Abstract
Hepatocellular cancer (HCC) accounts for approximately 90% of primary liver carcinoma and is a significant health threat worldwide. Circular RNA basonuclin 2 (circBNC2) is implicated with the progression of several cancers. However, its roles in carcinogenesis and glycolysis are still unclear in HCC. In this study, the levels of circBNC2 and high mobility group AT-hook 2 (HMGA2) were highly expressed, while these of miR-217 were poorly expressed in HCC tissues and cells. Upregulation of circBNC2 was related to poor prognosis and tumor node metastasis (TNM) stage. Knockdown of circBNC2 inhibited the HCC progression. Moreover, knockdown of circBNC2 suppressed the levels of Ras, ERK1/2, PCNA, HK2, and OCT4. Notably, circBNC2 functioned as a molecular sponge of microRNA 217 (miR-217) to upregulate the HMGA2 expression. The inhibitory effects of the circBNC2 silence on the growth and stemness of HCC cells, and levels of PCNA, HK2 and OCT4 were aggravated by the miR-217 overexpression, but neutralized by the HMGA2 overexpression. Besides, silencing of circBNC2 blocked the tumor growth through upregulating the expression of miR-217 and downregulating the levels of HMGA2, PCNA2, HK2 and OCT4 in vivo. Thus, the current data confirmed that circBNC2 sponged miR-217 to upregulate the HMGA2 level, thereby contributing to the HCC glycolysis and progression. These findings might present novel insight into the pathogenesis and treatment of HCC.
Keywords: Hepatocellular carcinoma, circBNC2, miR-217, HMGA2, Glycolysis
1. Introduction
As a common and aggressive cancer, liver cancer remains the sixth leading cause of cancer-related death globally [1]. Moreover, patients with liver cancer are generally diagnosed at advanced stage with the poor prognosis [2]. Hepatocellular carcinoma (HCC) represents over 90% of primary liver cancer, and chemotherapy and immunotherapy are the best options to extend survival before resistance arises [2]. Besides, immune checkpoint inhibitors alone or in combination with other agents are vital strategies for the treatment of HCC [[3], [4], [5]]. However, high metastasis and recurrence of HCC pose great challenges to the HCC treatment. A small population of cancer cells referred as cancer stem cells (CSCs) have the distinct metabolic phenotypes, such as high glycolytic activity [6,7]. CSCs are generally believed to take charge the tumor initiation, recurrence and therapy resistance [6,7]. In this case, it is urgent to identify the potential biomarkers for prognosis and develop the new therapeutic targets for HCC.
Emerging evidences have shown the involvement of circular RNAs (circRNAs) in the occurrence and development of cancers [6,7]. For instance, cSMARCA5, a circRNA originated from exons 15 and 16 of the SMARCA5 gene, suppressed tumor growth and metastasis in HCC, making it as a potential therapeutic target [8]. CircAKT3 enhanced cisplatin resistance in gastric cancer, which highlighted that is might be a potential therapeutic target for patients with gastric cancer receiving cisplatin therapy [9]. CircBNC2 (has_circ_0008732) is derived from the zinc-finger protein basonuclin 2 (BNC2) that is a susceptibility gene for epithelial ovarian cancer [10]. A previous study confirmed that circBNC2 was poorly expressed, thus acting as a promising diagnostic biomarker for epithelial ovarian cancer [11,12]. Furthermore, circBNC2 was dysregulated in patients with non-small cell lung cancer. However, the impact of circBNC2 on HCC is unknown.
CircRNA can affect the expression of target genes by serving as competing endogenous RNAs (ceRNAs) for microRNAs (miRNAs), thereby participating in the regulation of cancer development [13]. Dysregulation of miRNAs has been identified in various cancers, including HCC. For instance, miRNA-1179 inhibited the HCC metastasis by interacting with zinc-finger E-box-binding homeobox 2 (ZEB2). MiR-194 played a crucial role in the progression of HCC via inhibiting polypyrimidine tract-binding protein 1 (PTBP1)/cyclin D3 (CCND3) axis, acting as a novel therapeutic target for the HCC patients [14]. A previous study demonstrated that miRNA-448 served as a tumor suppressor by downregulating BCL-2 in HCC [15]. Overexpression of miR-140-5p was demonstrated to increase the sensitivity to sorafenib in HCC [16]. Notably, miR-217 functioned as a tumor suppressor. MiR-217 could act as a target of long non-coding RNA CRNDE to suppress the cell viability and motility by downregulating the level of mitogen-activated protein kinase 1 (MAPK1) in HCC [17]. Moreover, miR-217 suppressed the HCC development through targeting metadherin (MTDH) [18]. Downregulation of miR-217 could serve as a potential biomarker for the malignant progression and poor prognosis in HCC [19]. However, the interaction and regulatory relationship between circBNC2 and miR-217 have not been investigated.
Noticeably, the bioinformatics tool predicted that miR-217 acted as a target miRNA of circBNC2. Therefore, in this study, we hypothesized that circBNC2 might regulate the carcinogenesis and glycolysis in HCC by mediating the miR-217. Herein, we sought to investigate the function of circBNC2 in cell growth, glycolysis and stemness in HCC. The involvement of miR-217/high mobility group AT-hook 2 (HMGA2) was also explored during above processes. Significantly, the current findings confirmed that circBNC2 sponged miR-217 to upregulate the HMGA2 level, which contributed to the HCC glycolysis and progression, indicating a promising therapeutic approach for HCC.
2. Materials and methods
2.1. HCC tumor samples and ethics statement
HCC and adjacent normal tissues (n = 54) were obtained from the Chongqing Cancer Hospital and promptly frozen in liquid nitrogen for the use. All patient signed the informed consent. This research was approved by the Ethics Review Committees of Chongqing Cancer Hospital (No. 2020-LS-036) and conducted according to the Declaration of Helsinki.
2.1.1. Cell culture
The cell lines (L02, Huh7, HCCLM3 and Hep3B) were obtained from Procell (Wuhan, China), and maintained in Dulbecco's modified Eagle's medium (DMEM) with fetal bovine serum (FBS) (10%) and penicillin/streptomycin (100 U/mL, Gibco, Grand Island, NY, USA) under 5% carbon dioxide (CO2) at 37 °C.
2.2. Real-time quantitative polymerase chain reaction (RT-qPCR)
In brief, total RNA from tissues and cells was isolated and purified using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The reversed transcription was conducted with RT-PCR kit (Invitrogen), and 2 μl of cDNA template was applied for the RT-qPCR assays on AB7300 thermo-recycler purchased from Applied Biosystems (Foster City, CA, USA) using SYBR Green. The internal references were GAPDH and U6. The PCR amplification reaction was denatured at 95 °C for 10 min followed by a thermocycling step of 40 cycles (95 °C for 10 s; 60 °C for 35 s). The 2−ΔΔCt method was used to calculate the relative expression levels of genes. The sequences were listed in Table 1.
Table 1.
The sequences of primers used in the present study.
| Name | Forward primer (5ʹ-3ʹ) | Reverse primer (5ʹ-3ʹ) |
|---|---|---|
| circBNC2 | GCAGTTCGGAACCAGAACGAC | ATGCTGGCCAGTCTTGCTCAC |
| miR-217 | ACACTCCAGCTGGGTACTGCATCAGGAACTG | TGGTGTCGTGGAGTCG |
| HMGA2 | GGGCGCCGACATTCAAT | ACTGCAGTGTCTTCTCCCTTCAA |
| GAPDH | GGGAAACTGTGGCGTGAT | GAGTGGGTGTCGCTGTTGA |
| U6 | CGCTTCGGCAGCACATATACTAAAATTGGAAC | GCTTCACGAATTTGCGTGTCATCCTTGC |
2.2.1. RNase R treatment
Approximately 10 μg of RNA was extracted from Huh7 cells and cultured at 37 °C for 1 h with or without 4 U/μg of RNase R (Epicentre Biotechnologies). Finally, the relative messenger RNAs (mRNAs) levels of circBNC2 and GAPDH were measured by RT-qPCR.
2.2.2. Cell transfection
Small interfering ribonucleic acid (siRNA) against circBNC2 (si-circBNC2), short hairpin RNA (shRNA) targeting circ-BNC2 (sh-circBNC2), and their negative control (si-NC, and sh-NC) were constructed by Sangon Biotechnology (Shanghai, China). MiR-217 mimic (miR-217), miR-217 inhibitor, HMGA2 mimic (HMGA2), and their negative control (miR-NC, NC inhibitor, and vector) were purchased from GenePharma (Shanghai, China). Cell transfection was conducted as previously described [20]. Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was applied for the transfection assay.
2.2.3. Cell counting Kit-8 (CCK-8) assay
The cell viability of Huh7 and Hep3B was performed by CCK-8 assay [14]. Cells were seeded in 96-well plates and incubated at 37 °C for 24 h. At 0 h, 24 h, 48 h and 72 h, 10 μL of CCK-8 solution (Beyotime, Shanghai, China) was added into each well, respectively. Four hours later, absorbance at 450 nm was recorded by a Bio-Rad microplate reader (Hercules, CA, USA).
2.2.4. 5-Ethynyl-2′-deoxyuridine (EdU) assay
The cell proliferation ability was also evaluated using the EdU assay [14]. In brief, HCC cells (1 × 105) under various treatments were seeded in 96-well plates and treated with EdU labelling medium for 2 h. Then, the labeled cells were fixed with paraformaldehyde (4%) for 15 min and cultured with TritonX-100 (0.5%) in PBS for 15 min. Next, cells together with Apollo dye solution (100 μL) were incubated at room temperature without light for 30 min. Finally, the cells were stained with DAPI for 5 min, and visualized using a microscope. ImageJ software was applied to calculate the percentage of EdU positive cells.
2.2.5. Glucose consumption, lactate production and ATP level detection
Based on the manufacturer's instructions, the levels of glucose uptake, lactate and ATP in HCC cells were determined by Glucose Assay Kit purchased from Shanghai Abcam (ab65333, China), Lactate Assay kit purchased from Sigma St (MAK064, Louis, MO, USA) and ATP Assay Kit purchased from Abcam (ab83355), respectively. For the examination of the levels of glucose uptake, cells (2 × 106) were harvested washed with cold PBS, and then resuspended into 100 μL of Assay Buffer. The resuspended cells were homogenized quickly by pipetting up and down a few times, and centrifuged with 2 min at 4 °C at top speed in a cold microcentrifuge to remove any insoluble material. A PCA/KOH deproteinization step was performed before glucose assay. The levels of glucose uptake were determined by colorimetric assay based on the protocol described. For the examination of the levels of lactate, cells (1 × 104) were harvested washed with cold PBS, and then resuspended into the extracting solution. Then, cells were subjected to the ultrasonication, and centrifuged with 12000 g for 10 min at 4 °C. The levels of lactate were determined by colorimetric assay based on the protocol described. For the examination of the levels of ATP, cells (1 × 106) were harvested washed with cold PBS, and then resuspended into 100 μL of ATP Assay Buffer. The resuspended cells were homogenized quickly by pipetting up and down a few times, and centrifuged with 13000 g for 5 min at 4 °C in a cold microcentrifuge to remove any insoluble material. A PCA/KOH deproteinization step was performed before glucose assay. The levels of ATP were determined by colorimetric assay based on the protocol described.
2.2.6. Tumor sphere formation
Tumor sphere formation was performed as previously described [14]. Briefly, 1 × 103 cells were placed into 6-well plates and kept in DMEM supplemented with epidermal growth factor (EGF, 20 ng/ml), B27 (2%) and fibroblast growth factor (FGF, 10 ng/ml) at room temperature for 7 days. After the transfection with si-NC, si-circBNC2, si-circBNC2+miR-217 or si-circBNC2+HMGA2 for 72 h, spheres were photographed and counted by a microscope.
2.3. Western blot analysis
The extraction of proteins from tissues or cells was implemented via radioimmunoprecipitation assay (RIPA, Sangon Biotech, Shanghai, China). Then, the protein samples were subjected to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (ThermoFisher, Waltham, USA). After the block by 5% non-fat milk, Western blot analysis was applied to determine the protein expression [14]. The membranes were incubated with the diluted primary antibodies (1:2000) and secondary antibodies (1:5000), respectively. Finally, the protein signals were achieved using an enhanced chemiluminescence (Amersham Pharmacia, Piscataway, USA). Primary antibodies against Ras (1:1,000, ab180772), phosphor-ERK1/2 (p-ERK1/2, 1:1,000, ab47339), ERK1/2 (t-ERK1/2, 1:3,000, ab196883), PCNA (1:2,000, ab18197), OCT4 (1:2,000, ab109183), hexokinase 2 (HK2) (1:2,000, ab227198), HMGA2 (1:2,000, ab97276), β-actin (1:2,000, ab8227), GAPDH (1:2,500, ab9485) and secondary antibodies (1:2,000, ab205718) were all obtained from Abcam. ImageJ software was used to evaluate the band density by densitometric analysis.
2.3.1. Flow cytometry
The cell apoptosis was determined through the flow cytometry assay. Briefly, cells were resuspended into 1 ml of binding buffer after cells were rinsed with PBS. Then, cells were stained with propidium iodide (PI) and FITC-Annexin V (Thermo Fisher Scientific), separately. Apoptotic cells were detected on a FACScan flow cytometry (BD Biosciences, NJ, USA) and analyzed by BD CellQuest Pro software (version 5.1, BD Biosciences). Besides, the cell cycle was also determined by the flow cytometry assay. Following the wash with PBS, cells were fixed with ice-cold 100% ethanol (E7023, Sigma) at 4 °C for 60 min, and then washed with PBS thrice. Then, cells were incubated with RNase A (100 μg/ml, R1030, Solarbio) at 37 °C for 60 min, and with propidium iodide (PI, 1000 μg/ml, P3566, Invitrogen) for 30 min without the light. The cell population was detected by the FACScan flow cytometry.
2.3.2. Dual-luciferase reporter assay
Dual-luciferase reporter assay was performed as previous description. The wild type sequences of circBNC2 (circBNC2-WT), mutant type sequences of circBNC2 (circBNC2-MUT), wild type 3′Untranslated Region (UTR) sequences of HMGA2 (HMGA2-WT) or mutant type 3′UTR sequences of HMGA2 (HMGA2-MUT) were obtained and inserted into pGL-3 vector (Promega, Madison, USA). Huh7 cells were co-transfected with luciferase reporter plus miR-217 or miR-NC using Lipofectamine 3000. Luciferase intensity was measured by Dual-Luciferase Assay Kit of Promega after 48 h.
2.3.3. RNA pull-down assay
RNA pull-down analysis was conducted as a previous report [14]. Briefly, wild-type miR-217 and its negative control (miR-NC) were labeled with biotin and incubated with streptavidin beads (Biolinkedin Biotech, Shanghai, China) for 24 h at 4 °C. Huh7 cells were lysed and cultured with bead-biotin complex for 2 h. Subsequently, the bounded RNA was isolated and subjected to qRT-PCR assay for the quantitative analysis of the circBNC2 level.
2.3.4. Tumor xenograft assay
The 5-weeks-old nude mice (BALB/c) were used to perform xenograft assay. 1 × 107 Huh7 cells transfected with sh-circBNC2 or sh-NC were subcutaneously inoculated into the mice. After 7 days, tumor volume was measured every 7 d. All mice were sacrificed by inhaling excess CO2 on day 35 after the inoculation. The dissected tumors were weighted. The mice experiment was approved by the Animal Ethics Committee of Chongqing University Cancer Hospital.
2.3.5. Enzyme-linked immunosorbent assay (ELISA)
Serum of mice was collected and the levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were measured using ELISA kits (Beyotime, PV963). All protocols were carried out according to the manufacturer's recommendations.
2.4. Hematoxylin and eosin (H&E) staining
The collected tumor and liver tissues were fixed with paraformaldehyde (4%) for 24 h and then embedded in paraffin. The tissue samples were sectioned into slices with the 5 μm thickness and the sections were stained with hematoxylin and eosin.
2.4.1. Immunohistochemistry (IHC)
The sections of tumor samples were rehydrated for IHC [21]. After the antigen retrieval, the sections were incubated with hydrogen peroxide (3%) for 20 min to block endogenous peroxidase. The sections were sealed with normal goat serum (5%) for 30 min, followed by the incubation with antibodies against HMGA2 (ab97276, Abcam), PCNA (ab18197, Abcam), HK2 (ab227198, Abcam) and OCT4 (ab109183, Abcam) overnight at 4 °C. Subsequently, the sections were incubated with secondary antibodies at 37 °C for 1 h, followed by stained with DAB. Finally, the sections were counter-stained with hematoxylin and observed under an Olympus light microscope (400× magnification).
2.5. Statistical analysis
All data were analyzed using GraphPad Prism 7 and showed as mean ± standard deviation (SD). Difference between groups was calculated by Student's t-test or one-way ANOVA with Tukey's post-hoc test. Overall survival (OS) was generated by the Kaplan-Meier plot, and the difference was assessed by log-rank test. The relationship of circBNC2 level with clinical pathological features was analyzed by chi-square test. *p < 0.05 was regarded as the statistical significance.
3. Results
Herein, we hypothesized that circBNC2 might exert the potential roles in the carcinogenesis and glycolysis in HCC by mediating the miR-217/HMGA2 axis. Therefore, in the current study, we detected the effects of circBNC2 knockdown on cell growth, glycolysis and stemness in HCC, as well as the role of miR-217/HMGA2 involved in the above processes. As expected, circBNC2 knockdown could inhibit HCC cell growth, glycolysis and stemness by sponging miR-217 to up-regulate the HMGA2 level, suggesting a promising target for the treatment of HCC.
3.1. Overexpression of circBNC2 in HCC tumor tissues and cells
Previous study identified circBNC2 as a promising diagnostic biomarker for epithelial ovarian cancer [11,12]. Before the investigation of its role in HCC, we first conducted the in silico analysis based on the GSE164803 database and found that the circBNC2 level was higher in 6 HCC tumor tissues compared with that in the matched adjacent normal tissues (p < 0.01; Fig. 1A). As shown in Fig. 1B, the circBNC2 abundance was higher in HCC tumor tissues compared with that in the matched adjacent tissues (p < 0.001). Moreover, expression of circBNC2 was negative correlated with the OS (p < 0.01; Fig. 1C) and recurrence-free survival (p < 0.05; Fig. 1D). Besides, the circBNC2 level was higher in all HCC cells than that in L02 cells (p < 0.001; Fig. 1E). More importantly, the mRNA level of GAPDH in Huh7 cells was significantly declined after RNase R disposition, while that of circBNC2 was unaffected (Fig. 1F), implying a resistance of circBNC2 to RNase R digestion. Thus, circBNC2 acted as a stable circRNA that involved in the HCC progression. CircBNC2 expression was higher in tumors metastasis (p = 0.006) and advanced-stage tumors (p = 0.013) (Table 1). There is no significant association of circBNC2 expression with gender (p = 0.413), age (p = 0.785), tumor size (p = 0.172), or microvascular invasion (p = 0.586) (Table 2). Taken together, the level of circBNC2 was overexpressed in HCC tumor and cells, which was related to the poor survival.
Fig. 1.
CircBNC2 was highly expressed in HCC tissues and cells. (A) Compared with the adjacent normal tissues (n = 6), circBNC2 was upregulated in HCC tumor tissues (n = 6) according to GSE164803 database. **p < 0.01. (B) The expression of circBNC2 in our cohort of 54 pairs of HCC tumor tissues and adjacent normal tissues was detected by RT-qPCR. ***p < 0.001. (C) HCC patients (n = 54) were divided into circBNC2 high expression group and low expression based on the median value of circBNC2, and the survival curve of HCC patients in two groups was assessed by log-rank test. (D) The correlation between CircBNC2 expression and recurrence-free survival of HCC patients. (E) CircBNC2 expression in HCC cell lines (Huh7, HCCLM3 and Hep3B) and a normal liver cell line (L02) were assessed by RT-qPCR. ***p < 0.001 vs. L02. (F) Expression of circBNC2 and GAPDH in Huh7 cell treated with RNase R were assessed by RT-qPCR. ***p < 0.001 vs. Mock.
Table 2.
Correlation between circBNC2 expression and the clinical pathological features of 54 HCC patients.
| Characteristic | All cases | circBNC2 expression |
p-value | |
|---|---|---|---|---|
| High (n = 27) | Low (n = 27) | |||
| Gender | 0.413 | |||
| male | 29 | 16 | 13 | |
| female | 25 | 11 | 14 | |
| Age (years) | 0.785 | |||
| <60 | 29 | 14 | 15 | |
| ≥60 | 25 | 13 | 12 | |
| Tumor size (cm) | 0.172 | |||
| ≥5 | 25 | 10 | 15 | |
| <5 | 29 | 17 | 12 | |
| Tumor metastasis | 0.006* | |||
| Present | 26 | 18 | 8 | |
| Absent | 28 | 9 | 19 | |
| Microvascular invasion | 0.586 | |||
| Absent | 26 | 12 | 14 | |
| Present | 28 | 15 | 13 | |
| TNM Stages | 0.013* | |||
| Ⅰ/Ⅱ | 23 | 7 | 16 | |
| Ⅲ/Ⅳ | 31 | 20 | 11 | |
*p < 0.05.
3.2. Knockdown of circBNC2 inhibited HCC cell growth, glycolysis and stemness
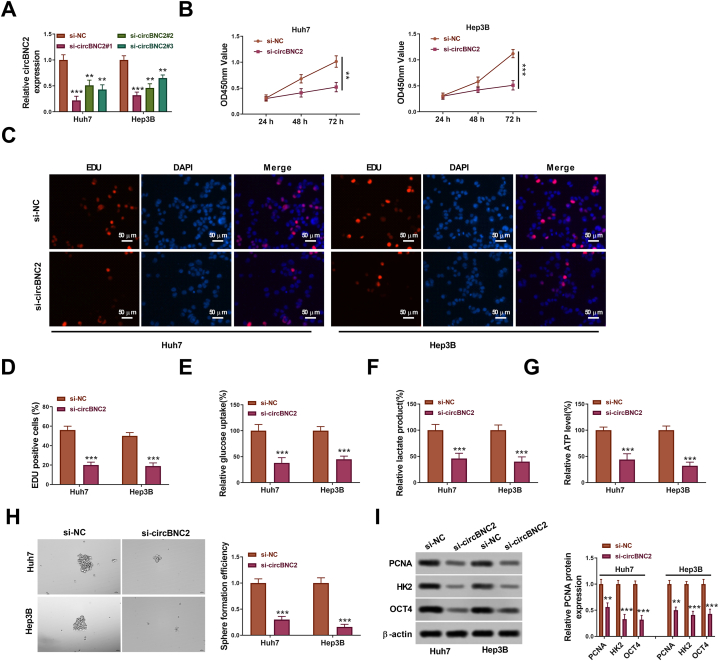
To investigate the roles of circBNC2 in HCC cells, loss-of-function experiment was performed in HCC cells (Huh7 and Hep3B). As shown in Fig. 2A, the knockdown efficiency of si-circBNC2 #1 was higher than that of si-circBNC2 #2 and #3 (p < 0.01). Thus, the si-circBNC2 #1 was named as si-circBNC2 and used for the following experiments.
Fig. 2.
Function of circBNC2 on proliferation, glycolysis and stemness of HCC cells. (A) The knockdown efficiency of si-circBNC2#1, si-circBNC2#2 and si-circBNC2#3 in Huh7 and Hep3B cells was detected by qRT-PCR. After Huh7 and Hep3B cells transfected with si-NC or si-circBNC2, the proliferation, relative glucose uptake, relative lactate production, relative ATP level, sphere formation, and levels of PCNA, HK2 and OCT4 protein were assessed by CCK-8 assay (B), EdU assay(C-D), Glucose Assay Kit (E), Lactate Assay Kit (F), ATP Assay Kit (G), sphere-forming assay(H), Western blot assay (I), respectively. The original images of gels can be found in Supplementary material 2. **p < 0.01, ***p < 0.001 vs. si-NC.
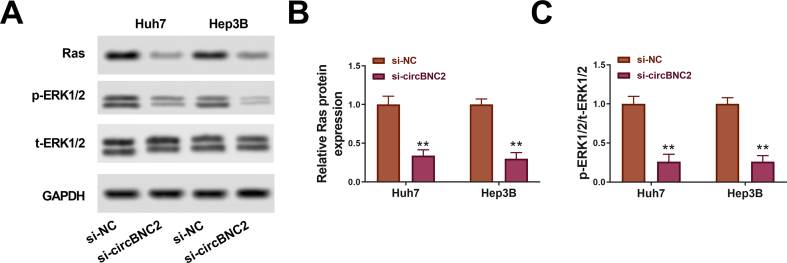
CCK-8 assays indicated that knockdown of circBNC2 inhibited cell proliferation (Fig. 2B). EdU assays also showed that the circBNC2 knockdown significantly suppressed the cell proliferation by 40% in Huh7 cells and 35% in Hep3B cells compared with the si-NC group (p < 0.001; Fig. 2C and D). Besides, the circBNC2 downregulation inhibited glucose uptake, lactate production and ATP level relative to the si-NC group (Fig. 2E–G). Moreover, the circBNC2 knockdown significantly reduced the number of sphere formation (Fig. 2H). Furthermore, the levels of survival markers (Ras and ERK), proliferation marker PCNA, glycolysis-related factor HK2, and stem-cell marker OCT4 were decreased in Huh7 and Hep3B cells transfected with si-circBNC2 (Fig. S1 A-C, Fig. 2I). Overall, these findings implied that deficiency of circBNC2 restrained cell viability, glycolysis and stemness in HCC cells.
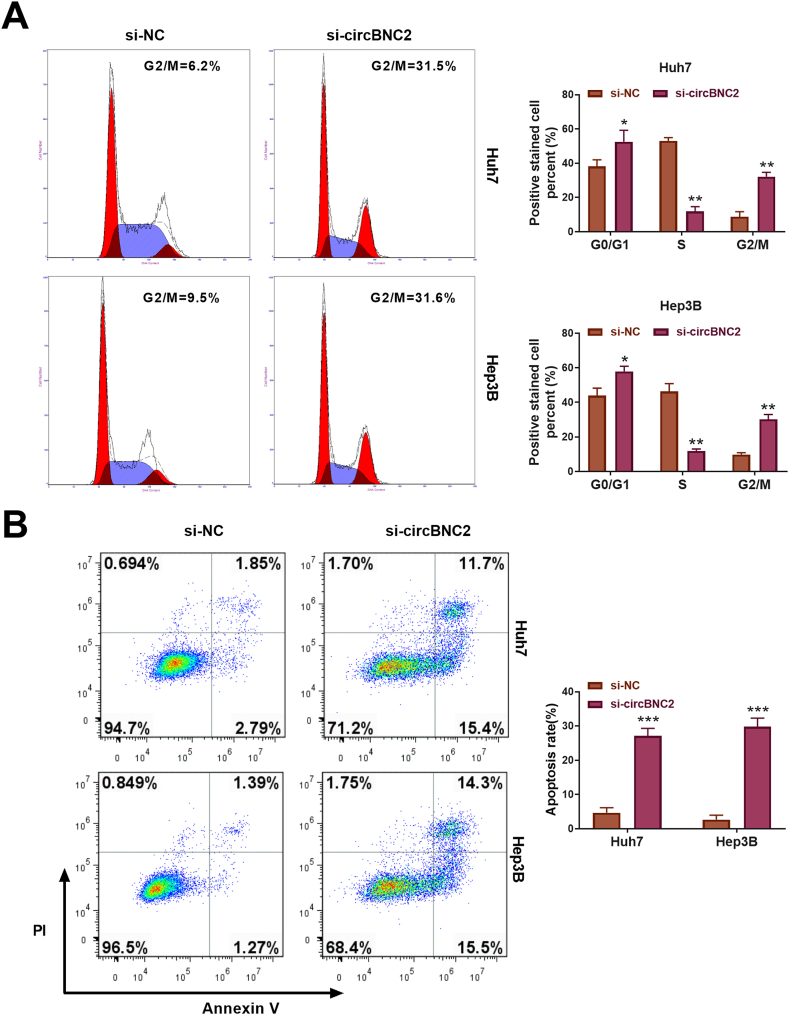
3.3. Knockdown of circBNC2 induced HCC cell G2/M phase arrest and apoptosis
To investigate the role of circBNC2 in the cell arrest and apoptosis of HCC cells, si-circBNC2 was transfected into Huh7 and Hep3B cells. Downregulation of circBNC2 prominently induced G2/M phase arrest in both Huh7 and Hep3B cells (p < 0.01; Fig. 3A). Besides, knockdown of circBNC2 significantly enhanced the apoptosis rate of both Huh7 and Hep3B cells (p < 0.001; Fig. 3B). Thus, these results indicated that silencing of circBNC2 promoted cell cycle arrest and apoptosis in HCC cells.
Fig. 3.
Knockdown of circBNC2 induced HCC cell S phase arrest and apoptosis. (A) The cell cycle of Huh7 and Hep3B cells was determined by the flow cytometry. (B) The cell apoptosis of Huh7 and Hep3B cells was analyzed by the flow cytometry. *p < 0.05, **p < 0.01 and ***p < 0.001 vs. si-NC.
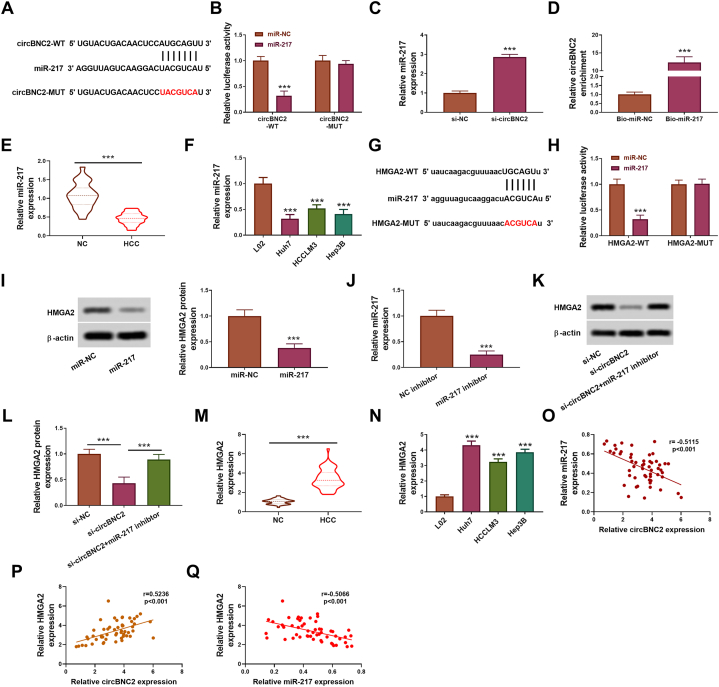
3.4. CircBNC2 up-regulated the HMGA2 expression by absorbing miR-217
According to bioinformatics analysis by circinteractome, miR-217 was predicted to be a potential target miRNA of circBNC2 (Fig. 4A). MiR-217 overexpression significantly suppressed the luciferase activity of circBNC2-WT in Huh7 cells but did not influence the luciferase activity of circBNC2-MUT (Fig. 4B). Moreover, transfection of si-circBNC2 increased the expression of miR-217 in Huh7 cells (p < 0.001; Fig. 4C). CircBNC2 was significantly enriched in biotinylated-miR-217 in Huh7 cells (p < 0.001; Fig. 4D). Besides, the miR-217 level was lower in HCC tumor tissues and cell lines (Huh7, HCCLM3 and Hep3B cells) (p < 0.001; Fig. 4E and F).
Fig. 4.
CircBNC2 increased HMGA2 expression by absorbing miR-217. (A) The potential binding sites between circBNC2 and miR-217 was predicted by circinteractome. (B) The luciferase activity in Huh7 cells co-transfected with circBNC2-WT/circBNC2-MUT and miR-217/miR-NC was examined by dual-luciferase reporter assay. ***p < 0.001 vs. miR-NC. (C) After Huh7 cells transfected with si-NC or si-circBNC2, miR-217 expression was determined by RT-qPCR assay. ***p < 0.001 vs. si-NC. (D) The level of circBNC2 in Huh7 cells after RNA pull-down assay using Bio-miR-NC probe or Bio-miR-217 probe was determined by RT-qPCR. ***p < 0.001 vs. Bio-miR-NC. (E) MiR-217 expression in HCC tumor tissues (n = 54) and adjacent normal tissues (n = 54) was detected by RT-qPCR. ***p < 0.001. (F) MiR-217 expression in HCC cell lines (Huh7, HCCLM3 and Hep3B) and a normal liver cell line (L02) were assessed by RT-qPCR. ***p < 0.001 vs. L02. (G) The potential binding sites between miR-217 and HMGA2 was predicted by starBase. (H) After co-transfected with HMGA2-WT/HMGA2-MUT and miR-217/miR-NC, the luciferase activity in Huh7 cells was examined. ***p < 0.001 vs. miR-NC. (I) After Huh7 cells transfected with miR-NC or miR-217 vector, HMGA2 protein expression was detected by Western blot assay. The original images of gels can be found in Supplementary material 2. ***p < 0.001 vs. miR-NC. (J) After Huh7 cells transfected with miR-217 inhibitor or NC inhibitor, miR-217 expression was detected by RT-qPCR assay. ***p < 0.001 vs. NC inhibitor. (K–L) After Huh7 cells transfected with si-NC, si-BNC2 or si-circBNC2+miR-217 inhibitor, the levels of PCNA, HK2 and OCT4 in Huh7 cells were assessed by Western blot assay. The original images of gels can be found in Supplementary material 2. ***p < 0.001. (M) HMGA2 expression in HCC tumor tissues (n = 54) and adjacent normal tissues (n = 54) was detected by RT-qPCR. ***p < 0.001. (N) HMGA2 expression in HCC cell lines (Huh7, HCCLM3 and Hep3B) and normal liver cell line (L02) was assessed by RT-qPCR. ***p < 0.001 vs. L02. (O–Q) The correlations between circBNC2 and miR-217 and HMGA2 were analyzed in HCC tissues.
By searching via starBase, we found that miR-217 contained the binding sites of HMGA2 (Fig. 4G). The luciferase activity was observably inhibited in Huh7 cells co-transfected with HMGA2-WT plus miR-217 (p < 0.001; Fig. 4H), indicating the interaction between HMGA2 and miR-217. The relative protein expression of HMGA2 was downregulated in Huh7 cells transfected with miR-217 (p < 0.001; Fig. 4I). RT-qPCR assay demonstrated that the miR-217 expression was downregulated after the transfection with miR-217 inhibitor (p < 0.001; Fig. 4J). The HMGA2 protein expression was inhibited in Huh7 cells transfected with si-circBNC2 (p < 0.001; Fig. 4K and L). However, the inhibition was abolished in Huh7 cell co-transfected with si-circBNC2 plus miR-217 inhibitor (p < 0.001; Fig. 4K and L). Essentially, the miR-217 inhibitor could neutralize the repression of si-circBNC2 on Huh7 cells. Besides, the HMGA2 level was upregulated in HCC tumor tissues and cell lines (Huh7, HCCLM3 and Hep3B cells) (p < 0.001; Fig. 4M and N). Additionally, we analyzed the correlation between circBNC2 and miR-217 and HMGA2 and found that there was a negative correlation between circBNC2 and miR-217 (p < 0.001; Fig. 4O) and a positive correlation between circBNC2 and HMGA2 (p < 0.001; Fig. 4P) in HCC tissues. Furthermore, a negative correlation between the expression of miR-217 and HMGA2 (p < 0.001; Fig. 4Q).
Analyses of the associations of the miR-217 and HMGA2 expression with clinical pathological features of 54 HCC patients were performed. Significant correlation was observed in miR-217 and HMGA2 expression with tumors metastasis. There is no significant association of miR-217 and HMGA2 expression with gender, age, tumor size, microvascular invasion, or TNM stage (Table 3, Table 4).
Table 3.
Correlation between miR-217 expression and the clinical pathological features of 54 HCC patients.
| Characteristic | All cases | miR-217 expression |
p-value | |
|---|---|---|---|---|
| High (n = 27) | Low (n = 27) | |||
| Gender | 0.056 | |||
| male | 29 | 11 | 18 | |
| female | 25 | 16 | 9 | |
| Age (years) | 0.413 | |||
| <60 | 29 | 16 | 13 | |
| ≥60 | 25 | 11 | 14 | |
| Tumor size (cm) | 0.413 | |||
| ≥5 | 25 | 13 | 16 | |
| <5 | 29 | 14 | 11 | |
| Tumor metastasis | 0.029* | |||
| Absent | 26 | 17 | 9 | |
| Present | 28 | 10 | 18 | |
| Microvascular invasion | 0.278 | |||
| Absent | 26 | 11 | 15 | |
| Present | 28 | 16 | 12 | |
| TNM Stages | 0.054 | |||
| Ⅰ/II | 23 | 15 | 8 | |
| Ⅲ/IV | 31 | 12 | 19 | |
*p < 0.05.
Table 4.
Correlation between HMGA2 expression and the clinical pathological features of 54 HCC patients.
| Characteristic | All cases | HMGA2 expression |
p-value | |
|---|---|---|---|---|
| High (n = 27) | Low (n = 27) | |||
| Gender | 0.172 | |||
| male | 29 | 12 | 17 | |
| female | 25 | 15 | 10 | |
| Age (years) | 0.413 | |||
| <60 | 29 | 13 | 16 | |
| ≥60 | 25 | 14 | 11 | |
| Tumor size (cm) | 0.413 | |||
| ≥5 | 25 | 11 | 14 | |
| <5 | 29 | 16 | 13 | |
| Tumor metastasis | 0.006* | |||
| Absent | 26 | 8 | 18 | |
| Present | 28 | 19 | 9 | |
| Microvascular invasion | 0.586 | |||
| Absent | 26 | 14 | 12 | |
| Present | 28 | 13 | 15 | |
| TNM Stages | 0.054 | |||
| Ⅰ/II | 23 | 8 | 15 | |
| Ⅲ/IV | 31 | 19 | 12 | |
*p < 0.05.
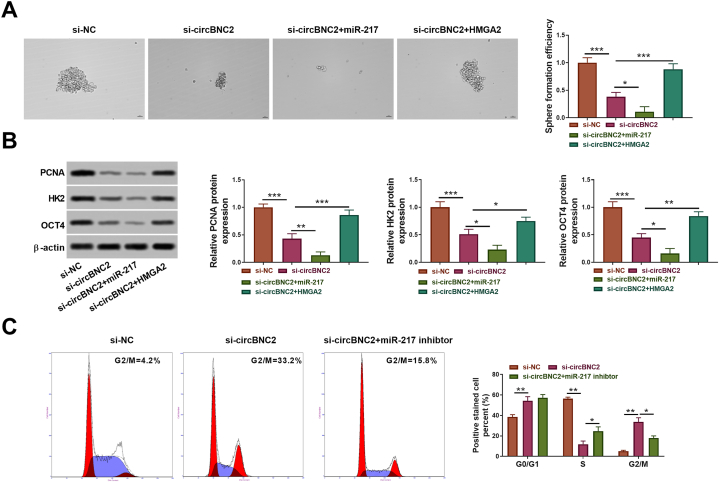
3.5. CircBNC2 regulated HCC cell proliferation, glycolysis and stemness through miR-217/HMGA2 axis
To investigate whether miR-217/HMGA2 axis contributed to the circBNC2-mediated HCC development, we performed experiments by restoring the miR-217 or HMGA2 expression. Transfection of the HMGA2 vector significantly increased its expression (p < 0.001; Fig. 5A). MiR-217 overexpression significantly promoted the inhibitory effect of circBNC2 silence on HCC cell growth based on the CCK-8 and EdU assays (p < 0.05; Fig. 5B–D). Conversely, HMGA2 overexpression significantly reversed the decrease of circBNC2 knockdown on HCC cell growth (p < 0.01; Fig. 5B–D). Moreover, circBNC2 silencing-mediated inhibitory effects on glucose uptake, lactate production and ATP level of Huh7 cells were enhanced by the miR-217 overexpression or neutralized by the HMGA2 overexpression (p < 0.05; Fig. 5E–G).
Fig. 5.
CircBNC2/miR-217/HMGA2 regulated HCC cell proliferation and glycolysis . (A) After transfection with the empty vector or HMGA2 vector, HMGA2 protein expression was detected by Western blot. The original images of gels can be found in Supplementary material 2. ***p < 0.001 vs. vector. After Huh7 cells transfected with si-NC, si-circBNC2, si-circBNC2+miR-217, or si-circBNC2+HMGA2, the proliferation, relative glucose uptake, relative lactate production, relative ATP level, sphere formation, and levels of PCNA, HK2 and OCT4 protein were assessed by CCK-8 assay (B), EdU assay (C and D), Glucose Assay Kit (E), Lactate Assay Kit (F), ATP Assay Kit (G), respectively. *p < 0.05, **p < 0.01, ***p < 0.001.
The sphere-forming assay showed that the circBNC2 silencing-mediated inhibitory effects on sphere formation (p < 0.05; Fig. 6A) and protein expression of PCNA, HK2 and OCT4 (p < 0.05; Fig. 6B) could also be facilitated by the miR-217 elevation or reversed by the HMGA2 overexpression. Besides, downregulation of circBNC2 prominently induced G2/M phase arrest of both Huh7 and Hep3B cells (p < 0.05; Fig. 6C). However, administration of miR-217 inhibitor had a restoring effect on the circBNC2 silencing-mediated inhibitory role in G2/M phase (p > 0.05; Fig. 6C). Collectively, these results suggested that miR-217/HMGA2 axis involved in circBNC2-mediated HCC cell proliferation, cell cycle arrest, glycolysis and stemness.
Fig. 6.
CircBNC2/miR-217/HMGA2 regulated HCC cell stemness and cycle. (A) After Huh7 cells transfected with si-NC, si-circBNC2, si-circBNC2+miR-217, or si-circBNC2+HMGA2, the sphere formation, and levels of PCNA, HK2 and OCT4 protein were assessed by sphere-forming assay(A) and Western blot assay (B), respectively. The original images of gels can be found in Supplementary material 2. (C) The cell cycle was determined by the flow cytometry. *p < 0.05, **p < 0.01, ***p < 0.001.
3.6. CircBNC2 knockdown inhibited HCC cell growth in vivo
There was a significant reduction of tumor volume and weight in sh-circBNC2 group compared with that in sh-NC group (p < 0.01; Fig. 7A and C), but no significant difference in mice weight between sh-circBNC2 group and sh-NC group was observed (p > 0.05; Fig. 7B). The level of circBNC2 was declined, whereas that of miR-217 was elevated in sh-circBNC2 group (p < 0.001; Fig. 7D). However, there was no significant differences in serum levels of ALT and AST between sh-circBNC2 group and sh-NC group (p > 0.05; Fig. 7E and F). Besides, H&E staining showed improvement of liver damage in sh-circBNC2 group (Fig. 7G), and the IHC staining rates of HMGA2, PCNA, HK2 and OCT4 were reduced in sh-circBNC2 group (p < 0.001; Fig. 7G). Therefore, these results indicated that circBNC2 knockdown inhibited HCC cell growth in vivo.
Fig. 7.
CircBNC2 inhibition reduced HCC growth in vivo. (A) The growth curve of xenograft tumor in the sh-circBNC2 and sh-NC groups. Mice weight (B) and tumor weight (C) were evaluated in the sh-circBNC2 and sh-NC groups. (D) The expression of circBNC2 and miR-217 in the sh-circBNC2 and sh-NC groups were examined by qRT-PCR. The serum levels of ALT (E) and AST (F) in the sh-circBNC2 and sh-NC groups were examined by ELISA. (G) Representative H&E stain images of tumor foci of liver sample and positive staining rates of HMGA2, PCNA, HK2 and OCT4 in the sh-circBNC2 and sh-NC groups were examined by immunohistochemistry. N = 6 per group. **p < 0.01, ***p < 0.001 vs. sh-NC.
4. Discussion
Currently, glycolysis has been implicated with the progression of malignant tumors, including HCC, and the abnormal glucose metabolism may contribute to tumor growth and malignant behavior of HCC [22]. In this study, we identified a glycolysis-associated circRNA, circ-BNC2, with a significant upregulation in HCC tissues and cell lines, which was closely related to aggressive clinical pathological features as well as poor prognosis. Functional assays indicated that circBNC2 was a driver of HCC cell viability and glycolysis both in vitro and in vivo. Notably, circBNC2 could elevate the HMGA2 expression by sponging miR-217. Our findings reveal the essential biological effect of circBNC2 in HCC and suggest that deregulation of circBNC2 may be responsible for the malignant progression of HCC.
Accumulating evidences have supported that circRNAs act as the important regulators in glycolysis and progression of HCC. For instance, exosome circFBLIM1 promoted HCC progression and glycolysis via regulating the miR-338/LRP6 axis [23]. Circ_0004913 could inhibit cell growth, metastasis, and glycolysis in HCC via regulating the miR-184/HAMP axis [24]. Fang et al. [25] demonstrated that circ_0046599 knockdown suppressed HCC cell viability, motion and glycolysis by absorbing miR-1258 to regulate RPN2. Circ-PRMT5 silence could repress viability and glycolysis of HCC cells by modulating the miR-18-5p and HK2 axis [26]. These findings indicate that circRNAs play the critical roles in tumor growth and glycolysis in HCC.
The previous study found that circBNC2 was significantly downregulated in the epithelial ovarian cancer and could be a potential diagnostic biomarker for epithelial ovarian cancer [11,12]. Herein, we confirmed that the level of circBNC2 was elevated in HCC, and high level of circBNC2 was associated with poor prognosis and advanced-stage, indicating that circBNC2 might function as a prognostic biomarker for HCC. ERK1 and ERK2 are the downstream components of a phosphorelay pathway that conveys growth and mitogenic signals largely channelled by the small RAS GTPases [27]. PCNA is one of the key viability-related proteins in cancers [28]. In this study, we demonstrated an inhibitory effect of circBNC2 silence on cell viability and tumor growth. As an important feature of cancers, glycolysis contributes to the development of HCC. Glucose uptake, lactate production and ATP level are the three indicators of glycolysis, and HK2 is an important enzyme in the glycolysis pathway [29]. In this study, circBNC2 knockdown retarded glycolysis by downregulating the levels of glucose uptake, lactate production and ATP. Besides, OCT4 is one of the stemness-associated transcription factors in cancers [30]. Through detecting sphere-forming ability and the OCT4 expression, the current findings revealed that circBNC2 down-regulation restrained the HCC stemness. In addition, knockdown of circBNC2 induced HCC cell cycle arrest and apoptosis. More importantly, a significant reduction of tumor volume and weight was observed in sh-circBNC2 mice. Collectively, circBNC2 knockdown served as an anti-cancer role in HCC in vitro and in vivo, and this was the first report of carcinogenicity of circBNC2 in HCC.
Emerging evidences have confirmed that circRNAs can regulate cell function and pathological responses by acting as the miRNA sponges to regulate the gene expression. For instance, circRNA-5692 could inhibit the progression of HCC by modulating the miR-328-5p/DAB2IP axis [31]. CircMTO1 weakened HCC cell proliferation, invasion and tumor growth by sponging miR-9 to promote p21 expression [32]. CircSETD3 acted as a sponge for micR-421 and negatively modulated miR-421 to alleviate the HCC growth [33]. Herein, we demonstrated that circBNC2 up-regulated the HMGA2 expression via sponging miR-217. MiR-217 has been validated to poorly express in many cancers and plays an anti-cancer role in the progression of cancers [34]. For example, a previous study confirmed that miR-217 weakened thyroid cancer cell proliferation and motility via inhibiting AKT3 expression [35]. Furthermore, miR-217 overexpression inhibited cell viability, motion and increased cell apoptosis in glioma [34]. More importantly, miR-217 was downregulated in HCC, and its upregulation could inhibit invasion of HCC cells [36]. Therefore, there is a reason to believe that miR-217 may function an anti-cancer role in the process of HCC. Similar with the previous study [17,18], this study demonstrated a low expression of miR-217 in HCC. Importantly, miR-217 expression was negatively regulated by circBNC2 in vitro and in vivo in HCC. Moreover, overexpression of miR-217 reversed the inhibitory effects of circBNC2 silence on HCC cell growth, glycolysis, stemness and levels of PCNA, HK2 and OCT4, implying that circBNC2 regulated the HCC development by sponging miR-217.
HMGA2 is a member of HMGA family that binds to AT-rich regions in DNA, altering the chromatin architecture [37]. HMGA2 is usually highly expressed and associated with the malignant progression of cancers. For example, HMGA2 expression was increased in lung cancer, indicating a promising molecular marker for lung cancer [38]. For research in cervical cancer, HMGA2 silencing suppressed the epithelial-mesenchymal transition and lymph node metastasis [39]. Overexpression of HMGA2 was closely related to occurrence of epithelial-mesenchymal transition in bladder cancer [40]. Moreover, induction of the HMGA2 expression by circSHKBP1 facilitated the cell proliferation, invasion, angiogenesis and stem cell-like properties in laryngeal squamous cell carcinoma [41]. Notably, Xu et al. [42] demonstrated that HMGA2 was up-regulated in HCC, and it could be involved in the inhibition of cell proliferation, glycolysis and tumor growth regulated by the circZFR/miR-375 axis. Therefore, HMGA2 always served as an oncogenic factor to promote the tumor progression. Our study presented that HMGA2 was upregulated in HCC, which analogously with a previous report [42]. Importantly, HMGA2 was predicted and confirmed to be a target of miR-217. Overexpression of HMGA2 overturned the inhibitory effect of circBNC2 knockdown on the HCC development and levels of PCNA, HK2 and OCT4. These findings suggested that circBNC2 could promote the HMGA2 expression via sponging miR-217.
5. Conclusions
Together, we first identified circBNC2 as an oncogene in HCC. Importantly, circBNC2 could sponge miR-217 to up-regulate the HMGA2 expression, thereby driving the HCC stemness, malignancy and glycolysis. This study indicates that circBNC2 may become a promising prognostic biomarker and therapeutic target for HCC. Notably, the characterization of the circBNC2/miR-217/HMGA2 axis provides an important insight for the HCC stemness and tumorigenesis. However, the effect of circBNC2 on other cellular functions need to be investigated, such as cell motility, oxidative stress, ferroptosis or immune response. Besides, this study only explored the effect of circBNC2 knockdown on HCC in animal experiments, the role of circBNC2/miR-217/HMGA2 axis in HCC development need to be further addressed in vivo.
Author contribution statement
Yan Feng: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Shufeng Xia: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Junlan Hui: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yan Xu: Conceived and designed the experiments.
Funding statement
This study did not receive any specific funding or grant.
Data availability statement
Data will be made available on request.
Statement
The preprint of this article can be found in Researchsquare (https://www.researchsquare.com/article/rs-2810839/v1).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgment
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17120.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- 1.Villanueva A. Hepatocellular carcinoma. N. Engl. J. Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Anwanwan D., Singh S.K., Singh S., Saikam V., Singh R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Canc. 2020;1873 doi: 10.1016/j.bbcan.2019.188314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viscardi G., Tralongo A.C., Massari F., Lambertini M., Mollica V., Rizzo A., Comito F., Di Liello R., Alfieri S., Imbimbo M., Della Corte C.M., Morgillo F., Simeon V., Lo Russo G., Proto C., Prelaj A., De Toma A., Galli G., Signorelli D., Ciardiello F., Remon J., Chaput N., Besse B., de Braud F., Garassino M.C., Torri V., Cinquini M., Ferrara R. Comparative assessment of early mortality risk upon immune checkpoint inhibitors alone or in combination with other agents across solid malignancies: a systematic review and meta-analysis. Eur. J. Cancer. 2022;177:175–185. doi: 10.1016/j.ejca.2022.09.031. [DOI] [PubMed] [Google Scholar]
- 4.Rizzo A., Cusmai A., Gadaleta-Caldarola G., Palmiotti G. Which role for predictors of response to immune checkpoint inhibitors in hepatocellular carcinoma? Expet Rev. Gastroenterol. Hepatol. 2022;16:333–339. doi: 10.1080/17474124.2022.2064273. [DOI] [PubMed] [Google Scholar]
- 5.Rizzo A., Ricci A.D., Di Federico A., Frega G., Palloni A., Tavolari S., Brandi G. Predictive biomarkers for checkpoint inhibitor-based immunotherapy in hepatocellular carcinoma: where do we stand? Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.803133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee E.C.S., Elhassan S.A.M., Lim G.P.L., Kok W.H., Tan S.W., Leong E.N., Tan S.H., Chan E.W.L., Bhattamisra S.K., Rajendran R., Candasamy M. The roles of circular RNAs in human development and diseases. Biomed. Pharmacother. 2019;111:198–208. doi: 10.1016/j.biopha.2018.12.052. [DOI] [PubMed] [Google Scholar]
- 7.Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., Wu Y.M., Dhanasekaran S.M., Engelke C.G., Cao X., Robinson D.R., Nesvizhskii A.I., Chinnaiyan A.M. The landscape of circular RNA in cancer. Cell. 2019;176:869–881 e813. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J., Xu Q.G., Wang Z.G., Yang Y., Zhang L., Ma J.Z., Sun S.H., Yang F., Zhou W.P. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018;68:1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Huang X., Li Z., Zhang Q., Wang W., Li B., Wang L., Xu Z., Zeng A., Zhang X., Zhang X., He Z., Li Q., Sun G., Wang S., Li Q., Wang L., Zhang L., Xu H., Xu Z. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer. 2019;18:71. doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winham S.J., Armasu S.M., Cicek M.S., Larson M.C., Cunningham J.M., Kalli K.R., Fridley B.L., Goode E.L. Genome-wide investigation of regional blood-based DNA methylation adjusted for complete blood counts implicates BNC2 in ovarian cancer. Genet. Epidemiol. 2014;38:457–466. doi: 10.1002/gepi.21815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ning L., Long B., Zhang W., Yu M., Wang S., Cao D., Yang J., Shen K., Huang Y., Lang J. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int. J. Oncol. 2018;53:2637–2646. doi: 10.3892/ijo.2018.4566. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y., Zhu Y., Zhang W., Lang J., Ning L. Utility of plasma circBNC2 as A diagnostic biomarker in epithelial ovarian cancer. OncoTargets Ther. 2019;12:9715–9723. doi: 10.2147/OTT.S211413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong Y., Du Y., Yang X., Mo Y., Fan C., Xiong F., Ren D., Ye X., Li C., Wang Y., Wei F., Guo C., Wu X., Li X., Li Y., Li G., Zeng Z., Xiong W. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang H., Heo S., Shin J.J., Ji E., Tak H., Ahn S., Lee K.J., Lee E.K., Kim W. A miR-194/PTBP1/CCND3 axis regulates tumor growth in human hepatocellular carcinoma. J. Pathol. 2019;249:395–408. doi: 10.1002/path.5325. [DOI] [PubMed] [Google Scholar]
- 15.Liao Z.B., Tan X.L., Dong K.S., Zhang H.W., Chen X.P., Chu L., Zhang B.X. miRNA-448 inhibits cell growth by targeting BCL-2 in hepatocellular carcinoma. Dig. Liver Dis. 2019;51:703–711. doi: 10.1016/j.dld.2018.09.021. [DOI] [PubMed] [Google Scholar]
- 16.Ye J., Zhang R., Du X., Chai W., Zhou Q. Long noncoding RNA SNHG16 induces sorafenib resistance in hepatocellular carcinoma cells through sponging miR-140-5p. OncoTargets Ther. 2019;12:415–422. doi: 10.2147/OTT.S175176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H., Ke J., Guo Q., Barnabo Nampoukime K.P., Yang P., Ma K. Long non-coding RNA CRNDE promotes the proliferation, migration and invasion of hepatocellular carcinoma cells through miR-217/MAPK1 axis. J. Cell Mol. Med. 2018;22:5862–5876. doi: 10.1111/jcmm.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M., Li M., Li N., Zhang Z., Liu N., Han X., Liu Q., Liao C. miR-217 suppresses proliferation, migration, and invasion promoting apoptosis via targeting MTDH in hepatocellular carcinoma. Oncol. Rep. 2017;37:1772–1778. doi: 10.3892/or.2017.5401. [DOI] [PubMed] [Google Scholar]
- 19.Tian Y.W., Shen Q., Jiang Q.F., Wang Y.X., Li K., Xue H.Z. Decreased levels of miR-34a and miR-217 act as predictor biomarkers of aggressive progression and poor prognosis in hepatocellular carcinoma. Minerva Med. 2017;108:108–113. doi: 10.23736/S0026-4806.16.04616-4. [DOI] [PubMed] [Google Scholar]
- 20.Gao H.B., Gao F.Z., Chen X.F. MiRNA-1179 suppresses the metastasis of hepatocellular carcinoma by interacting with ZEB2. Eur. Rev. Med. Pharmacol. Sci. 2019;23:5149–5157. doi: 10.26355/eurrev_201906_18179. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z., Zhou Y., Liang G., Ling Y., Tan W., Tan L., Andrews R., Zhong W., Zhang X., Song E., Gong C. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10:55. doi: 10.1038/s41419-018-1287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wani N.A., Zhang B., Teng K.Y., Barajas J.M., Motiwala T., Hu P., Yu L., Bruschweiler R., Ghoshal K., Jacob S.T. Reprograming of glucose metabolism by zerumbone suppresses hepatocarcinogenesis. Mol. Cancer Res. 2018;16:256–268. doi: 10.1158/1541-7786.MCR-17-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai Z., Wei T., Li Q., Wang X., Zhang Y., Zhang S. Exosomal circFBLIM1 promotes hepatocellular carcinoma progression and glycolysis by regulating the miR-338/LRP6 axis. Cancer Biother. Radiopharm. 2020 doi: 10.1089/cbr.2020.3564. [DOI] [PubMed] [Google Scholar]
- 24.Wu M., Sun T., Xing L. Circ_0004913 inhibits cell growth, metastasis, and glycolysis by absorbing miR-184 to regulate HAMP in hepatocellular carcinoma. Cancer Biother. Radiopharm. 2020 doi: 10.1089/cbr.2020.3779. [DOI] [PubMed] [Google Scholar]
- 25.Fang Q., Liu H., Zhou A., Zhou H., Zhang Z. Circ_0046599 promotes the development of hepatocellular carcinoma by regulating the miR-1258/RPN2 network. Cancer Manag. Res. 2020;12:6849–6860. doi: 10.2147/CMAR.S253510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding Z., Guo L., Deng Z., Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann. Hepatol. 2020;19:269–279. doi: 10.1016/j.aohep.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Lavoie H., Gagnon J., Therrien M. ERK signalling: a master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 2020;21:607–632. doi: 10.1038/s41580-020-0255-7. [DOI] [PubMed] [Google Scholar]
- 28.Jurikova M., Danihel L., Polak S., Varga I. Ki67, PCNA, and MCM proteins: markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Sheng H., Tang W. Glycolysis inhibitors for anticancer therapy: a review of recent patents. Recent Pat. Anti-Cancer Drug Discov. 2016;11:297–308. doi: 10.2174/1574892811666160415160104. [DOI] [PubMed] [Google Scholar]
- 30.Gkountela S., Castro-Giner F., Szczerba B.M., Vetter M., Landin J., Scherrer R., Krol I., Scheidmann M.C., Beisel C., Stirnimann C.U., Kurzeder C., Heinzelmann-Schwarz V., Rochlitz C., Weber W.P., Aceto N. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176:98–112 e114. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao W., Zhang X., Yang W., Dou D., Zhang H., Tang Y., Zhong W., Meng J., Bai Y., Liu Y., Yang L., Chen S., Liu H., Yang C., Sun T. Prim-O-glucosylcimifugin enhances the antitumour effect of PD-1 inhibition by targeting myeloid-derived suppressor cells. J Immunother Cancer. 2019;7:231. doi: 10.1186/s40425-019-0676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M., Li N., Zhou W., Yu Y., Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 33.Xu L., Feng X., Hao X., Wang P., Zhang Y., Zheng X., Li L., Ren S., Zhang M., Xu M. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J. Exp. Clin. Cancer Res. 2019;38:98. doi: 10.1186/s13046-019-1041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng J., Liu X., Xue Y., Gong W., Ma J., Xi Z., Que Z., Liu Y. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J. Hematol. Oncol. 2017;10:52. doi: 10.1186/s13045-017-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y., Cheng K., Wang T., Xie Q., Chen M., Chen Q., Wen Q. miR-217 inhibits proliferation, migration, and invasion via targeting AKT3 in thyroid cancer. Biomed. Pharmacother. 2017;95:1718–1724. doi: 10.1016/j.biopha.2017.09.074. [DOI] [PubMed] [Google Scholar]
- 36.Su J., Wang Q., Liu Y., Zhong M. miR-217 inhibits invasion of hepatocellular carcinoma cells through direct suppression of E2F3. Mol. Cell. Biochem. 2014;392:289–296. doi: 10.1007/s11010-014-2039-x. [DOI] [PubMed] [Google Scholar]
- 37.Fusco A., Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 38.Meyer B., Loeschke S., Schultze A., Weigel T., Sandkamp M., Goldmann T., Vollmer E., Bullerdiek J. HMGA2 overexpression in non-small cell lung cancer. Mol. Carcinog. 2007;46:503–511. doi: 10.1002/mc.20235. [DOI] [PubMed] [Google Scholar]
- 39.Wang W.Y., Cao Y.X., Zhou X., Wei B., Zhan L., Fu L.T. HMGA2 gene silencing reduces epithelial-mesenchymal transition and lymph node metastasis in cervical cancer through inhibiting the ATR/Chk1 signaling pathway. Am. J. Transl. Res. 2018;10:3036–3052. [PMC free article] [PubMed] [Google Scholar]
- 40.Ding X., Wang Y., Ma X., Guo H., Yan X., Chi Q., Li J., Hou Y., Wang C. Expression of HMGA2 in bladder cancer and its association with epithelial-to-mesenchymal transition. Cell Prolif. 2014;47:146–151. doi: 10.1111/cpr.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mansoori B., Duijf P.H.G., Mohammadi A., Najafi S., Roshani E., Shanehbandi D., Hajiasgharzadeh K., Shirjang S., Ditzel H.J., Kazemi T., Mokhtarzadeh A., Gjerstorff M.F., Baradaran B. Overexpression of HMGA2 in breast cancer promotes cell proliferation, migration, invasion and stemness. Expert Opin. Ther. Targets. 2020:1–11. doi: 10.1080/14728222.2020.1736559. [DOI] [PubMed] [Google Scholar]
- 42.Xu R., Yin S., Zheng M., Pei X., Ji X. Circular RNA circZFR promotes hepatocellular carcinoma progression by regulating miR-375/HMGA2 Axis. Dig. Dis. Sci. 2021 doi: 10.1007/s10620-020-06805-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.