Abstract
Background
Low back pain is a major contributor to disability worldwide and generates a tremendous socioeconomic impact. The degenerative intervertebral disc (IVD) has been hypothesized to contribute to discogenic pain by sensitizing nociceptive neurons innervating the disc to stimuli that is nonpainful in healthy patients. Previously, we demonstrated the ability of degenerative IVDs to sensitize neurons to mechanical stimuli; however, elucidation of degenerative IVDs discogenic pain mechanisms is required to develop therapeutic strategies that directly target these mechanisms.
Aims
In this study, we utilized CRISPR epigenome editing of nociceptive neurons to identify mechanisms of degenerative IVD‐induced changes to mechanical nociception and demonstrated the ability of multiplex CRISPR epigenome editing of nociceptive neurons to modulate inflammation‐induced mechanical nociception.
Methods and Results
Utilizing an in vitro model, we demonstrated degenerative IVD‐produced IL‐6‐induced increases in nociceptive neuron activity in response to mechanical stimuli, mediated by TRPA1, ASIC3, and Piezo2 ion channel activity. Once these ion channels were identified as mediators of degenerative IVD‐induced mechanical nociception, we developed singleplex and multiplex CRISPR epigenome editing vectors that modulate endogenous expression of TRPA1, ASIC3, and Piezo2 via targeted gene promoter histone methylation. When delivered to nociceptive neurons, the multiplex CRISPR epigenome editing vectors abolished degenerative IVD‐induced mechanical nociception while preserving nonpathologic neuron activity.
Conclusion
This work demonstrates the potential of multiplex CRISPR epigenome editing as a highly targeted gene‐based neuromodulation strategy for the treatment of discogenic pain, specifically; and, for the treatment of inflammatory chronic pain conditions, more broadly.
Keywords: back pain, discogenic pain, inflammation, mechanical nociception
Multiplex CRISPR epigenome editing of ion channel expression in nociceptive neurons abolished degenerative IVD‐induced mechanical nociception in an in vitro model of discogenic back pain. These results demonstrate multiplex epigenome editing of nociceptive neurons as a potential neuromodulation strategy for discogenic back pain and other chronic pain conditions.

1. INTRODUCTION
Low back pain is amongst the most prevalent global maladies, ranking as the leading cause of disability worldwide, 1 third in disease burden according to disease‐adjusted life years, 2 and generating an enormous socioeconomic cost estimated at $100 billion annually in the United States alone. 3 While the prevalence of low back pain and its significant impact on society have been established, the underlying mechanisms of back pain remain nebulous and treatment strategies are limited.
Disc degeneration, commonly associated with low back pain, 4 , 5 , 6 , 7 , 8 is characterized by pathological changes in the intervertebral disc (IVD) including decreased disc height, 9 extracellular matrix deterioration, 10 , 11 elevated inflammatory cytokines levels, 12 , 13 , 14 , 15 , 16 , 17 , 18 and altered disc innervation. 19 , 20 , 21 , 22 While these changes to the IVD have been linked with discogenic pain, 23 , 24 , 25 the interactions between nociceptive neurons and degenerative IVD tissue, as well as the mechanisms by which these interactions contribute to discogenic pain, have yet to be elucidated.
Healthy IVDs are innervated in the peripheral lamella of the annulus fibrosus (AF) by neurons whose soma resides in the dorsal root ganglia (DRG). 26 , 27 , 28 Most of these neurons are nociceptors that express the neuropeptide calcitonin gene‐related peptide (CGRP), 29 , 30 , 31 , 32 , 33 as well as multiple mechano‐sensitive ion channels, such as Piezo1 34 and Piezo2, 35 indicating the disc may be sensitive to mechanical stimuli. Degenerative IVDs exhibit multiple changes in innervation including an increased nociceptive neuron denisty 36 and neo‐innervation of typically aneural regions of the disc with CGRP 32 , 37 expressing nociceptive neurons. 19 , 20 , 26 , 32 , 37 , 38 In addition, neurons innervating the degenerative IVD are exposed to pathologically high levels of interleukin 6(IL‐6), tumor necrosis factor‐alpha (TNF‐α), and interleukin 1 beta (IL‐1β), 13 , 14 , 39 , 40 , 41 as well as other inflammatory cytokines. TNF‐α, IL‐1β, and IL‐6/IL‐6st have been demonstrated to sensitize peripheral nerves and to induce mechanical allodynia 42 , 43 , 44 , 45 , 46 in peripheral neuropathy. Moreover, degenerative IVD tissue has been demonstrated to directly sensitize neurons to mechanical stimuli, by altering the activation thresholds of mechanically sensitive ion channels. 47 Together, these results demonstrate the degenerative IVD contains multiple factors that may initiate discogenic pain by sensitizing nociceptive neurons mechanical stimuli that are nonpainful in healthy patients.
CRISPR‐based epigenome editing facilitates stable, long‐term, highly site‐specific 48 histone modifications to modulate gene expression. Briefly, CRISPR‐Cas9‐based epigenome editing vectors are comprised of three key components: a nuclease‐deficient Cas9 (dCas9), a synthetic guide RNA (sgRNA) to target specific DNA sequences, 49 , 50 , 51 and the effector molecule Kruppel‐associated box (KRAB). The fusion of KRAB to dCas9 produces localized, highly targeted H3K9 histone methylation, 52 , 53 , 54 , 55 which can be used to suppress endogenous gene expression. Previously, we have demonstrated the ability of CRISPR epigenome editing of nociceptive neurons to modulate degenerative IVD‐induced sensitization of neurons to thermal stimuli. 56 , 57 In this study, we propose direct regulation of mechanical nociception, via multiplex CIRPSR epigenome editing of ion channel expression, as a neuromodulation strategy for the treatment of discogenic back pain. Utilizing this therapeutic strategy, back pain symptoms may be treated by epigenome modifications of specific pain‐related genes (i.e., mechano‐sensitive ion channels) expressed in the nociceptive neurons that innervate the degenerative.
We hypothesize that pathologically high inflammatory cytokines levels present in the degenerative IVD contribute to discogenic pain by lowering the activation thresholds of nociceptive neurons in the degenerative IVD, resulting in neuron sensitivity to stimuli that are not painful in healthy patients. To test this hypothesis, we developed an in vitro model to investigate the interactions between human degenerative IVDs, nociceptive neurons, and cyclic mechanical loading of the IVD. Utilizing this model, we explored the ability of the degenerative IVDs to alter mechanical nociception, elucidated the specific ion channels involved in these interactions, and CRISPR‐based neuromodulation to regulate these mechanisms.
This work elucidates transient receptor potential ankyrin subtype 1 (TRPA1) protein, acid‐sensing ion channel 3 (ASIC3), and Piezo2 as mediators of degenerative IVD‐induced mechanical nociception and suggests multiplex epigenome regulation of these ion channels as a pain modulation strategy for the treatment of discogenic pain.
2. METHODS
2.1. Experimental overview
In this study, we conducted a set of experiments to elucidate the cytokine receptors and ion channels that mediate degenerative IVD‐induced enhanced mechanical nociception and to demonstrate the ability of singleplex and multiplex CRISPR epigenome editing of nociceptive neuron cytokine receptor and ion channel profiles to modulate mechanical nociception, in an in vitro model of discogenic back pain.
First, we investigated the potential of singleplex CRISPR epigenome editing of nociceptive neurons to mitigate degenerative IVD augmentation of mechanical nociception. Lentiviral CRISPR epigenome editing constructs that co‐express endonuclease deficient Cas 9(dCas9)‐Krüppel‐associated box domain (KRAB)‐T2A‐Green Fluorescent Protein (GFP) and guide RNAs (sgRNAs) targeting the IL‐6st, ASIC3, TRPA1, Piezo1, or Piezo2 gene promoter region were designed, constructed, and tested for their ability to downregulate target gene expression. Validated CRISPR IL‐6st, ASIC3, TRPA1, Piezo1, and Piezo2 epigenome editing lentiviral vectors were selected and delivered, individually, to DRG neurons, prior to neuron exposure to degenerative IVD‐conditioned media. Subsequently, the ability of epigenome editing of nociceptive neurons to modulate degenerative IVD‐induced mechanical nociception was tested, utilizing our in vitro model of discogenic back pain. 47
Once the ion channels ASIC3, TRPA1, and Piezo2 were implicated in degenerative IVD‐induced enhanced mechanical nociception, experiments in the multiplex epigenome‐edited neuron pathological AF tissue exposure section were conducted to investigate the potential combined/redundant roles of AISC3, TRPA1, and Piezo2 ion channel activity in degenerative IVD‐induced mechanical nociception and to demonstrate the ability of multiplex CRISPR epigenome editing to modulate degenerative IVD‐induced mechanical nociception.
2.2. Degenerative IVD‐conditioned media
All procedures were performed with the approval of the Institutional Review Board of the University of Utah (discarded surgical waste tissue was collected, as permissible under exemption 4). Degenerative IVD tissue was harvested from the lumbar IVDs of five patients (n = 5; two male patients age: 61 and 51; three female patients age: 38, 60, 76) undergoing surgical treatment for axial back pain, degenerative disc disease, and lumbar spondylosis. These surgical patients had demonstrated disc disease on magnetic resonance imaging, reported axial back pain, and disc degeneration was classified using the Pfirrmann method. 58 Degenerative IVD‐conditioned media was produced, via a previously reported method. 47 , 56 , 59
Briefly, surgical IVD tissue (containing both nucleus pulposus [NP] and AF tissue) was transferred into a glass petri dish, washed twice in washing medium (DMEM‐HG [Life Technologies] supplemented with 1% [v/v] gentamycin (Gibco), 1% [v/v] kanamycin (Sigma) and 1% [v/v] fungizone (Gibco)) and cut into small pieces (~3 mm2). Next, IVD tissue was weighed, and cultured in DMEM‐HG supplemented with 50 μg/mL ascorbic acid (Life Technologies), 5 μg/mL gentamicin, and 0.125 μg/mL fungizone at a media‐to‐tissue ratio of 3.5 mL/g for 48 h (37°C and 5% CO2). 18 , 47 Following culture, degenerative IVD conditioned media from each individual patient were collected and stored separately, by patient at −80°C, until time of experiments (up to 3 months).
2.3. Bovine caudal AF tissue culture
Bovine tails were ethically obtained from a local merchant. AF tissue was extracted from bovine caudal IVDs. Cutting between adjacent lamellae, the outermost lamellae of AF tissue was separated from tissue determined to be NP and transition zone tissue, via gross anatomical observation. AF tissue was washed twice with washing medium, wrapped in sterile PBS‐soaked sterile gauze, placed in 50 cc microcentrifuge tubes, and frozen at −20°C, until the time of experiments.
At time of experiments, AF tissue was transferred into a glass petri dish, washed twice in washing medium, 47 , 56 cut to size (30 mm × 15 mm × 5 mm) in circumferentially oriented samples, and cultured in washing media for 48 h (37°C and 5% CO2). Subsequently, media were removed, samples were coated with laminin, and incubated for 3 h (37°C and 5% CO2). Following incubation, media were replaced with washing media supplemented with 10% FBS (volume FBS/total media volume) and cultured for 48 h (37°C and 5% CO2) prior to DRG neuron seeding.
2.4. DRG neuron cell culture
All procedures were performed with the approval of the University of Utah Institutional Animal Care and Use Committee (IACUC, protocol #21‐11 006). Adult (200–250 g) male Sprague–Dawley rat DRG were harvested, cleaned, dissociated, and DRG neurons were isolated, utilizing a previously reported method. 47 , 56 , 59 In brief, rat DRGs were explanted and cleaned of connective tissue, and anterior and posterior roots. Ganglia were dissociated in 2 mL of L‐15 medium supplemented with 500 μL of collagenase IV (1.33%[v/v], Worthington Biochemical) for 30 min at 37°C. The cell suspension was centrifuged at 1000 RPM for 5 min after which the supernatant was aspirated and the cells were incubated in 4 mL of DMEM/F‐12 supplemented with 1 mL of 1% trypsin (Worthington Biochemical) plus 50 μL of 1%(v/v) DNase I (Worthington Biochemical) for 20 min at 37°C. Following incubation, 1 mL of soybean trypsin inhibitor (Worthington Biochemical) was added to the cell suspension and the cell suspension was centrifuged at 1000 RPM for 5 min. Next, the supernatant was aspirated, and cells were resuspended in 1 mL of DMEM/ F‐12 plus 50 μL of 1%(v/v) DNase and triturated through a fire‐polished Pasteur pipette.
Following rat DRG neuron isolation, DRG neurons were seeded onto healthy bovine AF tissue at a density of 25 000 cells/cm2, and cultured in Sato− media [DMEM/F12 supplemented with 2.2% [v/v] SATO‐mix, 1% [v/v] transferrin (Sigma), 2% [v/v] insulin (Sigma), 1% (v/v) Glutamax (Invitrogen)], 0.5% (v/v) gentamicin, and 2.5 S NGF (10 ng/mL, Worthington Biochemical) 47 , 56 , 59 for 3–4 days, until experiments.
2.5. Mechanical testing frame
All calcium transient imaging experiments were conducted using a custom‐built mechanical testing frame 47 mounted onto the stage of a multiphoton microscope (Bruker/Prairie view; see Scheme 1). Briefly, DRG neuron‐seeded AF tissue constructs (30 mm long; 15 mm wide; and 5 mm thick) we loaded into the mechanical testing frame, oriented with lamellae aligned parallel to the axis of strain application, and the samples were secured by clamping the ends of the AF tissue construct. AF tissue gauge length was standardized to a 20 mm clamp‐to‐clamp distance. The saline bath was filled with room‐temperature PBS until AF tissue samples were submerged.
SCHEME 1.

(A) Mechanical testing frame mounted onto multiphoton microscope. (B) Close‐up image of DRG neuron seeded AF tissues constructs loaded into the mechanical testing frame.
Application of cyclic tensile strain was controlled using a custom LabView (National Instruments) VI. For the duration of baseline calcium imaging, samples were maintained at 0% tissue. Following baseline imaging, samples were exposed to a single cycle (1 Hz; sinusoidal waveform) of tensile strain at 1%, 2%, 4%, and 6% tissue strain. Cycles of strain exposure were separated by 5 min periods during which samples were held at constant baseline (0%) strain.
Strain levels were selected to span the range of tissue strain levels neurons innervating the AF of the degenerative IVD would be subjected to during normal loading of the IVD. 60
2.6. Imaging of strain‐induced calcium transients in DRG neurons seeded upon AF tissue
In these experiments, DRG neuron activity was measured, via calcium imaging, during neuron exposure to cyclic tensile strain (0%–6% AF tensile strain, 1 Hz). 47 Strain levels were selected to span to a range of tissue strain levels neurons innervating the AF of the degenerative IVD would be subjected to during normal loading of the IVD62. Imaging of strain‐induced calcium transients was conducted using a previously reported method. 47
Briefly, 1 h prior to imaging, rat DRG neuron seeded upon AF tissue constructs were loaded with the calcium indicator dye (Rhod‐2 AM, 4 μM, Molecular Probes) and incubated in the dark at 37°C, until the time of experiments. At the time of experiments, AF tissue constructs seeded with DRG neurons were loaded into a custom built, multiphoton microscope stage‐mounted mechanical testing frame, and incubated at room temperature (20–25°C) for 15 min in order to establish a baseline calcium signal.
Following baseline calcium image acquisition, AF tissue samples were subjected to cyclic tensile strain (1 Hz; 0%, 1%, 2%, 4%, and 6%) during imaging of nociceptive neuron calcium transients. Samples were returned to baseline strain levels (0%) for 5 min between subsequent applications of cyclic strain. Fluorescent measurement of calcium was performed using a multiphoton microscope (Prairie View [Bruker], Ex. 1105 nm, Em. 585 nm, 1.0 Hz).
Image analysis was conducted using Fiji software. 61 For each imaging condition, 40 DRG neurons were analyzed. ROIs were drawn around DRG neurons cell bodies and neural processes, and the average fluorescent intensity was calculated. The background signal was subtracted from each cell and the mean baseline (F 0(t)) was calculated at the baseline strain:
| (1) |
ΔF/F (Equation 1) was calculated for each cell across the entire experiment. The baseline mean and standard deviation of ΔF/F were calculated for all cells at the baseline strain level. Neurons were considered to exhibit calcium transients in response to mechanical stimuli if the ΔF/F for the cell was 10 standard deviations greater than the mean baseline value 47 , 62 in response to mechanical stimulation.
2.7. Curve fitting of neuron calcium transients
Data from the singleplex epigenome‐edited neuron and multiplex epigenome‐edited neuron degenerative IVD conditioned media exposure experiments were fitted to the sigmoidal Boltzmann equation (Equation 2) with the percentage of neurons exhibiting mechanical strain‐induced calcium transients plotted as a function of strain:
| (2) |
Each trial was individually fit to produce values for S 50 and S max. S 50 was defined as the strain at which 50% of the maximum response occurs. The S max was defined as the maximum percentage of neurons exhibiting strain‐induced calcium transients as predicted by the curve fitting.
2.8. Singleplex epigenome‐edited neuron degenerative IVD conditioned media exposure
DRG neurons seeded on AF tissue were cotransduced with lentiviral constructs targeting the IL‐6st, ASIC3, TRAP1, Piezo1, or Piezo2 gene promoter region, or a nontarget sgRNA with no match to the rat genome, and a lentiviral CGRP‐α fluorescent reporter(utilized to identify CGRP positive neurons). 47 Following transduction, neurons were cultured in Sato− medium for 6 days, following removal of transduction media. Subsequently, Sato− media were removed and replaced with 2 mL of fresh Sato− media in combination with either 2 mL of degenerative IVD‐conditioned media or 2 mL of control (DMEM) media, and incubated for 24 h under standard cell culture conditions (37°C and 5% CO2). Fluorescent measurements of calcium transients were conducted as described in Section 2.6. Images of CGRP‐positive neurons were acquired (excitation 810 nm, emission 457 nm) and imaging was conducted as previously reported. 47
2.9. Multiplex epigenome‐edited neuron pathological AF tissue exposure
To allow for multiplex targeting of multiple ion channels simultaneously, we created an epigenome editing vector that co‐expresses dCas9‐KRAB‐T2A‐GFP and ASIC3, TRPA1, and Piezo2 sgRNAs, utilizing a previously reported method 63 (Supporting Information Methods). Efficacy of multiplex regulation of gene expression in DRG neurons transduced with multiplex epigenome editing vectors was confirmed via qPCR, as described in the qPCR Experiments section (Supporting Information Methods).
DRG neurons seeded upon AF tissue were co‐transduced with a lentiviral multiplex CRISPR epigenome editing vector targeting the ASIC3, TRPA1, and Piezo2 gene promoter regions and a lentiviral CGRP‐α fluorescent reporter, and cultured as described in Section 2.8.
Following incubation, transduced DRG neurons seeded upon AF tissue were loaded with the calcium indicator dye Rhod‐2 AM (4 μM; molecular probes) and incubated in the dark at 37°C for 1 h. Fluorescent measurements of calcium were performed as described in Section 2.6.
Images of CGRP‐positive neurons were acquired (excitation 810 nm, emission 457 nm) and imaging conducted as described above.
2.10. Statistical analysis
Singleplex epigenome‐edited neuron degenerative IVD conditioned media exposure and Multiplex epigenome‐edited neuron pathological AF tissue exposure experiment data were analyzed by two‐way analysis of variance (ANOVA) on repeated measures with Tukey's post hoc test, treating media condition and strain as factors. S 50 and S max data from all experiments were analyzed by one‐way ANOVA with Tukey's post hoc test, treating media condition as the factor. qPCR experiments were analyzed by one‐way ANOVA with Tukey's post hoc test, treating transduction vector as the factor. For all experiments, individual the patient conditioned media exposures were treated as replicates Significance was tested at α = 0.05 for all statistical analyses. All values are reported as mean ± standard deviation.
3. RESULTS
3.1. Singleplex epigenome editing of IL‐6st promoter in DRG neurons abolished degenerative IVD‐induced elevated mechanical nociception
DRG neurons seeded onto AF tissue constructs (15 000 cells/cm2) transduced with CRISPR epigenome editing vectors targeting IL‐6st (Figure S1A) demonstrated robustly downregulated gene expression of IL‐6st at the RNA level. Guide four demonstrated maximum downregulation of IL‐6st expression (97.7 ± 2.4%; Figure S1B) and was selected for subsequent functional experiments.
The percentage of neurons exhibiting strain‐induced Ca2+ transients in CGRP positive (CGRP+) neurons exposed to degenerative IVD‐conditioned media group was significantly elevated (p < 0.05) when compared to the percentage of neurons exhibiting strain‐induced Ca2+ transients in the control media group (Figure 1A); however, the percentage of IL‐6st epigenome‐edited CGRP+ neurons exhibiting strain‐induced Ca2+ transients when exposed to degenerative IVD‐conditioned media was returned to baseline levels (Figure 1A). In addition, the maximum calcium transients (ΔF/F) of CGRP+ neurons exposed to degenerative IVD‐conditioned media were significantly elevated over baseline levels (Figure 1B); although, the maximum calcium transients of IL‐6st epigenome‐edited CGRP+ neurons exposed to degenerative IVD‐conditioned media returned to baseline levels (Figure 1B). Furthermore, nontarget epigenome editing of CGRP+ neurons had no effect on neuronal responses of CGRP+ neurons subjected to cyclic strain in the presence of either degenerative IVD conditioned media or control media (Figure S2A,B).
FIGURE 1.

Epigenome editing of IL‐6st expression in CGRP‐positive nociceptive neurons abolishes degenerative IVD‐induced mechanical nociception. (A) The percentage of CGRP+ neurons exhibiting calcium transients as a function of strain was fitted to a Boltzmann equation for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, IL6st edited CGRP+ neuron control media, and IL6st edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). (B) The maximum calcium transient (ΔF/F) of CGRP+ neurons as a function of strain was fitted to a Boltzmann curve for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, IL6st edited CGRP+ neuron control media, and IL6st edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). The S 50 (C) and S max (D) generated from the dose–response curve fitting for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, IL6st edited CGRP+ neuron control media, and IL6st edited CGRP+ neuron degenerative IVD‐conditioned media groups. (A–D) *p < 0.05 when compared to the CGRP+ neuron control media group. Error bars are SD.
From the curve fitting, the S 50 of CGRP+ neurons in the degenerative IVD‐conditioned media group (1.11 ± 0.02) was significantly reduced when compared to the S 50 of CGRP+ neurons in the control group (3.99 ± 0.9; Figure 1C); however, IL‐6st epigenome editing of CGRP+ neurons in the degenerative IVD‐conditioned media group returned the S 50 (3.62 ± 0.49) to baseline (3.99 ± 0.9; Figure 1C). Similarly, the S max of CGRP+ neurons in the degenerative IVD‐conditioned media group (47.0 ± 2.1%) was significantly elevated (p < 0.05) over the S max of CGRP+ neurons in the control group (7.6 ± 3.2%; Figure 1C); nonetheless, the IL‐6st epigenome‐edited CGRP+ neurons exposed to degenerative IVD‐conditioned media displayed S max values (9.62 ± 3.0%) not significantly different from the S max values of neurons in the control media group (7.6 ± 3.2%). Moreover, IL‐6st epigenome editing of CGRP+ neurons returned the S 50 (4.2 ± 0.6; Figure 1C) and S max (7.8 ± 2.4%; Figure 1D) values of neurons exposed to degenerative IVD‐conditioned media to baseline control levels (Figure 1C,D). Nontarget epigenome editing of CGRP+ neurons had no effect on S 50 (Figure S2C) and S max values (Figure S2D).
3.2. Singleplex epigenome editing of ASIC3, TRPA1, and Piezo2 in CGRP+ nociceptive neurons reduced degenerative IVD‐enhanced mechanical nociception but did not eliminate it
CRISPR epigenome editing of ASIC3, TRPA1, Piezo1, and Piezo2 gene promoters in DRG neurons resulted in robust downregulation of target gene expression at the RNA level. Maximum downregulation of ASIC3 (95 ± 2%; Figure S1C), TRPA1 (99 ± 1%; Figure S1D), Piezo1 (79 ± 9%; Figure S1E), and Piezo2 (81 ± 9%; Figure S1F) gene expression were significantly reduced when compared to gene expression in nontarget epigenome‐edited neurons. In addition, immunostaining of epigenome‐edited DRG neurons exhibited significantly downregulated protein expression (Figure S3) of target gene, for all genes tested.
FIGURE 3.
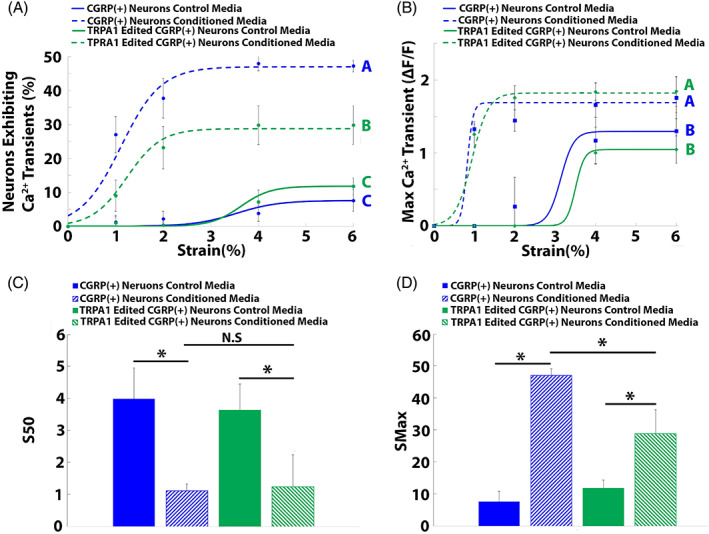
Epigenome editing of TRPA1 expression in CGRP+ DRG neurons reduced degenerative IVD‐induced sensitization of nociceptive neurons to mechanical stimuli. The percentage of CGRP+ neurons exhibiting calcium transients as a function of strain was fitted to a Boltzmann equation for the CGRP+ neuron control media, CGRP+ neuron degenerative‐IVD‐conditioned media, TRPA1 edited CGRP+ neuron control media, and TRPA1 edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). (B) The maximum calcium transient (ΔF/F) of CGRP+ neurons as a function of strain was fitted to a Boltzmann curve for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, TRPA1 edited CGRP+ neuron control media, and TRPA1 edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). The S 50 (C) and S max (D) were generated from the dose–response curve fitting for the control media, degenerative IVD‐conditioned media, TRPA1‐edited control media, and TRPA1‐edited degenerative IVD‐conditioned media groups. (A–B) Different letters indicate groups significantly (p < 0.05) different from one another. Error bars are SD. (C–D) *p < 0.05 when compared to the CGRP+ neuron control media group, #p = 0.05 when compared to the TRPA1 edited CGRP+ neuron control media group. Error bars are SD.
ASIC3 edited CGRP+ neurons exposed to degenerative IVD‐conditioned media exhibited significantly reduced percentages of neurons generating calcium transients, when compared to nonedited CGRP+ neurons exposed to conditioned media (Figure 2A); however, neuron activity remained significantly elevated (p < 0.05) over the neuron activity of both ASIC3 edited and nonedited CGRP+ neurons subjected to cyclic strain in the presence of control media (Figure 2A). Epigenome editing of ASIC3 had no effect on maximum calcium transients (Figure 2B).
FIGURE 2.

Epigenome editing of ASIC3 expression in CGRP+ DRG neurons reduced degenerative IVD‐induced mechanical nociception. The percentage of CGRP+ neurons exhibiting calcium transients as a function of strain was fitted to a Boltzmann equation for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, ASIC3 edited CGRP+ neuron control media, and ASIC3 edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). (B) The maximum calcium transient (ΔF/F) of CGRP+ neurons as a function of strain was fitted to a Boltzmann curve for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, ASIC3 edited CGRP+ neuron control media, and ASIC3 edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). The S 50 (C) and S max (D) were generated from the dose–response curve fitting for the control media, degenerative IVD‐conditioned media, ASIC3 edited control media, and ASIC3 edited degenerative IVD conditioned‐media groups. (A–D) *p < 0.05 when compared to the CGRP+ neuron control media group, #p = 0.05 when compared to the ASIC3 edited CGRP+ neuron control media group. Error bars are SD.
From the curve fitting, the S 50 values (1.37 ± 0.52) of ASIC3‐edited CGRP+ neurons exposed to degenerative IVD‐conditioned media were similar (p > 0.05) to those of nonedited CGRP+ neurons exposed to degenerative IVD‐conditioned media and significantly decreased compared to those of CGRP+ neurons exposed to control media (3.99 ± 0.9; Figure 2C). Additionally, the S max (25.3 ± 4.9%) of ASIC3‐edited CGRP+ neurons exposed to degenerative IVD‐conditioned media was significantly lower than the S max of nonedited CGRP+ neurons exposed to degenerative IVD‐conditioned media (47.0 ± 2.1%; Figure 2D); however, it remained significantly elevated over baseline (7.6 ± 3.2%; Figure 2D).
TRPA1 epigenome‐edited CGRP+ neurons exposed to cyclic strain in the presence of degenerative IVD‐conditioned media exhibited significantly reduced (p < 0.05) percentages of neurons exhibiting Ca2+ transients when compared to nonedited CGRP+ neurons exposed to cyclic strain in the presence of degenerative IVD‐conditioned media (Figure 3A); none the less, the percentage of TRPA1 edited neurons exhibiting Ca2+ transients remained significantly elevated over baseline (Figure 3A). In contrast, TRPA1 epigenome editing had no effect on the maximum calcium transients (Figure 3B).
TPRA1 epigenome editing had no effect on the S 50 values of neurons subjected to cyclic strain in the presence of either control (3.63 ± 0.81) or degenerative IVD‐conditioned media (1.23 ± 0.36; Figure 3C). Contrastingly, TRPA1‐edited neurons exposed to degenerative IVD‐conditioned media displayed significantly reduced S max (28.8 ± 7.5%) values when compared to the S max of nonedited neurons exposed to degenerative IVD‐conditioned media (47 ± 2.1%; Figure 3D); however, the S max of TRPA1‐edited neurons remained significantly elevated over baseline S max values (11.8 ± 2.5%; Figure 3D).
Piezo1 epigenome editing of CGRP+ neurons had no effect on degenerative IVD‐induced mechanical nociception, as the percentage of neurons exhibiting calcium transients (Figure 4A) and the maximum calcium transients (Figure 4B) of Piezo1 edited neurons exposed to degenerative IVD‐conditioned media were similar to those of nonedited neurons exposed to degenerative IVD‐conditioned media. In addition, the S 50s of Piezo1 edited neurons exposed to degenerative IVD‐conditioned media (1.21 ± 0.36) were not significantly different from the S 50s of nonedited neurons exposed to degenerative IVD‐conditioned media (1.10 ± 0.1; Figure 4C). Moreover, the S max of Piezo1 edited neurons exposed to degenerative IVD‐conditioned media (45.6 ± 8.1%) were similar to the S max of nonedited neurons exposed to degenerative IVD‐conditioned media (44.8 ± 3.8%; Figure 4D) and remained significantly elevated over the S max of neurons exposed to control media (13.4 ± 1.9%).
FIGURE 4.

Epigenome editing of Piezo1 expression in CGRP+ DRG neurons had no effect on degenerative IVD‐induced mechanical nociception. The percentage of CGRP+ neurons exhibiting calcium transients as a function of strain was fitted to a Boltzmann equation for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, Piezo1 edited CGRP+ neuron control media, and Piezo1 edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). (B) The maximum calcium transient (ΔF/F) of CGRP+ neurons as a function of strain was fitted to a Boltzmann curve for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, Piezo1 edited CGRP+ neuron control media, and Piezo1 edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). The S 50 (C) and S max (D) generated from the dose–response curve fitting for the control media, degenerative IVD‐conditioned media, Piezo1 edited control media, and Piezo1 edited degenerative IVD‐conditioned media groups. (A–B) Different letters indicate groups significantly (p < 0.05) different from one another. Error bars are SD. (C–D) *p < 0.05 when compared to the CGRP+ neuron control media group, #p = 0.05 when compared to the Piezo1 edited CGRP+ neuron control media group. Error bars are SD.
Piezo2‐edited neurons subjected to cyclic strain in the presence of degenerative IVD‐ conditioned media exhibited significantly reduced percentages (p < 0.05) of neurons demonstrating Ca2+ transients when compared to nonedited neurons subjected to cyclic strain in the presence of degenerative IVD‐conditioned media (Figure 5A); none the less, the percentage of Piezo2 edited neurons exhibiting Ca2+ transients remained significantly elevated over baseline (Figure 5A). In addition, Piezo2‐edited neurons subjected to cyclic strain in the presence of degenerative IVD‐conditioned media displayed significantly reduced maximum calcium transients when compared to nonedited neurons subjected to cyclic strain in the presence of degenerative IVD‐conditioned media (Figure 5B); yet, the maximum calcium transients of Piezo2‐edited neurons remained elevated over baseline levels.
FIGURE 5.
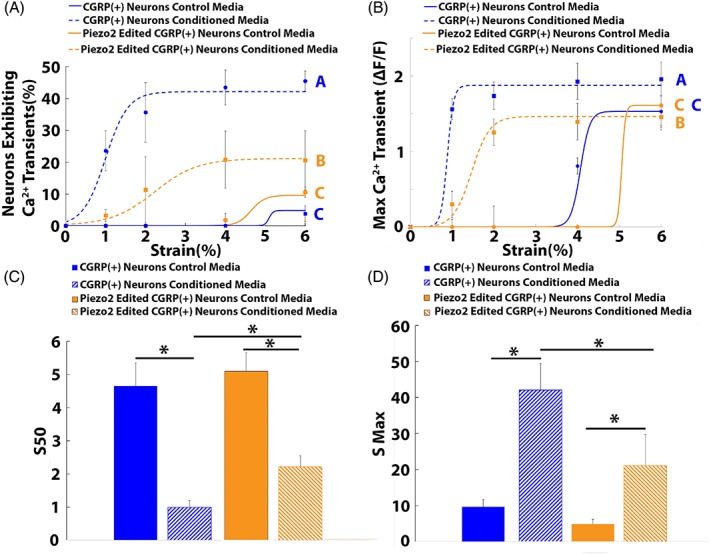
Epigenome editing of Piezo2 expression in CGRP+ DRG neurons reduced degenerative IVD‐induced mechanical nociception. The percentage of CGRP+ neurons exhibiting calcium transients as a function of strain was fitted to a Boltzmann equation for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD conditioned media, Piezo2‐edited CGRP+ neuron control media, and Piezo2‐edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). (B) The maximum calcium transient (ΔF/F) of CGRP+ neurons as a function of strain was fitted to a Boltzmann curve for the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, Piezo2‐edited CGRP+ neuron control media, and Piezo2‐edited CGRP+ neuron degenerative IVD‐conditioned media groups (n = 5 patients). The S 50 (C) and S max (D) generated from the dose–response curve fitting for the control media, degenerative IVD‐conditioned media, Piezo2‐edited control media, and Piezo2‐edited degenerative IVD‐conditioned media groups. (A–B) Different letters indicate groups significantly (p < 0.05) different from one another. Error bars are SD. (C–D) *p < 0.05 when compared to the CGRP+ neuron control media group, #p = 0.05 when compared the Piezo2 edited CGRP+ neuron control media group. Error bars are SD.
Piezo2‐edited neurons exposed to degenerative IVD‐conditioned media displayed significantly elevated S 50 values (2.22 ± 0.32) when compared to nonedited neurons exposed to degenerative IVD‐conditioned media (0.99 ± 0.2; Figure 5C) and significantly decreased when compared to baseline control S 50 values (4.65 ± 0.7; Figure 5C). Furthermore, the S max of Piezo2‐edited neurons exposed to degenerative IVD‐conditioned media (21.2 ± 8.6%) was significantly reduced when compared to the S max of nonedited neurons exposed to degenerative IVD‐conditioned media (42.1 ± 7.3%; Figure 5D), however; the S max of Piezo2‐edited neurons remained elevated over the S max of nonedited neurons exposed to control media (9.6 ± 2.1%; Figure 5D ).
3.3. Multiplex epigenome editing of ASIC3, TRPA1, and Piezo2 in CGRP+ nociceptive neurons abolished degenerative IVD‐enhanced mechanical nociception
DRG neurons transduced with the multiplex CRISPR epigenome editing vector (61 ± 5% transduction efficiency, n = 3 independent transduction; Figure 6C) demonstrated significantly downregulated expression of ASIC3 (70.5 ± 10.8%), TRPA1 (72.1 ± 11.4%), and Piezo2 (67 ± 13%; Figure 6B) when compared to neurons transduced with the nontargeting multiplex CRISRP epigenome editing vector.
FIGURE 6.

Multiplex epigenome editing of TRPA1, ASIC3, and Piezo2 gene promoter in DRG neurons robustly downregulated gene expression and abolished degenerative IVD‐induced mechanical nociception. (A) Vector map depicting TRPA1, ASIC3, and Piezo sgRNAs cloned into CRISRP multiplex epigenome editing vector co‐expressing dCASP‐KRAB‐T2A‐eGFP. (B) Multiplex epigenome editing of TRPA1, ASIC3, and Piezo2 in DRG neurons, *denotes p < 0.05 compared to nontarget epigenome edited DRG neurons, n = 3 independent transductions. Error bars are SD. (C) DRG neurons expressing CRISPR multiplex epigenome editing vectors (E) The percentage of neurons exhibiting strain‐induced calcium transients for neurons in the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, multiplex‐edited CGRP+ neuron control media, and multiplex‐edited CGRP+ neuron degenerative IVD‐conditioned media treatment groups (n = 5 patients). (E) The maximum strain‐induced calcium transient (ΔF/F) of neurons in the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, multiplex‐edited CGRP+ neuron control media, and multiplex edited CGRP+ neuron degenerative IVD‐conditioned media treatment groups (n = 5 patients). The S 50 (F) and S max (G) for neurons in the CGRP+ neuron control media, CGRP+ neuron degenerative IVD‐conditioned media, multiplex edited CGRP+ neuron control media, and multiplex edited CGRP+ neuron degenerative IVD‐conditioned media treatment groups (n = 5 patients). (D–E). Different letters indicate groups significantly (p < 0.05) different from one another. Error bars are SD. (F–G) *p < 0.05 when compared to the CGRP+ neuron control media group, #p = 0.05 when compared to the Multiplex edited CGRP+ neuron control media group. Error bars are SD.
Multiplex epigenome editing of ASIC3, TRPA1, and Piezo2 in CGRP+ neurons abolished degenerative IVD‐induced mechanical nociception and returned the percentages of neurons exhibiting calcium transients to baseline levels (p = 0.4089; Figure 6D). Similarly, multiplex‐edited neurons subjected to cyclic strain in the presence of degenerative IVD‐conditioned media exhibited maximum calcium transients that were not significantly different from the baseline level (p = 1; Figure 6E). Moreover, the S 50 (Figure 6F) and the S max (Figure 6G) of multiplex‐edited neurons subjected to cyclic strain in the presence of degenerative IVD‐conditioned media were returned to baseline levels.
4. DISCUSSION
In this study, we modeled and investigated the interactions between the degenerative IVD environment, nociceptive neurons, and cyclic mechanical loading to elucidate the potential mechanisms of discogenic back pain. Utilizing this model, we tested the ability of CRISPR epigenome editing of nociceptive neuron receptor profiles as a neuromodulation strategy for the treatment of discogenic back pain. Our data demonstrated that specific factors/cytokines produced from the degenerative IVD are capable of directly increasing nociceptive neuron activity in response to mechanical stimuli, suggesting the degenerative IVD environment directly induces elevated mechanical nociception.
In our in vitro model, rat DRG neurons were seeded directly upon bovine AF tissue and subjected to cyclic tensile strain to identify potential underlying mechanisms by which the degenerative IVD can sensitize nociceptive neurons to mechanical stimuli. One limitation of this model is the potential of reactions between the AF tissue and DRG neurons to mimic degenerative IVD‐conditioned media‐induced neuron sensitization to mechanical stimuli; however, the immuno‐privileged state of the healthy IVD minimizes these concerns. In addition, physiologically, nociceptive neurons innervating the IVD cell bodies reside in the DRG, and are not exposed to cyclic strain. Additional or different mechanisms by which degenerative IVDs can alter nociceptive neurons responses to mechanical stimuli may exist that are not accounted for in out model. Despite these limitations, the current study provides valuable information on the potential mechanisms of discogenic pain and demonstrates CRISPR epigenome editing of nociceptive neurons as a potential treatment strategy for discogenic pain.
Previously, utilizing this model, we demonstrated pathologically elevated levels of IL‐6 and sIL‐6R in degenerative IVD‐conditioned media, 59 and that treatment of degenerative IVD‐conditioned media with IL‐6 blocking antibody abolished elevated nociceptive neuron activity in response to mechanical stimuli, indicating its induction by IL‐6. In this study, we again demonstrated that degenerative IVD conditioned media sensitized neuronal responses to mechanical stimuli, indicating degenerative IVDs release factors capable of modulating neuron responses. Based on our previous results identifying the IL‐6 signaling pathways as a primary mediator of degenerative IVD‐induced neuron sensitization to mechanical stimuli, we tested the ability of singleplex CRISPR epigenome editing of IL‐6st expression in nociceptive neurons to regulate elevated neuron responses induced by degenerative IVDs. Our data demonstrated that epigenome editing of IL‐6st expression in rat nociceptive neurons abolished degenerative IVD‐induced elevated nociceptive neuron activity in response to cyclic strain, while these neurons maintained their baseline (nonpathological) activity. Together, these results confirm the results of our previous work, implicating activation of IL‐6 signaling pathways as the primary initiating factor for degenerative IVD‐induced neuron mechanical sensitization in our model.
Next, we investigated the specific mechano‐sensitive ion channels that mediate degenerative IVD‐induced mechanical nociception. While multiple ion channels have been implicated in pain, we focused our study on mechanosensitive ion channels (i.e., Piezo1, Peizo2, and TRPA1) and ASIC3, due to its implication in mechanical pain in models where neurons are directly exposed to degenerative IVD tissue. Our data demonstrated singleplex CRISPR epigenome editing of TRPA1, ASIC3, or Piezo2 expression in nociceptive neurons reduced degenerative IVD‐induced elevated nociceptive neuron activation in response to mechanical stimuli; however, neural activity remained elevated over baseline levels. Together, these results demonstrate that degenerative IVD‐induced sensitization of neurons to mechanical stimuli is induced by IL‐6 signaling, and mediated by the combined activation of TRPA1, ASIC3, and Piezo2 ion channels in our model system; however, the degenerative IVD‐induced mechanical sensitization cannot be attributed to the activity of a single ion channel.
To determine the combined contributions of TRPA1, ASIC3, and Piezo2 activity to degenerative IVD‐induced nociceptor neuron sensitization to mechanical stimuli, we developed a multiplex CRISPR epigenome editing vector that targets the TPRA1, ASIC3, and Piezo2 gene promoter regions, simultaneously, in nociceptive neurons and tested the ability of this vector to modulate degenerative IVD‐induced neuron responses to cyclic strain. Our data demonstrate that multiplex epigenome editing of TRPA1, ASIC3, and Piezo2 expression in nociceptive neurons abolished degenerative IVD‐induced elevated neuron activity in response to mechanical stimuli, while these multiplex‐edited neurons maintained baseline neuron activity. Together, these results implicate IL‐6‐induced activation of TRPA1, ASIC3, and Piezo2 ion channels as a potential underlying mechanism of degenerative IVD‐induced mechanical nociception, demonstrate singleplex CRISPR epigenome editing of IL‐6st and multiplex CRISPR epigenome editing of TRPA1, ASIC3, and Piezo2 expression in nociceptive neurons abolished degenerative IVD‐induced sensitization of neurons to mechanical stimuli, and establishes CRISPR multiplex epigenome editing of nociceptive neurons as a potential gene‐based neuromodulation strategy for the treatment of discogenic back pain.
We demonstrated that singleplex CRISPR epigenome editing of IL6‐st expression in nociceptive neurons abolished degenerative IVD‐induced elevated nociceptive neuron activity in response to mechanical stimuli. Previous studies have established the presence of pathologically elevated cytokine levels, such as TNF‐α, IL‐1β, and IL‐6, in painful IVDs 14 , 15 , 16 , 39 , 40 , 41 , 64 and the role of IL‐6 in radiculopathy and peripheral neuropathy. 42 , 65 , 66 , 67 Additionally, we have previously demonstrated the ability of IL‐6 and sIL‐6R produced by degenerative IVDs 59 to directly augment nociceptive neuron activity in response to mechanical stimulation. 47 Our data indicate that epigenome editing of IL‐6st expression in nociceptive neurons abolished degenerative IVD‐induced elevated neuron activation in response to mechanical stimuli while these neurons maintained baseline neuron activity. Notably, nociceptive neurons innervating the degenerate IVD, do not express membrane‐bound IL‐6 receptors. In these neurons, IL‐6 exhibits trans‐signaling that is facilitated though a complex formed by three components: IL‐6, soluble IL‐6 receptor (sIL‐6R), and IL‐6st. 44 , 65 Combined with our findings that demonstrate IL‐6 and sIL‐6r released from the degenerative IVD directly sensitize nociceptive neurons, these results indicate that targeting the IL‐6/sIL‐6R/IL‐6st signaling complex, in nociceptive neurons innervating the disc, could be a potential therapeutic development strategy for discogenic back. Recent clinical trials for the treatment of discogenic pain have focused on intradiscal injection of monoclonal antibodies targeting IL‐6 68 to disrupt degenerative IVD‐induced activation of the IL‐6 signaling pathway. While these treatments have been demonstrated to provide short‐term pain reduction, 68 they have been ineffective at providing long‐term pain relief. 68 In this study, we demonstrated that epigenome editing of IL‐6st in nociceptive neurons, a component of the IL‐6st signaling complex not targeted in these clinical trials, abolished degenerative IVD‐induced neuron sensitization. Together, these results demonstrate the ability of epigenome editing of IL‐6st expression in nociceptive neurons to abolish degenerative IVD‐induced mechanical sensitization of nociceptive neurons, establish IL‐6st as a potential target of epigenome editing‐based treatment strategies for discogenic pain, and suggests intradiscal delivery of gene‐based neuromodulation therapeutics targeting IL‐6st as a potential treatment strategy for discogenic pain specifically, and more broadly, other inflammatory chronic pain conditions.
Our data demonstrated that singleplex epigenome editing of Acid Sensing Ion Channel 3 (ASIC3) expression in nociceptive neurons reduced degenerative IVD‐induced sensitization of nociceptive neurons to mechanical stimuli; however, neuron activity remained elevated over baseline levels. ASIC3 is a proton‐gated ion channel widely expressed in neurons of the peripheral nervous system, such as the nociceptive neurons that innervate the IVD. 69 , 70 , 71 , 72 ASIC3 has been implicated in inflammatory acidosis‐induced nociception, 73 , 74 , 75 is upregulated in DRG neurons exposed to NP tissue, and has been demonstrated to regulate mechanical allodynia in NP placement models of radiculopathy. 76 , 77 , 78 While some studies have suggested a direct role for ASIC3 79 , 80 in mechanosensation, the exact mechanisms by which ASIC3 channels contribute to mechanical nociception, remain unknown. Together, these results implicate ASIC3 as a potential mediator of degenerative IVD‐induced mechanical nociception and establish ASIC3 as a potential target for epigenome editing‐based therapeutic development.
We demonstrated singleplex epigenome editing of TRPA1 expression in nociceptive neurons partially ameliorated degenerative IVD‐induced elevated nociceptive neuron activity in response to cyclic strain. TRPA1 is a polymodal ion channel, dominantly expressed in nociceptive neurons, 81 , 82 that can be activated by painful or potentially painful stimuli, such as inflammatory mediators 83 , 84 , 85 , 86 , 87 and has been implicated in mechanosensation, inflammation, and numerous chronic pain conditions. 87 , 88 , 89 In addition, TRPA1 signaling activates intracellular signaling pathways that produce upregulated TRPA1 and CGRP expression, 90 increased neuron activity, 91 and increased CGRP release. Together, these results implicate TRPA1 activation as a possible mediator of degenerative IVD‐induction of mechanical nociception, and establishes TRPA1 as a potential therapeutic target for the treatment of discogenic pain.
Our data demonstrated that singleplex epigenome editing of Piezo2 expression in nociceptive neurons significantly reduced degenerative IVD‐induced mechanical sensitization of nociceptive neurons. DRG neurons express the mechanical gate ion channel Piezo2, 92 , 93 , 94 , 95 , 96 a mediator of several mechano‐sensitive functions of neurons including discriminative touch, vibration detection, and proprioception, but not normal mechanical nociception. 92 , 93 , 97 , 98 , 99 , 100 , 101 , 102 , 103 However, Piezo2 has been implicated in the mediation of mechanical hyperalgesia and mechanical allodynia in multiple chronic pain conditions 35 , 103 , 104 , 105 , 106 , 107 with Piezo2 knockout mice exhibiting partially reduced sensitivity to mechanical pain. 103 , 107 Consistent with these findings, our data demonstrated epigenome editing of Piezo2 expression in nociceptive neurons reduced degenerative IVD‐induced mechanical sensitization of nociceptive neurons while having no effect on baseline neuron responses to mechanical stimuli. These results implicate Piezo2 as a potential mediator of degenerative IVD‐induced mechanical nociception.
In addition to Piezo2, DRG neurons express the mechanically gated ion channel Piezo1. Piezo1 has been implicated as a potential mediator of mechanical nociception 95 , 96 ; however, our data show Piezo1 is not involved in the degenerative IVD‐induced sensitization of nociceptive neurons to cyclic strain observed in this study. Several potential explanations exist for these findings. In prior work, Piezo1 has been activated by different modalities of mechanical stimuli from the narrow range of cyclic strain frequencies (1 Hz), cyclic strain levels (0%–6%), and strain modalities (tensile strain) tested in our in vitro model. In addition, the majority of Piezo1‐expressing nociceptive neurons (~60%) 95 co‐express Piezo2, suggesting potential redundant roles for Piezo1 and Piezo2 in the degenerative IVD‐induced sensitization of nociceptive neurons to mechanical stimuli. Furthermore, most neurons that express Piezo1 but not Piezo2 are nonpeptidergic nociceptive neurons. These neurons comprise a very small percentage (~3%) of neurons innervating the IVD; therefore, we limited our analysis to peptidergic (CGRP) nociceptors, as they are the most abundant neurons in IVD. Piezo1 may mediate degenerative IVD‐induced mechanical nociception in nonpeptidergic neurons not examined in this work.
CRISPR epigenome editing systems have tremendous therapeutic potential due to their ability to modulate specific cell functions in a pathological environment. Here, we demonstrated the ability of multiplex CRISPR epigenome editing‐based neuromodulation therapeutics to modulate nociceptive neuron ion channel expression profiles and regulate the response of those neurons to the degenerative IVD environment (Table 1). Our data demonstrate nociceptive neurons with multiplex epigenome modifications of the TRPA1, ASIC3, and Piezo2 promotor regions maintained normal sensitivity to mechanical stimuli under pathological conditions that induced elevated nociceptive neuron response to mechanical stimuli. These results demonstrate that CRISPR multiplex epigenome editing of ion channels in nociceptive neurons regulates degenerative IVD‐modulated mechanical sensitization of nociceptive neurons and establishes epigenome modification of sensory neurons as a potential therapeutic strategy for the treatment of back pain.
TABLE 1.
Summary of the effects of CRISPR epigenome editing (Target Gene) on degenerative IVD conditioned media sensitization of DRG neurons to mechanical stimuli.
| Target gene | Neurons exhibiting Ca2+ transients (%) | Max Ca2+ transient | S 50 | S max |
|---|---|---|---|---|
| IL‐6st | Abolished | Abolished | Abolished | Abolished |
| ASIC 3 | Reduced | No change | No change | Reduced |
| TRPA1 | Reduced | No change | No change | Reduced |
| Piezo1 | No change | No change | No change | No change |
| Piezo 2 | Reduced | Reduced | Reduced | Reduced |
| Multiplex | Abolished | Abolished | Abolished | Abolished |
In this study, we demonstrated that IL‐6 present in the degenerative IVD‐induced elevated nociceptive neurons responses to cyclic strain, at physiologically relevant strain levels, 60 , 108 via TRPA1, ASIC3, and Piezo2 ion channel activity. These results elucidate potential underlying mechanisms for discogenic pain generation by degenerative IVD modulation of nociceptive neuron activity. In addition, we demonstrated that multiplex epigenome editing of the gene promoter region of TRPA1, ASIC3, and Piezo2 in nociceptive neurons abolished degenerative IVD‐induced mechanical sensitization of nociceptive neurons. These results elucidate an underlying mechanism of discogenic pain, identify therapeutic targets for the treatment of back pain, and establish multiplex epigenome editing of nociceptive neurons as a potential neuromodulation strategy for the treatment of discogenic back pain.
AUTHOR CONTRIBUTIONS
Joshua D. Stover, Brandon Lawrence, and Robby D. Bowles. all contributed to research design. Joshua D. Stover, Matthew A. Trone, and Robby D. Bowles contributed to data acquisition, analysis, and interpretation. Joshua D. Stover, Matthew A. Trone, and Robby D. Bowles drafted and critically reviewed the manuscript. All authors have read and reviewed the final submitted manuscript.
CONFLICT OF INTEREST STATEMENT
Robby Bowles is an Editorial Board member of JOR Spine and a co‐author of this article. To minimize bias, they were excluded from all editorial decision‐making related to the acceptance of this article for publication. [Correction added on 22 June 2023, after first online publication: Conflict of Interest statement was revised]
Supporting information
Data S1: Supporting information.
Figure S1. qPCR results from singleplex epigenome editing of DRG neuron.
Figure S2. Nontarget epigenome editing of CGRP+ neurons has no effect on degenerative intervertebral disc‐induced mechanical nociception
Figure S3. Singleplex CRISPR epigenome editing downregulates target protein expression.
Table S1. Surgical patient data.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health under the award number R01AR074998. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We acknowledge Cell Imaging Core at the University of Utah for use of equipment (Bruker/Prairie View multiphoton microscope).
Stover, J. D. , Trone, M. A. , Lawrence, B. , & Bowles, R. D. (2023). Multiplex epigenome editing of ion channel expression in nociceptive neurons abolished degenerative IVD‐conditioned media‐induced mechanical sensitivity. JOR Spine, 6(2), e1253. 10.1002/jsp2.1253
REFERENCES
- 1. Vos T, Barber RM, Bell B, Bertozzi‐Villa A, Biryukov SBI, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386(9995):743‐800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murray CJL, Vos T, Lozano R, et al. Disability‐adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990‐2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2197‐2223. [DOI] [PubMed] [Google Scholar]
- 3. Katz JN. Lumbar disc disorders and low‐back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006;88(suppl 2):21‐24. [DOI] [PubMed] [Google Scholar]
- 4. Mayer JE, Iatridis JC, Chan D, Qureshi SA, Gottesman O, Hecht AC. Genetic polymorphisms associated with intervertebral disc degeneration. Spine J. 2013;13(3):299‐317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Williams FMK, Bansal AT, van Meurs JB, et al. Novel genetic variants associated with lumbar disc degeneration in northern Europeans: a meta‐analysis of 4600 subjects. Ann Rheum Dis. 2013;72(7):1141‐1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shirazi‐Adl A, Taheri M, Urban JPG. Analysis of cell viability in intervertebral disc: Effect of endplate permeability on cell population. J Biomech. 2010;43(7):1330‐1336. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y, Videman T, Battié MC. ISSLS prize winner: Lumbar vertebral endplate lesions: associations with disc degeneration and back pain history. Spine. 2012;37(17):1490‐1496. [DOI] [PubMed] [Google Scholar]
- 8. Adams MA, Roughley PJ. What is intervertebral disc degeneration, and what causes it? Spine. 2006;31(18):2151‐2161. [DOI] [PubMed] [Google Scholar]
- 9. Suthar P, Patel R, Mehta C, Patel N. MRI evaluation of lumbar disc degenerative disease. J Clin Diagn Res. 2015;9(4):TC04‐TC09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roberts S, Evans H, Trivedi J, Menage J. Histology and pathology of the human intervertebral disc. J Bone Jt Surg Am. 2006;88(Suppl 2):10‐14. [DOI] [PubMed] [Google Scholar]
- 11. Le Maitre CL, Pockert A, Buttle DJ, et al. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35(Pt 4):652‐655. [DOI] [PubMed] [Google Scholar]
- 12. Burke JG, Watson RWG, McCormack D, Dowling FE, Walsh MG, Fitzpatrick JM. Intervertebral discs which cause low back pain secrete high levels of proinflammatory mediators. J Bone Jt Surg Br. 2002;84(2):196‐201. [DOI] [PubMed] [Google Scholar]
- 13. Burke JG, Watson RWG, Conhyea D, et al. Human nucleus pulposis can respond to a pro‐inflammatory stimulus. Spine. 2003;28(24):2685‐2693. [DOI] [PubMed] [Google Scholar]
- 14. Shamji MF, Setton LA, Jarvis W, et al. Proinflammatory cytokine expression profile in degenerated and herniated human intervertebral disc tissues. Arthritis Rheum. 2010;62(7):1974‐1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kokubo Y, Uchida K, Kobayashi S, et al. Herniated and spondylotic intervertebral discs of the human cervical spine: histological and immunohistological findings in 500 en bloc surgical samples. Laboratory investigation. J Neurosurg Spine. 2008;9(3):285‐295. [DOI] [PubMed] [Google Scholar]
- 16. Specchia N, Pagnotta A, Toesca A, Greco F. Cytokines and growth factors in the protruded intervertebral disc of the lumbar spine. Eur Spine J. 2002;11(2):145‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vernon‐Roberts B, Moore RJ, Fraser RD. The natural history of age‐related disc degeneration: the pathology and sequelae of tears. Spine. 2007;32(25):2797‐2804. [DOI] [PubMed] [Google Scholar]
- 18. Stover JD, Farhang N, Berrett KC, Gertz J, Lawrence B, Bowles RD. CRISPR epigenome editing of AKAP150 in DRG neurons abolishes degenerative IVD‐induced neuronal activation. Mol Ther. 2017;25(9):2014‐2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Freemont AJ, Watkins A, Le Maitre C, et al. Current understanding of cellular and molecular events in intervertebral disc degeneration: implications for therapy. J Pathol. 2002;196(4):374‐379. [DOI] [PubMed] [Google Scholar]
- 20. Freemont AJ, Peacock TE, Goupille P, et al. Nerve ingrowth into diseased intervertebral disc in chronic back pain. Lancet. 1997;350(9072):178‐181. [DOI] [PubMed] [Google Scholar]
- 21. Coppes MH, Marani E, Thomeer RT, Groen GJ. Innervation of “painful” lumbar discs. Spine. 1997;22(20):2342‐2349. discussion 2349–50. [DOI] [PubMed] [Google Scholar]
- 22. Millecamps M, Stone LS. Delayed onset of persistent discogenic axial and radiating pain after a single‐level lumbar intervertebral disc injury in mice. Pain. 2018;159(9):1843‐1855. [DOI] [PubMed] [Google Scholar]
- 23. Kepler CK, Ponnappan RK, Tannoury CA, et al. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13(3):318‐330. [DOI] [PubMed] [Google Scholar]
- 24. Lotz JC, Ulrich JA. Innervation, inflammation, and hypermobility may characterize pathologic disc degeneration: review of animal model data. J Bone Joint Surg Am. 2006;88(Suppl 2):76‐82. [DOI] [PubMed] [Google Scholar]
- 25. García‐Cosamalón J, del Valle ME, Calavia MG, et al. Intervertebral disc, sensory nerves and neurotrophins: who is who in discogenic pain? J Anat. 2010;217(1):1‐15. doi: 10.1111/j.1469-7580.2010.01227.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coppes MH, Marani E, Thomeer RT, et al. Innervation of annulus fibrosis in low back pain. Lancet. 1990;336(8708):189‐190. [DOI] [PubMed] [Google Scholar]
- 27. Roberts S, Eisenstein SM, Menage J, et al. Mechanoreceptors in intervertebral discs. Morphology, distribution, and neuropeptides. Spine. 1995;20(24):2645‐2651. [DOI] [PubMed] [Google Scholar]
- 28. Ohtori S, Takahashi Y, Takahashi K, et al. Sensory innervation of the dorsal portion of the lumbar intervertebral disc in rats. Spine. 1999;24(22):2295‐2299. [DOI] [PubMed] [Google Scholar]
- 29. Aoki Y, Ohtori S, Ino H, et al. Disc inflammation potentially promotes axonal regeneration of dorsal root ganglion neurons innervating lumbar intervertebral disc in rats. Spine. 2004;29(23):2621‐2626. [DOI] [PubMed] [Google Scholar]
- 30. Aoki Y, Ohtori S, Takahashi K, et al. Expression and co‐expression of VR1, CGRP, and IB4‐binding glycoprotein in dorsal root ganglion neurons in rats: differences between the disc afferents and the cutaneous afferents. Spine. 2005;30(13):1496‐1500. [DOI] [PubMed] [Google Scholar]
- 31. Ohtori S, Takahashi K, Chiba T, Yamagata M, Sameda H, Moriya H. Substance P and calcitonin gene‐related peptide immunoreactive sensory DRG neurons innervating the lumbar intervertebral discs in rats. Ann Anat. 2002;184(3):235‐240. [DOI] [PubMed] [Google Scholar]
- 32. Ashton IK, Roberts S, Jaffray DC, Polak JM, Eisenstein SM. Neuropeptides in the human intervertebral disc. J Orthop Res. 1994;12(2):186‐192. [DOI] [PubMed] [Google Scholar]
- 33. Kestell GR, Anderson RL, Clarke JN, Haberberger RV, Gibbins IL. Primary afferent neurons containing calcitonin gene‐related peptide but not substance P in forepaw skin, dorsal root ganglia, and spinal cord of mice. J Comp Neurol. 2015;523:2555‐2569. [DOI] [PubMed] [Google Scholar]
- 34. Roh J, Hwang SM, Lee SH, Lee K, Kim YH. Functional Expression of Piezo1 in dorsal root ganglion (DRG) Neurons. Int J Mol Sci. 2020;21(11):3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dubin AE, Schmidt M, Mathur J, et al. Inflammatory signals enhance piezo2‐mediated mechanosensitive currents. Cell Rep. 2012;2(3):511‐517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson WE, Evans H, Menage J, et al. Immunohistochemical detection of Schwann cells in innervated and vascularized human intervertebral discs. Spine. 2001;26(23):2550‐2557. [DOI] [PubMed] [Google Scholar]
- 37. Brown MF, Hukkanen MV, McCarthy ID, et al. Sensory and sympathetic innervation of the vertebral endplate in patients with degenerative disc disease. J Bone Joint Surg. 1997;79(1):147‐153. [DOI] [PubMed] [Google Scholar]
- 38. Krock E, Rosenzweig DH, Chabot‐Doré A‐J, et al. Painful, degenerating intervertebral discs up‐regulate neurite sprouting and CGRP through nociceptive factors. J Cell Mol Med. 2014;18(6):1213‐1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Le Maitre CL, Hoyland JA, Freemont AJ. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL‐1beta and TNFalpha expression profile. Arthritis Res Ther. 2007;9(4):R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Le Maitre C, Freemont AJ, Hoyland J. The role of interleukin‐1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res Ther. 2005;7(4):R732‐R745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burke JG, Watson RWG, McCormack D, et al. Intervertebral discs which cause low back pain secrete high levels of proinflammatory mediators. J Bone Joint Surg. 2002;84(2):196‐201. [DOI] [PubMed] [Google Scholar]
- 42. Oprée A, Kress M. Involvement of the proinflammatory cytokines tumor necrosis factor‐alpha, IL‐1 beta, and IL‐6 but not IL‐8 in the development of heat hyperalgesia: effects on heat‐evoked calcitonin gene‐related peptide release from rat skin. J Neurosci. 2000;20(16):6289‐6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oka Y, Ibuki T, Matsumura K, et al. Interleukin‐6 is a candidate molecule that transmits inflammatory information to the CNS. Neuroscience. 2007;145(2):530‐538. [DOI] [PubMed] [Google Scholar]
- 44. Andratsch M, Mair N, Constantin CE, et al. A key role for gp130 expressed on peripheral sensory nerves in pathological pain. J Neurosci. 2009;29(43):13473‐13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fang D, Kong L‐Y, Cai J, et al. IL‐6‐mediated functional up‐regulation of TRPV1 receptors in DRG neurons via the activation of JAK/PI3K signaling pathway. Pain. 2015;156(6):1124‐1144. [DOI] [PubMed] [Google Scholar]
- 46. Bowles RD, Karikari IO, VanDerwerken DN, et al. In vivo luminescent imaging of NF‐κB activity and NF‐κB‐related serum cytokine levels predict pain sensitivities in a rodent model of peripheral neuropathy. Eur J Pain. 2016;20(3):365‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stover JD, Lawrence B, Bowles RD. Degenerative IVD conditioned media and acidic pH sensitize sensory neurons to cyclic tensile strain. J Orthop Res. 2021;39(6):1192‐1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13(2):127‐137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819‐823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957‐963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1‐demarcated microenvironments of heterochromatin required for KRAB zinc finger protein‐mediated transcriptional repression. Mol Cell Biol. 2006;26(22):8623‐8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Groner AC, Meylan S, Ciuffi A, et al. KRAB‐zinc finger proteins and KAP1 can mediate long‐range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6(3):e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krebs CJ, Schultz DC, Robins DM. The KRAB zinc finger protein RSL1 regulates sex‐ and tissue‐specific promoter methylation and dynamic hormone‐responsive chromatin configuration. Mol Cell Biol. 2012;32(18):3732‐3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reynolds N, Salmon‐Divon M, Dvinge H, et al. NuRD‐mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012;31(3):593‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stover JD, Farhang N, Lawrence B, Bowles RD. Multiplex epigenome editing of dorsal root ganglion neuron receptors abolishes redundant interleukin 6, tumor necrosis factor alpha, and interleukin 1b signaling by the degenerative intervertebral disc. 2019;30(9):1147‐1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Farhang N, Brunger JM, Stover JD, et al. CRISPR‐based epigenome editing of cytokine receptors for the promotion of cell survival and tissue deposition in inflammatory environments. Tissue Eng Part A. 2017;23(15–16):738‐749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pfirrmann CW, Metzdorf A, Zanetti M, et al. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26(17):1873‐1878. [DOI] [PubMed] [Google Scholar]
- 59. Stover JD, Farhang N, Lawrence B, Bowles RD. CRISPR epigenome editing of AKAP 150 in DRG neurons abolished degenerative IVD‐induced sensitization of sensory neurons. Mol Ther Rev. 2016;25:2014‐2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. O'Connell GD, Vresilovic EJ, Elliott DM. Human intervertebral disc internal strain in compression: the effect of disc region, loading position, and degeneration. J Orthop Res. 2011;29(4):547‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schindelin J, Arganda‐Carreras I, Frise E, et al. Fiji: an open‐source platform for biological‐image analysis. Nat Methods. 2012;9(7):676‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Greffrath W, Kirschstein T, Nawrath H, Treede R‐D. Changes in cytosolic calcium in response to noxious heat and their relationship to vanilloid receptors in rat dorsal root ganglion neurons. Neuroscience. 2001;104(2):539‐550. [DOI] [PubMed] [Google Scholar]
- 63. Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9‐based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42(19):e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kitano T, Zerwekh JE, Usui Y, et al. Biochemical changes associated with the symptomatic human intervertebral disk. Clin Orthop Relat Res. 1993;(293):372‐377. [PubMed] [Google Scholar]
- 65. Obreja O, Schmelz M, Poole S, Kress M. Interleukin‐6 in combination with its soluble IL‐6 receptor sensitises rat skin nociceptors to heat, in vivo. Pain. 2002;96(1–2):57‐62. [DOI] [PubMed] [Google Scholar]
- 66. Obreja O, Biasio W, Andratsch M, et al. Fast modulation of heat‐activated ionic current by proinflammatory interleukin 6 in rat sensory neurons. Brain. 2005;128(Pt 7):1634‐1641. [DOI] [PubMed] [Google Scholar]
- 67. Bowles RD, Mata BA, Bell RD, et al. 2014. In vivo luminescence imaging of NF‐κB activity and serum cytokine levels predict pain sensitivities in a rodent model of osteoarthritis. Arthritis Rheumatol.66(3):637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sainoh T, Orita S, Miyagi M, et al. Single intradiscal injection of the interleukin‐6 receptor antibody tocilizumab provides short‐term relief of discogenic low back pain; prospective comparative cohort study. J Orthop Sci. 2016;21(1):2‐6. [DOI] [PubMed] [Google Scholar]
- 69. Deval E, Noe J, Alloui A, et al. ASIC3, a sensor of acidic and primary. 2008;27(22):3047‐3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stephan G, Huang L, Tang Y, et al. The ASIC3/P2X3 cognate receptor is a pain‐relevant and ligand‐gated cationic channel. Nat Commun. 2018;9(1):1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Deval E, Lingueglia E. Acid‐Sensing Ion Channels and nociception in the peripheral and central nervous systems. Neuropharmacology. 2015;94:49‐57. [DOI] [PubMed] [Google Scholar]
- 72. Wemmie JA, Taugher RJ, Kreple CJ. Acid‐sensing ion channels in pain and disease. Nat Rev Neurosci. 2015;14(7):461‐471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wu J, Liu T‐T, Zhou Y‐M, et al. Sensitization of ASIC3 by proteinase‐activated receptor 2 signaling contributes to acidosis‐induced nociception. J Neuroinflammation. 2017;14(1):150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sluka KA, Price MP, Breese NM, Stucky CL, Wemmie JA, Welsh MJ. Chronic hyperalgesia induced by repeated acid injections in muscle is abolished by the loss of ASIC3, but not ASIC1. Pain. 2003;106(3):229‐239. [DOI] [PubMed] [Google Scholar]
- 75. Deval E, Noël J, Lay N, et al. ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J. 2008;27(22):3047‐3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ohtori S, Inoue G, Koshi T, et al. Up‐regulation of acid‐sensing ion channel 3 in dorsal root ganglion neurons following application of nucleus pulposus on nerve root in rats. Spine. 2006;31(18):2048‐2052. [DOI] [PubMed] [Google Scholar]
- 77. Wang D, Pan H, Zhu H, et al. Upregulation of nuclear factor‐κB and acid sensing ion channel 3 in dorsal root ganglion following application of nucleus pulposus onto the nerve root in rats. Mol Med Rep. 2017;16(4):4309‐4314. [DOI] [PubMed] [Google Scholar]
- 78. Kobayashi Y, Sekiguchi M, Konno S. Effect of an acid‐sensing ion channels inhibitor on pain‐related behavior by nucleus pulposus applied on the nerve root in rats. Spine. 2017;42(11):633‐641. [DOI] [PubMed] [Google Scholar]
- 79. Lin SH, Cheng YR, Banks R, et al. Evidence for the involvement of ASIC3 in sensory mechanotransduction in proprioceptors. Nat Commun. 2016;7:11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cheng YR, Jiang BYCC. Acid‐sensing ion channels: dual function proteins for chemo‐sensing and mechano‐sensing. J Biomed Sci. 2018;25(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Latorre R, Brauchi S, Orta G, et al. ThermoTRP channels as modular proteins with allosteric gating. Cell Calcium. 2007;42(4):427‐438. [DOI] [PubMed] [Google Scholar]
- 82. Story GM, Peier AM, Reeve AJ, et al. ANKTM1, a TRP‐like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell. 2003;112(6):819‐829. [DOI] [PubMed] [Google Scholar]
- 83. Nilius B, Prenen J, Owsianik G. Irritating channels: the case of TRPA1. J Physiol. 2011;589(Pt 7):1543‐1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nilius B, Appendino G, Owsianik G. The transient receptor potential channel TRPA1: from gene to pathophysiology. Eur J Physiol. 2012;464(5):425‐458. doi: 10.1007/s00424-012-1158-z [DOI] [PubMed] [Google Scholar]
- 85. Zygmunt PM, Högestätt ED. TRPA1 BT—Mammalian Transient Receptor Potential (TRP) Cation Channels: Volume I. Springer; 2014:583‐630. doi: 10.1007/978-3-642-54215-2_23 [DOI] [Google Scholar]
- 86. Chen J, Hackos DH. TRPA1 as a drug target—promise and challenges. Naunyn Schmiedebergs Arch Pharmacol. 2015;388(4):451‐463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Viana F. TRPA1 channels: molecular sentinels of cellular stress and tissue damage. J Physiol. 2016;594(15):4151‐4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bandell M, Story GM, Hwang SW, et al. Noxious Cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron. 2004;41(6):849‐857. [DOI] [PubMed] [Google Scholar]
- 89. Kwan KY, Allchorne AJ, Vollrath MA, et al. TRPA1 contributes to cold, mechanical, and chemical nociception but is not essential for hair‐cell transduction. Neuron. 2006;50(2):277‐289. [DOI] [PubMed] [Google Scholar]
- 90. Wang X‐L, Cui L‐W, Liu Z, et al. Effects of TRPA1 activation and inhibition on TRPA1 and CGRP expression in dorsal root ganglion neurons. Neural Regen Res. 2019;14(1):140‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ikeda‐Miyagawa Y, Kobayashi K, Yamanaka H, et al. Peripherally increased artemin is a key regulator of TRPA1/V1 expression in primary afferent neurons. Mol Pain. 2015;11(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Coste B, Mathur J, Schmidt M, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330(6000):55‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ranade SS, Woo S‐H, Dubin AE, et al. Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature. 2014;516(7529):121‐125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zheng W, Nikolaev YA, Gracheva EO, Bagriantsev SN. Piezo2 integrates mechanical and thermal cues in vertebrate mechanoreceptors. Proc Natl Acad Sci USA. 2019;116(35):17547‐17555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang J, La J‐H, Hamill OP. Piezo1 is selectively expressed in small diameter mouse drg neurons distinct from neurons strongly expressing TRPV1. Front Mol Neurosci. 2019;12:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mikhailov N, Leskinen J, Fagerlund I, et al. Mechanosensitive meningeal nociception via Piezo channels: Implications for pulsatile pain in migraine? Neuropharmacology. 2019;149:113‐123. [DOI] [PubMed] [Google Scholar]
- 97. Woo S‐H, Ranade S, Weyer AD, et al. Piezo2 is required for Merkel‐cell mechanotransduction. Nature. 2014;509(7502):622‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Woo S‐H, Lukacs V, de Nooij JC, et al. Piezo2 is the principal mechanotransduction channel for proprioception. Nat Neurosci. 2015;18(12):1756‐1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Florez‐Paz D, Bali KK, Kuner R, Gomis A. A critical role for Piezo2 channels in the mechanotransduction of mouse proprioceptive neurons. Sci Rep. 2016;6:25923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chesler AT, Szczot M, Bharucha‐Goebel D, et al. The role of Piezo2 in human mechanosensation. N Engl J Med. 2016;375(14):1355‐1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mahmud AA, Nahid NA, Nassif C, et al. Loss of the proprioception and touch sensation channel PIEZO2 in siblings with a progressive form of contractures. Clin Genet. 2017;91(3):470‐475. doi: 10.1111/cge.12850 [DOI] [PubMed] [Google Scholar]
- 102. Delle Vedove A, Storbeck M, Heller R, et al. Biallelic loss of proprioception‐related Piezo2 causes muscular atrophy with perinatal respiratory distress, arthrogryposis, and scoliosis. Am J Hum Genet. 2016;99(5):1206‐1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Szczot M, Liljencrantz J, Ghitani N, et al. Piezo2 mediates injury‐induced tactile pain in mice and humans. Sci Transl Med. 2018;10(462):eaat9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Eijkelkamp N, Linley JE, Torres JM, et al. A role for Piezo2 in EPAC1‐dependent mechanical allodynia. Nat Commun. 2013;4:1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Singhmar P, Huo X, Eijkelkamp N, et al. Critical role for Epac1 in inflammatory pain controlled by GRK2‐mediated phosphorylation of Epac1. Proc Natl Acad Sci USA. 2016;113(11):3036‐3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yang J, Zhang J, Yang H, Li K, Lei X, Xu C. The potential role of Piezo2 in the mediation of visceral sensation. Neurosci Lett. 2016;630:158‐163. [DOI] [PubMed] [Google Scholar]
- 107. Murthy SE, Loud MC, Daou I, et al. The mechanosensitive ion channel Piezo2 mediates sensitivity to mechanical pain in mice. Sci Transl Med. 2018;10(462):eaat9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. O'Connell GD, Johannessen W, Vresilovic EJ, Elliott DM. Human internal disc strains in axial compression measured noninvasively using magnetic resonance imaging. Spine. 2007;32(25):2860‐2868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Supporting information.
Figure S1. qPCR results from singleplex epigenome editing of DRG neuron.
Figure S2. Nontarget epigenome editing of CGRP+ neurons has no effect on degenerative intervertebral disc‐induced mechanical nociception
Figure S3. Singleplex CRISPR epigenome editing downregulates target protein expression.
Table S1. Surgical patient data.


