Abstract
Purpose
Retinitis pigmentosa (RP) is the most common cause of inherited blindness, with onset occurring as early as 4 years of age in certain rare but severe forms caused by mutations in the gamma subunit of phosphodiesterase 6 (PDE6). Studies in humans and mice have shown that RP pathology begins with progressive photoreceptor death, which then drives changes in downstream neurons, neighboring retinal pigment epithelium (RPE), and vasculature. Here, we present the first detailed analysis of RP disease progression in Pde6g-deficient mice.
Design
Experimental study of an RP mouse model.
Subjects
We studied Pde6g−/− and Pde6g+/− mice at the age of 7, 16, 30, 44, and 56 days with n = 2 to 5 per group and time point.
Methods
Photoreceptor degeneration and retinal remodeling were analyzed in retinal sections by immunofluorescence. Retinal blood vessel degradation was analyzed in flat-mounted retinas immunolabeled for isolectin GS-IB4. Protein expression was measured by immunoblot. Acellular capillaries were assessed in trypsin-digested and hematoxylin–eosin-stained retinas at postnatal day (P) 44. Retinal pigment epithelium cells were delineated in flat-mounted RPE-choroid-sclera by immunolabeling for the cell-adhesion protein β-catenin.
Main Outcome Measures
Immunofluorescence and morphometry (quantitative analysis of outer nuclear layer, dendrite area, vessel area, acellular vessels, RPE cell size, number of nuclei per RPE cell, RPE cell eccentricity, and RPE cell solidity).
Results
This novel RP model exhibits early onset and rapid rod degeneration, with the vast majority gone by P16. This pathology leads to retinal remodeling, including changes of inner retinal neurons, early activation of glia cells, degradation of retinal vasculature, and structural abnormalities of the RPE.
Conclusions
The pathology in our Pde6g−/− mouse model precisely mirrors human RP progression. The results demonstrate the significant role of the gamma subunit in maintaining phosphodiesterase activity and provide new insights into the disease progression due to Pde6g deficiency.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found after the references.
Keywords: Retinitis pigmentosa, Remodeling, Retinal vasculature, Retinal pigment epithelium (RPE), PDE6G
Retinitis pigmentosa (RP) is the most common inherited retinal dystrophy worldwide, with 1 case for every 4000 people.1 Retinitis pigmentosa is a progressive disease, with patients initially experiencing night blindness, followed by a gradual narrowing of the visual field (tunnel vision) and, ultimately, loss of daylight vision (color and fine acuity).2 This clinical progression is mirrored by progressive cellular degeneration—first of the mutant rod photoreceptors that mediate night vision, followed by the secondary death of cone photoreceptors that mediate daylight vision.3 This cell loss in the photoreceptor layer leads to remodeling in the neighboring inner retinal and retinal pigment epithelium (RPE) layers, as well as in blood vessels.4 For example, remodeling of neurons in the inner nuclear layer includes retraction of bipolar cell dendrites, formation of new processes, and migration of surviving bipolar and amacrine cells into the photoreceptor or ganglion cell layers. In addition, Müller glia cells undergo reactive gliosis, forming a glial seal between the remnant neural retina and the RPE. Typically, gliosis is characterized by a dramatic increase in expression of glial fibrillary acidic protein and pronounced hypertrophy of Müller cells. Alterations of the RPE cells (e.g., RPE atrophy or pigmented bone spicules) and attenuation of the retinal blood vessels are also common secondary changes in human RP.4,5
Rod-specific phosphodiesterase 6 (PDE6), composed of alpha and beta catalytic subunits and 2 identical inhibitory gamma subunits,6,7 is an essential component of the visual transduction cascade that, in response to light activation, regulates intracellular cyclic guanosine monophosphate levels by hydrolysis of cyclic guanosine monophosphate.8 Mutations in any of the 3 PDE6 subunits can cause autosomal recessive RP in humans,9 demonstrating that each subunit is essential for photoreceptor function and maintenance.9, 10, 11 Mutations in PDE6A and PDE6B are common, accounting for 5% to 8% of autosomal recessive RP cases.12, 13, 14 On the other hand, mutations in PDE6G are rare11 yet can lead to severe early-onset RP, with marked reduction in scotopic and photopic electroretinograms as early as 4 years of age.11
Loss of PDE6 function in mice also causes retinal degeneration that mimics the human phenotype, and numerous Pde6a- and Pde6b-deficient mouse models of RP have been used to study disease progression and remodeling as well as to develop therapies. In contrast, Pde6g-mutant mice (Pdegtm1/tm1, Del7C, Y84G, and W70A) have been mainly used to examine the effects of these mutations on photoreceptor degeneration and the activation and deactivation phases of phototransduction.15 Significantly, the retinal pathology in Pde6g-mutant mice has not been analyzed. Here, we characterize a new PDE6G-deficient mouse model, Pde6gCreERT2/CreERT2 (referred to as Pde6g−/−), and use it to study the effect of Pde6g gene disruption on retinal morphology. We demonstrate very early and rapid degeneration of rod photoreceptor cells, with only a single row of cone photoreceptor nuclei remaining by postnatal day (P) 16. We also observed remodeling of inner retinal neurons, glial cell activation, decreased retinal vascularization, and abnormal RPE morphology. Thus, our Pde6g−/− mouse model faithfully replicates the progressive structural pathologies of human RP. More specifically, with its early onset and rapid time course, these mice model the devastating PDE6G-driven forms of human RP.
Material and Methods
Animals
Animal experiments were performed according to the Association for Research in Vision and Ophthalmology statement for the use of animals in ophthalmic and vision research and were approved by the local authorities. Mice were kept at a 12-hour light/12-hour dark cycle with access to water and food ad libitum. The Pde6gCreERT2 mutation was generated in the Barbara & Donald Jonas Stem Cells Laboratory, Columbia University, by replacing exons 2 and 3 of the Pde6g gene with a CreERT2.16 In our study, mutant mice (Pde6gCreERT2/CreERT2, referred to as Pde6g−/−) and control mice (Pde6gCreERT2/+, referred to as Pde6g+/−) of both sexes were used. Polymerase chain reaction primers used for genotyping were as follows:
Pde6g forward: 5′-GGTCAGATTCCAGTGTGTGGG-3′
Pde6g internal: 5′-CTTAGGTGGTCCTTTCCTGGG-3′
Pde6g reverse: 5′-GTTTAGCTGGCCCAAATGTTG-3′
Eye Preparation
Eyes were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS; pH, 7.4) for 5 min. The cornea and lens were removed, followed by a 40-min incubation in 4% paraformaldehyde. For cryosections, eyecups were washed 3 times in PBS (pH, 7.4), cryoprotected overnight in 30% sucrose at 4°C, and embedded in Tissue-Tek optimum cutting temperature compound (Sakura). Eyecups were frozen and sectioned vertically at 10 μm using a Leica CM3050S cryostat, collected on Thermo Scientific SuperFrost Plus slides, and stored at −20°C. For flat-mounted retinal and RPE-choroid-sclera preparations, eyecups were washed 3 times in PBS (pH, 7.4). The retina was separated, and RPE was bleached in 10% H2O2 for 1.5 hours at 55°C. For immunoblotting, retinas were removed and immediately frozen in liquid nitrogen.
Immunofluorescence
Retinal sections were stained in 5% ChemiBlocker (MerckMillipore #2170), 0.3% Triton-X-100 in PBS (pH, 7.4) overnight at 4°C using anti-Pde6g/h (Santa Cruz #sc-166350), anticone arrestin (Sigma # AB15282), anti-Pde6b (Invitrogen #PA1722), antisecretagonin (gift from Prof. Dr Ludwig Wagner, University of Vienna, Austria), anticalbindin (Swant #300), antiprotein kinase c alpha (Santa Cruz #sc-8393), and antiglutamic-acid-rich protein (Sigma #MABN2429) primary antibodies. Flatmounts were stained in 5% ChemiBlocker, 3% dimethyl sulfoxide, and 0.3% Triton-X-100 in PBS (pH, 7.4) overnight at 4°C using anti-ionized calcium-binding adapter molecule 1 (Fujifilm Wako #019-19741) and anti-β-catenin (Cell Signaling #8480) primary antibodies or isolectin GS-B4 conjugated fluorescein (Sigma #L2895). Corresponding anti-Rabbit AF488 (Invitrogen #A-11070), anti-Rabbit AF647 (Invitrogen #A-21245), and anti-Mouse AF555 (Invitrogen #A-21425) secondary antibodies were incubated in 3% ChemiBlocker in PBS (pH, 7.4) for 1.5 hours at room temperature. Hoechst 33342 (Invitrogen #H1399) was used to stain nuclei. Samples were fixed with Aqua-Poly/Mount (Polysciences #18606) on Thermo Scientific SuperFrost Plus slides and stored at 4°C.
Trypsin Digestion and Hematoxylin–Eosin Staining
Retinal flatmounts were washed 5 times for 30 min in ddH2O and incubated overnight at 4°C, followed by digestion in 3% trypsin (Thermo #27250018) in 0.1M Tris buffer (pH, 7.8) for 90 min at 37°C. The inner limiting membrane was removed with scissors in ddH2O, and vasculature was cleaned by several washing steps. After drying, it was mounted on Thermo Scientific SuperFrost Plus slides and stained with the hematoxylin–eosin fast staining kit (Carl Roth #9194.1). Samples were fixed with Aqua-Poly/Mount.
Imaging and Quantification
Retinal sections were imaged with a Zeiss LSM710 confocal microscope. Images of retinal sections were taken in the ventral or dorsal area of the eye. Outer nuclear layer (ONL) thickness was measured 250 μm from the optic nerve using the Fiji software. Rod bipolar cell, horizontal cell, and cone bipolar cell dendrite areas close to the optic nerve were quantified as pixels in the outer plexiform layer using Fiji. Flatmounts were imaged as Z-Stack with a custom-made VisiScope CSU-X1 confocal system equipped with a high-resolution sCMOS camera (Visitron Systems) or with a Keyence BZ-X800 microscope using the sectioning tool. Relative vessel area was quantified using the AngioTool software.17 Acellular capillaries were manually counted in brightfield images with equal size using Fiji. Retinal pigment epithelium cell morphology parameters were measured in images with equal size using CellProfiler 4.0.7. Nuclei per RPE cell were manually counted. All data were plotted using GraphPad Prism 9.3. Data are expressed as mean ± standard error of mean using an unpaired t test. P ≤ 0.05 was considered statistically significant (∗P ≤ 0.05; ∗∗P ≤ 0.01; ∗∗∗P ≤ 0.001). The N values refer to the number of individual animals.
Immunoblot
Retinae were lysed in M-PER Mammalian Protein Extraction Reagent (Thermo #78503) containing protease inhibitor (Sigma #11697498001) with a Branson Sonifier W-450D at 40% amplitude. Protein concentration was determined using the Pierce Coomassie protein assay kit (Thermo #23200). Proteins (20 μg per sample) were separated by SDS-PAGE (Bio-Rad Mini Protean Tetra system) and transferred to a 0.2 μm polyvinylidene difluoride (PVDF) membrane (Sigma; GE10600021) for 2 hours at 120 V. Membranes were incubated in 5% nonfat dry milk in Tris-buffered saline with Tween20 for 1 hour at room temperature. Primary antibodies such as anti-Pde6g/h, antiglutamic-acid-rich protein, antiglial fibrillary acidic protein (Sigma #G3893), and anti-ß-actin-peroxidase (Sigma #A3854) were incubated in 5% nonfat dry milk overnight at 4°C. Corresponding anti-Mouse horseradish peroxidase (Santa Cruz # sc-516102) secondary antibody was incubated for 1 hour at room temperature. Proteins were detected using Immobilon Forte Western HRP substrate (Millipore #WBLUF0100) in a Bio-Rad ChemiDoc MP imager.
Results
Pde6g−/− Developing Mouse Retinas
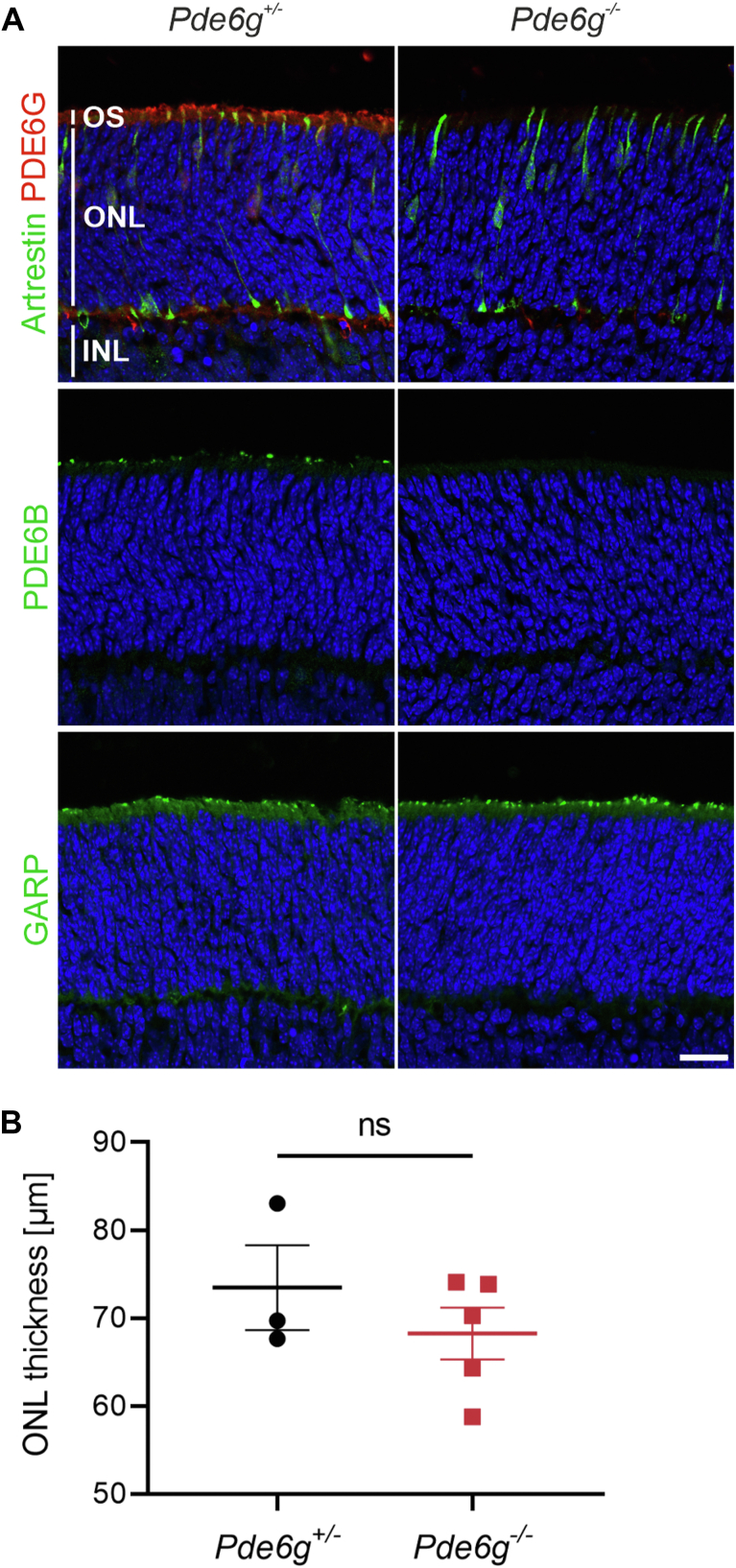
In our Pde6g−/− mice, Pde6g exons 2 and 3 are replaced by CreERT2 (Fig S1).16 Heterozygous Pde6g+/− mice are used to drive tamoxifen-inducible, rod-specific Cre recombinase in order to carry out deletions at specific sites in the DNA.16,18,19 To analyze the effect of PDE6G on postnatal development, we immunolabeled retinas from 7-day-old Pde6g−/− and Pde6g+/− (control) mice. It has already been shown that in P7 wild-type mice, photoreceptors are immature (i.e., rod and cone outer segments are very short) and programmed photoreceptor cell death is ongoing (i.e., the ONL is thicker than adult retina).20 We first immunolabeled sections for PDE6G and found, as expected, immunolabeling in outer segments of Pde6g+/− retinas but not Pde6g−/− (Fig 2A). Since loss of a PDE6 subunit leads to reduced expression of the other 2 subunits,21 we immunolabeled sections for PDE6B and found that, like PDE6G, PDE6B immunolabeling was not detectable in Pde6g−/− retinas (Fig 2A). To analyze rod outer segments and cones, sections were immunolabeled for N-terminal glutamic-acid-rich protein, which is a domain of rod cyclic nucleotide-gated beta1 channel subunit, and cone arrestin, respectively. Immunolabeling for both markers in Pde6g−/− and Pde6g+/− retinas was indistinguishable (Fig 2A). Rod and cone outer segments are still short as the first discs develop at approximately P7 in mice.22,23 Finally, the mean ONL thickness was not significantly different in Pde6g−/− retinas (vs. Pde6g+/−) (P = 0.4) (Fig 2B). These data suggest that, before P7, PDE6G has either no effect on gross morphological development or a very modest effect.
Figure 2.
Morphological development of photoreceptors before postnatal day (P) 7 is not affected by phosphodiesterase 6 (PDE6G) depletion. Retinas were dissected from P7 Pde6g+/− (control) and Pde6g−/− (mutant) mice, sectioned, and immunolabeled. A, Representative images of sections stained for PDE6G and cone arrestin, PDE6B, or glutamic-acid-rich protein (GARP), and counterstained with Hoechst. B, Outer nuclear layer (ONL) thickness. Circles and squares, individual mice. Pde6g+/− (n = 3); Pde6g−/− (n = 5). Data are presented as the mean ± standard error of the mean and group means compared by an unpaired t test. Scale bar, 20 μm. INL = inner nuclear layer; ns = not significant; OS = outer segment.
Progression of Photoreceptor Degeneration and Inner Retinal Remodeling in Pde6g−/− Retinas
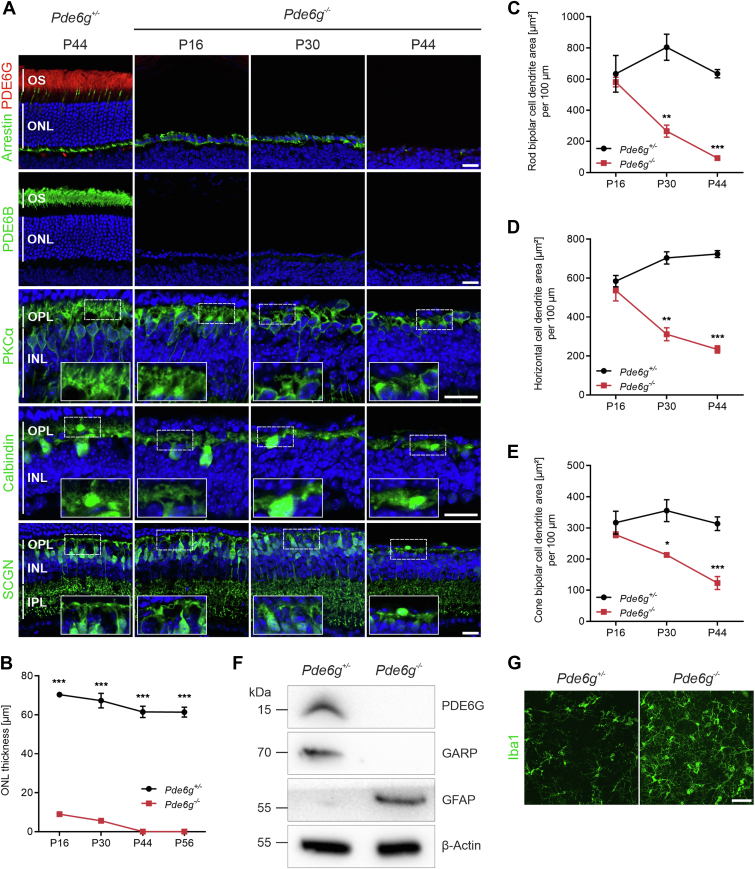
We next analyzed the effects of PDE6G loss at later time points—P16, P30, and P44. As expected, PDE6G and PDE6B immunolabeling was not detectable in Pde6g−/− retinas at all time points but was intense in outer segments of Pde6g+/− retinas at P44 and P30 (Fig 3A, data not shown). At P16, PDE6G expression was less intense due to shorter rod outer segments of immature photoreceptors (data not shown).22 In Pde6g−/− retinas, cones (arrestin immunolabeling) appeared as a single row of nuclei in the ONL at P16 as well as at P30 and were completely lost by P44 (Fig 3A). To quantify photoreceptor degeneration (rods and cones), ONL thickness was measured in retinal sections (Fig 3B). At P16 and P30, ONL thickness in Pde6g−/− retinas was already reduced by 87% and 92%, respectively, compared to age-matched Pde6g+/− retinas. By P44, all photoreceptor cells had died. In control Pde6g+/− retinas, the slight decrease in ONL thickness between P16 and P44 is due to normal programmed cell death of rods.24
Figure 3.
Loss of phosphodiesterase 6 (PDE6G) results in rapid photoreceptor degeneration and retinal remodeling. A, Representative images of retinal sections immunolabeled for PDE6G (rod outer segment [OS]), cone arrestin (cones), PDE6B (rod OS), protein kinase C alpha (PKCα) (rod bipolar cells), calbindin (horizontal cells), and secretagonin (SCGN) (cone bipolar cells) and counterstained with Hoechst. B, Outer nuclear layer (ONL) thickness of mice at postnatal day (P) 16 (n = 2–3), P30 (n = 3), P44 (n = 3), and P56 (n = 3–4). Circles and squares, individual mice. Data, presented as the mean ± standard error of the mean (SEM), were compared by unpaired t test. ∗∗∗P < 0.001. C–E, Quantification of dendrite area from rod bipolar cells (C), horizontal cells (D), and cone bipolar cells (E) at P16 (n = 2–3), P30 (n = 3), and P44 (n = 4), respectively. Circles and squares, individual mice. Data, presented as the mean ± SEM, were compared by unpaired t test. ∗P < 0.03; ∗∗P < 0.002; ∗∗∗P < 0.001. F, Representative immunoblot of P30 retinal lysates. β-actin, loading control. G, Representative images of microglia/macrophages in the center of P44 flat-mounted retinas immunostained for ionized calcium-binding adapter molecule 1 (Iba1). Scale bars, 10 μm. GARP = glutamic-acid-rich protein; GFAP = glial fibrillary acidic protein; INL = inner nuclear layer; IPL = inner plexiform layer; kDa = kilodalton; OPL = outer plexiform layer.
Photoreceptor degeneration is accompanied by changes in downstream neurons.25 To evaluate the time course of these changes in our Pde6g−/− mice, we visualized rod bipolar, horizontal, and cone bipolar neurons by immunostaining for protein kinase c alpha, calbindin, and secretagonin, respectively. In Pde6g−/− retinas, rod bipolar cell dendrites were shorter and less bushy along with thinning of the outer plexiform layer at P30, and at P44, most had disappeared. Horizontal cell processes underwent progressive retraction, which was evident at P30, and further progressed at P44. Cone bipolar cells and their dendrites (secretagonin) were shorter at P30; at P44, many cone bipolar cells had disappeared (Fig 3A). We next quantified the observed changes in rod bipolar cell dendrites, horizontal cell dendrites, and cone bipolar cell dendrites. At P16, there was no significant difference in the dendrite areas between Pde6g+/− and Pde6g−/− retinas. At P30, the dendritic areas were significantly reduced in Pde6g−/− compared with Pde6g+/− retinas. At P44, the dendritic areas were further decreased, reflecting the progressive retraction of dendrites (Fig 3C–E).
The absence of PDE6G and rod outer segments (glutamic-acid-rich protein) at P30 was further confirmed by immunoblotting (Figs 3F and S4). Because photoreceptor degeneration triggers reactive gliosis in Müller glial cells and chronic microglial activation, both were analyzed in our Pde6g−/− retinas. Reactive gliosis, analyzed by immunoblotting P30 retinal lysates, revealed dramatically increased glial fibrillary acidic protein expression in Pde6g−/− retinas (Fig 3F). Dramatic microglia/macrophage activation was revealed by increased ionized calcium-binding adapter molecule 1 immunolabeling in the center of P44 retinas (Fig 3G).
Thus, the loss of PDE6G leads to rapid degeneration of rod photoreceptor cells and secondary degeneration of cones (Fig 3A). The vast majority of rods degenerate between P7 (Fig 2B) and P16 (Fig 3A). At P30, rods are gone and few cones remain. At P44, all photoreceptors, including cones, have died (Fig 3A, B). Downstream neurons are already impacted by P16, with shorter and less complex processes (Fig 3A, C–E).
Retinal Blood Vessel Degeneration in Pde6g−/− Retinas
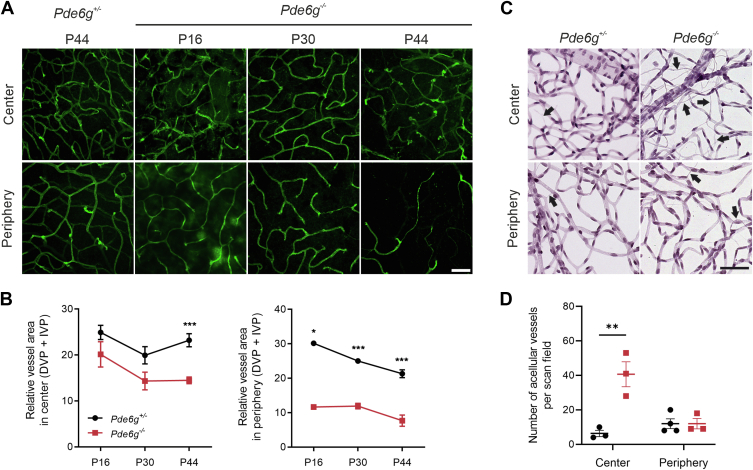
In mice, the retinal vasculature forms a trilaminar network of superficial, intermediate, and deep vascular plexuses. Following the loss of photoreceptors, this retinal vascular network decreases. In our Pde6g−/− mice (vs. Pde6g+/−), we analyzed vascular changes in flat-mounted P16, P30, and P44 retinas immunolabeled for isolectin GS-IB4, an endothelial cell marker. The mean vessel area in deep and intermediate vascular plexi was significantly reduced at all time points in peripheral retina. In central retinas, corresponding vessel area was again lower at all time points, but the difference was only significant at P44 (Fig 5A, B). In the superficial vascular plexus, vessel area was not changed at any time point in central or peripheral retinas (Fig S6).
Figure 5.
Retinal blood vessel degeneration in Pde6g−/− mice. A, Retinal flatmounts were immunostained for isolectin GS-IB4. B, Quantification of deep vascular plexus (DVP) + intermediate vascular plexus (IVP) blood vessel in retinal center and periphery: postnatal day (P) 16 (n = 2–3), P30 (n = 3–4), and P44 (n = 5). C, Retinas from P44 mice were flat-mounted, trypsin digested, and hematoxylin–eosin stained. Data presented as mean ± standard error of the mean. D, Quantification of acellular capillaries: Pde6g+/− (n = 3–4) and Pde6g−/− (n = 3). Circles and squares, individual mice. Pde6g−/− vs. Pde6g+/− age-matched group means were compared by unpaired t tests. ∗P 0.02; ∗∗P 0.01; ∗∗∗P < 0.001. Scale bars, 50 μm.
When endothelial cells die, the result is acellular capillaries—nonperfused basement membrane sleeves with no endothelial cell nuclei along their length and an important marker for monitoring progression of RP. We assessed acellular capillaries at P44 in retinal flatmounts. While there were only small numbers of acellular capillaries in central and peripheral Pde6g+/− retinas, the numbers were significantly increased in central Pde6g−/− retinas (Fig 5C, D).
RPE Dysmorphia in Pde6g−/− Retinas
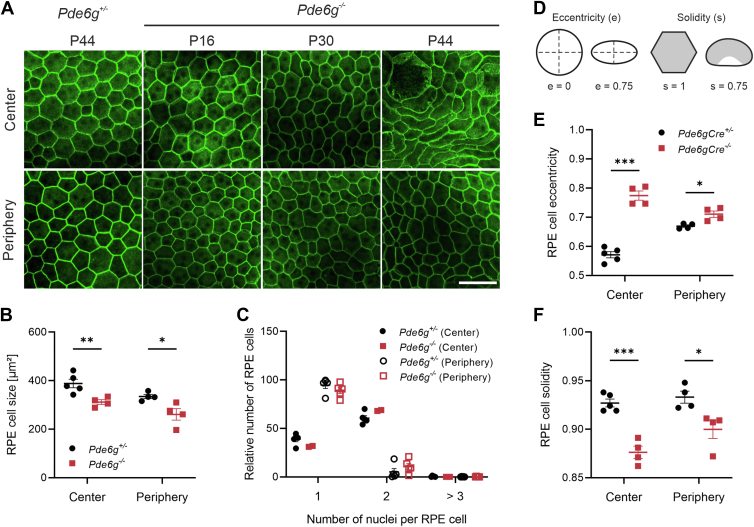
The RPE is a monolayer of columnar epithelial cells located between the retina and choroid, which is structurally and metabolically intertwined with the photoreceptor cell layer. Photoreceptor death in RP drives changes in the RPE.26 To characterize these changes in our Pde6g−/− mice (vs. Pde6g+/−), RPE cells were delineated in flat-mounted RPE-choroid-sclera by immunolabeling for the cell-adhesion protein β-catenin. As expected, RPE cells in Pde6g+/− mice were generally uniform in size and exhibited a polygonal (mostly hexagonal) shape (Fig 7A). Peripheral RPE from Pde6g−/− mice was generally indistinguishable from Pde6g+/− RPE, regardless of age (Fig 7A). In contrast, central RPE from Pde6g−/− mice was noticeably different from Pde6g+/− RPE—with minor morphological differences at P16 and P30 that were dramatic at P44. Essentially, the regular hexagonal geometry was replaced with polymorphous RPE of various, mostly elongated shapes. We next quantified these observations in RPE from P44 mice. The mean cell size was significantly smaller in Pde6g−/− (vs. Pde6g+/−) RPE: 312 versus 388 μm2 in the center and 261 versus 335 μm2 in the periphery (Fig 7B). All RPE cells from Pde6g−/− and Pde6g+/− mice had either 1 or 2 nuclei, and there was no significant difference in the frequency of either cell type (Fig 7C). The number of nuclei per RPE cell was not significantly different between the 2 groups (Fig 7C). We also analyzed cell eccentricity (shape/elongation) and solidity (proportion of RPE cell area filling a best-fit convex envelope) (Fig 7D). Compared to control, Pde6g−/− RPE cells had significantly greater eccentricity centrally and significantly reduced solidity centrally and peripherally (Fig 7E, F).
Figure 7.
Retinal pigment epithelium (RPE) remodeling in Pde6g−/− mice. RPE-choroid-sclera preparations from Pde6g+/− mice at postnatal day (P) 44 and Pde6g−/− mice at P16, P30, and P44 were flat-mounted, immunostained for β-catenin, images were taken in center and periphery, and Z-Stacks merged. A, Representative immunofluorescence images. Scale bar, 50 μm. B–F, Quantification of RPE cell parameters from P44 mice. Circles and squares, individual mice. N values for Pde6g+/− and Pde6g−/− mice, respectively: (B, D–F) 4–5 and 4; (C) 5 and 2–4. Data presented as mean ± standard error of the mean. Pde6g−/− vs. Pde6g+/− age-matched group means were compared by unpaired t tests. ∗P < 0.03; ∗∗P 0.01; ∗∗∗P < 0.001.
Discussion
Different variants in the same gene cause divergent RP disease progressions in humans and mice.27,28 For example, mice carrying different alleles of Pde6g resulted in retinal phenotypes ranging from biochemical defects with no photoreceptor loss to severe photoreceptor degeneration.15 In our Pde6g−/− mice, in which exons 2 and 3 of the Pde6g gene are replaced by CreERT2,16 we observed a very early disease onset, extremely rapid loss of photoreceptors (the vast majority of rod photoreceptors are lost at P16), and no detectable PDE6B immunolabeling. Interestingly, the Pde6gtm1/tm1 mouse line, in which the third exon was replaced with the bacterial neomycin resistance gene, shows a slightly later disease onset, a similarly rapid rate of photoreceptor degeneration (1 row of cone photoreceptors remaining at P21), and assembly, folding, and stability of the PDE6A/PDE6B complex does not appear to be affected.29 In a third Pde6g mutant, deletion of the last 7 amino acids (Del7C) led to photoreceptor degeneration that was approximately 1 week slower than Pdegtm1/tm1 mice, and expression of PDE6A and PDE6B was reduced by approximately 90%.30 Finally, mice carrying a point mutation in PDE6G at position 84 or 70 (Y84G or W70A, respectively) show no photoreceptor degeneration and normal expression of the 3 PDE6 subunits.31,32 Thus, observations in these 5 Pde6g mutant mice suggest that the onset and rate of photoreceptor loss correlates with the strength of the genetically induced insult. This is consistent with the demonstration that residual PDE6A protein slows photoreceptor degeneration.28
In this study, we analyzed secondary changes in inner retinal neurons, glia cells, blood vessels, and RPE in response to photoreceptor loss in Pde6g mutant mice. In our Pde6g−/− mice, photoreceptor loss led to progressive morphological changes in second-order neurons, which were evident at P16 and included dendritic retraction and dislocation of the cell body of horizontal and rod and cone bipolar cells. Dendritic retraction, mostly of rod bipolar cells, has been previously described in Pde6b mutant mice (e.g., rd1 and rd10)25,33,34 and in other retinal degenerative mouse models.35 Thus, it appears that photoreceptor loss, independent of the genetic cause, deafferents neural retina and induces negative plastic remodeling therein.4
Attenuation of retinal blood vessels is a characteristic clinical finding in both RP patients5 and mouse models.26,36,37 In our Pde6g−/− mice, we observed a significant decrease in vessel area in both center and periphery over time. The retinal vascular area in Pde6g+/− mice also becomes slightly reduced in the periphery which reflects remodeling processes that take place until adulthood (P120).38 In Pde6g−/− mice, a significant decrease in vessel area at P16 in deep and intermediate vascular plexi in the peripheral retina but not in the superficial vascular plexus was detected. Thus, the early onset photoreceptor degeneration observed in our model impacts the development of the deep and intermediate plexi that form in the second and third postnatal weeks but not the superficial plexus that forms from P0 to P10.39 In mutant rhodopsin Tg rat lines and rd1 mice that are characterized by partial loss of photoreceptor cells during development, partial failure of the deep vascular plexus development has been described.36,40 A suggested mechanism for this reduced vascular development is increased oxygen tension driven by photoreceptor loss.41,42 This idea is supported by research showing that developing retinal vasculature is reduced in hyperoxic conditions,43 the inner retina becomes relatively hyperoxic following photoreceptor cell loss,42 and the deep vascular plexus increases in an ambient hypoxic environment.44
We also analyzed secondary changes in RPE cells, which are structurally, functionally, and metabolically associated with photoreceptors.45,46 Thus, when photoreceptors are compromised, RPE cells are impaired and undergo significant morphological changes.26,47, 48, 49 This is the first study to examine correlations between RPE sheet morphology and photoreceptor degeneration in Pde6g mutant mice. In our Pde6g−/− mice, we observed minor changes in RPE at 16 or 30 days of age and dramatic abnormalities at 44 days, even though the vast majority of photoreceptors have degenerated at P16. In rd10 mutant mice, which carry a missense mutation (R560C) in Pde6b, rod degeneration begins around P18, peaks around P25, and at P45, RPE exhibits phenotypes that are not as severe as in our Pde6g−/− mice—compromised junctional complexes and both enlarged and abnormally small cells,48 but at P100, there are major changes in RPE cell morphometrics.50,51 Finally, in Pde6gstop/stop mice, which carry a stop cassette in the Pde6b gene, photoreceptor degeneration has a late onset (beginning around week 4) and is slow, and dramatic changes in RPE morphology are first apparent around week 24.26 These comparisons suggest that the age of onset for RPE remodeling is a function of the age of onset and speed of photoreceptor degeneration. These models also suggest that RPE remodeling appears sometime after photoreceptor loss, suggesting that bystander effects (e.g., oxidative stress, inflammation) require time to accumulate.52 Last but not least, the small and elongated RPE cells in the center at P44 indicate that the RPE cells undergo dedifferentiation and epithelial–mesenchymal transition through β-catenin induction of mesenchymal genes.53,54 The RPE dedifferentiation is a cardinal feature of the stress response55 and has been implicated in several retinal diseases, including proliferative vitreoretinopathy, diabetic retinopathy, and age-related macular degeneration.56 Thus, RPE cell survival in RP comes at the expense of epithelial attributes.
This preclinical mouse model and analysis describes the impact of a Pde6g mutation on RP disease progression and severity in multiple retinal layers. This study, in combination with analysis of other Pde6a and Pde6b mutants, as well as non-Pde6 mutants, reveals the common and unique features of these mutants. This information can be used to develop therapeutic strategies for RP while also identifying potential therapeutic targets.
Acknowledgments
The authors thank Prof. Dr Ludwig Wagner for sharing the secretagonin antibody.
Manuscript no. XOPS-D-22-00244.
Footnotes
Supplemental material available atwww.aaojournal.org.
Disclosures:
All authors have completed and submitted the ICMJE disclosures form.
The authors made the following disclosures:
S.H.T.: Support – Abeona Therapeutics, Inc, Emendo; Scientific advisory panel – Nanoscope Therapeutics, Medical Excellence Capital; Founder – Rejuvitas. The remaining authors have no proprietary or commercial interest in any materials discussed in this article.
This work was supported by the German Research Foundation [Emmy Noether grant 5719/1–1] and the Daimler and Benz Foundation to S.F.K. Jonas Children's Vision Care is supported by the National Institute of Health5P30CA013696, R01EY033770, R01EY018213, R01EY024698, R24EY027285, U01EY034590, U01EY030580, U54OD020351, R21AG050437, the Schneeweiss Stem Cell Fund, New York State [SDHDOH01-C32590GG-3450000] the Foundation Fighting BlindnessTA-GT-0321-0802-COLU-TRAP, Nancy & Kobi Karp, the Crowley Family Funds, The Rosenbaum Family Foundation, Alcon Research Institute, the Gebroe Family Foundation, the Research to Prevent Blindness (RPB) Physician-Scientist Award, unrestricted funds from RPB, New York, NY.
HUMAN SUBJECTS: Animal experiments were performed according to the Association for Research in Vision and Ophthalmology statement for the use of animals in ophthalmic and vision research and were approved by the local authorities.
Author Contributions:
Conception and design: Jentzsch, Tsang, Koch
Data collection: Jentzsch
Analysis and interpretation: Jentzsch, Koch; Obtained funding: N/A
Overall responsibility: Jentzsch, Tsang, Koch
Supplementary Data
References
- 1.Cross N., van Steen C., Zegaoui Y., et al. Current and future treatment of retinitis pigmentosa. Clin Ophthalmol. 2022;16:2909–2921. doi: 10.2147/OPTH.S370032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartong D.T., Berson E.L., Dryja T.P. Retinitis pigmentosa. Lancet. 2006;368(9549):1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 3.Hartong D.T., Berson E.L., Dryja T.P. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 4.Jones B.W., Pfeiffer R.L., Ferrell W.D., et al. Retinal remodeling in human retinitis pigmentosa. Exp Eye Res. 2016;150:149–165. doi: 10.1016/j.exer.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milam A.H., Li Z.Y., Fariss R.N. Histopathology of the human retina in retinitis pigmentosa. Prog Retin Eye Res. 1998;17:175–205. doi: 10.1016/s1350-9462(97)00012-8. [DOI] [PubMed] [Google Scholar]
- 6.Baehr W., Devlin M.J., Applebury M.L. Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J Biol Chem. 1979;254:11669–11677. [PubMed] [Google Scholar]
- 7.Hurley J.B., Stryer L. Purification and characterization of the gamma regulatory subunit of the cyclic GMP phosphodiesterase from retinal rod outer segments. J Biol Chem. 1982;257:11094–11099. [PubMed] [Google Scholar]
- 8.Cote R.H. Photoreceptor phosphodiesterase (PDE6): activation and inactivation mechanisms during visual transduction in rods and cones. Pflugers Arch. 2021;473:1377–1391. doi: 10.1007/s00424-021-02562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaughlin M.E., Ehrhart T.L., Berson E.L., Dryja T.P. Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci U S A. 1995;92:3249–3253. doi: 10.1073/pnas.92.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S.H., Pittler S.J., Huang X., et al. Autosomal recessive retinitis pigmentosa caused by mutations in the alpha subunit of rod cGMP phosphodiesterase. Nat Genet. 1995;11:468–471. doi: 10.1038/ng1295-468. [DOI] [PubMed] [Google Scholar]
- 11.Dvir L., Srour G., Abu-Ras R., et al. Autosomal-recessive early-onset retinitis pigmentosa caused by a mutation in PDE6G, the gene encoding the gamma subunit of rod cGMP phosphodiesterase. Am J Hum Genet. 2010;87:258–264. doi: 10.1016/j.ajhg.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dryja T.P., Rucinski D.E., Chen S.H., Berson E.L. Frequency of mutations in the gene encoding the alpha subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1999;40:1859–1865. [PubMed] [Google Scholar]
- 13.Kim M.S., Joo K., Seong M.W., et al. Genetic mutation profiles in Korean patients with inherited retinal diseases. J Korean Med Sci. 2019;34:e161. doi: 10.3346/jkms.2019.34.e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oishi M., Oishi A., Gotoh N., et al. Comprehensive molecular diagnosis of a large cohort of Japanese retinitis pigmentosa and Usher syndrome patients by next-generation sequencing. Invest Ophthalmol Vis Sci. 2014;55:7369–7375. doi: 10.1167/iovs.14-15458. [DOI] [PubMed] [Google Scholar]
- 15.Farber D.B., Tsang S.H. Stationary night blindness or progressive retinal degeneration in mice carrying different alleles of PDE gamma. Front Biosci. 2003;8:s666–s675. doi: 10.2741/1111. [DOI] [PubMed] [Google Scholar]
- 16.Koch S.F., Tsai Y.T., Duong J.K., et al. Halting progressive neurodegeneration in advanced retinitis pigmentosa. J Clin Invest. 2015;125:3704–3713. doi: 10.1172/JCI82462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamprecht M.R., Sabatini D.M., Carpenter A.E. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 18.Koch S.F., Duong J.K., Hsu C.W., et al. Genetic rescue models refute nonautonomous rod cell death in retinitis pigmentosa. Proc Natl Acad Sci U S A. 2017;114:5259–5264. doi: 10.1073/pnas.1615394114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Cui X., Jauregui R., et al. Genetic rescue reverses microglial activation in preclinical models of retinitis pigmentosa. Mol Ther. 2018;26:1953–1964. doi: 10.1016/j.ymthe.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma R.K., O'Leary T.E., Fields C.M., Johnson D.A. Development of the outer retina in the mouse. Brain Res Dev Brain Res. 2003;145:93–105. doi: 10.1016/s0165-3806(03)00217-7. [DOI] [PubMed] [Google Scholar]
- 21.Fu Y., Yau K.W. Phototransduction in mouse rods and cones. Pflugers Arch. 2007;454:805–819. doi: 10.1007/s00424-006-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spencer W.J., Lewis T.R., Phan S., et al. Photoreceptor disc membranes are formed through an Arp2/3-dependent lamellipodium-like mechanism. Proc Natl Acad Sci U S A. 2019;116:27043–27052. doi: 10.1073/pnas.1913518117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daum J.M., Keles O., Holwerda S.J., et al. The formation of the light-sensing compartment of cone photoreceptors coincides with a transcriptional switch. Elife. 2017;6 doi: 10.7554/eLife.31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young R.W. Cell death during differentiation of the retina in the mouse. J Comp Neurol. 1984;229:362–373. doi: 10.1002/cne.902290307. [DOI] [PubMed] [Google Scholar]
- 25.Strettoi E., Pignatelli V., Rossi C., et al. Remodeling of second-order neurons in the retina of rd/rd mutant mice. Vis Res. 2003;43:867–877. doi: 10.1016/s0042-6989(02)00594-1. [DOI] [PubMed] [Google Scholar]
- 26.Kajtna J., Tsang S.H., Koch S.F. Late-stage rescue of visually guided behavior in the context of a significantly remodeled retinitis pigmentosa mouse model. Cell Mol Life Sci. 2022;79:148. doi: 10.1007/s00018-022-04161-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y.N., Kim Y.J., Seol C.A., et al. Genetic profile and associated characteristics of 150 Korean patients with retinitis pigmentosa. J Ophthalmol. 2021;2021 doi: 10.1155/2021/5067271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sothilingam V., Garcia Garrido M., Jiao K., et al. Retinitis pigmentosa: impact of different Pde6a point mutations on the disease phenotype. Hum Mol Genet. 2015;24:5486–5499. doi: 10.1093/hmg/ddv275. [DOI] [PubMed] [Google Scholar]
- 29.Tsang S.H., Gouras P., Yamashita C.K., et al. Retinal degeneration in mice lacking the gamma subunit of the rod cGMP phosphodiesterase. Science. 1996;272:1026–1029. doi: 10.1126/science.272.5264.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang S.H., Yamashita C.K., Lee W.H., et al. The positive role of the carboxyl terminus of the gamma subunit of retinal cGMP-phosphodiesterase in maintaining phosphodiesterase activity in vivo. Vis Res. 2002;42:439–445. doi: 10.1016/s0042-6989(01)00213-9. [DOI] [PubMed] [Google Scholar]
- 31.Tsang S.H., Yamashita C.K., Doi K., et al. In vivo studies of the gamma subunit of retinal cGMP-phophodiesterase with a substitution of tyrosine-84. Biochem J. 2001;353:467–474. doi: 10.1042/0264-6021:3530467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsang S.H., Burns M.E., Calvert P.D., et al. Role for the target enzyme in deactivation of photoreceptor G protein in vivo. Science. 1998;282:117–121. doi: 10.1126/science.282.5386.117. [DOI] [PubMed] [Google Scholar]
- 33.Pang J.J., Dai X., Boye S.E., et al. Long-term retinal function and structure rescue using capsid mutant AAV8 vector in the rd10 mouse, a model of recessive retinitis pigmentosa. Mol Ther. 2011;19:234–242. doi: 10.1038/mt.2010.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strettoi E., Pignatelli V. Modifications of retinal neurons in a mouse model of retinitis pigmentosa. Proc Natl Acad Sci U S A. 2000;97:11020–11025. doi: 10.1073/pnas.190291097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baehr W., Frederick J.M. Naturally occurring animal models with outer retina phenotypes. Vis Res. 2009;49:2636–2652. doi: 10.1016/j.visres.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanks J.C., Johnson L.V. Vascular atrophy in the retinal degenerative rd mouse. J Comp Neurol. 1986;254:543–553. doi: 10.1002/cne.902540407. [DOI] [PubMed] [Google Scholar]
- 37.Hanna J., Yucel Y.H., Zhou X., et al. Progressive loss of retinal blood vessels in a live model of retinitis pigmentosa. Can J Ophthalmol. 2018;53:391–401. doi: 10.1016/j.jcjo.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 38.Rust R., Gronnert L., Dogancay B., Schwab M.E. A revised view on growth and remodeling in the retinal vasculature. Sci Rep. 2019;9:3263. doi: 10.1038/s41598-019-40135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorrell M.I., Aguilar E., Friedlander M. Retinal vascular development is mediated by endothelial filopodia, a preexisting astrocytic template and specific R-cadherin adhesion. Invest Ophthalmol Vis Sci. 2002;43:3500–3510. [PubMed] [Google Scholar]
- 40.Pennesi M.E., Nishikawa S., Matthes M.T., et al. The relationship of photoreceptor degeneration to retinal vascular development and loss in mutant rhodopsin transgenic and RCS rats. Exp Eye Res. 2008;87:561–570. doi: 10.1016/j.exer.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matthes M.T., Bok D. Blood vascular abnormalities in the degenerative mouse retina (C57BL/6J-rd le) Invest Ophthalmol Vis Sci. 1984;25:364–369. [PubMed] [Google Scholar]
- 42.de Gooyer T.E., Stevenson K.A., Humphries P., et al. Rod photoreceptor loss in Rho-/- mice reduces retinal hypoxia and hypoxia-regulated gene expression. Invest Ophthalmol Vis Sci. 2006;47:5553–5560. doi: 10.1167/iovs.06-0646. [DOI] [PubMed] [Google Scholar]
- 43.Ashton N. Some aspects of the comparative pathology of oxygen toxicity in the retina. Ophthalmologica. 1970;160:54–71. doi: 10.1159/000305969. [DOI] [PubMed] [Google Scholar]
- 44.Penn J.S., Li S., Naash M.I. Ambient hypoxia reverses retinal vascular attenuation in a transgenic mouse model of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:4007–4013. [PubMed] [Google Scholar]
- 45.Hurley J.B. Retina metabolism and metabolism in the pigmented epithelium: a busy intersection. Annu Rev Vis Sci. 2021;7:665–692. doi: 10.1146/annurev-vision-100419-115156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strauss O. The retinal pigment epitheliumin visual function. Physiol Rev. 2005;85(3):845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 47.Boatright J.H., Dalal N., Chrenek M.A., et al. Methodologies for analysis of patterning in the mouse RPE sheet. Mol Vis. 2015;21:40–60. [PMC free article] [PubMed] [Google Scholar]
- 48.Napoli D., Biagioni M., Billeri F., et al. Retinal pigment epithelium remodeling in mouse models of retinitis pigmentosa. Int J Mol Sci. 2021;22:5381. doi: 10.3390/ijms22105381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu D.M., Ji X., Ivanchenko M.V., et al. Nrf2 overexpression rescues the RPE in mouse models of retinitis pigmentosa. JCI Insight. 2021;6 doi: 10.1172/jci.insight.145029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chrenek M.A., Dalal N., Gardner C., et al. Analysis of the RPE sheet in the rd10 retinal degeneration model. Adv Exp Med Biol. 2012;723:641–647. doi: 10.1007/978-1-4614-0631-0_81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Y., Qi X., Chrenek M.A., et al. Functional principal component analysis reveals discriminating categories of retinal pigment epithelial morphology in mice. Invest Ophthalmol Vis Sci. 2013;54:7274–7283. doi: 10.1167/iovs.13-12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Datta S., Cano M., Ebrahimi K., et al. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog Retin Eye Res. 2017;60:201–218. doi: 10.1016/j.preteyeres.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou M., Geathers J.S., Grillo S.L., et al. Role of epithelial-mesenchymal transition in retinal pigment epithelium dysfunction. Front Cell Dev Biol. 2020;8:501. doi: 10.3389/fcell.2020.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J.W., Kang K.H., Burrola P., et al. Retinal degeneration triggered by inactivation of PTEN in the retinal pigment epithelium. Genes Dev. 2008;22:3147–3157. doi: 10.1101/gad.1700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao C., Yasumura D., Li X., et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sripathi S.R., Hu M.W., Turaga R.C., et al. Proteome landscape of epithelial-to-mesenchymal transition (EMT) of retinal pigment epithelium shares commonalities with malignancy-associated EMT. Mol Cell Proteomics. 2021;20 doi: 10.1016/j.mcpro.2021.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.