Abstract
Head and neck carcinoma (HNSC) is often diagnosed at advanced stage, incurring poor patient outcome. Despite of advances in chemoradiation and surgery approaches, limited improvements in survival rates of HNSC have been observed over the last decade. Accumulating evidences have demonstrated the importance of microRNAs (miRNAs) in carcinogenesis. In this context, we sought to identify a miRNA signature associated with the survival time in patients with HNSC. This study proposed a survival estimation method called HNSC-Sig that identified a miRNA signature consists of 25 miRNAs associated with the survival in 133 patients with HNSC. HNSC-Sig achieved 10-fold cross validation a mean correlation coefficient and a mean absolute error of 0.85 ± 0.01 and 0.46 ± 0.02 years, respectively, between actual and estimated survival times. The survival analysis revealed that five miRNAs, hsa-miR-3605-3p, hsa-miR-629-3p, hsa-miR-3127-5p, hsa-miR-497-5p, and hsa-miR-374a-5p, were significantly associated with prognosis in patients with HNSC. Comparing the relative expression difference of top 10 prioritized miRNAs, eight miRNAs, hsa-miR-629-3p, hsa-miR-3127-5p, hsa-miR-221-3p, hsa-miR-501-5p, hsa-miR-491-5p, hsa-miR-149-3p, hsa-miR-3934-5p, and hsa-miR-3170, were significantly expressed between cancer and normal groups. In addition, biological relevance, disease association, and target interactions of the miRNA signature were discussed. Our results suggest that identified miRNA signature have potential to serve as biomarker for diagnosis and clinical practice in HNSC.
Keywords: Machine learning, miRNA signature, Cancer survival, Head and neck cancer
1. Introduction
Head and neck squamous cell carcinomas (HNSCs) are a heterogenic malignancies that can involve multiple cellular origins and sites within the head and neck region. HNSCs are the most common malignancy accounting for nearly 430,000 deaths worldwide each year [1]. The major risk factors associated with HNSCs are including, tobacco, alcohol consumption [2] and human papillomavirus (HPV) infection [3]. Treatment of HNSCs involves chemotherapy, radiotherapy and surgical eradication. However, the heterogeneity of HNSCs, diversity of treatment approaches and individual patient response to these variables, make it difficult to define the outcome of the treatment. Though, advancements in the treatment strategies, concerning aged patients and individual response, these modalities affects quality of life [4,5] and radiotherapy resistance remains a major challenge for HNSC treatment. The toxic nature of treatment and poor survival rates, highlighting the need for effective therapies using the specific biomarkers to improve the treatment outcome.
MicroRNA (miRNA) brings new insights to cancer pathobiology and involves in the regulation of gene expression [6]. MiRNAs take part in many processes that are crucial for cancer progression, such as proliferation, migration and apoptosis. Several studies demonstrated that miRNA expression is dysregulated in human cancers [7,8]. There are various studies that have identified miRNAs for the prediction of progression in HNSC and explored the roles of miRNAs as biomarkers in cancer detection and prognosis. Upregulation of miR-21, miR-200c and miR-34a and downregulation of miR-375 was observed in tumors of HNSCs [9]. Inhibition of miR-16 suppress cell migration in laryngeal cancer cell line and targets zyxin and promotes cell mobility [10]. The expression of miR-375 is associated with localization of the tumors and alcohol consumption in patients with laryngeal squamous cell carcinoma [11]. Hsa-miR-21 expression was consistently reported to be significantly associated with poor survival in HNSCs [12]. A meta-analysis on HNSCs identified significant miRNAs that are up and downregulated in HNSCs and decreased expression of 26 miRNAs were associated with poor survival [13]. Decreased expression level of serum miR-9 was found to be associated with poor prognosis in patients with oral squamous cell carcinoma [14]. Altered expression of miRNAs were used to predict recurrence in patients with HNSC [15]. Expression levels of hsa-miR-205, hsa-let-7d and hsa-miR-616 were associated with poor survival in patients with HNSC [16,17]. Some miRNAs are identified as potential biomarkers in HNSC [18]. Additionally, there are some miRNAs that are associated with risk factors and treatment modalities of HNSCs, such as HPV associated miRNAs [19] and radiotherapy associated extra cellular miRNAs [20]. Therefore, miRNAs have been using as prognostic biomarker in HNSC.
Computational models have been using for prediction of prognosis in HNSCs. For instance, Bryce et al. developed an artificial neural network (ANN) model to predict survival in patients with HNSCs using clinical factors [21]. Recently, Jurmeister et al. developed machine learning-based models to distinguish lung cancer and HNSCs using DNA methylation data, in which support vector machine (SVM)-based model obtained a promising predictive accuracy for classifying HNSCs [22]. Reddy et al. utilized Random Forest, gradient boosted decision trees, and logistic regression models to predict the acute radiation toxicities in patients with HNSC [23]. Jiang et al. used ridge logistic regression to predict xerostomia after radiation therapy in patients with HNSC [24]. A set of machine learning models were used to detect the cancer in surgical specimens from 36 patients with HNSC [25]. A deep learning based model has been used to detect HNSC from computed tomography (CT) scan images [26]. A convolutional neural network-based frame work was utilized to predict patient outcome based on their CT images [27]. However, machine learning models based on miRNA expression profiles for estimating survival in HNSC are limited. Accordingly, our purpose is to utilize machine learning model to identify a miRNA signature that associated with survival patients with HNSC.
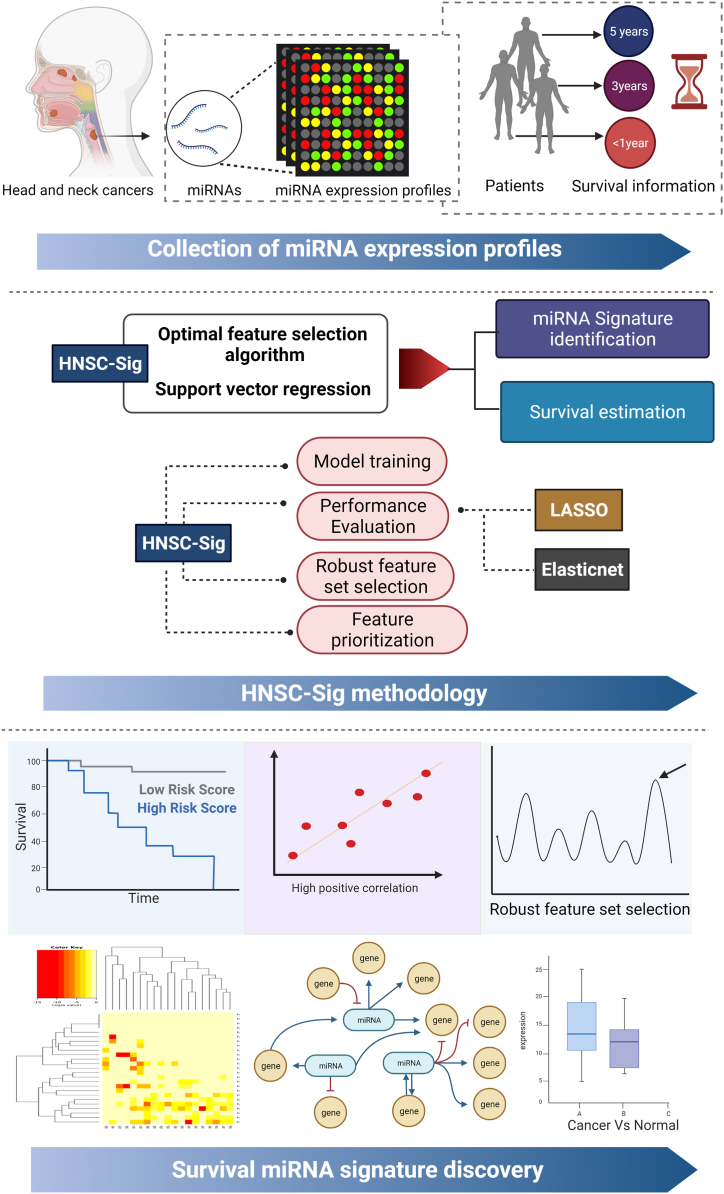
In this study, we utilized an optimized support vector regression (SVR) model incorporated with inheritable bi-objective combinatorial genetic algorithm (IBCGA) as a feature selection algorithm to estimate the survival time as well as identify a miRNA signature that is associated with survival in patients with HNSC. The optimization technique was adopted from our previous work [[28], [29], [30]]. The major contributions of this work includes identifying a set of survival associated miRNA signature; robust feature set selection using feature appearance score (FAS); survival validation of the identified miRNAs using Kaplan-Meier (KM) survival analysis; biological relevance of the miRNAs was analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) annotations; and relative miRNA expression difference and coexpression analysis of the miRNAs in HNSC and their importance in cancers were discussed further.
The manuscript is organized as follows: Section 1 provides background and related work, Section 2 describes the dataset, development of the HNSC-Sig method, and the bioinformatics tools employed in the study. Section 3 covers the identification of the miRNA signature for estimating survival time in HNSC patients, biological pathway analysis of the miRNA signature, gene ontology annotations, gene targets, miRNA disease association, and expression difference analysis. Section 4 discusses the roles of the identified miRNA signature in HNSC, the potential and limitations of utilizing miRNA signatures in cancer diagnosis and prognosis, and concludes with final remarks.
2. Materials and methods
2.1. Dataset
Initially, the dataset consisted of 523 HNSC samples were retrieved from TCGA database, and the miRNA profiling was performed using the Illumina HiSeq 2000 miRNA sequencing platform. We filtered the dataset based on the availability of survival information and miRNA expression profiling. Finally, the dataset consisted of 133 patients with 498 miRNA expression profiles of HNSC were further considered for the analysis.
2.2. HNSC-Sig
We establish the HNSC-Sig method based on SVR incorporated with optimal feature selection algorithm called IBCGA. The optimization technique has been successfully applied in various cancer survival estimations [28,[31], [32], [33], [34], [35]]. HNSC-Sig was designed to identify a survival associated miRNA signature as well as estimate the survival time in patients with HNSC. Recent years, SVMs playing increasingly prominent roles in solving several biological problems, especially in cancer prognosis and survival predictions [36]. The SVR implemented in this study was used from the LibSVM package [37].
2.2.1. An optimal feature selection method IBCGA
Feature selection algorithms are crucial in reducing dimensionality when working with datasets that have a large number of features. Gu et al. proposed a new method for transient stability assessment of power systems, combining kernelized fuzzy rough sets and the memetic algorithm [38]. Li et al. proposed a novel OS-extreme learning machine with binary Jaya-based feature selection [39]. In this study, we used optimal feature selection algorithm IBCGA, which is potential at solving large parameter optimization problems and can select minimum set of miRNAs as signature from a large number of miRNA expression profiles (n = 498) while maximizing the prediction performance. Here, we used correlation coefficient (CC) [Equation (1)] and mean absolute error (MAE) as the estimation measures to evaluate the prediction performance.
| (1) |
where xi and yi are actual and are the predicted survival times of the ith miRNA, and are the corresponding means, and N is the total number of patients in the cohort. The MAE is defined in Equation (2).
| (2) |
Here, we used genetic algorithm (GA) terminology to represents chromosomes and genes. The chromosome of IBCGA comprises 498 genes and three 4-bit genes for encoding, γ, C, and ν for the ν-SVR. The encoded chromosomes were designed as described in previous studies [36,40]. The IBCGA can simultaneously obtain a set of solutions, Xr, where r = rstart, rstart + 1 … rend in a single run. The detailed steps involved in IBCGA can be found in Supplementary method.
2.2.2. Robust feature set selection
The robust set of miRNA signature among 30 independent runs in HNSC-Sig has the highest FAS obtained using the following procedure [Equation (3)].
Step 1: Perform N independent runs of HNSC-Sig by maximizing accuracy of 10-CV for obtaining N miRNA signatures. There are features in the a-th signatures, a = 1, …, N.
- Step 2: FAS is calculated as follows:
-
a)Calculate the Feature appearance score f(a) for each feature ‘a’ that ever presents in the N sets of miRNAs.
-
b)Calculate the score Pa, t = 1, …, N where si is the i-th feature in the a-th solution:
-
a)
| (3) |
Step 3: Output the a-th feature set with the highest appearance score Fa.
2.3. Biological significance analysis
KEGG pathway GO annotation analysis was performed using DIANA tools. The DIANA-microT web server provided the predicted miRNA targets for the pathway analysis. The p-value threshold was set to 0.05 and Fishers’s exact test (hypergeometric distribution) was used for the enrichment analysis.
2.4. miRNA-target interaction network
We constructed the miRNA-target interaction network using Cytoscape v3.7.2. For the prediction of target genes miRTarBase and TargetScan predictions were used presented in the CyTargetLinker application of the Cytoscape. The interaction threshold was adjusted to two units.
3. Results
3.1. MiRNA signature selection and survival estimation
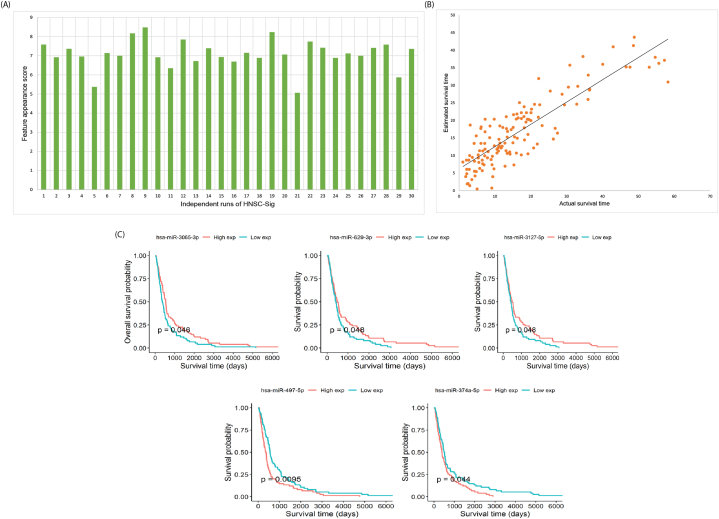
The dataset consisted of 133 patients with HNSC from The cancer genome atlas (TCGA) database. The optimized method HNSC-sig was incorporated with feature selection algorithm IBCGA to identify a set of survival miRNAs in patients with HNSC. HNSC-Sig identified an average 31 miRNAs and achieved a 10-fold cross-validation (10-CV) mean correlation coefficient and mean absolute error of 0.85 ± 0.01 and 0.46 ± 0.02 years, respectively, between actual and predicted survival time. We performed 30 independent runs of HNSC-sig to select the robust feature set. The system flow-chart of HNSC-Sig is depicted in Fig. 1. To select a robust feature set, FAS was measured for each independent run of the HNSC-Sig. The highest FAS indicates that frequency of these features are higher compared to the lower FAS. The highest FAS of HNSC-sig was 8.48 and the 25 miRNAs were selected as a signature. HNSC-sig with highest FAS obtained a correlation and a mean absolute error of 0.86 and 0.42, respectively, between actual and predicted survival time. The FAS for each independent run is shown in Fig. 2a.
Fig. 1.
System flow chart. Step 1: collection of miRNA expression data from TCGA data portal, Step: HNSC-Sig methodology, and Step 3: miRNA signature discovery by estimation of survival time, KM-survival curves, KEGG and GO annotation analysis, target network analysis, and expression difference analysis.
Fig. 2.
Robust miRNA signature identification. (A) Robust signature selection using Feature appearance score (FAS) measurement for each independent run of HNSC-Sig. The highest FAS obtained is 8.48 at the 9th independent run, (B) estimation performance of the robust miRNA signature, and (C) KM survival plots for hsa-miR-3605-3p, hsa-miR-629-3p, hsa-miR-3127-5p, hsa-miR-497-5p, and hsa-miR-374a-5p.
The prediction performance of HNSC-Sig was compared with some regression methods, such as the least absolute shrinkage and selection operator (LASSO) and elastic net. HNSC-Sig exhibited superior prediction performance compared to LAASO and elastic net methods. The comparison of prediction performance results are shown in Table 1. The prediction performance of HNSC-Sig is shown in Fig. 2b. Correlation plots for HNSC-sig, LASSO, and elastic net are shown in Supplementary Fig. S1.
Table 1.
HNSC-Sig prediction performance comparison.
| Method | Correlation coefficient | Mean absolute error in years | Features selected |
|---|---|---|---|
| LASSO | 0.62 | 0.69 | 22 |
| Elastic net | 0.82 | 0.53 | 55 |
| HNSC-Sig | 0.88 | 0.43 | 29 |
| HNSC-Sig-FAS | 0.86 | 0.42 | 25 |
| HNSC-Sig-Mean | 0.85 ± 0.01 | 0.46 ± 0.02 | 31.70 ± .75 |
The miRNAs of the signature were ranked based on main effect difference (MED) analysis [41]. The MED analysis gives score for each miRNA based on its estimation capability, hence, the miRNA with a highest MED score possess higher importance while estimating the survival time. The list of miRNAs of the signature and corresponding MED scores are shown in Supplementary Table S1.
3.2. Survival validation of the miRNAs
The miRNAs of the signature were validated using KM survival analysis. Survival outcome was explored considering p-values. A p-value < 0.05 was considered to be statistically significant. Five of the miRNA signature, hsa-miR-3605-3p, hsa-miR-629-3p, hsa-miR-3127-5p, hsa-miR-497-5p, and hsa-miR-374a-5p, were significantly associated with prognosis in patients with HNSC. These five miRNAs, hsa-miR-3605-3p, hsa-miR-629-3p, hsa-miR-3127-5p, hsa-miR-497-5p, and hsa-miR-374a-5p, obtained p-values of 0.046, 0.048, 0.048, 0.0095, and 0.044, respectively. KM survival curves for these five miRNAs are shown in Fig. 2c.
3.3. Biological pathway analysis of the miRNA signature
Biological significance of the identified miRNA signature was analyzed using KEGG pathways. The identified miRNA signature enriched in various pathways, the more significant enriched pathways (P < 0.005) including, fatty acid biosynthesis, lysine degradation, hippo signaling pathway, adherens junction, proteoglycans in cancer, fatty acid metabolism, ECM-receptor interaction, pathways in cancer, prostate cancer, glioma, steroid biosynthesis, and p53 signaling pathway. The identified miRNA signature involved in fatty acid biosynthesis (p < E−365), lysine degradation (p = 1.14E−13), hippo signaling pathway (p = 4.01E−13), adherens junction (p = 1.12E−12), proteoglycans in cancer (p = 1.98E−12), fatty acid metabolism (p = 8.20E−09), ECM-receptor interaction (p = 6.70E−08), pathways in cancer (p = 0.0006), prostate cancer (p = 0.0008), glioma (p = 0.0011), steroid biosynthesis (p = 0.0045), and p53 signaling pathway (p = 0.007) and targets 2, 20, 63, 38, 88, 9, 16, 124, 45, 28, 3, and 38 genes, respectively. The details of KEGG pathways and number of target genes are described in Table 2. The KEGG pathway enrichment analysis is depicted in Supplementary Fig. S2.
Table 2.
The miRNA signature involved in significant KEGG pathways.
| KEGG pathway | miRNAs | Target genes | p-value |
|---|---|---|---|
| Proteoglycans in cancer | 20 | 112 | 1.211E−16 |
| Adherens junction | 20 | 49 | 2.932E−12 |
| Viral carcinogenesis | 20 | 104 | 7.936E−09 |
| Hepatitis B | 19 | 78 | 2.281E−07 |
| Protein processing in endoplasmic reticulum | 21 | 95 | 8.18E−07 |
| TGF-beta signaling pathway | 17 | 47 | 9.288E−07 |
| Pancreatic cancer | 19 | 42 | 9.288E−07 |
| Bacterial invasion of epithelial cells | 20 | 48 | 9.288E−07 |
| Hippo signaling pathway | 21 | 72 | 1.983E−06 |
| Cell cycle | 21 | 70 | 4.562E−06 |
| Colorectal cancer | 20 | 41 | 6.81E−06 |
| Prostate cancer | 21 | 56 | 1.037E−05 |
| Estrogen signaling pathway | 20 | 55 | 1.306E−05 |
| Acute myeloid leukemia | 17 | 38 | 1.677E−05 |
| Glycosaminoglycan biosynthesis - keratan sulfate | 12 | 10 | 2.874E−05 |
| Pathways in cancer | 21 | 184 | 3.088E−05 |
| Focal adhesion | 21 | 108 | 3.515E−05 |
| Fatty acid biosynthesis | 9 | 5 | 4.006E−05 |
| Oocyte meiosis | 17 | 59 | 4.028E−05 |
| Glioma | 19 | 37 | 4.028E−05 |
| Endocytosis | 21 | 100 | 4.028E−05 |
| p53 signaling pathway | 19 | 43 | 6.164E−05 |
| N-Glycan biosynthesis | 14 | 27 | 6.176E−05 |
| Renal cell carcinoma | 20 | 43 | 7.457E−05 |
| Neurotrophin signaling pathway | 21 | 67 | 7.712E−05 |
| Chronic myeloid leukemia | 18 | 45 | 7.842E−05 |
| Shigellosis | 20 | 39 | 8.722E−05 |
| Prolactin signaling pathway | 18 | 42 | 0.0001111 |
| Non-small cell lung cancer | 17 | 33 | 0.0001449 |
| mTOR signaling pathway | 18 | 38 | 0.000156 |
| FoxO signaling pathway | 20 | 72 | 0.0001875 |
| Lysine degradation | 17 | 25 | 0.0001893 |
| Small cell lung cancer | 19 | 49 | 0.0003562 |
| Sphingolipid signaling pathway | 19 | 60 | 0.0003722 |
| Ubiquitin mediated proteolysis | 19 | 74 | 0.0003919 |
| Endometrial cancer | 19 | 32 | 0.0004231 |
| Insulin signaling pathway | 21 | 74 | 0.0004354 |
| AMPK signaling pathway | 20 | 67 | 0.0004477 |
| ErbB signaling pathway | 21 | 49 | 0.0004477 |
| HIF-1 signaling pathway | 20 | 58 | 0.0006434 |
| Thyroid cancer | 17 | 19 | 0.0007077 |
| Signaling pathways regulating pluripotency of stem cells | 20 | 72 | 0.0007951 |
3.4. Gene ontology annotations
GO annotations of the miRNA signature was analyzed in terms of biological processes, cellular components and molecular functions. The miRNA signature involved more significant biological pathways were epidermal growth factor receptor signaling pathway, fibroblast growth factor receptor signaling pathway, RNA metabolic process, DNA metabolic process, transcription, DNA-templated, mRNA metabolic process, immune system process, Fc-epsilon receptor signaling pathway, nucleobase-containing compound catabolic process, and Fc-gamma receptor signaling pathway involved in phagocytosis, by targeting 2120 genes.
The miRNA signature involved in more significant cellular components were protein complex, nucleoplasm, cytosol, organelle, microtubule organizing center, and focal adhesion, by targeting 6887 genes. The miRNA signature involved in various molecular functions, the most significant molecular functions includes, enzyme regulator activity, protein binding transcription factor activity, nucleic acid binding transcription factor activity, poly(A) RNA binding, enzyme binding, RNA binding, ion binding, cytoskeletal protein binding, transcription corepressor activity, and transcription factor binding, by targeting 5098 genes. The details of the miRNA signature involved in GO annotations are listed in Supplementary Table S2. The GO enrichment analysis is shown in Supplementary Fig. S3.
3.5. Identifying miRNA signature gene targets
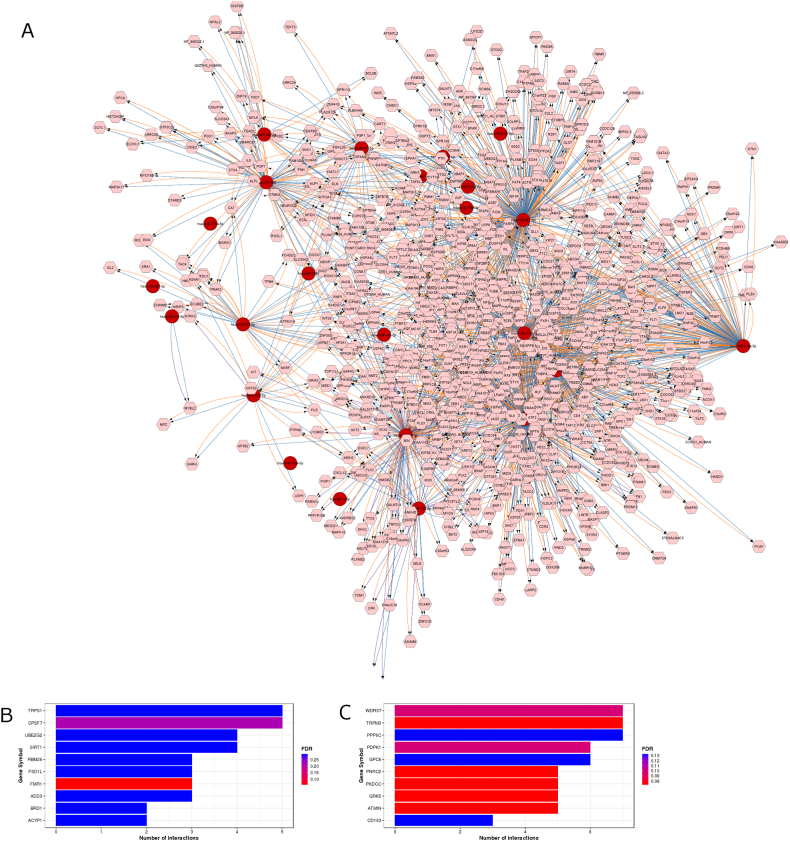
To investigate the possible genes targeted by the miRNA signature, we constructed a miRNA-target interaction network using Cytoscape v3.7.2. The miRNA-target network was constructed using predicted and validated targets by TargetScan Homo sapiens version 6.2 and miRTarBase v4.4 and MicroCosm v5. In which, miRTarBase collects target genes experimentally validated by western blot, microarrays, reporter assay, and next generation sequencing experiments [42]. TargetScan predicts biological targets of miRNAs [43], and MicroCosm contains computationally predicted targets for microRNAs, which are obtained from the miRBase sequence database and EnsEMBL.There are totally 20,832 target interactions in the network, in which TargetScan, miRTarBase and MicroCosm predicted and validated targets were 8313, 195, 12,324, respectively. For the better visualization overlap threshold was adjusted and in the constructed miRNA-target network (Fig. 3A), TargetScan, miRTarBase and MicroCosm targeted interactions were 1019, 107, and 981, respectively.
Fig. 3.
(A) miRNA-Target interaction using Cytoscape. The target genes of the miRNA signature were predicted using the miRTarBase, TargetScan, and MicroCosm. In this network, microRNAs and target genes are shown as red circles and pink rounded hexagons, respectively. The predicted microRNA–target interactions were visualized in orange, blue and violet using CyTargetLinker. (B) gene enrichment analysis predictions using miRTarBase, and (C) gene enrichment analysis predictions using TargetScan. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Additionally, statistical analysis for over-representation of miRNA-target interaction enrichment was performed using MIENTURNET [44]. The miRNA-target interaction enrichment analysis was performed for both predicted and validated interactions using TargetScan and miRTarBase. The bar chart representation of miRNA-target interaction enrichment is shown in Fig. 3B and C.
3.6. miRNA signature disease association
We predicted the miRNA-disease association of the miRNA signature using MISIM v2.0 [45], where miRNA functional similarity as weighting factors and calculates the frequencies of diseases. The miRNA signature and its weighted frequencies for HNSCs were measured and listed in Table 3.
Table 3.
The miRNA signature association with HNSCs.
| Rank | miRNA | Disease | Weighted frequency |
|---|---|---|---|
| 1 | hsa-miR-3065-3p | Head and Neck Neoplasms | 10.5936 |
| 2 | hsa-miR-629-3p | Head and Neck Neoplasms | 31.9993 |
| 3 | hsa-miR-3127-5p | Head and Neck Neoplasms | 18.8977 |
| 4 | hsa-miR-221-3p | Mouth neoplasms | 8.63 |
| 5 | hsa-miR-126-3p | Head and Neck Neoplasms | 39.1751 |
| 6 | hsa-miR-501-5p | Head and Neck Neoplasms | 20.6326 |
| 7 | hsa-miR-491-5p | Mouth Neoplasms | 6.491 |
| 8 | hsa-miR-149-3p | Mouth Neoplasms | 7.54158 |
| 9 | hsa-miR-3934-5p | NA | NA |
| 10 | hsa-miR-3170 | NA | NA |
| 11 | hsa-miR-378a-5p | Head and Neck Neoplasms | 33.9768 |
| 12 | hsa-let-7d-3p | Head and Neck Neoplasms | 36.8654 |
| 13 | hsa-miR-616-3p | Head and Neck Neoplasms | 20.5316 |
| 14 | hsa-miR-363-3p | Mouth Neoplasms | 5.50785 |
| 15 | hsa-miR-129-5p | Head and Neck Neoplasms | 37.1912 |
| 16 | hsa-miR-130a-5p | Head and Neck Neoplasms | 20.248 |
| 17 | hsa-miR-627-5p | Head and Neck Neoplasms | 21.3615 |
| 18 | hsa-miR-200c-3p | Head and Neck Neoplasms | 14.8893 |
| 19 | hsa-miR-497-5p | Head and Neck Neoplasms | 40.7023 |
| 20 | hsa-miR-141-5p | Mouth Neoplasms | 7.89423 |
| 21 | hsa-miR-374a-5p | Head and Neck Neoplasms | 28.7938 |
| 22 | hsa-miR-181c-5p | Head and Neck Neoplasms | 29.726 |
| 23 | hsa-miR-625-5p | Head and Neck Neoplasms | 37.3419 |
| 24 | hsa-miR-3193 | NA | NA |
| 25 | hsa-miR-760 | Head and Neck Neoplasms | 29.6151 |
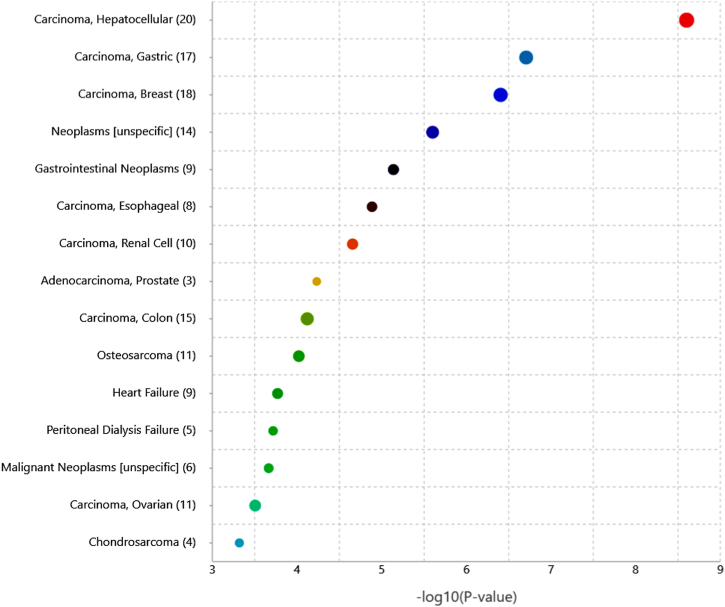
Additionally, the significant disease and functional association of the identified miRNA signature were analyzed using TAM 2.0 [46]. The top five significant associated with miRNA signature are hepatocellular carcinoma (p-value (−log10) = 8.60), gastric cancer (p-value (−log10) = 6.70), breast cancer (p-value (−log10) = 6.40), gastrointestinal neoplasms (p-value (−log10) = 5.13), and esophageal carcinoma (p-value (−log10) = 4.88). The significant disease association of the miRNA signature is shown in Fig. 4.
Fig. 4.
The miRNA signature association with various diseases. The size of bubble correlates with the count of input miRNAs.
Next, we investigated the functions of the identified miRNA signature in HNSC by examining scientific literature. Within the miRNA signature, 19 miRNAs were found to have various roles in the progression and survival of HNSC. The miRNAs and their respective roles are listed in Supplementary Table S3.
3.7. Expression difference in HNSC vs normal
The relative expression levels of the miRNA signature were compared between cancer and normal samples. Among top 10 ranked miRNAs, eight miRNAs, hsa-miR-629-3p, hsa-miR-3127-5p, hsa-miR-221-3p, hsa-miR-501-5p, hsa-miR-491-5p, hsa-miR-149-3p, hsa-miR-3934-5p, hsa-miR-3170 showed a significant difference between cancer and healthy groups with p-values of 2.99E−08, 4.47E−07, 1.07E−06, 5.97E−09, 5.34E−02, 3.60E−03, 1.62E−12, and 1.62E−12, respectively. Box plot representation of miRNA expression difference between cancer and normal groups are shown in Supplementary Fig. S4.
Further, we measured the relative expression difference of miRNA signature across different pathological stages of HNSCs. The significant expression of miRNAs were observed across different stages. The relative expression difference of the top 10 ranked miRNAs across stages of HNSC is shown in Supplementary Table S4.
4. Discussion
Identifying right biomarker could be aid in personalize and improve treatment strategies in patients with HNSC. An increasing number of studies have discovered the miRNAs as potential biomarkers for cancer prognosis, diagnosis and therapeutic targets. HNSCs are one of the major leading causes of deaths worldwide. The major complications in treating HNSC are, therapies, such as radiotherapy and chemotherapy affects the quality of life and also tumors relatively resistance to cytotoxic drugs [47]. Hence, treatment strategies has brought forward new therapeutic opportunities, where miRNAs are the key tools [48]. Identifying novel miRNA signature, and their biological functions and targets genes will advance our knowledge about the significance of miRNAs in tumorigenesis as well as developing the miRNA-based cancer prognosis and diagnosis. We utilized optimized survival estimation method HNSC-sig to identify a potential survival miRNA signature in patients with HNSCs.
HNSC-sig identified a miRNA signature consisted of 25 miRNAs that is associated with survival in patients with HNSC. The comparison of prediction performance results showed the estimation capability of HNSC-Sig was better when compared with some regression methods. We prioritize the miRNAs of the signature and analyzed further. The bioinformatics analysis revealed the miRNA signature involved in some significant pathways, biological process, cellular components, and molecular processes.
Next, we verified the roles of the top 10 ranked miRNAs using literature. The significant expression of hsa-miR-3605 was observed in laryngeal carcinoma, one of the commonly occurred HNSCs [49]. Expression difference of hsa-miR-3605-3p was observed when compared the lung squamous cell carcinomas, with a fold change of −1.9 [50]. A qRT-PCR study on laryngeal squamous cell carcinoma cells reported the up-regulation of hsa-miR-221 and its association with clinicopathelogical features of laryngeal squamous cell carcinoma [51]. Up-regulation of plasma miR-221 was also observed in larynx cancer [52]. Differential expression of hsa-miR-221 was observed in oral cancer, common type of HNSCs [53]. The increased expression level of hsa-miR-629-3p was associated with the poor survival in HNSC. A significant association between miR-126 and poor prognosis was observed in oral squamous cell carcinoma (OSCC) and decreased expression of miR-126 was correlated with disease-free survival [54]. The overexpression of miR-126 significantly regulate the expression of epidermal growth factor-like domain 7 in OSCC cell lines [55] and acts as a tumor suppressor in OSCC cells. A meta-analysis study on HNSC demonstrated the decreased expression of miR-126 is associated with poor prognosis [56]. Recent study on HNSC cells demonstrated that expression of miR-501-5p was up-regulated and targets calcium activated chloride channel A4 resulting poor outcome in patients with HNSC [57]. Hsa-miR-491 targets G-protein-coupled receptor kinase-interacting protein 1 and regulate migration and metastasis of OSCC cells, and lower expression of miR-419-5p was associated with poor survival in patients with OSCC [58]. A follow-up study on 97 laryngeal squamous cell carcinoma patients reported that lower expression of miR-149 was associated with the shorter survival [59]. The expression of miR-149 was found to be down-regulated and associated with poorer survival in patients with HNSC [60]. An array analysis study on laryngeal squamous cell carcinoma revealed an expression of miR-149a was up-regulated when compared to the healthy samples [61]. Hsa-miR-3170 is one of the novel seven-miRNA prognostic model in HNSC, which was identified by LASSO and Cox regression analyses.
Among top 10 ranked miRNAs, eight miRNAs (hsa-miR-3065, hsa-miR-221, hsa-miR-629, hsa-miR-126, hsa-miR-501, hsa-miR-491, hsa-miR-149, and hsa-miR-3170) were involved in HNSC as well as other cancers. Two miRNAs, hsa-miR-3127 and hsa-miR-3934 were not involved in HNSC, but they are actively expressed in other cancer types. For instance, hsa-miR-629 targets FOXO3 and promotes cancer progression in pancreatic carcinoma [62]. The expression of hsa-miR-3127 was upregulated in hepatocellular carcinoma cells and promotes tumorigenicity and proliferation in hepatocellular carcinoma cells [63]. The downregulation of hsa-miR-3934-5p regulated proliferation and apoptosis in non-small cell lung carcinoma cells [64]. Hence, the participation of these three miRNAs, hsa-miR-629, hsa-miR-3127, and hsa-miR-3934, were not previously reported in HNSC, we assume that these are the new targets to explore their roles in HNSC.
Future research on miRNA signatures in HNSC should focus on several promising directions to advance our understanding and clinical applications [[65], [66], [67]]. Firstly, there is a need to investigate novel miRNAs that have not been extensively studied in this context. Utilizing high-throughput sequencing technologies, researchers can profile the expression and functional significance of these less characterized miRNAs. Secondly, integrating miRNA data with other omics data, such as gene expression profiles, DNA methylation patterns, and proteomic data, can provide a comprehensive molecular landscape of HNSC, allowing for the identification of novel regulatory networks and key molecular players. Longitudinal studies tracking miRNA expression changes throughout the course of treatment and disease progression can offer insights into the dynamic nature of miRNA regulation and its association with treatment response, therapy resistance, and disease recurrence [68,69]. Furthermore, conducting functional characterization and mechanistic studies to unravel the biological mechanisms underlying miRNA-mediated effects in HNSC is essential. Another important direction is the development of non-invasive diagnostic tools utilizing miRNAs present in body fluids such as blood, saliva, and urine, which show potential as minimally invasive biomarkers for cancer detection and monitoring. Lastly, translating miRNA signatures into clinical practice requires conducting large-scale clinical trials to validate their prognostic value, assess predictive accuracy, and determine their impact on treatment decision-making and patient outcomes.
The use of miRNA signatures as prognostic markers for HNSC has shown promise in research studies. However, there are some limitations and challenges associated with their application, which need to be addressed for their effective utilization in clinical settings [70,71]. A key limitation is the inherent heterogeneity of miRNA expression within HNSC, which can vary across different subtypes and individual patients. This heterogeneity poses a challenge in developing an applicable miRNA signature that can accurately predict prognosis for all patients. The lack of standardized protocols for miRNA detection and quantification hampers the reproducibility and comparability of results across different studies. Future research should aim to establish standardized methodologies for sample collection, RNA isolation, miRNA profiling, and data analysis to ensure consistent and reliable results. Many miRNA signature studies in HNSC have been conducted with relatively small sample sizes, which limits the statistical power and generalizability of the findings. Future studies should involve larger cohorts and independent validation sets to ensure the robustness and reliability of miRNA signatures as prognostic markers. Addressing these limitations and challenges would enhance the potential of miRNA signatures as prognostic markers for HNSC patients.
Before miRNA signatures can be translated into clinical practice, several challenges related to clinical implementation need to be overcome. These challenges include the development of standardized assays, the establishment of cost-effectiveness, and the feasibility of incorporating miRNA signatures into routine diagnostic settings. Clinical trials and translational studies are necessary to evaluate the clinical utility, reliability, and potential impact of miRNA signatures as prognostic markers for HNSC patients. In conclusion, our findings provide an important resource for the exploration of survival specific transcripts that may guide target-based therapeutic strategies and future discoveries in HNSCs.
Author contributions
Srinivasulu Yerukala Sathipati (SYS) and Shinn-Ying Ho (SYH) designed the system, participated in manuscript preparation, and carried out the detailed study. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Availability of data and materials
All the data used in this analysis can be found at TCGA data portal [https://cancergenome.nih.gov/].
Funding
This work was supported by grants from National Science and Technology Council, Taiwan ((110-2221-E−A49-099-MY3–, 111-2740-B-400-002–), and was financially supported by the “Center For Intelligent Drug Systems and Smart Bio-devices (IDS2B)” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. This work was also supported in part by the Marshfield Clinic Research Institute, Marshfield, WI. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17218.
Contributor Information
Srinivasulu Yerukala Sathipati, Email: sathipathi.srinivasulu@marshfieldclinic.org.
Shinn-Ying Ho, Email: syho@nctu.edu.tw.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bray F., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Maier H., et al. Tobacco and alcohol and the risk of head and neck cancer. Clin. Invest. 1992;70(3–4):320–327. doi: 10.1007/BF00184668. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi A.K., et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 2011;29(32):4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.List M.A., Bilir S.P. Functional outcomes in head and neck cancer. Semin. Radiat. Oncol. 2004;14(2):178–189. doi: 10.1053/j.semradonc.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Bjordal K., Kaasa S. Psychological distress in head and neck cancer patients 7-11 years after curative treatment. Br. J. Cancer. 1995;71(3):592–597. doi: 10.1038/bjc.1995.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019;20(1):21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S., Gregory R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takamizawa J., et al. Reduced expression of the <strong><em>let-7</em></strong> MicroRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 9.Kalfert D., et al. MicroRNA profile in site-specific head and neck squamous cell cancer. Anticancer Res. 2015;35(4):2455–2463. [PubMed] [Google Scholar]
- 10.Wu H., et al. MicroRNA-16 targets zyxin and promotes cell motility in human laryngeal carcinoma cell line HEp-2. IUBMB Life. 2011;63(2):101–108. doi: 10.1002/iub.417. [DOI] [PubMed] [Google Scholar]
- 11.Hu A., et al. miR-21 and miR-375 microRNAs as candidate diagnostic biomarkers in squamous cell carcinoma of the larynx: association with patient survival. Am. J. Transl. Res. 2014;6(5):604–613. [PMC free article] [PubMed] [Google Scholar]
- 12.Avissar M., et al. MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis. 2009;30(12):2059–2063. doi: 10.1093/carcin/bgp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lubov J., et al. Meta-analysis of microRNAs expression in head and neck cancer: uncovering association with outcome and mechanisms. Oncotarget. 2017;8(33):55511–55524. doi: 10.18632/oncotarget.19224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun L., et al. Association of decreased expression of serum miR-9 with poor prognosis of oral squamous cell carcinoma patients. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2016;22:289–294. doi: 10.12659/MSM.895683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganci F., et al. Altered peritumoral microRNA expression predicts head and neck cancer patients with a high risk of recurrence. Mod. Pathol. 2017;30(10):1387–1401. doi: 10.1038/modpathol.2017.62. [DOI] [PubMed] [Google Scholar]
- 16.Childs G., et al. Low-level expression of MicroRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am. J. Pathol. 2009;174(3):736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vahid A., et al. 2023. PERK-Mediated Induction of miR-5p and miR-3p Arms of miR-616 Regulates Cell Growth by Targeting C-MYC. bioRxiv. 2023.03.06.531445. [Google Scholar]
- 18.Nagadia R., et al. miRNAs in head and neck cancer revisited. Cell. Oncol. (Dordr) 2013;36(1):1–7. doi: 10.1007/s13402-012-0122-4. [DOI] [PubMed] [Google Scholar]
- 19.Sannigrahi M.K., et al. Role of host miRNA hsa-miR-139-3p in HPV-16-Induced carcinomas. Clin. Cancer Res. 2017;23(14):3884–3895. doi: 10.1158/1078-0432.CCR-16-2936. [DOI] [PubMed] [Google Scholar]
- 20.Summerer I., et al. Changes in circulating microRNAs after radiochemotherapy in head and neck cancer patients. Radiat. Oncol. 2013;8:296. doi: 10.1186/1748-717X-8-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryce T.J., et al. Artificial neural network model of survival in patients treated with irradiation with and without concurrent chemotherapy for advanced carcinoma of the head and neck. Int. J. Radiat. Oncol. Biol. Phys. 1998;41(2):339–345. doi: 10.1016/s0360-3016(98)00016-9. [DOI] [PubMed] [Google Scholar]
- 22.Jurmeister P., et al. Machine learning analysis of DNA methylation profiles distinguishes primary lung squamous cell carcinomas from head and neck metastases. Sci. Transl. Med. 2019;11(509):eaaw8513. doi: 10.1126/scitranslmed.aaw8513. [DOI] [PubMed] [Google Scholar]
- 23.Reddy J.P., et al. Applying a machine learning approach to predict acute radiation toxicities for head and neck cancer patients. Int. J. Radiat. Oncol. Biol. Phys. 2019;105(1):S69. [Google Scholar]
- 24.Jiang W., et al. Machine learning methods uncover radiomorphologic dose patterns in salivary glands that predict xerostomia in patients with head and neck cancer. Adv. Radiat. Oncol. 2019;4(2):401–412. doi: 10.1016/j.adro.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu G., et al. Detection of head and neck cancer in surgical specimens using quantitative hyperspectral imaging. Clin. Cancer Res. 2017;23(18):5426–5436. doi: 10.1158/1078-0432.CCR-17-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta P., Malhi A.K. 2018. Using deep learning to enhance head and neck cancer diagnosis and classification. (2018 Ieee International Conference on System, Computation, Automation and Networking (Icscan)). [Google Scholar]
- 27.Diamant A., et al. Deep learning in head & neck cancer outcome prediction. Sci. Rep. 2019;9(1):2764. doi: 10.1038/s41598-019-39206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yerukala Sathipati S., Ho S.Y. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci. Rep. 2017;7(1):7507. doi: 10.1038/s41598-017-07739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yerukala Sathipati S., et al. MicroRNA signature for estimating the survival time in patients with bladder urothelial carcinoma. Sci. Rep. 2022;12(1):4141. doi: 10.1038/s41598-022-08082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sathipati S.Y., et al. Survival estimation in patients with stomach and esophageal carcinoma using miRNA expression profiles. Comput. Struct. Biotechnol. J. 2022;20:4490–4500. doi: 10.1016/j.csbj.2022.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yerukala Sathipati S., et al. MicroRNA signature for estimating the survival time in patients with bladder urothelial carcinoma. Sci. Rep. 2022;12(1):4141. doi: 10.1038/s41598-022-08082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yerukala Sathipati S., Ho S.-Y. Identifying a miRNA signature for predicting the stage of breast cancer. Sci. Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-34604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yerukala Sathipati S., Huang H.-L., Ho S.-Y. Estimating survival time of patients with glioblastoma multiforme and characterization of the identified microRNA signatures. BMC Genom. 2016;17(13):75–86. doi: 10.1186/s12864-016-3321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yerukala Sathipati S., Ho S.-Y. Novel miRNA signature for predicting the stage of hepatocellular carcinoma. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-71324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sathipati S.Y., et al. Artificial intelligence-driven pan-cancer analysis reveals miRNA signatures for cancer stage prediction. Hum. Genet. Genom. Adv. 2023;4(3) doi: 10.1016/j.xhgg.2023.100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yerukala Sathipati S., et al. Identification and characterization of the lncRNA signature associated with overall survival in patients with neuroblastoma. Sci. Rep. 2019;9(1):5125. doi: 10.1038/s41598-019-41553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang C.-C., Lin C.-J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. (TIST) 2011;2(3):1–27. [Google Scholar]
- 38.Gu X., Li Y., Jia J. Feature selection for transient stability assessment based on kernelized fuzzy rough sets and memetic algorithm. Int. J. Electr. Power Energy Syst. 2015;64:664–670. [Google Scholar]
- 39.Li Y., Yang Z. Application of EOS-ELM with binary jaya-based feature selection to real-time transient stability assessment using PMU data. IEEE Access. 2017;5:23092–23101. [Google Scholar]
- 40.Yerukala Sathipati S., Ho S.-Y. Identifying a miRNA signature for predicting the stage of breast cancer. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-34604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tung C.-W., Ho S.-Y. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinf. 2008;9(1):1–15. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang H.-Y., et al. miRTarBase 2020: updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2019;48(D1):D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agarwal V., et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Licursi V., et al. MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinf. 2019;20(1):545. doi: 10.1186/s12859-019-3105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., et al. MISIM v2.0: a web server for inferring microRNA functional similarity based on microRNA-disease associations. Nucleic Acids Res. 2019;47(W1):W536–W541. doi: 10.1093/nar/gkz328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., et al. TAM 2.0: tool for MicroRNA set analysis. Nucleic Acids Res. 2018;46(W1):W180–w185. doi: 10.1093/nar/gky509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 48.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 49.Lu Z.M., et al. Micro-ribonucleic acid expression profiling and bioinformatic target gene analyses in laryngeal carcinoma. OncoTargets Ther. 2014;7:525–533. doi: 10.2147/OTT.S59871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muñoz-Largacha J.A., et al. miRNA profiling of primary lung and head and neck squamous cell carcinomas: addressing a diagnostic dilemma. J. Thorac. Cardiovasc. Surg. 2017;154(2):714–727. doi: 10.1016/j.jtcvs.2017.02.071. [DOI] [PubMed] [Google Scholar]
- 51.Hussein S., et al. Up-regulated miR-221 expression as a molecular diagnostic marker in laryngeal squamous cell carcinoma and its correlation with Apaf-1 expression. Cancer Biomarkers. 2017;19(3):279–287. doi: 10.3233/CBM-160444. [DOI] [PubMed] [Google Scholar]
- 52.Yilmaz S.S., et al. MiR-221 as a pre- and postoperative plasma biomarker for larynx cancer patients. Laryngoscope. 2015;125(12):E377–E381. doi: 10.1002/lary.25332. [DOI] [PubMed] [Google Scholar]
- 53.Lopes C.B., et al. Differential expression of hsa-miR-221, hsa-miR-21, hsa-miR-135b, and hsa-miR-29c suggests a field effect in oral cancer. BMC Cancer. 2018;18(1):721. doi: 10.1186/s12885-018-4631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sasahira T., et al. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br. J. Cancer. 2012;107(4):700–706. doi: 10.1038/bjc.2012.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X., Wu H., Ling T. Suppressive effect of microRNA-126 on oral squamous cell carcinoma in vitro. Mol. Med. Rep. 2014;10(1):125–130. doi: 10.3892/mmr.2014.2171. [DOI] [PubMed] [Google Scholar]
- 56.Jamali Z., et al. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51(4):321–331. doi: 10.1016/j.oraloncology.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 57.Li B., et al. MiR-501-5p acts as an energetic regulator in head and neck squamous cell carcinoma cells growth and aggressiveness via reducing CLCA4. Mol. Biol. Rep. 2020;47(3):2181–2187. doi: 10.1007/s11033-020-05317-6. [DOI] [PubMed] [Google Scholar]
- 58.Huang W.C., et al. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014;74(3):751–764. doi: 10.1158/0008-5472.CAN-13-1297. [DOI] [PubMed] [Google Scholar]
- 59.Xu Y., et al. Clinical significance of miR-149 in the survival of patients with laryngeal squamous cell carcinoma. BioMed Res. Int. 2016;2016 doi: 10.1155/2016/8561251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tu H.F., et al. The association between genetic polymorphism and the processing efficiency of miR-149 affects the prognosis of patients with head and neck squamous cell carcinoma. PLoS One. 2012;7(12) doi: 10.1371/journal.pone.0051606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lucas Grzelczyk W., et al. Serum expression of selected miRNAs in patients with laryngeal squamous cell carcinoma (LSCC) Diagn. Pathol. 2019;14(1):49. doi: 10.1186/s13000-019-0823-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan H., et al. MiR-629 promotes human pancreatic cancer progression by targeting FOXO3. Cell Death Dis. 2017;8(10):e3154. doi: 10.1038/cddis.2017.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang J., et al. MicroRNA-3127 promotes cell proliferation and tumorigenicity in hepatocellular carcinoma by disrupting of PI3K/AKT negative regulation. Oncotarget. 2015;6(8):6359–6372. doi: 10.18632/oncotarget.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ren A., Wen Z., Zheng L. Downregulation of miR-3934-5p enhances A549 cell sensitivity to cisplatin by targeting TP53INP1. Exp. Ther. Med. 2019;18(3):1653–1660. doi: 10.3892/etm.2019.7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doghish A.S., et al. The role of miRNAs in liver diseases: potential therapeutic and clinical applications. Pathol. Res. Pract. 2023 doi: 10.1016/j.prp.2023.154375. [DOI] [PubMed] [Google Scholar]
- 66.El-Mahdy H.A., et al. miRNAs as potential game-changers in head and neck cancer: future clinical and medicinal uses. Pathol. Res. Pract. 2023 doi: 10.1016/j.prp.2023.154457. [DOI] [PubMed] [Google Scholar]
- 67.Tajik F., et al. MicroRNA-372 acts as a double-edged sword in human cancers. Heliyon. 2023;9(5):e15991. doi: 10.1016/j.heliyon.2023.e15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Losurdo P., et al. microRNAs combined to radiomic features as a predictor of complete clinical response after neoadjuvant radio-chemotherapy for locally advanced rectal cancer: a preliminary study. Surg. Endosc. 2023:1–8. doi: 10.1007/s00464-022-09851-1. [DOI] [PubMed] [Google Scholar]
- 69.Uzuner E., et al. The role of MiRNA in cancer: pathogenesis, diagnosis, and treatment. miRNomics: MicroRNA Biol. Comput. Anal. 2022:375–422. doi: 10.1007/978-1-0716-1170-8_18. [DOI] [PubMed] [Google Scholar]
- 70.Diener C., Keller A., Meese E. Emerging concepts of miRNA therapeutics: from cells to clinic. Trends Genet. 2022;38(6):613–626. doi: 10.1016/j.tig.2022.02.006. [DOI] [PubMed] [Google Scholar]
- 71.Ho P.T.B., Clark I.M., Le L.T.T. MicroRNA-based diagnosis and therapy. Int. J. Mol. Sci. 2022;23(13):7167. doi: 10.3390/ijms23137167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in this analysis can be found at TCGA data portal [https://cancergenome.nih.gov/].