Abstract
Interstitial fibrosis, tubular atrophy, and inflammation are major contributors to kidney allograft failure. Here we sought an objective, quantitative pathological assessment of these lesions to improve predictive utility and constructed a deep-learning–based pipeline recognizing normal vs. abnormal kidney tissue compartments and mononuclear leukocyte infiltrates. Periodic acid– Schiff stained slides of transplant biopsies (60 training and 33 testing) were used to quantify pathological lesions specific for interstitium, tubules and mononuclear leukocyte infiltration. The pipeline was applied to the whole slide images from 789 transplant biopsies (478 baseline [preimplantation] and 311 post-transplant 12-month protocol biopsies) in two independent cohorts (GoCAR: 404 patients, AUSCAD: 212 patients) of transplant recipients to correlate composite lesion features with graft loss. Our model accurately recognized kidney tissue compartments and mononuclear leukocytes. The digital features significantly correlated with revised Banff 2007 scores but were more sensitive to subtle pathological changes below the thresholds in the Banff scores. The Interstitial and Tubular Abnormality Score (ITAS) in baseline samples was highly predictive of one-year graft loss, while a Composite Damage Score in 12-month post-transplant protocol biopsies predicted later graft loss. ITASs and Composite Damage Scores outperformed Banff scores or clinical predictors with superior graft loss prediction accuracy. High/intermediate risk groups stratified by ITASs or Composite Damage Scores also demonstrated significantly higher incidence of estimated glomerular filtration rate decline and subsequent graft damage. Thus, our deep-learning approach accurately detected and quantified pathological lesions from baseline or post-transplant biopsies and demonstrated superior ability for prediction of post-transplant graft loss with potential application as a prevention, risk stratification or monitoring tool.
Keywords: deep learning, graft survival, kidney transplantation, renal pathology
Kidney transplantation is the treatment of choice for patients with end-stage renal disease (ESRD).1 Interstitial fibrosis and tubular atrophy and inflammation are considered major contributors to post-transplant kidney allograft failure irrespective of etiology of injury.2 Currently interstitial fibrosis and tubular atrophy and inflammation are graded by pathologic assessment of biopsies. While cumulative injury represented as categorical Banff scores have been associated with post-transplant graft function and survival, these have intermediate sensitivity for graft failure prediction in any given biopsy, due to interobserver and intraobserver variability.3 Prediction of long-term graft survival remains a major challenge. Post-transplant factors, such as the rate of decline of estimated glomerular filtration rate (eGFR) up to 2 years4,5 have shown predictive ability. However, factors obtained early post-transplantation that predict longer-term post-transplant course would offer distinct advantages for identifying patients at risk for graft loss and therefore, potentially guide subsequent patient management.
Recently, deep-learning–based approaches have been successfully applied to radiological medical images6,7 and histologically stained images,8,9 and studies in renal digital pathology have shown promise in detecting glomerular or interstitial abnormalities.10–15 Good prediction of kidney tissue compartments16–18 was obtained with pixel-level prediction algorithm U-Net.19 An instance-level object detection algorithm mask Region-based Convolution Neural Network (R-CNN)20 was developed recently with advantages of performing object localization, shape prediction, and object classification at the same time and that accurately distinguishes sclerotic from nonsclerotic glomeruli.21 We reasoned that these deep-learning–based approaches could be applied for observer-independent histopathologic assessment of transplant biopsies, offering distinct advantages for graft prognostication.
In this study, we first trained a deep-learning model based on both U-Net and mask R-CNN algorithms to accurately recognize normal and abnormal kidney tissue compartments and infiltrated mononuclear leukocytes (MNLs) from both baseline (pre-implantation) and post-transplant biopsies. We then extracted slide-wide features to ensure capture of abnormalities in the interstitium, tubules, and inflammation and investigated their association with Banff scores and post-transplant graft outcomes in 2 large, independent cohorts.
METHODS
Study cohorts and biopsy slides
The Genomics of Chronic Allograft Rejection (GoCAR)22 study is a prospective, multicenter study with patients that have been followed for a median of 5 years. Australian Chronic Allograft Dysfunction (AUSCAD) is an Australia transplant cohort from Westmead Hospital, University of Sydney with patients being followed for a median duration of 4.5 years. Living- and deceased-donor recipients between 18 and 75 years old were included and sensitized; patients with multiple organ transplants were excluded in this study. Blood, kidney biopsy specimens, and clinical data were collected at the time of post-transplantation visit. In GoCAR, 2 protocol biopsy cores were taken from baseline (pre-implantation) or various times (3, 12, and 24 months) post-transplantation. One formalin-fixed, paraffin-embedded corewas processed for histologic stains and scored centrally and agreed byat least 2 pathologists at Massachusetts General Hospital according to revised Banff 2007 classification for renal allograft pathology23 at the time of biopsy. AUSCAD biopsy samples were formalin-fixed and paraffin-embedded prior to routine histologic staining including periodic acid–Schiff (PAS). These biopsies were scored locally at Westmead according to the revised Banff 2007 classification for renal allograft pathology and reviewed by pathologists at Massachusetts General Hospital to ensure consistency in diagnosis between the 2 centers. GoCAR slides were scanned with Aperio CS scanner at ×20 objective with a ×2 magnifier; AUSCAD slides were scanned by scanner from Hamamatsu company with a ×20 objective.
PAS-stained slides in both cohorts were used in this study (Figure 1). To fully capture abnormalities regarding interstitium, tubules, and inflammation, the training set should incorporate all types of abnormal cases covering all 3 aspects. However, due to the various incidences of pathologic lesions observed in 1164 kidney biopsies taken at various time points in the entire GoCAR cohort, a random selection of a subset from these biopsies may miss certain types of abnormal instances. Therefore, we examined each PAS-stained slide and selected 93 slides that represented the spectrum of histologic lesions for model construction. Multiple selected regions covering glomeruli, interstitium, tubules, arteries, and MNL infiltration on these slides were annotated under the guidance of pathologists prior to model construction. Here abnormal tubules are defined as shrunken tubules with thickened and wrinkled membrane, while interstitium is defined as intertubular, nonglomerular space within tissue sections. Next, the established whole-slide image (WSI) investigation pipeline was applied to all available PAS-stained slides, including 478 slides at baseline (GoCAR, n = 317; AUSCAD, n = 161) and 311 slides at 12 months post-transplantation (GoCAR, n = 200; AUSCAD, n = 111) to extract digital features to be correlated with graft survival. These slides represented 404 patients from GoCAR cohort and 212 patients from the AUSCAD cohort.
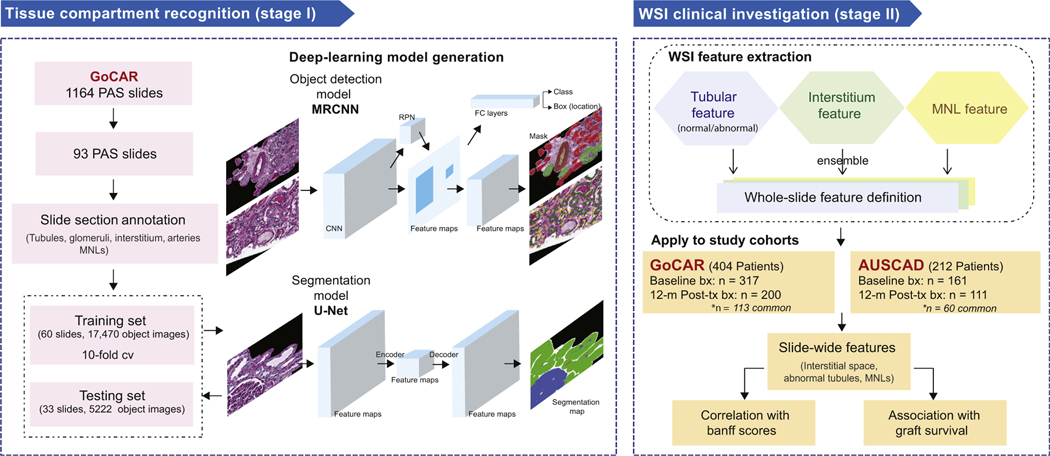
Figure 1 |. Study design.
This study consists of 2 major stages. Stage I is tissue compartment recognition. Ninety-three slides that represented the spectrum of histologic lesions were selected from the Genomics of Chronic Allograft Rejection (GoCAR) periodic acid–Schiff (PAS) slides and then randomly divided into the discovery set (n = 60) and the testing set (n = 33). The annotated sections of these slides were used for deep-learning model construction and evaluation. During the training process, we built the models based on 2 types of deep-learning structures for compartment or mononuclear leukocyte (MNL) detection (by mask region-base convolution neural network [MRCNN]) and tissue segmentation (by U-Net). Models were determined through evaluation with 10-fold cross-validation and finally applied to the testing set. Stage II is the whole-slide image (WSI) clinical investigation. Using the established deep-learning model, we processed 789 baseline and 12-month post-transplantation (post-tx) WSIs from 2 independent cohorts (GoCAR and Australian Chronic Allograft Dysfunction [AUSCAD]) and extracted a series of slide-wide digital features capturing the abnormalities in the interstitium and tubules, and MNL infiltration. These features were further examined through association with Banff scores and post-transplantation graft survival. bx, biopsies; FC, fully connected; RPN, region proposal network.
WSI deep-learning analysis
The WSI deep-learning analysis procedure was divided into 2 stages: stage I includes development of deep-leaning–based models for tissue compartment recognition; and stage II includes the application of pretrained models on WSI to extract slide-wide features to be correlated with graft outcomes (the details were depicted in Figure 1 and Supplementary Figure S1 and described in the Supplementary Methods). Briefly, at stage I, annotated PAS sections including most tissue compartments were preprocessed into 22,692 fixed-sized image tiles for model generation. The deep-learning model was tuned on training image tiles from 60 slides with 10-fold cross-validation to avoid overfitting and the established model was tested on independent image tile set from 33 slides for unbiased model evaluation. We constructed a compartment detection model and an MNL detection model using mask R-CNN20 and an interstitium estimation model using U-Net.19 Detailed hyperparameter settings are described in the Supplementary Methods. By comparing prediction results with ground truth annotations, the accuracies were measured by true positive rate, positive predictive value, and general Fβ score,24 where β = 2. At stage II, outputs from pretrained tissue compartment recognition models were first combined into whole-slide prediction image. Through use of a unit window scanning across the prediction image of entire slide, we defined interstitial or inflammatory regions of interest and slide-wide digital features capturing abnormalities in interstitium, tubules, and MNL infiltration, which were then summarized into composite features reflecting overall kidney damage. This WSI feature extraction process was applied to 2 independent transplant cohorts (GoCAR and AUSCAD) and estimated digital features were correlated with Banff scores and graft survival separately.
Statistical analysis
Quantitative outcomes such as Banff scores or eGFR were treated as continuous variables, and missing data were excluded for specific analyses. Association of digital features with Banff scores or eGFR were measured by Spearman’s correlation. Graft loss was defined as loss of graft function; association with graft loss was assessed by Cox proportional hazards regression; and multiple testing correction has been applied. Time-dependent area under the curve (AUC) values were estimated by R package “timeROC”.25 T denotes follow-up days of a patient. At certain time point t, a case is defined as patient lost graft at T ≤ t; a control is defined as patient survived through t (T > t). As for survival confounders adjustment, a series of clinical parameters including recipient age, sex, race, donor age, number of transplants, kidney diseases, living or deceased donor, human leukocyte antigen mismatch, cold ischemia time (CIT), induction type, baseline donor-specific antibodies, and delayed graft function were first evaluated with graft loss through univariate analysis. Significant parameters—living or deceased donor, CIT, baseline donor-specific antibodies, human leukocyte antigen mismatch, and induction type—were selected as confounders. Further investigations of graft loss and other graft outcomes among risk groups stratified by composite scores were evaluated by log-rank test and Fisher’s exact test, respectively.
RESULTS
Demographic and clinical characteristics of study cohorts
We applied artificial intelligence techniques to all available PAS-stained slides of kidney donor biopsies taken at baseline (pre-implantation) or 12 months post-transplantation in 404 patients from a multicenter international cohort (GoCAR)22 and 212 patients from an external Australian cohort (AUSCAD) (Figure 1). Among these patients, 113 patients in GoCAR and 60 patients in AUSCAD had biopsies taken at both time points, and others were biopsied at either baseline or 12 months. The 2 populations had similar sex distribution, age, and CIT, but they differed in ethnicity and clinical management protocols (Table 1). Patients from GoCAR had more diverse ethnic backgrounds including African American or Hispanic (25% vs. none in AUSCAD), whereas AUSCAD recorded more deceased donors (78.77% vs. 53.71% in GoCAR). All patients from AUSCAD received induction therapy predominantly with lymphocyte nondepleting agents (93.87%), while among 78.22% of recipients from GoCAR who received induction, lymphocyte-depleting agents (Thymoglobulin or Campath-1) were used in 39.36% and nondepleting agents in 38.86%. Overall, the AUSCAD cohort had a lower graft loss rate (4.72% vs. 12.13% in GoCAR) during slightly shorter follow-up period (median 4.5 years vs. 5 years in GoCAR).
Table 1 |.
Demographic and clinical characteristics in 2 independent kidney transplant cohorts
| Characteristics | GoCAR (n = 404) | AUSCADa (n = 212) | P valueb |
|---|---|---|---|
| Recipient age, yr | 49.38 ± 13.52 | 48.44 ± 12.11 | 0.381 |
| Recipient sex | 0.282 | ||
| Female | 129 (31.93) | 77 (36.32) | |
| Male | 275 (68.07) | 135 (63.68) | |
| Recipient race | 1.7e–19 | ||
| Caucasian | 261 (64.6) | 177 (83.49) | |
| Asian | 24 (5.94) | 27 (12.74) | |
| African American | 76 (18.81) | 0 (0) | |
| Hispanic | 25 (6.19) | 0 (0) | |
| Other | 18 (4.46) | 8 (3.77) | |
| Dialysis | 6.0e–04 | ||
| No | 89 (22.03) | 23 (10.95) | |
| Yes | 315 (77.97) | 187 (89.05) | |
| Kidney disease | 1.3e–05 | ||
| Diabetes mellitus | 139 (34.41) | 79 (37.98) | |
| Glomerulonephritis | 74 (18.32) | 66 (31.73) | |
| Hypertension | 77 (19.06) | 14 (6.73) | |
| Polycystic kidney disease | 41 (10.15) | 20 (9.62) | |
| Other | 73 (18.07) | 29 (13.94) | |
| Donor age, yr | 42.02 ± 15.51 | 45.43 ± 16.8 | 0.019 |
| Donor sex | 0.609 | ||
| Female | 197 (48.76) | 105 (50.97) | |
| Male | 207 (51.24) | 101 (49.03) | |
| Deceased donor | 6.5e-10 | ||
| No | 187 (46.29) | 45 (21.23) | |
| Yes | 217 (53.71) | 167 (78.77) | |
| CIT, min | 530.65 ± 494.21 | 501.06 ± 245.1 | 0.324 |
| HLA mismatch | 0.010 | ||
| 0 | 46 (11.39) | 12 (6.19) | |
| 1–2 | 55 (13.61) | 42 (21.65) | |
| 3–4 | 150 (37.13) | 58 (29.9) | |
| 5–6 | 153 (37.87) | 82 (42.27) | |
| Delayed graft function | 9.0e–04 | ||
| No | 334 (82.67) | 150 (70.75) | |
| Yes | 70 (17.33) | 62 (29.25) | |
| Induction type | 5.1e–46 | ||
| Lymphocyte nondepletion | 157 (38.86) | 199 (93.87) | |
| Lymphocyte depletion | 159 (39.36) | 13 (6.13) | |
| None | 88 (21.78) | 0 (0) | |
| Follow-up, d | 1776.98 ± 660.2 | 1637.39 ± 849.81 | 0.038 |
| Death-censored graft loss | 0.002 | ||
| No | 355 (87.87) | 202 (95.28) | |
| Yes | 49 (12.13) | 10 (4.72) |
AUSCAD, Australian Chronic Allograft Dysfunction; CIT, cold ischemia time; GoCAR, Genomics of Chronic Allograft Rejection; HLA, human leukocyte antigen.
Several variables in the AUSCAD cohort had missing values: donor age (n = 24), CIT (n = 8), dialysis (n = 2), kidney disease (n = 4), donor sex (n = 6), HLA mismatch (n = 18).
P values were calculated using Fisher’s exact test (categorical variables) or Student’s t test (continuous variables).
Values are median ± SD or n (%).
Deep-learning–based WSI investigation defined abnormality in interstitium or tubules and MNL infiltration
Our 2-stage study first generated a deep-learning model detecting tissue compartments and MNLs, and then defined slide-wide abnormality features to be correlated with Banff scores23 and graft outcomes (Figure 1, Supplementary Methods). In stage I, 3 types of models based on 2 deep-learning architectures were built on 17,470 images from 60 slides (training set) using 10-fold cross-validation. The models, respectively, identified tissue compartments (tubules, glomeruli, etc.), MNLs (mask R-CNN), and interstitial area (U-Net). The final model was tested on an independent set of 5,222 images from 33 slides, and accurately recognized 96% of glomeruli and 91% of tubules and differentiated normal and abnormal tubules at true positive rates of 81% and 84%, respectively. On the other hand, we were able to detect 90% of normal epithelial cells as well as 77% of MNLs at the individual nuclei level. The slightly lower accuracy of MNL detection was reflective of challenges in MNL annotation on PAS slides. Lastly, 85% and 96% of predicted interstitial area and area covered by arteries were correctly identified (Supplementary Table S1).
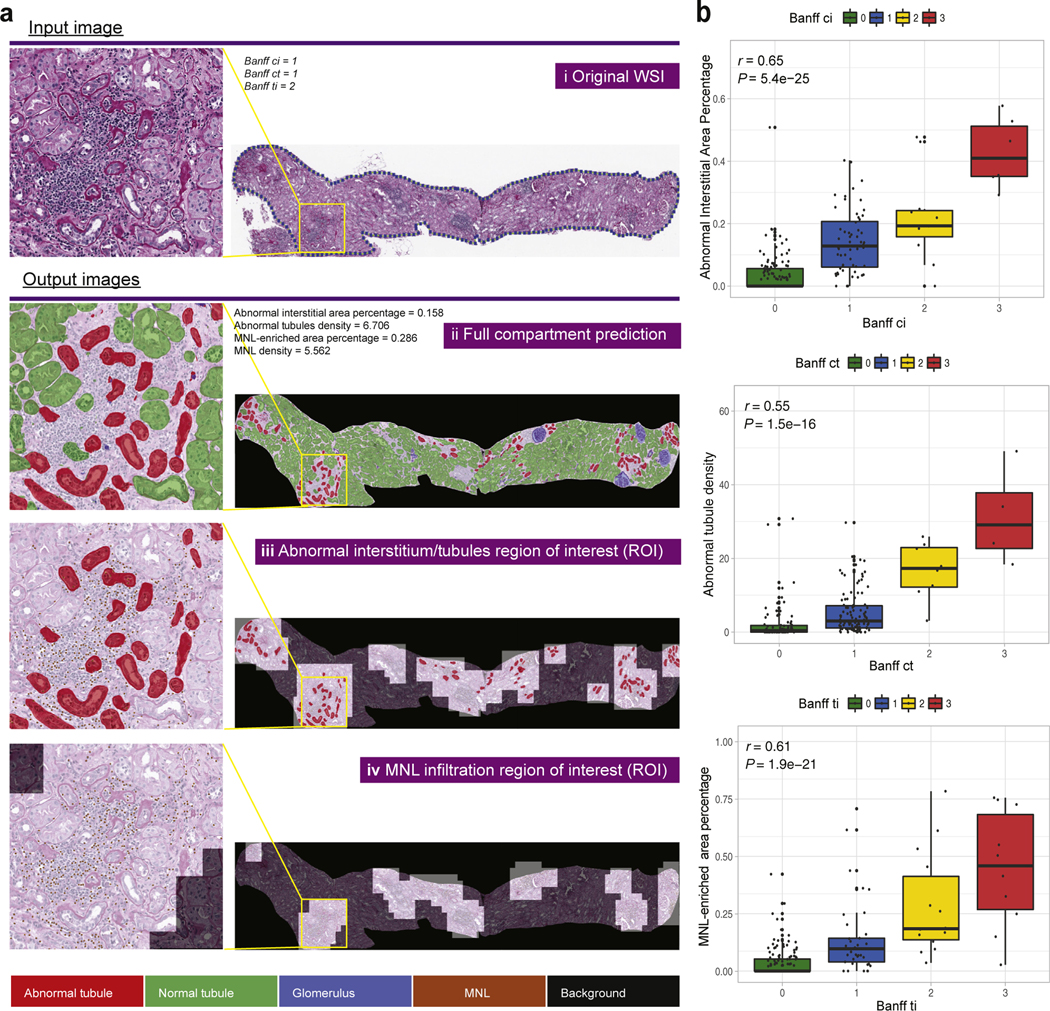
In stage II, we applied the pretrained tissue compartment recognition models to WSIs to extract a series of slide-wide digital features specifically capturing abnormalities within biopsies (Supplementary Figure S1, Supplementary Methods). For quantifying abnormalities in tubules and/or interstitium, we defined (i) abnormal interstitial area percentage, proportion of total abnormal interstitium area over WSI; (ii) standardized abnormal tubule density; (iii) Interstitial and Tubular Abnormality Score (ITAS), a composite score of (i) and (ii). To quantify inflammation in biopsies (i.e., MNL infiltration), we defined (iv) MNL-enriched area percentage, proportion of MNL infiltration area over WSI; (v) standardized MNL density; (vi) MNL Infiltration Score, a composite score of (iv) and (v). Lastly, a Composite Damage Score (CDS), integrating both ITAS and MNL Infiltration Score, was defined as the estimation of overall graft damage. Figure 2a demonstrates an example application of our pipeline to an abnormal case: (i) original WSI, (ii) whole-slide prediction, and the masks highlighting (iii) abnormal interstitium or tubule regions or (iv) MNL infiltration regions that agreed with assessment by pathologists.
Figure 2 |. Demonstration of slide-wide digital features and correlation with corresponding Banff scores.
(a) Demonstration of slidewide digital features from the whole-slide image (WSI) investigation by an example WSI: (i) original WSI; (ii) whole-slide prediction; (iii) predicted abnormal interstitium or tubules regions of interest (ROIs); (iv) predicted mononuclear leukocytes (MNLs) infiltrated ROIs. Left panel shows zoom-in inspections of 1 particular abnormal region within the yellow box on the WSI. (b) Correlation of digital features with Banff scores. Correlation of abnormal interstitial area percentage and Banff ci score (top), abnormal tubules density and Banff ct score (middle), MNL-enriched area percentage and Banff ti score (bottom) in the Genomics of Chronic Allograft Rejection (GoCAR) 12-month post-transplantation biopsy slides (n = 200). P values were calculated from Spearman’s correlation test. To optimize viewing of this image, please see the online version of this article at www.kidney-international.org.
Digital features were correlated with Banff scores
The Banff scores such as interstitial fibrosis (ci), tubular atrophy (ct), and total inflammation (ti) (graded by expert visual-assessment from different histologic stains) are similar in pathologic principle but different in quantification and technique to our PAS-based digital features (as illustrated in Supplementary Figure S1B). Here, we examined the relationship between these 2 methods. We performed a WSI investigation, extracting digital features in 789 WSIs from biopsies at baseline (n = 478) and 12 months post-transplantation (n = 311) in both the GoCAR and AUSCAD cohorts. Our data indicated that digital features (abnormal interstitial area percentage, abnormal tubules density, and MNL-enriched area percentage) were significantly correlated with respective Banff scores in GoCAR biopsies at baseline (Supplementary Figure S2A) and 12 months (Figure 2b). Similarly, the digital scores were correlated with Banff scores in AUSCAD (12 months) where i+t was used because of unavailability of ti score (Supplementary Figure S2B). Notably, although MNL detection at nuclei level yielded a lower accuracy compared to detection of glomeruli or tubules, MNL-derived digital inflammation feature was strongly correlated with ti score in GoCAR (P = 1.9e–21) and i+t in AUSCAD (P = 1.9e–05) at slide level.
Although highly correlated, we still identified discrepancies between the 2 scoring systems such as the case demonstrated in Supplementary Figure S3A: here, Banff assessment reported all zeros but digital features indicated abnormal scores (illustrated by small clusters of shrunken tubules and MNLs). We then inspected all 137 cases classified as normal by Banff criteria (ci, ct, i, t, ti, g, cv = 0) from baseline biopsies and identified 50 abnormal and 87 normal cases based on digital features. No graft loss by 1 year was observed in these cases, but the baseline digitally abnormal group, compared with the all-normal group, had higher subsequent Banff ci+ct scores early post-transplantation, which were especially significant within the first 3 months (Supplementary Figure S3B). Moreover, the digitally abnormal group had a worse subsequent graft function as measured by eGFR within 12 months post-transplantation (Supplementary Figure S3C).
Taken together, the above-mentioned data indicate that our digital features accurately reflect Banff scores and identified similar histologic lesions. Furthermore, it suggested that in cases of discrepancy, digital quantitative scores offer a more sensitive assessment of graft damage below the Banff threshold.
Baseline interstitial and tubular abnormality score predicted early graft damage and 1-year graft loss
The pathologic evaluation of baseline biopsies could reveal donor kidney quality. However, its utility in post-transplant prognosis has been debated.26 To explore a novel application for our digital features in baseline biopsies, we examined the association of individual or composite features with post-transplant graft failure and compared these with the performance of Banff-based scores. We also compared these with Kidney Donor Profile Index (KDPI), a composite demographic and clinical factor that is validated for deceased donors.27–29 In the GoCAR cohort (n=317) (Supplementary Figure S4A, Supplementary Table S2), we observed significant association of individual interstitial or tubular features and composite ITAS with death-censored graft loss (DCGL) in univariate or multivariate Cox models. In the AUSCAD cohort (n = 161) (Supplementary Table S3), the association with graft survival was not confirmed in DCGL, which could be because there were fewer DCGL cases.
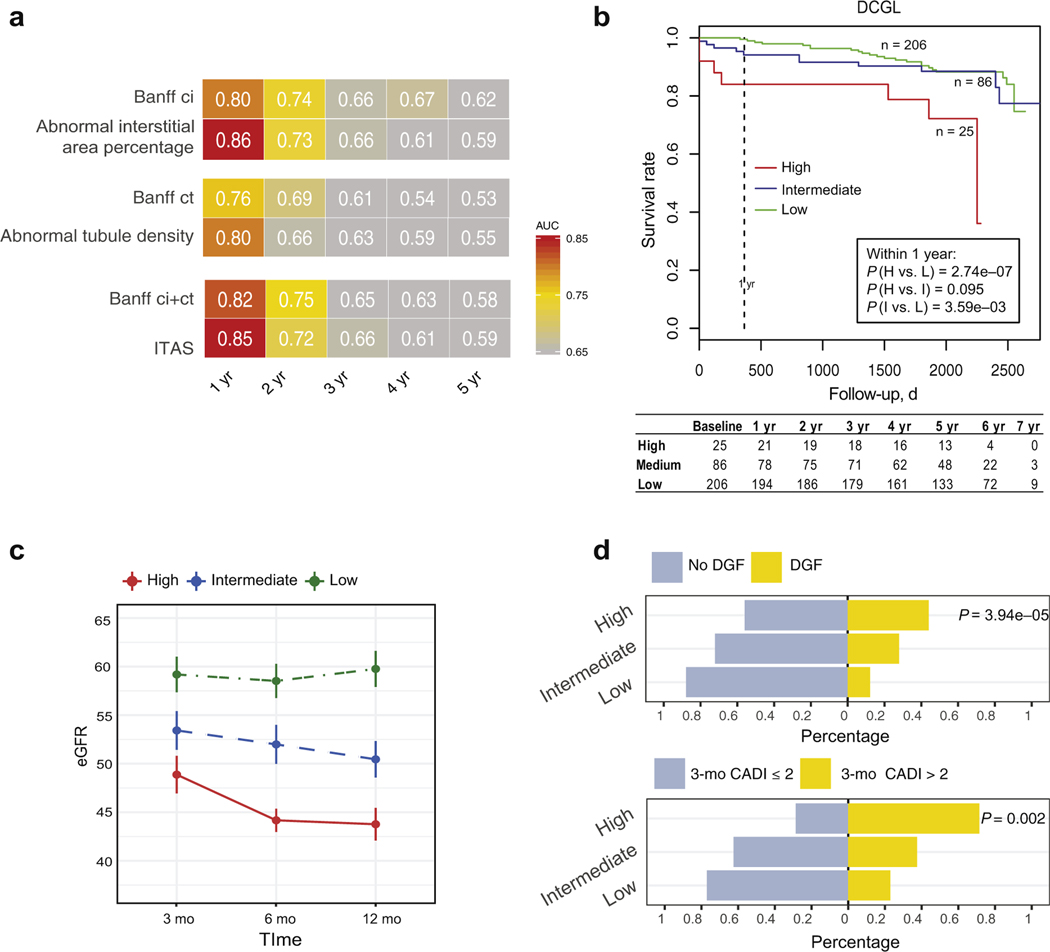
Time-dependent AUC estimation in GoCAR indicated that baseline individual or composite digital features outperformed individual Banff scores or ci+ct, respectively, in prediction of DCGL within 12 months (Figure 3a). Next, we divided baseline biopsies into 3 risk groups by composite feature ITAS: high (ITAS > 0.6), intermediate (0.1 ≤ ITAS ≤ 0.6), and low (ITAS < 0.1) risk. Threshold of high baseline ITAS was determined according to the percentile of baseline ci+ct > 1 in the GoCAR cohort. A second threshold for low risk was added to identify healthy donor kidneys with zero or extremely low ITAS (< 0.1). The high and intermediate ITAS risk groups exhibited significantly higher DCGL rates compared with those of the low ITAS risk group over the entire period of follow-up. These differences were most apparent in the first 12 months post-transplantation (P = 2.8e–07 for high vs. low and P = 3.6e–03 for intermediate vs. low) (Figure 3b) and in the deceased-donor subcohort (P = 5.3e–04 for high vs. low and P = 0.011 for intermediate vs. low) (Supplementary Figure S4B). ITAS was superior to ci+ct (P = 6.0e–04 for high vs. low and P = 0.197 for intermediate vs. low in the entire population) (Supplementary Figure S4C) and KDPI (P = 0.024 for high vs. low and P = 0.141 for intermediate vs. low, KDPI > 85%, 20% < KDPI ≤ 85%, KDPI ≤ 20% in deceased-donor population) (Supplementary Figure S4D) for risk stratification of DCGL. Of note, high and intermediate ITAS risk groups demonstrated a sustained decline in eGFR over the first 12 months post-transplantation (Figure 3c), which is consistent with incrementally significant correlation of ITAS with post-transplant eGFR at 3 months (P = 0.001), 6 months (P = 7.6e–05), 12 months (P = 1.5–05), and 24 months (P = 0.015). A significantly higher incidence of delayed graft function (P = 3.9e–05), and early (3 months post-transplantation) graft damage as measured by the Chronic Allograft Damage Index (CADI) score30 > 2 (P = 0.002) were observed in high and intermediate ITAS risk groups (Figure 3d). In the AUSCAD cohort (n = 161), the associations of ITAS risk groups with other clinical outcomes were demonstrated in Supplementary Figure S5. To summarize, donor baseline ITAS is strongly associated with early graft function within 1 year post-transplantation, but the degree of association weakens afterward, implicating that recipient factors and post-transplant conditions come into play.
Figure 3 |. Association of baseline digital features with post-transplant graft outcomes in the Genomics of Chronic Allograft Rejection (GoCAR) cohort.
(a) Heat map of time-dependent area under the curve (AUC) values in predicting death-censored graft loss (DCGL) by Banff scores and digital features at different time intervals in baseline biopsy slides (n = 317). Numbers and yellow-red color range of boxes represent AUC values at given time points. (b) Kaplan-Meier curves of DCGL in high, intermediate, and low risk groups stratified by the Interstitial and Tubular Abnormality Score (ITAS) from baseline biopsies (n = 317). Baseline ITAS groups are defined as high, ITAS > 0.6; intermediate, 0.1 ≤ ITAS ≤ 0.6; and low, ITAS < 0.1. P values are calculated by log-rank test. (c) Average estimated glomerular filtration rate (eGFR) values over time within 12-months post-transplantation per baseline ITAS risk group. Error bars represent ×0.1 SD from mean values. (d) Bar charts demonstrating proportions of delayed graft function (DGF) and no DGF (upper) and 3-month post-transplant Chronic Allograft Damage Index (CADI) >2 or ≤2 (lower) among 3 baseline ITAS risk groups. P values are calculated by Fisher’s exact test.
Twelve-month post-transplant composite damage score predicted long-term graft loss
Because our data showed that composite baseline digital score predicts early but not long-term DCGL, we examined longer-term subsequent graft survival using 12-month post-transplant biopsy slides in both cohorts. In GoCAR (n = 200) (Supplementary Figure S6A, Supplementary Table S4), digital interstitial and tubular features, which are superior to corresponding Banff ci and ct scores, were significantly associated with long-term DCGL with or without adjustment for clinical confounders (living or deceased donor, CIT, baseline donor-specific antibodies, human leukocyte antigen mismatch, and induction type), while the MNL feature was comparable to the Banff ti score in association with DCGL. The associations of 12-month digital features with long-term survival were validated in the AUSCAD cohort (n = 111) (Supplementary Table S5).
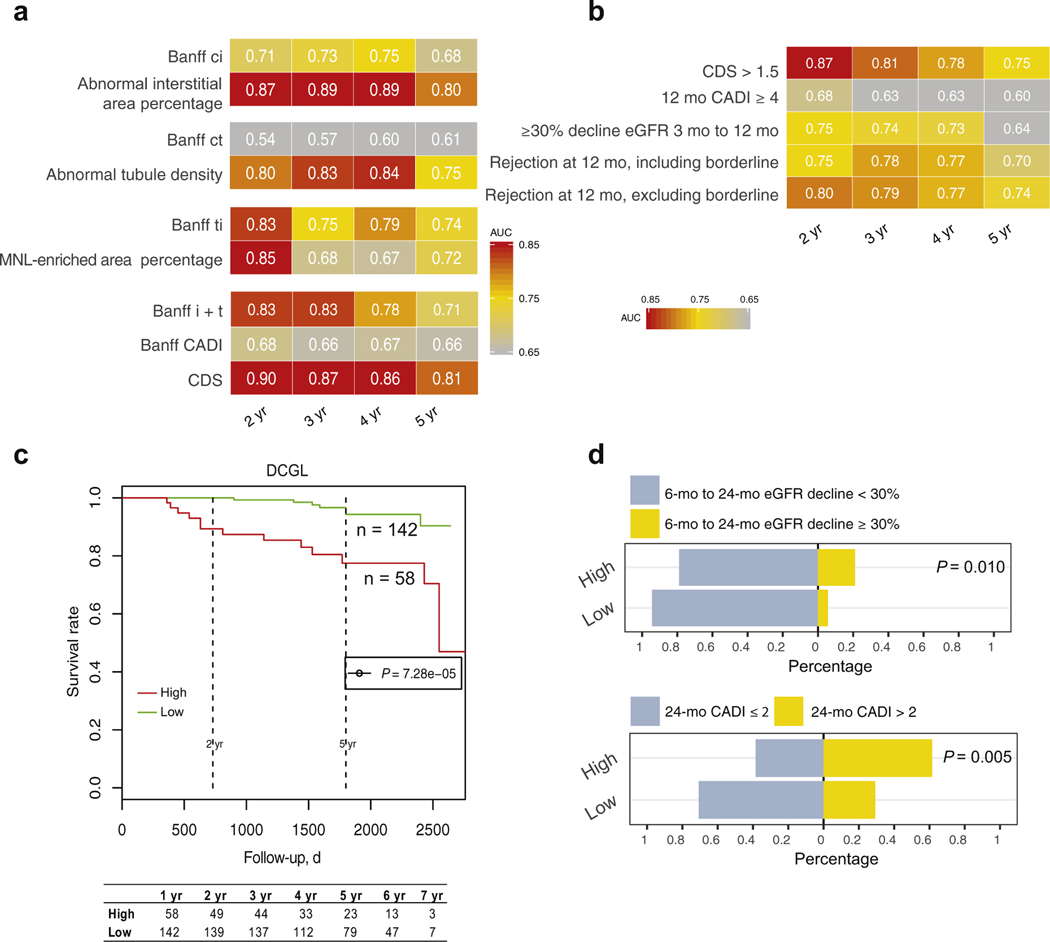
We observed that 12-month digital features outperformed corresponding Banff scores including CADI in predicting long-term graft loss with superior time-dependent AUCs in the GoCAR cohort (Figure 4a). We then used the CDS summarizing abnormalities detected in interstitium, tubules, and inflammation for graft loss risk stratification. We determined the threshold of 12-month CDS (> 1.5) according to the percentile of 12-month CADI ≥ 4 in the GoCAR cohort, as 1-year CADI ≥ 4 is considered a surrogate for high risk of graft loss in patients who received transplants.31 A 12-month CDS > 1.5 outperformed 12-month CADI ≥ 4, >30% 3-month to 12-month eGFR decline, and acute cellular rejection (including or excluding borderline cases) at 12 months in long-term survival prediction, especially for graft survival within 2 years post-transplantation (Figure 4b). Kaplan-Meier curves of DCGL (P = 7.3e–05) (Figure 4c) confirmed significantly lower survival rate in patients with high 12-month CDS. We also identified significant associations of 12-month CDS risk groups with other published surrogate outcomes including >30% 6-month to 24-month eGFR decline4,5 (P = 0.010) and progressive histologic damage (P = 0.005; 24-month CADI > 2) (Figure 4d). These analyses in the AUSCAD cohort (n = 111) also validated the predictive ability of 12-month CDS for long-term survival (Supplementary Figure S7). Thus, high 12-month CDS (> 1.5), obtained at 12 months post-transplantation, is an alternative surrogate for long-term graft loss.
Figure 4 |. Association of 12-month post-transplant digital features with post-transplant graft outcomes in the Genomics of Chronic Allograft Rejection (GoCAR) cohort.
(a) Heat map of time-dependent area under the curve (AUC) values in predicting death-censored graft loss (DCGL) by Banff scores and digital features at different time intervals in 12-month post-transplant biopsy slides (n = 200). Numbers and yellow-red color range of boxes represent AUC values at given time points. (b) Heat map of time-dependent AUCs in predicting DCGL by the 12-month Composite Damage Score ([CDS], capturing the interstitial and tubular abnormality and mononuclear leukocyte [MNL] infiltration) high or low group and other pathologic or clinical (or both) factors that were obtained prior to or at 12 months. The 12-month CDS groups are defined as high, CDS > 1.5, and low, CDS ≤ 1.5. (c) Kaplan-Meier curves of the DCGL in high and low risk groups stratified by the 12-month CDS. P value is calculated by log-rank test. (d) Bar charts demonstrating proportions of 6-month to 24-month estimated glomerular filtration rate (eGFR) decline ≥30% or <30% (upper) and the 24-month post-transplant Chronic Allograft Damage Index (CADI) >2 or ≤2 (lower) between 12-month CDS risk groups. P values are calculated by Fisher’s exact test.
DISCUSSION
We constructed a deep-learning–based histopathologic assessment model recognizing and quantifying interstitial, tubular, and inflammatory abnormalities in kidney transplant biopsies. WSI investigation of baseline and 12-month post-transplant biopsies validated these digital features and further explored potential applications of composite features in clinical practice. Our digital features not only exhibited strong correlation with relevant Banff scores, but they also detected subtle changes below the thresholds in Banff scores. Composite features of baseline ITAS and 12-month CDS were identified to be predictive of early and late graft outcomes, respectively, implying utility in transplant prognosis. To the best of our knowledge, this is the first study applying artificial intelligence techniques in identifying digital pathologic features associated with solid organ transplant survival from both baseline and post-transplant biopsies with validation in multiple prospective cohorts.
Compared to previous investigations in deep-learning–based kidney tissue compartment detection,16–18 our study advances the field in 4 ways: (i) Besides U-Net, we incorporated a mask R-CNN architecture for more efficient and accurate detection of the normal and abnormal compartments. (ii) As inflammation is another major contributor to graft failure, we added a mask R-CNN–based MNL detection model in post-transplant biopsy evaluations, improving graft loss predictive ability. (iii) The slide-wide pathologic lesions were quantified through definition of individual features in interstitium, tubules, and MNL infiltration, respectively, or composite features reflecting overall kidney damage. (iv) We explored a novel clinical application of developed digital features for graft survival prediction in 2 well-designed cohorts. Both GoCAR and AUSCAD are large prospective cohorts that collected protocol biopsies pre-implantation and at time points post-transplantation and followed up for medians of 4.5 to 5 years. GoCAR is a multicenter prospective (non-interventional) cohort involving 4 regions in the United States (New York, Michigan, Wisconsin, Illinois) and 1 region in Australia (Sydney). The patients who received kidney transplants are truly heterogeneous, coming from various race or ethnicity backgrounds and using different standard-of-care protocols at different sites. Therefore, the demographic, clinical, and pathologic data in GoCAR were reflective of heterogeneous patients who received transplants and “real-world” clinical management. The models developed from the GoCAR cohort have been validated in external AUSCAD cohort and are very likely to be applicable to other cohorts.
Although many attempts have been made, no consistent association has been established between baseline histologic findings and post-transplant outcomes among publications.26,32 Comparing our GoCAR cohort with previous studies, we obtained superior performance in predicting graft loss with baseline digital features as well as Banff scores, we consider the following reasons: (i) Our GoCAR biopsies were collected from multiple centers but were scored centrally by the pathology experts at Massachusetts General Hospital, which minimized the variation from pathology expertise from different centers. (ii) Our pre-implanted baseline biopsies were preserved through paraffin embedding rather than freezing procedure. It has been reported that frozen tissue stained with hematoxylin and eosin contain less contrast thus subtle lesions can easily be missed, and the artifacts in frozen sections often cause misdiagnoses,26 leading to poor association with post-transplant outcomes. (iii) Although controversial, a few studies have reported significant associations between interstitial fibrosis– and tubular atrophy–related pathologic features and graft function or survival.33–41 Taken together, the studies from our group and others proved the association of baseline pathologic features with transplant outcomes including graft survival. We particularly demonstrated a strong prediction power of short-term survival using baseline digital features. The major limitations of current approaches in pathologic evaluation for baseline biopsies are the variations from slide processing procedure and the expertise in transplant pathologic assessment.32,42 The Banff system itself has limitations by using categories rather than continuous variables.43 Our machine-based process overcomes these drawbacks by producing consistent and automated results within 30 minutes from scanned images. The ITAS at baseline was superior to Banff ci + ct and KDPI and demonstrated the ability of stratifying risk of early graft damage, thus providing early information with utility for post-transplant monitoring, risk stratification, or potential interventional trials.
We identified that the CDS from 12-month protocol biopsies predicted long-term graft survival, outperforming histology and clinical factors. Reporting longer-term hard outcomes from prospective trials has been an issue in kidney transplantation research.44 The identification of surrogate end points is a major unmet need that often prevents the design of adequately powered trials. Recent studies proposed using eGFR decline within 24 to 36 months as a long-term graft loss surrogate.4,5 However, such a surrogate has the following limitations: (i) Creatinine measurement is impacted by a number of factors including timing of collection in the day, diet, and interlaboratory variation.45,46 (ii) eGFR decline has low detection sensitivity because it requires multiple measurements during long-term follow-up, and the ≥ 40% decline from 6 to 24 months, as suggested by a prior study for graft loss prediction,5 only occurred in 4% of patients in the GoCAR cohort although rates of graft loss were 12% for DCGL. In contrast, 12-month CDS was able to detect 29% of GoCAR and 21% of AUSCAD populations as high risk as early as 12 months while still exhibiting optimal AUCs in long-term graft loss prediction.
This work focused on investigation of the digital features from protocol biopsies at baseline and 12-month post-transplantation with transplant graft outcomes (particularly graft loss) for prognosis purpose. However, we expect our tissue recognition model, which was built from protocol biopsies, works on for-cause biopsies as well, because the severity of histologic lesions (such as Banff ct, ti score) relies largely on the amount or density of individual abnormal objects and similarly our slide-wide digital features are summarized from detection of corresponding abnormal objects. Thus, with an accurate detection of individual abnormal objects, our slide-wide digital features would be expected to accurately reflect the pathologic lesions regarding interstitium, tubules, and MNL infiltration and correlate with Banff scores in both protocol and for-cause biopsies. As an ongoing project, we are collecting additional clinical outcomes such as treatment information for patients who had indication biopsy taken to extensively investigate the utility of the digital features from for-cause biopsies in clinical diagnosis or prognosis.
Our study has several limitations. First, the identification of microvascular inflammation (g and ptc) and arteritis (v) requires further refinement. Additionally, due to the challenges of distinguishing MNLs from epithelial cells in tubules, current MNL detection appears less accurate within tubules than in interstitium therefore current slide-wide inflammation feature focused on overall inflammation estimation (similar to Banff ti score). Second, its ability to diagnose and grade acute cellular rejection has not been demonstrated and it is unable to differentiate between antibody- and cell-mediated rejection. We aim to improve the MNL detection model in various compartments on PAS-stained slides in conjunction with CD3 staining to extend the capability of inflammation capture. In addition, further refinements are required to diagnose transplant glomerulopathy or de novo or recurrent glomerular diseases. Lastly, the thresholds of composite scores for risk stratification were determined based on the percentile of CADI and ci+ct in the GoCAR cohort. Nevertheless, these unsupervised risk stratifications clearly outperformed conventional multivariate risk calculators using clinical factors47–49 for predicting graft failure. However, expanded cohorts with sufficient graft loss cases are necessary to determine precise thresholds based on a supervised model by incorporating both digital features and clinical factors.
In summary, our deep-learning approach provided a reliable risk stratification of post-transplant graft survival using transplant biopsies at baseline and 12 months post-transplantation. This represents a novel and reproducible approach to facilitate early prevention, risk stratification, or post-transplant monitoring in clinical practice.
Supplementary Material
Figure S1. Illustration of slide-wide digital features extraction process and definition.
Figure S2. Correlation of digital features with corresponding Banff scores.
Figure S3. Discrepancy between digital features and Banff scores.
Figure S4. Association of baseline digital features with post-transplant graft outcomes in the Genomics of Chronic Allograft Rejection (GoCAR) cohort.
Figure S5. Association of baseline digital features with post-transplant graft outcomes in the Australian Chronic Allograft Dysfunction (AUSCAD) cohort.
Figure S6. Association of 12-month post-transplant digital features with post-transplant graft outcomes in the Genomics of Chronic Allograft Rejection (GoCAR) cohort.
Figure S7. Association of 12-month post-transplant digital features with post-transplant graft outcomes in the Australian Chronic Allograft Dysfunction (AUSCAD) cohort.
Table S1. Accuracy summary of kidney tissue compartment prediction model.
Table S2. Association of baseline Banff scores and digital features with graft loss in the Genomics of Chronic Allograft Rejection (GoCAR) cohort
Table S3. Association of baseline Banff scores and digital features with graft loss in the Australian Chronic Allograft Dysfunction (AUSCAD) cohort.
Table S4. Association of 12-month post-transplant Banff scores and digital features with graft loss in the Genomics of Chronic Allograft Rejection (GoCAR) cohort.
Table S5. Association of 12-month post-transplant Banff scores and digital features with graft loss in the Australian Chronic Allograft Dysfunction (AUSCAD) cohort.
Translational Statement.
This is the first study applying artificial intelligence techniques to both baseline (pre-implantation) and post-transplantation biopsies to identify quantitative digital features using 2 large prospective transplant cohorts. Our automated deep-learning approach showed accuracy in predicting early and long-term graft survival using baseline and 12-month transplantation biopsies, respectively. This approach represents a novel tool for risk stratification of allografts and post-transplantation monitoring in clinical practice.
ACKNOWLEDGMENTS
We thank Ms. Meyke Hermsen in Dr. Jeroen A.W.M. van der Laak’s lab in Department of Pathology of Radboud University Medical Center in Nijmegen for suggestions in kidney compartment annotation using ASAP (Automated Slide Analysis Platform) program. We thank the Scientific Computing Division at the Icahn School of Medicine at Mount Sinai for providing computational resources.
This work is a substudy of the Genomics of Chronic Renal Allograft Rejection (GoCAR) study sponsored by National Institutes of Health grant no. 5U01AI070107-03. The cost of clinical, histologic, and genomic experiments, as well as the authors’ effort involved in patient enrollment, data analysis, and manuscript preparation were paid by this grant. All the authors have reviewed the manuscript and agreed to submission.
DISCLOSURE
BM reports stock in RenalytixAI and VericiDx. WZ reports personal fees from VericiDx. BM and WZ report the following patents: (i) Patents US provisional patent application F&R ref 27527-0134P01, serial no. 61/951,651, filled March 2014: Method for identifying kidney allograft recipients at risk for chronic injury; (ii) US provisional patent application: Methods for diagnosing risk of renal allograft fibrosis and rejection (miRNA); (iii) US provisional patent application: Method for diagnosing subclinical acute rejection by RNA sequencing analysis of a predictive gene set; and (iv) US provisional patent application: Pretransplant prediction of posttransplant acute rejection. PJO’C is a consultant for CSL Behring and Vitaeris.
Footnotes
All the other authors declared no competing interests.
DATA AVAILABILITY STATEMENT
The coded and deidentified participant data will be made available to qualifying researchers by requesting the data from the corresponding authors. Proposals will be reviewed by the investigators and collaborators based on scientific merit. If the proposal is approved, the data will be shared through a secure data transfer site.
REFERENCES
- 1.Hunsicker LG. A survival advantage for renal transplantation. N Engl J Med. 1999;341:1762–1763. [DOI] [PubMed] [Google Scholar]
- 2.Parajuli S, Aziz F, Garg N, et al. Histopathological characteristics and causes of kidney graft failure in the current era of immunosuppression. World J Transplant. 2019;9:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furness PN, Taub N. Convergence of European Renal Transplant Pathology Assessment Procedures (CERTAP) Project. International variation in the interpretation of renal transplant biopsies: report of the CERTPAP project. Kidney Int. 2001;60:1998–2012. [DOI] [PubMed] [Google Scholar]
- 4.Clayton PA, Lim WH, Wong G, Chadban SJ. Relationship between eGFR decline and hard outcomes after kidney transplants. J Am Soc Nephrol. 2016;27:3440–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faddoul G, Nadkarni GN, Bridges ND, et al. CTOT-17 Consortium. Analysis of biomarkers within the initial 2 years posttransplant and 5-year kidney transplant outcomes: results from clinical trials in Organ Transplantation-17. Transplantation. 2018;102:673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29:102–127. [DOI] [PubMed] [Google Scholar]
- 7.Xue Y, Chen S, Qin J, et al. Application of deep learning in automated analysis of molecular images in cancer: a survey. Contrast Media Mol Imaging. 2017;2017:9512370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Yang DM, Rong R, et al. Pathology image analysis using segmentation deep learning algorithms. Am J Pathol. 2019;189:1686–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukowy JD, Dayton A, Cloutier D, et al. Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol. 2018;29:2081–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallego J, Pedraza A, Lopez S, et al. Glomerulus classification and detection based on convolutional neural networks. J Imaging. 2018;4:20. [Google Scholar]
- 12.Ginley B, Lutnick B, Jen K-Y, et al. Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol. 2019;30:1953–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kannan S, Morgan LA, Liang B, et al. Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Rep. 2019;4:955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh JN, Matlock MK, Kudose S, et al. Deep learning global glomerulosclerosis in transplant kidney frozen sections. IEEE Trans Med Imaging. 2018;37:2718–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ginley B, et al. Automated computational detection of interstitial fibrosis, tubular atrophy, and glomerulosclerosis. J Am Soc Nephrol. 2021;32:837–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouteldja N, Klinkhammer BM, Bülow RD, et al. Deep learning-based segmentation and quantification in experimental kidney histopathology. J Am Soc Nephrol. 2021;32:52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermsen M, de Bel T, den Boer M, et al. Deep learning–based histopathologic assessment of kidney tissue. J Am Soc Nephrol. 2019;30:1968–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayapandian CP, Chen Y, Janowczyk AR, et al. ; Nephrotic Syndrome Study Network (NEPTUNE). Development and evaluation of deep learning–based segmentation of histologic structures in the kidney cortex with multiple histologic stains. Kidney Int. 2021;99:86–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. 2015. arXiv:1505.04597. [Google Scholar]
- 20.Abdulla W Mask R-CNN for object detection and instance segmentation on Keras and TensorFlow. GitHub repository 2017. https://github.com/matterport/Mask_RCNN. Accessed March 31, 2019. [Google Scholar]
- 21.Altini N, Cascarano G, Brunetti A, et al. A deep learning instance segmentation approach for global glomerulosclerosis assessment in donor kidney biopsies. Electronics. 2020;9:1768. [Google Scholar]
- 22.O’Connell PJ, Zhang W, Menon MC, et al. Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: a multicentre, prospective study. Lancet. 2016;388:983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8:753–760. [DOI] [PubMed] [Google Scholar]
- 24.Van Rijsbergen CJ. Information Retrieval. 2nd ed. ButterworthHeinemann; 1979. [Google Scholar]
- 25.Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32:5381–5397. [DOI] [PubMed] [Google Scholar]
- 26.Naesens M Zero-time renal transplant biopsies: a comprehensive review. Transplantation. 2016;100:1425–1439. [DOI] [PubMed] [Google Scholar]
- 27.OPTN. A guide to calculating and interpreting the Kidney Donor Profile Index (KDPI). 2020. https://optn.transplant.hrsa.gov/media/1512/guide_to_calculating_interpreting_kdpi.pdf. Accessed June 5, 2020. [Google Scholar]
- 28.OPTN. KDRI to KDPI mapping table. 2018. https://optn.transplant.hrsa.gov/media/2974/kdpi_mapping_table_2018.pdf. Accessed June 5, 2020. [Google Scholar]
- 29.Rao PS, Schaubel DE, Guidinger MK, et al. A comprehensive risk quantification score for deceased donor kidneys: the kidney donor risk index. Transplantation. 2009;88:231–236. [DOI] [PubMed] [Google Scholar]
- 30.Helanterä I, Ortiz F, Koskinen P. Chronic Allograft Damage Index (CADI) as a biomarker in kidney transplantation. In: Patel VB, Preedy VR, eds. Biomarkers in Kidney Disease. Springer Netherlands; 2016:669–687. [Google Scholar]
- 31.Hayry P, Paavonen T, Taskinen E, et al. Protocol core needle biopsy and histological chronic allograft damage index as surrogate endpoint for long-term graft survival. Transplant Proc. 2004;36:89–91. [DOI] [PubMed] [Google Scholar]
- 32.Wang CJ, Wetmore JB, Crary GS, Kasiske BL. The donor kidney biopsy and its implications in predicting graft outcomes: a systematic review. Am J Transplant. 2015;15:1903–1914. [DOI] [PubMed] [Google Scholar]
- 33.Howie AJ, Ferreira MAS, Lipkin GW, Adu D. Measurement of chronic damage in the donor kidney and graft survival. Transplantation. 2004;77:1058–1065. [DOI] [PubMed] [Google Scholar]
- 34.De Vusser K, Lerut E, Kuypers D, et al. The predictive value of kidney allograft baseline biopsies for long-term graft survival. J Am Soc Nephrol. 2013;24:1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopes JA, Moreso F, Riera L, et al. Evaluation of pre-implantation kidney biopsies: comparison of Banff criteria to a morphometric approach. Kidney Int. 2005;67:1595–1600. [DOI] [PubMed] [Google Scholar]
- 36.Navarro MD, López-Andréu, Rodríguez-Benot A, et al. Significance of preimplantation analysis of kidney biopsies from expanded criteria donors in long-term outcome. Transplantation. 2011;91:432–439. [DOI] [PubMed] [Google Scholar]
- 37.Hofer J, Regele H, Böhmin GA, et al. Pre-implant biopsy predicts outcome of single-kidney transplantation independent of clinical donor variables. Transplantation. 2014;97:426–432. [DOI] [PubMed] [Google Scholar]
- 38.Losappio V, Stallone G, Infante B, et al. A single-center cohort study to define the role of pretransplant biopsy score in the long-term outcome of kidney transplantation. Transplantation. 2014;97:934–939. [DOI] [PubMed] [Google Scholar]
- 39.Kahu J, Kyllönen L, Räisänen-Sokolowski A, Salmela K. Donor risk score and baseline biopsy CADI value predict kidney graft outcome. Clin Transplant. 2011;25:E276–E283. [DOI] [PubMed] [Google Scholar]
- 40.Heilman RL, Smith ML, Smith BH, et al. Progression of interstitial fibrosis during the first year after deceased donor kidney transplantation among patients with and without delayed graft function. Clin J Am Soc Nephrol. 2016;11:2225–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arias LF, Blanco J, Sanchez-Fructuoso A, et al. Histologic assessment of donor kidneys and graft outcome: multivariate analyses. Transplant Proc. 2007;39:1368–1370. [DOI] [PubMed] [Google Scholar]
- 42.Singh P, Farber JL, Doria C, et al. Peritransplant kidney biopsies: comparison of pathologic interpretations and practice patterns of organ procurement organizations. Clin Transplant. 2012;26:E191–E199. [DOI] [PubMed] [Google Scholar]
- 43.Vasquez-Rios G, Menon MC. Kidney transplant rejection clusters and graft outcomes: revisiting Banff in the era of “big data.”. J Am Soc Nephrol. 2021;32:1009–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fergusson NA, Ramsay T, Chassé M, et al. Impact of using alternative graft function endpoints: a secondary analysis of a kidney transplant trial. Transplant Direct. 2019;5:e439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joffe M, Hsu C, Feldman HI, et al. Variability of creatinine measurements in clinical laboratories: results from the CRIC study. Am J Nephrol. 2010;31:426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delanaye P, Cavalier E, Pottel H. Serum creatinine: not so simple! Nephron. 2017;136:302–308. [DOI] [PubMed] [Google Scholar]
- 47.Haller MC, Wallisch C, Mjøen G, et al. Predicting donor, recipient and graft survival in living donor kidney transplantation to inform pretransplant counselling: the donor and recipient linked iPREDICTLIVING tool—a retrospective study. Transpl Int. 2020;33:729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irish WD, Ilsley JN, Schnitzler MA, et al. A risk prediction model for delayed graft function in the current era of deceased donor renal transplantation. Am J Transplant. 2010;10:2279–2286. [DOI] [PubMed] [Google Scholar]
- 49.Kasiske BL, Israni AK, Snyder JJ, et al. A simple tool to predict outcomes after kidney transplant. Am J Kidney Dis. 2010;56:947–960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Illustration of slide-wide digital features extraction process and definition.
Figure S2. Correlation of digital features with corresponding Banff scores.
Figure S3. Discrepancy between digital features and Banff scores.
Figure S4. Association of baseline digital features with post-transplant graft outcomes in the Genomics of Chronic Allograft Rejection (GoCAR) cohort.
Figure S5. Association of baseline digital features with post-transplant graft outcomes in the Australian Chronic Allograft Dysfunction (AUSCAD) cohort.
Figure S6. Association of 12-month post-transplant digital features with post-transplant graft outcomes in the Genomics of Chronic Allograft Rejection (GoCAR) cohort.
Figure S7. Association of 12-month post-transplant digital features with post-transplant graft outcomes in the Australian Chronic Allograft Dysfunction (AUSCAD) cohort.
Table S1. Accuracy summary of kidney tissue compartment prediction model.
Table S2. Association of baseline Banff scores and digital features with graft loss in the Genomics of Chronic Allograft Rejection (GoCAR) cohort
Table S3. Association of baseline Banff scores and digital features with graft loss in the Australian Chronic Allograft Dysfunction (AUSCAD) cohort.
Table S4. Association of 12-month post-transplant Banff scores and digital features with graft loss in the Genomics of Chronic Allograft Rejection (GoCAR) cohort.
Table S5. Association of 12-month post-transplant Banff scores and digital features with graft loss in the Australian Chronic Allograft Dysfunction (AUSCAD) cohort.
Data Availability Statement
The coded and deidentified participant data will be made available to qualifying researchers by requesting the data from the corresponding authors. Proposals will be reviewed by the investigators and collaborators based on scientific merit. If the proposal is approved, the data will be shared through a secure data transfer site.