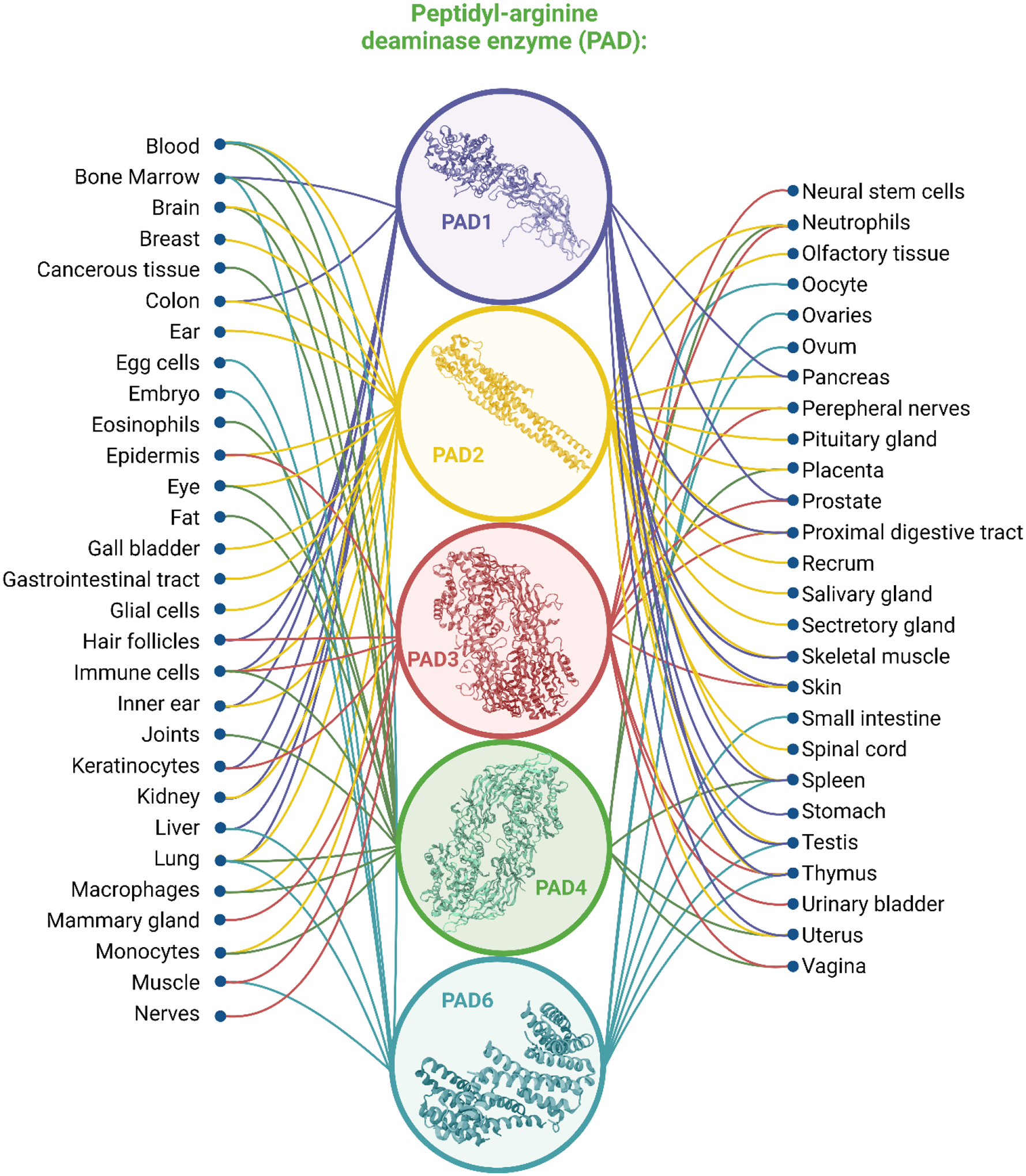
Five types of peptidylarginine deiminases (PADs) and their corresponding distribution.
The chemical mechanism of the citrullination reaction is characterized by the primary, positively-charged ketimine group of arginine undergoing replacement by a ketone group, forming neutrally-charged citrulline, a process mediated by PAD catalysis. The loss of a net charge directly impedes hydrogen bond formation and alters charge distribution, increasing the hydrophobicity of the protein and adversely affecting the protein stability (Figure 2) [26–28]. Therefore it is no surprise that many studies have defined connections between ubiquitous PAD expression, citrullination, and numerous diseases, thought to aid in the development and progression of rheumatoid arthritis (RA) [29,30], prion disease [31], psoriasis [32], Alzheimer’s disease (AD)[33–35], multiple sclerosis [36–39], various cancers [40], diabetes [41,42], and others [43]. While these connections between citrullination and disease conditions are intriguing, it should be noted that a major barrier to draw definitive conclusions is presented by the scale of the existing studies, the majority of which examined a limited number of specimens, ultimately identifying only a few citrullinated proteins without site-specific localization information.
Figure 2.
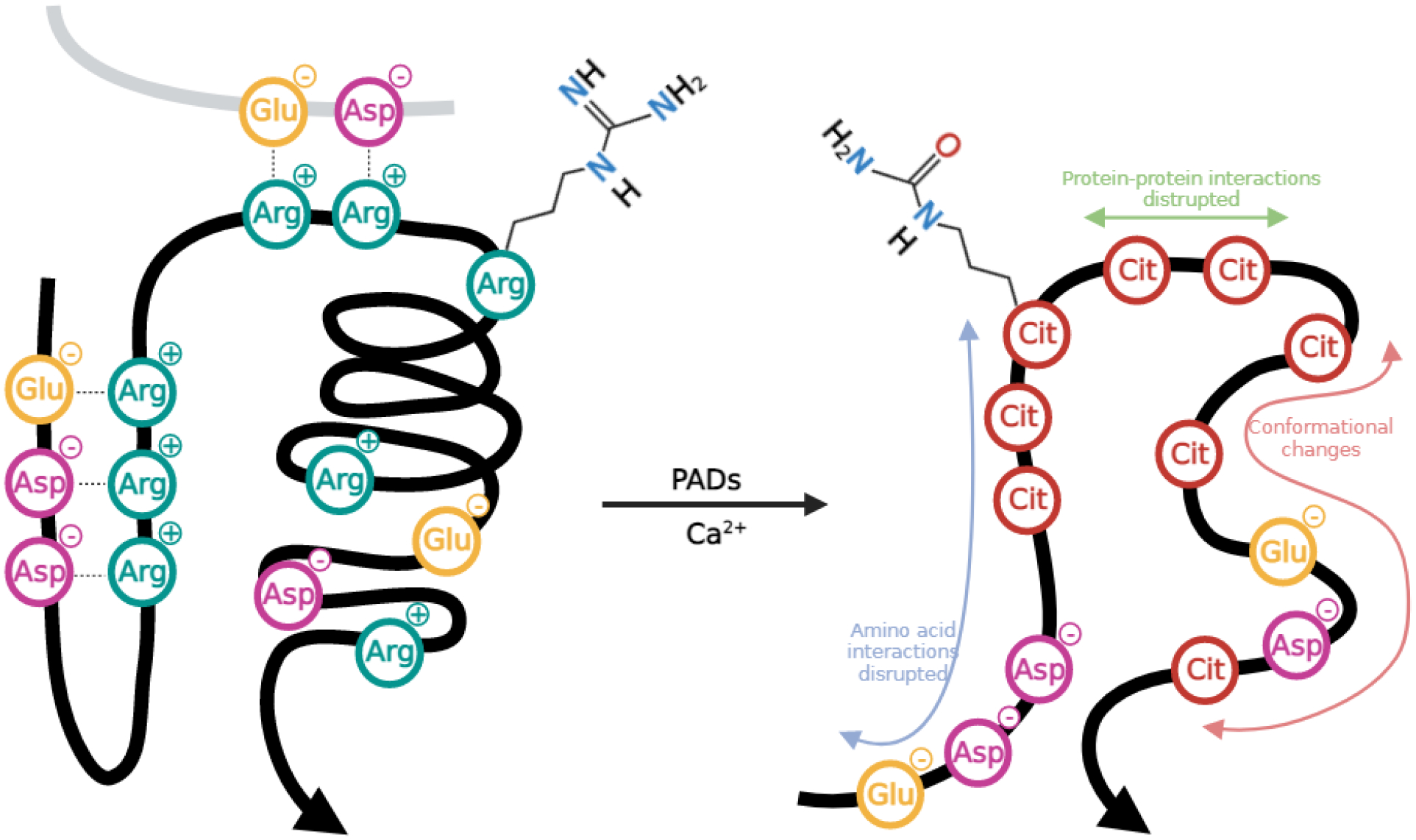
Citrullination of arginine by peptidylarginine deiminases and the subsequent impact on protein–protein interactions.
Antibody-based methods are currently the most popular, as these are well-established and documented, for detecting citrullination, though these methods are intrinsically incapable of large-scale analysis, and it is difficult to pinpoint the specific modification site [44–46]. Mass spectrometry (MS) presents a promising approach for citrullination site localization due to its excellent sensitivity, but even MS-based approaches for citrullination investigation are in dire need of optimization, as the small corresponding mass shift and low abundance of citrullinated proteins introduces significant obstacles [45,47]. Tremendous efforts have been made to overcome these challenges, working to optimize both data acquisition and data analysis processes. With respect to acquisition method optimization, methods for chemical derivatization [48,49] and enrichment [50–52] have been presented, while others have worked to generate novel algorithms for data analysis and database searching [53–58], in addition to approaches complemented with careful, manual examination of spectra [59,60]. These methods seek to overcome the intrinsic hurdles encountered with citrullination identification, including its small mass shift, by inducing a larger mass shift for citrullinated peptides compared to their non-modified counterparts for more confident identification; low abundance, by effective enrichment methods; and localization, by developing algorithms to increase statistical confidence of MS identification results. The aim of this review is to introduce and survey novel MS-based citrullination identification approaches and investigate the ways in which these methods can be integrated into citrullination investigations, motivated by network biology. The involvements of citrullination in the pathogenesis of various diseases are also discussed. A comprehensive overview of citrullination-associated diseases along with corresponding tissues, proteins, cell types, and PAD isozyme types is presented (Supplementary Table 1), as well as the utilization of MS techniques and methodologies that lead to these biological findings (Supplementary Table 2).
2. Strategies to identify citrullination sites and method integration
2.1. Antibody-based and probe-based detection of protein citrullination
Our knowledge of the citrullinome is still limited primarily due to the lack of effective and robust analytical tools, as many rely on low-throughput methods, such as immunodetection using available antibodies that recognize citrulline residues, identified through traditional Western blot or immunostaining procedures [46,61,62]. Two primary polyclonal antibodies are used for citrullination detection, histone H3, which recognizes citrulline residues at positions 2, 8, and 17, and histone H4, which recognizes these residues at position 3. Anti-peptidyl-citrulline, clone F95 antibody has also been validated for detection of peptidyl-citrulline [63,64]. Clonal F95 antibodies have been employed to recognize citrulline by staining the fibrinoid extracellular matrix of necrotic synovial tissue of RA patients, enabling visualization of citrullinated proteins in those areas. Another antibody-based method utilizes further modification of any present citrulline residues with 2, 3-butanedione monoxime and antipyrine in a strong acidic solution, which forms an ureido group adduct, detectable by Western blot with an anti-modified citrulline antibody [65]. Moreover, a citrulline-specific probe, rhodamine, is a sensitive fluorophore that specifically detects protein citrullination via a chemoselective reaction between the glyoxal group of the rhodamine and citrulline [64,66]. Although these methods are sufficient for some applications, they lack specificity, the sensitivity of the antibodies is less than ideal, they are incapable of profiling citrullinated proteins of low abundance, and they cannot provide localization of the citrullinated residue, rendering these approaches unfit for global citrullinome characterization.
2.2. Standard Mass Spectrometry Approaches
MS-based strategies are gaining popularity as powerful tools for large-scale characterization and localization of various PTMs due to the rapid development of MS sensitivity, accuracy, and compatibility, though its application to mapping citrullination is much less established [44,45,47,67,68]. As citrullinated proteins are often present in low abundance, corresponding signals can be largely suppressed by other molecules within the sample, with effective enrichment solutions lacking. In the event that the low abundance citrullinated protein is detected, the small mass shift induced by citrullination (+0.984 Da) can be easily confused with deamidation (+0.984 Da) and 13C isotopic peaks (+1.0033 Da). These hurdles result in poor-quality tandem MS spectra, presenting additional challenges in confident identification, localization, and quantification of citrullinated proteins. Standard MS analysis is possible, but often requires a mass spectrometer with high mass accuracy and time-consuming manual examination of the MS spectra [59,69,70]. Therefore, optimized strategies for citrullination sample preparation, MS parameters, and data searching have been developed to fill these gaps (Figure 3). These approaches and their effectiveness will be discussed in the rest of this section.
Figure 3.
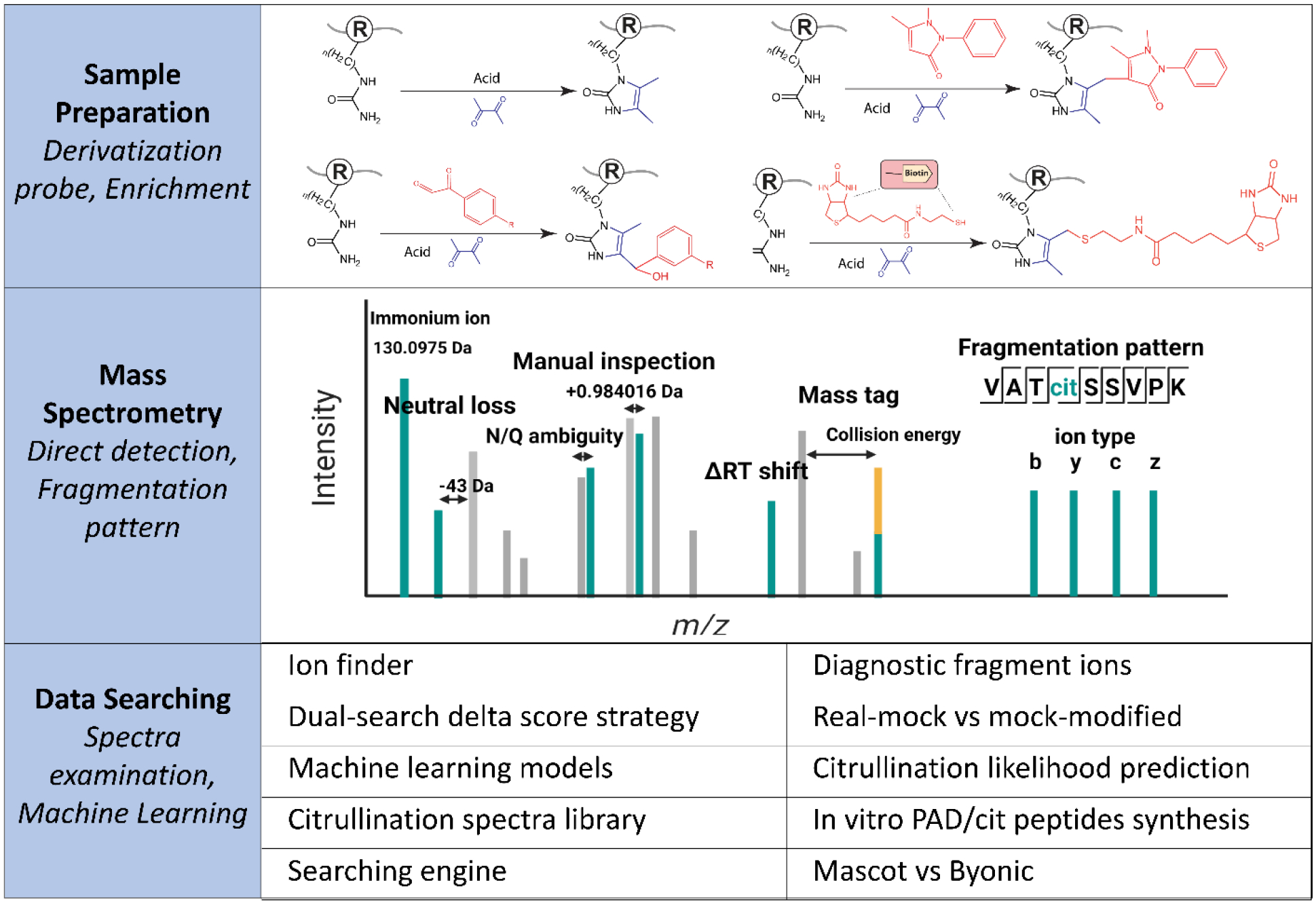
Mass spectrometry approaches to analyze citrullinated proteins.
2.3. Fragmentation characteristics of citrullinated peptides
To combat these issues, significant effort has been devoted to leverage the characteristics of fragmentation to aid in the efficiency and accuracy of citrullination site detection and localization. An abundant neutral loss of 43 Da from citrullinated peptide precursor ions was detected by collision-induced dissociation (CID), as the HNCO moiety, isocyanic acid, is eliminated from the citrulline ureido group. This loss was reportedly observed not only in multiple charge states of precursor ions, but also in b- and y-fragmented ions, providing a diagnostic marker for citrullination identification [71]. This observation became the foundation for confident citrullination discovery and profiling, and has been successfully adopted by numerous citrullination identification workflows and applications [51,52,72,73]. For example, Lee and colleagues made tremendous headway by mining the human tissue proteome for protein citrullination evidence from 30 human tissues by MS, using this foundational knowledge as a starting point. By combining fragmentation characteristics associated with the neutral loss of isocyanic acid with manual interpretation of the spectrum, coupled with the use of reference spectra for deiminated peptides or synthetically citrullinated peptides, database searching of 70 million tandem mass spectra yielded 13,000 candidate citrullinated spectra, which were further filtered by spectra quality metrics and the detection of the diagnostic ions from citrullinated peptides to reduce false positives. The team also synthesized about 2,200 citrullinated and 1,300 deamidated peptides to build a library of reference spectra, enabling the validation of 375 citrullination sites from 209 human proteins [59]. While this study expanded the human protein citrullination database, it also demonstrated a possible solution to increase the confidence of citrullination identification, providing a robust method that can be used to explore the biological roles of these modifications in the future. While these findings and the implementation of quality assurance methods are invaluable, it must be noted that the time devoted to the manual interpretation of the spectrum and the synthesis of thousands of peptides is simply unfeasible for global investigations where high-throughput procedures are a necessity. With this, Maurais and colleagues developed a streamlined data analysis pipeline for citrullination identification using an automated workflow to rigorously and rapidly mine proteomic datasets to identify the sites of citrullination from complex peptide mixtures via ionFinder and envoMatch software programs. Citrullination sites were classified with high confidence based on the presence of diagnostic fragment ions, specifically the neutral loss of isocyanic acid, as a marker for the diagnosis of protein citrullination. They used this method to map the sites of autocitrullination on purified PADs 1−4, as well as global citrullinome in a PAD2-overexpressing cell line, and finally used this framework to identify more than 350 unique citrullinated peptides from 220 proteins. Many of the proteins identified to be citrullinated were nuclear proteins, further supporting the nuclear localization of PAD2 [56]. With the fast search speed of automated software compared to manual inspection, this method significantly improved the efficiency and throughput of identification of citrullinated proteins. EnvoMatch and ionFinder are good complements to commercially available software for database searching, with the ability to increase the confidence of peptide-citrullination assignments. Automated annotation of diagnostic neutral loss species by standard database search algorithms is still a core process to verify the assignment of citrullination. In addition, the customizable nature of these post-processing tools has potential to benefit other proteomic analyses, by defining stringent criteria for modified peptide assignment, though it should be noted that the described automation program is not publicly available for others to consult. Despite these advancements, low-abundance proteins still pose a major challenge, as coverage of citrullination events in these scenarios is limited, generating a bias in terms of identification of citrullination sites from abundant proteins within the proteome. Selective enrichment of citrullinated peptides is thus a necessity for comprehensive citrullination site identifications.
2.4. Citrullination data integration with database analysis and machine learning
When abundance becomes a limiting factor in the identification of PTMs, many have turned from traditional data-dependent acquisition (DDA) methods to data-independent acquisition (DIA) methods [74–76]. While DDA methods operate so that the top n most abundant precursor ions are selected for fragmentation, biasing against ions of low abundance, DIA methods ensure the fragmentation of all precursors, in essence minimizing this biasing issue [77,78]. To accomplish this, the precursor m/z range is essentially divided into windows, of a selected m/z width, and all precursors within this window are subjected to fragmentation [78,79]. Though attractive for these applications, DIA methods can present tremendous challenges in data analysis, specifically with deconvolution [80].
Fert-Bober and colleagues developed a citrullination-targeted proteomic strategy using a DIA method to improve quantitative consistency and accuracy of identifications [28]. This method involved the development of a citrullinated peptide spectral library, used to parse identifications from complicated DIA output spectra, followed by downstream quantification. After validation using two-dimensional gel electrophoresis, 304 citrullinated sites were identified belonging to 145 proteins in human myocardium samples [28]. While these initial results were promising, identifications were limited to peptides already present within their spectral library. Aiming to adapt their method for larger-scale investigation, the same team generated a robust mouse hyper-citrullinated spectral library. In order to draw comparisons between modified and unmodified peptides, spectral matching of both forms was accomplished through the use of delta retention time shift (ΔRT) as a signature for citrullination [53]. Subsequent validation of findings was achieved through the detection of the neutral loss of isocyanic acid in peptides within the CID spectra using Skyline software. In total, they identified 3026 citrullinated peptides in 1037 citrullinated proteins, 90% of which were novel, from several mouse organs, probing their involvement in a variety of biological processes [53]. This work provides a rich resource of candidate citrullinated proteins and potential citrullinated biomarkers in clinical research. With several cases of citrullination library utilization [56,59], machine learning (ML) emerged as an attractive route to generate models capable of predicting citrullination position. Chaerkady and team generated a random forest ML prediction model to characterize citrullination sites in neutrophils and mast cells via integration of MS and ML [81]. Prior to ML, they performed preliminary data analysis using MaxQuant software with stringent, automatic filtering of peptides, obtained from neutrophil and mast cells treated with and without ionomycin, with the expected neutral loss signature from the MS/MS output files used to ensure the presence of at least one citrullination site in candidate peptides. Preliminary data analysis yielded a total of 833 validated citrullination sites on 395 proteins. Using this benchmark dataset, they developed random forest ML-based prediction models, one for each neutrophil and mast cell analyses, for identification of citrullinated protein sites according to motif presence [81]. Of note, one-fifth of PAD4 substrates contained an RG/RGG motif, which has been reported in previous studies [82], and Gly and Asp residues were largely found surrounding citrullinated sites within the ML work [81]. This study further reveals substrate motif preferences for PAD enzyme−substrate interactions and provides insight into citrullination of proteins in important biological processes. To date, this was the first report that integrated the use of ML in MS-based citrullination analyses and enabled the prediction of a catalytic motif for PAD2 and PAD4. These catalytic motifs need further experimental validation, which will improve the understanding of PAD catalysis as well as aid in future drug or inhibitor design.
2.5. Probe design and development for citrullination identification
As the small mass shift induced from citrullination can be easily misinterpreted as deamidation or another isotope with a similar mass, a chemical probe specifically designed to target citrullination sites would be advantageous, enabling an increase of the observed mass shift. De Ceuleneer and team developed a chemical tagging strategy, which allowed them to selectively pinpoint citrullinated peptides in a complex mixture with liquid chromatography-mass spectrometry (LC-MS) analysis [48]. Citrulline residues in the peptide mixture were able to covalently react with 2, 3-butanedione, which resulted in a 50 Da mass shift from singly charged ions. With this, citrullinated peptides in the mixture could be identified by comparison of the peptide mass fingerprint of a modified and an unmodified version of the same sample. The reaction conditions, including pH and incubation time, were then optimized, with synthetic peptides, and validated using a digest of citrullinated fibrinogen, a protein highly implicated in citrullination-mediated disease pathogenesis [83]. It should be emphasized that the use of an acidic reaction environment, specifically necessary for citrulline-containing residues, should be strictly controlled, as carrying out the reaction under neutral or basic conditions can cause arginine to adversely react. This was the first study to introduce a mass probe for easier detection of citrullinated residues and was also a milestone study, as it inspired further citrullination derivatization design [51,52,72,73].
As the abundance of citrullinated proteins is typically low in biological systems, an effective enrichment strategy is needed for MS analysis. Tutturen and team presented a technique for both specific modification and selective enrichment of citrullinated peptides from biological samples [51]. The technique relied on an induced reaction between a glyoxal derivative, 4-glyoxalbenzoic acid (GBA), and the ureido group of the citrulline residue under strong acidic conditions, with an additional biotin moiety attached to the GBA molecule, enabling further enrichment by streptavidin beads, producing biotinylated, citrullinated peptides upon biotin-PEG-GBA (BPG) modification [51]. Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) was conducted to analyze the impact of the enrichment strategy, revealing that the sensitivity of citrullination detection was greatly improved, though performance of the BPG modification in LC-MS for high-throughput citrullination profiling remains unknown. Lewallen and team developed a biotin-conjugated phenylglyoxal (biotin-PG) strategy to enrich, detect, and quantify protein citrullination [52]. This probe was shown to serve as an antibody surrogate, compatible with Western blotting, for the enrichment of citrullinated proteins from complex mixtures. Moreover, the probe was implemented for MS identification, serving as an enrichment tag to identify and quantify abundance of citrullinated proteins among different biological samples. More than 50 citrullinated proteins were identified and significantly enriched by at least 2-fold in a PAD2-overexpressing cell line compared to the controls using this probe and platform technology. More recently, Shi et al. reported the design and development of a biotin-thiol tag that enabled derivatization, enrichment, and confident identification of citrullination via MS with high specificity and efficiency [73]. They performed proof of principle experiments using recombinant human histone H3 protein with or without in vitro PAD. The team also explored different fragmentation techniques and enzymatic digestion methods for optimized citrullination analysis, with LysC/trypsin digestion in conjunction with stepped higher energy collision dissociation (HCD) fragmentation providing the best coverage of the citrullinome. In total, 691 citrullination sites from 432 proteins were globally mapped through large-scale citrullinome profiling of different mouse tissues [73]. This work exemplified an effective and greatly improved MS-based approach, with higher reproducibility and accuracy than previous methods. It should be mentioned that this work presented a new area in need of investigation, as fundamentally, citrulline is resistant to trypsin digestion, though their work identified peptides with C-terminal citrullination sites, which were treated as potential artificial identifications being removed from the citrullination identification list. However, these results and complementary experiments suggest that the cleavage might be protein-specific, or occur at a lower rate, which has been discussed in other studies [60,84]. More in-depth characterization might be needed to fully elucidate the underlying mechanism of the observation of C-terminal citrullination products.
Isobaric mass tagging strategies provide a unique avenue to achieve relative quantification, by enabling higher sample throughput and improved accuracy. Custom-developed N, N-dimethyl leucine (DiLeu) isobaric tags serve as a cost-effective alternative to expensive commercial isobaric tagging reagents, such as TMT, presenting a valuable tool for high-throughput MS-based quantitative proteomics [85,86]. Recent work has combined the aforementioned novel derivatization methods with DiLeu isobaric tags to achieve multiplexed quantitative analysis of citrullination from up to 12 samples [72]. This approach was applied to investigate citrullination alterations in response to DNA damage-induced stress using human cell lines. In total, they identified and quantified 78 citrullination sites from 63 citrullinated proteins in three DNA damage treatment groups and one control group [72]. This strategy combined the pipeline for isobaric labeling with previous methods for derivatization and enrichment of citrullinated peptides, enabling high-throughput quantitation of citrullinated proteins from complex biological samples with greatly improved accuracy. This approach provides a simple yet powerful tool for unambiguous, high-throughput identification and quantification of citrullinated peptides, which could be easily adopted by other researchers in the field. Clearly, the established target MS methodology coupled with a top-down proteomics approach is still needed to capture specific citrullinated proteins with unique properties, such as highly basic histone proteins [72].
Determination of sites citrullinated by PAD using 18O stable isotope labeling was previously conducted via MS [87]. In this method, an oxygen atom was incorporated into the citrulline residue from H2O during citrullination by PAD, peptides citrullinated in 50% H218O could be recognized through a characteristic isotope distribution which could be distinguished from its natural abundance, making the citrullinated sites easier to be detected. To demonstrate the proof of concept with the peptides citrullinated in 50% H218O, the authors conducted in vivo experiments of human fibrinogen citrullination identification by PAD4 treatment. The results showed complete agreement of identified citrullinated sites and equivalent sequence coverage with MS/MS spectra. We believe this methodology can be more readily adapted to in vitro protein citrullination identification and validation, though in analyses of a complex protein mixture in vivo, this method will still require time-consuming manual examination of the MS spectra given the small mass shift.
2.6. Top-down and bottom-up proteomics integration
With the development of advanced MS instrumentation and improved enrichment strategies, the integration of different proteomics methodologies can facilitate more comprehensive investigation into citrullination to answer specific questions. Top-down proteomics (TDP) employs MS to analyze intact proteins for effective characterization of PTMs, while bottom-up proteomics (BUP) examines digested protein-derived peptide sequences, localizing PTMs using tandem MS analyses. TDP enables the identification of novel protein isoforms and PTMs, characterization of sequence variations, and quantification of pathological alterations in a form highly similar to the protein’s native form [88]. However, few studies have explored citrullination profiles achieved with TDP, as these methods are considerably more challenging to analyze compared to BUP approaches, particularly due to the complexity of the generated data and instrumental limitations [89]. One TDP study presented an electrospray ionization (ESI) MS method to observe the structural alteration of recombinant human histone H2A/H2B dimers induced by PAD4 citrullination, finding that citrullination stabilizes the histone dimer association [90]. Citrullination-related PTM crosstalk is another important but complex biological event in need of more attention, as it influences complex phenotypic outcomes, thus inducing physiological and pathological alterations. The existence of crosstalk between citrullination of H3R26 and methylation of H3K27 has been reported [91]. We believe the combination of complementary information from BUP and TDP analyses will be beneficial to capture more crosstalk information.
2.7. Site-specific incorporation of citrulline into proteins
Despite recent advances for identifying citrullination sites, it still remains challenging to investigate the functional impact of citrullination with respect to a single residue. Mondal et al. recently reported a technology that enables the site-specific incorporation of citrulline into proteins in mammalian cells using an Escherichia coli-derived engineered leucyl tRNA synthetase-tRNA pair[92]. This approach incorporated a photocaged-citrulline (SM60) into proteins in response to a nonsense codon. SM60 can be further converted to citrulline with light in vitro and in living cells. To demonstrate proof of concept, they further characterized the effect of incorporating citrullination at two known auto-citrullination sites in PAD4 (R372 and R374) and showed that these two mutants were 181- and 9-fold less active than the wild-type enzyme, respectively. This work elucidated how these modifications could impact enzyme activity and indicated potential to decipher the biology of citrullination with this technology. This strategy enables the incorporation of citrullination on demand and mechanistically addresses how this PTM impacts fundamental biological processes and pathways, which will ultimately pave way for further functional validation of citrullination coupled with traditional knockout or knockdown experiments.
3. Networks of citrullinated proteins implicated in various disease phenotypes
With a growing number of studies implicating citrullination in disease pathology, this modification is now considered to be a hallmark of disease progression in many scenarios. In this section, we summarize the reported citrullinated proteins and corresponding autoantibodies that are responsible for disease pathogenesis and development including rheumatoid arthritis, neurodegenerative diseases, cancer, and diabetes (Figure 4). These relationships highlight the citrullinated hallmarks that may serve as probable targets for diagnosis and treatment solutions.
Figure 4.
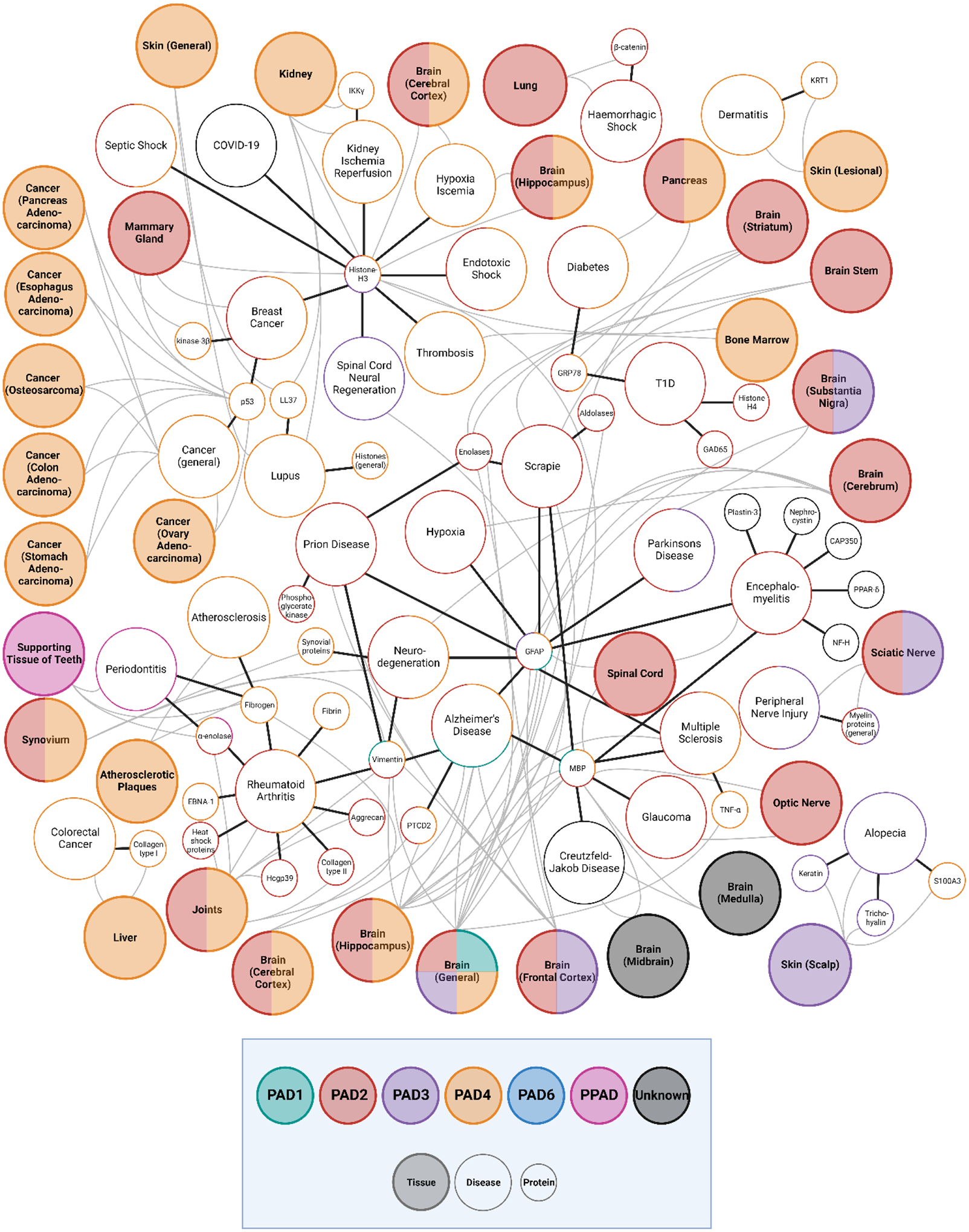
Network of protein, tissues, and diseases implicated with citrullination, with citrullinated proteins reported in small circles, diseases reported in larger circles, and tissues reported in the large shaded circles. The PAD associated with each protein, tissue, and disease are indicated by the outline of the corresponding circle.
3.1. Rheumatoid Arthritis (RA)
RA is a chronic, progressive autoimmune disease that primarily affects the lining of synovial joints and can lead to pain, functional disability, and premature death [93]. Citrullination is considered to be a major contributor to RA disease pathogenesis, with citrullinated proteins serving as major targets of antibodies in patients with RA. The participation of citrullination in RA pathogenesis has been observed in several contexts. Dysregulated PAD activity and aberrant expression of citrullinated proteins were found in the synovial fluid of RA patients, including citrullinated trichohyalin, filaggrin, histones, and transcription factors [29,30]. Furthermore, anti-citrullinated protein antibodies (ACPAs) have been found to be produced against various citrullinated protein antigens and have frequently been found in many affected individuals, with the presence of ACPAs in RA linked closely with disease severity [29,52,72,81,94]. With this, generation of ACPAs at early developmental stages can have a strong predictive value and be used for early RA diagnosis. The development of ACPAs is also considered critical for RA pathogenesis, with observation of autoantigens for ACPAs in the synovial fluid, including filaggrin, fibronectin, fibrinogen α- and β-chains, α-enolase, vimentin, type II collagen, immunoglobulin binding protein-BIP, tenascin-C, as well as microbial components VCP1 and VCP2 [95–99]. Another autoantibody discovered in RA patients is the rheumatoid factor (RF), which was determined to be directed against serum γ-globulins and have a strong association with RA [100]. RF has been found in multiple immunoglobulin isotypes, including IgM, IgG, and IgA; RF is capable of directly binding to the Fc portion of aggregated globulins. RF has been used as a biomarker for RA testing, with reported 60% to 90% sensitivity in patient blood samples [101]. PAD2 and PAD4 are the two main isozymes determined to mediate citrullination in RA, both of which have been localized in neutrophils [102]. Inflammation is one of the trademark characteristics of RA and results from atypical citrullination, which promotes the formation of NETs, a network of extracellular strings of DNA that bind pathogenic microbes [103,104]. It has also been found that citrullinated histones from neutrophils were routinely targeted by ACPAs, suggesting that increased histone citrullination could be linked to increased autoimmune response [105]. Several other citrullinated proteins reported to be in NETs include actin, actin related protein 2/3 complex subunit 1B, coronin, and leukocyte elastase inhibitor [106,107].
3.2. Neurodegenerative diseases
Alzheimer’s disease (AD) is the most common neurodegenerative disease (ND), characterized by the abnormal aggregation of misfolded proteins, such as amyloid beta (Aβ) and phosphorylated tau protein within the brain [108,109]. To date, the development of therapeutic approaches targeting these two biomarkers has been lacking. It has been suggested that excessive expression of PAD isozymes, specifically PAD2 and PAD4, with the PAD2 isoform considered to be predominantly expressed in the CNS while PAD4 has been observed in the hippocampus and cerebral cortex, can contribute to ND changes implicated in AD pathology [33,110,111]. Another study found an abnormal accumulation of citrullinated proteins and an increase of the PAD2 content in the hippocampus of AD patients [33]. Most recently, citrullination of Aβ in AD brains was detected by MS-based methods, concluding that a common mechanism for citrullination of Aβ exists in both the sporadic and familial representation of AD, and that citrullinated Aβ peptides could possibly trigger adverse immune response [112]. The most frequently reported citrullinated proteins associated with AD are often structural proteins, such as fibrillary acidic protein (GFAP), myelin basic protein (MBP), vimentin, and neurogranin (NRGN), with 12 citrullination sites on MBP, 2 citrullinated sites on vimentin, 4 citrullinated sites on GFAP, and 1 citrullinated site on NRGN detected by either MS-based methods or monoclonal antibody strategies [60]. GFAP, a cytoskeletal protein of astrocytes in the AD brain, is a substrate compatible with PAD2, indicating that citrullinated GFAP may function in the progression of AD [113]. The relationship between ND and citrullination is not limited to AD, as elevated citrullinated MBP levels have also been reported in multiple sclerosis patients, representing an important biochemical pathway in its pathogenesis [36]. Over 27% of MBP arginine residues are citrullinated in multiple sclerosis compared to healthy individuals [38]. Hypercitrullinated MBP has been shown to result in a partial conformational change, hypothesized to suppress the transition of lipid bilayers into compact multilayers, resulting in loss of proper nerve signal transduction [114].
3.3. Cancers
Citrullination has also been reported to participate in various processes associated with the pathogenesis of cancer and tumor biology. PAD is also an active area of cancer therapeutic and biomarker development. The pathway of this relationship is quickly becoming clearer, as new evidence shows that PAD2 and PAD4 enzymes are overexpressed in many types of neutrophils and tumor cells, such as adenocarcinoma and prostate cancer cells [102,115]. PAD4 has also been documented in association with gastric cancer, liver cancer, and ovarian cancer [116–119]. PAD4 levels were also found to be elevated in blood samples of patients with lung cancer [120]. Colorectal cancer liver metastatic growth was reported to be relevant to PAD4-driven citrullination of the extracellular matrix [14,121]. These findings suggest the important role of citrullination in metastasis and disease progression. Furthermore, PAD2 appears to play a tumorigenic role in breast cancer, digestive system cancers, and skin cancers [122]. A well-known tumor biomarker, cytokeratin, was found to be citrullinated, and as a result prevent caspase digestion, suggesting its citrullinated form may serve as a novel target for tumor treatment [120]. It was found that PAD4 and histone hypercitrullination are crucial for chromatin unfolding and the formation of NETs, as histone citrullination can not only regulate various cellular processes but also alter the formation of NETs. In addition, citrullinated histones are typically associated with the promotion of NETs-mediated inflammation by regulating the localization and activation of toll-like receptor 4 (TLR4) [123,124]. PADs and citrullination also regulate gene transcription and induce response to DNA damage. For example, most of the p53 target genes are tumor suppressor genes which regulate cell cycle arrest, DNA repair, metabolism, translation control, programed cell death, and autophagy [125]. The level of citrullinated arginine 3 residues of histone H4 is negatively correlated with p53 protein expression and tumor size in non-small cell lung cancer tissues [126]. Citrullination of H4R3 and Lamin C in response to DNA damage was demonstrated to induce nuclear fragmentation through the p53-PAD4 pathway [127]. Citrullination and methylation crosstalk on histone H3 are also reported to regulate ER-target gene transcription. H3K27 demethylases were able to activate transcription after H3Cit26 formation, which supports the existence of crosstalk between citrullination and methylation in the adjacent position [91]. It can be hypothesized that citrullination crosstalk may have a crucial role in carcinogenesis through the effect of histone modifications on aberrant tumor suppressor genes.
3.4. Type 1 diabetes (T1D)
T1D is an insulin-dependent glucose metabolic disorder caused by inflammation of pancreatic islet cells, specified as an organ-specific autoimmune disease induced by autoimmune response against pancreatic β cells. It was first reported that insulin B can be citrullinated in Porphyromonas gingivalis, with subsequent publications establishing the relationship between citrullinated β-cell proteins and coordinated response in T1D [128,129]. More recently, PADs and citrullination have emerged in additional T1D-associated pathways, coupled with several key proteins responsible for blood sugar regulation, encompassing glutamic acid decarboxylase 65 (GAD65), islet amyloid polypeptide (IAPP), islet antigen-2 (IA2), glucose-regulated protein 78 (GRP78), and islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP) [41,42]. In recent studies, citrullinated GRP78 and GAD65 were discovered as novel autoantigens when human islets were exposed to inflammatory stress induced by interleukin-1β, tumor necrosis factor-α, and interferon-γ, thus contributing to loss of immune self-tolerance toward β-cells in T1D [42]. The citrullination of glucokinase (GK) was also identified as a result of inflammation, triggering an autoimmune response. Citrullination of GK is also known to alter its biological activity and suppress glucose-stimulated insulin secretion [130]. Surmounting evidence has shown the citrullination participation of neutrophils and NETosis in the pathogenesis of T1D. Proteomic profiling has indicated that the level of PAD2 is higher in prediabetic nonobese diabetes (NOD) islets than the control group [131]. Inflammatory cytokines, which activate endoplasmic reticulum (ER) stress pathways, were shown to induce citrullination of β-cell proteins. These findings support a potential therapeutic strategy of inhibiting PAD enzymes and utilizing specific citrullinated autoantigens as drug targets for T1D.
Conclusions and future directions
This review presents a frame of reference for the role of citrullination concerning the pathogenesis of several diseases including RA, cancer, ND, and T1D. It also highlights novel MS techniques and their applications that facilitate profiling of citrullination sites with diagnostic and therapeutic significance. The precise identification and characterization of protein citrullination remains challenging and quantification is even more convoluted due to the limited approaches available. There are many technological gaps in this area, as well as many plausible hypotheses to investigate. Future integration of these methodologies with biological networks will promise to provide a foundation for the elucidation of these underlying mechanisms and disease pathologies. Some immediate areas of examination are necessary for the field to progress, namely, there are discrepancies in the relationship between trypsin cleavage and citrullination sites, as these sites should not be cleaved, though experimental evidence has reported C-terminal citrulline residues on a number of occasions [43,54,60,73]. Most importantly, future research efforts should not only develop robust and efficient methodologies to expand the “citrullinome” database, but also explain corresponding pathways to establish viable therapeutic targets and produce novel drugs for the treatment of citrullination-associated diseases.
Supplementary Material
Significance statement.
Citrullination is a key PTM that affects protein structure and functionality. It has been associated with the development of diverse pathological states and has raised much interest in recent decades, though advancement in this area has been hindered due to challenges associated with enrichment, detection, and localization of protein citrullination. This review summarizes recent advances in MS-based citrullination characterization approaches and methodology integration. The impact and association of protein citrullination on signaling networks in disease pathogenesis and progression is also highlighted in this review.
Acknowledgements
Preparation of this manuscript and some of the research cited was supported in part by grant funding from the NIH (R21AG060242, RF1AG052324, R21AG065728, and R01DK071801). L.L. acknowledges funding support of NIH shared instrument grants (NIH-NCRR S10RR029531, S10OD025084, and S10OD028473), a Vilas Distinguished Achievement Professorship and the Charles Melbourne Johnson Distinguished Chair Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy. L.F. was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number T32GM008505 (Chemistry–Biology Interface Training Program). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All figures were created with BioRender.com.
Abbreviations
- PTM
Post-translational modifications
- LC-MS
liquid chromatography-mass spectrometry
- PAD
peptidylarginine deiminase
- CID
collision-induced dissociation
- DDA
data-dependent acquisition
- DIA
data-independent acquisition
- ΔRT
delta retention time shift
- ML
machine learning
- MALDI-TOF
matrix-assisted laser desorption/ionization-time of flight
- HCD
higher energy collision dissociation
- RA
Rheumatoid Arthritis
- ACPA
Anti-citrullinated protein antibodies
- AD
Alzheimer’s disease
- RF
rheumatoid factor
- CNS
central nervous system
- T1D
Type 1 diabetes
- NETs
Neutrophil extracellular traps
- NOD
prediabetic nonobese diabetes
- BUP
bottom-up proteomics
- TDP
top-down proteomics
- ESI
electrospray ionization
- ER
endoplasmic reticulum
- GFAP
glial fibrillary acidic protein
- MBP
myelin basic protein
- NRGN
neurogranin
- ND
neurodegenerative disease
- TLR4
toll-like receptor 4
- GAD65
glutamic acid decarboxylase 65
- IGRP
islet-specific glucose-6-phosphate catalytic subunit-related protein
- GK
glucokinase
- DiLeu
N, N-dimethyl leucine
Biographies

Bin Wang received his bachelor’s degree in chemical engineering and master’s degree in biology from Tianjin University where he focused on the structural biology and protein engineering. He is currently pursuing a Ph.D. in Pharmaceutical Sciences under the supervision of Professor Lingjun Li at the University of Wisconsin-Madison. His Ph.D. dissertation work centers on developing novel mass spectrometry–based methods to characterize protein posttranslational modifications and exploring protein-protein interactions and structural alterations in neurodegenerative diseases. He is currently developing new methods to explore the role of protein citrullination in several disease models.

Lauren Fields received a B.S. in chemistry from the University of North Carolina Asheville in 2020 where she developed analytical methods to assess the mechanism of action of antibiotics. She is pursuing a Ph.D. in Analytical Chemistry under the supervision of Professor Lingjun Li at the University of Wisconsin-Madison and her present work involves improving the identification of neuropeptides. She is currently developing bioinformatics strategies to enable high-throughput, optimized identification and quantification of neuropeptides.

Dr. Lingjun Li is a Vilas Distinguished Achievement Professor and the Charles Melbourne Johnson Distinguished Chair Professor of Pharmaceutical Sciences and Chemistry at UW-Madison. Dr. Li received her Ph.D. degree in Chemistry from UIUC and did joint postdoctoral research at PNNL and Brandeis University prior to her faculty appointment. Dr. Li’s research interests include the development of novel MS-based tools for proteomics, peptidomics, and glycomics, and their applications in neuroscience and cancer research. She was recognized with ASMS Research Award, NSF CAREER Award, Sloan Fellowship, PittCon Achievement Award, and ASMS Biemann Medal. Dr. Li is an Associate Editor for JASMS.
Footnotes
Supporting Information can be found at journal website.
Supplemental Tables S1 and S2.
Declaration of interests
The authors have declared no conflict of interests.
References
- [1].Fuhrmann J, Clancy KW, & Thompson PR (2015). Chemical biology of protein arginine modifications in epigenetic regulation. Chem Rev, 115(11), 5413–5461. doi: 10.1021/acs.chemrev.5b00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fuhrmann J, & Thompson PR (2016). Protein Arginine Methylation and Citrullination in Epigenetic Regulation. ACS Chem Biol, 11(3), 654–668. doi: 10.1021/acschembio.5b00942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Witalison E, Thompson P, & Hofseth L (2015). Protein Arginine Deiminases and Associated Citrullination: Physiological Functions and Diseases Associated with Dysregulation. Current Drug Targets, 16(7), 700–710. doi: 10.2174/1389450116666150202160954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mondal S, & Thompson PR (2019). Protein Arginine Deiminases (PADs): Biochemistry and Chemical Biology of Protein Citrullination. Acc Chem Res, 52(3), 818–832. doi: 10.1021/acs.accounts.9b00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Abu Rus’d A, Ikejiri Y, Ono H, Yonekawa T, Shiraiwa M, Kawada A, & Takahara H (1999). Molecular cloning of cDNAs of mouse peptidylarginine deiminase type I, type III and type IV, and the expression pattern of type I in mouse. European Journal of Biochemistry, 259(3), 660–669. [DOI] [PubMed] [Google Scholar]
- [6].Guerrin M, Ishigami A, Mechin MC, Nachat R, Valmary S, Sebbag M, … Serre G (2003). cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochemical Journal, 370, 167–174. doi: Doi 10.1042/Bj20020870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Darrah E, Rosen A, Giles JT, & Andrade F (2012). Peptidylarginine deiminase 2, 3 and 4 have distinct specificities against cellular substrates: novel insights into autoantigen selection in rheumatoid arthritis. Annals of the Rheumatic Diseases, 71(1), 92–98. doi: 10.1136/ard.2011.151712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Watanabe K, & Senshu T (1989). Isolation and Characterization of Cdna Clones Encoding Rat Skeletal-Muscle Peptidylarginine Deiminase. Journal of Biological Chemistry, 264(26), 15255–15260. [PubMed] [Google Scholar]
- [9].Watanabe K, Akiyama K, Hikichi K, Ohtsuka R, Okuyama A, & Senshu T (1988). Combined Biochemical and Immunochemical Comparison of Peptidylarginine Deiminases Present in Various Tissues. Biochimica Et Biophysica Acta, 966(3), 375–383. doi: Doi 10.1016/0304-4165(88)90088-8 [DOI] [PubMed] [Google Scholar]
- [10].Lamensa JWE, & Moscarello MA (1993). Deimination of Human Myelin Basic-Protein by a Peptidylarginine Deiminase from Bovine Brain. Journal of Neurochemistry, 61(3), 987–996. doi: DOI 10.1111/j.1471-4159.1993.tb03612.x [DOI] [PubMed] [Google Scholar]
- [11].Senshu T, Kan SH, Ogawa H, Manabe M, & Asaga H (1996). Preferential deimination of keratin K1 and filaggrin during the terminal differentiation of human epidermis. Biochemical and Biophysical Research Communications, 225(3), 712–719. doi: DOI 10.1006/bbrc.1996.1240 [DOI] [PubMed] [Google Scholar]
- [12].Nachat R, Mechin MC, Charveron M, Serre G, Constans J, & Simon M (2005). Peptidylarginine deiminase isoforms are differentially expressed in the anagen hair follicles and other human skin appendages. Journal of Investigative Dermatology, 125(6), A15–A15. [DOI] [PubMed] [Google Scholar]
- [13].Wang S, & Wang YM (2013). Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms, 1829(10), 1126–1135. doi: 10.1016/j.bbagrm.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chang XT, Han JX, Pang L, Zhao Y, Yang Y, & Shen ZL (2009). Increased PADI4 expression in blood and tissues of patients with malignant tumors. Bmc Cancer, 9. doi: Artn 40 10.1186/1471-2407-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vossenaar ER, Radstake TRD, van der Heijden A, van Mansum MAM, Dieteren C, de Rooij DJ, … van Venrooij WJ (2004). Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Annals of the Rheumatic Diseases, 63(4), 373–381. doi: 10.1136/ard.2003.012211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chavanas S, Mechin C, Takahara H, Kawada A, Nachat R, Serre G, & Simon M (2004). Comparative analysis of the mouse and human peptidylarginine deiminase gene (PADI) clusters reveals highly conserved non-coding segments and a new human gene, PAD16. Journal of Investigative Dermatology, 123(6), A105–A105. [DOI] [PubMed] [Google Scholar]
- [17].Kan R, Yurttas P, Kim B, Jin M, Wo L, Lee B, … Coonrod SA (2011). Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Developmental Biology, 350(2), 311–322. doi: 10.1016/j.ydbio.2010.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Esposito G, Vitale AM, Leijten FPJ, Strik AM, Koonen-Reemst AMCB, Yurttas P, … Gossen JA (2007). Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Molecular and Cellular Endocrinology, 273(1–2), 25–31. doi: 10.1016/j.mce.2007.05.005 [DOI] [PubMed] [Google Scholar]
- [19].Liu YL, Tsai IC, Chang CW, Liao YF, Liu GY, & Hung HC (2013). Functional Roles of the Non-Catalytic Calcium-Binding Sites in the N-Terminal Domain of Human Peptidylarginine Deiminase 4. PLoS One, 8(1). doi: ARTN e51660 10.1371/journal.pone.0051660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhou J, Biesterveld BE, Li YQ, Wu ZY, Tian YZ, Williams AM, … Alam HB (2020). Peptidylarginine Deiminase 2 Knockout Improves Survival in Hemorrhagic Shock. Shock, 54(4), 458–463. doi: 10.1097/Shk.0000000000001489 [DOI] [PubMed] [Google Scholar]
- [21].Raijmakers R, Vogelzangs J, Raats J, Panzenbeck M, Corby M, Jiang HP, … Werneburg B (2006). Experimental autoimmune encephalomyelitis induction in peptidylarginine deiminase 2 knockout mice. Journal of Comparative Neurology, 498(2), 217–226. doi: 10.1002/cne.21055 [DOI] [PubMed] [Google Scholar]
- [22].Li PX, Li M, Lindberg MR, Kennett MJ, Xiong N, & Wang YM (2010). PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. Journal of Experimental Medicine, 207(9), 1853–1862. doi: 10.1084/jem.20100239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Suzuki A, Kochi Y, Shoda H, Seri Y, Fujio K, Sawada T, … Yamamoto K (2016). Decreased severity of experimental autoimmune arthritis in peptidylarginine deiminase type 4 knockout mice. Bmc Musculoskeletal Disorders, 17. doi: ARTN 205 10.1186/s12891-016-1055-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gyorgy B, Toth E, Tarcsa E, Falus A, & Buzas EI (2006). Citrullination: A posttranslational modification in health and disease. International Journal of Biochemistry & Cell Biology, 38(10), 1662–1677. doi: 10.1016/j.biocel.2006.03.008 [DOI] [PubMed] [Google Scholar]
- [25].Witalison EE, Thompson PR, & Hofseth LJ (2015). Protein Arginine Deiminases and Associated Citrullination: Physiological Functions and Diseases Associated with Dysregulation. Current Drug Targets, 16(7), 700–710. doi: Doi 10.2174/1389450116666150202160954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Orgovan G, & Noszal B (2011). The complete microspeciation of arginine and citrulline. Journal of Pharmaceutical and Biomedical Analysis, 54(5), 965–971. doi: 10.1016/j.jpba.2010.11.023 [DOI] [PubMed] [Google Scholar]
- [27].Ordonez A, Martinez-Martinez I, Corrales FJ, Miqueo C, Minano A, Vicente V, & Corral J (2009). Effect of citrullination on the function and conformation of antithrombin. Febs Journal, 276(22), 6763–6772. doi: 10.1111/j.1742-4658.2009.07391.x [DOI] [PubMed] [Google Scholar]
- [28].Ciesielski O, Biesiekierska M, Panthu B, Soszynski M, Pirola L, & Balcerczyk A (2022). Citrullination in the pathology of inflammatory and autoimmune disorders: recent advances and future perspectives. Cell Mol Life Sci, 79(2), 94. doi: 10.1007/s00018-022-04126-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Maloley P, Ryan E, Aripova N, Duryee M, England B, Mikuls T, & Thiele G (2021). Citrullination Drives the Expression of Pro-fibrotic Genes in Rheumatoid Arthritis Fibroblast-like Cells. Arthritis & Rheumatology, 73, 27–30. [Google Scholar]
- [30].Darrah E, & Andrade F (2018). Rheumatoid arthritis and citrullination. Current Opinion in Rheumatology, 30(1), 72–78. doi: 10.1097/Bor.0000000000000452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jang B, Kim E, Choi JK, Jin JK, Kim JI, Ishigami A, … Choi EK (2008). Accumulation of citrullinated proteins by up-regulated peptidylarginine deiminase 2 in brains of scrapie-infected mice: a possible role in pathogenesis. Am J Pathol, 173(4), 1129–1142. doi: 10.2353/ajpath.2008.080388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ishida-Yamamoto A, Senshu T, Takahashi H, Akiyama K, Nomura K, & Iizuka H (2000). Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. J Invest Dermatol, 114(4), 701–705. doi: 10.1046/j.1523-1747.2000.00936.x [DOI] [PubMed] [Google Scholar]
- [33].Ishigami A, Ohsawa T, Hiratsuka M, Taguchi H, Kobayashi S, Saito Y, … Maruyama N (2005). Abnormal accumulation of citrullinated proteins catalyzed by peptidylarginine deiminase in hippocampal extracts from patients with Alzheimer’s disease. Journal of Neuroscience Research, 80(1), 120–128. doi: 10.1002/jnr.20431 [DOI] [PubMed] [Google Scholar]
- [34].Acharya NK, Nagele EP, Han M, Coretti NJ, DeMarshall C, Kosciuk MC, … Nagele RG (2012). Neuronal PAD4 expression and protein citrullination: Possible role in production of autoantibodies associated with neurodegenerative disease. Journal of Autoimmunity, 38(4), 369–380. doi: 10.1016/j.jaut.2012.03.004 [DOI] [PubMed] [Google Scholar]
- [35].Ishigami A, Masutomi H, Handa S, Nakamura M, Nakaya S, Uchida Y, … Toda T (2015). Mass Spectrometric Identification of Citrullination Sites and Immunohistochemical Detection of Citrullinated Glial Fibrillary Acidic Protein in Alzheimer’s Disease Brains. Journal of Neuroscience Research, 93(11), 1664–1674. doi: 10.1002/jnr.23620 [DOI] [PubMed] [Google Scholar]
- [36].Moscarello MA, Mastronardi FG, & Wood DD (2007). The role of citrullinated proteins suggests a novel mechanism in the pathogenesis of multiple sclerosis. Neurochemical Research, 32(2), 251–256. doi: 10.1007/s11064-006-9144-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yang L, Tan DW, & Piao H (2016). Myelin Basic Protein Citrullination in Multiple Sclerosis: A Potential Therapeutic Target for the Pathology. Neurochemical Research, 41(8), 1845–1856. doi: 10.1007/s11064-016-1920-2 [DOI] [PubMed] [Google Scholar]
- [38].Gs Chirivi R (2013). Citrullination: A Target for Disease Intervention in Multiple Sclerosis and other Inflammatory Diseases? Journal of Clinical & Cellular Immunology, 04(03). doi: 10.4172/2155-9899.1000146 [DOI] [Google Scholar]
- [39].Bradford CM, Ramos I, Cross AK, Haddock G, McQuaid S, Nicholas AP, & Woodroofe MN (2014). Localisation of citrullinated proteins in normal appearing white matter and lesions in the central nervous system in multiple sclerosis. J Neuroimmunol, 273(1–2), 85–95. doi: 10.1016/j.jneuroim.2014.05.007 [DOI] [PubMed] [Google Scholar]
- [40].Yuzhalin AE (2019). Citrullination in Cancer. Cancer Research, 79(7), 1274–1284. doi: 10.1158/0008-5472.Can-18-2797 [DOI] [PubMed] [Google Scholar]
- [41].Yang ML, Sodre FMC, Mamula MJ, & Overbergh L (2021). Citrullination and PAD Enzyme Biology in Type 1 Diabetes - Regulators of Inflammation, Autoimmunity, and Pathology. Frontiers in Immunology, 12. doi: ARTN 678953 10.3389/fimmu.2021.678953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nguyen H, & James EA (2016). Immune recognition of citrullinated epitopes. Immunology, 149(2), 131–138. doi: 10.1111/imm.12640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yu K, & Proost P (2022). Insights into peptidylarginine deiminase expression and citrullination pathways. Trends Cell Biol, 32(9), 746–761. doi: 10.1016/j.tcb.2022.01.014 [DOI] [PubMed] [Google Scholar]
- [44].Hensen SM, & Pruijn GJ (2014). Methods for the detection of peptidylarginine deiminase (PAD) activity and protein citrullination. Mol Cell Proteomics, 13(2), 388–396. doi: 10.1074/mcp.R113.033746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Clancy KW, Weerapana E, & Thompson PR (2016). Detection and identification of protein citrullination in complex biological systems. Curr Opin Chem Biol, 30, 1–6. doi: 10.1016/j.cbpa.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Moelants EA, Van Damme J, & Proost P (2011). Detection and quantification of citrullinated chemokines. PLoS One, 6(12), e28976. doi: 10.1371/journal.pone.0028976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Verheul MK, van Veelen PA, van Delft MAM, de Ru A, Janssen GMC, Rispens T, … Trouw LA (2018). Pitfalls in the detection of citrullination and carbamylation. Autoimmun Rev, 17(2), 136–141. doi: 10.1016/j.autrev.2017.11.017 [DOI] [PubMed] [Google Scholar]
- [48].De Ceuleneer M, De Wit V, Van Steendam K, Van Nieuwerburgh F, Tilleman K, & Deforce D (2011). Modification of citrulline residues with 2,3-butanedione facilitates their detection by liquid chromatography/mass spectrometry. Rapid Communications in Mass Spectrometry, 25(11), 1536–1542. doi: 10.1002/rcm.5015 [DOI] [PubMed] [Google Scholar]
- [49].Stensland M, Holm A, Kiehne A, & Fleckenstein B (2009). Targeted analysis of protein citrullination using chemical modification and tandem mass spectrometry. Rapid Communications in Mass Spectrometry, 23(17), 2754–2762. doi: 10.1002/rcm.4185 [DOI] [PubMed] [Google Scholar]
- [50].Tutturen AE, Fleckenstein B, & de Souza GA (2014). Assessing the citrullinome in rheumatoid arthritis synovial fluid with and without enrichment of citrullinated peptides. J Proteome Res, 13(6), 2867–2873. doi: 10.1021/pr500030x [DOI] [PubMed] [Google Scholar]
- [51].Tutturen AEV, Holm A, & Fleckenstein B (2013). Specific biotinylation and sensitive enrichment of citrullinated peptides. Analytical and Bioanalytical Chemistry, 405(29), 9321–9331. doi: 10.1007/s00216-013-7376-1 [DOI] [PubMed] [Google Scholar]
- [52].Lewallen DM, Bicker KL, Subramanian V, Clancy KW, Slade DJ, Martell J, … Thompson PR (2015). Chemical Proteomic Platform To Identify Citrullinated Proteins. Acs Chemical Biology, 10(11), 2520–2528. doi: 10.1021/acschembio.5b00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fert-Bober J, Venkatraman V, Hunter CL, Liu RN, Crowgey EL, Pandey R, … Van Eyk JE (2019). Mapping Citrullinated Sites in Multiple Organs of Mice Using Hypercitrullinated Library. Journal of Proteome Research, 18(5), 2270–2278. doi: 10.1021/acs.jproteome.9b00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang X, Swensen AC, Zhang T, Piehowski PD, Gaffrey MJ, Monroe ME, … Qian WJ (2020). Accurate Identification of Deamidation and Citrullination from Global Shotgun Proteomics Data Using a Dual-Search Delta Score Strategy. Journal of Proteome Research, 19(4), 1863–1872. doi: 10.1021/acs.jproteome.9b00766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Villacres C, Spicer V, & Krokhin OV (2021). Confident Identification of Citrullination and Carbamylation Assisted by Peptide Retention Time Prediction. Journal of Proteome Research, 20(3), 1571–1581. doi: 10.1021/acs.jproteome.0c00775 [DOI] [PubMed] [Google Scholar]
- [56].Maurais AJ, Salinger AJ, Tobin M, Shaffer SA, Weerapana E, & Thompson PR (2021). A Streamlined Data Analysis Pipeline for the Identification of Sites of Citrullination. Biochemistry. doi: 10.1021/acs.biochem.1c00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Huh S, Hwang D, & Kim MS (2020). Statistical Modeling for Enhancing the Discovery Power of Citrullination from Tandem Mass Spectrometry Data. Anal Chem, 92(19), 12975–12986. doi: 10.1021/acs.analchem.0c01687 [DOI] [PubMed] [Google Scholar]
- [58].De Ceuleneer M, Van Steendam K, Dhaenens M, Elewaut D, & Deforce D (2012). Quantification of citrullination by means of skewed isotope distribution pattern. J Proteome Res, 11(11), 5245–5251. doi: 10.1021/pr3004453 [DOI] [PubMed] [Google Scholar]
- [59].Lee CY, Wang DX, Wilhelm M, Zolg DP, Schmidt T, Schnatbaum K, … Kuster B (2018). Mining the Human Tissue Proteome for Protein Citrullination. Molecular & Cellular Proteomics, 17(7), 1378–1391. doi: 10.1074/mcp.RA118.000696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jin ZC, Fu ZM, Yang J, Troncosco J, Everett AD, & Van Eyk JE (2013). Identification and characterization of citrulline-modified brain proteins by combining HCD and CID fragmentation. Proteomics, 13(17), 2682–2691. doi: 10.1002/pmic.201300064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Senshu T, S. T, Inoue T, Akiyama K, Asaga H. (1992). Detection of citrulline residues in deiminated proteins on polyvinylidene difluoride membrane. Anal Biochem, 203(1), 94–100. [DOI] [PubMed] [Google Scholar]
- [62].Nicholas AP, King JL, Sambandam T, Echols JD, Gupta KB, McInnis C, & Whitaker JN (2003). Immunohistochemical localization of citrullinated proteins in adult rat brain. J Comp Neurol, 459(3), 251–266. doi: 10.1002/cne.10607 [DOI] [PubMed] [Google Scholar]
- [63].Holmes CL, Shim D, Kernien J, Johnson CJ, Nett JE, & Shelef MA (2019). Insight into Neutrophil Extracellular Traps through Systematic Evaluation of Citrullination and Peptidylarginine Deiminases. Journal of Immunology Research, 2019. doi: Artn 2160192 10.1155/2019/2160192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bawadekar M, Gendron-Fitzpatrick A, Rebernick R, Shim D, Warner TF, Nicholas AP, … Shelef MA (2016). Tumor necrosis factor alpha, citrullination, and peptidylarginine deiminase 4 in lung and joint inflammation. Arthritis Research & Therapy, 18. doi: ARTN 173 10.1186/s13075-016-1068-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bawadekar M, Shim D, Johnson CJ, Warner TE, Rebernick R, Damgaard D, … Shelef MA (2017). Peptidylarginine deiminase 2 is required for tumor necrosis factor alpha-induced citrullination and arthritis, but not neutrophil extracellular trap formation. Journal of Autoimmunity, 80, 39–47. doi: 10.1016/j.jaut.2017.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bicker KL, Subramanian V, Chumanevich AA, Hofseth LJ, & Thompson PR (2012). Seeing Citrulline: Development of a Phenylglyoxal-Based Probe To Visualize Protein Citrullination. Journal of the American Chemical Society, 134(41), 17015–17018. doi: 10.1021/ja308871v [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Vitorino R, Guedes S, Vitorino C, Ferreira R, Amado F, & Van Eyk JE (2020). Elucidating Citrullination by Mass Spectrometry and Its Role in Disease Pathogenesis. J Proteome Res. doi: 10.1021/acs.jproteome.0c00474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].De Ceuleneer M, Van Steendam K, Dhaenens M, & Deforce D (2012). In vivo relevance of citrullinated proteins and the challenges in their detection. Proteomics, 12(6), 752–760. doi: 10.1002/pmic.201100478 [DOI] [PubMed] [Google Scholar]
- [69].Raijmakers R, van Beers JJBC, El-Azzouny M, Visser NFC, Bozic B, Pruijn GJM, & Heck AJR (2012). Elevated levels of fibrinogen-derived endogenous citrullinated peptides in synovial fluid of rheumatoid arthritis patients. Arthritis Research & Therapy, 14(3). doi: ARTN R114 10.1186/ar3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Stobernack T, Glasner C, Junker S, Gabarrini G, de Smit M, de Jong A, … van Dijl JM (2016). Extracellular Proteome and Citrullinome of the Oral Pathogen Porphyromonas gingivalis. J Proteome Res, 15(12), 4532–4543. doi: 10.1021/acs.jproteome.6b00634 [DOI] [PubMed] [Google Scholar]
- [71].Hao G, Wang D, Gu J, Shen Q, Gross SS, & Wang Y (2009). Neutral loss of isocyanic acid in peptide CID spectra: a novel diagnostic marker for mass spectrometric identification of protein citrullination. J Am Soc Mass Spectrom, 20(4), 723–727. doi: 10.1016/j.jasms.2008.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Li ZH, Wang B, Yu QY, Shi YT, & Li LJ (2022). 12-Plex DiLeu Isobaric Labeling Enabled High-Throughput Investigation of Citrullination Alterations in the DNA Damage Response. Analytical Chemistry, 94(7), 3074–3081. doi: 10.1021/acs.analchem.1c04073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shi Y, Li Z, Wang B, Shi X, Ye H, Delafield DG, Lv L, Ye Z, Chen Z, Ma F, & Li L (2022). Enabling global analysis of protein citrullination via biotin thiol tag-assisted mass spectrometry. Anal. Chem doi: 10.1021/acs.analchem.2c03844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Demichev V, Szyrwiel L, Yu FC, Teo GC, Rosenberger G, Niewienda A, … Ralser M (2022). dia-PASEF data analysis using FragPipe and DIA-NN for deep proteomics of low sample amounts. Nature communications, 13(1). doi: ARTN 3944 10.1038/s41467-022-31492-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lou R, Liu W, Li R, Li S, He X, & Shui W (2021). DeepPhospho accelerates DIA phosphoproteome profiling through in silico library generation. Nat Commun, 12(1), 6685. doi: 10.1038/s41467-021-26979-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Dong MM, Lih TSM, Ao MH, Hu YW, Chen SY, Eguez RV, & Zhang H (2021). Data-Independent Acquisition-Based Mass Spectrometry (DIA-MS) for Quantitative Analysis of Intact N-Linked Glycopeptides. Analytical chemistry, 93(41), 13774–13782. doi: 10.1021/acs.analchem.1c01659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bruderer R, Bernhardt OM, Gandhi T, Xuan Y, Sondermann J, Schmidt M, … Reiter L (2017). Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility of Results. Molecular & Cellular Proteomics, 16(12), 2296–2309. doi: 10.1074/mcp.RA117.000314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].DeLaney K, & Li LJ (2019). Data Independent Acquisition Mass Spectrometry Method for Improved Neuropeptidomic Coverage in Crustacean Neural Tissue Extracts. Analytical chemistry, 91(8), 5150–5158. doi: 10.1021/acs.analchem.8b05734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Demichev V, Messner CB, Vernardis SI, Lilley KS, & Ralser M (2020). DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nature Methods, 17(1), 41–+. doi: 10.1038/s41592-019-0638-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hu A, Noble WS, & Wolf-Yadlin A (2016). Technical advances in proteomics: new developments in data-independent acquisition. F1000Res, 5. doi: 10.12688/f1000research.7042.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Chaerkady R, Zhou YB, Delmar JA, Weng SHS, Wang JM, Awasthi S, … Hess S (2021). Characterization of Citrullination Sites in Neutrophils and Mast Cells Activated by Ionomycin via Integration of Mass Spectrometry and Machine Learning. Journal of Proteome Research, 20(6), 3150–3164. doi: 10.1021/acs.jproteome.1c00028 [DOI] [PubMed] [Google Scholar]
- [82].Tanikawa C, Ueda K, Suzuki A, Iida A, Nakamura R, Atsuta N, … Matsuda K (2018). Citrullination of RGG Motifs in FET Proteins by PAD4 Regulates Protein Aggregation and ALS Susceptibility. Cell Reports, 22(6), 1473–1483. doi: 10.1016/j.celrep.2018.01.031 [DOI] [PubMed] [Google Scholar]
- [83].Kim JS, Choi M, Choi JY, Kim JY, Kim JY, Song JS, … Lee EY (2020). Implication of the Association of Fibrinogen Citrullination and Osteoclastogenesis in Bone Destruction in Rheumatoid Arthritis. Cells, 9(12). doi: ARTN 2720 10.3390/cells9122720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bradford CM, Ramos I, Cross AK, Haddock G, McQuaid S, Nicholas AP, & Woodroofe MN (2014). Localisation of citrullinated proteins in normal appearing white matter and lesions in the central nervous system in multiple sclerosis. Journal of Neuroimmunology, 273(1–2), 85–95. doi: 10.1016/j.jneuroim.2014.05.007 [DOI] [PubMed] [Google Scholar]
- [85].Frost DC, Greer T, & Li LJ (2015). High-Resolution Enabled 12-Plex DiLeu Isobaric Tags for Quantitative Proteomics. Analytical chemistry, 87(3), 1646–1654. doi: 10.1021/ac503276z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Greer T, Hao L, Nechyporenko A, Lee S, Vezina CM, Ricke WA, … Li LJ (2015). Custom 4-Plex DiLeu Isobaric Labels Enable Relative Quantification of Urinary Proteins in Men with Lower Urinary Tract Symptoms (LUTS). PLoS One, 10(8). doi: ARTN e0135415 10.1371/journal.pone.0135415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kubota K, Yoneyama-Takazawa T, & Ichikawa K (2005). Determination of sites citrullinated by peptidylarginine deiminase using O-18 stable isotope labeling and mass spectrometry. Rapid Communications in Mass Spectrometry, 19(5), 683–688. doi: 10.1002/rcm.1842 [DOI] [PubMed] [Google Scholar]
- [88].Cai WX, Tucholski TM, Gregorich ZR, & Ge Y (2016). Top-down Proteomics: Technology Advancements and Applications to Heart Diseases. Expert Review of Proteomics, 13(8), 717–730. doi: 10.1080/14789450.2016.1209414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Smith LM, Kelleher NL, & Proteomics CTD (2013). Proteoform: a single term describing protein complexity. Nature Methods, 10(3), 186–187. doi: 10.1038/nmeth.2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Shimoyama S, Nagadoi A, Tachiwana H, Yamada M, Sato M, Kurumizaka H, … Akashi S (2010). Deimination stabilizes histone H2A/H2B dimers as revealed by electrospray ionization mass spectrometry (vol 45, pg 900, 2010). Journal of Mass Spectrometry, 45(10), 1232–1232. doi: 10.1002/jms.1853 [DOI] [PubMed] [Google Scholar]
- [91].Clancy KW, Russell AM, Subramanian V, Nguyen H, Qian YW, Campbell RM, & Thompson PR (2017). Citrullination/Methylation Crosstalk on Histone H3 Regulates ER-Target Gene Transcription. Acs Chemical Biology, 12(6), 1691–1702. doi: 10.1021/acschembio.7b00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Mondal S, Wang S, Zheng YA, Sen S, Chatterjee A, & Thompson PR (2021). Site-specific incorporation of citrulline into proteins in mammalian cells. Nature communications, 12(1). doi: ARTN 45 10.1038/s41467-020-20279-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Guo Q, Wang YX, Xu D, Nossent J, Pavlos NJ, & Xu JK (2018). Rheumatoid arthritis: pathological mechanisms and modern pharmacologic therapies. Bone Research, 6. doi: ARTN 15 10.1038/s41413-018-0016-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Tilvawala R, Nguyen SH, Maurais AJ, Nemmara VV, Nagar M, Salinger AJ, … Thompson PR (2018). The Rheumatoid Arthritis-Associated Citrullinome. Cell Chemical Biology, 25(6), 691–+. doi: 10.1016/j.chembiol.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ioan-Facsinay A, Willemze A, Robinson DB, Peschken CA, Markland J, van der Woude D, … El-Gabalawy HS (2008). Marked Differences in Fine Specificity and Isotype Usage of the Anti-Citrullinated Protein Antibody in Health and Disease. Arthritis and Rheumatism, 58(10), 3000–3008. doi: 10.1002/art.23763 [DOI] [PubMed] [Google Scholar]
- [96].Snir O, Widhe M, von Spee C, Lindberg J, Padyukov L, Lundberg K, … Malmstrom V (2009). Multiple antibody reactivities to citrullinated antigens in sera from patients with rheumatoid arthritis: association with HLA-DRB1 alleles. Annals of the Rheumatic Diseases, 68(5), 736–743. doi: 10.1136/ard.2008.091355 [DOI] [PubMed] [Google Scholar]
- [97].Schwenzer A, Jiang X, Mikuls TR, Payne JB, Sayles HR, Quirke AM, … Midwood KS (2016). Identification of an immunodominant peptide from citrullinated tenascin-C as a major target for autoantibodies in rheumatoid arthritis. Annals of the Rheumatic Diseases, 75(10), 1876–1883. doi: 10.1136/annrheumdis-2015-208495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Johansson L, Pratesi F, Brink M, Arlestig L, D’Amato C, Bartaloni D, … Rantapaa-Dahlqvist S (2016). Antibodies directed against endogenous and exogenous citrullinated antigens pre-date the onset of rheumatoid arthritis. Arthritis Research & Therapy, 18. doi: ARTN 127 10.1186/s13075-016-1031-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Shoda H, Fujio K, Shibuya M, Okamura T, Sumitomo S, Okamoto A, … Yamamoto K (2011). Detection of autoantibodies to citrullinated BiP in rheumatoid arthritis patients and pro-inflammatory role of citrullinated BiP in collagen-induced arthritis. Arthritis Research & Therapy, 13(6). doi: ARTN R191 10.1186/ar3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Trela M, Perera S, Sheeran T, Rylance P, Nelson PN, & Attridge K (2019). Citrullination facilitates cross-reactivity of rheumatoid factor with non-IgG1 Fc epitopes in rheumatoid arthritis. Scientific Reports, 9. doi: ARTN 12068 10.1038/s41598-019-48176-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Ingegnoli F, Castelli R, & Gualtierotti R (2013). Rheumatoid Factors: Clinical Applications. Disease Markers, 2013, 727–734. doi: 10.1155/2013/726598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Zhou YB, Chen B, Mittereder N, Chaerkady R, Strain M, An LL, … Sims GP (2017). Spontaneous Secretion of the Citrullination Enzyme PAD2 and Cell Surface Exposure Of PAD4 by Neutrophils. Frontiers in Immunology, 8. doi: ARTN 1200 10.3389/fimmu.2017.01200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Song WP, Ye J, Pan NF, Tan CY, & Herrmann M (2021). Neutrophil Extracellular Traps Tied to Rheumatoid Arthritis: Points to Ponder. Frontiers in Immunology, 11. doi: ARTN 578129 10.3389/fimmu.2020.578129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Maronek M, & Gardlik R (2022). The Citrullination-Neutrophil Extracellular Trap Axis in Chronic Diseases. Journal of Innate Immunity. doi: 10.1159/000522331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Sohn DH, Rhodes C, Onuma K, Zhao X, Sharpe O, Gazitt T, … Sokolove J (2015). Local Joint inflammation and histone citrullination in a murine model of the transition from preclinical autoimmunity to inflammatory arthritis. Arthritis & Rheumatology, 67(11), 2877–2887. doi: 10.1002/art.39283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Carmona-Rivera C, Carlucci M, Moore E, Lingampalli N, Uchtenhagen H, James E, … Kaplan MJ (2020). Synovial fibroblast-neutrophil interactions promote pathogenic adaptive immunity in rheumatoid arthritis (vol 5, pg 43, 2019). Science Immunology, 5(43). doi: ARTN eaaz9319 10.1126/sciimmunol.aaz9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Spengler J, Lugonja B, Ytterberg AJ, Zubarev RA, Creese AJ, Pearson MJ, … Scheel-Toellner D (2015). Release of Active Peptidyl Arginine Deiminases by Neutrophils Can Explain Production of Extracellular Citrullinated Autoantigens in Rheumatoid Arthritis Synovial Fluid. Arthritis & Rheumatology, 67(12), 3135–3145. doi: 10.1002/art.39313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Hardy J, & Selkoe DJ (2002). Medicine - The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science, 297(5580), 353–356. doi: DOI 10.1126/science.1072994 [DOI] [PubMed] [Google Scholar]
- [109].Citron M (2010). Alzheimer’s disease: strategies for disease modification. Nature Reviews Drug Discovery, 9(5), 387–398. doi: 10.1038/nrd2896 [DOI] [PubMed] [Google Scholar]
- [110].Wang L, Chen HY, Tang J, Guo ZW, & Wang YM (2022). Peptidylarginine Deiminase and Alzheimer’s Disease. Journal of Alzheimers Disease, 85(2), 473–484. doi: 10.3233/Jad-215302 [DOI] [PubMed] [Google Scholar]
- [111].Roth EB, Theander E, Londos E, Sandberg-Wollheim M, Larsson A, Sjoberg K, & Stenberg P (2008). Pathogenesis of autoimmune diseases: Antibodies against transglutaminase, peptidylarginine deiminase and protein-bound citrulline in primary Sjogren’s syndrome, multiple sclerosis and Alzheimer’s disease. Scandinavian Journal of Immunology, 67(6), 626–631. doi: 10.1111/j.1365-3083.2008.02115.x [DOI] [PubMed] [Google Scholar]
- [112].Mukherjee S, Perez KA, Dubois C, Nisbet RM, Li QX, Varghese S, … Masters CL (2021). Citrullination of Amyloid-beta Peptides in Alzheimer’s Disease. ACS Chem Neurosci, 12(19), 3719–3732. doi: 10.1021/acschemneuro.1c00474 [DOI] [PubMed] [Google Scholar]
- [113].Kim SE, Park JW, Kim MJ, Jang B, Jeon YC, Kim HJ, … Jang MK (2018). Accumulation of citrullinated glial fibrillary acidic protein in a mouse model of bile duct ligation-induced hepatic fibrosis. PLoS One, 13(8). doi: ARTN e0201744 10.1371/journal.pone.0201744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Moscarello MA, Lei H, Mastronardi FG, Winer S, Tsui H, Li Z, … Wood DD (2013). Inhibition of peptidyl-arginine deiminases reverses protein-hypercitrullination and disease in mouse models of multiple sclerosis. Disease Models & Mechanisms, 6(2), 467–478. doi: 10.1242/dmm.010520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kholia S, Jorfi S, Thompson PR, Causey CP, Nicholas AP, Inal JM, & Lange S (2015). A novel role for peptidylarginine deiminases in microvesicle release reveals therapeutic potential of PAD inhibition in sensitizing prostate cancer cells to chemotherapy. Journal of Extracellular Vesicles, 4. doi: ARTN 26192 10.3402/jev.v4.26192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Chang XT, Wu H, Li HL, Li HL, & Zheng YB (2022). PADI4 promotes epithelial-mesenchymal transition(EMT) in gastric cancer via the upregulation of interleukin 8. Bmc Gastroenterology, 22(1). doi: ARTN 25 10.1186/s12876-022-02097-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Wang Y, Chen R, Gan Y, & Ying S (2021). The roles of PAD2- and PAD4-mediated protein citrullination catalysis in cancers. Int J Cancer, 148(2), 267–276. doi: 10.1002/ijc.33205 [DOI] [PubMed] [Google Scholar]
- [118].Wang L, Chang XT, Yuan GY, Zhao Y, & Wang PC (2010). Expression of Peptidylarginine Deiminase Type 4 in Ovarian Tumors. International Journal of Biological Sciences, 6(5), 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Chang XT, & Han JX (2006). Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Molecular Carcinogenesis, 45(3), 183–196. doi: 10.1002/mc.20169 [DOI] [PubMed] [Google Scholar]
- [120].Baka Z, Barta P, Losonczy G, Krenacs T, Papay J, Szarka E, … Nagy G (2011). Specific expression of PAD4 and citrullinated proteins in lung cancer is not associated with anti-CCP antibody production. International Immunology, 23(6), 405–414. doi: 10.1093/intimm/dxr026 [DOI] [PubMed] [Google Scholar]
- [121].Yuzhalin AE, Gordon-Weeks AN, Tognoli ML, Jones K, Markelc B, Konietzny R, … Muschel RJ (2018). Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nature communications, 9. doi: ARTN 4783 10.1038/s41467-018-07306-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Nagai T, Matsueda Y, Tomita T, Yoshikawa H, & Hirohata S (2018). The expression of mRNA for peptidylarginine deiminase type 2 and type 4 in bone marrow CD34+cells in rheumatoid arthritis. Clinical and Experimental Rheumatology, 36(2), 248–253. [PubMed] [Google Scholar]
- [123].Tsourouktsoglou TD, Warnatsch A, Ioannou M, Hoving D, Wang Q, & Papayannopoulos V (2020). Histones, DNA, and Citrullination Promote Neutrophil Extracellular Trap Inflammation by Regulating the Localization and Activation of TLR4. Cell Reports, 31(5). doi: ARTN 107602 10.1016/j.celrep.2020.107602 [DOI] [PubMed] [Google Scholar]
- [124].Zhu DW, Zhang Y, & Wang SJ (2021). Histone citrullination: a new target for tumors. Molecular Cancer, 20(1). doi: ARTN 90 10.1186/s12943-021-01373-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Chen JD (2016). The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harbor Perspectives in Medicine, 6(3). doi: ARTN a026104 10.1101/cshperspect.a026104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Li PX, Yao HJ, Zhang ZQ, Li M, Luo Y, Thompson PR, … Wang YM (2008). Regulation of p53 target gene expression by peptidylarginine deiminase 4. Molecular and Cellular Biology, 28(15), 4745–4758. doi: 10.1128/Mcb.01747-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Tanikawa C, Espinosa M, Suzuki A, Masuda K, Yamamoto K, Tsuchiya E, … Matsuda K (2012). Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nature communications, 3. doi: ARTN 676 10.1038/ncomms1676 [DOI] [PubMed] [Google Scholar]
- [128].Hayashi H, Morioka M, Ichimiya S, Yamato K, Hinode D, Nagata A, & Nakamura R (1993). Participation of an Arginyl Residue of Insulin Chain-B in the Inhibition of Hemagglutination by Porphyromonas-Gingivalis. Oral Microbiology and Immunology, 8(6), 386–389. doi: DOI 10.1111/j.1399-302X.1993.tb00616.x [DOI] [PubMed] [Google Scholar]
- [129].Sherina N, de Vries C, Kharlamova N, Sippl N, Jiang X, Brynedal B, … Lundberg K (2022). Antibodies to a Citrullinated Porphyromonas gingivalis Epitope Are Increased in Early Rheumatoid Arthritis, and Can Be Produced by Gingival Tissue B Cells: Implications for a Bacterial Origin in RA Etiology. Frontiers in Immunology, 13. doi: ARTN 804822 10.3389/fimmu.2022.804822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Yang ML, Horstman S, Gee R, Guyer P, Lam TT, Kanyo J, … Callebaut N (2022). Citrullination of glucokinase is linked to autoimmune diabetes. Nature Communications, 13(1). doi: ARTN 1870 10.1038/s41467-022-29512-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Petrelli A, Popp SK, Fukuda R, Parish CR, Bosi E, & Simeonovic CJ (2022). The Contribution of Neutrophils and NETs to the Development of Type 1 Diabetes. Frontiers in Immunology, 13. doi: ARTN 930553 10.3389/fimmu.2022.930553 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


