Abstract
Background
Clear cell renal cell carcinoma (ccRCC) is a common urinary cancer. Although diagnostic and therapeutic approaches for ccRCC have been improved, the survival outcomes of patients with advanced ccRCC remain unsatisfactory. Fatty acid metabolism (FAM) has been increasingly recognized as a critical modulator of cancer development. However, the significance of the FAM in ccRCC remains unclear. Herein, we explored the function of a FAM-related risk score in the stratification and prediction of treatment responses in patients with ccRCC.
Methods
First, we applied an unsupervised clustering method to categorize patients from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) datasets into subtypes and retrieved FAM-related genes from the MSigDB database. We discern differentially expressed genes (DEGs) among different subtypes. Then, we applied univariate Cox regression analysis followed by least absolute shrinkage and selection operator (LASSO) linear regression based on DEGs expression to establish a FAM-related risk score for ccRCC.
Results
We stratified the three ccRCC subtypes based on FAM-related genes with distinct overall survival (OS), clinical features, immune infiltration patterns, and treatment sensitivities. We screened nine genes from the FAM-related DEGs in the three subtypes to establish a risk prediction model for ccRCC. Nine FAM-related genes were differentially expressed in the ccRCC cell line ACHN compared to the normal kidney cell line HK2. High-risk patients had worse OS, higher genomic heterogeneity, a more complex tumor microenvironment (TME), and elevated expression of immune checkpoints. This phenomenon was validated in the ICGC cohort.
Conclusion
We constructed a FAM-related risk score that predicts the prognosis and therapeutic response of ccRCC. The close association between FAM and ccRCC progression lays a foundation for further exploring FAM-related functions in ccRCC.
Keywords: Fatty acid metabolism, ccRCC, Immune features, Prognostic model, Treatment
1. Introduction
Clear cell renal cell carcinoma (ccRCC) accounts for approximately 70–80% of all renal malignancies and is one of the most prevalent urinary tumors [[1], [2], [3], [4]]. More than 30% of ccRCC patients have disease metastases at diagnosis, and the 5-year survival rate of metastatic ccRCC is only 10–20% [[5], [6], [7]]. Although targeted therapies focused on the vascular epithelial growth factor (VEGF) pathway, such as sunitinib, axitinib, and pazopanib, have improved the survival of patients with ccRCC, acquired resistance and side effects remain a challenge during ccRCC treatment [[8], [9], [10], [11]]. Recent studies have shown that immune checkpoint inhibitor therapies, including pembrolizumab, ipilimumab, and nivolumab, significantly prolong the overall survival (OS) and disease-free survival (DFS) in patients with localized ccRCC [[12], [13], [14]]. However, high recurrence rates after immunotherapy and the lack of reliable biomarkers to predict therapeutic effects still hinder the development of ccRCC treatments [12]. Therefore, building a powerful model to precisely predict ccRCC characteristics, patient clinical stage, and drug response is essential for formulating personalized treatment regimens and improving overall patient survival.
Although cancer types and underlying etiologies greatly differ, dysregulation of cellular metabolism is a common feature of tumorigenesis [[15], [16], [17]]. Aerobic glycolysis is a classic example of a metabolic perturbation, altering glucose metabolism to aerobic glycolysis, called the Warburg effect [[18], [19], [20]]. Abnormal glucose metabolism satisfies the metabolic necessities for cell growth and supports high levels of glycolytic intermediates for various biosynthetic pathways in cancer progression [21,22]. Additionally, aerobic glycolysis establishes a suppressive immune microenvironment that triggers immunosuppression in the tumor microenvironment (TME) [[23], [24], [25]]. Reprogramming fatty acid metabolism (FAM) is another characteristic of cancer. Multiple critical enzymes involved in FAM are dysregulated in several cancers [[26], [27], [28], [29]]. Many studies have demonstrated that abnormally activated fatty acid (FA) oxidation and de novo FA synthesis provide metabolic energy, cell membranes, and signaling molecules for carcinogenesis [[30], [31], [32], [33], [34]]. Most understanding of FAM indicates that limiting the FA supply may be a therapeutic strategy for cancer [[35], [36], [37]].
FAs in the TME influence the role and infiltration of immune cells [[38], [39], [40], [41], [42]]. Metabolic reprogramming is intricately linked to antitumor immunity [[43], [44], [45], [46]]. ccRCC cells are characterized by abundant glycogen and lipid accumulation [47,48] and exhibit diverse remodeling of cellular metabolism, including glucose, FA, and tricarboxylic acid cycle [[49], [50], [51], [52]]. Tumor-specific metabolic changes in ccRCC, including FAM, are associated with poor patient outcomes [53,54]. Moreover, enhanced lipid import and droplet formation benefit ccRCC tumorigenesis [55]. Multiple FAM enzymes, including anabolic enzymes such as fatty acid synthase (FASN), and catabolic enzymes, such as carnitine palmitoyltransferase I (CPT1A), hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha (HADHA) and beta (HADHB), and acetyl-CoA acetyltransferase 1 (ACAT1), have been considered potential prognostic biomarkers for ccRCC [[56], [57], [58]]. The von Hippel–Lindau (VHL) inactivation, commonly present in ccRCC, stabilized the hypoxia-inducible factor (HIF)1α to further repress FAM by inhibiting CPT1A and inducing lipid deposition [59]. Mechanistic investigations showed that the metabolic vulnerabilities of key enzymes involved in FA metabolism are novel therapeutic targets for ccRCC [50]. Therefore, an in-depth exploration of FAM may uncover the mechanisms underlying ccRCC progression and new therapeutic strategies.
In this study, we explored the potential roles of key FAM-related genes in ccRCC and provided a theoretical foundation for future ccRCC research. We developed and validated a novel FAM-related risk score using The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) datasets. The FAM-related risk score predicted clinical features, TME characteristics, and therapeutic efficacy in patients with ccRCC. In summary, our results provide new insights into FAM-related mechanisms in ccRCC. The established FAM-related risk score is reliable for predicting OS and treatment response in patients with ccRCC.
2. Materials and methods
2.1. Data acquisition and preprocessing
We retrieved RNA sequencing (RNA-Seq) profiles and clinical information from TCGA (https://tcga‐data.nci.nih.gov/tcga/) and ICGC databases. TCGA-KIRC cohort with 513 samples was used as the training set, and the ICGC (RECA-EU) dataset with 91 patients was used as the validation set. Samples with incomplete expression profiles, clinical follow-up data, survival time, or statuses were excluded. OS is the period from the day of diagnosis to death from any cause or the last follow-up and expressed in years. We converted the ensemble gene symbols to gene symbols for subsequent analyses and used the mean as the gene expression value when several probes were matched to the same gene. We retrieved 309 FAM-related genes from three FAM single-sample gene set, including the hallmark, the Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome, from the molecular signature database (MSigDB) (http://www.broadinstitute.org/gsea/msigdb/).
2.2. Enrichment analysis and correlations between FAM and clinicopathological features
To explore whether FAM was involved in ccRCC pathogenesis, we performed sample gene set enrichment analysis (ssGSEA) using the “GSVA” and “clusterProfiler” R packages to compute FAM-related enrichment scores and enrichment levels of FAM pathways, respectively, between tumor and para-cancerous tissues in TCGA cohort. Furthermore, we analyzed the association between FAM and clinicopathological features of patients with ccRCC.
2.3. Molecular subtypes based on FAM-related genes
We conducted the consensus clustering of TCGA cohort based on FAM levels using the “ConsensusClusterPlus” R package [60]. We varied the number of clusters k from 2 to 9 to determine the optimal k value for stratifying the appropriate subtypes based on the delta area, cumulative distribution function (CDF), and consensus matrix. We applied the Spearman's rank correlation coefficient as the distance measurement and completed 500 repetitions to ensure cluster stability. We then performed principal component analysis (PCA) to demonstrate the typing effect. We constructed Kaplan–Meier (K–M) survival curves to assess the OS between molecular subtypes using the “survival” R package [61]. Finally, we evaluated the distribution of clinicopathological features and differences in FAM between the different subtypes.
2.4. Gene mutational landscape of FAM-related subtypes
Tumor mutation characteristics are closely related to tumor progression and immunotherapeutic efficacy. Therefore, we investigated mutational characteristics, including genetic mutations and the mutant-allele tumor heterogeneity (MATH) score of FAM-related subtypes using the “maftools” R package. MATH is a tumor-specific score used to measure intratumoral genetic heterogeneity based on variations in the variant allele frequency of all mutations in the tumor. TCGA mutation data were retrieved from Genomic Data Commons (GDC) (https://portal.gdc.cancer.gov/). Furthermore, we estimated the association of our subtypes with six previously published immune subtypes (IFN-γ dominant, inflammatory, lymphocyte-depleted, immunologically quiet, wound healing, and TGF-β dominant) using a pan-cancer analysis [62].
2.5. Functional enrichment analysis of ccRCC FAM-related subtypes
To identify the underlying characteristics of FAM-related subtypes in ccRCC development, we performed GSEA using KEGG gene signatures to explore the enriched pathways among different FAM-related subtypes. We then conducted the enrichment score of FAM genes of the hallmarks gene signatures to quantify the enrichment levels of different pathways among FAM-related subtypes.
2.6. Immune features and pathway enrichment of FAM-related subtypes
To investigate the immune microenvironment of FAM-related subtypes in ccRCC, we used the “ESTIMATE” R package to evaluate the immune status of patients with ccRCC [63]. The “CIBERSORT” R package was used to measure the infiltration of immune cells [64].
2.7. Therapeutic differences among FAM-related subtypes
Based on the immune characteristics among different FAM-related subtypes, we explored the response of different molecular subtypes to conventional ccRCC chemotherapy agents by the IC50 value measurement using “pRRophetic” R package, including cisplatin, bortezomib, temsirolimus, axitinib, gefitinib, sunitinib, sorafenib, and vinblastine. As immune checkpoint inhibitors (ICIs) have revolutionized cancer immunotherapy, we used a heatmap to compare the distribution of the published 45 immune coinhibitory/costimulatory genes among FAM-related subtypes [65].
2.8. Differentially expressed genes (DEGs) and relevant function analysis of FAM-related subtypes
To classify key FAM genes, we conducted pairwise comparisons of DEGs among FAM-related subtypes using the “Limma” R package and exhibited the results in heatmaps [66]. Significant DEGs were determined by false discovery rate (FDR) < 0.05, and |log2 [Fold Change (FC)] | > 1. Subsequently, KEGG and Gene Ontology (GO) analyses were performed using the “webgestalt” R package to evaluate pathway differences for the key FAM genes identified [67].
2.9. Establishment of a prognostic FAM-related risk score
We applied univariate Cox regression analysis to preliminarily screen crucial prognostic genes from the DEGs identified preliminarily [68]. Then, we conducted LASSO analysis using the “glmnet” R package to select the crucial genes in TCGA cohort [69]. We used 10-time cross-validation to estimate the confidence interval for each lambda and subsequently identified the optimal lambda with the lowest average error. In addition, we conducted multivariate Cox regression in a backward stepwise fashion to curtail the gene number and acquire the regression coefficient for every gene. We assumed that p-selected genes are input into a prognostic prediction model, denoted as (x1, …,xp). The risk score was a weighted sum of genes whose weights reflect the degree of association and defined as [70]. Then, patients were divided into high- and low-risk groups based on the cutoff value determined by the “survminer” R package (http://www.sthda.com/english/rpkgs/survminer). Survival curves were constructed using the K–M to compare OS between the different risk groups. The ICGC cohort was used for the external validation to verify the robustness of the established prediction model. Finally, we examined the relationships between risk scores and clinical characteristics to elucidate their potential for clinical application. We also evaluated the differences in risk scores among subtypes.
2.10. Immune status between different risk groups
We performed GSEA to compare the differences in enrichment scores of FAM pathways between groups using genes selected from MSigDB, hallmark, KEGG, and Reactome. We then applied the ESTIMATE R package to explore the TME features between groups. We used CIBERSORT to analyze the relative abundance of tumor-infiltrating immune cells between the two groups. Thereafter, we collected 29 functional gene expression signatures (FGES) previously published associated with the TME in different risk groups [71]. Additionally, we obtained previously published gene signatures from 15 pathways characterized by immune, stromal, DNA repair, and oncogenic signatures to investigate the heterogeneity of the immune microenvironment between different risk groups [72]. We also assessed the correlation of the risk score with the FAM pathways, 29 gene expression signatures, and gene signatures from 15 pathways using Pearson’s correlation.
2.11. The predictive performance of the risk score on ccRCC therapy
First, we evaluated drug sensitivity using IC50 for common chemotherapy agents in TCGA risk groups. We then analyzed whether immunotherapy sensitivity varied between risk groups. We analyzed the differences in MATH scores using the Wilcoxon test to investigate genomic heterogeneity between groups. Given the close relationship between tumor immune regulation and immune checkpoint-related genes (ICGs), we explored the relationship between the risk score and ICGs and the expression profiles of ICGs between groups.
2.12. Expression validation of the selected genes in FAM-related prognostic model
To further explore the FAM-related genes selected from the constructed prognostic model at the mRNA level in ccRCC and normal renal cells, we performed cell culture and real-time quantitative PCR (RT–qPCR) [73]. Human renal tubular epithelial HK2 and ccRCC ACHN cells were cultured in Dulbecco’s modified eagle medium (DMEM, Gibco, Invitrogen, Carlsbad, CA, USA). Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), following the standard protocol. cDNA was synthesized using a PrimeScript™ RT reagent kit (Takara, Shiga, Japan). RT–qPCR was conducted using an ABI7300 real-time PCR system (Applied Biosystems) to validate the expression of the selected FAM-related genes from the constructed prognostic model, following the manufacturer’s instructions. The amplification conditions for qRT-PCR were 95 °C for 30 s, 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. The primers used are listed in Supplementary Table 1. Each mRNA expression was normalized to GAPDH expression according to the 2−ΔΔCt method, and all reactions were performed thrice in triplicate to calculate the average cycle threshold (Ct).
2.13. Statistical analysis
Statistical analyses were conducted using R (version 3.4.3) and GraphPad Prism (version 8.4.2) software. To compare clinical features between different groups, we applied the Wilcoxon test to evaluate two independent non-parametric samples and the Kruskal–Wallis test to compare three or more samples. Continuous variables with a normal distribution are presented as means ± standard deviation. Pearson’s or Spearman’s rank correlation coefficients were used to explore the correlations between variables. Multivariate Cox regression analysis was performed to explore the independence of the constructed FAM-related model for prognostic performance. p-value < 0.05 indicated statistical significance [74].
3. Results
3.1. Influence of FAM in ccRCC
To elucidate the mechanisms of FAM in ccRCC progression, we found that the enrichment scores of the genes in FAM pathways in TCGA cohort were higher in para-cancer tissues than in ccRCC tissues (p < 0.05) (Fig. S1A). We comprehensively assessed the association between FAM and clinicopathological features. As expected, higher clinical stages resulted in lower FAM scores, and women had more active FAM than men (p < 0.05) (Fig. S1B).
3.2. Identification of ccRCC subtypes associated with FAMGs
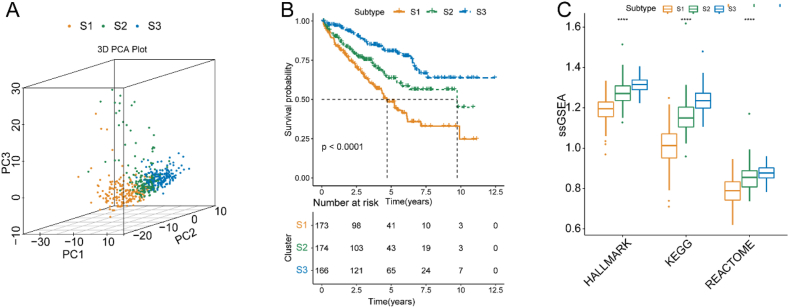
To explore the carcinogenic mechanisms of FAMGs in ccRCC, we performed a consensus cluster analysis based on the expression of FAMGs in TCGA cohort. Based on the relative CDF and delta area changes, (the number of clusters) achieved the flattest middle portion of the CDF curve and a definitive boundary for the consensus matrix (Figs. S2A–C). Therefore, we classified patients in TCGA cohort into subtypes: subtype 1, subtype 2, and subtype 3, named S1, S2, and S3, respectively. Furthermore, PCA analysis also proved the typing results with distinct differences among the three subtypes (p < 0.05) (Fig. 1A). K–M survival curves showed that ccRCC patients in S1 had shorter OS than those in S2 and S3 in both TCGA and ICGC cohorts (p < 0.05) (Fig. 1B). Additionally, we found clear differences in the enrichment levels of FAM among the S1, S2, and S3 subtypes, with the highest FAM scores in S3 and the lowest in S1 (p < 0.05) (Fig. 1C). Next, we evaluated the differences in clinicopathological features between the S1, S2, and S3 subtypes in TCGA cohort and found that S1 patients with poorer prognoses had higher TNM and clinical stages and grades (p < 0.05) (Fig. S3).
Fig. 1.
Consensus clustering of FAMGs of ccRCC in TCGA cohort. (A) PCA plot of the three FAM-related subtypes. (B) Kaplan–Meier survival curves for OS of the S1, S2, and S3 subtypes. (C) Comparison of FAM of the S1, S2, and S3 subtypes. Abbreviations: ccRCC, clear cell renal cell carcinoma; CDF, cumulative distribution function; FAMGs, fatty acid metabolism genes; KEGG, the Kyoto Encyclopedia of Genes and Genomes; OS, overall survival; PCA, principal component analysis; ssGSEA, sample gene set enrichment analysis; TCGA, The Cancer Genome Atlas.
3.3. Characteristics of gene mutations among FAMG-related subtypes
First, we found that VHL, PBRM1, TTN, SETD2, and BAP1 mutations were commonly found in TCGA cohort and were associated with FAMG-related subtypes (Fig. S4A). To understand the influence of FAM-related gene mutations in ccRCC, we observed that the S1, S2, and S3 subtypes exhibited substantial differences in tumor heterogeneity, and S3 had the lowest MATH scores (p < 0.05) (Fig. S4B). Finally, we found that S3 patients were mostly matched to the widely recognized immune subtype S3 (p < 0.05) (Fig. S4C).
3.4. Enriched pathways among FAMG-related subtypes
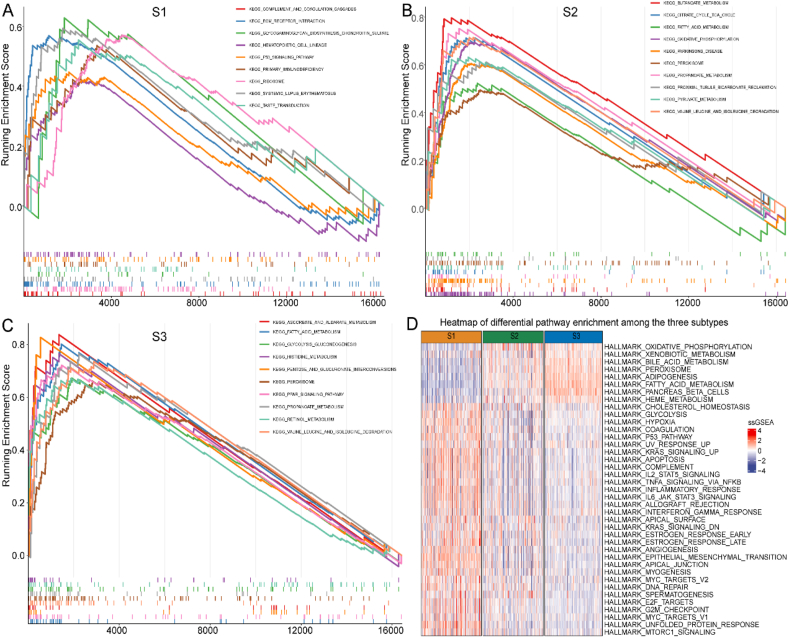
Furthermore, the GSEA results identified distinct enriched pathways in the three ccRCC subtypes. For example, S1 was largely enriched in pathways correlated to inflammation and immune response, including the “complement and coagulation cascades,” “primary immunodeficiency,” “systemic lupus erythematosus,” and “P53 signaling pathway” (Fig. 2A). Besides “FAM pathways,” other metabolism-related pathways such as “butanoate,” “propanoate,” and “pyruvate” were enriched in S2 (Fig. 2B). S3 was also enriched in several metabolism-related pathways, including “FAM,” “pentose and glucuronate interconversions,” and “ascorbate and aldarate metabolism” (Fig. 2C). S1 was enriched in several pathways, including immune-, cell cycle-, and cancer-related pathways. S3 was enriched for some metabolism-related pathways such as “FAM,” “oxidative phosphorylation,” “xenobiotic metabolism,” and “bile acid metabolism” (Fig. 2D).
Fig. 2.
Pathway enrichment analysis among three subtypes in TCGA cohort. (A) S1, (B) S2, and (C) S3. (D) Heatmap of differential pathway enrichment among the three subtypes. Abbreviations: KEGG, the Kyoto Encyclopedia of Genes and Genomes; TCGA, The Cancer Genome Atlas.
3.5. Immune features of FAMG-related subtypes
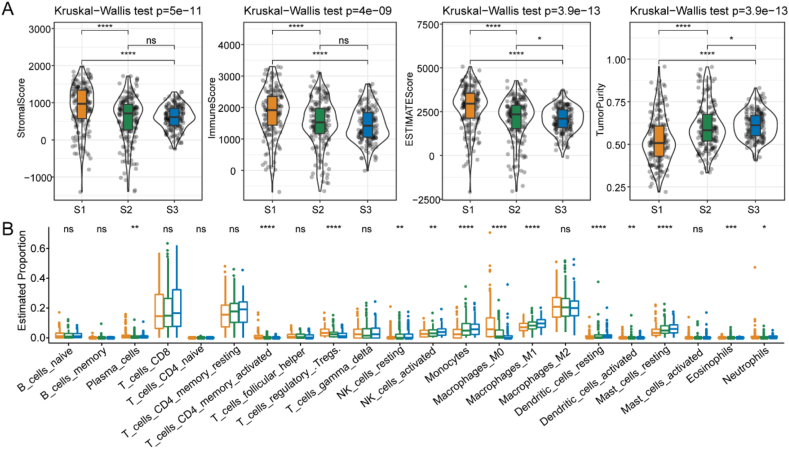
To evaluate the TME components among different FAMG-related subtypes, we used ESTIMATE to assess the immune cell status. We found that S1 had higher stromal, immune, and ESTIMATE scores than the other two subtypes and a lower tumor purity, suggesting that S1 had a high immune infiltration density and heterogeneity (p < 0.05) (Fig. 3A). We further used CIBERSORT to determine the abundance of 22 immune cells among the FAMG-related subtypes. S1 showed an increased proportion of M0 macrophages, regulatory T, and Treg cells (p < 0.05) (Fig. 3B).
Fig. 3.
Associations between the immune signatures of TME and FAM-related subtypes in TCGA cohort. (A) Comparison of ESTIMATE scores in the S1, S2, and S3 subtypes. (B) Comparison of the immune cell distribution in the S1, S2, and S3 subtypes. Abbreviations: CD4, cluster of differentiation 4; CD8, cluster of differentiation 8; FAM, fatty acid metabolism; NK, natural killer; TCGA, The Cancer Genome Atlas; TME, tumor microenvironment.
3.6. Differences in cancer therapy response among FAMG-related subtypes
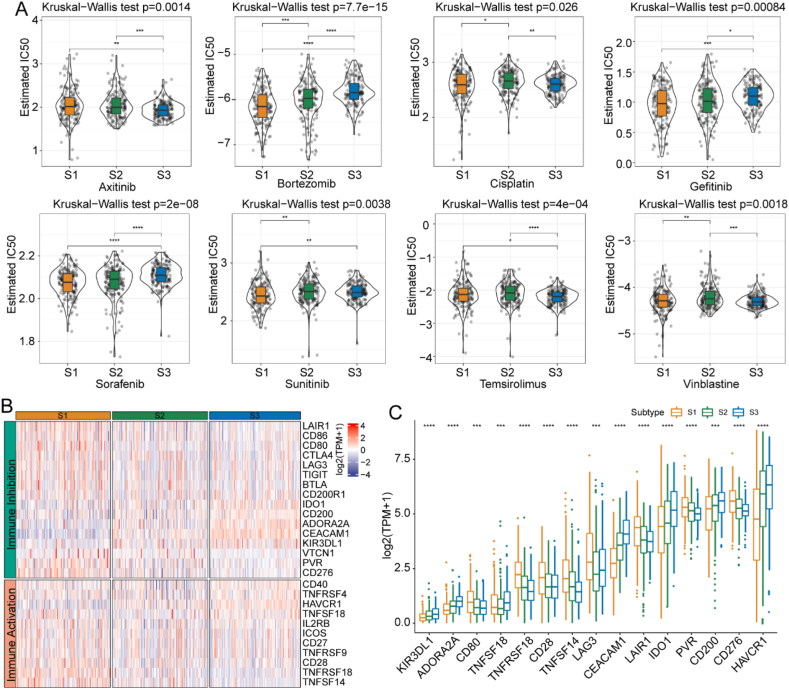
Next, we examined whether the FAMG-related subtypes significantly influenced cancer chemotherapy and immunotherapy. We analyzed the response of the subtypes to conventional chemotherapy agents and found that S1 was more sensitive to bortezomib and sunitinib, whereas S3 was more sensitive to axitinib in TCGA cohort (p < 0.05) (Fig. 4A). Given the promising applications of immunotherapy with checkpoint inhibitors, we also tested the levels of several representative immune checkpoints in the three FAMG-related subtypes. The results revealed that most immune checkpoint genes, characterized by “immune activation” and “immune inhibition, were highly expressed in the S1 stage (Fig. 4B). Furthermore, we identified 15 significantly differentially expressed ICGs among FAMG-related subtypes: KIR3DL1, ADORA2A, CD80, TNFSF18, TNFRSF18, CD28, TNFSF14, LAG3, CEACAM1, LAIR1, IDO1, PVR, CD200, CD276, and HAVCR1 (p < 0.05) (Fig. 4C).
Fig. 4.
Estimation of chemotherapy and immunotherapy response of the S1, S2, and S3 subtypes in TCGA cohort. (A) Estimated IC50 for commonly used chemotherapy drugs in the S1, S2, and S3 subtypes. (B) Expression of ICGs in the S1, S2, and S3 subtypes. (C) Expression of 15 differentially expressed ICGs in the S1, S2, and S3 subtypes. Abbreviations: ICGs, immune checkpoint-related genes; IC50, half maximal inhibitory concentration; TCGA, The Cancer Genome Atlas.
3.7. Differential gene screening among FAMG-related subtypes
After establishing three distinct FAMG subtypes in ccRCC, we compared their gene expression levels to determine the co-expressed differential genes. We found 321 DEGs between the S1 and S2 subtypes, 146 upregulated and 175 downregulated genes in S1, 973 DEGs between the S1 and S3 subtypes, 305 upregulated and 668 downregulated genes in S1, 151 DEGs between the S2 and S3 subtypes, and 29 upregulated and 122 downregulated genes in S2 (p < 0.05) (Figs. S5A–B). We found that 86 DEGs overlapped among the three FAMG-related subtypes: SLC27A2, CYP4A11, FMO1, CYP4A22, HAO2, CYP2J2, HMGCC2, and CA4 (Fig. S5C). Moreover, we performed a functional enrichment analysis of 305 DEGs between S1 and S3 and found that these DEGs were mainly mapped to stroma-related signaling pathways (Figs. S5D–G).
3.8. Construction of a FAMG-related risk prediction model
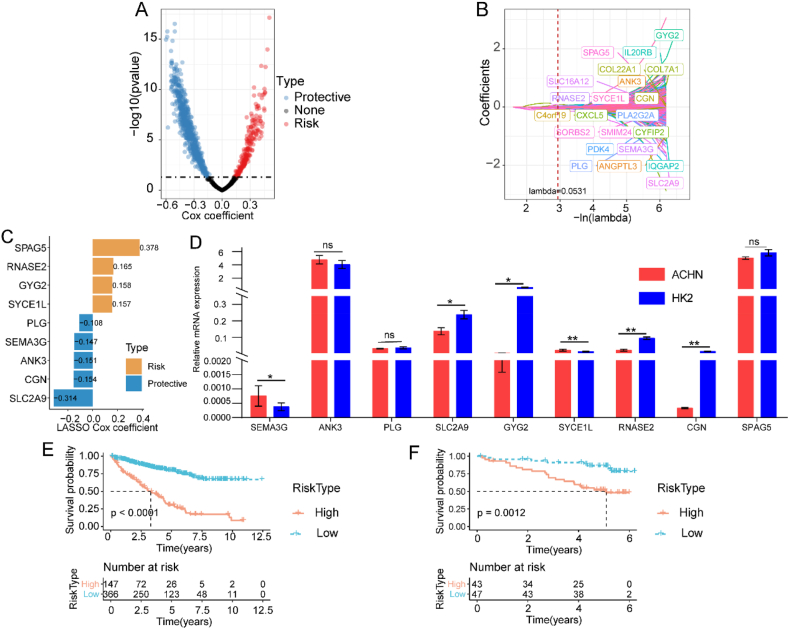
To evaluate the relationship between FAMGs and the prognosis of ccRCC patients, we used univariate Cox regression analysis and verified 852 DEGs with prognostic values; 214 were risk genes and 638 were protective genes (p < 0.05) (Fig. 5A). Next, we incorporated the 852 prognostic genes to establish a prognostic risk prediction model using LASSO regression. First, regression coefficients approached zero and gradually increased with an increase in lambda. The optimal lambda was set to 0.0531 based on a 10-time validation, and 22 genes were finally included in the risk score (Fig. 5B). Subsequently, multivariate stepwise linear regression analysis verified the final nine prognostic genes and determined the coefficient for each targeted gene (Fig. 5C). The following formula was established using the mRNA level of each risk gene and coefficients: . Moreover, we validated the expression of nine genes in the HK2 and ACHN cell lines. Compared to HK2 cells, the ccRCC cell line ACHN had higher SEMA3G and SYCE1L mRNA levels and lower CGN, GYG2, RNASE2, and SLC2A9 mRNA levels (p < 0.05) (Fig. 5D). With the cutoff value of −0.199807 set by the “survminer” R package, we allocated patients to the high- and low-risk groups in TCGA cohort. We constructed K–M survival curves and found that high-risk patients had significantly poorer prognoses (p < 0.05) (Fig. 5E). To verify the robustness of risk scores, the same model was applied to the ICGC cohort for external validation. High-risk patients showed shorter OS than low-risk ones in the ICGC cohort (p < 0.05) (Fig. 5F). These results demonstrated that our risk score was predictive of OS in different cohorts, suggesting robust performance of the constructed nine-gene risk score. We also investigated the distribution of risk scores among clinical characteristics (i.e., cancer grade, stage, and TNM) and found that the more advanced the ccRCC clinical stage, the greater the risk score (p < 0.05). In addition, the distribution of risk scores exhibited significant differences among the three FAMG-related subtypes (p < 0.05) (Fig. S6).
Fig. 5.
Construction and validation of FAM-related risk score for ccRCC in TCGA and ICGC cohorts. (A) Univariate Cox regression analysis of DEGs identifying OS-related FAMGs. (B) Partial likelihood deviance of LASSO coefficient profiles. (C) Multivariate Cox regression analysis for nine selected FAMGs of the risk score. (D) The mRNA expression levels of the nine selected FAMGs by RT–qPCR. ROC curve of the FAM-related risk score in the ICGC cohort. (E) Kaplan–Meier curves of the FAM-related risk score in TCGA cohort. (F) Kaplan–Meier curves of the FAM-related risk score in the ICGC cohort. Abbreviations: ccRCC, clear cell renal cell carcinoma; DEGs, differentially expressed genes; FAM, fatty acid metabolism; FAMGs, fatty acid metabolism genes; ICGC, International Cancer Genome Consortium; LASSO, least absolute shrinkage and selection operator; ROC, receiver operating characteristic; RT–qPCR, real-time quantitative PCR; TCGA, The Cancer Genome Atlas.
3.9. Associations between the risk score and immune characteristics
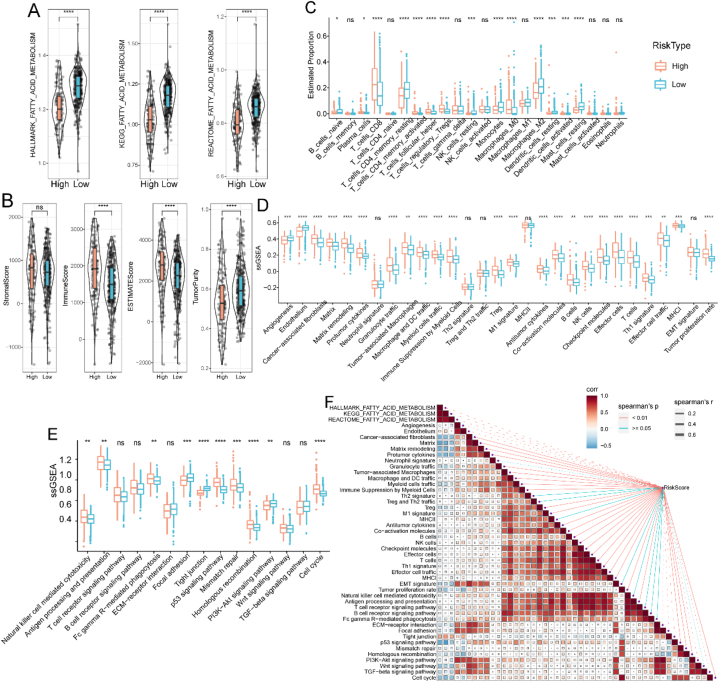
We first evaluated the enrichment of FAM pathways between low- and high-risk groups using the Hallmark, KEGG, and Reactome databases. Low-risk patients had higher FAM enrichment scores than high-risk patients (p < 0.05) (Fig. 6A). To further illustrate the differences in the TME between the different risk groups, we used ESTIMATE to obtain tumor stroma and immune cell infiltration. The immune, stromal, and ESTIMATE scores were higher in the high-risk group than in the low-risk group. In contrast, the tumor purity was higher in the low-risk group than in the high-risk one (p < 0.05) (Fig. 6B). We also compared the levels of 22 immune cell types between the risk groups and found a remarkable difference in the proportion of 22 immune cell types between the groups (p < 0.05) (Fig. 6C). Moreover, the enrichment scores between the high- and low-risk groups differed significantly for 29 FGES. The FGES associated with tumor stroma and promotion accounted for most of the enrichment in the high-risk group (Fig. 6D). Compared to the low-risk group, several pathways correlated with DNA damage repair were differentially enriched in the high-risk group based on the 15 pathway-related enrichment score distributions (Fig. 6E). Additionally, we explored the relationship between the risk score and the FAM pathways, 29 FGES, and 15 pathways. The risk score exhibited a negative correlation with FAM, a positive correlation with FGES associated with tumor stroma and promotion, and pathways correlated with DNA damage repair (Fig. 6F).
Fig. 6.
Relationship between immune infiltration features and the risk score in TCGA cohort. (A) FAM enrichment scores in high- and low-risk groups. (B) Immune, stromal, estimate, and tumor purity scores in high- and low-risk groups. (C) Distribution of 22 immune cells in high- and low-risk groups. (D) TME-related 29 gene signatures in high- and low-risk groups. (E) Scores of 15 pathways in high- and low-risk groups. (F) Correlation of risk score with TME-related gene signatures and 15 pathways. Abbreviations: ssGSEA, sample gene set enrichment analysis; TCGA, The Cancer Genome Atlas; TME, tumor microenvironment.
3.10. Comparisons of therapeutic responses between groups
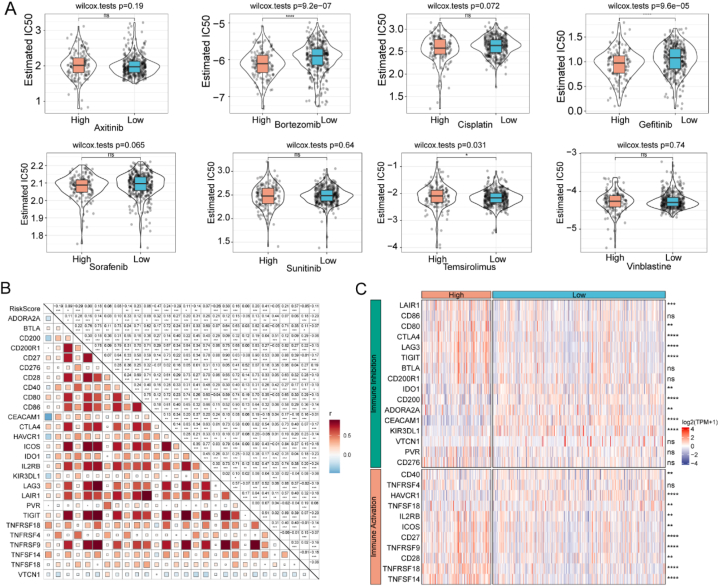
For drug sensitivity analysis, we compared the IC50 of several commonly used chemotherapeutic agents between the low- and high-risk groups, including bortezomib, axitinib, cisplatin, sorafenib, temsirolimus, sunitinib, gefitinib, and vinblastine. The high-risk group exhibited greater sensitivity to bortezomib, gefitinib, and temsirolimus than the low-risk group (p < 0.05) (Fig. 7A). Further correlation analysis revealed that the risk score was significantly correlated with the expression of several ICGs, including CTLA4, IDO1, and ICOS (p < 0.05) (Fig. 7B). Many ICGs were significantly overexpressed in the high-risk group, including LAG3, IDO1, and CTLA4 (p < 0.05) (Fig. 7C).
Fig. 7.
Risk score predictions for ccRCC treatment sensitivity in TCGA cohort. (A) Violin plots of the estimated IC50 for chemotherapy agents in high- and low-risk groups. (B) Association of risk score with ICGs. (C) Heatmap of differentially expressed ICGs in high- and low-risk groups. ccRCC, clear cell renal cell carcinoma; ICGs, International Cancer Genome Consortium; TCGA, The Cancer Genome Atlas.
4. Discussion
Metabolic and epigenetic studies in the last decade have revealed a new relationship between metabolic alterations and cancer progression [[75], [76], [77], [78]]. Metabolic modifications trigger oncogenic transformations and are hallmarks of cancer. For example, upregulated glycolytic metabolism contributes to energy production and essential metabolic intermediates in most solid tumors [[79], [80], [81]]. The metabolic properties of tumors induce immunosuppression in the TME and constrain cancer immunotherapy [[82], [83], [84]]. Although most studies have focused on the influence of FAM on cell physiological functions and biological behaviors, the effects of FAM on ccRCC remain unclear [[85], [86], [87]]. FAM is dysregulated in ccRCC and correlated with a poor prognosis of patients. Various key enzymes related to FAM have been found to participate in the malignance of ccRCC. However, the pathogenic mechanism of FAM in ccRCC has not been elucidated [53]. In addition, the prognostic performance of the previously published FAM-related risk scores and gene signatures requires further clinical verification [88,89]. The immune features and therapeutic responses of FAM-related genes have also been inadequately illustrated in the constructed models. Therefore, we constructed a novel FAM-related risk score to explore the regulatory mechanisms of FAM in ccRCC progression and identify high-risk patients with poor prognoses to guide personalized treatment strategies.
FAM reprogramming is a ubiquitous metabolic process in cancer [[90], [91], [92]]. Several key enzymes of the FAM play a critical role in cancer cell survival and progression and are currently attracting considerable attention as new therapeutic targets [93,94]. Emerging omics techniques, including genomics, epigenomics, transcriptomics, proteomics, and metabolomics, have shown that ccRCC characteristically exhibits high lipid accumulation and abnormal FAM [53,55,95,96]. In this study, we found that the FAM scores differed between the primary tumor and para-tumorous normal tissues. We also found a correlation between the FAM and the clinicopathological features of patients with ccRCC. A higher clinical stage was associated with lower FAM scores.
The three molecular subtypes S1, S2, and S3, categorized by FAM features using consensus cluster analysis, exhibited significant differences in prognosis, clinical grade, tumor genomic heterogeneity, biological pathways, TME, and response to common treatments. Thus, to better understand the functions of FAM-related genes in ccRCC, we constructed a predictive risk score to discern different degrees of risk in patients with ccRCC. We showed that the FAM-related risk score predicted OS efficiently in both TCGA and ICGC cohorts. Consistent with previous studies, high-risk patients had worse OS than low-risk patients and were susceptible to posterior clinical staging, suggesting that the FAM-related risk score can be used to identify high-risk patients and provide personalized treatment to improve outcomes [[97], [98], [99], [100]].
Previous studies demonstrated that ccRCC is highly infiltrated by different immune cells and has high immune heterogeneity [98,[101], [102], [103]]. We found that high-risk patients had more plentiful immune and stromal components, lower tumor purity in the TME, and more abundant immunosuppressive cells. As expected, the high-risk group had elevated enrichment scores for cancer-associated fibroblasts (CAFs), tumor- and matrix-associated macrophages, and tumor proliferation rates. Additionally, pathways involved in oncogenesis, such as the DNA damage repair pathway, were identified in high-risk groups.
Chemotherapy has remained the most effective treatment for advanced ccRCC for decades [104,105]. Herein, we found that high-risk patients were more sensitive to bortezomib, gefitinib, and temsirolimus than other treatments, indicating that they might benefit from chemotherapy. The significant genomic heterogeneity differences between the groups reflect features of tumor promotion and treatment resistance in the high-risk group, highlighting the importance of immunotherapy in ccRCC [106]. Therefore, we explored immunotherapy responses between the low- and high-risk groups and found that the characteristics reflecting the immunotherapy response were increased in the high-risk group compared to the low-risk group. Thus, high-risk patients may show an improved immunotherapy response. In summary, these results demonstrate that the constructed FAM-related risk score exhibits a promising prognostic performance for patients with ccRCC and provides insights into the function of FAMGs in ccRCC.
This study has some limitations. First, we used multiple online databases and bioinformatics methods without prospective clinical validation. Thus, we will conduct an additional validation cohort study at our center to confirm the predictive values of the constructed nine-gene risk score for ccRCC. Moreover, we validated the mRNA levels of the nine genes in the risk prediction model. Therefore, future studies should investigate the underlying mechanisms of the nine FAMGs in ccRCC progression.
In conclusion, we successfully developed and verified a robust risk score associated with the FAM in ccRCC that can be applied to predict the prognosis of patients with ccRCC. The FAM-related risk score described the clinicopathological features of the patients and predicted their susceptibility to chemotherapy and immunotherapy. The FAM-related risk score has contributed to research on promising ccRCC prognostic predictors and future individualized treatments for ccRCC.
Author contribution statement
Conceived and designed the experiments: Qinfan Yao, Dajin Chen, and Jianghua Chen.
Performed the experiments: Xiuyuan Zhang, Chunchun Wei, Qiannan Xu, and Hongjun Chen.
Analyzed and interpreted the data: Qinfan Yao, Chunchun Wei, and Xiuyuan Zhang.
Contributed reagents, materials, analysis tools or data: Qiannan Xu, and Jianghua Chen.
Wrote the paper: Qinfan Yao, Dajin Chen, and Xiuyuan Zhang.
Funding statement
This work was funded by the National Natural Science Foundation of China (81802085), and the Natural Science Foundation of Zhejiang (LY21H160033).
Data availability statement
The datasets generated in the current study are available from the corresponding author.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17224.
Contributor Information
Jianghua Chen, Email: chenjianghua@zju.edu.cn.
Dajin Chen, Email: zju2001@zju.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Ljungberg B., Albiges L., Abu-Ghanem Y., et al. European association of urology guidelines on renal cell carcinoma: the 2022 update. Eur. Urol. 2022;82:399–410. doi: 10.1016/j.eururo.2022.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Jonasch E., Gao J., Rathmell W.K. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuch B., Amin A., Armstrong A.J., et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur. Urol. 2015;67:85–97. doi: 10.1016/j.eururo.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 5.Li Y., Lih T.M., Dhanasekaran S.M., et al. Histopathologic and proteogenomic heterogeneity reveals features of clear cell renal cell carcinoma aggressiveness. Cancer Cell. 2023;41:139–163.e117. doi: 10.1016/j.ccell.2022.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J., Zhu L., Zheng L.P., et al. Overexpression of ULK1 represents a potential diagnostic marker for clear cell renal carcinoma and the antitumor effects of SBI-0206965. EBioMedicine. 2018;34:85–93. doi: 10.1016/j.ebiom.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroeder A., Heller D.A., Winslow M.M., et al. Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer. 2011;12:39–50. doi: 10.1038/nrc3180. [DOI] [PubMed] [Google Scholar]
- 8.Motzer R.J., Hutson T.E., Cella D., et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N. Engl. J. Med. 2013;369:722–731. doi: 10.1056/NEJMoa1303989. [DOI] [PubMed] [Google Scholar]
- 9.Motzer R.J., Hutson T.E., Tomczak P., et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N. Engl. J. Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 10.Choueiri T.K., Motzer R.J. Systemic therapy for metastatic renal-cell carcinoma. N. Engl. J. Med. 2017;376:354–366. doi: 10.1056/NEJMra1601333. [DOI] [PubMed] [Google Scholar]
- 11.Buchler T., Bortlicek Z., Poprach A., et al. Outcomes for patients with metastatic renal cell carcinoma achieving a complete response on targeted therapy: a registry-based analysis. Eur. Urol. 2016;70:469–475. doi: 10.1016/j.eururo.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 12.Bedke J., Albiges L., Capitanio U., et al. The 2022 updated European association of urology guidelines on the use of adjuvant immune checkpoint inhibitor therapy for renal cell carcinoma. Eur. Urol. 2023;83:10–14. doi: 10.1016/j.eururo.2022.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Motzer R.J., Escudier B., McDermott D.F., et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motzer R.J., Escudier B., George S., et al. Nivolumab versus everolimus in patients with advanced renal cell carcinoma: updated results with long-term follow-up of the randomized, open-label, phase 3 CheckMate 025 trial. Cancer. 2020;126:4156–4167. doi: 10.1002/cncr.33033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Raggi C., Taddei M.L., Rae C., Braconi C., Marra F. Metabolic reprogramming in cholangiocarcinoma. J. Hepatol. 2022;77:849–864. doi: 10.1016/j.jhep.2022.04.038. [DOI] [PubMed] [Google Scholar]
- 18.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 19.Bononi G., Masoni S., Di Bussolo V., Tuccinardi T., Granchi C., Minutolo F. Historical perspective of tumor glycolysis: a century with Otto Warburg. Semin. Cancer Biol. 2022;86:325–333. doi: 10.1016/j.semcancer.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Stancliffe E., Fowle-Grider R., et al. Saturation of the mitochondrial NADH shuttles drives aerobic glycolysis in proliferating cells. Mol. Cell. 2022;82:3270–3283.e3279. doi: 10.1016/j.molcel.2022.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J., Kim K., Kwon I.C., Lee K.Y. Intracellular glucose-depriving polymer micelles for anti-glycolytic cancer treatment. Adv. Mater. 2022 doi: 10.1002/adma.202207342. [DOI] [PubMed] [Google Scholar]
- 22.Paul S., Ghosh S., Kumar S. Tumor glycolysis, an essential sweet tooth of tumor cells. Semin. Cancer Biol. 2022;86:1216–1230. doi: 10.1016/j.semcancer.2022.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Guo D., Tong Y., Jiang X., et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IκBα. Cell Metabol. 2022;34:1312–1324.e1316. doi: 10.1016/j.cmet.2022.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y. Targeting glycolysis for cancer therapy using drug delivery systems. J. Contr. Release. 2022;353:650–662. doi: 10.1016/j.jconrel.2022.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Lin J., Liu G., Chen L., Kwok H.F., Lin Y. Targeting lactate-related cell cycle activities for cancer therapy. Semin. Cancer Biol. 2022;86:1231–1243. doi: 10.1016/j.semcancer.2022.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Nieman K.M., Kenny H.A., Penicka C.V., et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beckers A., Organe S., Timmermans L., et al. Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 2007;67:8180–8187. doi: 10.1158/0008-5472.Can-07-0389. [DOI] [PubMed] [Google Scholar]
- 28.Boroughs L.K., DeBerardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brusselmans K., De Schrijver E., Verhoeven G., Swinnen J.V. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res. 2005;65:6719–6725. doi: 10.1158/0008-5472.Can-05-0571. [DOI] [PubMed] [Google Scholar]
- 30.Lee H., Woo S.M., Jang H., Kang M., Kim S.Y. Cancer depends on fatty acids for ATP production: a possible link between cancer and obesity. Semin. Cancer Biol. 2022;86:347–357. doi: 10.1016/j.semcancer.2022.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Currie E., Schulze A., Zechner R., Walther T.C., Farese R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metabol. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snaebjornsson M.T., Janaki-Raman S., Schulze A. Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer. Cell Metabol. 2020;31:62–76. doi: 10.1016/j.cmet.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Ackerman D., Simon M.C. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24:472–478. doi: 10.1016/j.tcb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Röhrig F., Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X., Huang F., Ma G., Wei W., Wu N., Liu Z. Dysregulated ceramides metabolism by fatty acid 2-hydroxylase exposes a metabolic vulnerability to target cancer metastasis. Signal Transduct. Targeted Ther. 2022;7:370. doi: 10.1038/s41392-022-01199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T., Yuan Z., Lei Z., et al. Hippocalcin-Like 1 blunts liver lipid metabolism to suppress tumorigenesis via directly targeting RUVBL1-mTOR signaling. Theranostics. 2022;12:7450–7464. doi: 10.7150/thno.75936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Wang Y., Ren Y., Zhang Q., Yi P., Cheng C. Metabolic modulation of immune checkpoints and novel therapeutic strategies in cancer. Semin. Cancer Biol. 2022;86:542–565. doi: 10.1016/j.semcancer.2022.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Abrego J., Sanford-Crane H., Oon C., et al. A cancer cell-intrinsic GOT2-PPARδ Axis suppresses antitumor immunity. Cancer Discov. 2022;12:2414–2433. doi: 10.1158/2159-8290.Cd-22-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su P., Wang Q., Bi E., et al. Enhanced lipid accumulation and metabolism are required for the differentiation and activation of tumor-associated macrophages. Cancer Res. 2020;80:1438–1450. doi: 10.1158/0008-5472.Can-19-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corn K.C., Windham M.A., Rafat M. Lipids in the tumor microenvironment: from cancer progression to treatment. Prog. Lipid Res. 2020;80 doi: 10.1016/j.plipres.2020.101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu L., Zhang X., Zheng L., et al. RIPK3 orchestrates fatty acid metabolism in tumor-associated macrophages and hepatocarcinogenesis. Cancer Immunol Res. 2020;8:710–721. doi: 10.1158/2326-6066.Cir-19-0261. [DOI] [PubMed] [Google Scholar]
- 42.Liu S., Zhang H., Li Y., et al. S100A4 enhances protumor macrophage polarization by control of PPAR-γ-dependent induction of fatty acid oxidation. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2021-002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galván-Peña S., Carroll R.G., Newman C., et al. Malonylation of GAPDH is an inflammatory signal in macrophages. Nat. Commun. 2019;10:338. doi: 10.1038/s41467-018-08187-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho P.C., Bihuniak J.D., Macintyre A.N., et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W., Bai Y., Xiong Y., et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature. 2016;531:651–655. doi: 10.1038/nature17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu B., Ackerman D., Sanchez D.J., et al. HIF2α-Dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.Cd-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walther T.C., Farese R.V., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan S.K., Hougen H.Y., Merchan J.R., Gonzalgo M.L., Welford S.M. Fatty acid metabolism reprogramming in ccRCC: mechanisms and potential targets. Nat. Rev. Urol. 2023;20:48–60. doi: 10.1038/s41585-022-00654-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hakimi A.A., Reznik E., Lee C.H., et al. An integrated metabolic Atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanderson S.M., Locasale J.W. Revisiting the Warburg effect: some tumors hold their breath. Cell Metabol. 2018;28:669–670. doi: 10.1016/j.cmet.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wettersten H.I., Aboud O.A., Lara P.N., Jr., Weiss R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017;13:410–419. doi: 10.1038/nrneph.2017.59. [DOI] [PubMed] [Google Scholar]
- 54.Yong C., Stewart G.D., Frezza C. Oncometabolites in renal cancer. Nat. Rev. Nephrol. 2020;16:156–172. doi: 10.1038/s41581-019-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakraborty S., Balan M., Sabarwal A., Choueiri T.K., Pal S. Metabolic reprogramming in renal cancer: events of a metabolic disease. Biochim. Biophys. Acta Rev. Canc. 2021;1876 doi: 10.1016/j.bbcan.2021.188559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan Y., Yang X., Li Y., et al. Expression and prognostic significance of fatty acid synthase in clear cell renal cell carcinoma. Pathol. Res. Pract. 2020;216 doi: 10.1016/j.prp.2020.153227. [DOI] [PubMed] [Google Scholar]
- 57.Du S., Hu W., Zhao Y., et al. Long non-coding RNA MAGI2-AS3 inhibits breast cancer cell migration and invasion via sponging microRNA-374a. Cancer Biomarkers. 2019;24:269–277. doi: 10.3233/cbm-182216. [DOI] [PubMed] [Google Scholar]
- 58.Liu S., Liu X., Wu F., et al. HADHA overexpression disrupts lipid metabolism and inhibits tumor growth in clear cell renal cell carcinoma. Exp. Cell Res. 2019;384 doi: 10.1016/j.yexcr.2019.111558. [DOI] [PubMed] [Google Scholar]
- 59.Du W., Zhang L., Brett-Morris A., et al. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat. Commun. 2017;8:1769. doi: 10.1038/s41467-017-01965-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X., Yuan X., Ni J., et al. MAGI2-AS3 inhibits breast cancer by downregulating DNA methylation of MAGI2. J. Cell. Physiol. 2021;236:1116–1130. doi: 10.1002/jcp.29922. [DOI] [PubMed] [Google Scholar]
- 62.Thorsson V., Gibbs D.L., Brown S.D., et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshihara K., Shahmoradgoli M., Martínez E., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Auslander N., Zhang G., Lee J.S., et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 2018;24:1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 67.Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emura T., Matsui S., Chen H.Y. compoundCox: Univariate feature selection and compound covariate for predicting survival. Comput. Methods Progr. Biomed. 2019;168:21–37. doi: 10.1016/j.cmpb.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 69.Witten D.M., Tibshirani R. Survival analysis with high-dimensional covariates. Stat. Methods Med. Res. 2010;19:29–51. doi: 10.1177/0962280209105024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeh C.T., Liao G.Y., Emura T. Sensitivity analysis for survival prognostic prediction with gene selection: a copula method for dependent censoring. Biomedicines. 2023;11 doi: 10.3390/biomedicines11030797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bagaev A., Kotlov N., Nomie K., et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. 2021;39:845–865.e847. doi: 10.1016/j.ccell.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 72.Li L., Wang X. Identification of gastric cancer subtypes based on pathway clustering. NPJ Precis. Oncol. 2021;5:46. doi: 10.1038/s41698-021-00186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harshitha R., Arunraj D.R. Real-time quantitative PCR: a tool for absolute and relative quantification. Biochem. Mol. Biol. Educ. 2021;49:800–812. doi: 10.1002/bmb.21552. [DOI] [PubMed] [Google Scholar]
- 74.Durślewicz J., Klimaszewska-Wiśniewska A., Antosik P., Grzanka D. Low expression of MATR3 is associated with poor survival in clear cell renal cell carcinoma. Biomedicines. 2023;11 doi: 10.3390/biomedicines11020326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu T.J., Ma D., Liu Y.Y., et al. Bulk and single-cell transcriptome profiling reveal the metabolic heterogeneity in human breast cancers. Mol. Ther. 2021;29:2350–2365. doi: 10.1016/j.ymthe.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hosios A.M., Manning B.D. Cancer signaling drives cancer metabolism: AKT and the Warburg effect. Cancer Res. 2021;81:4896–4898. doi: 10.1158/0008-5472.Can-21-2647. [DOI] [PubMed] [Google Scholar]
- 77.Thakur C., Chen F. Connections between metabolism and epigenetics in cancers. Semin. Cancer Biol. 2019;57:52–58. doi: 10.1016/j.semcancer.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sosa V., Moliné T., Somoza R., Paciucci R., Kondoh H., Me L.L. Oxidative stress and cancer: an overview. Ageing Res. Rev. 2013;12:376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 79.Icard P., Shulman S., Farhat D., Steyaert J.M., Alifano M., Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist. Updates. 2018;38:1–11. doi: 10.1016/j.drup.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 80.Dang C.V., Le A., Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.Ccr-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abdel-Wahab A.F., Mahmoud W., Al-Harizy R.M. Targeting glucose metabolism to suppress cancer progression: prospective of anti-glycolytic cancer therapy. Pharmacol. Res. 2019;150 doi: 10.1016/j.phrs.2019.104511. [DOI] [PubMed] [Google Scholar]
- 82.Leone R.D., Zhao L., Englert J.M., et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366:1013–1021. doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z.H., Peng W.B., Zhang P., Yang X.P., Zhou Q. Lactate in the tumour microenvironment: from immune modulation to therapy. EBioMedicine. 2021;73 doi: 10.1016/j.ebiom.2021.103627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ganapathy-Kanniappan S., Geschwind J.F. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol. Cancer. 2013;12:152. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rolver M.G., Holland L.K.K., Ponniah M., et al. Chronic acidosis rewires cancer cell metabolism through PPARα signaling. Int. J. Cancer. 2022 doi: 10.1002/ijc.34404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chu Q., An J., Liu P., et al. Repurposing a tricyclic antidepressant in tumor and metabolism disease treatment through fatty acid uptake inhibition. J. Exp. Med. 2023;220 doi: 10.1084/jem.20221316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ramos Meyers G., Samouda H., Bohn T. Short chain fatty acid metabolism in relation to gut microbiota and genetic variability. Nutrients. 2022;14 doi: 10.3390/nu14245361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang H., Zhang D., Hu X. A potential fatty acid metabolism-related gene signature for prognosis in clear cell renal cell carcinoma. Cancers (Basel) 2022;14 doi: 10.3390/cancers14194943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang R., Shen J., Chen Y., Gao J., Yao J. Fatty acid metabolism-related signature predicts survival in patients with clear cell renal carcinoma. Aging (Albany NY) 2022;14:9969–9979. doi: 10.18632/aging.204433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baron A., Migita T., Tang D., Loda M. Fatty acid synthase: a metabolic oncogene in prostate cancer? J. Cell. Biochem. 2004;91:47–53. doi: 10.1002/jcb.10708. [DOI] [PubMed] [Google Scholar]
- 91.Menendez J.A., Decker J.P., Lupu R. In support of fatty acid synthase (FAS) as a metabolic oncogene: extracellular acidosis acts in an epigenetic fashion activating FAS gene expression in cancer cells. J. Cell. Biochem. 2005;94:1–4. doi: 10.1002/jcb.20310. [DOI] [PubMed] [Google Scholar]
- 92.Zhao S., Cheng L., Shi Y., Li J., Yun Q., Yang H. MIEF2 reprograms lipid metabolism to drive progression of ovarian cancer through ROS/AKT/mTOR signaling pathway. Cell Death Dis. 2021;12:18. doi: 10.1038/s41419-020-03336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feng K., Ma C., Liu Y., et al. Encapsulation of LXR ligand by D-Nap-GFFY hydrogel enhances anti-tumorigenic actions of LXR and removes LXR-induced lipogenesis. Theranostics. 2021;11:2634–2654. doi: 10.7150/thno.53139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hoxha M., Zappacosta B. A review on the role of fatty acids in colorectal cancer progression. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1032806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wettersten H.I., Hakimi A.A., Morin D., et al. Grade-Dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. 2015;75:2541–2552. doi: 10.1158/0008-5472.Can-14-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weiss R.H. Metabolomics and metabolic reprogramming in kidney cancer. Semin. Nephrol. 2018;38:175–182. doi: 10.1016/j.semnephrol.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin X., Wang Z., Wang J., Xu Y., Kong W., Zhang J. Development of a novel gene signature to predict prognosis and response to PD-1 blockade in clear cell renal cell carcinoma. OncoImmunology. 2021;10 doi: 10.1080/2162402x.2021.1933332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Q., Tang H., Luo X., et al. Immune-associated gene signatures serve as a promising biomarker of immunotherapeutic prognosis for renal clear cell carcinoma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.890150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gui C.P., Wei J.H., Chen Y.H., et al. A new thinking: extended application of genomic selection to screen multiomics data for development of novel hypoxia-immune biomarkers and target therapy of clear cell renal cell carcinoma. Briefings Bioinf. 2021;22 doi: 10.1093/bib/bbab173. [DOI] [PubMed] [Google Scholar]
- 100.Ding R., Wei H., Jiang X., Wei L., Deng M., Yuan H. Prognosis and pain dissection of novel signatures in kidney renal clear cell carcinoma based on fatty acid metabolism-related genes. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.1094657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu C., Wang Y., Nie L., et al. Comprehensive analysis of cellular senescence-related genes in the prognosis, tumor microenvironment, and immunotherapy/chemotherapy of clear cell renal cell carcinoma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.934243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krishna C., DiNatale R.G., Kuo F., et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy. Cancer Cell. 2021;39:662–677.e666. doi: 10.1016/j.ccell.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vuong L., Kotecha R.R., Voss M.H., Hakimi A.A. Tumor microenvironment dynamics in clear-cell renal cell carcinoma. Cancer Discov. 2019;9:1349–1357. doi: 10.1158/2159-8290.Cd-19-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Atkins M.B., Tannir N.M. Current and emerging therapies for first-line treatment of metastatic clear cell renal cell carcinoma. Cancer Treat Rev. 2018;70:127–137. doi: 10.1016/j.ctrv.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 105.Rathmell W.K., Rumble R.B., Van Veldhuizen P.J., et al. Management of metastatic clear cell renal cell carcinoma: ASCO guideline. J. Clin. Oncol. 2022;40:2957–2995. doi: 10.1200/jco.22.00868. [DOI] [PubMed] [Google Scholar]
- 106.Dionellis V.S., Norkin M., Karamichali A., et al. Genomic instability profiles at the single cell level in mouse colorectal cancers of defined genotypes. Cancers (Basel) 2021;13 doi: 10.3390/cancers13061267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in the current study are available from the corresponding author.